Abstract
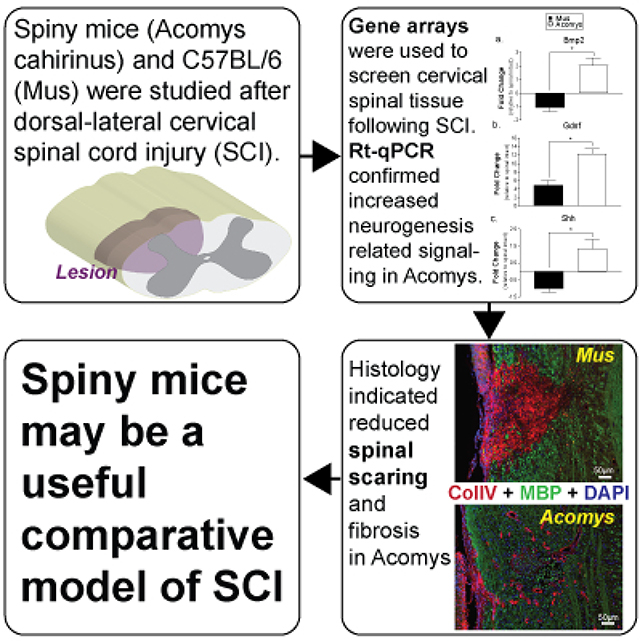
The spiny mouse (Acomys cahirinus) appears to be unique among mammals by showing little scarring or fibrosis after skin or muscle injury, but the Acomys response to spinal cord injury (SCI) is unknown. We tested the hypothesis that Acomys would have molecular and immunohistochemical evidence of reduced spinal inflammation and fibrosis following SCI as compared to C57BL/6 mice (Mus), which similar to all mammals studied to date exhibits spinal scarring following SCI. Initial experiments used two pathway-focused RT-PCR gene arrays (“wound healing” and “neurogenesis”) to evaluate tissue samples from the C2-C6 spinal cord 3-days after a C3/C4 hemi-crush injury (C3Hc). Based on the gene array results, specific genes were selected for RT-qPCR evaluation using species-specific primers. The results supported our hypothesis by showing increased inflammation and fibrosis related gene expression (Serpine 1, Plau, Timp1) in Mus as compared to Acomys (P<0.05). RT-qPCR also showed enhanced stem cell and axonal guidance related gene expression (Bmp2, GDNF, Shh) in Acomys compared to Mus (P<0.05). Immunohistochemical evaluation of the spinal lesion at 4-wks post-injury indicated reduced collagen IV immunostaining in Acomys (P<0.05). Glial fibrillary acidic protein (GFAP) and ionized calcium binding adaptor molecule 1(IBA1) immunostaining indicated morphological differences in the appearance of astrocytes and macrophages/microglia in Acomys. Collectively, the molecular and histologic results support the hypothesis that Acomys has reduced spinal inflammation and fibrosis following SCI. We suggest that Acomys may be a useful comparative model to study adaptive responses to SCI.
Keywords: Spiny mice, spinal injury, RRID: AB_2572352, RRID: AB_839504, RRID: AB_2572267, RRID: AB_2109953, RRID: AB_305584
Graphical Abstract
We tested the hypothesis that the spiny mouse (Acomys cahirinus) would have molecular and immunohistochemical evidence of reduced spinal inflammation and fibrosis following spinal cord injury (SCI) as compared to C57BL/6 mice (Mus). Based on initial gene array results of the local response to SCI, specific genes were selected for RT-qPCR using species-specific primers. The results showed increased inflammation and fibrosis related gene expression in Acomys as compared to Mus. Immunohistochemical evaluation of the spinal lesion indicated reduced collagen IV immunostaining in Acomys. Collectively, the results support the hypothesis that Acomys has reduced spinal inflammation and fibrosis following SCI. Acomys may be a useful comparative model to study adaptive responses to SCI.
Introduction
A fundamental goal of regenerative medicine is to develop therapies to reduce inflammation, fibrosis, scarring, or other detrimental outcomes in damaged or diseased mammalian tissues. Spinal cord injury (SCI) is one condition in which inflammation, scarring, and limited regeneration has severe functional consequences. Immediately after SCI an inflammatory response is induced involving activation of resident immune cells (e.g. microglia), and recruitment of monocyte/macrophages and neutrophils to the site of the injury (Carlson, Parrish, Springer, Doty, & Dossett, 1998; Hawthorne & Popovich, 2011). At later time points, a glial/fibrotic scar forms and provides a barrier that prevents axonal regeneration (Tran, Warren, & Silver, 2018). Current regenerative therapies in SCI include delivery of exogenous stem cells (Pereira, Marote, Salgado, & Silva, 2019) and/or the incorporation of biomolecular scaffolds into the damaged spinal cord (Kim, Park, & Choi, 2014). However, regenerative failure remains a consistent feature of SCI in animal models and humans.
One contributing factor to the lack of progress in identifying therapeutic targets has been the absence of a relevant model system, namely an adult mammal with high regenerative capacity of spinal tissue. In this regard, we recently discovered that the adult spiny mouse (Acomys cahirinus) can regenerate a remarkable range of tissues following injury including dermis, hair follicles, sebaceous glands, smooth muscle, skeletal muscle, adipose cells and cartilage (Brant, Yoon, Polvadore, Barbazuk, & Maden, 2016; Seifert et al., 2012). Acomys does not undergo fibrosis in response to damage in any organ examined thus far, and presents a unique opportunity to interrogate the mechanisms involved in mammalian tissue regeneration (Pinheiro, Prata, Araújo, & Tiscornia, 2018). A mammalian model of improved regenerative capacity after SCI would provide an opportunity for comprehensive examination of the mechanisms contributing to this response. Towards this goal, we evaluated the acute molecular and long-term immunohistochemical impact of SCI in Acomys. This response was compared to Mus, which displays signatures of the “typical” mammalian response to spinal injury (i.e. inflammation, scarring, limited regeneration). The overall hypothesis was that the injured spinal cord in adult Acomys would have a molecular and immunohistochemical signature consistent with reduced fibrosis and improved regenerative capacity as compared to Mus.
Materials and methods
Animals
All experiments were conducted with adult, male C57/BL6 (n=16; 29.8 ± 2.8g) mice (Mus; Jackson Laboratory) and male, spiny mice (n=15; 45.1 ± 6.4g) (Acomys cahirinus; in-house colony at the University of Florida). Descriptions of skin and muscle wound healing using spiny mice from this colony have been published (Brant, Lopez, Baker, Barbazuk, & Maden, 2015; Brant et al., 2016; Maden et al., 2018). Mice were housed in a controlled environment (12h light/dark cycles) with food and water ad libitum. All experimental protocols were approved by the Institutional Animal Care and Use Committee at the University of Florida.
Spinal Cord Injury
Anesthesia was induced by placing mice in a chamber flushed with 3% isoflurane mixed with 100% O2. Anesthetized mice were transferred to a heated surgical station and core body temperature was maintained at 37.5 ± 0.5°C (model 700 TC-1000, CWE). During surgery, anesthesia was maintained by having the mice breathe 1.5–2% isoflurane through a nose cone with 100% O2. The surgical area was cleaned with three, alternating rounds of betadine surgical scrub followed by 70% ethanol. A dorsal incision was then made over the spinal midline from the base of the skull to the fifth cervical segment. A C3 laminectomy was performed to expose the spinal cord, and a C3 lateral, dorsal crush injury was induced similar to previous report (Hilton et al., 2013). A sterilized pair of fine tipped Dumont #5 forceps was marked 1mm from the tip with a black sharpie marker. To induce the injury, one prong of the forceps was inserted at the spinal midline at a depth of 1mm (identified by the black sharpie mark) while the other prong remained outside the lateral edge of the spinal cord. The forceps were held closed for 15 seconds. The forceps were removed, reinserted for a second time and closed again for 15 seconds. Following the injury, the overlying muscles were sutured with 4–0 Vicryl suture and the skin was closed with sterile wound clips. Mice received an analgesic, buprenorphine (0.03 mg/kg, s.q.) and nonsteroidal anti-inflammatory medication meloxicam (2.0 mg/kg, s.q.) for the initial 48 hours post-injury. Mice also received Lactated Ringer’s solution (2 ml/day, s.q.) and oral Nutri-cal supplements (0.5–1ml, Webster Veterinary, MA, USA) until adequate volitional eating and drinking resumed. Manual bladder expression was performed twice daily until voluntary micturition was apparent. The bladder size was estimated by palpating the lower abdomen using two fingers (thumb and index finger) and was then expressed by gently applying pressure until the bladder was completely emptied. Body temperature was assessed daily for two weeks post-SCI using a digital mouse rectal thermometer (model 700 TC-1000, CWE) and Vaseline.
