Abstract
Background
Although clinical trials testing immunotherapies in glioblastoma (GBM) have yielded mixed results, new strategies targeting tumor-specific somatic coding mutations, termed “neoantigens,” represent promising therapeutic approaches. We characterized the microenvironment and neoantigen landscape of the aggressive CT2A GBM model in order to develop a platform to test combination checkpoint blockade and neoantigen vaccination.
Methods
Flow cytometric analysis was performed on intracranial CT2A and GL261 tumor-infiltrating lymphocytes (TILs). Whole-exome DNA and RNA sequencing of the CT2A murine GBM was employed to identify expressed, somatic mutations. Predicted neoantigens were identified using the pVAC-seq software suite, and top-ranking candidates were screened for reactivity by interferon-gamma enzyme linked immunospot assays. Survival analysis was performed comparing neoantigen vaccination, anti-programmed cell death ligand 1 (αPD-L1), or combination therapy.
Results
Compared with the GL261 model, CT2A exhibited immunologic features consistent with human GBM including reduced αPD-L1 sensitivity and hypofunctional TILs. Of the 29 CT2A neoantigens screened, we identified neoantigen-specific CD8+ T-cell responses in the intracranial TIL and draining lymph nodes to two H2-Kb restricted (Epb4H471L and Pomgnt1R497L) and one H2-Db restricted neoantigen (Plin2G332R). Survival analysis showed that therapeutic neoantigen vaccination with Epb4H471L, Pomgnt1R497L, and Plin2G332R, in combination with αPD-L1 treatment was superior to αPD-L1 alone.
Conclusions
We identified endogenous neoantigen specific CD8+ T cells within an αPD-L1 resistant murine GBM and show that neoantigen vaccination significantly augments survival benefit in combination with αPD-L1 treatment. These observations provide important preclinical correlates for GBM immunotherapy trials and support further investigation into the effects of multimodal immunotherapeutic interventions on antiglioma immunity.
Key Points
1. Neoantigen vaccines combined with checkpoint blockade may be promising treatments.
2. CT2A tumors exhibit features of human GBM microenvironments.
3. Differential scanning fluorimetry assays may complement in silico neoantigen prediction tools.
Keywords: glioblastoma, immunogenomics, neoantigen , personalized vaccination
Importance of the Study.
To date, GBM clinical trials testing standard immune-checkpoint blocking antibodies have yielded mixed results. However, new strategies aimed at promoting immune recognition of tumor-specific neoantigens offer promising approaches to augment antitumor responses to immune-checkpoint blocking therapy. Thus, the characterization of checkpoint blockade–resistant GBM models that can enable combination neoantigen vaccine studies has been a major focus of the field. In this study, we show that unlike other syngeneic murine GBMs, the CT2A model exhibits significant resistance to immune-checkpoint blocking treatment. We characterize the immunogenicity of predicted CT2A neoantigens and perform survival studies in CT2A-bearing mice, showing that polyvalent neoantigen vaccination combined with anti–PD-L1 blockade provides synergistic survival benefit compared with either modality alone. This work reveals new insights into a clinically relevant model of GBM and provides a proof of concept for a combination immunotherapy approach that is the basis of an ongoing clinical trial by our group here at Washington University.
The treatment of glioblastoma (GBM) remains a major challenge. With over 13 000 new cases diagnosed annually, GBM is the most common and lethal malignancy of the central nervous system (CNS) in adults. Despite multimodality treatment that includes maximal surgical resection, radiotherapy, and temozolomide chemotherapy, nearly all patients will die from their diseases. Although there have been significant advances in understanding the molecular and genomic characteristics of GBM in order to guide new treatment approaches, median patient survival has not increased significantly.1 Thus, there remains a clear need to develop more effective treatments for this disease. To this end, the success of immunotherapies in other cancers has stimulated a search for effective immune-based treatments for GBM patients. However, to date the results of these efforts have been mixed. In newly diagnosed GBM, the dendritic cell–based tumor lysate vaccination, DC-Vax-L, appeared to improve survival in a subset of patients,2 whereas the anti–epidermal growth factor receptor variant III peptide vaccine, Rintega, did not improve survival over the control group.3 Moreover, anti–programmed cell death 1 (PD-1) immunotherapy did not improve survival as monotherapy in unselected recurrent GBM patients.4
Of the myriad barriers to effective brain tumor immunotherapy, one potentially critical consideration is that a more systematic approach to tumor-specific antigen identification and targeting is necessary to potently stimulate and direct T cells to treat GBM.5 To this end, the “cancer immunogenomics” concept represents an approach in which bioinformatics tools are used to predict candidate T-cell neoantigens from cancer genomics data. We have applied the cancer immunogenomics approach to neoantigen discovery in preclinical models6 and to the development of personalized polyvalent therapeutic cancer vaccines for patients with GBM.7–9 We previously employed a cancer immunogenomics approach to identify endogenous neoantigens in 2 orthotopic mouse GBM models, SMA-560 and GL261.6 Using a screening approach that combined functional interferon-gamma enzyme linked immunospot (IFN-γ ELISPOT) assays and tetramer-based fluorescence activated cell sorting (FACS) analysis, we detected endogenous CD8+ T-cell responses to 3 neoantigens derived from orthotopically transplanted, syngeneic tumors. In addition, we and others recently demonstrated that personalized cancer vaccines targeting predicted neoantigens in GBM patients can elicit tumor-specific immune responses.7–9 Thus, cancer immunogenomics-mediated neoantigen targeting represents a compelling form of precision immunotherapy in GBM that merits further study.
To investigate neoantigen vaccine approaches in GBM, we leveraged our previous work in preclinical models to study therapeutic neoantigen targeting. Although the GL261 and SMA-560 models have provided key proof of concept that neoantigens can be identified and targeted, their sensitivity to checkpoint blockade immunotherapy10–12 does not recapitulate the lack of response observed in human GBM checkpoint blockade clinical trials. Thus, we sought to employ a preclinical model that is less sensitive to checkpoint blockade treatment to determine if precision targeting of neoantigens could augment responsiveness to checkpoint blockade. We applied a cancer immunogenomics approach to the highly aggressive CT2A GBM model to identify neoantigens specific to the tumor. We confirmed the reduced sensitivity of intracranial CT2A to checkpoint blockade and found that these tumors harbor a larger fraction of tumor-associated macrophages and hypofunctional T cells compared with GL261 tumors. To identify CT2A neoantigen candidates for vaccination, DNA whole-exome and RNA sequencing were performed, and the pVAC-seq software suite13 was used to identify high-affinity H2-Db and H2-Kb restricted candidate neoantigens. Using IFN-γ ELISPOT assays, we demonstrated that 13 of the 29 top ranked neoantigen candidates elicited CD8+ splenocyte responses in mice vaccinated with mutant peptide. Of those immunogenic candidates, we identified endogenously primed neoantigen-specific CD8+ T-cell responses within tumors and draining lymph nodes to two H2-Kb restricted neoantigens, Epb4H471L and Pomgnt1R497L, and one H2-Db restricted neoantigen, Plin2G332R. These neoantigens formed stable complexes with their respective major histocompatibility complex (MHC) molecules based on differential Scanning fluorimetry (DSF). Finally, we showed that multivalent therapeutic neoantigen vaccination with Epb4H471L, Pomgnt1R497L, and Plin2G332R in combination with anti–PD-L1 treatment was superior to either vaccine or anti–PD-L1 treatment alone, suggesting that combination vaccine and checkpoint blockade treatments may represent a therapeutic pathway in GBM and, potentially, in other cancers. By applying a cancer immunogenomics approach to identify endogenous neoantigen reactivity in an aggressive and treatment-resistant murine GBM, we provide a preclinical framework to investigate the effects of multimodality immunotherapeutic interventions in antiglioma immunity.
