Abstract
Phosphoribosyl-pyrophosphate synthetase 2 (PRPS2) is a rate-limiting enzyme and plays an important role in purine and pyrimidine nucleotide synthesis. Recent studies report that PRPS2 is involved in male infertility. However, the role of PRPS2 in hypospermatogenesis is unknown. In this study, the relationship of PRPS2 with hypospermatogenesis and spermatogenic cell apoptosis was investigated. The results showed that PRPS2 depletion increased the number of apoptotic spermatogenic cells in vitro. PRPS2 was downregulated in a mouse model of hypospermatogenesis. When PRPS2 expression was knocked down in mouse testes, hypospermatogenesis and accelerated apoptosis of spermatogenic cells were noted. E2F transcription factor 1 (E2F1) was confirmed as the target gene of PRPS2 and played a key role in cell apoptosis by regulating the P53/Bcl-xl/Bcl-2/Caspase 6/Caspase 9 apoptosis pathway. Therefore, these data indicate that PRPS2 depletion contributes to the apoptosis of spermatogenic cells and is associated with hypospermatogenesis, which may be helpful for the diagnosis of male infertility.
Keywords: hypospermatogenesis, male infertility, molecular marker, phosphoribosyl-pyrophosphate synthetase 2, spermatogenesis
INTRODUCTION
Hypospermatogenesis is the most prevalent histological type in azoospermic patients,1,2 which is characterized by a low production of spermatozoa and is associated with male infertility.3,4 However, the etiology and the molecular basis of hypospermatogenesis remain unknown.
During spermatogenesis, germ cell apoptosis maintains the normal development and output of germ cells.5 Increased apoptosis of germ cells in the testis is implicated in hypospermatogenesis and male infertility.6,7,8,9 Accelerated apoptosis of spermatogenic cells is frequently observed in azoospermic patients with hypospermatogenesis.10,11,12 Thus, studies in germ cell apoptosis may be helpful in clarifying the molecular basis of hypospermatogenesis.
Phosphoribosyl-pyrophosphate synthetase 2 (PRPS2) belongs to the family of phosphoribosylpyrophosphate synthetases (PRS) and plays an important role in purine and pyrimidine nucleotide synthesis.13 PRPS2 is located in the p22 region of the X chromosome and has high homology with PRPS1.14 Although both the PRPS1 and PRPS2 genes are highly expressed in the thymus, adipose, and testes,15,16 they display different biological functions. Compared with PRPS2, PRPS1 plays an important role in the nucleotide metabolism of normal cells.13,17,18 PRPS2, but not PRPS1, is reported to be associated with tumor formation and development.18–20 Previous studies have revealed that PRPS2 expression inhibits the apoptosis of Sertoli cells and is correlated with Sertoli cell-only syndrome (SCOS).21,22 However, the effect of PRPS2 expression on spermatogenic cell apoptosis is unknown. Furthermore, the correlation between PRPS2 expression and hypospermatogenesis is unclear. In this study, we investigated the effect of PRPS2 expression on the apoptosis of spermatogenic cells and evaluated the correlation between PRPS2 expression and hypospermatogenesis.
MATERIALS AND METHODS
Cell culture and transfection
The mouse-derived spermatogonial cell line, GC1-spg, and the spermatocyte cell line, GC2-spg, were purchased from the American Type Culture Collection (Manassas, VA, USA) and cultured with Dulbecco's Modified Eagle Medium (DMEM; Gibco, Utah, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) at 37°C in a 5% CO2 incubator. PRPS2 knockdown and overexpression in the GC1 and GC2 cells were performed by murine lentivirus vectors supplied by GenePharma Biomedical Company (Shanghai, China). The sequence of the murine PRPS2-specific shRNA was CCACCAAAGTTTATGCTAT. PRPS2 overexpression was achieved by a recombinant lentivirus containing murine PRPS2. The negative control (NC) was transfected with the empty lentivirus vector. All experimental methods were carried out in accordance with the approved guidelines of the Southern Medical University Research Center, Guangzhou, China. All the experimental protocols were approved by the Institutional Animal Care and Use Licensing Committee of the Southern Medical University (No. S3077).
Animal models
All C57 male mice were supplied by the Southern Medical University Animal Center. This study was supported by the university's institutional animal care and use committee. The mouse testis tissues were collected at different postpartum times (P4-P10 weeks) for detecting PRPS2 expression. A mouse model of hypospermatogenesis was obtained as previously described.23 Briefly, a total of 12 C57 male mice that were 8 weeks old were purchased and raised in the Southern Medical University Animal Center. All mice were randomly assigned to an experimental group or a negative control group. Those in the experimental group (n = 6) were treated with busulfan and dimethyl sulfoxide (DMSO; Biyuntian, Shanghai, China) solution (30 mg kg−1 body weight) by intraperitoneal injection. Mice in the control group (n = 6) were treated with the same volume of DMSO. Two weeks after treatment, the histopathological changes of the testis tissues were evaluated using hematoxylin–eosin (H and E; Yuanye, Shanghai, China) staining. Then, PRPS2 expression was quantified in the mice with hypospermatogenesis.
PRPS2-specific shRNA lentivirus treatment
Eighteen C57 male mice that were 8 weeks old were randomly assigned to an experimental group, a NC group, or a blank group. Under anesthesia, the murine PRPS2-specific short-hairpin RNA lentivirus (10 μl) was injected into both testes of six mice in the experimental group by a microsyringe (size = 0.45 mm). Six mice in the NC group were treated with the same volume of empty lentivirus (10 μl), whereas the mice in the blank group (n = 6) did not receive any treatment. One week after the treatment, the mice were killed by ketamine (IsoReag, Shanghai, China), and the testes were collected for further investigation. The murine PRPS2-specific shRNA lentivirus and the empty lentivirus were supplied by GeneChem bioMedical Biotechnology (Shanghai, China).
Immunohistochemical (IHC) analysis
The procedure was performed as previously described.23,24 Briefly, the testis tissues were fixed with Bouin solution (Meilun, Dalian, China) for 6 h and dehydrated and embedded in paraffin. Antigen retrieval was performed with 0.01 mol l−1 sodium citrate buffer (pH 6.0; Biyuntian). The sections were incubated with a polyclonal PRPS2 antibody (0.5 mg ml−1, dilution 1: 150, product No. PA5-42007, Invitrogen, Carlsbad, CA, USA), a polyclonal Caspase 3 antibody (0.5 mg ml−1, dilution 1: 100, product No. BS90186, Bioworld Technology, Minneapolis, MN, USA), a polyclonal Caspase 6 antibody (0.5 mg ml−1, dilution 1: 100, product No. BS90188, Bioworld Technology), and a polyclonal Caspase 9 antibody (0.5 mg ml−1, dilution 1: 100, product No. BS1731, Bioworld Technology) at room temperature for 2 h. The secondary antibody (Biyuntian) was incubated at room temperature for 30 min. Then, the sections were visualized by diaminobenzidine (DAB; Biyuntian) and counterstained by hematoxylin. Fetal bovine serum (FBS; Gibco) replaced the primary antibody was used as negative control. Those sections with confirmed positive expression of PRPS2, Caspase 3, Caspase 6, and Caspase 9 were treated as positive controls.