Gene Arrays
Three days after SCI, mice (Mus: n=6, Acomys: n=5) were anesthetized by breathing 3% isoflurane mixed into 100% O2. An age matched, spinal-intact comparison group did not undergo surgery or receive an injury (Mus: n=4, Acomys: n=5). After isoflurane anesthesia, mice received an i.p. injection of beuthanasia solution (150mg/kg). The depth of anesthesia was confirmed via lack of foot withdrawal to toe pinch, and the cervical spinal cord from C2 to C6 was quickly extracted, placed in an ice cold RNA free dish, cut sagittally to isolate the ipsilateral (same side as injury) spinal cord and immediately placed into 1.5mL of RNALater (Qiagen Cat. 76104). Spinal cord samples were stored at 4°C for 24 hours and then maintained at −80°C. Tissues were subsequently thawed at 4°C, washed in RNase-free water and homogenized using a rotor stator type tissue homogenizer (ProScientific Bio-Gen PRO200 Homogenizer; Multi-Gen 7XL Generator Probes) in RLT Buffer and processed using the Rneasy Plus Mini Kit (Qiagen Cat. 74134). RNA quality was assessed using an Agilent 2200 TapeStation (Agilent, Andover, MA). All samples had a RIN score > 7.0. For the wound healing pathway-focused RT-PCR array (Qiagen PAMM-121Z) and the neurogenesis pathway-focused RT-PCR array (Qiagen PAMM-404Z) cDNA was generated using the RT2 PreAMP cDNA Synthesis Kit (Qiagen 330451), using array specific primers (Qiagen 330241) followed by the RT2 First Strand Kit (Qiagen 33041). Arrays were run using RT2 SYBR Green qPCR Mastermix (Qiagen 330502). For RT-qPCR with species-specific primers cDNA was generated SuperScript™ IV VILO Reverse Transcriptase (Invitrogen 11756050) following the manufacturer’s protocol. Real-Time PCR was performed using Sso-Fast™ EvaGreen® Supermix (Bio-Rad 172–5200) on a Bio-Rad C1000 Touch™ Thermal Cycler. The fold change in gene expression was calculated using the ΔΔCt relative expression method (Livak & Schmittgen, 2001) using Gapdh as the reference gene. Sequence of species-specific PCR primers can be found in Supplemental Table 1. All reactions were run with an annealing temperature of 60°C.
Antibody Selection
Antibody name, source, catalog number, concentration, immunogen, and research resource identifiers (RRID) for each primary antibody used in the study are provided in Table 1. The polyclonal, Collagen IV antibody (Abcam, Cat#ab6586; RRID: AB_305584; sequence available here: https://www.uniprot.org/uniprot/P02462) was validated by the manufacturer and binds to native collagen epitopes composed of multiple subunit strands and has negligible cross-reactivity with Type I, II, III, V or VI collagens. Specifically, Collagen IV has been shown to accumulate within the scar following SCI (Liesi & Kauppila, 2002). This antibody detects a single band ~250kDa on western blots in baby hamster kidney fibroblasts (Abcam, 2012), and has an expression pattern following SCI in mouse (Vangansewinkel et al., 2019) and rat (Tuinstra et al., 2014) similar to that reported herein.
Table 1:
Antibody name, source, catalog number, concentration, immunogen, and research resource identifiers (RRID) for each primary antibody used in the study.
| Antibody | Source | Catalog # | Concentration | Immunogen | RRID |
|---|---|---|---|---|---|
| Collagen IV | Abcam | ab6586 | 1:250 | Full length native protein (purified) corresponding to Collagen IV from human and bovine placenta | AB_305584 |
| MBP | Encor Biotechnology | CPCA-MBP | 1:500 | Purified myelin basic protein isolated from bovine brain | AB_2572352 |
| Iba1 | Wako Chemicals | 019–19741 | 1:300 | Synthetic peptide (C-terminal of Iba1) | AB_839504 |
| NeuN | Encor Biotechnology | MCA-1B7 | 1:1000 | N-terminal 99 amino acids of human FOX3 expressed in and purified from E. coli | AB_2572267 |
| GFAP | Encor Biotechnology | CPCA-GFAP | 1:1000 | Recombinant full length human GFAP isotype 1 expressed in and purified from E. coli. | AB_2109953 |
The polyclonal, Myelin Basic Protein (MBP) antibody (EnCor Biotechnology, Cat#CPCA-MBP; RRID: AB_2572352; sequence available here: http://encorbio.com/Alignments/MBP%20isoforms.pdf binds to all four gene products from the single mammalian MBP gene which is observed by four bands between ~14–21.5 kDa in western blots in rodent spinal tissue (EnCor Biotechnology). The Iba-1 antibody (Wako Chemicals, Cat#019–19741; RRID: AB_839504) recognizes a calcium-binding protein with a molecular weight of 17 kDa specifically expressed in macrophage/microglia (Ito et al., 1998) and has been validated for immunohistochemistry in rodent, human, dog, cat, pig, marmoset, and zebrafish (Ahn et al., 2012; Fantin et al., 2010; Gaige et al., 2013; Ide, Uchida, Tamura, & Nakayama, 2010; Rodriguez-Callejas, Fuchs, & Perez-Cruz, 2016; S. M. Turner et al., 2016). The Neu-N antibody (EnCor Biotechnology, Cat#MCA-1B7; RRID: AB_2572267; sequence available here: https://www.uniprot.org/uniprot/A6NFN3) is a reliable neuronal marker, binding to neurons in all vertebrates (Mullen, Buck, & Smith, 1992). We have previously used the polyclonal, Glial Fibrillary Acidic Protein (GFAP) antibody (EnCor Biotechnology, Cat#CPCA-GFAP; RRID: AB_2109953; sequence available here: https://www.uniprot.org/uniprot/P14136) to stain reactive astrocytes in mouse brain and spinal tissue during CNS disease (S. M. Turner et al., 2016; Turner, Falk, Byrne, & Fuller, 2016). Similar to other reports (Bovolenta, Wandosell, & Nieto-Sampedro, 1992; Cregg et al., 2014), GFAP is a major constituent of the astro-glial scar in most mammals which forms following SCI. Western blot analysis of whole brain lysates indicates GFAP antibody shows a single, strong band at ~50kDa.
Immunohistochemistry
A separate cohort of mice that recovered for four weeks following SCI mice (Mus: n=6, Acomys: n=5) were anesthetized by breathing 3% isoflurane mixed into 100% O2 followed by an i.p. injection of urethane solution (1.0–1.6g/kg in distilled water). After the depth of anesthesia was verified, mice were transcardially perfused with ice cold saline (1ml per gram body weight) followed by 4% paraformaldehyde (PFA; 1ml/g) in 1X Dulbecco’s phosphate buffered saline (DPBS, Mediatech, Inc., Cat#21–030-CV). The cervical spinal cord (C2-C6) was harvested, post-fixed in 4% PFA overnight at 4°C and placed in 70% ethanol at room temperature for at least 48 hrs. The spinal cord was then paraffin embedded and stored at room temperature. Paraffin embedded spinal cords were cut longitudinally at 7 μm on a Leica Biosystems microtome and mounted on glass slides (Fisher, Superfrost Plus). Alternate slides were then used for immunostaining, and this enabled each stain to be evaluated at 35μm increments (i.e., every 5th slide was stained).
Prior to immunochemistry, tissue sections were deparaffinized and rehydrated through xylenes followed by a graded series of ethanol exposures (100%, 95% and 70%). An antigen retrieval procedure was done using 0.1M Citrate solution, pH 6.0 for 25 minutes (collagen IV/MBP staining; Abcam) or Trilogy reagent (IBA1/GFAP/NeuN staining; Cell Marque) at 95 °C for 15 min. Slides were blocked in 2% normal horse serum (Vector Labs) in 1X TBS with 0.2% Triton-X for 1hr at room temperature. Spinal sections were then incubated for two days at 4°C in the primary antibody solution (Table1): rabbit anti-collagen IV (1:250; RRID: AB_305584), chicken anti-MBP (1:500; RRID: AB_2572352), rabbit anti-Iba-1 (1:300; RRID: AB_839504), mouse anti-NeuN (1:1,000; RRID: AB_2572267), chicken anti-GFAP (1:1,000; RRID: AB_2109953), in antibody diluent (ThermoFisher Scientific, #003118). Immunoreactivity was detected using Alexa Fluor® secondary antibodies (all 1:500; ThermoFisher Scientific) donkey anti-mouse 488 (Cat#A-21202), donkey anti-rabbit 594 (Cat#A-21207), goat anti-chicken 488 (Cat#A-11039), and goat anti-chicken 594 (Cat#A-11042) in 2% normal horse serum in 1X TBS with 0.2% Triton-X for 1hr at room temperature. Positive control tissues and negative control tissue concentration matched Ig controls were included with each immunoassay. Negative controls resulted in no staining. Slides were coverslipped with VectaShield antifade mounting medium containing DAPI (Vector Labs, Cat#H-1200). Slides were air-dried and stored at 4°C.
All fluorescent images were captured with a BZ-X series all-in-one fluorescence microscope (Keyence Corporation) at 10, 20 and 40X and stitched using BZ-X Analyzer Software. Representative images in Figure 6 and 7 were edited to enhance NeuN signal. The tonal range for the input levels of the green channel was adjusted to 6–180 for Figure 6 and 12–140 for Figure 7 and the brightness was increased to 30. These changes were only performed on representative images (not those used for quantification) and were performed equally for all Mus and Acomys images.