Materials and Methods
Animals and Cells
Animal studies were approved by the Institutional Animal Care and Use Committee at Washington University. C57BL/6 mice were purchased from Taconic Biosciences. Mice were housed in specific pathogen-free conditions. GL261 was obtained from the National Cancer Institute Tumor Repository. CT2A was obtained from Dr Peter Fecci (Duke University). Either 1 × 106 (subcutaneous) or 5 × 104 (intracranial) GL261 or CT2A cells were implanted into 6- to 10-week-old naïve syngeneic C57BL/6 mice. For intracranial experiments, tumor cells were resuspended in 5 µL phosphate buffered saline and injected into the right striatum of anesthetized syngeneic mice in a stereotactic frame. Subcutaneously implanted tumors were harvested when approximately 10 mm in greatest diameter (~2 wk for CT2A). Intracranially implanted tumors were harvested when mice became moribund.
Lymphocyte Isolation
Subcutaneous tumor-infiltrating lymphocytes (TILs) were isolated as previously described.6 Intracranial TILs were harvested by mechanical dissociation, and a Percoll density gradient was used to removed myelin and cellular debris. Draining lymph nodes and spleens were mechanically dissociated and filtered through a 70-micron cell strainer. Mononuclear cells were isolated from spleens by Ficoll-Paque PLUS density gradient (GE Healthcare Life Sciences).
DNA Whole-Exome and RNA Sequencing
Libraries were captured using the Agilent Mouse Exome reagent. Sequencing was performed on an Illumina HiSeq2000. Sequence coverage was as follows: C57BL/6 normal (92.1x) and CT2A tumor (82.4x). Data were aligned to reference sequence using Burrows–Wheeler Aligner v0.5.9, then merged and deduplicated using Picard v1.46 (https://broadinstitute.github.io/picard/). Single nucleotide variations (SNVs) were detected using the union of 3 callers: (i) Samtools vr96314 intersected with Somatic Sniper v1.2.15 and processed through false-positive filter v1; (ii) VarScan v2.2.616 filtered by VarScan high-confidence filter v1 and processed through false-positive filter v1; and (iii) Strelka v0.4.6.2.17 Insertions/deletions (indels) were detected using the union of 3 callers: (i) Genome Analysis Toolkit18 somatic-indel v5336 Pindel v0.5 filtered with Pindel false-positive and variant allele frequency (VAF) filters (params: --variant-freq-cutoff = 0.08); (ii) VarScan16 v2.2.6 filtered by VarScan high-confidence-indel version v1; and (iii) Strelka17 v0.4.6.2. SNVs and Indels were further filtered using a Bayesian classifier (https://github.com/genome/genome/blob/master/lib/perl/Genome/ Model/Tools/Validation/IdentifyOutliers.pm), retaining variants classified as somatic with a binomial log-likelihood of at least 3. Results were filtered to require expression of the mutant allele (fragments per kilobase of exon per million fragments mapped [FPKM] >1 and at least one variant-supporting read in the RNA) and VAF >1%.
MHC Class I Binding Predictions
The potential for CT2A nonsynonymous missense mutations to bind to H2-Db or H2-Kb molecules was predicted using the pVAC-seq pipeline (http://www.pvactools.org/).13 Predicted binding affinity of half-maximal inhibitory concentration (IC50) <500 nM by at least 1 of 6 prediction algorithms and a gene FPKM >1 were used at cutoffs for candidate neoantigens. Selection of candidates for screening was based on median predicted H2-Db or H2-Kb binding affinity and RNA-seq expression level (gene FPKM).
Peptide Immunization
For neoantigen immunogenicity screening, candidate neoantigens were divided into groups of 5 and administered as peptide pools (Peptide 2.0). Subcutaneous injections of pooled peptides (50 µg each) and 100 µg of polyinosinic-polycytidylic (polyIC) adjuvant were administered on days −10, −3, +4, +11, with contralateral subcutaneous CT2A tumor implantation on day 0. Spleens and tumors were harvested 2 weeks after implantation for ELISPOT assays. For survival experiments, synthetic long peptides were administered as subcutaneous injections (50 µg each) and 100 µg of polyIC adjuvant on days 3, 6, and 9, after intracranial tumor implantation.
(mEpb4: ELEQFESTIGFKLPNLRAAKRLWK; mPomgnt1: ECIIPDVSLSYHFGIVGLNMNGYF; mPlin2: QQLQTTCQTVLV NAQRLPQNIQDQA)
Enzyme Linked Immunospot Assays
Naïve splenocytes were plated at a concentration of 125 000 cells/well in 100 µL serum-free Cellular Technology Limited (CTL) media on precoated murine IFN-γ ELISPOT plates (CTL). CD8+ cells were isolated from spleens or tumors using magnetic bead-based positive selection kits (Stemcell Technologies). Effector cells were added to a final concentration of 25 000 CD8+ TILs/well, 400 000 CD8+ splenocytes/well, or 200 000 cells/well for draining lymph nodes, for a total volume of 200 µL/well with peptide (10 µM) (Peptide 2.0). Concanavalin A (1 µg/well) was used as a positive control. Plates were incubated overnight at 37oC and analyzed using the CTL ImmunoSpot kit. Positive responses were determined when a given neoantigen elicited a statistically significant CD8+ T cell response compared with media controls.
Flow Cytometry and Cytokine Detection
Flow cytometric analysis of lymphoid and myeloid populations was performed on TILs using multicolor staining panels comprising antibodies purchased from BioLegend (myeloid: Ly6C: HK1.4, F4/80: BM8, Ly6G: 1A8, B220: RA3–6B2, CD11b: M1/70, CD4 GK1.5, I-ab: 25–9–17, NK1.1: PK136, CD11c: N418, CD8: 53–6.7, CD3: 17A2, CD45: 30-F11, PD-L1: 10F.9G2; lymphoid: Lag-3: C9B7W, TIM-3: RMT3–23, CTLA-4: UC10–4B9, ICOS: C398.4A, FOXP3: MF-14, CD4: GK1.5, CD8: 53–6.7, TIGIT: 1G9, CD3: 145–2C11, Ox-40: OX-86, PD-1: 29F.1A12, CD19: 6D5, NK1.1: PK136, CD45: 104, 4–1BB: 17B5). Intracellular staining was performed using the eBioscience Fixation and Permeabilization Buffer kit. For intracellular cytokine staining, cells were stimulated with 50 nmol/L phorbol 12-myristate 13-acetate (PMA) and 500 nmol/L ionomycin for 4 to 6 hours at 37°C, 5% CO2, in the presence of 1 μg/mL brefeldin A (BD Biosciences). Surface staining was performed, and cells were fixed and permeabilized with the eBioscience Fixation and Permeabilization Buffer kit and stained for intraceullar IFN-γ, tumor necrosis factor alpha (TNF-α), and interleukin (IL)-2 (eBioscience Intracellular Fixation and Permeabilization Buffer Set, 88–8824–00). Analysis was performed on a BD Fortessa X-20 flow cytometer using FACSDiva software.