Quantitative real-time polymerase chain reaction (qRT-PCR)
The procedure was performed as previously described.24 Total RNA was extracted from the testes and cells with Trizol reagent (TAKARA, Dalian, China) in accordance with the manufacturer's protocol. RNA was reverse transcribed into cDNA by the Reverse Transcription System (TAKARA). Real-time PCR was used to measure and compare the expression of the genes. The sense sequences of genes are described in Supplementary Table 1. The mRNA level of each sample was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression.
Supplementary Table 1.
The sense sequences of genes were as follow
| Name | Sequece |
|---|---|
| PRPS2-F-Human | 5’- AGCTCGCATCAGGACCTGT-3’ |
| PRPS2-R-Human | 5’- ACGCTTTCACCAATCTCCACG-3’ |
| PRPS2-F-Mouse | 5’- ATGCTGGAGGAGCCAAAAG-3’ |
| PRPS2-R-Mouse | 5’- ACATCACCCACGAGAACCAT-3’ |
| SRA1-F-Mouse | 5’- ACGACCCGCCACAATTCTC-3’ |
| SRA1-R-Mouse | 5’- CTGGAAGCCTTACTTGAAGGAG-3’ |
| PHLDL3-F-Mouse | 5’- CCGTGGAGTGCGTAGAGAG-3’ |
| PHLDA3-R-Mouse | 5’- TCTGGATGGCCTGTTGATTCT-3’ |
| E2F1-F-Mouse | 5’- CTCGACTCCTCGCAGATCG-3’ |
| E2F1-R-Mouse | 5’- GATCCAGCCTCCGTTTCACC-3’ |
| BRE-F-Mouse | 5’- GGTGGACTACGCGGAGAAC-3’ |
| BRE-R-Mouse | 5’- CACCTGACTCAACCGCTCTTT-3’ |
| Sirt6-F-Mouse | 5’- ATGTCGGTGAATTATGCAGCA-3’ |
| Sirt6-R-Mouse | 5’- GCTGGAGGACTGCCACATTA-3’ |
| Egr1-F-Mouse | 5’- TCGGCTCCTTTCCTCACTCA-3’ |
| Egr1-R-Mouse | 5’- CTCATAGGGTTGTTCGCTCGG-3’ |
| Celsr2-F-Mouse | 5’- CACGATGGCCTGAGGGTTT-3’ |
| Celsr2-R-Mouse | CCTTGTGGAGAAAGGTGTCCT-3’ |
| Polg-F-Mouse | 5’- GAGCCTGCCTTACTTGGAGG-3’ |
| Polg-R-Mouse | 5’- GGCTGCACCAGGAATACCA-3’ |
| Spata2l-F-Mouse | 5’- ATGTGCTGAAGGGTGTACTCT-3’ |
| Spata2l-R-Mouse | 5’- GGGGTGGTCACCATTAGGC-3’ |
| GAPDH-F-Human | 5’- CTGAACGGGAAGCTCACTGG-3 |
| GAPDH-R-Human | 5’- TGAGGTCCACCACCCTGTTG-3’ |
| GAPDH-F-Mouse | 5’- AGGTCGGTGTGAACGGATTTG-3’ |
Western blot
The procedure was performed as previously described.24 The total protein was extracted from the testicular tissue and cells by radioimmunoprecipitation assay (RIPA) buffer and was quantified by the bicinchoninic acid method (Biyuntian). The protein lysates were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes (Biosharp, Waltham, MA, USA). After being blocked in phosphate-buffered saline with Tween 20 (PBST) solution for 1.5 h, the membranes were incubated with polyclonal PRPS2 antibody (0.1 mg ml−1, dilution 1: 1000, product No. PA5-42007, Invitrogen), polyclonal E2F1 antibody (0.2 mg ml−1, dilution 1: 2000, product No. sc-137059, Santa Cruz Corp., Santa Cruz, CA, USA), polyclonal P53 antibody (0.2 mg ml−1, dilution 1: 2000, product No. LBP62662, Immunoway Corp., Plano, TX, USA), polyclonal Bcl-xl antibody (0.1 mg ml−1, dilution: 1: 500, product No. A10753, Abclonal Technology, Boston, MA, USA), polyclonal Bcl-2 antibody (0.2 mg ml−1, dilution 1: 500, product No. A11025, Abclonal Technology), polyclonal Caspase 6 antibody (0.1 mg ml−1, dilution 1: 500, product No. BS90188, Bioworld Technology), polyclonal Caspase 9 antibody (0.1 mg ml−1, dilution 1: 500, product No. BS1731, Bioworld Technology), β-actin antibody (0.1 mg ml−1, dilution 1: 2000, product No. 3700T, Cell Signaling Technology, Beverly, MA, USA), and monoclonal Tubulin antibody (0.1 mg ml−1, dilution 1: 2000, product No. 2146s, Cell Signaling Technology). Then, the membranes were incubated with a secondary antibody (Biyuntian) at room temperature for 30 min and developed by an enhanced chemiluminescence detection kit (Alpha Innotech, San Leandro, CA, USA).
Immunofluorescence
Cells were plated on sterile glass sections, fixed with 95% ethanol for 30 min, and treated with 0.4% Triton X-100 (Kaiji, Guangzhou, China) for 15 min. The testicular sections were dehydrated, dewaxed, and underwent antigen retrieval. After being blocked in 10% FBS at room temperature for 20 min, the sections were incubated with PRPS2 (0.5 mg ml−1, dilution 1: 150, product No. PA5-42007, Invitrogen) and E2F1 (0.5 mg ml−1, dilution: 1: 200, product No. sc-137059, Santa Cruz Corp.) antibodies at 4°C overnight. Then, the sections were incubated with fluorescein isothiocyanate (FITC)-conjugated and Texas Red (TR)-conjugated antibodies (Zsbio Commerce Store, Beijing, China) at room temperature for 60 min and stained with 6-diamidino-2-phenylindole (Invitrogen). Finally, the immunofluorescence images were captured in a fluorescence microscope (Olympus, Tokyo, Japan). FBS replaced the primary antibody in the negative controls. Tissues with positive expression were used as positive controls. Sections with confirmed positive expression of PRPS2 and E2F1 were treated as positive controls.
Cell apoptosis
Apoptosis in the GC1 and GC2 cells was determined as previously described.22 Cells (2 × 105) were harvested and incubated with specific binding of propidium iodide and annexin V-FITC (BestBio, Shanghai, China). Cell apoptosis was examined by flow cytometry. Experiments were repeated three times in the same conditions.
Gene expression analysis
Whole mouse gene expression was detected using the Agilent Mouse Gene Expression Microarray (Agilent Technologies, Palo Alto, CA, USA), which contains more than 39 000 well-characterized mouse genes and transcripts. The quality control, GeneChip hybridization, and data acquisition were performed by KangChen Bio-tech Company (Shanghai, China). The data were analyzed using Agilent feature extraction software, and the data were normalized using Agilent GeneSpring GX version 12.1 software (Agilent Technologies). With the Agilent GeneSpring algorithms, the gene expression was measured in GC1/shPRPS2, GC1/PRPS2, and GC1/NC cells. The differential genes (P < 0.05) were selected and further analyzed by Gene Ontology and Pathway Analyses.