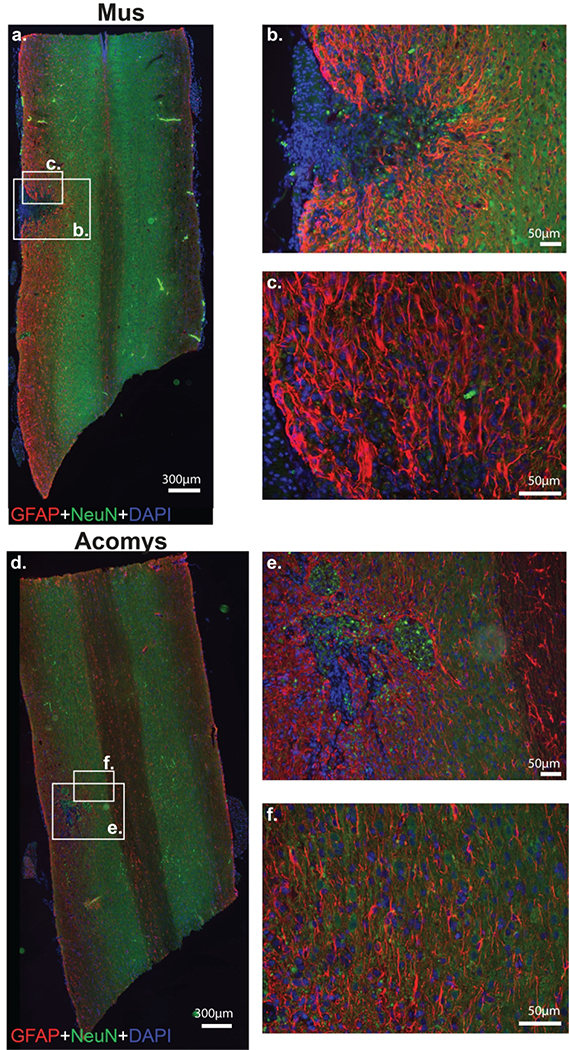
Figure 6:
Photomicrographs of GFAP (red), NeuN (green) and DAPI (blue) staining 4 weeks post-SCI in Mus (a-c) and Acomys (d-f). Representative longitudinal cervical spinal images (C2-C6) depicting astrocytes (GFAP), neurons (NeuN), and nuclei (DAPI) in and around the spinal lesion in Mus (a) and Acomys (d). High magnification images of the lesion epicenter shown in panel a for Mus (b,c) and panel d for Acomys (e,f). Note: linear adjustments were performed on representative images (see Materials and Methods section for details).
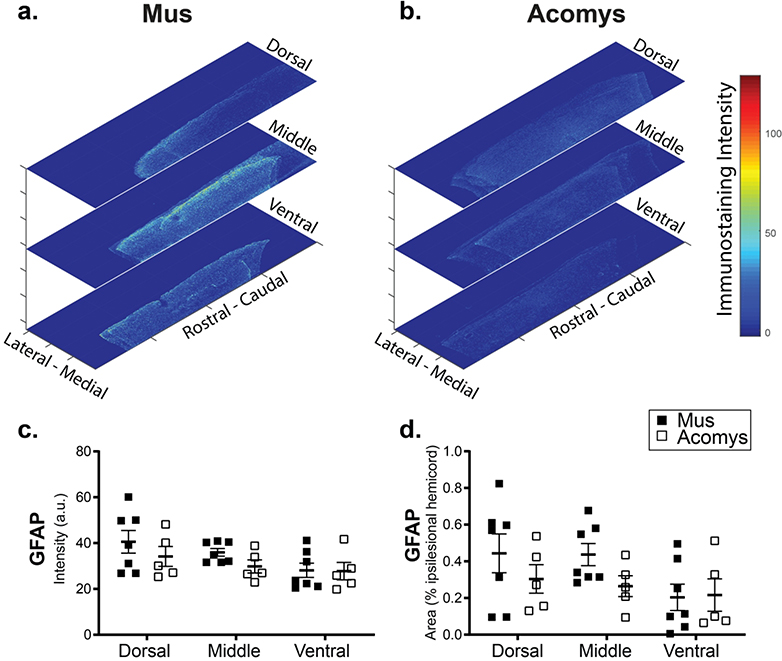
Figure 7:
Representative heat maps of the intensity of GFAP staining at the epicenter for Mus (a) and Acomys (b). Note: the size of heat map images are the ROI that were used for the analysis. Quantification of the intensity of GFAP staining (arbitrary units, c) and normalized to the size of the ROI (% ipsilesional hemicord, d) within the dorsal, middle, and ventral spinal regions.
Data and Statistical Analyses
Gene expression 3 days post-SCI was compared to spinal intact controls (which did not undergo surgery or receive an injury) within each species and genes with a fold change >1.5 are provided in Supplemental Table 2. Individual one tailed t-tests were used for analysis of species-specific primer results. For immunohistochemistry analysis, images captured at 10X were analyzed using MATLAB (The MathWorks R2015a). Briefly, MATLAB code was written to 1) detect the total number of pixels in the section, 2) remove the background (determined empirically based on a blinded subset of images), and 3) calculate the average fluorescent intensity and number of the remaining pixels. Quantification of all immunohistochemistry was performed on every third stained spinal cord section (Mus: n=11 ± 0.2; Acomys: n=14 ± 0.6 sections). This represented 105μm increments from the dorsal surface of the spinal cord. For GFAP and IBA1 staining, quantification was performed on the entire ipsilateral spinal cord. Collagen IV and MBP staining was quantified at the site of lesion. For quantification, an area of interest that was 4.7% of the tissue section area was positioned to encompass all the collagen IV staining. This value was empirically determined by assessing the spinal lesion from all animals based on the distribution of collagen IV staining. Immunostaining for GFAP and IBA1 was normalized to the size of the ipsilateral (to SCI) tissue section (% positive). To assess immunostaining according to the location of the section within the dorsal-ventral neuroaxis, the total number of imaged sections for each animal was divided into three equal groups representing ventral, middle, and dorsal spinal regions. The average number of sections in each region was similar: ventral: Mus: n=3.6 ± 0.2; Acomys: n=4.6 ± 0.2 sections, middle: Mus: n=3.4 ± 0.2; Acomys: n=4.6 ± 0.2 sections, and dorsal: Mus: n=3.6 ± 0.2; Acomys: n=4.6 ± 0.2 sections. Quantitation of the staining intensity for various makers of the SCI response (e.g., collagen IV, MBP, GFAP, IBA1) was done by evaluating both the raw signal intensity captured via fluorescence microscopy (arbitrary units) and also by normalization relative to the total amount of positive staining that could detected. Statistical analyses were performed in GraphPad Prism 8 (GraphPad Software, Inc). Means are presented with standard error.
Data availability statement
The data reported herein are available from the corresponding author (DDF) upon reasonable request.
Results
Behavioral observations
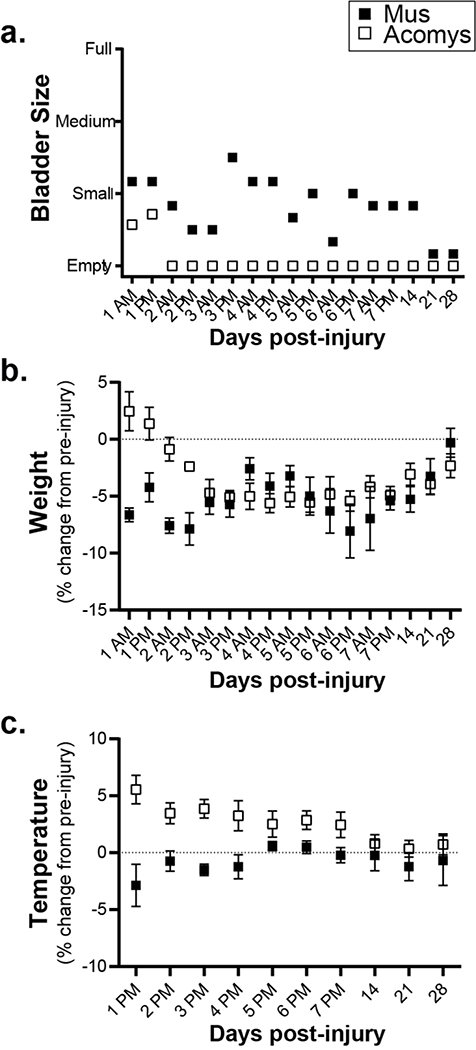
Several relevant observations were made during the routine care of the animals following SCI (Figure 1). First, Acomys resumed spontaneous bladder voiding earlier than Mus (Figure 1a). Acomys regained the ability to void the bladder by two days post-injury whereas Mus required manual bladder expression for more than two weeks. Second, both species showed a transient weight loss after the injury, but with a different time course as illustrated in Figure 1b. Mus showed an immediate drop in body mass, whereas Acomys did not drop body mass until day three. Prior to injury, body temperature was different between Mus (n=6; 37.6±0.1°) and Acomys (n=7; 35.7±0.2°, P<0.0001). Differences were also noted in rectal temperature over the first few days following the injury. Acomys showed an increase of approximately 1°C during days 1–7 post-injury whereas Mus slightly dropped temperature (Figure 1c).
Figure 1:
Behavioral observations for 28 days post-SCI. a. Bladder size in Mus (shaded squares) and Acomys (open squares) measured by manually palpating the lower abdomen. b. Weight (as a percent change from pre-injury values) assessed twice a day for the first 7 days and at 2, 3, and 4 weeks post-injury. c. Rectal temperature (as a percent change from pre-injury values) assessed once a day for the first 7 days and at 2, 3, and 4 weeks post-injury. Data shown are means ± standard error.
