Abstract
Motivation
Reliable identification of expressed somatic insertions/deletions (indels) is an unmet need due to artifacts generated in PCR-based RNA-Seq library preparation and the lack of normal RNA-Seq data, presenting analytical challenges for discovery of somatic indels in tumor transcriptome.
Results
We present RNAIndel, a tool for predicting somatic, germline and artifact indels from tumor RNA-Seq data. RNAIndel leverages features derived from indel sequence context and biological effect in a machine-learning framework. Except for tumor samples with microsatellite instability, RNAIndel robustly predicts 88–100% of somatic indels in five diverse test datasets of pediatric and adult cancers, even recovering subclonal (VAF range 0.01–0.15) driver indels missed by targeted deep-sequencing, outperforming the current best-practice for RNA-Seq variant calling which had 57% sensitivity but with 14 times more false positives.
Availability and implementation
RNAIndel is freely available at https://github.com/stjude/RNAIndel.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Transcriptome sequencing (RNA-Seq) is a versatile platform for performing a multitude of cancer genomic analyses such as gene expression profiling, allele specific expression, alternative splicing and fusion transcript detection. However, variant identification in RNA-Seq is not a common practice due to the presence of artifacts introduced in library preparation, the intrinsic complexity of splicing and RNA editing (Piskol et al., 2013). RNA-Seq data are predominantly generated from tumor-only samples as acquisition of a normal tissue with a comparable transcriptome is a rare practice. This lack of matching normal data further complicates somatic variant discovery in RNA-Seq. Despite these challenges, there are compelling reasons to explore RNA-Seq data for variant detection: (i) RNA variants are expressed and therefore more interpretable to cancer phenotype and clinical actionability than DNA variants; and (ii) Some studies only analyze tumor specimen by RNA-Seq, and performing variant detection in RNA-Seq will make full use of the available data resources. Thus, successful development of RNA-Seq variant calling tools will make this platform an interpretable and cost-effective alternative to DNA-based whole-genome or whole-exome sequencing (DNA-Seq), the current standard platform for somatic variant detection.
Various single nucleotide variant (SNV) detection tools dedicated to RNA-Seq have been developed. SNPiR (Piskol et al., 2013) calls true RNA-Seq SNVs by hard-filtering calls in repetitive and low-quality regions, around splice sites and at known RNA-editing sites. RVboost (Wang et al., 2014) is a machine-learning method to prioritize true SNVs trained on common SNPs in the input RNA-Seq data. eSNV-Detect (Tang et al., 2014) incorporates results generated from two mappers to confidently call expressed SNVs by removing mapping artifacts from individual mappers. Opossum (Oikkonen and Lise, 2017) preprocesses RNA-Seq reads for SNV calling by splitting intron-spanning reads and removing spurious reads. By contrast, indel detection in transcriptome is more challenging and has been largely unexplored (Sun et al., 2017). Even in DNA-Seq, indel discovery suffers from a high false discovery rate (Fang et al., 2014). In RNA-Seq, in addition to the artifacts from library preparation due to polymerase chain reaction (PCR), misalignment of spliced reads can introduce mapping artifacts. Indels are less common than SNVs in the genome with the ratio 1:7 and even more so in coding regions with 1: 43 (Ng et al., 2008). This low prevalence poses additional challenges for developing a robust indel detection method that optimizes sensitivity and specificity. In cancer studies, somatic indels should also be distinguished from germline indels. Therefore, somatic indel identification in tumor RNA-Seq can be formulated as a three-class classification problem where somatic, germline and artifact indels must be considered.
Here, we introduce RNAIndel, a novel tool that takes a tumor RNA-Seq BAM file as input, calls and annotates coding indels, and classifies them into somatic, germline and artifacts by supervised learning. RNAIndel was developed by using 765 475 indels collected from 330 pediatric tumor transcriptomes. To test the generality of the model, we tested RNAIndel against five RNA-Seq datasets, two from pediatric cancers and three from adult cancers, which are comprised of 547 samples analyzed by different RNA-Seq protocols on different NGS platforms. RNAIndel predicted 88‒100% of known somatic indels with 4‒16 false positives out of 500‒5000 RNA-Seq indels per sample. RNAIndel is also flexibly designed to allow researchers to import data from their own variant caller rather than the built-in caller and the model can be retrained using the user data. With its high sensitivity on somatic indel prediction, we anticipate that RNAIndel will augment the range of RNA-Seq applications and facilitate the investigation of expressed somatic variants.
2 Overview
The RNAIndel software (Fig. 1A) requires a RNA-Seq BAM file mapped by STAR (Dobin et al., 2013) as the input. Indel calling can be performed by the built-in Bambino (Edmonson et al., 2011) caller using parameters optimized for RNA-Seq indel calling, or by supplying variants in the Variant Call Format (VCF) (Danecek et al., 2011) generated by a user-preferred caller. Indels are annotated using all RefSeq (Pruitt et al., 2004) isoforms containing coding exons; indels within a coding exon or in an intron region within 10 bases of a splice site (splice region) are considered coding indels and subjected to further analysis. For each coding indel, RNAlndel extracts reads covering the indel locus to retrieve the actual alignment pileup. This process also enables the incorporation of additional variations such as polymorphisms near the indel into the feature calculation. RNAIndel queries a custom germline database, which is described in detail below, for matches to the indel, with the query result being used as a feature. The database query looks for both exact matches and equivalent matches (Supplementary Fig. S1). For indels supported by ≥2 unique reads, prediction is made by classifiers specifically trained based on their size (i.e. single-nucleotide [s-indel] or multi-nucleotide [m-indel]), which consist of an ensemble of random forest (Breiman, 2001) models. RNAIndel generates a VCF file where indel entries are parsimonious and left-aligned (Tan et al., 2015) to unify equivalent alignments of supporting reads (Supplementary Fig. S1). Predicted class and probability are reported in the VCF INFO field as well as calculated feature values and other annotations.
Fig. 1.