Dual-Luciferase Reporter Assays
The coding region of the E2F1 gene was amplified by polymerase chain reaction (PCR) and inserted into the GV238 vector. The E2F1 plasmid and the PRPS2 lentivirus vectors were cotransfected into the cells using Lipofectamine 2000 (Invitrogen). After 48 h, luciferase activity was measured by the Dual-Luciferase Reporter Assay System (Promega, Madison, CA, USA). Each assay was done in triplicate.
Transferase-mediated deoxyuridine triphosphate-biotin nick end labeling (TUNEL) assay
The TUNEL assay was performed as previously described,24 according to the manufacturer's instructions (Promega). Briefly, the tissue sections were dehydrated, dewaxed, and quenched with 1% proteinase K (20 mg ml−1) at 37°C for 30 min. The sections were incubated with the TUNEL mix (1.0 μl biotin-11-dUTP, 45 μl equilibration buffer, 4.0 μl TdT enzyme, and 50 μl reaction buffer) at 37°C for 60 min. After staining with 6-diamidino-2-phenylindole, the apoptotic cells became red and were counted by fluorescence microscopy at 570 nm.
Statistical analysis
All the data were recorded as mean ± standard deviation and analyzed by SPSS software (version 18.0; SPSS, Chicago, IL, USA). Independent-samples t- test was used to compare the difference after normality tests. P < 0.05 was regarded as statistical significance.
RESULTS
PRPS2 depletion and apoptosis of spermatogenic cells
To evaluate the effect of PRPS2 expression on the apoptosis of spermatogenic cells, flow cytometry was performed on GC1 and GC2 cells. First, the lentivirus vectors were transfected into GC1 and GC2 cells (Supplementary Figure 1 (203.2KB, tif) ). Then, qRT-PCR and Western blot analysis confirmed that PRPS2 expression was successfully downregulated by shRNA gene silencing and upregulated when overexpressed in both GC1 and GC2 cells (Figure 1a). The apoptotic rate in the GC1/shPRPS2 group was significantly higher (16.1% ± 3.0%, P < 0.01) than that in the GC1/NC group (2.8% ± 0.2%), whereas the apoptotic rate in the GC1/PRPS2 group was significantly reduced (0.5% ± 0.3%, P < 0.01) (Figure 1b). In addition, the apoptotic rates (mean±s.d.) in GC2/NC, GC2/shPRPS2, and GC2/PRPS2 groups were 4.0% ± 0.2%, 18.7% ± 1.9%, and 1.9% ± 0.2%, respectively (Figure 1c). Compared with GC2/NC group, apoptosis in GC2/shPRPS2 was significantly increased (P < 0.01), while significantly decreased in GC2/PRPS2 (P < 0.01).
Figure 1.
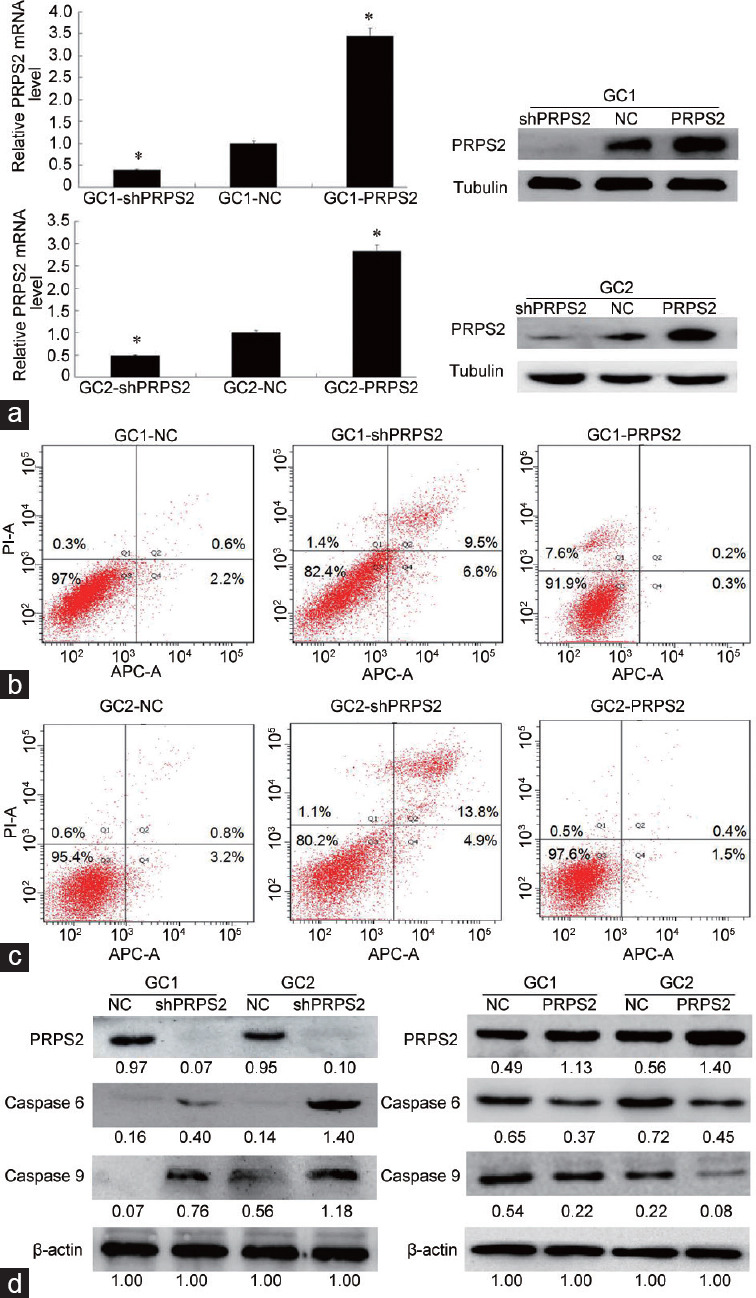
PRPS2 depletion and apoptosis of spermatogenic cells. (a) qRT-PCR and Western blot analysis confirmed that PRPS2 was successfully downregulated by gene silencing and upregulated by PRPRS2 overexpression in both GC1 and GC2 cells. (b) Flow cytometry analysis shows that PRPS2 depletion promotes the apoptosis of GC1 cells. (c) Flow cytometry analysis shows that PRPS2 depletion promotes the apoptosis of GC2 cells (abscissa: cell count; ordinate: the fluorescence intensity). (d) Western blot analysis indicates that PRPS2 depletion activates the expression of apoptotic proteins (values beneath blots are relative to the control). *Significantly different compared with negative control, P < 0.05 by independent-samples t-test. shPRPS2: small-hairpin RNA gene silencer; NC: negative control; PRPS2: overexpression; PRPS2: phosphoribosyl-pyrophosphate synthetase 2; qRT-PCR: quantitative real-time polymerase chain reaction; APC-A: Allophycocyanin-A.
Subsequently, the expression levels of Caspase 6 and Caspase 9 were measured by Western blotting. The results revealed that the expression levels of Caspase 6 and Caspase 9 were significantly increased when PRPS2 expression was downregulated in the GC1 and GC2 cells (Figure 1d). PRPS2 overexpression suppressed the expression of Caspase 6 and Caspase 9.