RT-PCR arrays
For an initial comparison of the molecular genetic response to SCI in Mus vs. Acomys, we examined the expression profile of the ipsilesional cervical spinal cord (C2-C6) at three days post-SCI (relative to non-injured controls) using two different pathway-focused RT-PCR arrays. These initial experiments were intended to identify genes of interest and individual genes were subsequently validated with RT-qPCR quantitation using both Mus-specific and Acomys-specific primers (next section). The first gene array targeted 84 genes involved in wound healing (e.g., inflammation, granulation, tissue remodeling) and the second array evaluated 84 genes associated with neurogenesis (e.g., proliferation, differentiation, motility, migration) and stem cell differentiation.
The results of the wound healing array and neurogenesis array are presented in Supplemental Table 2. The wound healing array indicated Mus had a stronger wounding response compared to Acomys. Specifically, Mus had 16 upregulated genes (>1.5 fold) and 21 downregulated compared to expression in the non-injured spinal cord; and Acomys had three upregulated and 24 downregulated genes (Supplemental Table 2a). The upregulated genes in Mus can be associated into four groups: 1) pro-inflammatory cytokines involved in the immune response to wounding (Il6, Cxcl3, Ccl12, Ccl7, Tnf, and Il1b); 2) Extracellular matrix and its remodeling (Timp1, Itga5, Plaur, Mmp1a, Col5a3, and Plau); 3) Tgfβ1 signaling and fibrosis (Serpine1, Tagln and Tgfb1); and 4) a growth factor (Hgf). In contrast, upregulated genes in Acomys consisted of one inflammatory response gene (Tnf) and two genes associated with the ECM and cell surface (Col 5a1 and Cadherin1). The majority of gene changes in Acomys were in the negative direction including extracellular matrix constituents (Vtn and Clo4a3), a vitronectin receptor (Itgav), inflammatory cytokines (Cxcl3 and Il2) and receptor (Il6st), and two integrins (Itgb5 and Itgb6).
In stark contrast to the wounding arrays, the majority of the upregulated genes in neurogenesis arrays were observed in Acomys. Specifically, there were 22 upregulated genes and two downregulated in Acomys, whereas in Mus there were 7 upregulated genes and 37 downregulated (Supplemental Table 2b). The upregulated genes in Mus were Tgfb1, two growth factors (Gdnf and Fgf2), a transcriptional activator of growth factors (Stat3), a neuregulin receptor (Erbb2), and two genes involved in the regulation of actin cytoskeleton and cell motility (S100a6 and Flna). Four of these seven were also upregulated in Acomys. The 22 upregulated genes in the Acomys spinal cord included: 1) growth factors/transcriptional activator of growth factors (Bdnf, Gdnf, Ptn, Stat3); 2) WNT signaling molecules (Dvl3, Ndp, Shh); 3) neural stem cell genes/transcription factors (Notch1, Notch2, Creb1, Sox2, Ascl1, Heyl and Mef2c); 4) genes involved in axonal guidance (Ntn1, Robo1, and Efnb1); 5) Tgfβ1 signaling molecules (Tgfb1, Bmp2); and 6) a cell death gene (Bcl2).
RT-qPCR of select genes using species-specific primers
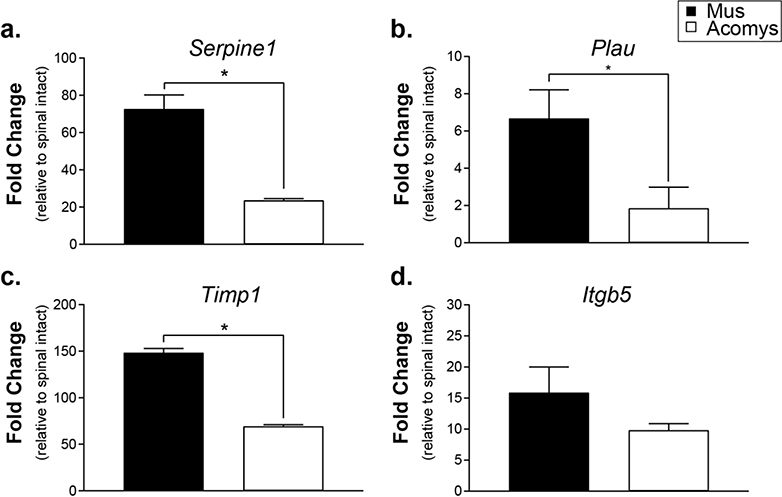
Based on the results of the gene arrays, we selected and verified the expression of four genes associated with wound healing and fibrosis in Mus and Acomys following SCI: Serpine1, Plau, Timp1, and Itgb5 (Figure 2). The levels of mRNA for each gene are expressed relative to non-injured spinal tissue of each species. Serpine 1, a major physiological regulator of the plasmin-based cascade involved in fibrotic disorders and pro-inflammatory molecule (Gupta, Xu, Castellino, & Ploplis, 2016) was upregulated post-SCI to a greater extent in Mus than Acomys (t(4)=6.441, P=0.0015). Plau, another plasminogen activator involved in the fibrotic pathway (He, Tsou, Khanna, & Sawalha, 2018) was also upregulated in Mus (t(4)=2.516, P=0.0328; Figure 2b). Analysis of Timp1, an extracellular matrix enzyme which functions to inhibit the action of the matrix metalloproteases was highly upregulated in Mus (t(4)=27.75, P<0.0001; Figure 2c.). Itgb5, an integrin that interacts with the matrix and intracellular compartments was similar between Mus and Acomys, which was supported by statistical analysis (t(4)=1.405, P=0.1163; Figure 2d).
Figure 2:
Gene expression levels at 3 days post-SCI expressed as fold change compared to spinal intact controls determined by RT-qPCR using species-specific primers. a. Serpine1 (plasminogen activator inhibitor 1). b. Plau (plasminogen activator urokinase). c. Timp1 (tissue inhibitor of metalloprotease 1). d. Itgb5 (integrin β5). Data shown are means ± standard error. * = P < 0.05.
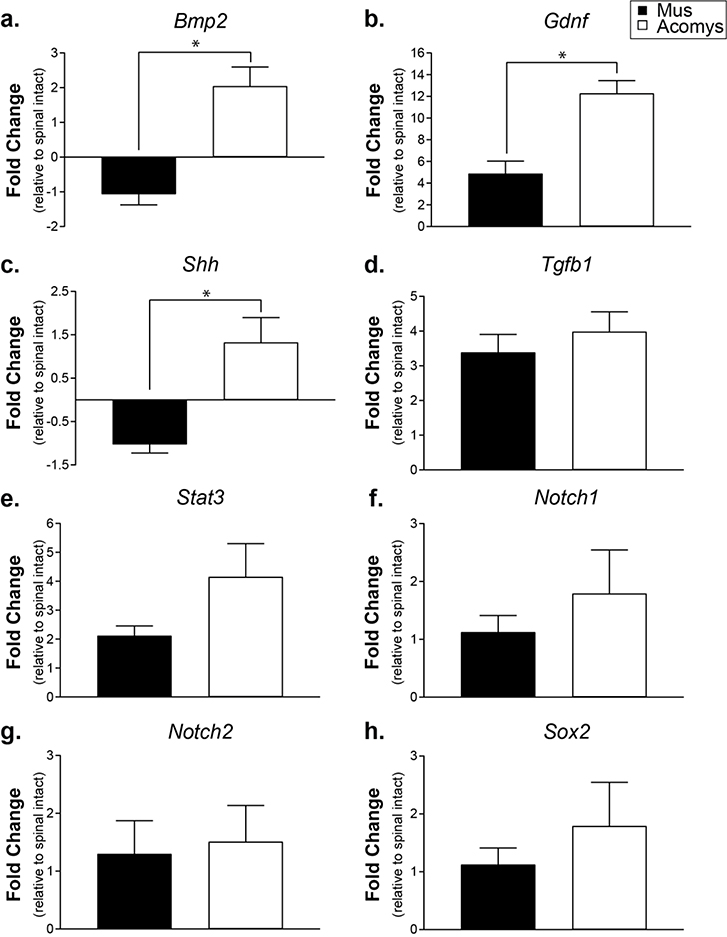
We also selected eight growth factor, neural stem cell, and signaling molecules genes identified by the neurogenesis array and verified their expression in both Mus and Acomys following SCI: Bmp2, GDNF, Shh, Tgfβ1, Stat3, Notch1, Notch2, and Sox2 (Figure 3). Bmp2 and GDNF, which regulate cellular differentiation (Xiao, Du, Wu, & Yip, 2010) and growth (Rosich, Hanna, Ibrahim, Hellenbrand, & Hanna, 2017) were upregulated in Acomys compared to Mus ((t(4)=4.936, P=0.0039; Figure 3a) and (t(4)=4.458, P=0.0056; Figure 3b)). Shh, a morphogen that aids in progenitor proliferation (Bambakidis, Wang, Franic, & Miller, 2003) and myelination (Thomas & Shea, 2013) was also upregulated in Acomys (t(4)=3.883, P=0.0089; Figure 3c). As observed in both gene arrays, levels of Tgfβ1 were similar between Mus and Acomys (t(4)=0.7728, P=0.2414; Figure 3d). Although a trend for upregulation, there was no difference in the transcription factor Stat3 (t(4)=1.696, P=0.0826; Figure 3e). Notch signaling, known to regulate neural progenitor cell fate (Cardozo, Mysiak, Becker, & Becker, 2017) was similar between Mus and Acomys (Notch1: (t(4)=0.8279, P=0.2271; Figure 3f); Notch2: (t(4)=0.2466, P=0.4087; Figure 3g)). Sox2, a transcription factor related to neuronal differentiation was not different between species (t(4)=1.373, P=0.1208; Figure 3g).
