Abstract
Recent findings demonstrate the origin of the plasmid-mediated colistin resistance gene mcr-3 from aeromonads. The present study aimed to screen for plasmid-mediated colistin resistance among 30 clinical multidrug-resistant (MDR) Aeromonas spp. PCR was used to screen for the presence of mcr-1, mcr-2, mcr-3 and mcr-4, which revealed mcr-3 in a colistin-susceptible isolate (FC951). All other isolates were negative for mcr. Sequencing of FC951 revealed that the mcr-3 (mcr-3.30) identified was different from previously reported variants and had 95.62 and 95.28 % nucleotide similarity with mcr-3.3 and mcr-3.10. Hybrid assembly using IonTorrent and MinION reads revealed structural genetic information for mcr-3.30 with an insertion of ISAs18 within the gene. Due to this, mcr-3.30 was non-expressive, which makes FC951 susceptible to colistin. Further, in silico sequence and protein structural analysis confirmed the new variant. To the best of our knowledge, this is the first report on a novel mcr-3 variant from India. The significant role of mcr-like genes in different Aeromonas species remains unknown and requires additional investigation to obtains insights into the mechanism of colistin resistance.
Keywords: mcr-3, colistin, Aeromonas, ISAs18, blaOXA-12, blaCEPH-A3
Introduction
Aeromonas spp. are ubiquitous and are known to cause gastroenteritis, wound infections and septicaemia; they are commonly known as ‘jacks of all trades’. Aeromonads are universally resistant to the penicillin group of antibiotics (penicillin, ampicillin, carbenicillin and ticarcillin) and are generally susceptible to tetracyclines and quinolones [1]. Recently, increasing resistance to third-generation cephalosporins and carbapenems has been reported [2, 3].
The recent discovery of plasmid-mediated colistin resistance genes (mcr) has attracted global attention. A study reported that the mcr-3 identified in Escherichia coli is similar to the gene present in Aeromonas species and suggested that it might have originated from aeromonads [4]. It should be noted that most Aeromonas species are susceptible to colistin, whereas Aeromonas jandaei and Aeromonas hydrophila have been reported to be intrinsically resistant to polymyxins [4]. However, the role of mcr-like genes in different Aeromonas species is not clearly understood and requires further investigation.
This study examined the presence of plasmid-mediated colistin resistance among Aeromonas spp. using PCR and structural analysis with next-generation sequencing. The nucleotide sequences obtained using experimental methods were translated into a protein sequence, and the 3D structure was modelled using in silico approaches to understand the structural changes in the different variants of mcr.
Methods
Isolates and identification
A total of 30 Aeromonas spp. isolated from stool specimens collected from January to December 2017 from symptomatic patients attending the Christian Medical College, Vellore were included in the study. Isolation and identification of the genus and species were carried out using a standard culture and biochemical tests [5].
Antimicrobial susceptibility testing (AST)
Disc diffusion
AST testing was carried out using the Kirby–Bauer disk diffusion method. The antimicrobial agents tested were trimethoprim/sulfamethoxazole (1.25/23.75 µg), tetracycline (30 µg), ciprofloxacin (5 µg), cefotaxime (30 µg), imipenem (10 µg) and meropenem (10 µg) (Oxoid, UK). Quality control (QC) strains ( Klebsiella pneumoniae ATCC 700603, Pseudomonas aeruginosa ATCC 27853 and E. coli ATCC 25922) were included in all batches, as recommended by the Clinical and Laboratory Standards Institute (CLSI-M45) [6].
Minimum inhibitory concentration (MIC)
The colistin MIC was determined for the studied isolates by broth microdilution and interpreted using the CLSI 2017 breakpoint recommendation [7]. mcr-1-positive E. coli with the expected range 4–8 µg ml−1, E. coli ATCC 25922 (0.25–2 µg ml−1) and P. aeruginosa ATCC 27853 (0.5–4 µg ml−1) were used as QC) strains for colistin MIC determination.
Screening of mcr genes by PCR
The presence of mcr-1, mcr-2, mcr-3 and mcr-4 encoding for plasmid-mediated colistin resistance was screened by PCR as described previously [4, 6–8].