PRPS2 depletion and hypospermatogenesis
To investigate the correlation between PRPS2 expression and hypospermatogenesis, a mouse model of hypospermatogenesis was used. Compared with the normal controls (Figure 2a and 2b), a significant decrease in spermatogenic cells and epididymal spermatozoa was observed in the mouse model of hypospermatogenesis (Figure 2c and 2d). Compared with that in the normal controls (Figure 2e), PRPS2 expression in the spermatogenic cells was obviously reduced in the mouse model of hypospermatogenesis (Figure 2f) by IHC. The Western blot (Figure 2g) and qRT-PCR (Figure 2h) results also revealed that the PRPS2 expression level was significantly decreased in the mouse model of hypospermatogenesis.
Figure 2.
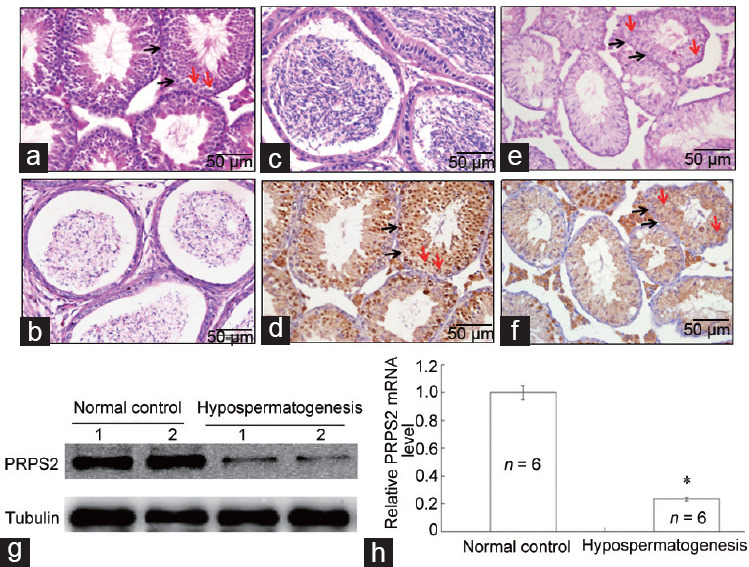
Correlation of PRPS2 expression with hypospermatogenesis. (a) H and E staining of the testis with normal spermatogenesis. (b) H and E staining of normal cauda epididymidis. (c) H and E staining of a testis with hypospermatogenesis. (d) H and E staining of cauda epididymidis in hypospermatogenesis. (e) PRPS2 expression was detected in normal testis by IHC. (f) PRPS2 expression was detected in a testis with hypospermatogenesis by IHC. Scale bars = 50 μm. (g) PRPS2 expression was detected in testis with normal spermatogenesis and hypospermatogenesis by Western blot. (h) PRPS2 expression was detected in testis tissues with normal spermatogenesis and hypospermatogenesis by qRT-PCR. *Hypospermatogenesis group compared with normal control, P < 0.05 by independent-samples t-test. Red arrow: spermatogonia; Black arrow: spermatocyte. PRPS2: phosphoribosyl-pyrophosphate synthetase 2; H and E: hematoxylinand and eosin; IHC: immunohistochemical; qRT-PCR: quantitative real-time polymerase chain reaction.
To further investigate the correlation between PRPS2 and hypospermatogenesis, a PRPS2-specific shRNA lentivirus was used to knock down PRPS2 expression in the mouse testis. The endogenous PRPS2 expression in the test is gradually increased from 4 to 10 weeks (Figure 3a and 3b). After treatment with the shPRPS2 lentivirus, the endogenous testicular PRPS2 expression in the testis at 9 weeks was successfully knocked down compared with that in the negative and blank controls (Figure 3c). The H and E staining revealed that shPRPS2 lentivirus treatment caused serious damage to many seminiferous tubules, while few seminiferous tubules were damaged in the negative controls (Figure 3d). The number of spermatogenic cells in seminiferous tubules was significantly decreased in the mice treated with the shPRPS2 lentivirus, which was similar to the observations in the mouse model of hypospermatogenesis (Figure 3d).
Figure 3.
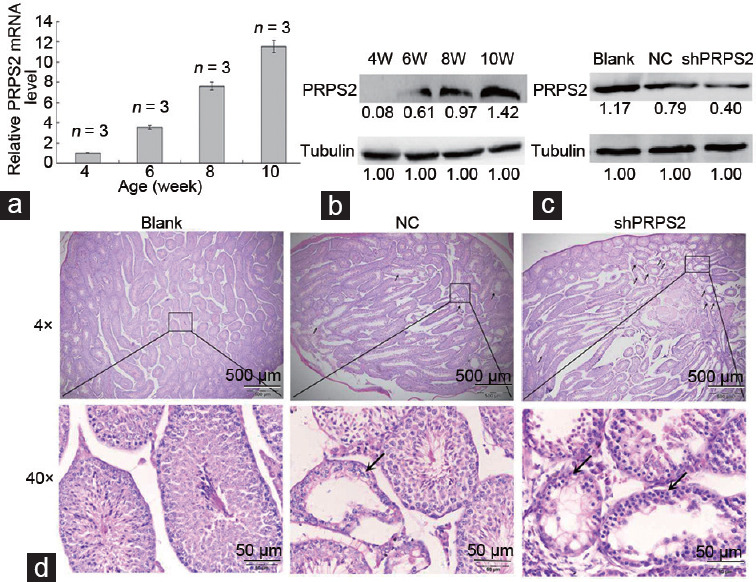
PRPS2 depletion and hypospermatogenesis. (a) PRPS2 mRNA levels were increased with age in the testis. (b) PRPS2 protein level increased with age in the testis. (c) When treated with murine PRPS2-specific shRNA lentivirus, PRPS2 downregulation was observed in testis tissues at 9 weeks of age. (d) When treated with murine PRPS2-specific shRNA lentivirus, hypospermatogenesis was observed in the testis. Upper row: low-power magnification, scale bars=500 μm. Lower row: high-power magnification, scale bars=50 μm. Black arrow: damaged seminiferous tubules. PRPS2: phosphoribosyl-pyrophosphate synthetase 2; NC: negative control; W: weeks.
Next, the TUNEL assay was performed to evaluate the relationship of PRPS2 with spermatogenic cell apoptosis and hypospermatogenesis. A significant increase in the number of apoptotic cells in the seminiferous tubules was observed in the mice treated with the shPRPS2 lentivirus, whereas apoptotic cells were hardly detected in the negative and blank controls (n = 6, P < 0.05, Figure 4). The expression levels of Caspase 3, Caspase 6, and Caspase 9 were significantly elevated in the mice treated with the shPRPS2 lentivirus (Supplementary Figure 2 (196.7KB, tif) ).
Figure 4.