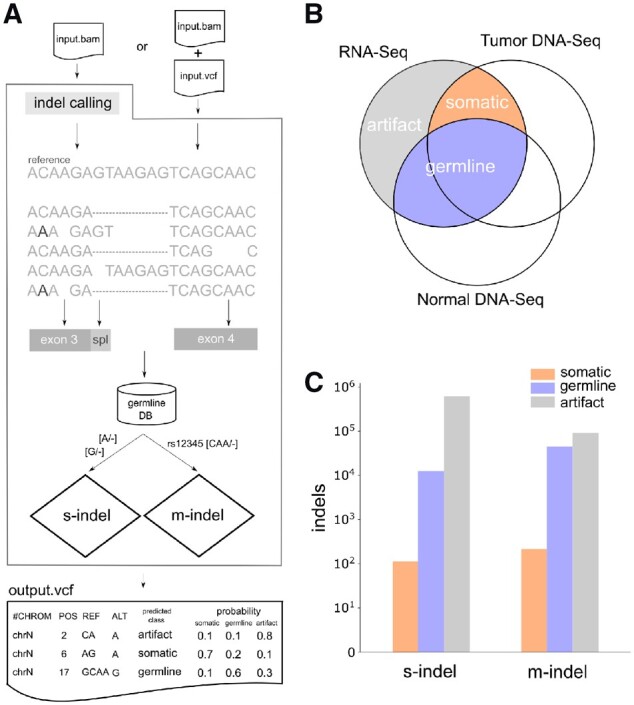
Computational framework and training dataset construction. (A) Workflow of RNAIndel. A tumor RNA-Seq BAM file is a required input. If an optional VCF file from user’s variant caller is supplied, indel calls in the file are used for prediction. Otherwise, indel calling is performed on the input BAM file using the built-in caller. Features are calculated using alignment pileup, transcript structure, and database. Alignments are spliced (dashed) and may contain non-reference variations which alter the indel flanking sequence (C > A at the 2nd base). Indels are annotated for coding exon (grey box) and splice region (light-grey box), defined as an intronic region within 10 bp of the exon boundary. After annotating a germline database membership, single-nucleotide (s-indel) and multi-nucleotide (m-indel) indels are separately predicted using random forest classifiers specifically trained for each type. Predicted class is based on the highest probability of being somatic, germine or artifact. RNAIndel outputs an annotated VCF file. (B) Training set generated from 330 cases. Indel calls in RNA-Seq were classified somatic, germline and artifact by matching with T/N-paired WES and WGS (DNA-Seq) data. (C) The s-indel and m- indel distribution in the categories of somatic, germline and artifacts. The class distribution of each dataset is shown in logarithm scale
3 Indel classifiers
3.1 Training set
Each case in the training set (N = 330) was sequenced by tumor RNA-Seq, and paired tumor (T)/normal (N) whole exome sequencing (WES) and PCR-free paired T/N whole genome sequencing (WGS) (Section 6). Coding indels in the training RNA-Seq dataset were labelled somatic, germline or artifact based on the paired T/N WES and WGS analysis (Fig. 1B). Specifically, an indel in RNA-Seq was labelled somatic if it matched to a somatic indel identified by the paired T/N DNA-Seq analysis. Expressed germline indels were defined if they were supported by the normal WGS and WES. The remaining indels, present in RNA-Seq but absent in WGS and WES, were labelled as artifacts. RNA-Seq indels with < 10× coverage in WGS or WES were excluded as ambiguous unless identified as somatic in the T/N-paired WGS/WES analysis. The resulting training set, comprised of single-nucleotide (s-indels) and multi-nucleotide (m-indels) indels, showed distinct distributions in the three categories of somatic, germline and artifact (Fig. 1C) where s-indels were highly enriched in artifacts. Specifically, s-indels accounted for 115, 12 529 and 616 121 of somatic, germline and artifact loci, respectively while m-indels accounted for 213, 45 098 and 91 399 of somatic, germline and artifact loci, respectively. The T/N-paired DNA-Seq analysis identified 959 somatic coding indels, 35.2% of which were expressed. Each sample harbored, on average, 0.88 ± 1.19 s.d. expressed somatic indels, ranging from 0 to 6. The somatic indels in the training set did not recur except for known somatic hotspots: NPM1 W288fs (3 acute myeloid leukemia (AML) samples), FLT3 I836_M837>M (2 B-lineage acute lymphoblastic leukemia samples) and WT1 V362fs (2 AML samples). The top 10 genes with most frequent indel mutations were ETV6 (11 samples), CCND3 (6), GATA1 (6), NOTCH1 (6), PTCH1 (6), BCOR (5), SETD2 (5), XBP1 (5), PTEN (4) and WT1 (4), highlighting the role of expressed somatic indels in tumorigenesis.
3.2 Features
We developed a total of 31 features in the following three categories: (i) sequence and alignment, (ii) effect on transcription and protein coding and (iii) match to a germline database (Table 1 and Supplementary Methods). In the first category, several features were selected based on the strand-slippage hypothesis, a widely accepted model for explaining the mechanism by which indels are generated in the process of DNA replication (Garcia-Diaz and Kunkel, 2006). Under this model, a DNA polymerase pauses synthesis in repetitive regions, and this delay in replication allows unpaired nucleotides to transiently anneal, leading to a misaligned replication. Thus, features governing sequence complexity (feature 1‒3) which include ‘repeat’ (feature 1) for quantifying homopolymer and simple tandem repeat (STR) content, as well as annealing temperature (feature 4‒7) are expected to be important parameters of this model. By contrast, insertions or deletions that are dissimilar to the flanking sequences are unlikely to be caused by strand slippage (feature 8). Cancer-associated indels can be complex (Supplementary Fig. S2) (Ye et al., 2016), so we define indel complexity based on misalignments near the indel site. Indel size (feature 10) is negatively correlated to prevalence, and insertions (feature 11) are rarer than deletions in the human genome (Zhang and Gerstein, 2003). Polymers of adenine or thymine, i.e. polyA or polyT, are known to be more error-prone than polyG or polyC (feature 12‒15) (Fang et al., 2014). Indels with high read support are more likely to be true (feature 16‒18). Mapping artifacts which stem from ambiguous mapping (feature 19) or difficulty in mapping spliced reads may cause false positives (feature 20). Equivalent indels are alternative alignments of the identical indel sequence, a type of mapping artefact which may confound indel detection (feature 21). PCR-based genotyping can frequently create false multiallelic indels (feature 22) (Weber et al., 2002).
Table 1.
RNAIndel features
| Feature identifiera | Feature description | Selection statusb | ||
|---|---|---|---|---|
| 1 | Sequence/Alignment | repeat | Count of repeat unit including homopolymers and STRs in indel flanking region | s, m |
| 2 | lc (linguistic complexity) | Diversity of k-mers in flanking 50-bp region | ||
| 3 | local_lc | Diversity of k-mers in flanking 6-bp region | s, m | |
| 4 | gc | GC content in flanking 50-bp region | ||
| 5 | local_gc | GC content in flanking 6-bp region | ||
| 6 | strength | DNA pair-bond strength of 2-mers in flanking 50-bp region | m | |
| 7 | local_strength | DNA pair-bond strength of 2-mers in flanking 6-bp region | s | |
| 8 | dissimilarity** | Edit distance between indel and flanking sequences | m | |
| 9 | indel_complexity | Mismatches around the indel measured by edit distance | s | |
| 10 | indel_size** | Length of inserted or deleted nucleotides | m | |
| 11 | is_ins | True for insertions | m | |
| 12 | is_at_ins* | True for ‘A’ or ‘T’ insertions | s | |
| 13 | is_at_del* | True for ‘A’ or ‘T’ deletions | ||
| 14 | is_gc_ins* | True for ‘G’ or ‘C’ insertions | ||
| 15 | is_gc_del* | True for ‘G’ or ‘C’ deletions | s | |
| 16 | ref_count | Count of RNA-Seq reads representing the reference allele | s, m | |
| 17 | alt_count | Count of RNA-Seq reads representing the indel allele | s, m | |
| 18 | is_bidirectional | True if an indel is supported by forward and reverse reads | s | |
| 19 | is_uniq_mapped | True if an indel is supported by uniquely mapped reads | s, m | |
| 20 | is_near_exon_boundary | True if an indel is within exon but on the exon boundary | s, m | |
| 21 | equivalence exists | True if alternative indel alignments are observe | s, m | |
| 22 | is_multiallelic | True if multiple indels are observed at the locus | s, m | |
| 23 | Transcript/Protein | is_inframe** | True if an indel is in-frame | |
| 24 | is_splice | True if an indel is in an intronic region within 10-bp to exon | ||
| 25 | is_truncating | True if an indel causes frame-shift, or stop gain, or destroys splice motif | ||
| 26 | is_in_cdd** | True if an indel is located in conserved domain | ||
| 27 | indel_location | Relative indel location in coding region | s | |
| 28 | is_nmd_insensitive | True if nonsense-mediate decay insensitive | ||
| 29 | indels_per_gene | Number of indels detected in the gene in the sample | ||
| 30 | cds_length | Length of the coding region | s | |
| 31 | DB | is_on_db | True if indel is present in the default germline database | s, m |
Note: A total of 31 features related to sequence/alignment, biological effect on transcription and protein coding, and match to germline variant database are examined.