Differential Scanning Fluorimetry
H2-Kb and -Db heavy chains and β2-microglobulin (β2m) were each expressed in Escherichia coli strain BL21 and refolded from inclusion bodies, together with an ultraviolet (UV)-photocleavable peptide (FAPGNYPJL, where J represents the cleavable 3-amino-3-(2-nitro) phenyl-propionic acid), as described previously.19 UV-mediated peptide-exchange reactions were performed in 96-well plates (CellTreat Scientific Products, #229190), as described previously19 to obtain peptide/H2-Kb and H2-Db complexes of interest. DSF was performed using a QuantStudio 7 Flex System real-time PCR machine (Applied Biosystems), 384-well Microamp optical plates (Applied Biosystems, #4309849), and Microamp adhesive film (Applied Biosystems, #4311971), as described previously,19 for all peptide/H2-Kb and H2-Db complexes. H2-Db and H2-Kb heavy chains and β2m were each expressed in E. coli strain BL21 and refolded from inclusion bodies as described for human leukocyte antigen A*02:01.19 Each 20 µL DSF reaction consisted of 2.5 µL of 40x SYPRO Orange dye (Invitrogen), 8.5 µL molecular biology grade water, 5 µL assay buffer (defined below), and the 4 µL UV-exchanged complexes. SYPRO Orange dye binds to hydrophobic regions of proteins that are exposed upon denaturation, resulting in fluorescence enhancement. DSF assay buffer consisted of 40 mM HEPES, 600 mM NaCl, and 12 mM EDTA for most experiments at a pH of 7.4. The real-time PCR machine was programmed for a temperature ramp rate of 1°C per minute from 25° to 99°C. Triplicate data points for each peptide/MHC were analyzed using OriginPro 2018 software. Several peptides were unable to be examined as complexes due to either solubility or stability issues.
Statistical Analysis
For comparisons of intergroup differences in mean number of spots on ELISPOT and for comparisons of melting temperature (Tm) values between immunogenic and non-immunogenic neoantigens, a two-tailed Student’s t-test was used to determine significance, with P < 0.05 as statistically significant (GraphPad Prism).
Results
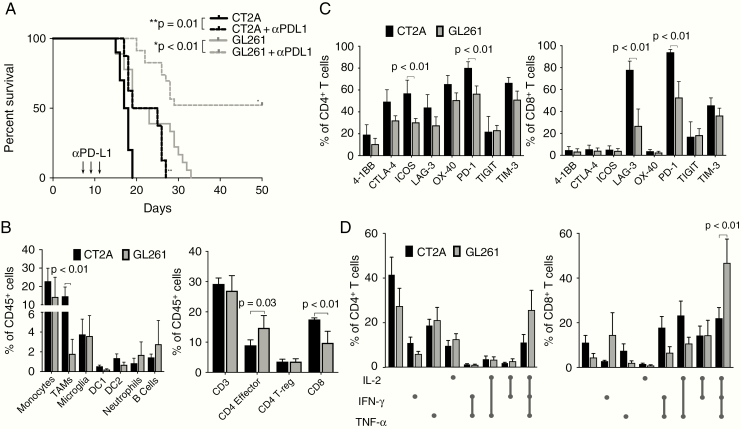
To determine the relative immunologic responses to checkpoint blockade in mice bearing GL261 or CT2A tumors, mice harboring either intracranial GL261 or CT2A GBMs were treated with anti–PD-L1 and monitored for overall survival. Although previous work has shown that a high percentage of GL261-bearing mice can reject tumors following checkpoint blockade treatment10–12 and that CT2A-bearing mice may be more resistant,20 we sought to compare responses between the 2 preclinical models directly. Treatment with anti–PD-L1 was started 7 days after intracranial tumor implantation and repeated every 2 days for a total of 3 treatments. Whereas untreated GL261-bearing mice exhibited a median survival of 21 days, 50% of GL261-bearing mice survived long term following treatment with anti–PD-L1 (Fig. 1A). In contrast, whereas untreated CT2A tumor–bearing mice experienced a median survival of 17.5 days, anti–PD-L1 treatment only modestly improved median survival to 22 days and did not lead to long-term survival.
Fig. 1.
Checkpoint blockade sensitivity and tumor-infiltrating immune cell distributions of GL261 and CT2A orthotopic murine GBMs. (A) Kaplan–Meier survival curves following immune checkpoint blockade using monoclonal antibodies against PD-L1 in mice bearing intracranial GL261 or CT2A tumors (n = 15 per cohort). (B) Immune profiling of predominant myeloid and lymphoid populations present in progressively growing intracranial GL261 and CT2A tumors (n = 5 per cohort). (C) Analysis of CD8+ and CD4+ T-cell expression of immune checkpoint molecules (n = 5 per cohort). (D) Intracellular cytokine analysis of GL261 and CT2A tumor-infiltrating CD8+ and CD4+ T cells. Boolean gating was performed to determine coexpression of IL-2, IFN-γ, and TNF-α.
To determine whether there were differences in the frequencies or phenotypes of immune cell populations that correlated with checkpoint blockade sensitivity, we assessed the composition and features of immune cells within the tumor microenvironments of mice with progressively growing GL261 or CT2A tumors by flow cytometry (Supplementary Figures 1–4). Although there was a trend toward increased absolute numbers of immune cells infiltrating into CT2A tumors, this finding was not statistically significant (data not shown). In the innate immune compartment, CD11b+ F4/80+ tumor-associated macrophages comprised a significantly greater proportion of CD45+ cells within CT2A tumors compared with GL261 tumors (Fig. 1B, left). There were no statistically significant differences between tumor-infiltrating monocytes, microglia, DC1 or DC2 dendritic cell subsets, neutrophils, or B cells between GL261 and CT2A. Furthermore, while percentages of total CD3+ T cells and regulatory T cells were comparable between GL261 and CT2A, CD4+/CD8+ T-cell ratios were decreased in CT2A tumors (Fig. 1B, right). Although CD3+ T cells made up similar proportions of the overall immune infiltrate in each tumor type, both CD4+ and CD8+ T cells exhibited greater expression of a panel of immune checkpoint molecules in CT2A tumors relative to GL261 tumors (Fig. 1C). Both inducible co-stimulator (ICOS) and PD-1 were significantly overexpressed within the CT2A CD4+ population, whereas lymphocyte-activation gene 3 (LAG-3) and PD-1 were the 2 predominant checkpoint molecules overexpressed by CT2A CD8+ T cells compared with those in GL261 tumors. Specifically, LAG-3 and PD-1 expression were identified in 80% and 95% of CD8+ T cells, respectively. In addition, we observed a trend of increased expression of 4-1BB, CTLA-4, LAG-3, OX-40, and T-cell immunoglobulin and mucin-domain containing 3 (TIM-3) within CD4+ T cells harvested from CT2A compared with GL261 tumors. Thus, the T cells within CT2A tumors express higher levels of regulatory checkpoint molecules compared with T cells within GL261.
Having evaluated the checkpoint molecule phenotypes of T cells in CT2A and GL261, we next evaluated the endogenous intracellular cytokine production profiles of tumor-infiltrating CD8+ and CD4+ T cells in GL261 and CT2A as a measure of TIL immune function immediately prior to tumor explant. TILs were isolated from GL261 and CT2A tumors once mice became moribund at approximately day 16 after intracranial tumor implantation, stimulated with PMA/ionomycin, and assessed for intracellular IFN-γ, IL-2, and TNF-α production by intracellular flow cytometry (Supplementary Figure 5). All combinations of cytokine production profiles were identified, and Boolean gating was performed to identify “triple positive” and “triple negative” cell populations, representing T cells staining positive or negative for all 3 cytokines, respectively. CT2A TILs contained significantly fewer triple-positive CD8+ T cells and trended toward a greater percentage of triple-negative CD8+ T cells compared with GL261 TIL (Fig. 1D), although not statistically significant. These same patterns between models were observed among CD4+ T-cell populations, although the differences did not reach statistical significance.