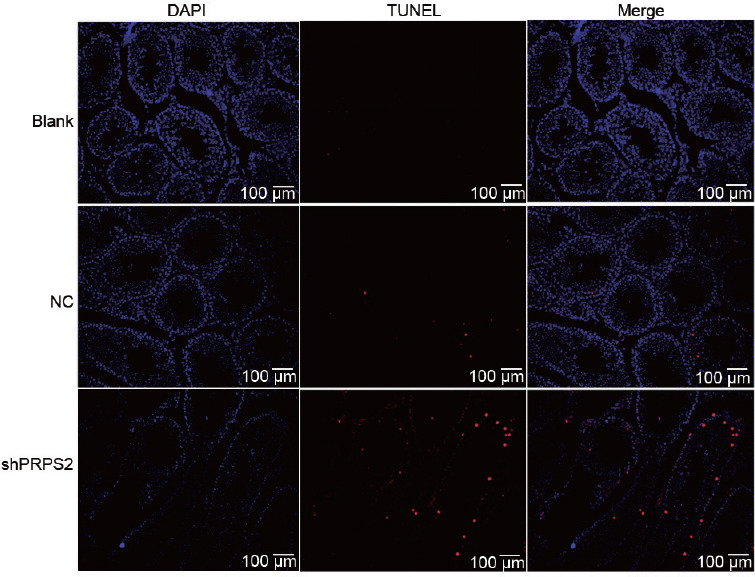
Apoptotic cell number visualized by the TUNEL assay. Representative micrographs indicating an increase in number after treatment with murine PRPS2-specific shRNA lentivirus. PRPS2: phosphoribosyl-pyrophosphate synthetase 2; TUNEL: transferase-mediated deoxyuridine triphosphate-biotin nick end labeling; DAPI: 4',6-diamidino-2-phenylindole.
PRPS2, E2F1, and activation of the P53/Bcl-xl/Bcl-2 signal pathway
To identify the potential signaling pathway associated with PRPS2, a whole-genome expression microarray was performed in GC1/NC, GC1/shPRPS2, and GC1/PRPS2 cells. A total of 1571 upregulated genes and 972 downregulated genes were recorded in GC1/PRPS2 cells compared with GC1/NC cells, which were implicated in the cytoskeleton, differentiation, cell cycle, cell adhesion, cell junction, and apoptosis pathways (P < 0.05, Figure 5a and 5b). A total of 1582 upregulated genes and 1549 downregulated genes were observed in GC1/shPRPS2 cells compared with GC1/NC cells, which were also connected with the cytoskeleton, cell cycle, differentiation, apoptosis, and cell adhesion pathways (P < 0.05, Figure 5c and 5d). To identify potential PRPS2-targeted genes, nine genes associated with the cell apoptosis signaling pathway were selected for further validation by qRT-PCR (Figure 5e). E2F1, Sirt6, and Egr1 were confirmed to be significantly upregulated in GC1/PRPS2 cells and were significantly downregulated in GC1/shPRPS2 cells compared with GC1/NC cells. A luciferase reporter assay was used to evaluate the correlation between PRPS2 and E2F1. The luciferase activity of E2F1 was significantly increased in GC1/PRPS2 and GC2/PRPS2 cells compared with that in the negative controls (P < 0.05; (Figure 5f). Furthermore, double immunofluorescent staining revealed that PRPS2 and E2F1 were colocated in GC1 and GC2 cells and normal testicular tissue (Figure 6a). PRPS2 downregulation decreased the expression of E2F1 in GC1 and GC2 cells (Figure 6b). In addition, when E2F1 and shPRPS2 vectors were cotransfected in GC1 and GC2 cells, the percentage of apoptotic cells was significantly reduced (Supplementary Figure 3 (314.8KB, tif) , P < 0.05).
Figure 5.
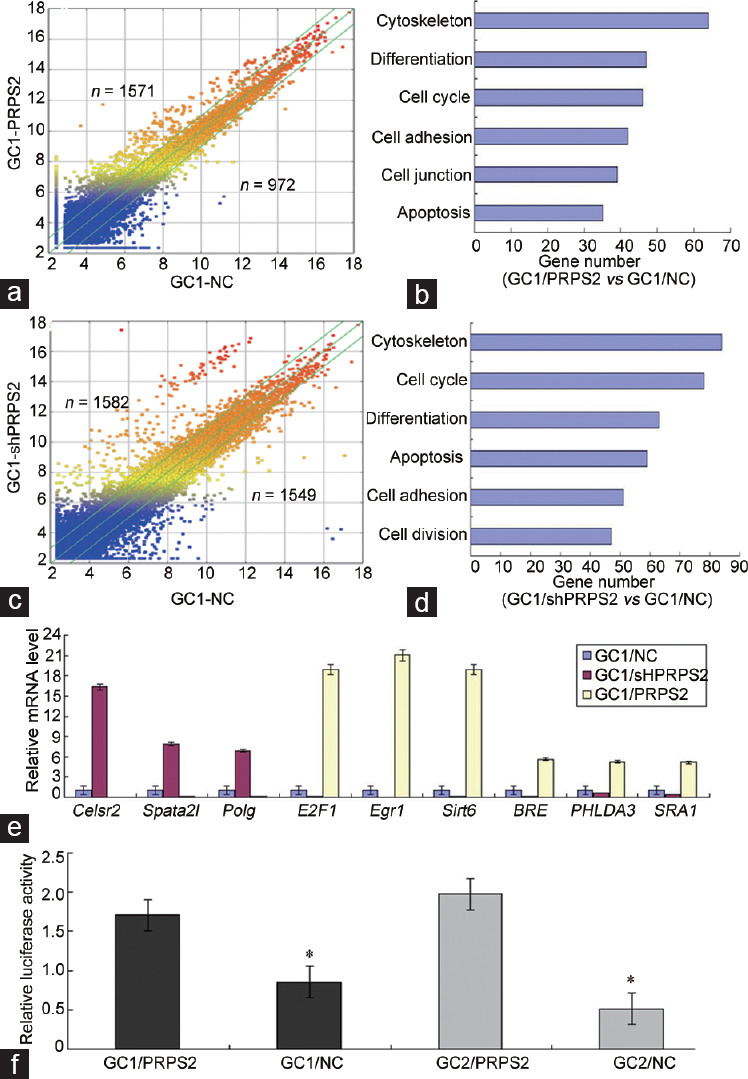
The potential genes and pathways regulated by PRPS2 were identified in GC1 cells. (a) Scatter diagram of the differential genes in GC1/PRPS2 and GC1/NC cells. (b) Potential signaling pathways of the differential genes in GC1/PRPS2 and GC1/NC cells. (c) Scatter diagram of the differential genes in GC1/shPRPS2 and GC1/NC cells. (d) Potential signaling pathways of the differential genes in GC1/shPRPS2 and GC1/NC cells. (e) Nine genes associated with cell apoptosis signal were confirmed in GC1 cells by qRT-PCR. (f) Transcriptional activity of E2F1 was measured by Luciferase Reporter Assay. *GC1/NC versus GC1/PRPS2 and GC2/NC versus GC2/PRPS2, P < 0.05 by independent-samples t-test. PRPS2: phosphoribosyl-pyrophosphate synthetase 2; NC: negative control; qRT-PCR: quantitative real-time polymerase chain reaction; Polg: polymerase (DNA directed), gamma; E2F1: E2F transcription factor 1; Egr1: early growth response 1; BRE: brain and reproductive organ-expressed; PHLDA3: pleckstrin homology-like domain family A member 3; SRA1: steroid receptor RNA activator 1.
Figure 6.