Figure 3:
Gene expression levels at 3 days post-SCI expressed as fold change compared to spinal intact controls determined by RT-qPCR using species-specific primers. a. Bmp2 (bone morphogenetic protein 2). b. Gdnf (glial derived neurotrophic factor). c. Shh (sonic hedgehog). d. Tgfb1 (transforming growth factor β1). e. Stat3 (signal transducer and activator of transcription 3). f. Notch1 (Notch homolog 1, translocation-associated). g. Notch2 (Notch homolog 2, translocation-associated). h. Sox2 (SRY (sex determining region Y)-box 2). Data shown are means ± standard error. * = P < 0.05.
Spinal cord immunohistochemistry
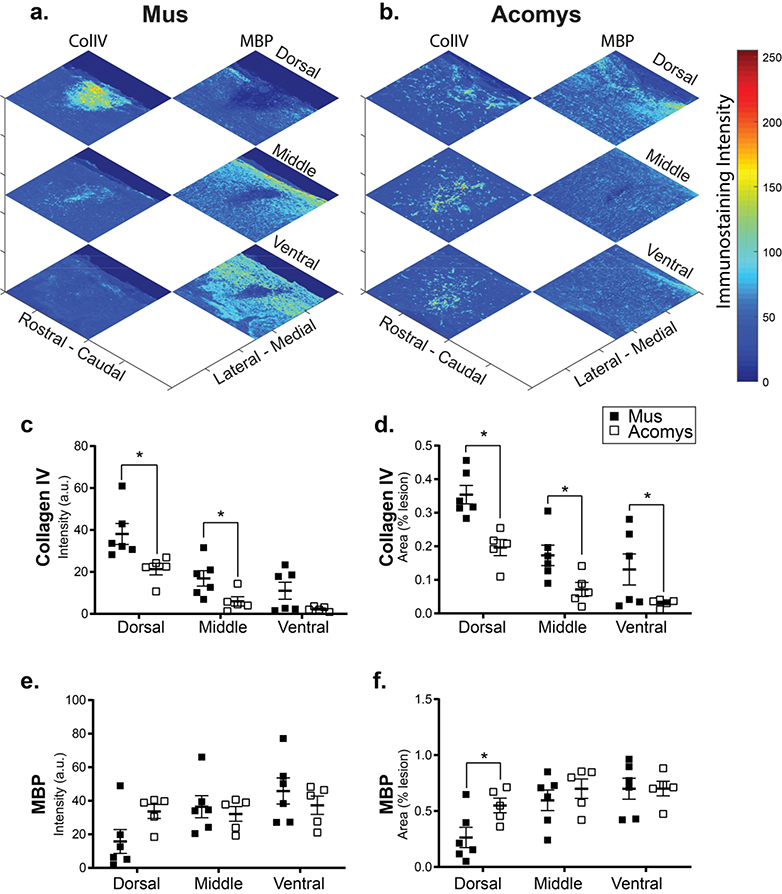
Representative photomicrographs of lesioned spinal cords stained for collagen IV and MBP are provided in Figure 4. Example “heat maps” which illustrate the intensity and density of these markers and quantification are shown in Figure 5. Evaluation of the immunohistochemical staining indicated a reduction in the density of collagen IV staining within the spinal lesion in Acomys, and this conclusion is supported by the statistical analyses. Evaluation of signal intensity (a.u.) produced low P-values for both species (Mus vs. Acomys, F(1,27) = 17.11, P=0.0003) and location (i.e., dorsal, middle, ventral, F(2,27) = 22.87, P<0.0001). Evaluation of normalized staining (% of the lesion area that stained positive for collagen IV) also produced low P-values: species: F(1,27)=23.37, P<0.0001; location: F(2,27)=22.70, P<0.0001. Post-hoc evaluation of the normalized data confirms a difference between Acomys and Mus in the dorsal (P=0.001), middle (P=0.025) and ventral (P=0.028) spinal regions (Figure 5d).
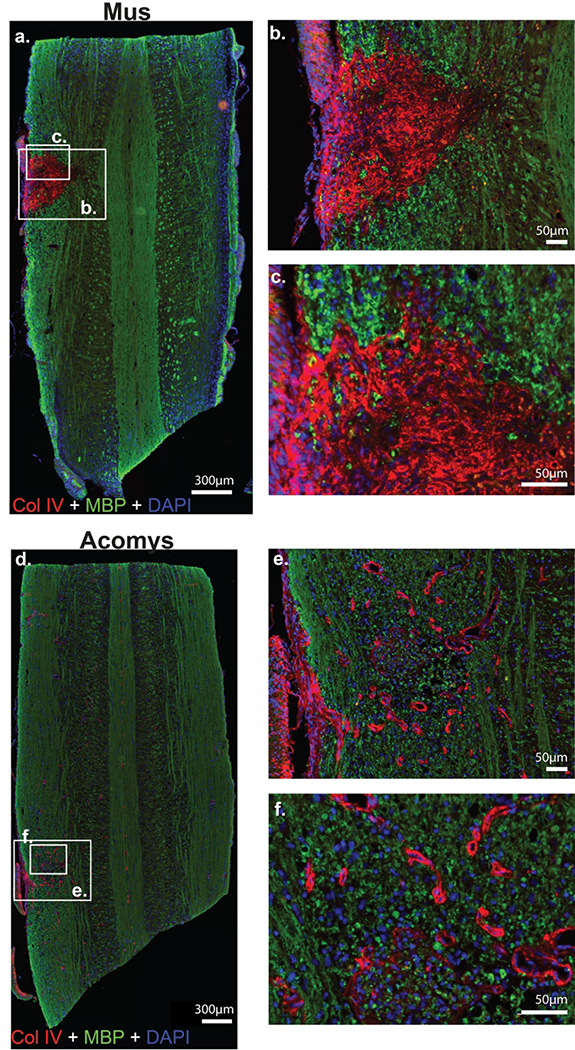
Figure 4:
Photomicrographs of collagen IV (red), myelin basic protein (green) and DAPI (blue) staining 4 weeks post-SCI in Mus (a-c) and Acomys (d-f). Representative longitudinal cervical spinal images (C2-C6) depicting collagen IV (Col IV) deposition, myelin (MBP), and nuclei (DAPI) in and around the spinal lesion in Mus (a) and Acomys (d). High magnification images of the lesion epicenter shown in panel a for Mus (b,c) and panel d for Acomys (e,f).
Figure 5:
Representative heat maps of the intensity of collagen IV and myelin basic protein staining at the lesion epicenter for Mus (a) and Acomys (b). Note: the size of heat map images are the ROI that were used for the analysis. Quantification of the intensity of collagen IV staining (arbitrary units, c) and normalized to the size of the ROI (% lesion, d) within the dorsal, middle, and ventral spinal regions. Quantification of the intensity of myelin basic protein staining (arbitrary units, e) and normalized to the size of the lesion (%lesion, f) within the dorsal, middle, and ventral spinal regions. Data shown are means ± standard error. * = P < 0.05.
In contrast to collagen IV, the density of MBP within the spinal lesion appeared greater in Acomys. The intensity of MBP staining (a.u.) was similar between species (F(1,27)=0.105, P=0.748), but different across location (F(2,27)=3.52, P=0.0438). Inspection of the data distribution indicated a possible difference in the dorsal spinal cord (Figure 5e–f). This is more evident when MBP is expressed relative to the lesion area (Figure 5f), as follows: location, F(2,27)=6.644, P=0.005; species, F(1,27)=3.474, P=0.073. Post hoc tests indicate differences in the dorsal spinal cord with greater staining in Acomys as compared to Mus (P=0.026; Figure 5f).
Photomicrographs of GFAP immunostaining in Acomys and Mus are provided in Figure 6 with heat maps and quantification of intensity shown in Figure 7. Evaluation of the immunohistochemical sections indicated differences between the two groups. Specifically, in Mus, GFAP positive astrocytes had a hypertrophied morphology and were concentrated at the lesion epicenter (Figure 6b), characteristic of reactive gliosis (Wilhelmsson et al., 2004). In Acomys GFAP positive astrocytes were organized in a network near the spinal lesion (Figure 6e). The distribution of GFAP staining intensity across all spinal cords is shown in Figure 57. In the middle of the spinal cord, the non-normalized (% of total ipsilesional cord area) GFAP staining intensity suggest a different distribution in Acomys. The 2-way ANOVA values for non-normalized data were as follows: species, F(1,30)=2.038, P=0.164; location, F(2,30)=3.251, P=0.053). The 2-way ANOVA values for normalized data were: species, F(1,30)=2.213, P=0.147; location, F(2,30)=2.332, P=0.0115.
