Abstract
Background
Systemic infiltration of the brain by tumor cells is a hallmark of glioma pathogenesis which may cause disturbances in functional connectivity. We hypothesized that aggressive high-grade tumors cause more damage to functional connectivity than low-grade tumors.
Methods
We designed an imaging tool based on resting-state functional (f)MRI to individually quantify abnormality of functional connectivity and tested it in a prospective cohort of patients with newly diagnosed glioma.
Results
Thirty-four patients were analyzed (World Health Organization [WHO] grade II, n = 13; grade III, n = 6; grade IV, n = 15; mean age, 48.7 y). Connectivity abnormality could be observed not only in the lesioned brain area but also in the contralateral hemisphere with a close correlation between connectivity abnormality and aggressiveness of the tumor as indicated by WHO grade. Isocitrate dehydrogenase 1 (IDH1) mutation status was also associated with abnormal connectivity, with more alterations in IDH1 wildtype tumors independent of tumor size. Finally, deficits in neuropsychological performance were correlated with connectivity abnormality.
Conclusion
Here, we suggested an individually applicable resting-state fMRI marker in glioma patients. Analysis of the functional connectome using this marker revealed that abnormalities of functional connectivity could be detected not only adjacent to the visible lesion but also in distant brain tissue, even in the contralesional hemisphere. These changes were associated with tumor biology and cognitive function. The ability of our novel method to capture tumor effects in nonlesional brain suggests a potential clinical value for both individualizing and monitoring glioma therapy.
Keywords: functional connectivity , functional MRI , glioblastoma , glioma, resting state
Key Points.
1. Functional MRI showed that functional connectivity is affected by glioma throughout the whole brain, even in the nonlesional hemisphere.
2. Functional MRI provides information about the individual glioma disease burden in the whole brain.
Importance of the Study.
Glioma is a systemic disease because tumor cells can spread throughout the whole brain. Current imaging tools, however, provide information only about the main tumor and its immediate surroundings. Hence, we developed a method which uses fMRI to provide information about individual disease burden in the whole patient brain. Using fMRI, we assessed whole brain connectivity in glioma patients and found that connectivity is impaired throughout the brain, even in sites far from the main tumor. Importantly, connectivity is more disturbed in patients with more aggressive tumors. This finding might open up the possibility of designing treatment concepts which are tailored to the individual disease severity and disease distribution in the patient brain, possibly making existing and future treatment regimens more effective.
Glioma constitutes a systemic disease of the brain with tumor cells spreading far beyond the macroscopically visible lesion and forming networks throughout the whole brain.1,2 The World Health Organization (WHO) grading system reflects the clinical prognosis, with worst prognosis in grade IV gliomas (glioblastoma).3 Assessing the status of nonlesional brain in glioma patients could provide crucial information about the aggressiveness of the disease in the individual patient,4 which will enable risk stratification on an individual basis and allow for personalized treatment strategies, tailored to the individual disease burden. To date, however, there is no imaging modality able to capture the full extent of how glioma affects the whole brain. Conventional MRI and PET provide only structural and metabolic information about the visible lesion but neglect the nonlesional brain. An imaging modality that provides information about nonlesional brain tissue in glioma patients could thus represent a major advance in the management of these patients. A potential tool for this purpose is resting-state functional MRI (rsfMRI), which assesses functional connectivity in the whole brain by tracking intrinsic fluctuations of the blood oxygenation level dependent (BOLD) signal.5,6
There are two potential mechanisms of how glioma cells might alter functional connectivity. First, on a microscopic level, glioma cells have been shown to migrate far beyond the tumor itself. This can lead to a breakdown of functional connectivity, as was recently shown in an animal model of glioblastoma.7 We therefore suspected that invasion of glioma cells into the human brain compromises the integrity of neuronal connections and that the aggressiveness of the tumor and consecutively the disease severity is correlated with the extent of the damage to neuronal connections. Second, the ability of the brain vasculature to respond to neuronal activation with increased flow of oxygenated blood is severely compromised in glioma patients. In the healthy brain, astrocytes regulate blood flow in response to neuronal activation.8 There is an extensive body of evidence which shows that invading glioma cells displace astrocytic processes from the perivascular space.9,10 This leads to a loss of the ability of astrocytes to induce vasoconstriction or vasodilation in response to neuronal activity in glioma patients.11 It is thus possible that functional connectivity, measured by the BOLD signal, is disturbed in brain areas with glioma cell invasion.
In order to test our hypothesis, we designed a novel rsfMRI-based imaging tool and applied it in a prospective cohort of patients with de novo glioma. Our imaging tool, called the Abnormality Index (ABI), quantifies damage to functional connectivity in glioma patients by comparing the strength of functional connectivity in each voxel with a reference cohort of 1000 healthy controls. We found that ABI is positively correlated with WHO tumor grade, which in turn is closely correlated with invasiveness of tumor cells.4 Furthermore, there was an association with isocitrate dehydrogenase (IDH) mutation status, with higher ABI observed in IDH wildtype gliomas, which are known to be more invasive than IDH mutated gliomas.12 Clinical features of glioma patients, like neurocognitive performance, were inversely correlated with ABI.
Materials and Methods
Patients and Study Design
Inclusion criteria were: suspected, newly diagnosed glioma and age over 18 years. Exclusion criteria were: previous cranial surgery, more than one epileptic seizure, anti-epileptic medication, neuropsychiatric comorbidities, and any contraindications to MR scanning such as metal implants. Patients with epilepsy or with long-term use of anti-epileptic drugs (AEDs) were excluded because of the well-documented effects of seizures and AEDs on functional connectivity.13,14 Histological confirmation of the diagnosis was obtained either by stereotactic biopsy or open resection. Diagnosis was made according to the criteria of the WHO classification system in the revised version of 2016.3 Patients were followed with routine clinical visits after initiation of therapy. In a subgroup of patients (n = 13), neuropsychological testing was conducted prior to the first scan using the Montreal Cognitive Assessment (MOCA) test.15 The study was reviewed and approved by the local institutional review board of Ludwig Maximilians University Munich. All study procedures were in accordance with the Declaration of Helsinki. Written informed consent was obtained from all patients.