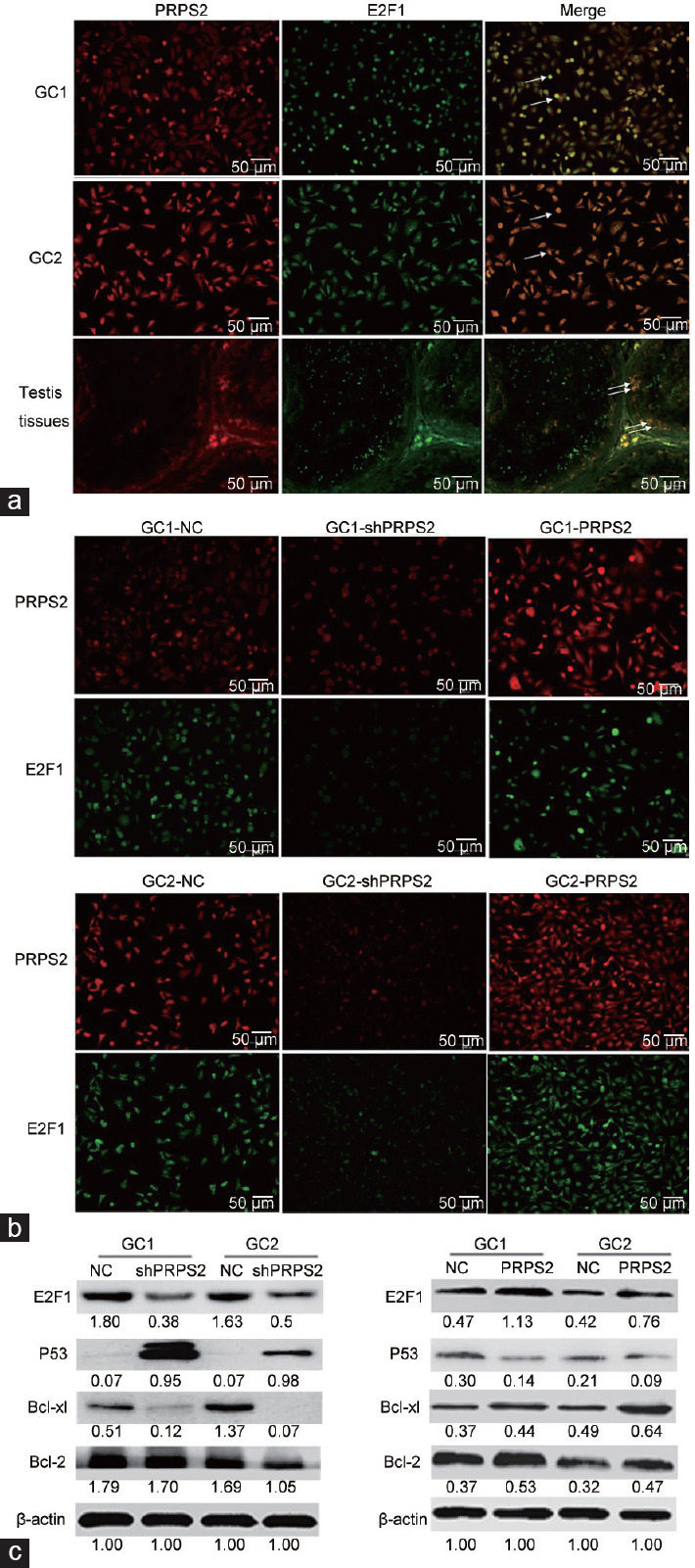
PRPS2 targetes E2F1 and activation of the P53/Bcl-xl/Bcl-2 signal pathway. (a) Colocation of PRPS2 and E2F1 in GC1 and GC2 cells and normal testis tissue was observed by double immunofluorescent staining. (b) After transfection, the expression levels of PRPS2 and E2F1 were detected by double immunofluorescent staining. (c) Western blot was used to measure the expression levels of the E2F1/P53/Bcl-xl/Bcl-2 signaling pathway. Values beneath the blots are relative to the control. White arrow: colocation. PRPS2: phosphoribosyl-pyrophosphate synthetase 2; NC: negative control; E2F1: E2F transcription factor 1.
PRPS2 downregulation decreased the expression of E2F1, Bcl-2, and Bcl-xl, but elevated the expression of P53. PRPS2 overexpression induced the increase of E2F1, Bcl-2, and Bcl-xl, but inhibited the expression of P53 (Figure 6c).
DISCUSSION
PRPS2, a rate-limiting enzyme, is crucial for purine and pyrimidine nucleotide synthesis. In a previous study, endogenous PRPS2 protein was found to be positively expressed in spermatogenic cells, Sertoli cells, and Leydig cells, and the abnormally high expression of PRPS2 in Sertoli cells was associated with the formation of SCOS.22 However, the relationship of PRPS2 with spermatogenic cell apoptosis and hypospermatogenesis is unclear. It is well known that increased apoptosis of germ cells contributes to the occurrence of hypospermatogenesis.6–9 Thus, we hypothesized that PRPS2 is associated with hypospermatogenesis by regulating germ cell apoptosis.
PRPS2 expression was confirmed to be associated with cell apoptosis in the present study, which is consistent with previous studies.18,22 PRPS2 depletion contributed to the apoptosis of spermatogenic cells in vitro. PRPS2 expression in mouse testis progressively increased from 4 to 10 weeks, indicating that PRPS2 expression is associated with testicular development. PRPS2 downregulation was observed in the mouse hypospermatogenesis model. When PRPS2 expression was knocked down in the mouse testis tissues, hypospermatogenesis and spermatogenic cell apoptosis were observed. These observations suggest that PRPS2 depletion contributes to apoptosis in spermatogenic cells and is associated with hypospermatogenesis.
The apoptosis signaling pathway was screened with a whole-genome expression microarray. Only E2F1 was identified as the targeted gene of PRPS2 and played a key role in cell apoptosis. Current studies report that E2F1 is a key regulator of cell cycle progression and correlates with germ cell apoptosis.25,26,27,28 E2F1 depletion and overexpression induce the increase of apoptosis in a P53-dependent or -independent manner.25,26,29 P53, a tumor suppressor, plays an important role in spermatogenesis and spermatogonial cell apoptosis.30,31 The activation of P53 might trigger apoptosis by inducing the synthesis of the Bcl-2 family of pro-apoptosis proteins.32,33,34 Moreover, the inhibition of Bcl-2 and Bcl-xl might activate Caspases to induce intrinsic apoptosis.5,35 The P53/Bcl-2/Caspase signaling pathway is activated by PRPS2 in TM4 cells.22 However, whether there is a correlation between PRPS2 and P53/Bcl-2/Caspases signal pathway in spermatogenic cells is unclear. In this study, the results showed that PRPS2 downregulation decreased the expression of E2F1, Bcl-2, and Bcl-xl, but elevated the expression of P53, Caspase 6, and Caspase 9 in spermatogenic cells. PRPS2 overexpression had reversed the effect. These data indicate that the effect of PRPS2 on cell apoptosis is correlated with the E2F1/P53/Bcl-xl/Bcl-2/Caspase 6/Caspase 9 signal pathway, which contributes to hypospermatogenesis.