Next-generation sequencing
The isolate that was positive for mcr was selected for next-generation sequencing to analyse colistin resistance determinants and other genetic factors. A QIAamp DNA Mini kit (Qiagen, Hilden, Germany) was used for genomic DNA extraction. Whole-genome sequencing (WGS) of the isolate was performed with 400 bp read chemistry using an IonTorrent Personal Genome Machine (PGM) (Life Technologies, Carlsbad, CA, USA) as per the manufacturer’s instructions. Data were assembled de novo using Assembler SPAdes v.5.0.0.0 embedded in Torrent Suite Server v.5.0.3. Sequence annotation was performed using PATRIC, the bacterial bioinformatics database and analysis resource (http://www.patricbrc.org), and the National Center for Biotechnology Information’s (NCBI’s) Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP, http://www.ncbi.nlm.nih.gov/genomes/static/Pipeline.html).
The CGE server (http://www.cbs.dtu.dk/services) and PATRIC [8] were employed for downstream analysis. ResFinder 2.1 (https://cge.cbs.dtu.dk//services/ResFinder/) was used to analyse the resistance gene profile [9]. Antimicrobial resistance genes were also screened using the Antibiotic Resistance Genes Database (ARDB) and the Comprehensive Antibiotic Resistance Database (CARD) through PATRIC. PlasmidFinder 1.3 (https://cge.cbs.dtu.dk//services/PlasmidFinder/) was used to screen for the presence of plasmids [10]. The clustered regularly interspaced short palindromic repeats (CRISPR) and Cas genes was identified from the genome using the CRISPRFinder tool [11]. The MultiLocus Sequence Typing (MLST) 1.8 tool was employed for sequence type analysis (https://cge.cbs.dtu.dk//services/MLST/) [12]. The genome was screened for insertion sequence elements using ISFinder (https://www-is.biotoul.fr/blast.php) [13].
MinION Oxford Nanopore sequencing
DNA library preparation and sequencing was prepared using SQK-LSK108 Kit R9 version (Oxford Nanopore Technologies, Oxford, UK) using a 1D sequencing method according to the manufacturer’s protocol. Sequencing was performed using the FLO-MIN106 R9 flow cell in the MinION Mk 1B sequencer. MinKNOW software v 1.15.1 (Oxford Nanopore Technologies, Oxford, UK) was employed in a Windows platform to perform sequencing and raw data (fast5 files) were obtained.
MinION sequence analysis
The fast5 files were generated from MinION sequencing and the reads were base-called with Albacore 2.0.1 (https://nanoporetech.com/about-us/news/new-basecaller-now-performs-raw-basecalling-improved-sequencing-accuracy). Furthermore, the adapters were trimmed using Porechop (https://github.com/rrwick/Porechop). Canu 1.7 [14] was used for MinION error correction and assembly with a genome size of 5.0 m as input. After de novo assembly, the contigs were polished with Nanopolish 0.10.1 (https://github.com/jts/nanopolish).
Hybrid assembly using IonTorrent and MinION reads
To increase the accuracy and completeness of he genome, a hybrid assembly using both IonTorrent and MinION reads with Unicycler (v0.4.6) was performed [15]. By default, Unicycler utilizes SPAdes [16] to assemble the short reads with different k-mers and filter out the low-depth regions. Subsequently, it trims and generates the short-read assembly graph. In addition, it uses Miniasm [17] and Racon [18] to assemble the MinION long reads and further the reads were bridged to determine all the genome repeats and produce complete genome assembly. The short reads were also polished with multiple rounds of Pilon [19] to reduce the base-level errors. After assembly, the assembly statistics and average nucleotide identity of different assemblies were evaluated using the Quast [20] and OrthoANI 0.93.1 tools [21], respectively.
In silico sequence analysis
The mcr nucleotide sequences obtained from the experimental techniques were translated to a protein sequence using the online Expasy Translate tool (https://web.expasy.org/translate/). The percentage identity of known MCR variants in comparison with novel protein was identified using blast search. The conserved amino acid region from the closely related variants was determined using Clustal Omega.
In silico structure analysis
The sequences of the MCR variant was used to model the 3D structure of the proteins. The 3D structures of the variants were modelled using Swiss-Model [22]. The translated variant sequences were given as the input for the 3D variant modelling. The rampage server was used to evaluate the quality of the modelled variants [23]. Finally, the MetaPocket server was used to predict the active pocket of the novel variant [24]. The structure visualization was performed using PyMOL.
Results
Antimicrobial susceptibility
The resistance profiles for the studied Aeromonas isolates are presented in Table 1. The colistin MIC was determined for all the isolates and this showed 30 % of the isolates were resistant to colistin. The MIC was identified to be 0.5 µg ml−1 (susceptible) for the isolate that was positive for mcr-3.
Table 1.