Scanning Procedure Functional MRI
Resting-state functional (f)MRI data were collected on a 3T Skyra scanner (Siemens Medical Solutions) using a 20-channel head and neck coil. Subjects were instructed to stay awake and keep their eyes closed; no other task instruction was provided. Functional images were acquired using an echo planar imaging (EPI) pulse sequence (repetition time [TR] = 3000 ms, echo time [TE] = 30 ms, flip angle = 90°, 3 × 3 × 3 mm3 voxels, 120 timepoints per run). Each participant underwent 2 resting-state runs of 6 minutes each, resulting in a total of 240 timepoints. Structural MRI scans were acquired using a sagittal 3D T1-weighted sequence (radiofrequency pulses and magnetization prepared rapid acquisition gradient echo [MPRAGE] sequence, 1 × 1 × 1 mm3 voxels).
18F-FET-PET Scanning Procedure
In a subset of 15 patients, 18F-fluoro-ethyl-tyrosine (FET)-PET was performed on a Biograph 64 PET/CT scanner (Siemens Medical Solutions). After injection of approximately 180 mega-becquerel FET, images were acquired for 40 minutes, and 20–40 minutes summation images were used for static evaluation. The maximal standardized uptake value (SUVmax) and the tumor-to-background ratios (TBRs) were assessed as described previously.16 Patients with a TBR of <1.6 compared with the visually unaffected contralateral hemisphere were considered FET negative.
Genomics Superstruct Project Sample
The reference sample consisted of 1000 healthy subjects (mean age, 21.3 y; range, 18–35; ~43 % male). The data were collected as part of the Brain Genomics Superstruct Project of Harvard University and Massachusetts General Hospital.17 This sample has been described in detail in prior publications.18–20 Resting-state fMRI data were collected on one of 3 different matched 3 tesla TimTrio scanners (Siemens Medical Solutions) using a 12-channel head and neck coil. Subjects were instructed to remain still, stay awake, and keep their eyes open. Functional images were acquired using an EPI pulse sequence (TR = 3000 ms, TE = 30 ms, flip angle = 85°, 3 × 3 × 3 mm3 voxels). Each participant underwent one or two resting-state runs (average of 1.7 runs) of 6.2 minutes each, resulting in 124 timepoints per run. Structural MRI scans were acquired using a sagittal 3D T1-weighted sequence (radiofrequency pulses and MPRAGE, 1 × 1 × 1 mm3 voxels).
Preprocessing
Resting-state fMRI data were processed using previously described procedures.18,21,22 Preprocessing included (i) slice timing correction (SPM2 software, Wellcome Department of Cognitive Neurology), (ii) rigid body correction for head motion with the FSL package,23,24 (iii) normalization for global mean signal intensity across runs, and (iv) low-pass temporal filtering, head motion regression, and ventricular, white matter, and whole brain signal regression. Whole brain signal regression was also included in the processing stream, which can improve the correction of motion-related artifacts.25,26 All subjects included in this study had met the quality control criteria of slice-based temporal signal-to-noise ratio (tSNR) >100 and of mean relative motion <0.05 mm.20 Structural data were processed using the FreeSurfer v5.3.0 software package (http://surfer.nmr.mgh.harvard.edu). To lower computation burden, we uniformly subsampled the cerebrum in the FreeSurfer nonlinear volumetric space (voxel size 8 × 8 × 8 mm3, non-gap), resulting in 1635 isometric voxels in the left hemisphere, 1479 isometric voxels in the right hemisphere, and a total of 3114 voxels. Alignment of functional and structural images was carried out using FsFast v4.5 (http://surfer.nmr.mgh.harvard.edu).
Calculation of the Abnormality Index in Glioma Patients
In order to detect and quantify altered functional connections in individual patients, we compared the connection strength of each link in an individual patient with the mean and standard deviation of the connection strength of this link derived from 1000 healthy subjects. Specifically, functional connectivity was assessed at the whole brain level by taking each of the 3114 voxels in the cerebrum as a seed and calculating its connectivity strength (defined as the correlation strength r of the BOLD signal time course) to all other voxels. The connectivity map based on each seed voxel i can be denoted as a vector Fi,
where i = 1, 2, …, 3114, representing the 3114 voxels in the cerebrum.
For each vector Fi we calculated the mean and standard deviation of correlation values r across 1000 subjects, resulting in a vector called MEANi and a vector called STDi.
We then compared each patient’s vector Fi(p), where P = 1, 2, …, 34, representing 34 patients, with the mean and standard deviation across the 1000 healthy subjects, resulting in an abnormality score (ABS) for each connection involving seed voxel i:
where j = 1, 2, …, 3114, and j ≠ i, representing all voxels in the cerebrum other than the seed voxel.
In order to quantify abnormality for each voxel, the connections whose absolute ABS exceeded a certain threshold z were counted, resulting in an abnormality count (ABC) per voxel:
Connections with z > 4.0 (corresponding to an uncorrected P-value of 0.0001) were considered significantly abnormal.
In order to quantify abnormality for each hemisphere, the ABC was summarized in the lesional hemisphere (LE) and in the contralesional hemisphere (CL), and normalized by the number of voxels in the respective hemisphere, resulting in an abnormality index (ABI):
Where x is the number of voxels in the lesional hemisphere
where y is the number of voxels in the contralesional hemisphere
Visualization
For each patient, the ABC of each voxel was assigned to the respective voxel of the FreeSurfer nonlinear volumetric template (voxel size 2 × 2 × 2 mm3). The resulting maps were smoothed using a Gaussian kernel (sigma = 2.3, mean filtering, kernel weighted). Smoothed maps were then registered back to the individual patient’s anatomy using nonlinear registration (Functional Magnetic Resonance Imaging of the Brain’s [FMRIB] nonlinear image registration tool24) as implemented in FSL (the FMRIB software library). Fig. 1 shows example maps of individual patients.