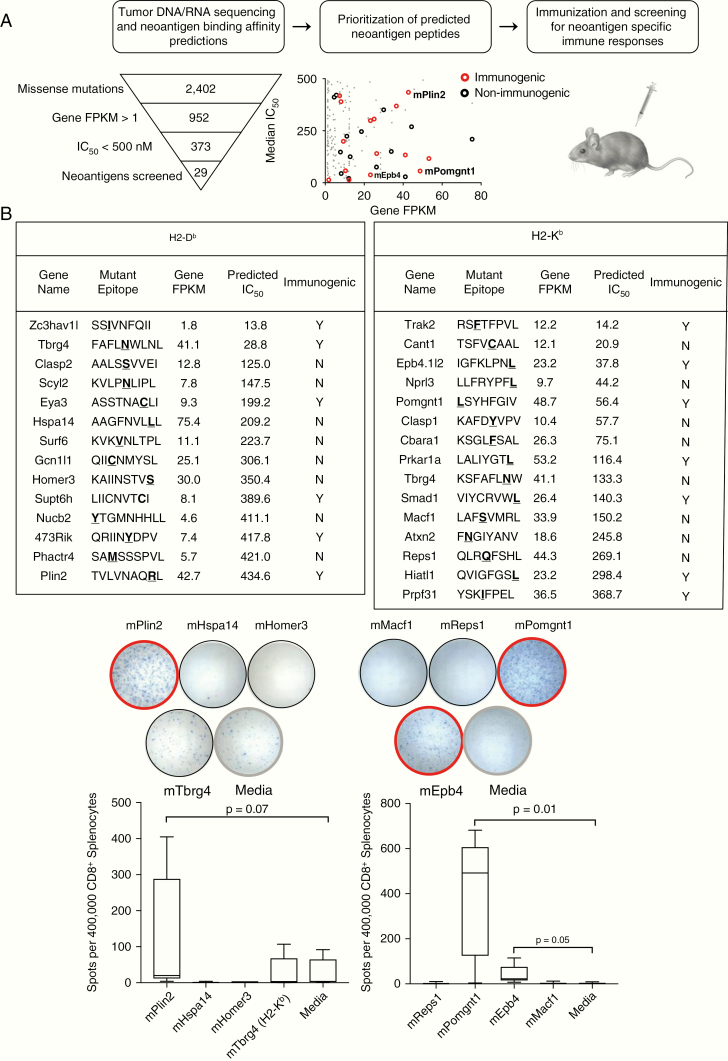
Because CT2A tumors were less responsive to anti–PD-L1 treatment, exhibited evidence of increased checkpoint regulatory expression, and harbored T cells with decreased capacity for cytokine secretion, we wished to determine whether T cells within CT2A tumors had the ability to recognize CT2A-specific tumor antigens. To determine the identities of potential neoantigen targets recognized by the host immune system, we used a cancer immunogenomics approach to characterize the somatic mutation burden and subsequent expressed candidate neoantigen landscape in CT2A, similar to previous work in the GL261 and SMA-560 preclinical GBM models.6 To this end, we employed DNA whole-exome sequencing and RNA sequencing to detect expressed, tumor-specific missense SNVs and frameshift mutations. This analysis revealed that CT2A harbored 2402 missense and 96 frameshift mutations, of which 952 were expressed as confirmed by RNA sequencing (gene FPKM >1) (Fig. 2A, Supplementary Table 1). We then employed the pVAC-seq bioinformatic pipeline13 to identify putative neoantigens predicted to bind class I MHC, H2-Db and H2-Kb. To determine candidate neoantigens, we prioritized candidates with median MHC H2-Db or H2-Kb binding affinities <500 nM and gene FPKM >1. Using these filters, we identified 193 H2-Db‒ and 180 H2-Kb‒restricted high-affinity neoepitopes in CT2A.
Fig. 2.
Cancer immunogenomics neoantigen discovery workflow. (A) Schematic of CT2A immunogenomic analysis and approach to identifying immunogenic neoantigens. Open circles represent top-ranking neoantigens that were selected for validation (red = immunogenic, black = non-immunogenic). (B) Left: Top 29 in silico H2-Db and H2-Kb candidate CT2A-derived neoantigens selected for immunological validation with corresponding amino acid sequence, gene FPKM, median IC50, and immunogenicity. Right: Representative wells from IFN-γ ELISPOT of CD8+ spleens stimulated with candidate neoantigens. Mice were immunized with pools of neoantigen candidates (n = 3 per cohort). Red outline = immunogenic neoantigens, black outline = non-immunogenic neoantigens.
To determine which of the predicted neoantigens could be recognized by the immune system, we characterized the immunogenicity of the top 14 H2-Db and 15 H2-Kb candidate neoantigen peptides that were prioritized by median IC50 and evidence of expression (Fig. 2B). To assess local and systemic T-cell responses to candidate neoantigens, mice bearing subcutaneous CT2A tumors were vaccinated with pools of neoantigen peptides, and both splenocytes and TILs were harvested. Of note, none of the combinations of pooled peptide vaccinations induced rejection of the subcutaneous tumors. Positively selected CD8+ T cells from harvested spleen and TIL population were tested for reactivity to individual neoantigen peptides and irrelevant control peptides by IFN-γ ELISPOT assays. Of the 29 candidates screened, 13 elicited CD8+ T-cell responses in the spleens of mice vaccinated with their mutant peptides (Fig. 2B, left). In addition, for 10 of the 13 candidates where immune responses were observed in the spleen, we also detected CD8+ T-cell responses in the TILs of vaccinated mice (Supplementary Figure 6). Representative ELISPOT data are shown for immunogenic neoantigens mPlin2, mEpb4, and mPomgnt1 (red circles) and non-immunogenic neoantigens (black circles) (Fig. 2B, right).
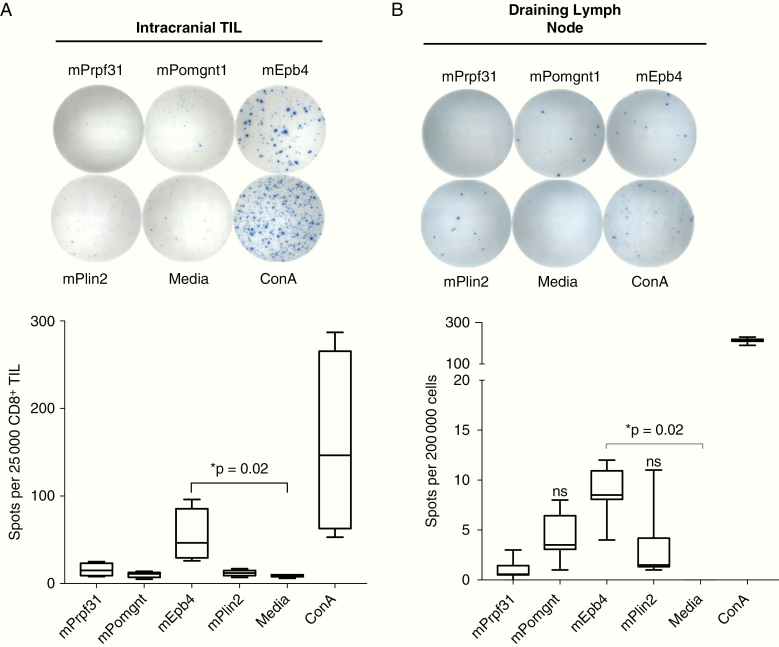
Having demonstrated that 13 of 29 tested neoantigens could elicit CD8+ T cells in response to vaccination, we next asked whether we could detect evidence of endogenously primed immune responses to immunogenic neoantigens following orthotopic tumor implantation. For these experiments, we prioritized screening neoantigens where T-cell reactivity had been detected in both the spleen and TILs of immunized mice, and selected 9 candidates for evaluation of endogenous immune responses. In non-immunized mice, CD8+ TILs isolated from orthotopically transplanted CT2A brain tumors demonstrated increased IFN-γ production following stimulation with the H2-Kb restricted neoantigen, Epb4H471L (Fig. 3A). Within cervical draining lymph nodes, we also detected IFN-γ activation in response to Epb4H471L as well as to the H2-Db Plin2G332R and H2-Kb Pomgnt1R497L neoantigens (Fig. 3B). No endogenous immune responses were detected against the other 6 candidates tested. Reactivity to all 3 neoantigens was also detected in the spleens of intracranial tumor–bearing mice, albeit at a significantly lower frequency than was found in the TILs or lymph nodes (data not shown).