Features marked with * were used only for training of single-nucleotide indel model while those marked with ** were used only for training of multi-nucleotide indel model.
Features selected by the single-nucleotide or the multi-nucleotide model are marked as s and m, respectively.
In the second category, indels were also characterized in terms of variant effect on gene transcription and protein coding. In-frame indels are non-truncating unless they create a de novo stop codon (stop gain). Indels in splicing regions may not affect splicing unless they destroy the splice motif (feature 23‒25). Further, in-frame indels may be less deleterious if they occur outside of conserved domains (feature 26). The relative location of indels (feature 27) within proteins are bimodal around the N and C-termini (Ng et al., 2008). It has been postulated that transcripts with N-terminal indels can be rescued by an alternative start codon downstream, while a subset of C-terminus truncations may retain the all functional domains. Indels in the first and last exons are therefore known to be less sensitive to nonsense mediated decay (NMD) (feature 28) (Popp and Maquat, 2016). We also hypothesized that the number of true somatic indels per gene is expected to be few. The number of indels within a gene was normalized by the length of the coding region (feature 29‒30).
The third category has a single feature (feature 31) which describes the membership of an indel to a custom germline indel database constructed by combining common indels from the dbSNP database (build 151) (Sherry et al., 2001) and gnomAD database (ver.2.1.1) (Karczewski et al., 2019) with > 0.0001 allele frequency in non-cancer populations. Indels curated as ‘Pathogenic’ or ‘Likely Pathogenic’ by ClinVar (ver.20180603) (Landrum et al., 2014) were subtracted from this custom germline database. A user can also supply their own germline database for this feature.
3.3 Training
RNAIndel random forest model training consists of the following three steps: down-sampling, feature selection and model optimization. Each step is performed on -fold cross-validation to maximize a generalized F-score () with being a user-configurable weight for true positive rate (TPR) over precision (Section 6). In most applications, the somatic class will represent a minority as in our training set (Fig. 1C) and users are generally interested in sensitive detection of somatic indels. Thus, we expect a relatively large to be used for training. Here, we trained the models to maximize for somatic prediction on a 5-fold cross-validation. Selected features and performance metric are summarized in Table 1 and Table 2, respectively.
Table 2.
Performance of the trained model for s-indel and m-indel prediction
| Indel* | TPR | FPR | Fβ | Hand-Till | |
|---|---|---|---|---|---|
| somatic | s | 0.887 | 0.005 | 0.789 | 0.971 |
| m | 0.953 | 0.032 | 0.873 | 0.985 | |
| germline | s | 0.932 | 0.004 | 0.92 | 0.979 |
| m | 0.925 | 0.005 | 0.956 | 0.989 | |
| artifact | s | 0.994 | 0.021 | 0.997 | 0.991 |
| m | 0.978 | 0.007 | 0.984 | 0.995 |
Note: TPR (sensitivity): true positive rate; FPR (): false positive rate; : generalized -score. = 15 for somatic, 1 otherwise. Hand-Till: Hand-Till’s measure, a generalization of AUC to multi-class problem (Section 6). *s for single-nucleotide indel, m for multi-nucleotide indel.
The selected features generated a distinct distribution of somatic, germline and artifact indels. For example, the majority of somatic s-indels occur in regions with ≤2 homopolyers while the artifacts occur mostly in regions with ≥3 homopolymers (Fig. 2) accompanied by lower DNA-bond strength, a strong predictor of annealing temperature (Khandelwal and Bhyravabhotla, 2010; see Supplementary Fig. S3B). Related to this, m-indels with sequences dissimilar to the flanking region are prevalent in somatic events but rare in germline or artifacts. Complex indels were uncommon in general and they occur almost exclusively as somatic s- or m-indels (Fig. 2). When considering indel size as the feature of interest, germline m-indels exhibit distinct peaks where the indel size is a multiple of three, indicating an enrichment for in-frame indels (Fig. 2). Indeed, germline indels showed a deficiency of protein truncation events and an enrichment for events that exhibited insensitivity to NMD compared to somatic indels or artifacts (Supplementary Fig. S3A). As expected, 85% of the germline indels were present in the germline database constructed for training both s-indel and m-indel (Supplementary Fig. S3A).
Fig. 2.
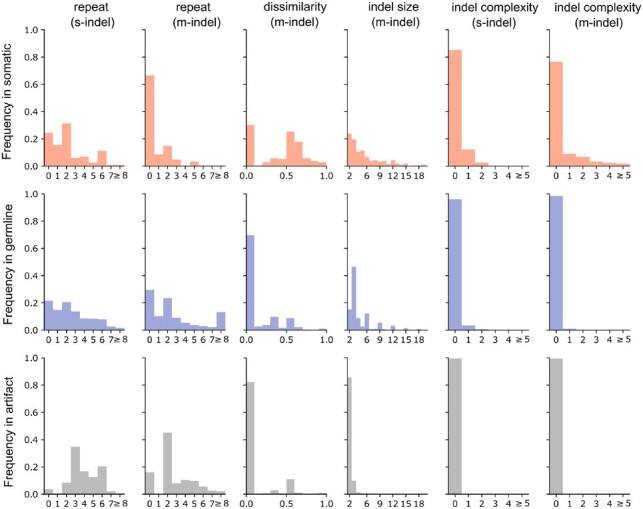
Distribution of somatic, germline and artifact indels based on features selected in the trained model. The complexity feature was not selected for m-indel (likely due to the overlapping with the dissimilarity feature) but shown for comparison with s-indel. Distribution of somatic (top panel, red), germline line (middle panel, blue) and artifacts (bottom panel, gray) are shown in histogram for each feature
4 Performance
4.1 Somatic indel discovery from tumor RNA-Seq data
The training set was generated from 330 pediatric cancer samples from 17 types; tumor transcriptomes were analyzed on Illumina HiSeq 4000 using total RNA libraries with 125-bp read length (Supplementary Table S1). To evaluate the broad applicability of the trained model, we tested the performance of RNAIndel in two additional pediatric cancer datasets (TestSet 1 and 2, Supplementary Tables S2 and S3) and three adult cancer datasets (TestSet 3‒5, Supplementary Tables S4–S6). Altogether, the five test datasets represent a broad spectrum of mutational processes in cancer genomes and technical variability in transcriptome profiling (Table 3). Published somatic DNA indels of these five test datasets, which were derived by various approaches (Section 6), were used as the truth datasets for evaluation.