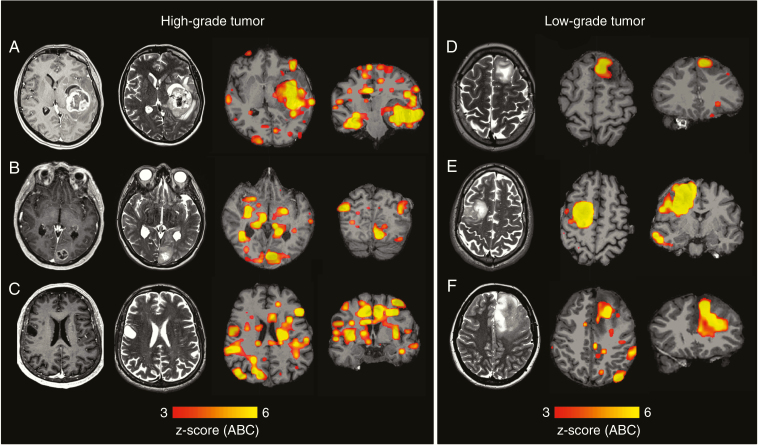
Fig. 1.
Examples of individual abnormality maps show clinical value of examining the connectome in glioma patients. (A) Structural scans from patient 25 (IDH wildtype glioblastoma WHO grade IV) with gadolinium-enhanced T1 on the far left and T2 images second from left. On the right, maps with individual functional data are shown. Functional maps depict the z-score of the voxel-wise abnormality count (ABC). Severe damage to functional connectivity is observed in both the lesional and the non-lesional hemisphere. In a similar fashion, (B) (patient 9; IDH wildtype glioblastoma WHO grade IV) shows extensive damage to functional connectivity in both hemispheres. (C) Structural and functional scans of patient 12 (IDH1 mutated glioblastoma, WHO grade IV) are shown. On gadolinium-enhanced T1 images (far left) no contrast enhancement by the tumor is noted, making the structural scans suggestive of a low-grade lesion. The functional scans (second from right and far right) with widespread damage to functional connectivity are suggestive of a high-grade tumor which is consistent with the final diagnosis. (D) T2 images of patient 23 (IDH-mutated oligodendroglioma, WHO grade II) as well as functional scans. Here the disturbances in functional connectivity are limited to the area of the tumor, the contralateral hemisphere appears intact. (E, F) Similarly (patient 15 and 34), large tumors which exhibit favorable histopathological and molecular features (IDH-mutated oligodendroglioma WHO grade II) are shown. Here, an area with high abnormality in the location of the tumor is seen while very little abnormality is observed in the rest of the brain.
Association with WHO Grade, IDH Mutation Status, Metabolic Information from PET, and Cognition
Associations of the ABI with WHO grade, MOCA scores, and SUV scores were established using Spearman rank correlation. Specifically, analyses were conducted using “partial correlation coefficients adjusted for internal variables” as implemented in MATLAB (https://www.mathworks.com/help/stats/partialcorri.html). Variables that were adjusted for included age (categorical), tSNR of the resting-state runs, and tumor volume.
Tumor volume was manually segmented by consensus reading (one radiologist, one neurosurgeon) on contrast-enhanced T1-weighted images and on non-enhanced T2-weighted images using Osirix v9.4. In non-enhancing tumors the T2-positive volume was segmented, while in enhancing tumors both the enhancing volume as well as non-enhancing tumor visible on T2-weighted images were segmented. The hereby defined volume will be referred to as the lesion throughout the paper. Furthermore, tumor location was recorded as a categorical variable (left/right and lobe, eg, right frontal) and was evaluated separately for influence on ABI using regression analysis. Group comparisons of the ABI in patients with IDH wildtype versus IDH mutated tumors were conducted for the lesional and contralesional hemisphere using one-way ANOVA as implemented in MATLAB (https://de.mathworks.com/help/stats/anova1.html).
Reliability of the Abnormality Index
In order to assess test-retest reproducibility of the ABI in tumor patients, we applied a so-called split-session approach that we have tested and validated in previous studies.27,28 Briefly, the raw data from the fMRI resting-state session were split into two halves and treated as test-retest data. The ABI of the lesional and contralesional hemisphere was then calculated based solely on the information of the first or second half of the rsfMRI session, respectively. Reproducibility was then estimated using Pearson’s correlation coefficient.
Data Availability
Genome Sequencing Program data are publicly available.17 Patient data and the code of the ABI are available from the corresponding authors upon request.
Results
Histology, Molecular Markers, and Clinical Course
Thirty-eight patients (mean age 48.7 ± 16.9 y, 14 female) were prospectively included between September 2015 and July 2017. After quality control of the data, 4 patients were excluded because of motion artifacts. Thirty-four glioma patients were retained for subsequent analyses. Table 1 shows a summary of patient characteristics. Detailed histological and molecular characteristics for each individual patient can be found in Supplementary Table 1. Eight patients died during the follow-up period (last follow-up July 2018, mean follow-up 15 mo across all 34 patients; range, 1‒32 mo). Median overall survival was therefore not reached. Sixteen patients underwent primary resection while 18 received a stereotactic biopsy. An overview of treatment data, follow-up, and overall survival for each patient is given in Supplementary Table 2.
Table 1.