Fig. 3.
Identification of neoantigen-reactivity in intracranial TILs and draining cervical lymph nodes in CT2A. (A) Representative images from IFN-γ ELISPOT of intracranial CT2A CD8+ TILs stimulated with candidate neoantigens. TILs were isolated from intracranial tumors at day 16 and used the same day for experiments. Twenty-five thousand CD8+ TILs/well were incubated overnight with indicated peptide (10 µM) and assessed for IFN-γ production by ELISPOT. (B) Representative images from IFN-γ ELISPOT of draining cervical lymph nodes (200 000 cells/well) from mice bearing intracranial CT2A tumors. Presented data depict pooled results from 3 experiments with 2–3 mice per experiment.
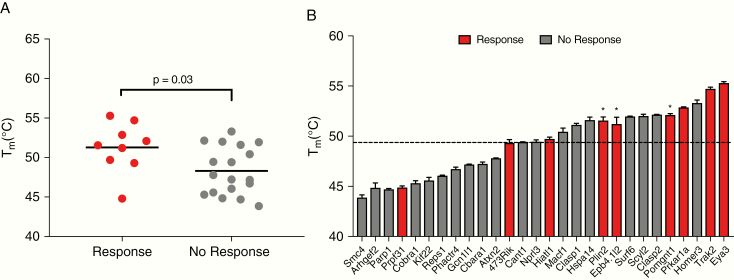
We next explored the correlation between neoantigen immunogenicity and peptide-MHC stability for the CT2A candidate neoantigens. Of 29 neoantigens tested for immunogenicity, 13 generated immune responses, while endogenous CD8+ T cell responses were detected against 3 of those epitopes. However, these findings did not correlate with median predicted binding affinity or gene expression (Fig. 2A and data not shown). Prior work21,22 has suggested that peptide-MHC stability, rather than binding affinity, may represent an improved predictor of immunogenicity. Furthermore, recent studies have shown that peptide-MHC stability can be used to accurately identify immunogenic neoantigens across multiple patient samples.19,21 Therefore, to assess the role of peptide-MHC stabilities in identifying immunogenic neoantigens, we employed a DSF assay19 to determine the thermal stability (Tm) of selected neoantigen/MHC. DSF analysis revealed that immunogenic CT2A neoantigens demonstrated significantly higher Tm values compared with CT2A neoantigens for which no T-cell response was detected (Fig. 4A). Moreover, the 3 neoantigens that elicited endogenous immune responses—mEpb4, mPomgnt1, and mPlin2—all harbored Tm values above the average (49.3°C) for all CT2A neoantigens (Fig. 4B). Thus, increased stability of the neoantigen peptide-MHCs correlated with observed immunogenicity.
Fig. 4.
Peptide-MHC stability predicts neoantigen immunogenicity. Experimentally determined thermal stabilities (Tm) of peptide-MHCs as measured by DSF for CT2A neoantigens that did or did not induce a T-cell response. (B) Red bars represent predicted CT2A neoantigens that were shown to be immunogenic; gray bars represent neoantigens for which no T-cell responses were detected. Values in (A) and (B) represent means of triplicates. The dotted line represents the average Tm for tested peptides.
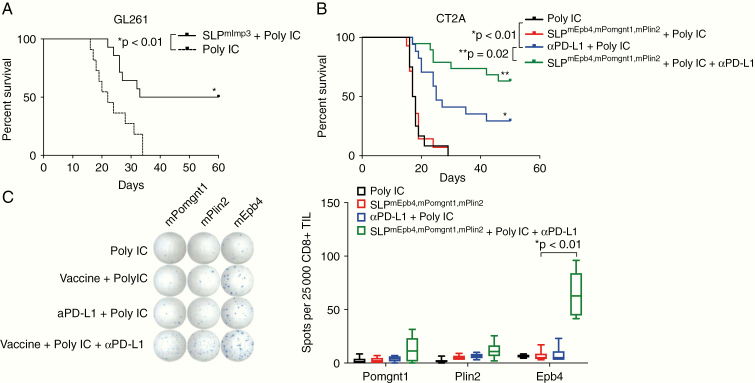
Finally, given the presence of spontaneously arising neoantigen-specific immune responses with the CNS and secondary lymphoid organs of glioma bearing mice detected in both GL2616 and in CT2A, we tested the effect of therapeutically targeting neoantigens using personalized vaccines. Because GL261-bearing mice exhibited responses to anti–PD-L1 (Fig. 1A10,12), we determined whether a vaccine targeting the mutant Imp3 neoantigen, to which we have previously identified spontaneous immune responses using immunogenomics approaches6 could substitute for this treatment. Whereas GL261-bearing mice treated with polyIC adjuvant alone exhibited a median survival of 22 days, vaccination with the mImp3 synthetic long peptide (SLP) and polyIC adjuvant on days 3, 6, and 9 after tumor implantation resulted in a significant survival benefit and extended median survival to 46.5 days. We then applied this approach to the highly aggressive CT2A tumor. We focused on creating a vaccine that targeted the neoantigens to which we observed endogenous immune responses. Thus, we generated the SLPmEbp4,mPomgnt1,mPlin2 vaccine comprising 27-mer SLPs targeting the mutant Plin2G332R, Pomgnt1R497L, and Epb4H471L neoepitopes as well as polyIC adjuvant. To determine the clinical efficacy of neoantigen vaccination, CT2A-bearing mice were treated with the adjuvant polyIC alone, neoantigen vaccine and polyIC, combination anti–PD-L1 + polyIC, or combination of therapeutic vaccine with adjuvant and anti–PD-L1 blockade. Vaccinated mice were treated on days 3, 6, and 9 after tumor implantation, and mice treated with anti–PD-L1 were treated on days 7, 9, and 11. Mice treated either with adjuvant alone or with SLPmEbp4,mPomgnt1,mPlin2 vaccine with adjuvant exhibited a median overall survival of 17.5 days, and mice treated with anti–PD-L1 with adjuvant exhibited a median overall survival of 25 days (Fig. 4B). In contrast, 60% of mice treated with combination SLPmEbp4,mPomgnt1,mPlin2” vaccine and anti–PD-L1 blockade demonstrated long-term survival. To determine if vaccination increased the presence of neoantigen-specific T cells within TILs, we performed ELISPOT analysis on TILs from CT2A-bearing mice treated with polyIC alone, neoantigen vaccine and poly IC, combination anti–PD-L1 + polyIC, or combination of therapeutic neoantigen vaccine with adjuvant and anti–PD-L1 blockade (Fig. 5C). Compared with other treatment regimens, there was a statistically significant increase in tumor-infiltrating mEpb4-specific CD8+ TIL as well as a trend toward increased numbers of mPomgnt1- and mPlin2-specific T cells in response to the combination of checkpoint blockade and neoantigen vaccine. Taken together, combination neoantigen vaccination and checkpoint blockade treatment improved the overall survival of mice harboring the aggressive CT2A tumor.