Table 3.
Performance of RNAIndel on two pediatric and three adult cancer datasets
| Tumor | N | Library | ReadLen | Sequencer | SomaticIndels* | TPR | Median #FP/Sample^ |
Median indels per sample | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | ||||||||||
| 1 | Pediatric | 77 | TotalRNA | 100 | HiSeq2000or 2500 | s | 17 | 0.882 | 3 | 3 | 2318 | |
| m | 40 | 0.975 | 4 | 3 | 311 | |||||||
| 2 | AML | 158 | Poly-A | 75 | HiSeq2000 | s | 22 | 0.954 | 2 | 1 | 1036 | |
| m | 61 | 0.984 | 2 | 2 | 202 | |||||||
| 3 | NSCLC | 90 | Poly-A | 100 | HiSeq1500 | s | 97 | 0.887 | 6 | 3 | 4 | 3171 |
| m | 68 | 0.941 | 7 | 4 | 4 | 394 | ||||||
| 4 | RCC | 91 | Poly-A | 50 | HiSeq2000 | s | 130 | 0.877 | 8 | 5 | 5 | 4303 |
| m | 81 | 0.889 | 8 | 2 | 2 | 510 | ||||||
| 5 | COAD (Hyper) | 29 | Poly-A | 75 | GAIIX | s | 120 | 0.392 | 20 | 20 | 999 | |
| m | 53 | 0.953 | 11 | 10 | 141 | |||||||
| COAD (NonHyper) | 102 | s | 30 | 0.9 | 4 | 3 | 466 | |||||
| m | 14 | 1.000 | 4 | 2 | 128 | |||||||
Note: TestSet 1 consists of 77 samples from 20 types of pediatric cancers (Supplementary Table S2). The tumor type for TestSet 2–5 uses the following abbreviation: AML for acute myeloid leukemia; NSCLC for non-small cell lung cancer; RCC for renal cell carcinoma; COAD for colon adenocarcinoma with hypermutator phenotype (Hyper) or without hypermutator phenotype (NonHyper). *s for single-nucleotide indel, m for multi-nucleotide indel. ^Median number of false positives (FP) in somatic prediction per sample is shown for the default (column A), filtered with normal RNA-Seq data (column B, available only for TestSet 3 and 4) and filtered with cohort recurrence in RNA-Seq (Supplementary Fig. S4).
While RNAIndel robustly predicted somatic indels at a high sensitivity with a true positive rate (TPR) of 0.88‒1.0 in all five test sets except for single-nucleotide indels (s-indels) in a subset of colon adenocarcinoma (COAD) with a hypermutated phenotype caused by microsatellite instability (MSI) (TestSet 5). The low sensitivity (TPR of 0.392) for s-indel prediction in hypermutated COAD was caused by misclassifying MSI-induced s-indels, mostly located in homopolyers, as artifacts. Excluding the COAD hypermutators, RNAIndel was able to achieve a high sensitivity of 0.73 (45/62) even for subclonal variants with VAF < 0.1. In TestSets 2 (AML) and 5 (lung cancer), we were able to find DNA support for 10 pathogenic indels predicted as somatic by RNAIndel but absent from the truth dataset, by manually reviewing targeted capture exome sequencing or WGS for evidence (Supplementary Table S7): EP300 Y207fs (VAF: 0.1), CEBPA P23fs (0.17), RAD21 D543fs (0.02), KIT Y418_D419>Y (0.15) and CREBBP S1767fs (0.01) in AML; EGFR K745_R748>K (0.64), EGFR E746_K754>RSNISESQQ (0.68), TP53 P72_A76fs (0.57), TP53 S313fs (0.15) and TP53 V122_T123fs (0.46) in lung cancer. This indicates that the overall accuracy of RNAIndel could be higher than what was calculated using the existing truth datasets.
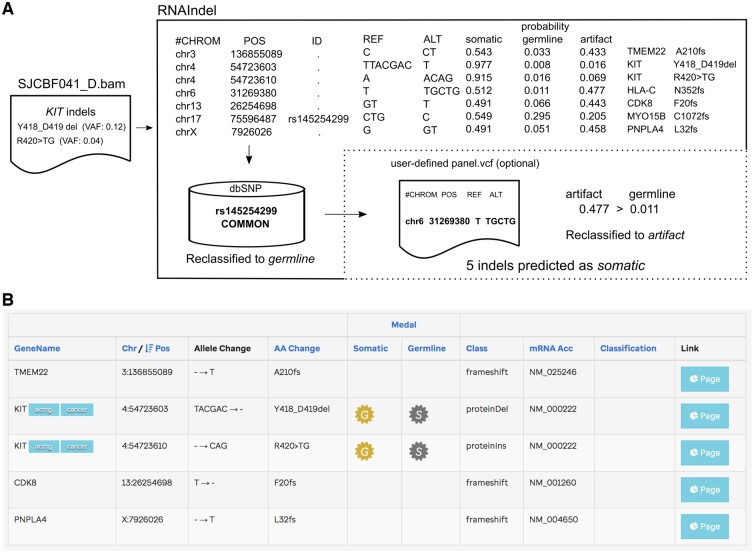
A refinement process can be applied to re-assign predicted somatic variants as non-somatic if they match custom databases that include common artifacts (Section 6, Supplementary Fig. S4). This can lead to great reduction in the false positive prediction per case especially when matched normal RNA-Seq is available e.g. TestSet 3 of lung cancer and TestSet 4 of renal cell carcinoma in Table 3). Users can also prioritize the RNAIndel prediction for variant pathogenicity by uploading the output VCF file to the St Jude Cloud tool PeCanPIE (https://platform.stjude.cloud/tools/pecan_pie) (Edmonson et al., 2019), which ranks variant pathogenicity into four tiers: gold, silver, bronze, or no medal with ‘gold’ being most likely to be pathogenic. In the example shown in Figure 3, two out the five predicted somatic indels are assigned a medal (both are gold), both are in-frame indels in KIT, a driver gene in AML while the remaining three genes are not driver genes in AML. This pathogenicity annotation can result in elimination of false positives or passenger events.
Fig. 3.