Antimicrobial susceptibility of the selected Aeromonas spp.
|
Sample no. |
Sample ID |
Age/sex |
Organism |
Resistance pattern (disc diffusion) |
Colistin MIC (µg ml−1) |
|---|---|---|---|---|---|
|
1 |
FC3340 |
29M |
Aeromonas spp. |
AMP-SXT-TAX |
4R |
|
2 |
FC193 |
0M |
AMP-IMI-MEM |
≥64R |
|
|
3 |
FC199 |
76M |
AMP |
1S |
|
|
4 |
FC284 |
75M |
Aeromonas spp. |
AMP-IMI-MEM |
16R |
|
5 |
FC728 |
71M |
AMP-TET-TAX-IMI-MEM-CIP |
0.5S |
|
|
6 |
FC729 |
66M |
AMP-TET-SXT-TAX-IMI-MEM-CIP |
0.5S |
|
|
7 |
FC715 |
34F |
AMP |
>32R |
|
|
8 |
FC850 |
27M |
AMP-TET-SXT-IMI-MEM-CIP |
1S |
|
|
9 |
*FC951 |
55M |
AMP-TET-IMI-MEM |
0.5S |
|
|
10 |
FC1239 |
21F |
AMP-TAX-IMI-MEM |
≥64R |
|
|
11 |
FC1520 |
45F |
AMP |
>32R |
|
|
12 |
FC1169 |
0M |
AMP |
2S |
|
|
13 |
FC599 |
57F |
AMP-SXT |
2S |
|
|
14 |
FC814 |
55F |
AMP-IMI |
>32R |
|
|
15 |
FC2457 |
0M |
AMP-TET-SXT |
2S |
|
|
16 |
FC538 |
58M |
AMP-IMP |
2S |
|
|
17 |
FC771 |
1M |
AMP-TAX-CIP |
4R |
|
|
18 |
FC578 |
26F |
AMP |
1S |
|
|
19 |
FC377 |
22F |
AMP-SXT-IMI-MEM |
1S |
|
|
20 |
FC1245 |
42F |
AMP-IMI |
2S |
|
|
21 |
FC788 |
0M |
AMP-TAX-IMI |
8R |
|
|
22 |
FC157 |
58M |
AMP |
0.5S |
|
|
23 |
FC390 |
9F |
AMP |
1S |
|
|
24 |
FC1411 |
5M |
AMP-IMI |
2S |
|
|
25 |
FC523 |
1M |
AMP-TET |
1S |
|
|
26 |
FC1999 |
54F |
AMP-IMI-MEM |
0.5S |
|
|
27 |
FC2051 |
28M |
AMP |
1S |
|
|
28 |
FC2019 |
60M |
AMP |
1S |
|
|
29 |
FC906 |
55M |
AMP-TET |
0.5S |
|
|
30 |
FC1435 |
73F |
AMP |
>32R |
*Isolate sequenced: R, resistant; S, susceptible; AMP, ampicillin; TET, tetracycline; SXT, trimethoprim/sulfamethoxazole; CIP, ciprofloxacin; TAX, cefotaxime; IMI, imipenem; MEM, meropenem.
Screening of mcr genes
Of the 30 isolates screened for mcr-1, mcr-2, mcr-3 and mcr-4, only one isolate (FC951) was positive for mcr-3.
Next-generation sequencing
The A. veronii (FC951) hat was positive for mcr-3 by PCR was sequenced using IonTorrent PGM. Analysis of mcr-3 revealed only 95.6 % identity against the reference sequences in the database (henceforth termed mcr-3.30). Further, ISFinder revealed that ISAs18 belongs to the IS4 family next to eptA, aka mcr-3.
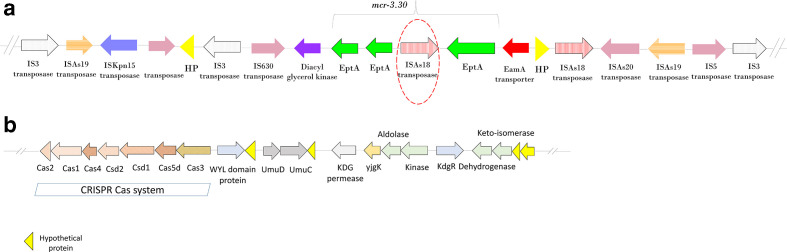
However, the mcr-3.30 was split in to two contigs (IonTorrent). Generally, the IonTorrent assembly is highly accurate, but the assembly had too many fragments. The long-read sequencing of FC951 using MinION resulted in a complete genome (a chromosome and a plasmid), but with errors. Hybrid assembly using IonTorrent and MinION reads in Unicycler resulted a complete chromosomal contig for FC951 with increased accuracy and few errors. Interestingly, analysis of the complete genome revealed mcr-3.30 integrated in the chromosome along with the insertion of ISAs18 within mcr-3.30. The structure of the genetic environment of mcr-3.30 is shown in Fig. 1a.