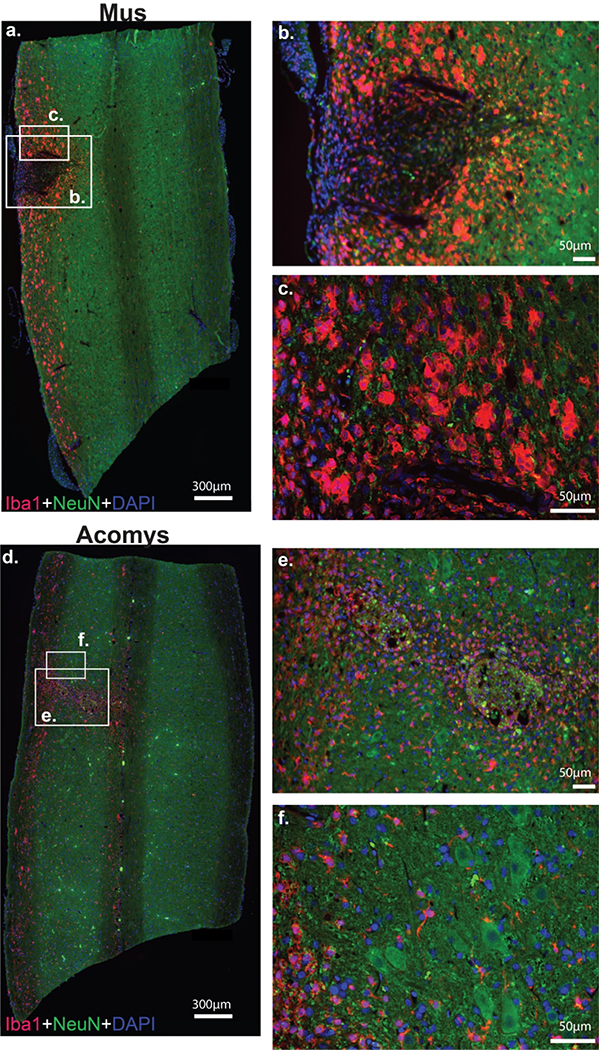
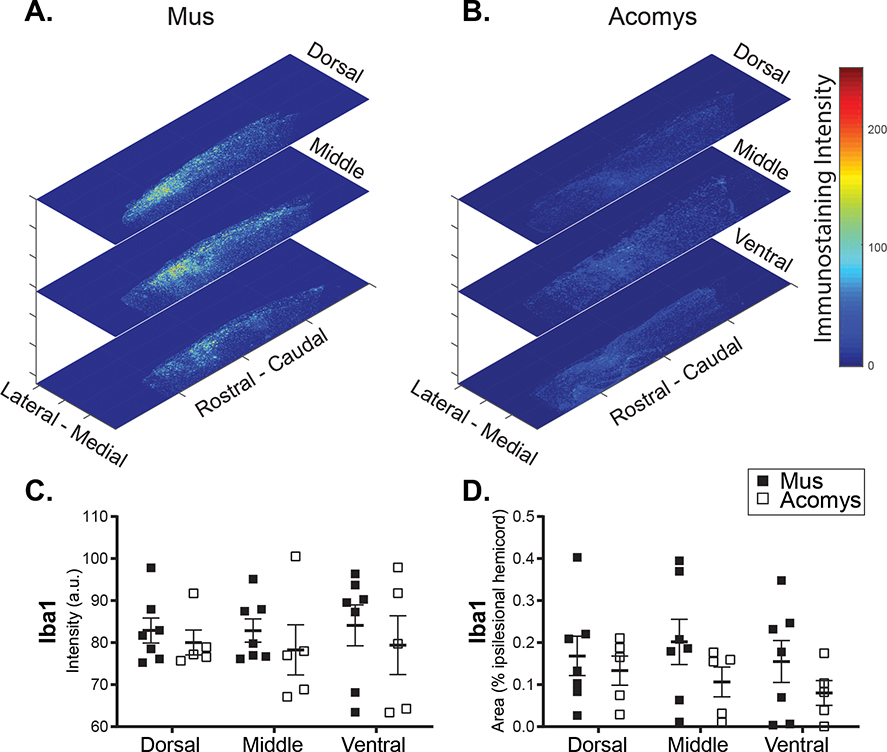
Photomicrographs depicting IBA1 immunostaining and associated heat maps and quantification are provided in Figure 8 and 9. Visual inspection suggested morphological differences between groups. Specifically, in Mus spinal cords, IBA1 positive cells demonstrated a large, rounded appearance (Figure 8a–c) that is consistent with activated macrophages/microglia (Wu et al., 2005). In Acomys, IBA1 positive cells were smaller as shown in Figure 8d–f. In addition, IBA1 staining tended to be distributed along the length of the cervical cord in Mus, while it was more localized to the injury in Acomys (Figure 8a and d). The dorsal-ventral distribution of staining intensity was generally similar between the species, with possible suggestion of reduced IBA1 intensity in the middle spinal cord sections of Acomys as compared to Mus. The 2-way ANOVA values for non-normalized data were as follows: species, F(1,30)=1.208, P=0.281; location, F(2,30)=0.0362, P=0.965. For the normalized data (% ipsilesional hemicord), the values were: species, F(1,30)=3.17, p=0.085; location, F(2,30)=0.374, P=0.691.
Figure 8:
Photomicrographs of IBA1 (red), NeuN (green) and DAPI (blue) staining 4 weeks post-SCI in Mus (a-c) and Acomys (d-f). Representative longitudinal cervical spinal images (C2-C6) depicting macrophages/microglia (IBA1), neurons (NeuN), and nuclei (DAPI) in and around the spinal lesion in Mus (a) and Acomys (d). High magnification images of the lesion epicenter shown in panel a for Mus (b,c) and panel d for Acomys (e,f). Note: linear adjustments were performed on representative images (see Materials and Methods section for details).
Figure 9:
Representative heat maps of the intensity of IBA1 staining at the epicenter for Mus (a) and Acomys (b). Note: the size of heat map images are the ROI that were used for the analysis. Quantification of the intensity of IBA1 staining (arbitrary units, c) and normalized to the size of the ROI (% ipsilesional hemicord, d) within the dorsal, middle, and ventral spinal regions.
DISCUSSION
These experiments indicate that the African spiny mouse, Acomys, has a unique molecular and immunohistochemical response to SCI. In specific, three times the number of neurogenesis related genes were induced in Acomys as compared to Mus and these results stand in sharp contrast to the wound healing arrays where many pro-inflammatory and fibrosis related genes were induced only in Mus. The Acomys reaction to SCI is consistent with a reduced inflammation and fibrosis observed in our studies on skin regeneration using wound healing arrays (Brant et al., 2015), antibody arrays (Brant et al., 2016) and transcriptome analyses (Brant et al., 2019). Thus, we conclude that the blunted immune response and lack of fibrosis in Acomys also extends to the injured spinal cord and may be a feature of any tissue damage inflicted upon this species. In contrast, the Mus response is typical of wounding in most mammals, namely the generation of a pro-inflammatory environment by the production of cytokines secreted by leukocytes, neutrophils, macrophages, microglia, and fibroblasts. These cytokines attract additional blood cells and macrophages to the wound site and can assist with the induction of neurotrophins (Fan et al., 2018), but excess levels cause scarring (Werner & Grose, 2003). This self-reinforcing system can induce fibrosis and subsequent inhibition of axonal regrowth across a spinal lesion. Conversely, in the absence of this “cytokine storm”, as it has been referred to (Tisoncik et al., 2012), the spinal environment in Acomys may be more conducive to regeneration.
In addition to the induction of pro-inflammatory cytokines, the majority of the remaining genes in Mus can be linked together through transforming growth factor-β (TGFβ) and the extracellular matrix (ECM) in the development of a fibrotic response. One of the most highly upregulated genes in Mus was Timp1, which functions to inhibit the action of the matrix metalloproteases. The same phenomenon was seen in Mus skin wound arrays (Brant et al., 2016), suggesting that the fibrotic response of Mus to wounding involves the inhibition of MMPs which may generate a more rigid ECM. The interaction of this rigid matrix with the intracellular compartment occurs via the integrins and the upregulation of integrins (e.g. Itgα5, along with Itgα1 – a receptor for fibronectin and fibrillin-1), could signal a fibrotic response to cells at the wound site. Activation of the TGFβ signaling pathway is also a key event in fibrosis (Schachtrup et al., 2010). Serpine1 expression is promoted by TGFβ1 (Honda et al., 2017; Samarakoon & Higgins, 2008) and it is the major physiological regulator of the plasmin based cascade which is involved in fibrotic disorders of the vascular system and several organs such as skin, liver, lung and kidney. Serpine1 is also itself a pro-inflammatory molecule as it activates macrophages (Gupta et al., 2016). Another plasminogen activator involved in the fibrotic pathway (He et al., 2018) upregulated following SCI in Mus was Plau, which could supplement the effects of Serpine1 on fibrosis. In addition to the role these TGFβ1 induced genes play in fibrosis, the activity of plasmin generated by their proteolytic activity is also involved in proteolysis of the ECM which itself could release TGFβ (Deryugina & Quigley, 2012). Interestingly, the TGFβ1 gene was induced to the same degree in the spinal tissue of Mus and Acomys in both the wound and neurogenesis arrays, and also in our skin regeneration experiments (Brant et al., 2015). This observation suggests that posttranscriptional mechanisms are involved in TGFβ1 and fibrotic responses such as its release from the matrix by biomechanical forces or the induction of downstream targets.