Pathogenic indel discovery by RNAIndel. (A) An example workflow is shown with a leukemia sample SJCBF041_D which harbors two distinct subclonal indels in the KIT oncogene based on WGS and WES analysis: Y418_D419del (VAF: 0.12) and R420>TG (0.04). Of the 7 somatic indels predicted by RNAIndel, only five remains after the refinement step. (B) By uploading the output VCF file to St Jude Cloud tool PeCanPIE, users can prioritize indels by pathogenicity. The KIT indels in somatic context were prioritized with the highest pathogenicity rank ‘gold’ (the ‘G’ symbol)
4.2 Comparison with paired analysis
While RNAIndel predicts somatic indels from tumor data alone, somatic variants are commonly discovered by T/N-paired analysis where systematic errors common to the tumor and the normal data would be subtracted along with germline variants. The Panel of Normals (PON) filtering supported by Mutect2 (Cibulskis et al, 2013) enhances this subtraction by collecting common variants found in a technically similar dataset of healthy samples. Performance of these two approaches was compared in an ideal situation where the matched normal controls and a dataset for PON creation are available using the non-small cell lung cancer (NSCLC) dataset (TestSet 3) (Section 6). While RNAIndel does not benefit from error subtraction by data pairing, it outperformed Mutect2’s paired analysis in both TPR and error rates (Table 4). The PON created from 100 healthy blood samples with similar technical specifications almost halved errors in the Mutect2 paired analysis with two true somatic single-nucleotide indels also filtered, even though no indels equivalent to them were found in the panel. RNAIndel optionally accepts a user-defined panel to refine its somatic prediction (Fig. 3). When the PON was used as the input panel, only a marginal improvement was seen in RNAIndel’s results, suggesting that indels found in the panel were already predicted as artifact or germline.
Table 4.
Performance of Mutect2 paired analysis and RNAIndel
| TPR |
#False Positives |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Mutect2 |
RNAIndel |
Mutect2 |
RNAIndel |
||||||
| PON | + | − | + | − | + | − | + | − | |
| NSCLC (n=90) | s | 0.464 | 0.485 | 0.887 | 0.887 | 94 | 50 | 6 | 5 |
| m | 0.603 | 0.603 | 0.941 | 0.941 | 26 | 16 | 7 | 7 | |
Note: Analysis was performed with (+) and without (−) Panel of Normals (PON) for non-small cell lung cancer (NSCLC) dataset (TestSet 3). s indicates single-nucleotide indel, m indicates multi-nucleotide indel.
4.3 Working with an external variant caller—an example using GATK-HaplotypeCaller
In addition to calling indels from a BAM file, RNAIndel can accept a VCF file made by an external variant caller (Fig. 1A). We chose Genome Analysis Toolkit-HaplotypeCaller (GATK-HC) (DePristo et al., 2011) to illustrate this feature for two reasons. First, GATK Best Practice of RNA-Seq variant calling has been documented in detail for GATK-HC with STAR (Section 6). Second, the STAR/GATK-HC pipeline showed best performance in a recent study where a variety of combinations of an RNA-Seq mapper and a variant caller were tested for detecting known somatic EGFR indels in lung cancer (Sun et al., 2017). Following this procedure, we evaluated the performance of three approaches for detecting expressed pathogenic indels: RNAIndel with the built-in caller, RNAIndel with GATK-HC, and the Best Practice-based approach without RNAIndel (Fig. 4A). We used TestSet 1 which contained 23 pathogenic indels and the RNA-Seq data were analyzed by the three approaches followed by automated pathogenicity classification (Fig. 3B). RNAIndel with the built-in caller achieved the highest sensitivity (22/23) with one somatic indel misclassified as artifact. The prediction from the combination of RNAIndel and GATK-HC had the fewest false positives (a total of 5 artifacts were predicted), but this was achieved at a cost of sensitivity: only 14 of the 23 pathogenic indels were predicted as somatic (the remaining 9 indels were not detected by GATK-HC) (Fig. 4B and C). The Best Practice prediction, i.e. the STAR/GATK-HC pipeline, was the noisiest with 442 artifacts, which is 14 times higher than the first two approaches. This approach also has the lowest sensitivity: only 13 of the 23 true somatic indels were correctly predicted with 1 removed by the Best Practice variant filtration and 9 undetected (Fig. 4B and C). The RNAIndel-based approaches predicted 2 germline indels as somatic that were curated as cancer predisposition mutations on ClinVar: RAD50 K994_E995fs (rs587780154) and BRCA2 P1062_Q1063 (rs80359374).
Fig. 4.
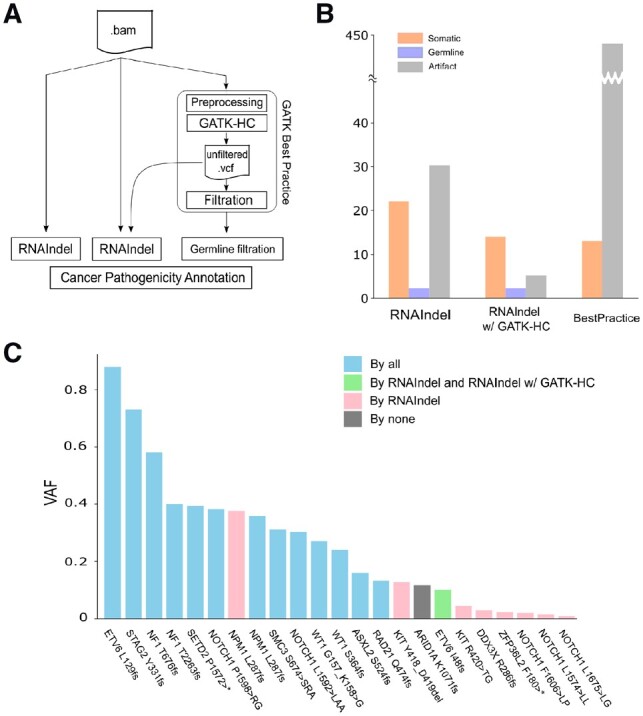
Working with an external variant caller. RNAIndel is flexibly designed to work with an external variant caller via a user-provided VCF file. GATK-HC was used as an example. (A) TestSet 1 (n = 77) was screened for pathogenic somatic indels by three approaches: 1) RNAIndel with built-in caller (left) using a RNA-Seq BAM file as input; 2) RNAIndel with GATK-HC (middle) using the VCF file from GATK-HC and the RNA-Seq BAM file as input; and 3) Best Practice-based approach (right). (B) Comparison of predicted somatic indels by the three approaches with the truth datasets. (C) Performance of the three methods on 23 known pathogenic somatic indels
5 Discussion
We developed RNAIndel, a machine-learning based method to classify coding indels derived from RNA-Seq data. To construct a high-quality training set, we used indel variants derived from three-platform sequencing of WGS, WES and RNA-Seq with WGS data generated from a PCR-free library protocol (Rusch et al., 2018). Previously, we have shown that exonic somatic indels that are validated by both WGS and WES and passed human review could achieve a positive predictive rate of 98.8% and a sensitivity of 94.3%, assuring the quality of this training set (Rusch et al., 2018). We focused on small indels with indel size < 23-bp as larger indels cannot be mapped accurately with the popular mapping algorithms and can be detected more effectively using software tools designed for structural variation analysis. For training the model, we excluded two high-grade glioma hypermutators to avoid feature selection bias; therefore, the current model is not able to predict indels caused by microsatellite instability (MSI) at high sensitivity. The low sensitivity of predicting s-indel in a subset of colon adenocarcinomas with hypermutator phenotype [TestSet 5 COAD (Hyper) in Table 3] reflects this deficiency in the training model. The vast majority of MSI-induced indels are s-indels in polyA or polyT regions (The Cancer Genome Atlas Network, 2012) and 81% of the missed indels in TestSet 5 occurred in polyA or polyT regions. MSI-induced hypermutation is rare in pediatric cancer but relatively common in a subset of adult cancers such as uterine corpus carcinoma (28.3%), stomach (21.9%) and colon adenocarcinomas (16.6%) (Cortes-Ciriano et al., 2017). We recommend users retrain the model for these cancer types.