Fig. 1.
(a) Genetic environment of mcr-3.30 with an insertion of ISAs18 transposase (1141 bp) leading to disruption of mcr-3.30 function. (b) CRISPR Cas system identified in FC951 and arrangement of Cas genes.
The Quast analysis showed the N50 and N75 value of the hybrid assembly to be 4 660 178, which is approximately 96 % of the total assembly length. In addition, ANI is calculated using different assembly methods. It is evident that the closeness between IonTorrent and hybrid assembly is about 99.92 %, which represents the high accuracy of the hybrid assembly. The hybrid assembly generated a 4.66 Mbp chromosome length single contig. In contrast, the IonTorrent-only assembly produced an assembly with more than 300 contigs and only 139 contigs >1000 bp. It was clear that hybrid assembly has its own advantages, with improved accuracy and a reduced error rate with genome completeness compared to MinION-only or IonTorrent-only assembly.
Moreover, annotation of the extra-chromosomal sequences from MinION could not be designated as a complete plasmid, although it showed 21 % similarity with a previously reported Xanthomonas citri plasmid (CP020883.1).
Further analysis of resistance genes using ResFinder revealed the presence of bla OXA12 and bla CEPH-A3 genes. A CRISPR Cas system was identified in FC951 and the arrangements of the genes were as shown in Fig. 1b. Notably, the sequence type of FC951 was identified to be novel, ST-515.
This complete genome project has been deposited at GenBank under the accession number CP032839 (plasmid accession number CP032840).
In silico sequence analysis
The mcr-3 nucleotide and protein sequence identified in this study was compared with the previously reported variants mcr-3.1–mcr-3.10 using blast search (KY924928.1, NMWW01000143.1, MF495680, NQCO01000074.1, MF489760, MF598076.1, MF598077.1, MF598078.1, MF598080.1 and MG214531) and identified to be a novel variant (mcr-3.30). The nucleotide percentage identity matrix for the mcr gene variants was as given in Table 2. From the analysis, mcr-3.3 and mcr-3.10 were found to be highly identical (95.62 and 95.28 %, respectively. The most conserved amino acid region among the three variants (mcr-3.3, mcr-3.10 and mcr-3.30) was identified using Clustal Omega. The region of amino acids from LEU-359 to ILE-427 was found to be the largest conserved sequence among them.
Table 2.
Percentage identity matrix of mcr-3 variants in comparison with FC951 mcr-3.30
|
mcr-3.30 (FC951) |
mcr-3.6 |
mcr-3.3 |
mcr-3.8 |
mcr-3.7 |
mcr-3.5 |
mcr-3.4 |
mcr-3.1 |
mcr-3.2 |
mcr-3.9 |
mcr-3.10 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
|
mcr-3.30 (FC951) |
100 |
||||||||||
|
mcr-3.6 |
94.07 |
100 |
|||||||||
|
mcr-3.3 |
95.62 |
98.15 |
100 |
||||||||
|
mcr-3.8 |
95.01 |
98.64 |
98.95 |
100 |
|||||||
|
mcr-3.7 |
92.6 |
94.33 |
94.09 |
93.97 |
100 |
||||||
|
mcr-3.5 |
94.07 |
93.72 |
95.32 |
94.22 |
95.69 |
100 |
|||||
|
mcr-3.4 |
94.13 |
93.72 |
95.32 |
94.22 |
95.82 |
99.75 |
100 |
||||
|
mcr-3.1 |
94.2 |
93.78 |
95.38 |
94.28 |
95.88 |
99.82 |
99.94 |
100 |
|||
|
mcr-3.2 |
94.2 |
93.84 |
95.44 |
94.34 |
95.82 |
99.88 |
99.88 |
99.94 |
100 |
||
|
mcr-3.9 |
94.54 |
94.39 |
95.75 |
94.9 |
96.62 |
97.72 |
97.85 |
97.91 |
97.85 |
100 |
|
|
mcr-3.10 |
95.28 |
94.7 |
96.55 |
95.33 |
96.56 |
98.65 |
98.77 |
98.83 |
98.77 |
99.08 |
100 |
In silico structure analysis
The 3D structures were modelled using the sequences for the variants mcr-3.1–mcr-3.10 using the Swiss-Model server. The template used and its respective PDB ID, coverage range and coverage identity are tabulated in Table 3. Further, the quality of the modelled variants was evaluated using the rampage server. The percentage of the amino acids in the favoured region, the percentage of amino acids in the allowed region and the percentage of amino acids in the outlier region are tabulated in Table 3. Superimposed structural evaluation of MCR variant 3.30 against the closely related MCR variant 3.3 and MCR variant 3.10 was visualized using PyMOL (Fig. 2). In addition, LEU53, ILE164, ALA57, TYR175, GLN186, ILE189, VAL176, GLY179, ALA192, PHE32, LEU50, VAL61, ARG180, VAL178, ASN182, LEU185, PHE46, LEU36, SER183, VAL29, GLY28, PRO51, LEU54, ASN25, ALA21, TRP26, GLU188, LEU24, LEU58, PRO191, ASN193, VAL195, PHE65 and ASN196 were identified as the active site of the MCR-3.30 variant using the Meta Pocket server (Fig. S1, available in the online version of this article).