Patient characteristics (N = 34)
| Age, y, mean ± SD (range) | 48.7 ± 16.9 (21–80) |
| Sex, n | |
| Female | 14 |
| Male | 20 |
| Tumor histology, n | |
| Oligodendroglioma | 8 |
| Diffuse astrocytoma | 5 |
| Anaplastic oligodendroglioma | 1 |
| Anaplastic astrocytoma | 5 |
| Glioblastoma | 15 |
| WHO grade, n | |
| II | 13 |
| III | 6 |
| IV | 15 |
| IDH1/2 mutation status, n | |
| Wildtype | 20 |
| Mutated | 14 |
| MOCA (mean ± SD, n = 14) | 22.1 ± 5.76 |
| Overall survival, mo, range, n = 8 | 7–17 |
The Abnormality Score Not Only Detects the Tumor but also Functional Changes in Nonlesional Brain Tissue
When the voxelwise ABC was projected to the individual anatomical space, it overlapped with the macroscopic tumor, as depicted on T2-weighted images and contrast-enhanced T1-weighted images (Fig. 1). However, voxels with a high ABC were detected not only in the lesional tissue, but also in nonlesional brain tissue in the lesional and contralesional hemisphere. For example, Fig. 1A shows a glioblastoma in the left temporal lobe that is well detected by the ABC. Similarly, Fig. 1B shows a tumor in the left occipital lobe which is also well detected by individual analysis of the patient connectome. In both cases, there are several clusters of increased ABC that spread beyond the tumor itself into the nonlesional brain tissue of the tumor-bearing and the contralesional hemisphere. Interestingly, the abnormality count did not appear to be simply driven by tumor size (see analyses below for more details about the effect of tumor volume). Fig. 1C shows a small T2-hyperintense tumor without contrast enhancement. According to structural MRI diagnostic criteria, this lesion would be suspected to be a low-grade tumor. However, this small tumor caused extensive damage to functional connectivity in both hemispheres and the final histopathological diagnosis indicated glioblastoma.
The involvement of nonlesional brain tissue was much less pronounced in low-grade tumors (Fig. 1D). Again, this was observed independent of tumor size (Fig. 1E, F). Here, large low-grade tumors are shown, which, in spite of the size of the tumor, cause disturbances in functional connectivity largely confined to the tumor with almost intact connectivity in the nonlesional hemisphere.
Tumor location was not found to influence ABI in a statistically significant manner on regression analysis (P > 0.05).
The Abnormality Index Is Significantly Associated with WHO Grade
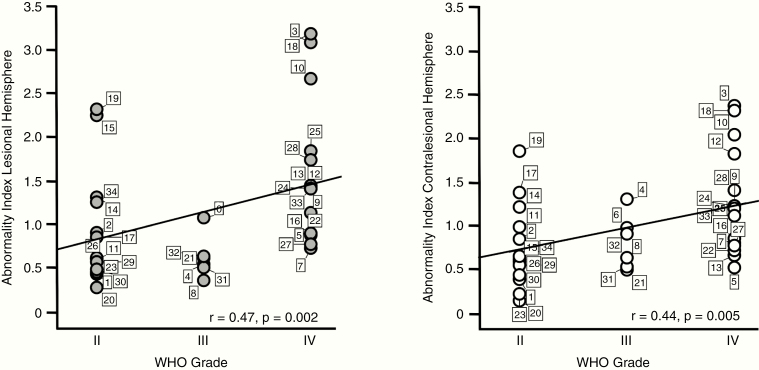
Quantitative analyses indicated that the ABI was significantly associated with WHO grade of the patients, when calculated based both on the lesional hemisphere (r = 0.47, P = 0.002) as well as on the nonlesional hemisphere (r = 0.44, P = 0.005; Fig. 2). In a next step, we aimed at assessing the impact of covariates like age, tumor volume, and data quality on this association. We found that age was significantly correlated with the ABI of both the lesional (r = 0.73, P < .0001) and the contralesional (r = 0.67, P < .0001) hemisphere. Interestingly, tumor volume was only correlated with the ABI of the lesional hemisphere (r = 0.334, p = 0.027), but not with the ABI of the contralesional hemisphere (r = 0.029, P = 0.871). Temporal SNR of the fMRI data was not significantly correlated with the ABI of the lesional or contralesional hemisphere (P > 0.05, respectively). To account for the impact of age and tumor volume, and the potential influence of tSNR variation, these covariates were included in a partial correlation analysis of ABI and WHO grade. The association remained significant, even when controlling for these covariates, for the lesional (r = 0.33, P = 0.04) and more importantly the nonlesional (r = 0.32, P = 0.04) hemisphere. This indicates that independently of age, tumor size, and tSNR, there was still a significant association between WHO grade and ABI.
Fig. 2.
The abnormality index is significantly associated with WHO grade. Scatter plots show the ABI of each individual patient (total n = 34) in the lesional hemisphere (left panel, gray dots) and contralesional (right panel, white dots) hemisphere. Patients are grouped according to WHO grade and are marked with their individual patient number (see Supplementary Tables 1 and 2 for patient characteristics). The ABI of both the lesional and contralesional hemisphere was significantly correlated with WHO grade.
The Abnormality Index Is Significantly Elevated in Patients with IDH Wildtype Tumors
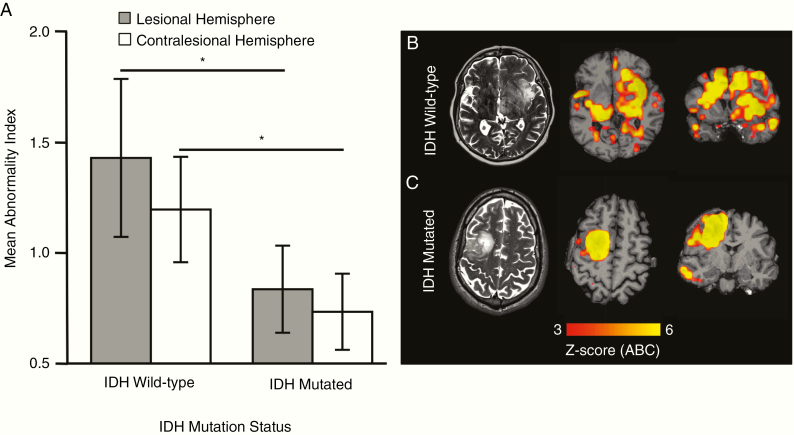
When patients were grouped according to IDH mutation status, the ABI was significantly elevated in patients with IDH wildtype tumors compared with patients with IDH mutant tumors. This could be shown with the ABI calculated based on the nonlesional and the lesional hemisphere (P = 0.016 and P = 0.010, respectively; Fig. 3A). Regarding IDH-mutation status, Fig. 3B shows a tumor which was histologically classified as low grade (diffuse astrocytoma) but showed genetic aberrations indicative of a glioblastoma-like lesion (IDH wildtype, telomerase reverse transcriptase [TERT]) mutation). Accordingly, connectome analysis showed widespread damage in both hemispheres, which is typical for high-grade tumors. In contrast, Fig. 3C shows an IDH-mutated low-grade glioma. Although the tumor volume was high, connectivity changes were mostly confined to the tumor, with high ABI in the lesional hemisphere, accounting for the large tumor volume, but low ABI in the contralesional hemisphere. For results of analyses of receiver operating characteristic curve showing the diagnostic value of ABI to predict IDH mutation status, refer to Supplementary Figure 4.