In addition to reduced fibrosis-related signaling, following SCI Acomys exhibit an upregulation of molecules associated with neurogenesis and regeneration including Bmp2, Shh, GDNF, and a tendency for elevated Notch 1 and Stat3 levels. This response involves signaling pathways identified in other model systems such as salamanders and zebrafish which successfully regenerate spinal tissue after injury. Specifically, Wnt, BMP and Hh pathways as well as several growth factors have been implicated in regulating the regenerative capacity of the spinal cord (Vergara, Arsenijevic, & Del Rio-Tsonis, 2005). In mammals, these molecules are known to play important roles during development by establishing signaling gradients which control cellular differentiation, axonal guidance, and neurogenesis (Cardozo et al., 2017).
Consistent with the molecular signature of the acutely injured Acomys spinal cord, we found a substantial reduction in staining for collagen IV in the vicinity of the spinal lesion after chronic injury in Acomys. Collagen is a major component of the fibrous scar that forms in the injured spinal cord (Hermanns, Klapka, & Müller, 2001; Stichel & Müller, 1998). Collagen can be expressed by multiple cells types, including endothelial cells (Schwab, Beschorner, Nguyen, Meyermann, & Schluesener, 2001), astrocytes (Liesi & Kauppila, 2002) and fibroblasts (Berry et al., 1983), and is generally considered to be part of the barrier to growth of axonal projections after SCI (Klapka & Müller, 2006). Another contribution to the glial scar comes from reactive astrocytes that express GFAP (Bovolenta et al., 1992). Our evaluation of GFAP intensity in the chronically injured Acomys spinal cord did not demonstrate a clear difference as compared to Mus, although a tendency for reduced staining can be seen in the data. Qualitative differences in the pattern and morphology of GFAP staining around the lesion epicenter are consistent with robust gliosis in Mus, which was not as pronounced in Acomys. Microglia are also activated after SCI and can be found in increased numbers near the interface of astrocytes and infiltrating leukocytes (Bellver-Landete et al., 2019; Hawthorne & Popovich, 2011). Recent evidence indicates that this microglial/macrophage response may in fact be beneficial to the injured spinal cord (Bellver-Landete et al., 2019; Gensel & Zhang, 2015). In our study, there was no suggestion of a difference in IBA1 staining intensity between the chronically injured Acomys and Mus spinal cords, which labels both macrophages and microglia (Imai, Ibata, Ito, Ohsawa, & Kohsaka, 1996). However, visual comparison of the IBA1 positive cells reveals Acomys have fewer, large, round IBA1 positive cells consistent with phagocytic morphology characteristic of activated microglia/macrophages (Wu et al., 2005).
In summary, the molecular and histologic data presented here are consistent with a growing literature that indicates that Acomys has a tissue response to injury that may be relatively unique among mammals. Prior work definitively establishes that Acomys does not undergo fibrosis and has remarkable regenerative capacity in dermis, smooth muscle, skeletal muscle, and other tissue following injury (Brant et al., 2019; Brant et al., 2016; Jiang, Harn, Ou, Lei, & Chuong, 2019; Seifert et al., 2012). Our current data support the hypothesis that this unique response to injury extends to the spinal cord. Based on the reduced inflammatory and fibrotic response and indication of enhanced regenerative capacity, we suggest that Acomys merits further study as comparative model to study adaptive responses to SCI.
Supplementary Material
Highlights.
Spiny mice (Acomys cahirinus) and C57BL/6 (Mus) were studied after spinal injury
RT-PCR gene arrays suggested different molecular response in Acomys
RTq-PCR with species-specific primers showed increased neurogenesis-related signaling in Acomys
Histology indicates reduced scarring and fibrosis in Acomys
Acomys may be a useful comparative model to study SCI
Acknowledgements
This work was supported by funding from the National Institute of Health, grant numbers: K99 HL143207-01 (KS), 1F32NS095620-01 (KS), 1R01NS080180-01A1 (DF), T32-ND043730 (MS) and F31HL145831 (MS), The Keck Foundation (MM) and R21OD023210 (MM and JOB). We thank Dr. Lila Wollman and Shreya Patel for their assistance with surgical procedures. We also thank Marda Jorgensen for excellent histological and immunochemical work, and Ethan Benevides for imaging work.
Footnotes
The authors declare no competing financial interests
REFERENCES
- Ahn JH, Choi JH, Park JH, Yan BC, Kim IH, Lee JC, … Won MH (2012). Comparison of alpha-synuclein immunoreactivity in the spinal cord between the adult and aged beagle dog. Lab Anim Res, 28(3), 165–170. doi: 10.5625/lar.2012.28.3.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bambakidis NC, Wang RZ, Franic L, & Miller RH (2003). Sonic hedgehog-induced neural precursor proliferation after adult rodent spinal cord injury. J Neurosurg, 99(1 Suppl), 70–75. doi: 10.3171/spi.2003.99.1.0070 [DOI] [PubMed] [Google Scholar]
- Bellver-Landete V, Bretheau F, Mailhot B, Vallières N, Lessard M, Janelle ME, … Lacroix S (2019). Microglia are an essential component of the neuroprotective scar that forms after spinal cord injury. Nat Commun, 10(1), 518. doi: 10.1038/s41467-019-08446-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry M, Maxwell WL, Logan A, Mathewson A, McConnell P, Ashhurst DE, & Thomas GH (1983). Deposition of scar tissue in the central nervous system. Acta Neurochir Suppl (Wien), 32, 31–53. [DOI] [PubMed] [Google Scholar]
- Bovolenta P, Wandosell F, & Nieto-Sampedro M (1992). CNS glial scar tissue: a source of molecules which inhibit central neurite outgrowth. Prog Brain Res, 94, 367–379. [DOI] [PubMed] [Google Scholar]
- Brant JO, Boatwright JL, Davenport R, Sandoval AGW, Maden M, & Barbazuk WB (2019). Comparative transcriptomic analysis of dermal wound healing reveals de novo skeletal muscle regeneration in Acomys cahirinus. PLoS One, 14(5), e0216228. doi: 10.1371/journal.pone.0216228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brant JO, Lopez MC, Baker HV, Barbazuk WB, & Maden M (2015). A Comparative Analysis of Gene Expression Profiles during Skin Regeneration in Mus and Acomys. PLoS One, 10(11), e0142931. doi: 10.1371/journal.pone.0142931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brant JO, Yoon JH, Polvadore T, Barbazuk WB, & Maden M (2016). Cellular events during scar-free skin regeneration in the spiny mouse, Acomys. Wound Repair Regen, 24(1), 75–88. doi: 10.1111/wrr.12385 [DOI] [PubMed] [Google Scholar]
- Cardozo MJ, Mysiak KS, Becker T, & Becker CG (2017). Reduce, reuse, recycle - Developmental signals in spinal cord regeneration. Dev Biol, 432(1), 53–62. doi: 10.1016/j.ydbio.2017.05.011 [DOI] [PubMed] [Google Scholar]
- Carlson SL, Parrish ME, Springer JE, Doty K, & Dossett L (1998). Acute inflammatory response in spinal cord following impact injury. Exp Neurol, 151(1), 77–88. doi: 10.1006/exnr.1998.6785 [DOI] [PubMed] [Google Scholar]
- Cregg JM, DePaul MA, Filous AR, Lang BT, Tran A, & Silver J (2014). Functional regeneration beyond the glial scar. Exp Neurol, 253, 197–207. doi: 10.1016/j.expneurol.2013.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deryugina EI, & Quigley JP (2012). Cell surface remodeling by plasmin: a new function for an old enzyme. J Biomed Biotechnol, 2012, 564259. doi: 10.1155/2012/564259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan B, Wei Z, Yao X, Shi G, Cheng X, Zhou X, … Feng S (2018). Microenvironment Imbalance of Spinal Cord Injury. Cell Transplant, 27(6), 853–866. doi: 10.1177/0963689718755778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantin A, Vieira JM, Gestri G, Denti L, Schwarz Q, Prykhozhij S, … Ruhrberg C (2010). Tissue macrophages act as cellular chaperones for vascular anastomosis downstream of VEGF-mediated endothelial tip cell induction. Blood, 116(5), 829–840. doi: 10.1182/blood-2009-12-257832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaige S, Bonnet MS, Tardivel C, Pinton P, Trouslard J, Jean A, … Dallaporta M (2013). c-Fos immunoreactivity in the pig brain following deoxynivalenol intoxication: focus on NUCB2/nesfatin-1 expressing neurons. Neurotoxicology, 34, 135–149. doi: 10.1016/j.neuro.2012.10.020 [DOI] [PubMed] [Google Scholar]
- Gensel JC, & Zhang B (2015). Macrophage activation and its role in repair and pathology after spinal cord injury. Brain Res, 1619, 1–11. doi: 10.1016/j.brainres.2014.12.045 [DOI] [PubMed] [Google Scholar]
- Gupta KK, Xu Z, Castellino FJ, & Ploplis VA (2016). Plasminogen activator inhibitor-1 stimulates macrophage activation through Toll-like Receptor-4. Biochem Biophys Res Commun, 477(3), 503–508. doi: 10.1016/j.bbrc.2016.06.065 [DOI] [PubMed] [Google Scholar]