Table 3.
List of parameters considered for modelling the variants using Swiss-Model and Ramachandran plot evaluation of the structures using rampage
|
Variants of mcr |
Template PDB |
Coverage range |
Coverage |
Identity |
% of amino acids in the favoured region |
% of amino acids in the allowed region |
% of amino acids in the outlier region |
|---|---|---|---|---|---|---|---|
|
3.1 |
5FGN |
6–544 |
0.98 |
37.55 |
94.2 % |
4.9 % |
0.9 % |
|
3.2 |
5FGN |
6–545 |
0.98 |
37.55 |
94.2 % |
4.9 % |
0.9 % |
|
3.3 |
5FGN |
1–546 |
0.99 |
35.82 |
94.0 % |
5.1 % |
0.9 % |
|
3.4 |
5FGN |
6–544 |
0.98 |
37.55 |
94.4 % |
4.7 % |
0.9 % |
|
3.5 |
5FGN |
6–546 |
0.97 |
40.27 |
95.9 % |
3.6 % |
0.6 % |
|
3.6 |
5FGN |
6–546 |
0.97 |
39.69 |
95.7 % |
3.6 % |
0.8 % |
|
3.7 |
5FGN |
6–546 |
0.97 |
40.08 |
95.1 % |
4.1 % |
0.8 % |
|
3.8 |
5FGN |
6–546 |
0.97 |
39.69 |
95.7 % |
3.6 % |
80.0 % |
|
3.9 |
5FGN |
6–546 |
0.97 |
40.08 |
95.3 % |
4.0 % |
80.0 % |
|
3.10 |
5FGN |
6–544 |
0.97 |
37.74 |
94.6 % |
4.5 % |
90.0 % |
|
3.30 |
5FGN |
6–469 |
0.98 |
36.24 |
93.7 % |
4.6 % |
1.7 % |
Fig. 2.
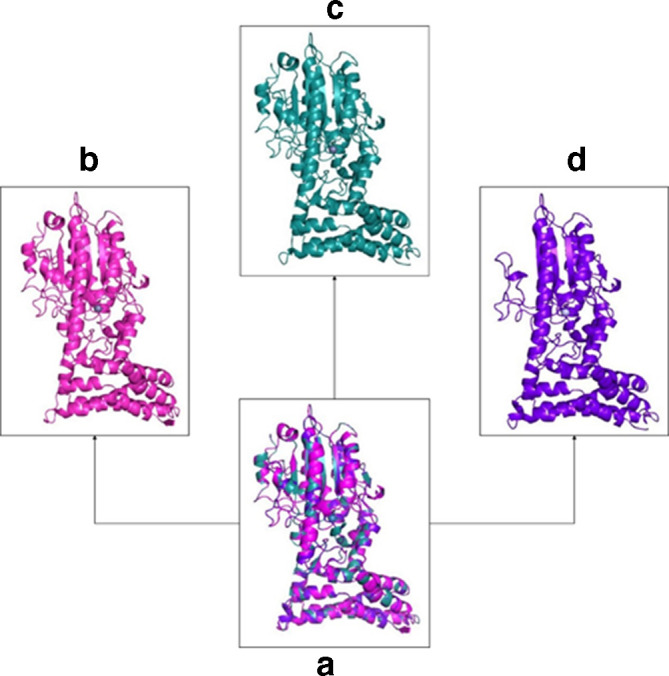
(a) Superimposed structural evaluation of MCR variant 3.30 against the closely related MCR variant 3.3 and MCR variant 3.10, (b) 3D structure of MCR variant 3.3, (c) 3D structure of MCR variant 3.10 and (d) 3D structure of MCR variant 3.30.