Artifact indels mostly occurred with a low variant allele frequency (VAF) of < 0.1; however more than 20% of somatic indels in the training dataset were in the same low VAF range (Supplementary Fig. S3C). VAF was initially used as a feature for model training but was abandoned after we recognized that this feature would lead to loss of true subclonal somatic indels. The combined results obtained from the five test datasets showed 11% of the somatic indels have < 0.1 VAF, confirming that RNAIndel is able to distinguish somatic indels of low VAF from those of artifacts. For the five test datasets, we used the published data as the ground truth for measuring the accuracy of RNAIndel. The real accuracy of RNAIndel may be higher in some cases—by manual review of WGS or deep capture sequencing, we were able to find DNA support for predicted somatic indels which are absent in the ground truth data (Supplementary Table S8).
Features assigned to variants by the software are used to inform its classification logic and at times may unveil interesting biological insights. For example, the strand-slippage model predicts a simple indel with the flanking sequence inserted or deleted. Two of our assigned features capture this state; the feature ‘dissimilarity’ is set to zero if the indel matches its flanking sequence, and the feature ‘indel complexity’ is set to zero for simple indels. Interestingly, somatic indels frequently deviated from the slippage model, while germline and artifact indels were generally consistent with it (Fig. 2). This suggests that the mechanisms of somatic indel acquisition differ from that of germline due to the instability of cancer genome. Alternatively, one may speculate that a high proportion of indels compatible with the slippage model tend to be under weaker selection pressure and may hence appear in germline. For example, a triplet indel in a tandem tri-nucleotide repeat region is a pattern typical of germline indels and can be explained by strand slippage. This type of indel may have limited impact on protein function due to the possible redundancy of the repetitive amino acid molecules.
Like other variant discovery tools, RNAIndel expects users to review the predicted outputs. To facilitate this process, we developed a refinement step by removing non-somatic indels using custom files (Supplementary Fig. S4), variant pathogenicity classification (Fig. 3), or using an alternative indel caller (Fig. 4) to ensure consensus. By combining automated classification with manual curation, RNAIndel enables an unbiased screening of somatic indels from tumor RNA-Seq data alone; an application of RNA-Seq data which has not been attempted effectively due to the lack of suitable tools.
6 Materials and methods
6.1 Datasets
All RNA-Seq datasets in this study, which consisted of a training set and five test sets, were mapped by STAR in 2-pass mode to GRCh38 (Supplementary Methods).
The training set was comprised of paired tumor (T)/normal (N) WGS and WES, and tumor RNA-Seq generated from 330 pediatric cancer patients from 17 major cancer types (Supplementary Table S1). In this dataset, somatic mutations in different cancer types were known to be acquired through diverse mutagenesis mechanisms including APOBEC, Reactive Oxidative Stress (ROS), Homologous Recombination (HR) deficiency and UV-light (Ma et al., 2018). Importantly, the WGS libraries were prepared by a PCR-free protocol to minimize PCR-artifacts. We did not include two high grade glioma samples with hypermutator phenotype to avoid bias in feature selection caused by the overwhelming number of s-indel in homopolymer regions.
The truth dataset used for training was comprised of expressed DNA indels that were validated by WGS and WES and passed manual review of variant quality and alignment by human analysts. Somatic exonic indels derived by this approach have a 98.8% validation rate based on capture sequencing (Rusch et al., 2018). A minimum of 10× coverage in tumor and normal WGS or WES is required for considering whether an RNA-Seq indel that does not match a known somatic DNA indel should be classified as germline (i.e. present in normal DNA) or artifact (absent in both tumor and normal DNA). On average, the WGS and WES of the training samples had 98.2 and 99.6% of the coding regions with ≥ 10× read coverage, respectively, showing that our overall classification of the training dataset was unbiased across the entire coding regions. The details of nucleic acid extraction, library preparation, sequencing, and variant detection and validation were previously described (Rusch et al., 2018).
Five public datasets were used as test sets. The first set (TestSet 1) was comprised of 77 RNA-Seq samples of 20 tumor types (EGAS00001002217) (Rusch et al., 2018). The ground-truth somatic indels in TestSet1 were based on paired T/N DNA-Seq analysis performed on the samples. The annotation of somatic, germline and artifacts presented in Figure 4 was determined using the same coverage criteria as the training dataset. To confirm the annotation derived by the Illumina DNA-Seq cross-platform validation with an orthogonal sequencing method, we selected 5 somatic, 2 germline and 4 artifact indels with a median VAF of 0.33 (range: 0.14‒0.79), for Sanger validation. The results were 100% concordant with NGS-based annotation (Supplementary Fig. S5). The second test set (TestSet 2) included 158 acute myeloid leukaemia samples by NCI TARGET project (dbGaP study identifier phs000465) (https://ocg.cancer.gov/programs/target). The ground-truth somatic indels in TestSet 2 were compiled from the paired-T/N WGS analysis by the Complete Genomics, Inc. (CGI) Cancer Sequencing service pipeline version 2 (Ma et al., 2018) and targeted capture sequencing analysis (Bolouri et al., 2018) by Strelka (Saunders et al., 2012). The third set (TestSet 3) was a non-small cell lung cancer dataset from 90 patients (Wang et al., 2018) (EGAD00001004071). The ground-truth somatic indels were called by Mutect2 (Cibulskis et al., 2013). The fourth (TestSet 4) and fifth (TestSet 5) datasets were TCGA datasets for renal cell carcinoma and colon adenocarcinoma (https://portal.gdc.cancer.gov). The ground-truths of these test sets were obtained from Broad GDAC Firehose (http://gdac.broadinstitute.org). In TestSet 5, the hypermutated samples were identified by the previous study (The Cancer Genome Atlas Network, 2012).
6.2 Performance metrics
As performance metrics for overall classification are sensitive to class imbalance, we used class-specific metrics for evaluation. For class , true positive (TP), true negative (TN), false positive (FP) and false negative (FN) are defined by:
where | | denotes the set size. True positive rate (TPR), false positive rate (FPR) and false discovery rate (FDR) are defined by TP/(TP+FN), FP/(FP+TN) and FP/(FP+TP), respectively.