Fig. 3.
The abnormality index is significantly elevated in patients with IDH wildtype tumors. (A) The ABI of both the lesional hemisphere (indicated in gray, mean ABI ± 2 standard deviations) and the contralesional hemisphere (indicated in white, mean ABI ± 2 standard deviations) was significantly higher in IDH wildtype glioma patients than in IDH-mutated glioma patients (P < 0.05, respectively, two-tailed t-test). (B) The individual connectome map (z-score of voxelwise abnormality count) of a patient with an IDH wildtype, TERT-mutated glioma with bilateral severe abnormality (patient 19). (C) The connectome map of a patient with an IDH1-mutated grade II oligodendroglioma with connectivity changes largely confined to the tumor itself.
The Abnormality Index Is Significantly Associated with Neurocognition
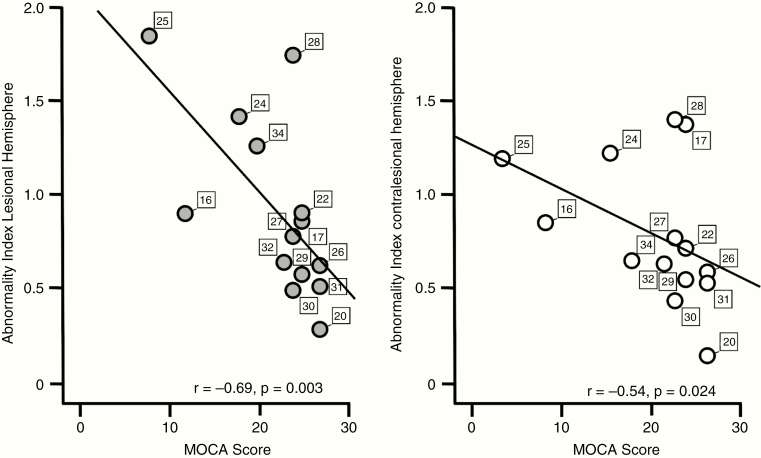
The results of neurocognitive testing using the MOCA test were inversely correlated with the ABI. We found a significant association of MOCA score with the ABI of the lesional hemisphere (r = −0.69, P = 0.003; Fig. 4A) and with the ABI of the contralesional hemisphere (r = −0.54, P = 0.024; Fig. 4B), ie, low MOCA scores were associated with high ABI. As MOCA scores were significantly correlated with rsfMRI data quality as indicated by SNR (r = −0.55, P = 0.04), we accounted for its potential impact in a partial correlation analysis and found that the correlation remained significant for both the lesional (r = −0.56, P = 0.022) and the contralesional (r = −0.64, P = 0.009) hemisphere. Age and tumor volume were not significantly correlated with the patients’ performance in the MOCA test (P > 0.05, respectively).
Fig. 4.
The abnormality index is significantly associated with neurocognition. Neurocognitive performance measured by the MOCA score was inversely correlated with the ABI score of both the lesional hemisphere (left panel) and the contralesional hemisphere (right panel). Patients are marked with their individual patient number.
The Abnormality Index Is Associated with Metabolic Information Obtained by PET
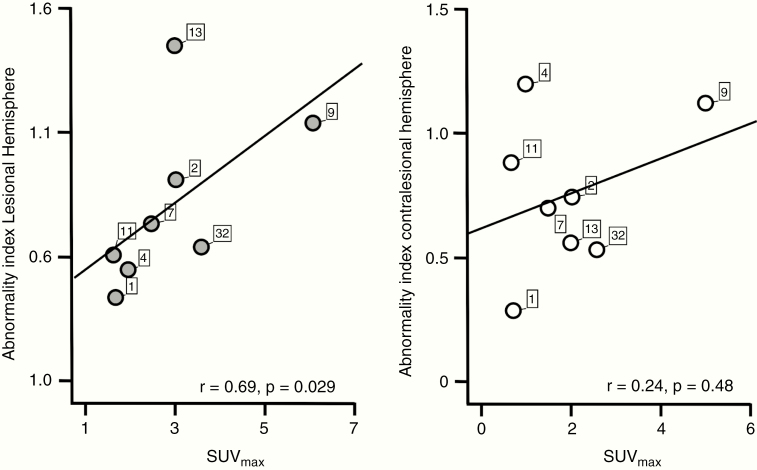
Prior to any therapy, FET-PET scans were obtained in 15 patients (grade II, n = 6; grade III, n = 5; and grade IV, n = 4). In 7 cases, no FET enhancement was noted. In 8 cases, ipsilateral PET-positive findings were reported. In none of these patients was FET enhancement noted in the contralesional hemisphere. In FET-PET positive patients, we found a significant positive correlation between SUVmax and the ABI of the lesional hemisphere (r = 0.69, P = 0.03; Fig. 5) but not the contralesional hemisphere (r = 0.24, P = 0.48). There was no significant correlation between SUVmax and age, tumor volume, or tSNR of the fMRI data (P > 0.05, respectively).