Discussion
Resistance to colistin is mainly mediated via alteration of the lipopolysaccharides (LPS) of the bacterial outer membrane. The alterations include mutations in lipid A-modifying genes. The most commonly reported mutations are in the mgrB gene and are therefore not transferable through horizontal gene transfer [25]. However, in 2015, the first plasmid-mediated colistin resistance gene (mcr-1) was reported [26], which belongs to the phosphoethanolamine transferase enzyme family (EptA). mcr-1 was identified in E. coli from human patients and animals in China. In 2016, another study reported the mobilizable colistin resistance gene, mcr-2, from porcine and bovine E. coli isolates in Belgium [27]. Further, mcr-3 and mcr-4 were identified in E. coli, Klebsiella spp. and Salmonella spp. [4, 28]. Recently, mcr-5 was identified in Salmonella enterica subsp. enterica serovar Paratyphi B isolated from poultry in Denmark [29].
Several mobile colistin resistance genes have been identified, but only mcr-1 and mcr-3 have been reported with a high number of variants in GenBank database. A recent study highlighted the importance of a third mobile colistin resistance gene, mcr-3 in Aeromonas salmonicida , due to its resemblance to various other phosphoethanolamine transferases in Enterobacteriaceae and also suggested that this resistance gene might have already been widely disseminated [4]. Here we discuss a novel variant of mcr-3 identified in A. veronii isolated from a clinical specimen.
The novel mcr-3 variant identified in this study exhibited ≤95 % nucleotide sequence similarity to all other previously reported mcr-3 variants, and is henceforth named mcr-3.30. In silico protein sequence comparison revealed the novelty of MCR-3.30. The superimposed protein structure comparison with MCR-3.3 and MCR-3.10 further confirmed the MCR-3.30 variant. Major structural changes were observed in domain 2. Similar comparisons of MCR-3 and MCR-1 protein structures were previously reported by [4]. Knowledge of the 3D structure of proteins is now in great demand in the field of computer-aided drug discovery (CADD). It helps researchers in identifying new drugs. In this study, we have used homology modelling technique to model the 3D structures of MCR variants.
mcr-3 was first identified in a 261 kb IncHI2-type plasmid pWJ1 from E. coli [4]. However, initially, known plasmid replicons were not identified in FC951 harbouring mcr-3.30 via PlasmidFinder. Later, sequence assembly and alignment with pWJ1 revealed an Inc-W like replicase gene in the extrachromosomal region, along with TraI and TrbN genes responsible for multi-functional conjugation and conjugal transfer proteins.
There had been a previous report on the chromosome integration of the mcr-3 variant in A. veronii [30]. In this study, a major insertion of ISAs18 (1141 bp) belonging to the IS4 family of insertional elements was found within mcr-3.30. ISAs18 had previously only been reported in A. salmonicida as a transposase [31]. The entire mcr-3.30 region, including the insertion, was flanked by eamA and dgkA. These genes encode a metabolite transporter and a diacylglycerol kinase, respectively [32]. The mcr-3.30 genetic environment also had other insertional elements, such as ISAs19, ISAs20, IS630, ISKpn15, IS5 and IS3 transposase.
However, mcr-3.30 disruption caused by the insertion of ISAs18 has rendered it non-expressive. Accordingly, the isolate FC951, in spite of harbouring mcr-3.30, was phenotypically susceptible to colistin. Similarly, a previous study by Pham Thanh et al. identified a deactivated mcr-1 due to the disruption of the gene by a 22 bp duplication in colistin-susceptible Shigella sonnei [33]. The gene was found to be reactivated by conjugation experiments. resulting in a colistin-resistant phenotype. Another study by Liassine et al. reported mcr-1 in susceptible E. coli with an unknown cause of susceptibility [34]. A recent study showed the presence of mcr-1 in susceptible E. coli due to the insertion of 1329 bp transposon IS10R and found that this could not be reactivated. As shown from these studies, the inactivated gene can be restored upon colistin exposure, particularly in settings where colistin use is high. This phenomenon emphasizes the importance of phenotypic confirmation despite detection of the gene in molecular screening [33, 35]. In contrast, the isolates of the present study that were resistant to colistin by MIC were negative for screened mcr; this could be due to other chromosomal mechanisms that need to be explored.