- Hawthorne AL, & Popovich PG (2011). Emerging concepts in myeloid cell biology after spinal cord injury. Neurotherapeutics, 8(2), 252–261. doi: 10.1007/s13311-011-0032-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Tsou PS, Khanna D, & Sawalha AH (2018). Methyl-CpG-binding protein 2 mediates antifibrotic effects in scleroderma fibroblasts. Ann Rheum Dis, 77(8), 1208–1218. doi: 10.1136/annrheumdis-2018-213022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanns S, Klapka N, & Müller HW (2001). The collagenous lesion scar--an obstacle for axonal regeneration in brain and spinal cord injury. Restor Neurol Neurosci, 19(1–2), 139–148. [PubMed] [Google Scholar]
- Hilton BJ, Assinck P, Duncan GJ, Lu D, Lo S, & Tetzlaff W (2013). Dorsolateral funiculus lesioning of the mouse cervical spinal cord at C4 but not at C6 results in sustained forelimb motor deficits. J Neurotrauma, 30(12), 1070–1083. doi: 10.1089/neu.2012.2734 [DOI] [PubMed] [Google Scholar]
- Honda H, Fujimoto M, Serada S, Urushima H, Mishima T, Lee H, … Naka T (2017). Leucine-rich. Physiol Rep, 5(24). doi: 10.14814/phy2.13556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide T, Uchida K, Tamura S, & Nakayama H (2010). Histiocytic sarcoma in the brain of a cat. J Vet Med Sci, 72(1), 99–102. doi: 10.1292/jvms.09-0312 [DOI] [PubMed] [Google Scholar]
- Imai Y, Ibata I, Ito D, Ohsawa K, & Kohsaka S (1996). A novel gene iba1 in the major histocompatibility complex class III region encoding an EF hand protein expressed in a monocytic lineage. Biochem Biophys Res Commun, 224(3), 855–862. doi: 10.1006/bbrc.1996.1112 [DOI] [PubMed] [Google Scholar]
- Ito D, Imai Y, Ohsawa K, Nakajima K, Fukuuchi Y, & Kohsaka S (1998). Microglia-specific localisation of a novel calcium binding protein, Iba1. Brain Res Mol Brain Res, 57(1), 1–9. doi: 10.1016/s0169-328x(98)00040-0 [DOI] [PubMed] [Google Scholar]
- Jiang TX, Harn HI, Ou KL, Lei M, & Chuong CM (2019). Comparative regenerative biology of spiny (Acomys cahirinus) and laboratory (Mus musculus) mouse skin. Exp Dermatol, 28(4), 442–449. doi: 10.1111/exd.13899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Park SR, & Choi BH (2014). Biomaterial scaffolds used for the regeneration of spinal cord injury (SCI). Histol Histopathol, 29(11), 1395–1408. doi: 10.14670/HH-29.1395 [DOI] [PubMed] [Google Scholar]
- Klapka N, & Müller HW (2006). Collagen matrix in spinal cord injury. J Neurotrauma, 23(3–4), 422–435. doi: 10.1089/neu.2006.23.422 [DOI] [PubMed] [Google Scholar]
- Liesi P, & Kauppila T (2002). Induction of type IV collagen and other basement-membrane-associated proteins after spinal cord injury of the adult rat may participate in formation of the glial scar. Exp Neurol, 173(1), 31–45. doi: 10.1006/exnr.2001.7800 [DOI] [PubMed] [Google Scholar]
- Livak KJ, & Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 25(4), 402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Maden M, Brant JO, Rubiano A, Sandoval AGW, Simmons C, Mitchell R, … Patel K (2018). Perfect chronic skeletal muscle regeneration in adult spiny mice, Acomys cahirinus. Sci Rep, 8(1), 8920. doi: 10.1038/s41598-018-27178-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen RJ, Buck CR, & Smith AM (1992). NeuN, a neuronal specific nuclear protein in vertebrates. Development, 116(1), 201–211. [DOI] [PubMed] [Google Scholar]
- Pereira IM, Marote A, Salgado AJ, & Silva NA (2019). Filling the Gap: Neural Stem Cells as A Promising Therapy for Spinal Cord Injury. Pharmaceuticals (Basel), 12(2). doi: 10.3390/ph12020065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro G, Prata DF, Araújo IM, & Tiscornia G (2018). The African spiny mouse ( Acomys spp.) as an emerging model for development and regeneration. Lab Anim, 52(6), 565–576. doi: 10.1177/0023677218769921 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Callejas JD, Fuchs E, & Perez-Cruz C (2016). Evidence of Tau Hyperphosphorylation and Dystrophic Microglia in the Common Marmoset. Front Aging Neurosci, 8, 315. doi: 10.3389/fnagi.2016.00315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosich K, Hanna BF, Ibrahim RK, Hellenbrand DJ, & Hanna A (2017). The Effects of Glial Cell Line-Derived Neurotrophic Factor after Spinal Cord Injury. J Neurotrauma, 34(24), 3311–3325. doi: 10.1089/neu.2017.5175 [DOI] [PubMed] [Google Scholar]
- Samarakoon R, & Higgins PJ (2008). Integration of non-SMAD and SMAD signaling in TGF-beta1-induced plasminogen activator inhibitor type-1 gene expression in vascular smooth muscle cells. Thromb Haemost, 100(6), 976–983. [PMC free article] [PubMed] [Google Scholar]
- Schachtrup C, Ryu JK, Helmrick MJ, Vagena E, Galanakis DK, Degen JL, … Akassoglou K (2010). Fibrinogen triggers astrocyte scar formation by promoting the availability of active TGF-beta after vascular damage. J Neurosci, 30(17), 5843–5854. doi: 10.1523/JNEUROSCI.0137-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab JM, Beschorner R, Nguyen TD, Meyermann R, & Schluesener HJ (2001). Differential cellular accumulation of connective tissue growth factor defines a subset of reactive astrocytes, invading fibroblasts, and endothelial cells following central nervous system injury in rats and humans. J Neurotrauma, 18(4), 377–388. doi: 10.1089/089771501750170930 [DOI] [PubMed] [Google Scholar]
- Seifert AW, Kiama SG, Seifert MG, Goheen JR, Palmer TM, & Maden M (2012). Skin shedding and tissue regeneration in African spiny mice (Acomys). Nature, 489(7417), 561–565. doi: 10.1038/nature11499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stichel CC, & Müller HW (1998). The CNS lesion scar: new vistas on an old regeneration barrier. Cell Tissue Res, 294(1), 1–9. [DOI] [PubMed] [Google Scholar]
- Thomas AM, & Shea LD (2013). Polysaccharide-modified scaffolds for controlled lentivirus delivery in vitro and after spinal cord injury. J Control Release, 170(3), 421–429. doi: 10.1016/j.jconrel.2013.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, & Katze MG (2012). Into the eye of the cytokine storm. Microbiol Mol Biol Rev, 76(1), 16–32. doi: 10.1128/MMBR.05015-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran AP, Warren PM, & Silver J (2018). The Biology of Regeneration Failure and Success After Spinal Cord Injury. Physiol Rev, 98(2), 881–917. doi: 10.1152/physrev.00017.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuinstra HM, Margul DJ, Goodman AG, Boehler RM, Holland SJ, Zelivyanskaya ML, … Shea LD (2014). Long-term characterization of axon regeneration and matrix changes using multiple channel bridges for spinal cord regeneration. Tissue Eng Part A, 20(5–6), 1027–1037. doi: 10.1089/ten.TEA.2013.0111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SM, Hoyt AK, ElMallah MK, Falk DJ, Byrne BJ, & Fuller DD (2016). Neuropathology in respiratory-related motoneurons in young Pompe (Gaa(−/−)) mice. Respir Physiol Neurobiol, 227, 48–55. doi: 10.1016/j.resp.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SMF, Falk DJ, Byrne BJ, & Fuller DD (2016). Transcriptome assessment of the Pompe (Gaa−/−) mouse spinal cord indicates widespread neuropathology. Physiol Genomics, 48(11), 785–794. doi: 10.1152/physiolgenomics.00075.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangansewinkel T, Lemmens S, Geurts N, Quanten K, Dooley D, Pejler G, & Hendrix S (2019). Mouse mast cell protease 4 suppresses scar formation after traumatic spinal cord injury. Sci Rep, 9(1), 3715. doi: 10.1038/s41598-019-39551-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara MN, Arsenijevic Y, & Del Rio-Tsonis K (2005). CNS regeneration: a morphogen’s tale. J Neurobiol, 64(4), 491–507. doi: 10.1002/neu.20158 [DOI] [PubMed] [Google Scholar]
- Werner S, & Grose R (2003). Regulation of wound healing by growth factors and cytokines. Physiol Rev, 83(3), 835–870. doi: 10.1152/physrev.2003.83.3.835 [DOI] [PubMed] [Google Scholar]
- Wilhelmsson U, Li L, Pekna M, Berthold CH, Blom S, Eliasson C, … Pekny M (2004). Absence of glial fibrillary acidic protein and vimentin prevents hypertrophy of astrocytic processes and improves post-traumatic regeneration. J Neurosci, 24(21), 5016–5021. doi: 10.1523/JNEUROSCI.0820-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Miyamoto O, Shibuya S, Okada M, Igawa H, Janjua NA, … Itano T (2005). Different expression of macrophages and microglia in rat spinal cord contusion injury model at morphological and regional levels. Acta Med Okayama, 59(4), 121–127. doi: 10.18926/AMO/31950 [DOI] [PubMed] [Google Scholar]
- Xiao Q, Du Y, Wu W, & Yip HK (2010). Bone morphogenetic proteins mediate cellular response and, together with Noggin, regulate astrocyte differentiation after spinal cord injury. Exp Neurol, 221(2), 353–366. doi: 10.1016/j.expneurol.2009.12.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported herein are available from the corresponding author (DDF) upon reasonable request.