Fig. 5.
The abnormality index is associated with metabolic information obtained by PET. X-axes of scatter plots show the maximal standardized uptake value (SUVmax) of tumors that showed increased uptake on FET-PET (n = 8, out of 15 patients who underwent FET-PET scanning). Y-axes indicate each patient’s ABI of the lesional (left panel, gray dots) and contralesional (right panel, white dots) hemisphere. ABI of the lesional hemisphere was significantly associated with SUVmax (P < 0.05), while the association of SUVmax and the ABI of the contralesional hemisphere failed to reach statistical significance (P > 0.05).
The Abnormality Index Shows High Test-Retest Reliability
In order to estimate reliability of the ABI in tumor patients, we applied a so-called split-session approach, which we have tested and validated in previous studies.27,28 We found that ABI scores were highly reproducible, with a strong correlation between test and retest results in the lesional (r = 0.83, P < 0.0001) and contralesional (r = 0.80, P < 0.0001) hemisphere.
Discussion
It was the aim of our study to investigate whether diffuse glioma of the brain leads to disturbances in whole brain functional connectivity and whether the extent of impaired functional connectivity can reflect disease severity in individual patients. For this purpose, a potential imaging marker (ABI) was developed and tested in glioma patients. We found that the ABI is associated with WHO grade (ie, high ABI in high-grade gliomas), IDH mutation status (ie, high ABI in IDH wildtype tumors), and neurocognitive performance (ie, low performance when ABI was high). These findings are significant in a number of ways. The current imaging standard of care for patients with suspected brain neoplasms is contrast-enhanced MRI, sometimes supplemented by special MRI studies like spectroscopy or fiber tract imaging. In addition to MRI studies, PET imaging using amino acid tracers provides additional information about extent and biological aggressiveness of the tumor.29 These imaging modalities are able to localize macroscopically visible tumor parts and characterize them further. They, however, do not provide information about the nonlesional brain. Our imaging marker, on the other hand, is based on a global analysis of functional connectivity which is not restricted to single functional networks and therefore captures tumor effects on the entire brain.
The abnormality in functional connectivity observed in our patient cohort may be attributed to 2 main causes. First, the tumor causes disruption in neuronal networks, affecting neuronal projections and therefore functional connectivity in all brain regions neuronally linked to the site of the tumor. This notion is supported by our results, because we found that tumor volume was moderately correlated with the ABI in the lesional hemisphere. An additional effect on functional connectivity in the lesional hemisphere, especially in the vicinity of the tumor, might arise from potential neuroplastic effects, as it has been observed in serial intraoperative functional mappings in glioma patients.30 In high-grade tumors, however, we observed significant functional abnormality in the nonlesional hemisphere, which cannot readily be explained by the effects of the main tumor alone, as the ABI in the nonlesional hemisphere was independent of tumor volume. An example of tumors with similar volumes but very differential impacts on the connectome is given in Fig. 3B, C. We thus suspect there is a second mechanism by which glioma causes damage to functional connectivity. There is significant tumor cell invasion in high-grade glioma4 and we observed high ABI in these patients. It therefore seems possible that abnormality in functional connectivity, especially in the nonlesional hemisphere, might not only be caused by disruption of large-scale functional networks involving 2 hemispheres but could also be related to tumor cell invasion. Although, at this point one can only speculate that microscopic tumor invasion contributes to the connectivity changes in macroscopically “normal” brain tissue as reported in this study, this interpretation is supported by the literature reporting the formation of widely distributed tumor cell networks via microtubes in glioblastoma,2 and changes at the synaptic and transmitter levels leading to altered neuronal activity that further fosters the formation of such glioma cell networks.31
The assumption that tumor cell invasion contributes to disturbances in functional connectivity detected here is further strengthened by the fact that glioma cell invasion is linked to dysregulation of cerebral blood flow. It has been shown that invading glioma cells cause the loss of physiological astrocyte function in regulating blood flow in the brain, especially in response to neuronal activation.8,11 This phenomenon has been described previously in brain tumor patients undergoing fMRI.32 As stated above, the BOLD signal has its basis in blood flow fluctuations in the brain which are caused by neuronal activation. Therefore, disruptions in cerebral blood flow which are caused by other factors than disturbances in neuronal connections are also picked up by techniques which rely on the BOLD signal. The damage to functional connectivity, which we have observed in glioma patients, might consecutively be representative not only for damage to neuronal connections but also for tumor-related disturbances in brain homeostasis in general. This is supported by our finding that high metabolic activity of the tumor on FET-PET imaging was correlated with ABI.
Recently, genetic characterization of brain tumors has gained increased clinical importance. There is strong evidence showing that molecular alterations in gliomas indicate the clinical course of the disease and are now integrated into treatment algorithms.33,34 Among the molecular markers studied, IDH mutation status has emerged as one of the most powerful predictors of patient outcome. In light of this development, there have been efforts to find imaging markers for non-invasive determination of IDH mutation status in glioma patients. So far, promising results have been published using specialized MRI sequences, some of them requiring equipment which is not widely available, such as 7T MRI scanners.35,36 There is still, however, no clinically established imaging marker for IDH mutation status. We found that patients with IDH wildtype glioma had significantly higher abnormality of functional connectivity, irrespective of histopathological grade. Furthermore, it was recently shown that IDH wildtype tumors might be more invasive than IDH mutant ones.37 Taken together, this too indicates that the ABI may reflect changes associated with glioma cell invasion. This is further supported by a recent study by Kesler et al which showed increased damage to the structural connectome in patients with IDH wildtype astrocytomas.38
We would like to point out, however, that it was not the aim of our study to find an imaging marker for histology or genetic characteristics of a tumor. Invasive measures are and will be necessary to determine these factors. Rather, our novel measure of abnormality in the functional connectome could serve as a potential additional source of information for the clinician when trying to determine how severely an individual patient is affected by the disease. We believe that the abnormality score can serve as a marker for (whole brain) disease severity in glioma patients by gaining additional information from nonlesional tissue. The overall aim of this study was to validate this novel measure, by demonstrating that it is associated with known markers of disease severity in glioma patients like WHO grade and IDH status.