There are no reports of mcr variants other than mcr-1 from India. Various Indian centres have reported colistin-resistant strains in hospitalized patients. So far, there are only published reports of colistin resistance in Enterobacteriaceae and non-fermenters such as E. coli , K. pneumoniae and A. baumannii from India.
Mutation in the mgrB gene is the most common resistance mechanism in K. pneumoniae and mutation in the lpxA/D, lpsB and pmrB genes is responsible for resistance in A. baumannii. On the other hand, the presence of plasmid-mediated mcr confers resistance in E. coli [25, 36, 37]
Apart from the colistin resistance gene, the genome analysis revealed the presence of other resistance genes, such as bla OXA-12, belonging to class D beta-lactamase in FC951, which confers resistance to ampicillin and is known to be naturally produced by Aeromonas jandaei and has strong activity against oxacillin [38]. The isolate also harboured bla CEPH-A3, which is the most common metallo-beta-lactamase (MBL) produced by Aeromonas species responsible for carbapenem resistance.
Conclusion
To the best of our knowledge, this is the first report on a novel mcr-3 variant (mcr-3.30) at the structural level, in comparison with the known variants (MCR-3.3–MCR-3.10). The mcr-3.30 identified in this study is non-functional for colistin resistance due to the insertion of ISAs18 within the gene. Further, this is the first complete genome sequence of A. veronii from India, and the first hybrid genome of A. veronii globally. These findings support extended screening of known and further exploration of unknown colistin resistance mechanisms in this pathogen as well as in other Gram-negative pathogens.
Supplementary Data
Funding information
The authors received no specific grant from any funding agency.
Acknowledgements
The authors gratefully acknowledge Christian Medical College, Vellore for providing laboratory space and facilities.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
The study was approved by the Institutional Review Board of the Christian Medical College, Vellore (83-i/11/13)
Footnotes
Abbreviations: ARDB, antibiotic resistance genes database; AST, antimicrobial susceptibility testing; CARD, comprehensive antibiotic resistance database; CRISPR, clustered regularly interspaced short palindromic repeats; MDR, multidrug resistant; MIC, minimum inhibitory concentration; MLST, multilocus sequence typing; WGS, whole genome sequencing.
One supplementary figure is available with the online version of this article.
References
- 1.Batra P, Mathur P, Misra MC. Aeromonas spp.: an emerging nosocomial pathogen. J Lab Physicians. 2016;8:1. doi: 10.4103/0974-2727.176234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhaskar M, Dinoop KP, Mandal J. Characterization of ceftriaxone-resistant Aeromonas spp. isolates from stool samples of both children and adults in southern India. J Health Popul Nutr. 2015;33:26. doi: 10.1186/s41043-015-0036-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anandan S, Gopi R, Devanga Ragupathi NK, Muthuirulandi Sethuvel DP, Gunasekaran P, et al. First report of blaOXA-181-mediated carbapenem resistance in Aeromonas caviae in association with pKP3-A: Threat for rapid dissemination. J Glob Antimicrob Resist. 2017;10:310–314. doi: 10.1016/j.jgar.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Yin W, Li H, Shen Y, Liu Z, Wang S, et al. Novel Plasmid-Mediated Colistin Resistance Gene mcr-3 in Escherichia coli . MBio. 2017;8:e00543–17. doi: 10.1128/mBio.00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abbott SL, Cheung WKW, Janda JM. The genus Aeromonas: biochemical characteristics, atypical reactions, and phenotypic identification schemes. J Clin Microbiol. 2003;41:2348–2357. doi: 10.1128/JCM.41.6.2348-2357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute (CLSI 2016 Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria. Document M45. 3rd ed. Wayne, PA: CLSI; [DOI] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute (CLSI 2017 Performance standards for antimicrobial susceptibility testing. Twenty-seventh Informational Supplement M100-S27. Wayne, PA: CLSI; [Google Scholar]