It is accepted that the pathological mechanism between hypospermatogenesis and SCOS is unclear. Hypospermatogenesis is characterized by a low production of spermatozoa and a decreased number of spermatogenic cells in the seminiferous tubules.3,4,36 However, SCOS is characterized only by Sertoli cells and a complete lack of spermatogenic cells in the seminiferous tubules.37,38 In a previous study, we reported that PRPS2 expression inhibited the apoptosis of TM4 Sertoli cells and was correlated with SCOS.22 In the present study, PRPS2 depletion contributed to apoptosis in spermatogenic cells and was associated with hypospermatogenesis. Thus, these data indicate that PRPS2 affects apoptosis in spermatogenic cells and Sertoli cells, but different expression levels of PRPS2 may result in different types of pathological changes in the testis. These data suggest that hypospermatogenesis might be associated with the formation of SCOS.
CONCLUSION
Our findings indicate that PRPS2 depletion contributes to apoptosis in spermatogenic cells and is associated with hypospermatogenesis, which may be helpful for the diagnosis of male infertility. Meanwhile, PRPS2 may serve as a potential biomarker associated with male infertility.
AUTHOR CONTRIBUTIONS
XMM and BL conceived and designed the experiments. BL and FPS performed the experiments. LXX, BW, and SBZ analyzed the data. LRZ and XMZ collected and checked the data. BL, LXX, and XMM wrote and revised the manuscript. All authors read and approved the final manuscript.
COMPETING INTERESTS
All authors declared no competing interests.
Lentivirus vectors were transfected into GC1 and GC2 cells to downregulate and upregulate the expression of PRPS2, respectively.
IHC detection indicated that the expression levels of Caspase 3, Caspase 6, and Caspase 9 were significantly higher in the mice with shPRPS2 lentivirus treatment compared with those in the blank and negative controls.
Flow cytometry analysis showed that cell apoptosis was significantly inhibited in GC1/shPRPS2/E2F1 cells compared with that in GC1/shPRPS2 cells.
ACKNOWLEDGMENTS
We thank the staff of the Department of Pathology, Nanfang Hospital and Peking University Shenzhen Hospital. This study was supported by the Science and Technology Projects of Shenzhen (No. JCYJ20160428173152329), the Fundamental Research Funds for the Central Universities (No. 21617316), the Guangdong Medical Research Fund (No. A2019553), and the National Natural Science Foundation of China (No. 81773277).
Supplementary Information is linked to the online version of the paper on the Asian Journal of Andrology website.
REFERENCES
- 1.Robin G, Boitrelle F, Leroy X, Peers MC, Marcelli F, et al. Assessment of azoospermia and histological evaluation of spermatogenesis. Ann Pathol. 2010;30:182–95. doi: 10.1016/j.annpat.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 2.Vogelweid CM, Verina T, Norton J, Harruff R, Ghetti B. Hypospermatogenesis is the cause of infertility in the male weaver mutant mouse. J Neurogenet. 1993;9:89–104. doi: 10.3109/01677069309083452. [DOI] [PubMed] [Google Scholar]
- 3.Sugimoto K, Koh E, Iijima M, Taya M, Maeda Y, et al. Aberrant methylation of the TDMR of the GTF2A1L promoter does not affect fertilisation rates via TESE in patients with hypospermatogenesis. Asian J Androl. 2013;15:634–9. doi: 10.1038/aja.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeshima T, Yumura Y, Iwasaki A, Noguchi K. Clinical review of hypospermatogenesis in patients with a previous episode of mumps orchitis. Hinyokika Kiyo. 2015;61:227–33. Article in Japanese. [PubMed] [Google Scholar]
- 5.Almeida C, Correi S, Rocha E, Alves A, Ferraz L, et al. Caspase signalling pathways in human spermatogenesis. J Assist Reprod Genet. 2013;30:487–95. doi: 10.1007/s10815-013-9938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kandirali E, Cayan S, Armagan A, Erol B, Kadioglu A. Does the testicular apoptotic index vary with serum gonadotropins and testicular histopathology in infertile men? Urol Int. 2009;83:349–53. doi: 10.1159/000241681. [DOI] [PubMed] [Google Scholar]
- 7.Shukla KK, Mahdi AA, Rajender S. Apoptosis, spermatogenesis and male infertility. Front Biosci (Elite Ed) 2012;4:746–54. doi: 10.2741/415. [DOI] [PubMed] [Google Scholar]
- 8.Almeida C, Sousa M, Barros A. Phosphatidylserine translocation in human from impaired spermatogenesis. Reprod Biomed Online. 2009;19:770–7. doi: 10.1016/j.rbmo.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Yao B, Yi N, Zhou S, OuYang W, Xu H, et al. The effect of induced anti-follicle-stimulating hormone autoantibody on serum hormone level and apoptosis in rat testis. Life Sci. 2012;91:83–8. doi: 10.1016/j.lfs.2012.04.026. [DOI] [PubMed] [Google Scholar]
- 10.Takagi S, Itoh N, Kimura M, Sasao T, Tsukamoto T. Spermatogonial proliferation and apoptosis in hypospermatogenesis associated with nonobstructive azoospermia. Fertil Steril. 2001;76:901–7. doi: 10.1016/s0015-0282(01)02732-7. [DOI] [PubMed] [Google Scholar]
- 11.Lin WW, Lamb DJ, Wheeler TM, Lipshultz LI, Kim ED. In situ end-labeling of human testicular tissue demonstrates increased apoptosis in conditions of abnormal spermatogenesis. Fertil Steril. 1997;68:1065–9. doi: 10.1016/s0015-0282(97)00372-5. [DOI] [PubMed] [Google Scholar]
- 12.Jaiswal DS, Trivedi S, Agrawal NK, Singh K. Dysregulation of apoptotic pathway candidate genes and proteins in infertile azoospermia patients. Fertil Steril. 2015;104:736–43. doi: 10.1016/j.fertnstert.2015.05.029. [DOI] [PubMed] [Google Scholar]
- 13.Taira M, Kudoh J, Minoshima S, Iizasa T, Shimada H, et al. Localization of human phosphoribosylpyrophosphate synthetase subunit I and II genes (PRPS1 and PRPS2) to different regions of the X chromosome and assignment of two PRPS1-related genes to autosomes. Somat Cell Mol Gene. 1989;15:29–37. doi: 10.1007/BF01534667. [DOI] [PubMed] [Google Scholar]
- 14.Taira M, Iizasa T, Yamada K, Shimada H, Tatibana M. Tissue-differential expression of two distinct genes for phosphoribosyl pyrophosphate synthetase and existence of the testis-specific transcript. Biochim Biophys Acta. 1989;1007:203–8. doi: 10.1016/0167-4781(89)90040-7. [DOI] [PubMed] [Google Scholar]
- 15.Ishizuka T, Iizasa T, Taira M, Ishijima S, Sonoda T, et al. Promoter regions of the human X-linked housekeeping genes PRPS1 and PRPS2 encoding phosphoribosylpyrophosphate synthetase subunit I and II isoforms. Biochim Biophys Acta. 1992;1130:139–48. doi: 10.1016/0167-4781(92)90521-z. [DOI] [PubMed] [Google Scholar]