If damage to functional connectivity is indeed an indicator of tumor cell invasion, this hypothesis needs to be verified with histological techniques. At this point, however, biopsies guided by ABI, especially of the contralateral hemisphere, were not felt to be ethically justified. Should it turn out, however, that the ABI derived from rsfMRI provides information about the extent of tumor invasion in the nonlesional hemisphere, this might fuel discussions about whether patients with IDH wildtype tumors and massive impairment of contralateral connectivity are suitable candidates for local therapies like open tumor resection at all.
Our findings suggest that deficits in functional connectivity could also be related to the clinical course of the patients. We found that neurocognitive function is negatively correlated with the ABI, especially in the lesional hemisphere, suggesting a strong influence of the main tumor on cognitive performance. This is in line with a recently published meta-analysis of studies on neurocognitive performance in glioma patients which showed a strong influence of the tumor on neurocognitive performance.39 Additionally, we found that there is a trend toward association of the ABI with overall survival, even though the significance of this finding might be limited by the small number of events in the overall survival analysis and by the heterogeneity of treatment in our cohort (Supplementary Figure 1). Regardless, this finding is another indicator that damage to functional connectivity might also be viewed as predictive of the clinical course of glioma patients. Further studies to validate this finding are necessary, however.
Clinical implementation of the ABI might be facilitated by the fact that special equipment is not required. Rather, the necessary sequences can be tacked onto a clinical routine MRI scan without major modification to the scanning procedure. The post analysis of imaging data can be done automatically and can be easily performed as a clinical routine. Furthermore, the ABI is an objective parameter, which is largely independent of patient cooperation, as its acquisition is task independent. Thus, it is also not dependent on cognitive status and language function of the patient, further easing clinical use. Additionally, the possibility to generate individual abnormality maps might prove useful in clinical practice. Pending further clinical validation, these maps might be used for delineating target volumes for therapies such as radiation or for therapy monitoring (eg, during the course of chemotherapy). As an example, Supplementary Figure 2 shows a low-grade glioma at baseline before temozolomide chemotherapy and after 3 and 6 cycles. There is a marked decrease in functional abnormality, which might indicate tumor response as well as the capability to restore impaired connectivity.
In summary, we propose a potential imaging marker in individual glioma patients based on whole brain functional connectivity. This imaging signature correlates with tumor-specific features such as WHO grade and molecular status. Furthermore, we found an association with neurocognitive performance. This technique has the potential to enhance the information which can be gained from conventional structural MRI, perhaps allowing for a more holistic assessment of disease burden in the individual glioma patient.
Ultimately, histological correlations from autopsy and/or biopsy studies are necessary to validate this imaging marker in glioma patients. Longitudinal patient studies are furthermore warranted to further investigate therapy effects on ABI and to test if ABI changes precede tumor progression or recurrence.
Supplementary Material
Funding
This work was supported by National Natural Science Foundation of China grants no. 81790650 and 81790652, all to H.L.; National Institutes of Health grants 1R01NS091604, 1R01DC017991, and 1R21MH121831; the Beijing Municipal Science & Technology Commission grant no. Z161100002616009; and postdoctoral fellowship grant no. 57370322 by the Dr Mildred Scheel Cancer Foundation in cooperation with the Deutscher Akademischer Austauschdienst (DAAD) to F.G., a career development grant by Ludwig Maximilians University to S.S., and a grant by Friedrich-Baur-Foundation to V.S.
Conflict of interest statement. The authors report no competing interests.
Authorship statement. Conception and design: VMS, SS, JCT, HL. Development of methodology: VMS, SS, FG, HL. Acquisition of data: VMS, SS, JR, MU, NLA, FWK, NT, TL, BEW. Analysis and interpretation of data: VMS, SS, FG, JR, JCT, HL. Writing, review, and/or revision of the manuscript: VMS, SS, FG, JR, MU, NLA, FWK, NT, TL, BEW, JCT, HL.