- 8.Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014;42:D581–D591. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, et al. Identification of acquired antimicrobial resistance genes. Journal of Antimicrobial Chemotherapy. 2012;67:2640–2644. doi: 10.1093/jac/dks261. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carattoli A, Zankari E, Garcìa-Fernandez A, Larsen MV, Lund O, et al. And pMLST: in silico detection and typing of plasmids. Antimicrob Agents Chemother. 2014:AAC-02412. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siguier P, Pérochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, et al. Canu: scalable and accurate long-read assembly via adaptive k -mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.XP L, Fang LX, Song JQ, Xia J, Huo W, et al. Clonal spread of mcr-1 in PMQR-carrying ST34 Salmonella isolates from animals in China. Sci Rep. 2016;6:38511. doi: 10.1038/srep38511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaser R, Sović I, Nagarajan N, Šikić M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017;27:737–746. doi: 10.1101/gr.214270.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee I, Ouk Kim Y, Park S-C, Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 22.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lovell SC, Davis IW, Arendall WB, de Bakker PI, Word JM, et al. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Z, Li Y, Lin B, Schroeder M, Huang B. Identification of cavities on protein surface using multiple computational approaches for drug binding site prediction. Bioinformatics. 2011;27:2083–2088. doi: 10.1093/bioinformatics/btr331. [DOI] [PubMed] [Google Scholar]
- 25.Pragasam AK, Shankar C, Veeraraghavan B, Biswas I, Nabarro LEB, et al. Molecular mechanisms of colistin resistance in Klebsiella pneumoniae causing bacteremia from India—A first report. Front Microbiol. 2017;7:2135. doi: 10.3389/fmicb.2016.02135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 27.Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Eurosurveillance. 2001;6:21. doi: 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 28.Carattoli A, Villa L, Feudi C, Curcio L, Orsini S, et al. Novel plasmid-mediated colistin resistance mcr-4 g ene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Eurosurveillance. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.31.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, et al. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar paratyphi B. J Antimicrob Agents Chemother. 2017;72:3317–3324. doi: 10.1093/jac/dkx327. [DOI] [PubMed] [Google Scholar]
- 30.Ling Z, Yin W, Li H, Zhang Q, Wang X, et al. Chromosome-Mediated mcr-3 Variants in Aeromonas veronii from Chicken Meat. Antimicrob Agents Chemother. 2017;61:e01272–17. doi: 10.1128/AAC.01272-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfeiffer F, Zamora-Lagos M-A, Blettinger M, Yeroslaviz A, Dahl A, et al. The complete and fully assembled genome sequence of Aeromonas salmonicida subsp. pectinolytica and its comparative analysis with other Aeromonas species: investigation of the mobilome in environmental and pathogenic strains. BMC Genomics. 2018;19:20. doi: 10.1186/s12864-017-4301-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kieffer N, Nordmann P, Moreno AM, Zanolli Moreno L, Chaby R, et al. Genetic and Functional Characterization of an MCR-3-Like Enzyme-Producing Escherichia coli Isolate Recovered from Swine in Brazil. Antimicrob Agents Chemother. 2018;62:e00278–18. doi: 10.1128/AAC.00278-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pham Thanh D, Thanh Tuyen H. Nguyen THI Nguyen T, Chung the H, wick Rr, Thwaites Ge, baker S, Holt Ke. inducible colistin resistance via a disrupted plasmid-borne mcr-1 gene in a 2008 Vietnamese Shigella sonnei isolate. J Antimicrob Agents Chemother. 2016;71:2314–2317. doi: 10.1093/jac/dkw173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liassine N, Assouvie L, Descombes M-C, Tendon VD, Kieffer N, et al. Very low prevalence of MCR-1/MCR-2 plasmid-mediated colistin resistance in urinary tract Enterobacteriaceae in Switzerland. Int J Infect Dis. 2016;51:4–5. doi: 10.1016/j.ijid.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 35.Terveer EM, Nijhuis RHT, Crobach MJT, Knetsch CW, Veldkamp KE, et al. Prevalence of colistin resistance gene (mcr-1) containing Enterobacteriaceae in feces of patients attending a tertiary care hospital and detection of a mcr-1 containing, colistin susceptible E. coli. PLoS One. 2017;12:e0178598. doi: 10.1371/journal.pone.0178598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghafur A, Shankar C, GnanaSoundari P, Venkatesan M, Mani D, et al. Detection of chromosomal and plasmid-mediated mechanisms of colistin resistance in Escherichia coli and Klebsiella pneumoniae from Indian food samples. J Glob Antimicrob Resist. 2019;16:48–52. doi: 10.1016/j.jgar.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 37.Vijayakumar S, S BA, Kanthan K, Veeraraghavan B. Whole-Genome shotgun sequences of seven colistin-resistant Acinetobacter baumannii isolates from bacteraemia. J Glob Antimicrob Resist. 2018;12:155–156. doi: 10.1016/j.jgar.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 38.Poirel L, Naas T, Nordmann P. Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrob Agents Chemother. 2010;54:24–38. doi: 10.1128/AAC.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.