A generalized -score () is formulated as:
where is a weighting parameter for TP. The Hand-Till measure (M) is a generalization of the area under the curve (AUC) to multi-class classification (Hand and Till, 2001). This quantifies the separation of classes in terms of prediction probability. For three classes and , the Hand-Till measure for class is defined:
where is the probability that a randomly chosen instance from class has a higher prediction probability of being in class than a randomly chosen class instance’s probability of being class .
6.3 Training by cross validation
Models are evaluated in the actual imbalanced distribution by -fold cross-validation (-fold CV), whose value is user-configurable (default 5). In the first fold, % of the data is held out un-sampled as a validation set and the training set is down-sampled from the remaining%. Trained models are evaluated using the un-sampled validation set. This process is repeated to the -th fold by rotating the validation set portion.
Training is carried by the following three steps: down-sampling, feature selection and parameter tuning. At each step, an optimal value or a subset of features is determined to maximizefor somatic prediction on -fold CV. In the down-sampling step, the training set is down-sampled with the following ratio: somatic: germline: artifact = . The ratio is searched within the range of for a maximum for somatic prediction using all features on -fold CV. Next, features are selected in a greedy best-first search. The search procedure begins with an empty set of selected features and a set of candidate features initially containing all features. In the first iteration, each feature is evaluated separately, and the one that achieves the maximum is added to the selected set and removed from the candidate feature set. For features tied for the highest value, the one with the minimum FDR is chosen. The second iteration examines the combination of the feature in the selected set and one of the remaining candidate features. The highest feature is similarly added to the selected set and removed from the candidate set. This procedure continues until the candidate set becomes empty and a subset of features with the highest is selected. Finally, the maximum number of features used in splitting a node during the tree construction is searched over from 1 to the number of features selected from the previous step. The value maximizing is chosen.
6.4 Prediction refinement
RNAIndel refines the somatic prediction using databases and the prediction probability. Common polymorphisms are reclassified to germline if predicted as somatic. Putative somatic indels matching a non-somatic indel panel, which is user-definable, are assigned to germline or artifact, whichever has the higher probability (Supplementary Fig. S4). This reclassification rule obviates the need to label non-somatic indels as germline or artifact when compiling a panel, which can be difficult in the absence of the DNA evidence. At default, RNAIndel uses a panel made from a reviewed list of non-somatic indels that were predicted somatic three time or more in the training set cross-validation. An ideal source of such non-somatic indels would be technically and biologically similar normal RNA-Seq data. RNAIndel can compile a non-somatic panel from normal VCF files. To avoid potential somatic variant contamination, the panel requires variants appear times in the VCF file set and absence in the COSMIC (Tate et al., 2019) database (Supplementary Fig. S4A). Such panels were created using the matched normal RNA-Seq data in TestSet 3 and 4 with, and successfully applied without affecting the TPR (Table 3). While a suitable normal dataset may not be available, it is a common scenario that multiple RNA-Seq samples are generated in a study. Putative somatic indels recurring in the output VCF files from the study cohort are possibly frequent artifacts or somatic hotspots as common polymorphisms are reclassified to germline if predicted as somatic. RNAIndel annotates recurrent indels that are absent in COSMIC in the output VCF INFO field (Supplementary Fig. S4B). Filtering non-COSMIC recurrent indels was also as effective in refining the somatic prediction, but, in the five test sets, one true somatic indel was also removed: the ACTB N111>MN insertion in TestSet 2 (Table 3).
6.5 Paired RNA-Seq analysis with panel of normals (PON)
While the expression difference between tissues is a limitation of applying PON to the RNA context, we selected a peripheral blood dataset from Dutch 500FG cohort as input for PON creation due to the limited availability of RNA-Seq dataset from healthy sources and the technical similarity to TestSet 3 (Table 3); the dataset was prepared by poly-A enriched library protocol and sequenced with 100-bp read length on Illumina HiSeq 2500, a higher-throughput version of HiSeq 1500. The FASTQ files (n = 100) (PRJNA553703) were downloaded and STAR 2-pass mapped against GRCh38. BAM files from this cohort and TestSet 3 were preprocessed by reassigning STAR’s MAPQ to 60 and GATK’s ‘SplitNCigarReads’ command. The PON was created by following (https://software.broadinstitute.org/gatk/documentation/tooldocs/4.0.2.0/org_broadinstitute_hellbender_tools_walkers_mutect_CreateSomaticPanelOfNormals.php). For each tumor and matched normal BAM file pair, Mutect2 in GATK 4.0.2.1 was run with and without the PON. The output VCF files were filtered using the ‘FilterMutectCalls’ command, and coding indels with ‘PASS’ status were considered putative somatic.
6.6 RNA-Seq variant calling by GATK-HC
Variant calling of BAM files were performed by GATK-HC in GATK 4.0.2.1. The work flow closely following the GATK Best Practice for calling variants in RNA-Seq (https://software.broadinstitute.org/gatk/documentation/article.php?id=3891). The Best Practice protocol filters spurious calls but does not distinguish somatic and germline calls. For a fair comparison with RNAIndel, which distinguishes somatic, germline and artifact calls, germline calls in the Best Practice protocol were filtered by matching the normal DNA-Seq data (Germline filtration in Fig. 4A). Indels passed these filters were considered putative somatic. Of those, indels annotated pathogenic by PeCanPIE were used for performance comparison.
6.7 Indel recovery
RNAIndel uses actual indel alignments in the BAM file for feature calculation. For this calculation, indels reported from the caller are searched in the BAM file. RNAIndel first searches for indels equivalent to the reported indel and merges them (Supplementary Fig. S1). If no equivalent indels are found, the nearest indel from the reported locus within a widow of ±5-bp is used for analysis as proxy. These substituted cases are labelled as such in the output VCF file. Typically, 100% of indels reported from the built-in Bambino caller are successfully recovered.
When GATK-HC is used as an external caller, ∼90% of indels are recovered from the alignment pileup while the remaining are missing. We evaluated the distribution of missing GATK indels in TestSet 1 and found that 48.6% were located in reads spanning intron-exon boundaries and 12.5% are at the human leukocyte antigen (HLA) loci, suggesting the missing indels may be caused by a combination of mapping artifacts and anomalies caused by GATK-HC locally assembled haplotypes.
For further investigation, we randomly selected 100 missing indels for manual curation. As shown in Supplementary Table S9, 47 cases were in intron-exon boundaries, 14 at HLA loci and 39 non-HLA exon cases. Remapping to the genome by the BLAT algorithm (Kent, 2002) confirmed that 44 of the 47 intronic indels were caused by mis-alignments over splice junctions. The remaining 3 cases had low quality mappings. The high degree of genetic diversity at HLA loci likely explains the missing indels at the HLA loci. For the remaining 39 non-HLA exonic indels, 17 were caused by mis-mappings, 15 were in highly-repetitive regions, 3 were caused by mis-alignment of SNVs in the neighborhood and 4 were in non-repetitive regions. Given that 96% of the missed indels can be attributed to artifacts, the impact of the underreporting is marginal for our analysis of somatic prediction by GATK. Indeed, none of the GATK indels that match the ground truth are in this category.