References
- 1. Sahm F, Capper D, Jeibmann A, et al. Addressing diffuse glioma as a systemic brain disease with single-cell analysis. Arch Neurol. 2012;69(4):523–526. [DOI] [PubMed] [Google Scholar]
- 2. Osswald M, Jung E, Sahm F, et al. Brain tumour cells interconnect to a functional and resistant network. Nature. 2015;528(7580):93–98. [DOI] [PubMed] [Google Scholar]
- 3. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 4. Palfi S, Swanson KR, De Boüard S, et al. Correlation of in vitro infiltration with glioma histological type in organotypic brain slices. Br J Cancer. 2004;91(4):745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34(4):537–541. [DOI] [PubMed] [Google Scholar]
- 6. Smith SM, Fox PT, Miller KL, et al. Correspondence of the brain’s functional architecture during activation and rest. Proc Natl Acad Sci U S A. 2009;106(31):13040–13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hadjiabadi DH, Pung L, Zhang J, et al. Brain tumors disrupt the resting-state connectome. Neuroimage Clin. 2018;18:279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Attwell D, Buchan AM, Charpak S, Lauritzen M, Macvicar BA, Newman EA. Glial and neuronal control of brain blood flow. Nature. 2010;468(7321):232–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Farin A, Suzuki SO, Weiker M, Goldman JE, Bruce JN, Canoll P. Transplanted glioma cells migrate and proliferate on host brain vasculature: a dynamic analysis. Glia. 2006;53(8):799–808. [DOI] [PubMed] [Google Scholar]
- 10. Cuddapah VA, Robel S, Watkins S, Sontheimer H. A neurocentric perspective on glioma invasion. Nat Rev Neurosci. 2014;15(7): 455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Watkins S, Robel S, Kimbrough IF, Robert SM, Ellis-Davies G, Sontheimer H. Disruption of astrocyte-vascular coupling and the blood-brain barrier by invading glioma cells. Nat Commun. 2014;5:4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Price SJ, Allinson K, Liu H, et al. Less invasive phenotype found in isocitrate dehydrogenase-mutated glioblastomas than in isocitrate dehydrogenase wild-type glioblastomas: a diffusion-tensor imaging study. Radiology. 2017;283(1):215–221. [DOI] [PubMed] [Google Scholar]
- 13. Xiao F, Caciagli L, Wandschneider B, et al. Effects of carbamazepine and lamotrigine on functional magnetic resonance imaging cognitive networks. Epilepsia. 2018;59(7):1362–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Luo C, Li Q, Lai Y, et al. Altered functional connectivity in default mode network in absence epilepsy: a resting-state fMRI study. Hum Brain Mapp. 2011;32(3):438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nasreddine ZS, Phillips NA, Bédirian V, et al. The Montreal Cognitive Assessment, MoCA: a brief screening tool for mild cognitive impairment. J Am Geriatr Soc. 2005;53(4):695–699. [DOI] [PubMed] [Google Scholar]
- 16. Suchorska B, Jansen NL, Linn J, et al. ; German Glioma Network Biological tumor volume in 18FET-PET before radiochemotherapy correlates with survival in GBM. Neurology. 2015;84(7):710–719. [DOI] [PubMed] [Google Scholar]
- 17. Holmes AJ, Hollinshead MO, O’Keefe TM, et al. Brain Genomics Superstruct Project initial data release with structural, functional, and behavioral measures. Sci Data. 2015;2:150031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yeo BT, Krienen FM, Sepulcre J, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106(3):1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Buckner RL, Krienen FM, Castellanos A, Diaz JC, Yeo BT. The organization of the human cerebellum estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106(5):2322–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Van Dijk KR, Sabuncu MR, Buckner RL. The influence of head motion on intrinsic functional connectivity MRI. Neuroimage. 2012;59(1):431–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mueller S, Wang D, Fox MD, et al. Individual variability in functional connectivity architecture of the human brain. Neuron. 2013;77(3):586–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Van Dijk KR, Hedden T, Venkataraman A, Evans KC, Lazar SW, Buckner RL. Intrinsic functional connectivity as a tool for human connectomics: theory, properties, and optimization. J Neurophysiol. 2010;103(1):297–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17(2):825–841. [DOI] [PubMed] [Google Scholar]
- 24. Smith SM, Jenkinson M, Woolrich MW, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(Suppl 1):S208–S219. [DOI] [PubMed] [Google Scholar]
- 25. Yan CG, Cheung B, Kelly C, et al. A comprehensive assessment of regional variation in the impact of head micromovements on functional connectomics. Neuroimage. 2013;76:183–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Satterthwaite TD, Elliott MA, Gerraty RT, et al. An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. Neuroimage. 2013;64:240–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mueller S, Wang D, Fox MD, et al. Reliability correction for functional connectivity: theory and implementation. Hum Brain Mapp. 2015;36(11):4664–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stoecklein S, Hilgendorff A, Li M, et al. Variable functional connectivity architecture of the preterm human brain: impact of developmental cortical expansion and maturation. Proc Natl Acad Sci U S A. 2020;117(2):1201–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Albert NL, Weller M, Suchorska B, et al. Response Assessment in Neuro-Oncology working group and European Association for Neuro-Oncology recommendations for the clinical use of PET imaging in gliomas. Neuro Oncol. 2016;18(9):1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Duffau H. The huge plastic potential of adult brain and the role of connectomics: new insights provided by serial mappings in glioma surgery. Cortex. 2014;58:325–337. [DOI] [PubMed] [Google Scholar]
- 31. Venkataramani V, Tanev DI, Strahle C, et al. Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature. 2019;573(7775):532–538. [DOI] [PubMed] [Google Scholar]
- 32. Pak RW, Hadjiabadi DH, Senarathna J, et al. Implications of neurovascular uncoupling in functional magnetic resonance imaging (fMRI) of brain tumors. J Cereb Blood Flow Metab. 2017;37(11): 3475–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eckel-Passow JE, Lachance DH, Molinaro AM, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Weller M, van den Bent M, Tonn JC, et al. ; European Association for Neuro-Oncology (EANO) Task Force on Gliomas European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017;18(6):e315–e329. [DOI] [PubMed] [Google Scholar]
- 35. Kickingereder P, Sahm F, Radbruch A, et al. IDH mutation status is associated with a distinct hypoxia/angiogenesis transcriptome signature which is non-invasively predictable with rCBV imaging in human glioma. Sci Rep. 2015;5:16238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Paech D, Windschuh J, Oberhollenzer J, et al. Assessing the predictability of IDH mutation and MGMT methylation status in glioma patients using relaxation-compensated multipool CEST MRI at 7.0 T. Neuro Oncol. 2018;20(12):1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cui D, Ren J, Shi J, et al. R132H mutation in IDH1 gene reduces proliferation, cell survival and invasion of human glioma by downregulating Wnt/β-catenin signaling. Int J Biochem Cell Biol. 2016;73:72–81. [DOI] [PubMed] [Google Scholar]
- 38. Kesler SR, Noll K, Cahill DP, Rao G, Wefel JS. The effect of IDH1 mutation on the structural connectome in malignant astrocytoma. J Neurooncol. 2017;131(3):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van Kessel E, Baumfalk AE, van Zandvoort MJE, Robe PA, Snijders TJ. Tumor-related neurocognitive dysfunction in patients with diffuse glioma: a systematic review of neurocognitive functioning prior to anti-tumor treatment. J Neurooncol. 2017;134(1):9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome Sequencing Program data are publicly available.17 Patient data and the code of the ABI are available from the corresponding authors upon request.