Fig. 5.
Polyvalent neoantigen vaccination provides synergistic protection when combined with anti–PD-L1 checkpoint blockade therapy in CT2A. (A) Treatment of intracranial GL261-bearing mice with mImp3 SLP neoantigen vaccine and polyIC adjuvant on days 3, 6, and 9 post tumor implantation. (B) Intracranial CT2A-bearing mice were treated with a polyvalent vaccine comprising the mEbp4, mPomgnt1, and mPlin2 neoantigens (days 3, 6, and 9); anti–PD-L1 therapy (days 7, 9, and 11); polyIC adjuvant alone (days 3, 6, and 9); or combination polyvalent neoantigen vaccination plus anti–PD-L1 treatment. (C) ELISPOT analysis of TILs from intracranial CT2A-bearing mice treated with a polyvalent vaccine comprising the mEbp4, mPomgnt1, and mPlin2 neoantigens (days 3, 6, and 9); anti–PD-L1 therapy (days 7, 9, and 11); polyIC adjuvant alone (days 3, 6, and 9); or combination polyvalent neoantigen vaccination plus anti–PD-L1 treatment. Shown are raw ELISPOT data (left) and quantitation (right).
Discussion
In this study, we characterized the immunogenomic landscape of the aggressive orthotopic GBM CT2A model and, using this information, tested the efficacy of treatment of tumor-bearing mice with a CT2A-specific neoantigen vaccine combined with anti–PD-L1 blockade combination. We found that the orthotopic GBM model, CT2A, exhibited several immunologic features consistent with human GBM, including high tumor infiltrate of macrophages and evidence of hypofunctional T cells.22–24 In addition, we compared the sensitivity to checkpoint blockade of CT2A and GL261. While anti–PD-L1 monotherapy induced survival in a subset of GL261-bearing mice consistent with previous studies,10,12 treatment provided only a limited benefit to CT2A-bearing mice. In order to identify CT2A-specific neoantigens with which to study endogenous antitumor immune responses as well as to develop a therapeutic vaccine, we applied a cancer immunogenomics methodology to predict expressed neoantigen candidates. We detected endogenous immune responses to mutant Plin2G332R, Pomgnt1R497L, and Epb4H471L neoantigens and then showed for the first time that combining a neoantigen vaccine targeting these epitopes with anti–PD-L1 was more effective than either treatment as monotherapy.
Although the therapeutic vaccination of cancer has a long history,25 limited success was observed, likely due to the nature of antigens targeted as well as the lack, at the time, of other immune stimulating combination agents such as checkpoint inhibitors. Of the classes of cancer antigens that have been identified, neoantigens represent compelling therapeutic targets due to (i) their cancer-specific expression, (ii) the potential lack of tolerance to them due to their absence during immunologic development, and (iii) feasibility of candidate identification using widely employed genomics approaches. Since the conception of their systematic identification,26 numerous studies have demonstrated that at least a subset of genomically identifiable candidate neoantigens could be recognized by the immune system both in preclinical models and in patients. In addition, our group has previously applied a cancer immunogenomics approach to show that endogenous immune responses can be generated to GBM neoantigens within the brain.6 Recently, personalized neoantigen vaccine trials have been developed against a growing number of cancer types, including GBM, following the initial reports of experiences in melanoma.27–29 For patients with GBM, 3 recent studies showed that the neoantigen vaccines in GBM patients are safe and feasible and can generate GBM-specific immune responses.7–9 Ongoing studies (www.clinicaltrials.gov, NCT03422094) combine GBM neoantigens with various combinations of checkpoint inhibitors to determine how to optimize the generation of GBM-specific immune responses.
Thus, the work described with the CT2A model is highly relevant to current efforts within the clinical trial landscape in patients with GBM. The observation that combination neoantigen vaccine and anti–PD-L1 treatment significantly augments the effect of either treatment alone points to another use for adjuvant checkpoint blockade in GBM beyond its use as a monotherapy to date. This study describes the first proof of concept in a preclinical model that combining a personalized vaccine with checkpoint blockade may represent an effective therapeutic strategy which may be relevant to other cancers in addition to GBM. Although checkpoint blockade has revolutionized the treatment of a growing number of advanced malignancies,30 recent studies in newly diagnosed and recurrent GBM4 have not demonstrated clinical efficacy of anti–PD-1 treatment as a monotherapy in these settings. However, the recent observation of clinical benefit in a subset of GBM patients treated with anti–PD-1 in the neoadjuvant setting provides evidence that checkpoint blockade may be beneficial in the appropriate context but insufficient to drive rejection.31 Given the complex ability of GBM to abrogate host antitumor immune responses, targeting T-cell inhibitory receptors alone may be insufficient in the presence of weak preexisting antitumor immune responses. Thus, therapeutic efforts aimed at driving effector immune cells and stimulating their activation in the GBM microenvironment coupled with strategies to alleviate tumor immune suppression represent a rational combinatorial treatment approach. In GBM and in other cancers, clinical benefit with checkpoint blockade is likely to be augmented through combination approaches. In the study by Ott et al,28 2 patients with metastatic melanoma treated with a personalized neoantigen vaccine demonstrated clinical responses after progression as well as an expansion of neoantigen-specific T-cell responses when anti–PD-1 treatment was added to the therapeutic vaccine. Thus, while there are important immunologic insights to be gained from other preclinical GBM models, the CT2A model represents a platform that models the response of most human GBM diseases to single anti–PD-1/PD-L1 blockade and enables the development of strategies to enhance responsiveness or to explore alternative immune treatments.
Additional studies of the CT2A tumor revealed important features of its associated microenvironment. In order to gain a deeper understanding of the infiltrating immune cell distribution in brain tumors exhibiting distinct responses to treatment, we used high dimensional flow cytometry analysis to compare the phenotypes and functions of freshly isolated immune cells from orthotopic CT2A and GL261 tumors. Characterizations of human GBMs show that up to 30% of the tumor mass can comprise tumor-associated macrophages (TAMs), a cell type that has been linked to accelerated disease progression and poor outcomes in cancer patients.24,25 In our studies, we found evidence of significant accumulation of TAMs within the CT2A GBM environment. Furthermore, the markers of T-cell exhaustion were more highly expressed in the CT2A model, with CD8+ T cells exhibiting higher levels of LAG-3 and PD-1 and CD4+ T cells exhibiting higher levels of ICOS and PD-1. In particular, the observation of elevated LAG-3 and TIM-3 expression among CT2A CD8+ TILs represents a compelling finding, as upregulation of these alternative immune checkpoints has been implicated in mechanisms conferring resistance to conventional checkpoint blockade24,32 and antagonistic antibodies have been developed to inhibit their function. Similar to results of other published studies in CT2A, we observed that higher expression of phenotypic markers were associated with T-cell exhaustion and greater reductions in T-cell cytokine output.24 Collectively, these findings of local tumor immune dysregulation in an anti–PD-L1 resistant murine GBM support the use of CT2A as a platform to evaluate targeted, combination immune treatment strategies in GBM. Given the distinct immunophenotypic profile of CT2A TILs, additional studies are warranted to better understand transcriptional programs associated with immune therapy resistance.