6.8 Sanger validation
Sanger validation was performed on samples for which DNA was available. Using PrimerTK, PCR primers were designed to include the regions to validate and generate amplicons between 200 bp and 350 bp in length (Supplementary Table S10). The PCR was performed using Amplitaq Gold 360 Master Mix (Thermo Fisher Scientific) in a 25 µl reaction with 50 ng of genomic DNA, 400 nM of each amplification primer and 1ul of 360 GC enhancer. The PCR was performed on a Veriti thermo cycler (Life Technologies) with a 10 min activation step at 95 °C followed by 35 cycles of amplification (94 °C 30 s, 59 °C 30 s, 72 °C 30 s), and a final extension for 7 min at 72 °C. Prior to sequencing, the PCR reactions were cleaned up using the Minelute PCR Purification Kit (Qiagen). Sequencing data was generated using an ABI3730 DNA Sequencer (Applied Biosystems).
Supplementary Material
Acknowledgements
We thank Mr James McMurry for downloading the adult cancer TCGA data for this study.
Funding
This work was funded by the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital. J.Z. and K.H. were also supported in part by grant from National Institute of General Medical Sciences (P50GM115279-03).
Conflict of Interest: none declared.
Contributor Information
Kohei Hagiwara, Computational Biology, St Jude Children’s Research Hospital, Memphis, TN 38105, USA.
Liang Ding, Computational Biology, St Jude Children’s Research Hospital, Memphis, TN 38105, USA.
Michael N Edmonson, Computational Biology, St Jude Children’s Research Hospital, Memphis, TN 38105, USA.
Stephen V Rice, Computational Biology, St Jude Children’s Research Hospital, Memphis, TN 38105, USA.
Scott Newman, Computational Biology, St Jude Children’s Research Hospital, Memphis, TN 38105, USA.
John Easton, Computational Biology, St Jude Children’s Research Hospital, Memphis, TN 38105, USA.
Juncheng Dai, Department of Epidemiology, Nanjing Medical University School of Public Health, Jiangning District, Nanjing, 211166, People’s Republic of China.
Soheil Meshinchi, Clinical Research Division, Fred Hutchinson Cancer Research Center, Seattle, WA 98109, USA.
Rhonda E Ries, Clinical Research Division, Fred Hutchinson Cancer Research Center, Seattle, WA 98109, USA.
Michael Rusch, Computational Biology, St Jude Children’s Research Hospital, Memphis, TN 38105, USA.
Jinghui Zhang, Computational Biology, St Jude Children’s Research Hospital, Memphis, TN 38105, USA.
References
- Bolouri H. et al. (2018) The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat. Med., 1, 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. (2001) Random forests. Mach. Learn., 45, 5–32. [Google Scholar]
- Cibulskis K. et al. (2013) Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol., 31, 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes-Ciriano I. et al. (2017) A molecular portrait of microsatellite instability across multiple cancers. Nat. Commun., 8, doi: 10.1038/ncomms15180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P. et al. (2011) The variant call format and VCFtools. Bioinformatics, 27, 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M.A. et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet, 43, 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A. et al. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonson M.N. et al. (2011) Bambino: a variant detector and alignment viewer for next-generation sequencing data in the SAM/BAM format. Bioinformatics, 27, 865–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonson M.N. et al. (2019) Pediatric cancer variant pathogenicity information exchange (PeCanPIE): a cloud-based platform for curating and classifying germline variants. Genome Res., doi: 10.1101/gr.250357.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H. et al. (2014) Reducing INDEL calling errors in whole genome and exome sequencing data. Genome Med., 6, doi: 10.1186/s13073-014-0089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Diaz M., Kunkel T.A. (2006) Mechanism of a genetic glissando: structural biology of indel mutations. Trends Biochem. Sci., 31, 206–214. [DOI] [PubMed] [Google Scholar]
- Hand D.J., Till R.J. (2001) A simple generalisation of the area under the ROC curve for multiple class classification problems. Mach. Learn., 45, 171–186. [Google Scholar]
- Karczewski K.J. et al. (2019) Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv 531210. doi: 10.1101/531210.
- Kent W.J. (2002) BLAT–the BLAST-like alignment tool. Genome Res., 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelwal G., Bhyravabhotla J. (2010) A phenomenological model for predicting melting temperatures of DNA sequences. PLoS One, 5, e12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum M.J. et al. (2014) ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res, 42, D980–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X. et al. (2018) Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature, 555, 371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P.C. et al. (2008) Genetic variation in an individual human exome. PLoS Genet., 4, e1000160.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikkonen L., Lise S. (2017) Making the most of RNA-seq: pre-processing sequencing data with Opossum for reliable SNP variant detection. Wellcome Open Res., doi: 10.12688/wellcomeopenres.10501.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskol R. et al. (2013) Reliable identification of genomic variants from RNA-Seq data. Am. J. Hum. Genet., 93, 641–‒651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp M.W., Maquat L.E. (2016) Leveraging rules of nonsense-mediated mRNA decay for genome engineering and personalized medicine. Cell, 165, 1319–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K.D. et al. (2004) NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res., 33, D501–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch M. et al. (2018) Clinical cancer genomic profiling by three-platform sequencing of whole genome, whole exome and transcriptome. Nat. Commun., 9, doi: 10.1038/s41467-018-06485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders C.T. et al. (2012) Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics, 28, 1811–1817. [DOI] [PubMed] [Google Scholar]
- Sherry S.T. et al. (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res., 29, 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z. et al. (2017) Indel detection from RNA-seq data: tool evaluation and strategies for accurate detection of actionable mutations. Brief. Bioinform., 18, 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan A. et al. (2015) Unified representation of genetic variants. Bioinformatics, 31, 2202–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X. et al. (2014) The eSNV-detect: a computational system to identify expressed single nucleotide variants from transcriptome sequencing data. Nucleic Acids Res., 42, e172.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J.G. et al. (2019) COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res., 47, D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network. (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature, 487, 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. et al. (2014) RVboost: RNA-seq variant prioritization using a boosting method. Bioinformatics, 30, 3414–3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. et al. (2018) Whole-genome sequencing reveals genomic signatures associated with the inflammatory microenvironments in Chinese NSCLC patients. Nat. Commun., doi: 10.1038/s41467-018-04492-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J.L. et al. (2002) Human diallelic insertion/deletion polymorphisms. Am. J. Hum. Genet., 71, 854–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K. et al. (2016) Systematic discovery of complex insertions and deletions in human cancers. Nat. Med., 22, 97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Gerstein M. (2003) Patterns of nucleotide substitution, insertion and deletion in the human genome inferred from pseudogenes. Nucleic Acids Res., 15, 5338–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.