Ultraviolet (UV) radiation is a major environmental mutagen. Exposure to UV leads to a sharp peak of γH2AX, the phosphorylated form of the histone variant H2AX, in the S phase within an asynchronous population of cells. γH2AX is often considered a definitive marker of DNA damage inside a cell. In this report, we show that γH2AX in the S-phase cells after UV irradiation reports neither on the extent of primary DNA damage in the form of cyclobutane pyrimidine dimers nor on the extent of its secondary manifestations in the form of DNA double-strand breaks or in the inhibition of global transcription.
KEYWORDS: γH2AX, cell cycle, replication, S phase, UV, DNA damage, DNA damage response (DDR), DNA repair, DNA replication
ABSTRACT
Ultraviolet (UV) radiation is a major environmental mutagen. Exposure to UV leads to a sharp peak of γH2AX, the phosphorylated form of the histone variant H2AX, in the S phase within an asynchronous population of cells. γH2AX is often considered a definitive marker of DNA damage inside a cell. In this report, we show that γH2AX in the S-phase cells after UV irradiation reports neither on the extent of primary DNA damage in the form of cyclobutane pyrimidine dimers nor on the extent of its secondary manifestations in the form of DNA double-strand breaks or in the inhibition of global transcription. Instead, γH2AX in the S phase corresponds to the sites of active replication at the time of UV irradiation. This accumulation of γH2AX at replication sites slows down the replication. However, the cells do complete the replication of their genomes and arrest within the G2 phase. Our study suggests that it is not DNA damage, but the response elicited, which peaks in the S phase upon UV irradiation.
INTRODUCTION
Chromatin, a complex of DNA and protein, transforms DNA inside a nucleus into a functional genome. Histones make up the majority of the protein content of chromatin. Their modifications, in the form of histone methylation, acetylation, or phosphorylation, are major regulators of nuclear processes, such as DNA transcription, replication, or repair (1, 2). One such histone modification is γH2AX, the phosphorylated form of the variant histone H2AX at serine 139, observed during a variety of cellular stresses (3–5). Genotoxic stress, in particular, leads to the accumulation of γH2AX at the sites of DNA damage within just a few minutes of damage, which at later time points starts to spread across millions of base pairs from those sites of damage (6). This phosphorylation of H2AX at serine 139 is mediated directly by the phosphatidylinositol 3-kinase-related kinases (PIKK) ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3-related (ATR)—the master transducers of DNA signals (7). This has established γH2AX as a standard, direct and faithful marker of DNA damage inside a cell (6). Using the levels of γH2AX as a measure of DNA damage, in particular that of DNA double-strand breaks (DSBs), previous studies have shown that DNA damage peaks in the S phase of the cell cycle with a variety of genotoxic treatments (8–11). These observations conform with the idea that S-phase cells within a population are most vulnerable to DNA damage owing to the active replication of their DNA, which requires permissible chromatin states that render the DNA more susceptible to DNA damage (12), or due to the fact that single-strand breaks (SSBs) formed as repair intermediates may be converted to DSBs upon unchecked replication (13). This notion, though, has been challenged by a relatively recent study where it was shown that only a minority of γH2AX foci in the S phase after UV irradiation represent DSBs (14). The study, while addressing many important aspects of UV-induced DNA damage, does not comment on the nature of γH2AX in the S-phase cells apart from the observation that it does not mark DSBs.
Here we fill the above lacuna by studying cell cycle-dependent DNA damage responses (DDR) using an improved imaging-based cell cycle staging developed previously (8). This helped us study not only the levels of different DDR proteins at a single-cell resolution but also their localization across the cell cycle. In particular, we show that the γH2AX peak in the S phase after UV irradiation reflects the colocalization of γH2AX with the active replication forks at the time of irradiation and reports neither on the extent of primary DNA damage, as measured by the induction of UV adducts, nor on that of the secondary damage in the form of DSBs. Moreover, we show that this colocalization of γH2AX with the sites of active replication does not entirely halt replication and that most of the S-phase cells at the time of UV irradiation complete the replication of their DNA and are later arrested in the G2 phase of the cell cycle. This further proves that cells in all the cell cycle phases are equally vulnerable to DNA damage from external sources, but it is the S-phase cells which show the highest levels of response in terms of γH2AX induction, owing to active DNA replication in those cells.
RESULTS
Cell cycle-dependent DNA damage responses.