Our genomic analysis of the GL26133 and CT2A models revealed that both tumors harbor tumor mutational burden (TMB) significantly greater than that typically observed in primary, newly diagnosed human GBM. Indeed, there are several scenarios in GBM in which a higher TMB state is observed. First, up to 20–30% of GBMs that recur following temozolomide chemotherapy exhibit the hypermutated genotype, due in large part to DNA base mispairing caused by alkylation which is propagated in the setting of mutations in DNA mismatch repair genes.34–38 Moreover, low-grade gliomas treated with temozolomide can also recur at a higher grade with the hypermutated genotype.36,39 Secondly, a rare subset of newly diagnosed GBM patients, in both the pediatric40 and adult settings,6 can also present with tumors harboring the hypermutated genotype. Thus, given the mutational burden in the GL261 and CT2A models, it is conceivable that they could represent valuable systems for studying the immune responses to the high TMB GBM state, their distinct responses to checkpoint blockade notwithstanding. Indeed, a number of studies have demonstrated a strong association between increased mutational burden and response to checkpoint blockade therapy in non-small-cell lung cancer,41,42 melanoma,43–45 bladder cancer,46 and others.47–49 This association has also been observed in anecdotal studies in adult6 and pediatric40 GBM. Interestingly, we showed that CT2A-bearing mice exhibited increased survival when treated with a combination of checkpoint blockade and the polyIC adjuvant, albeit not as significant as with additional neoantigen vaccine treatment. It is possible that, in high TMB settings, GBM may be susceptible to combinations of immunotherapies that bypass the need for personalized immunogenomics. In particular, polyIC is a toll-like receptor 3 agonist that activates dendritic cells in draining lymph nodes and within the tumor microenvironment. In combination with anti–PD-L1, this adjuvant may stimulate an immune response to CT2A that is augmented with directed neoantigen targeting. These compounds, as well as other potent immunostimulatory adjuvants, merit further study both alone and in combination with other immunotherapies.
Ultimately, the mutational burden in these cell lines reflects their genesis by means of chemical carcinogenesis rather than tumor evolution through treatment or germline disposition, and thus there are acknowledged differences between these preclinical models and some features of human disease. Additional models, such as the checkpoint blockade–resistant SB28 C57BL/6 model derived from in vivo genetic targeting using the sleeping beauty transposon system,50 harbor a lower mutational burden closer to the absolute number of exome-wide mutations (108)11 typically seen in adult GBM and therefore may also model relevant parameters of human GBM. Nevertheless, both the GL261 and CT2A cell lines remain models for neoantigen discovery and for studying the immune response to CNS tumors. Importantly, the higher the mutational burden, the more difficult it is to identify candidate neoantigens to which an immune response can be detected. Thus, the immunologic validation of predicted neoantigens in higher TMB models is a particularly strong confirmation of the in silico process we and others have employed for immunogenomics-drive discovery.
Our findings provide further validation that cancer immunogenomics approaches can be successfully employed to identify immunogenic neoantigens in murine brain tumor models, extending results we previously obtained in 2 other GBM models.33 Our sequencing and epitope prediction algorithms were similar to those used previously; we identified somatic missense mutations through exome sequencing of tumor and normal tissue, followed by mutation expression analysis by RNA sequencing. While a number of studies have reported the use of internally developed immunogenicity scores to improve neoantigen identification, these metrics have not been well validated. We employed a neoantigen discovery approach that prioritized predicted high-binding affinity candidates that were also transcriptionally present at high levels within the tumor. Our results indicate that CD8+ T-cell responses can be detected against neoantigens spanning both a range of predicted binding affinities within the IC50 < 500 nM limit as well as normalized gene expression scores. While the consideration of gene expression from RNA sequencing data improved our overall ability to identify immunogenic neoantigens, further enhancements to in silico neoantigen selection will be critical given increasing interest in the use of neoantigen-directed therapies. Indeed, we found that no predicted affinity or genomic expression metric was able to distinguish immunogenic neoantigens from those in which no T-cell response could be detected. Further work will also be directed to understanding the role of other sources of neoantigens such as posttranslational modifications and noncoding sequences.51
Some studies have demonstrated that peptide-MHC stability is a more accurate predictor of immunogenicity than binding affinity.52,53 To test the added value of peptide-MHC stability in identifying relevant neoantigens, we employed a DSF assay19 using temperature denaturation to measure thermal stability of peptide-MHC for CT2A neoantigens. DSF analysis of H2-Kb–restricted CT2A neoantigens revealed that higher Tm values were more predictive of T-cell reactivity than the conventional NetMHC54 peptide binding affinities. Given that testing the immunogenicity of all predicted peptides can be a slow and costly process, high-throughput evaluation of peptide-MHC stability may represent an additional efficient and orthogonal method of identifying immunogenic neoantigens where patient samples are limited or in instances of large tumor neoantigen burdens. Furthermore, once the factors influencing peptide-MHC interactions are more fully understood, it should be possible to develop algorithms that improve the success rate of neoantigen-based vaccines.
Taken together, this study provides novel preclinical evidence to suggest that combining vaccine efforts to clonally expand neoantigen-specific T cells with checkpoint blockade could represent a therapeutic approach to GBM. Additional work is needed to understand how best to vaccinate against identified targets, which combination treatments may be most effective, and how to rationally address the intratumoral heterogeneity in GBM with spatially targeted polyvalent methods. Moreover, although we have focused on CD8+ T cells in this study, further studies on the role of CD4+ T cells in anti-GBM responses will be critical to understand their role in therapeutic optimization.
Funding
This work was supported by National Institutes of Health grants K08NS092912 and R01NS112712 (G.P.D.), R01CA178844 (D.M.K.); the Damon Runyon Cancer Research Foundation (G.P.D.); and the Howard Hughes Medical Institute (C.J.L.).
Supplementary Material
Acknowledgments
We would like to thank Dan Yoakum and Abigail Brown for assistance with the DSF assays.
Conflict of interest statement
G.P.D. is a co-founder of Immunovalent, which has no bearing on this paper.
Authorship statement
Experimental design and execution: CJL, MS, DTB, JAB, AL, DKK, DB, DMK, TMJ, GPD. Data analysis and interpretation: CJL, MS, DTB, JAB, DB, CAM, DKK, TMJ, GPD. Manuscript writing: CJL, DTB, DKK, GPD. Manuscript review: CJL, MS, DTB, JAB, DKK, DB, DKK, TMJ, GPD.