- 16.Lee K, Keitz B, Taira M, Chapman VM. Linkage of phosphoribosylpyrophosphate synthetases 1 and 2, Prps1 and Prps2, on the mouse X chromosome. Mamm Genome. 1994;5:612–5. doi: 10.1007/BF00411455. [DOI] [PubMed] [Google Scholar]
- 17.Wang JC, Passage MB, Ellison J, Becker MA, Yen PH, et al. Physical mapping of loci in the distal half of the short arm of the human X chromosome: implications for the spreading of X-chromosome inactivation. Somat Cell Mol Genet. 1992;18:195–200. doi: 10.1007/BF01233165. [DOI] [PubMed] [Google Scholar]
- 18.Cunningham JT, Moreno MV, Lodi A, Ronen SM, Ruggero D. Protein and nucleotide biosynthesis are coupled by a single rate-limiting enzyme, PRPS2, to drive cancer. Cell. 2014;157:1088–103. doi: 10.1016/j.cell.2014.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mannava S, Grachtchouk V, Wheeler LJ, Im M, Zhuang D, et al. Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells. Cell Cycle. 2008;7:2392–400. doi: 10.4161/cc.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minchenko OH, Garmash IA, Kovalevska OV, Tsymbal DO, Minchenko DO. Expression of phosphoribosyl pyrophosphate synthetase genes in U87 glioma cells with ERN1 knockdown: effect of hypoxia and endoplasmic reticulum stress. Ukr Biochem J. 2014;86:74–83. [PubMed] [Google Scholar]
- 21.Li J, Guo W, Li F, He J, Yu Q, et al. HnRNPL as a key factor in spermatogenesis: lesson from functional proteomic studies of azoospermia patients with sertoli cell only syndrome. J Proteomics. 2012;75:2879–91. doi: 10.1016/j.jprot.2011.12.040. [DOI] [PubMed] [Google Scholar]
- 22.Lei B, Wan B, Peng J, Yang Y, Lv D, et al. PRPS2 expression correlates with sertoli-cell only syndrome and inhibits the apoptosis of TM4 Sertoli cells. J Urol. 2015;194:1491–7. doi: 10.1016/j.juro.2015.04.116. [DOI] [PubMed] [Google Scholar]
- 23.Zhang S, Zhao Y, Lei B, Li C, Mao X. PGAM1 is involved in spermatogenic dysfunction and affects cell proliferation, apoptosis, and migration. Reprod Sci. 2015;22:1236–42. doi: 10.1177/1933719115572485. [DOI] [PubMed] [Google Scholar]
- 24.Lei B, Zhou X, Lv D, Wan B, Wu H, et al. Apoptotic and nonapoptotic function of caspase 7 in spermatogenesis. Asian J Androl. 2017;19:47–51. doi: 10.4103/1008-682X.169563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rotgers E, Nurmio M, Pietila E, Cisneros-Montalvo S, Toppari J. E2F1 controls germ cell apoptosis during the first wave of spermatogenesis. Andrology. 2015;3:1000–14. doi: 10.1111/andr.12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agger K, Santoni-Rugiu E, Holmberg C, Karlstrom O, Helin K. Conditional E2F1 activation in transgenic mice causes testicular atrophy and dysplasia mimicking human CIS. Oncogene. 2005;24:780–9. doi: 10.1038/sj.onc.1208248. [DOI] [PubMed] [Google Scholar]
- 27.Al-Khalaf HH, Colak D, Al-Saif M, Al-Bakheet A, Hendrayani SF, et al. p16INK4a positively regulates cyclin D1 and E2F1 through negative control of AUF1. PLoS One. 2011;6:e21111. doi: 10.1371/journal.pone.0021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mann DJ, Jones NC. E2F-1 but not E2F-4 can overcome p16-induced G1 cell-cycle arrest. Curr Biol. 1996;6:474–83. doi: 10.1016/s0960-9822(02)00515-8. [DOI] [PubMed] [Google Scholar]
- 29.Hoja MR, Liu JG, Mohammadieh M, Kvist U, Yuan L. E2F1 deficiency impairs murine spermatogenesis and augments testicular degeneration in SCP3-nullizygous mice. Cell Death Differ. 2004;11:354–6. doi: 10.1038/sj.cdd.4401362. [DOI] [PubMed] [Google Scholar]
- 30.Li M, Yu M, Liu C, Zhu H, He X, et al. miR-34c works downstream of p53 leading to dairy goat male germline stem-cell (mGSCs) apoptosis. Cell Prolif. 2013;46:223–31. doi: 10.1111/cpr.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beumer TL, Roepers-Gajadien HL, Gademan IS, van Buul PP, Gil-Gomez G, et al. The role of the tumor suppressor p53 in spermatogenesis. Cell Death Differ. 1998;5:669–77. doi: 10.1038/sj.cdd.4400396. [DOI] [PubMed] [Google Scholar]
- 32.Bredow S, Juri DE, Cardon K, Tesfaigzi Y. Identification of a novel Bcl-2 promoter region that counteracts in a p53-dependent manner the inhibitory P2 region. Gene. 2007;404:110–6. doi: 10.1016/j.gene.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jana K, Jana N, De DK, Guha SK. Ethanol induces mouse spermatogenic cell apoptosis in vivo through over-expression of Fas/Fas-L, p53, and caspase-3 along with cytochrome c translocation and glutathione depletion. Mol Reprod Dev. 2010;77:820–33. doi: 10.1002/mrd.21227. [DOI] [PubMed] [Google Scholar]
- 34.Yeh YC, Liu TJ, Wang LC, Lee HW, Ting CT, et al. A standardized extract of Ginkgo biloba suppresses doxorubicin-induced oxidative stress and p53-mediated mitochondrial apoptosis in rat testes. Br J Pharmacol. 2009;156:48–61. doi: 10.1111/j.1476-5381.2008.00042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gunes S, Al-Sadaan M, Agarwal A. Spermatogenesis, DNA damage and DNA repair mechanisms in male infertility. Reprod Biomed Online. 2015;31:309–19. doi: 10.1016/j.rbmo.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 36.Lei B, Lv D, Zhou X, Zhang S, Shu F, et al. Biochemical hormone parameters in seminal and blood plasma samples correlate with histopathologic properties of testicular biopsy in azoospermic patients. Urology. 2015;85:1074–8. doi: 10.1016/j.urology.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 37.Gu U, Turunc T, Haydardedeoglu B, Yaycioglu O, Kuzgunbay B, et al. Sperm retrieval and live birth rates in presumed sertoli-cell-only syndrome in testis biopsy: a single centre experience. Andrology. 2013;1:47–51. doi: 10.1111/j.2047-2927.2012.00003.x. [DOI] [PubMed] [Google Scholar]
- 38.Stouffs K, Gheldof A, Tournaye H, Vandermaelen D, Bonduelle M, et al. Sertoli cell-only syndrome: behind the genetic scenes. Biomed Res Int. 2016;2016:6191307. doi: 10.1155/2016/6191307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lentivirus vectors were transfected into GC1 and GC2 cells to downregulate and upregulate the expression of PRPS2, respectively.
IHC detection indicated that the expression levels of Caspase 3, Caspase 6, and Caspase 9 were significantly higher in the mice with shPRPS2 lentivirus treatment compared with those in the blank and negative controls.
Flow cytometry analysis showed that cell apoptosis was significantly inhibited in GC1/shPRPS2/E2F1 cells compared with that in GC1/shPRPS2 cells.