Cell cycle is the process by which a cell divides into two daughter cells. Starting from the first gap phase, G1, the cell moves onto the S phase, where it replicates its DNA, and ends up in the second gap phase, G2, which then is followed by the cell division in the mitosis or M phase. Each cell cycle phase is marked by a distinct chromatin state facilitating the structure, function, and expression of the genome in that particular phase of the cell cycle (2, 15, 16). S phase, in particular, requires an open, more permissive chromatin state to aid DNA replication. This opened-up chromatin state, it is believed, renders the S-phase cells more vulnerable to DNA damage (12). Here we investigated the above hypothesis by studying cell cycle-dependent DNA damage responses (DDR) to UV and neocarzinostatin (NCS) treatments. UV is known to cause replication stress by forming cyclobutane-pyrimidine dimers and 6-4 pyrimidine-pyrimidone photoproducts that are repaired by nucleotide excision repair (NER). NCS—a radiomimetic drug—causes DSBs within just a few minutes of treatment (17) that may be repaired by homologous recombination (HR) or nonhomologous end joining (NHEJ) (18, 19). The aim was to study differences and similarities between the responses to the two treatments, especially in the S phase of the cell cycle.
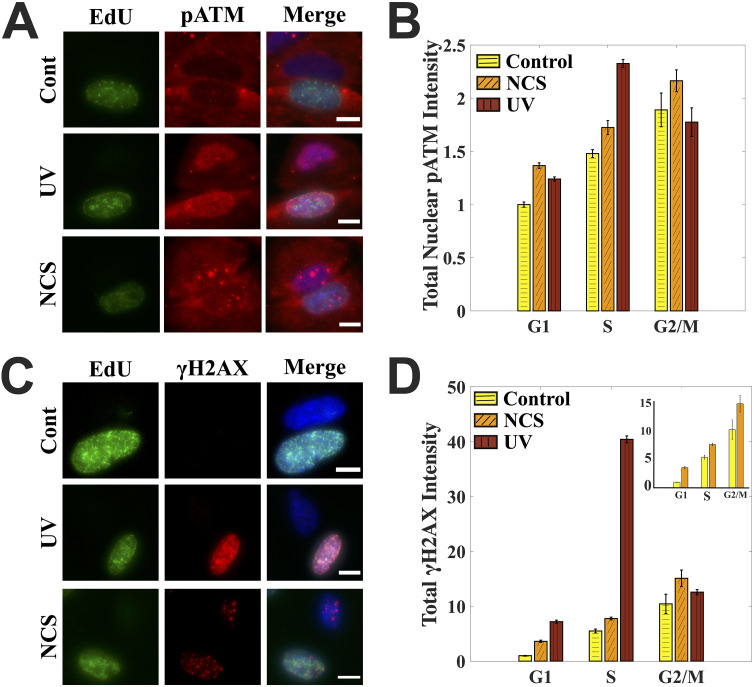
Cell cycle staging from just the DNA content may lead to mislabeling of early-S-phase cells as G1 or late-S-phase cells as G2, depending on their degree of progress inside the S phase. Hence, to identify true S-phase cells in the population, we labeled them with thymidine analogue 5-ethynyl-2’-deoxyuridine (EdU). EdU gets incorporated into the actively replicating DNA, which can later be detected via click chemistry (20). By combining EdU-based labeling of the S-phase cells with a previously developed microscopy-based cell cycle staging from DNA content (8), we were able to identify true S-phase cells, some of which were previously mislabeled as G1- or G2-phase cells when identified based on their DNA content alone (see Fig. S1A and B in the supplemental material). Once standardized, we used the technique to first study the DDR in S-phase cells treated with UV and NCS in terms of the levels of phosphorylation of the important DDR proteins ATM and H2AX as measured by quantifying total nuclear intensities of the proteins from the respective channels.
ATM is an important mediator of DNA damage response in its active form, which is marked by its phosphorylation at serine 1981 (pATM). It is activated during both NCS (which causes DSBs) and UV (which causes replication stress) treatments but via different mechanisms: DSBs directly recruit ATM via the Mre11-Rad50-Nbs1 (MRN) complex, which leads to autophosphorylation of ATM, while UV causes ATM activation in an ATR-dependent manner (21). Even though the phosphorylation of ATM in response to UV occurs later in the DDR cascade, it can be observed at a 30- to 60-min time point, as also reported previously (11, 21). Once activated, ATM also can phosphorylate H2AX at serine 139—a modification known by the name γH2AX and considered a standard DNA damage marker.
We observed that for the cells treated with NCS, the mean levels of γH2AX and nuclear pATM increased with the increase in DNA content through the cell cycle. In contrast, for the cells treated with UV the mean levels of both γH2AX and nuclear pATM peaked in the S phase of the cell cycle, with the γH2AX peak being much more pronounced than the pATM peak (Fig. 1D and F). We also observed that pATM and γH2AX foci counts followed different cell cycle trends in NCS-treated cells (Fig. S1C and D). The above observations pose the question: is it DNA damage or the response elicited which is cell cycle dependent? While it is well-known that the treatment with a radiomimetic drug or ionizing radiation leads to a clear accumulation of γH2AX at the sites of DSBs and that γH2AX foci in such cases clearly mark DSBS (17, 22–25), the role of γH2AX in the S-phase cells post-UV irradiation is not well understood. Hence, we decided to begin by investigating the cell cycle profile of UV-induced primary DNA lesions.