References
- 1. Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 2. Liau LM, Ashkan K, Tran DD, et al. First results on survival from a large phase 3 clinical trial of an autologous dendritic cell vaccine in newly diagnosed glioblastoma. J Transl Med. 2018;16(1):142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weller M, Butowski N, Tran DD, et al. ; ACT IV trial investigators Rindopepimut with temozolomide for patients with newly diagnosed, EGFRvIII-expressing glioblastoma (ACT IV): a randomised, double-blind, international phase 3 trial. Lancet Oncol. 2017;18(10):1373–1385. [DOI] [PubMed] [Google Scholar]
- 4. Omuro A, Vlahovic G, Lim M, et al. Nivolumab with or without ipilimumab in patients with recurrent glioblastoma: results from exploratory phase I cohorts of CheckMate 143. Neuro Oncol. 2018;20(5):674–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gubin MM, Artyomov MN, Mardis ER, Schreiber RD. Tumor neoantigens: building a framework for personalized cancer immunotherapy. J Clin Invest. 2015;125(9):3413–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Johanns TM, Miller CA, Dorward IG, et al. Immunogenomics of hypermutated glioblastoma: a patient with germline POLE deficiency treated with checkpoint blockade immunotherapy. Cancer Discov. 2016;6(11):1230–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johanns TM, Miller CA, Liu CJ, et al. Detection of neoantigen-specific T cells following a personalized vaccine in a patient with glioblastoma. Oncoimmunology. 2019;8(4):e1561106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Keskin DB, Anandappa AJ, Sun J, et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature. 2019;565(7738):234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hilf N, Kuttruff-Coqui S, Frenzel K, et al. Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature. 2019;565(7738):240–245. [DOI] [PubMed] [Google Scholar]
- 10. Reardon DA, Gokhale PC, Klein SR, et al. Glioblastoma eradication following immune checkpoint blockade in an orthotopic, immunocompetent model. Cancer Immunol Res. 2016;4(2):124–135. [DOI] [PubMed] [Google Scholar]
- 11. Genoud V, Marinari E, Nikolaev SI, et al. Responsiveness to anti-PD-1 and anti-CTLA-4 immune checkpoint blockade in SB28 and GL261 mouse glioma models. Oncoimmunology. 2018;7(12):e1501137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zeng J, See AP, Phallen J, et al. Anti-PD-1 blockade and stereotactic radiation produce long-term survival in mice with intracranial gliomas. Int J Radiat Oncol Biol Phys. 2013;86(2):343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hundal J, Carreno BM, Petti AA, et al. pVAC-Seq: a genome-guided in silico approach to identifying tumor neoantigens. Genome Med. 2016;8(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li H, Handsaker B, Wysoker A, et al. ; 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Larson DE, Harris CC, Chen K, et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28(3):311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28(14):1811–1817. [DOI] [PubMed] [Google Scholar]
- 18. McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Blaha DT, Anderson SD, Yoakum DM, et al. High-throughput stability screening of neoantigen/HLA complexes improves immunogenicity predictions. Cancer Immunol Res. 2019;7(1):50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nakashima H, Alayo QA, Penaloza-MacMaster P, et al. Modeling tumor immunity of mouse glioblastoma by exhausted CD8+ T cells. Sci Rep. 2018;8(1):208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Strønen E, Toebes M, Kelderman S, et al. Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science. 2016;352(6291):1337–1341. [DOI] [PubMed] [Google Scholar]
- 22. Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4(1):71–78. [DOI] [PubMed] [Google Scholar]
- 23. Bloch O, Crane CA, Kaur R, Safaee M, Rutkowski MJ, Parsa AT. Gliomas promote immunosuppression through induction of B7-H1 expression in tumor-associated macrophages. Clin Cancer Res. 2013;19(12):3165–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Woroniecka K, Chongsathidkiet P, Rhodin K, et al. T-cell exhaustion signatures vary with tumor type and are severe in glioblastoma. Clin Cancer Res. 2018;24(17):4175–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rosenberg SA, Yang JC, Restifo NP. Cancer immunotherapy: moving beyond current vaccines. Nat Med. 2004;10(9):909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Segal NH, Parsons DW, Peggs KS, et al. Epitope landscape in breast and colorectal cancer. Cancer Res. 2008;68(3):889–892. [DOI] [PubMed] [Google Scholar]
- 27. Carreno BM, Magrini V, Becker-Hapak M, et al. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science. 2015;348(6236):803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ott PA, Hu Z, Keskin DB, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547(7662):217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sahin U, Derhovanessian E, Miller M, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547(7662):222–226. [DOI] [PubMed] [Google Scholar]
- 30. Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018;359(6382):1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cloughesy TF, Mochizuki AY, Orpilla JR, et al. Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat Med. 2019;25(3):477–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Williams JB, Horton BL, Zheng Y, Duan Y, Powell JD, Gajewski TF. The EGR2 targets LAG-3 and 4-1BB describe and regulate dysfunctional antigen-specific CD8+ T cells in the tumor microenvironment. J Exp Med. 2017;214(2):381–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Johanns TM, Ward JP, Miller CA, et al. Endogenous neoantigen-specific CD8 T cells identified in two glioblastoma models using a cancer immunogenomics approach. Cancer Immunol Res. 2016;4(12):1007–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hunter C, Smith R, Cahill DP, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66(8):3987–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. TCGA Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Johnson BE, Mazor T, Hong C, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim J, Lee IH, Cho HJ, et al. Spatiotemporal evolution of the primary glioblastoma genome. Cancer Cell. 2015;28(3):318–328. [DOI] [PubMed] [Google Scholar]
- 38. Wang J, Cazzato E, Ladewig E, et al. Clonal evolution of glioblastoma under therapy. Nat Genet. 2016;48(7):768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van Thuijl HF, Mazor T, Johnson BE, et al. Evolution of DNA repair defects during malignant progression of low-grade gliomas after temozolomide treatment. Acta Neuropathol. 2015;129(4):597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bouffet E, Larouche V, Campbell BB, et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J Clin Oncol. 2016;34(19):2206–2211. [DOI] [PubMed] [Google Scholar]
- 41. Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hellmann MD, Nathanson T, Rizvi H, et al. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell. 2018;33(5):843–852 e844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350(6257):207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Johnson DB, Frampton GM, Rioth MJ, et al. Targeted next generation sequencing identifies markers of response to PD-1 blockade. Cancer Immunol Res. 2016;4(11):959–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rosenberg JE, Hoffman-Censits J, Powles T, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387(10031):1909–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Goodman AM, Sokol ES, Frampton GM, Lippman SM, Kurzrock R. Microsatellite-stable tumors with high mutational burden benefit from immunotherapy. Cancer Immunol Res. 2019;7(10):1570–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yarchoan M, Johnson BA 3rd, Lutz ER, Laheru DA, Jaffee EM. Targeting neoantigens to augment antitumour immunity. Nat Rev Cancer. 2017;17(4):209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chan TA, Yarchoan M, Jaffee E, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30(1):44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kosaka A, Ohkuri T, Okada H. Combination of an agonistic anti-CD40 monoclonal antibody and the COX-2 inhibitor celecoxib induces anti-glioma effects by promotion of type-1 immunity in myeloid cells and T-cells. Cancer Immunol Immunother. 2014;63(8):847–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Laumont CM, Vincent K, Hesnard L, et al. Noncoding regions are the main source of targetable tumor-specific antigens. Sci Transl Med. 2018;10(470):eaau5516.. [DOI] [PubMed] [Google Scholar]
- 52. Harndahl M, Rasmussen M, Roder G, et al. Peptide-MHC class I stability is a better predictor than peptide affinity of CTL immunogenicity. Eur J Immunol. 2012;42(6):1405–1416. [DOI] [PubMed] [Google Scholar]
- 53. van der Burg SH, Visseren MJ, Brandt RM, Kast WM, Melief CJ. Immunogenicity of peptides bound to MHC class I molecules depends on the MHC-peptide complex stability. J Immunol. 1996;156(9):3308–3314. [PubMed] [Google Scholar]
- 54. Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics. 2016;32(4):511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.