FIG 1.
Cell cycle-dependent DNA damage responses. (A) Cell cycle-dependent induction of pATM after UV and NCS treatment. Each row has two cells: one S phase (EdU positive) and the other non-S phase. Merged images also have a DAPI channel shown here in blue. Images were processed the same to aid the comparison. (B) Quantification for panel A. UV-treated cells have a pATM peak in the S phase, while NCS-treated cells have pATM levels that increase with the increase in DNA content through the cell cycle. (C) Cell cycle-dependent induction of γH2AX after UV and NCS treatment. Each row has two cells: one S phase and the other non-S phase. Merged images also have a DAPI channel shown here in blue. Images were processed with the same contrast adjustments to aid the comparison. (D) Quantification for panel C. UV-treated cells have a sharp γH2AX peak in the S phase, while NCS-treated cells have pATM levels that increase with the increase in DNA content through the cell cycle. The inset compares γH2AX levels just for the control and NCS-treated cells. DNA damage in cells was caused with either 1.6 μg/ml NCS for 2 min or 10 J/m2 UV, after which cells were left to recover for 60 min. Control cells were fixed right after EdU labeling. Bar graphs are normalized with respect to the mean value for G1 phase in control cells across the populations. Error bars are standard errors of the mean. Scale bars: 10 μm. See also Fig. S1 in the supplemental material.
UV-induced primary DNA lesions do not peak in the S phase, unlike γH2AX.
UV irradiation induces direct photodamage to DNA in the form of cyclobutane pyrimidine dimers (CPDs) and 6-4 pyrimidine-pyrimidone photoproducts, of which CPDs constitute up to 80% of the total photoproducts (26). We stained cells for CPDs along with γH2AX to measure the extent of primary DNA damage along with that of the response elicited on a cell-by-cell basis in the context of the cell cycle (Fig. 2A). The levels of CPDs, as well as γH2AX and EdU, were measured by quantifying the total nuclear intensities of the labeling from their respective channels.
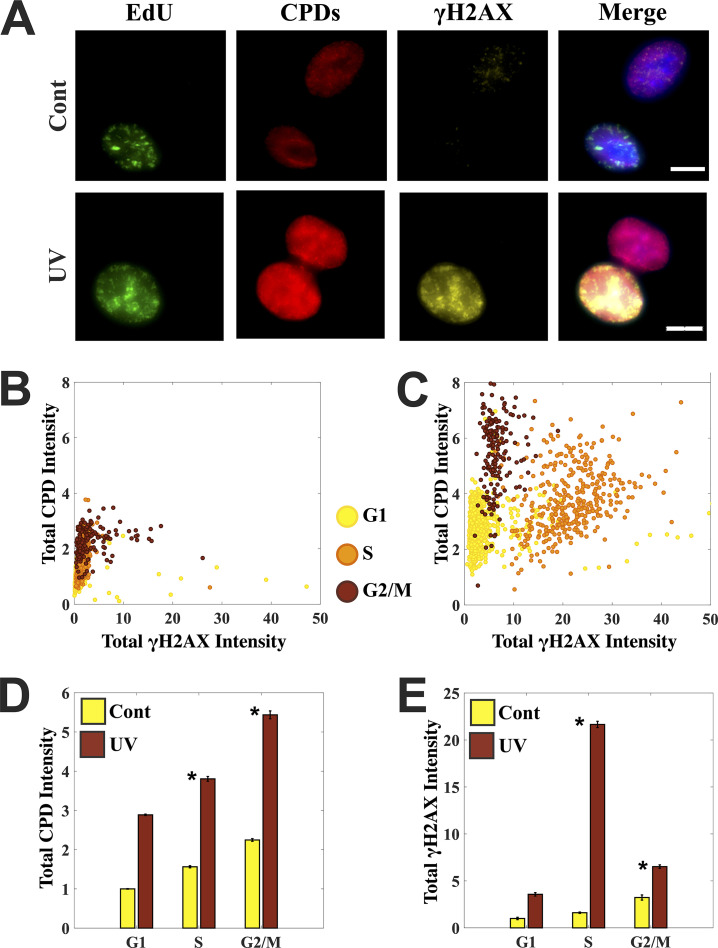
FIG 2.
UV-induced primary DNA lesions do not peak in the S phase, unlike the DNA damage response elicited in terms of γH2AX. (A) Control and UV-treated cells were stained simultaneously for cyclobutane pyrimidine dimers (CPDs) and γH2AX to measure the extent of primary DNA damage (CPDs) and compare the response (via γH2AX) in S- and non-S-phase cells. Each row has two cells: one S phase (EdU positive) and the other non-S phase. Merged images also have a DAPI channel shown here in blue. Images were processed with the same contrast adjustments to aid the comparison. (B and C) Scatterplots for control and UV-treated cells marking the levels of primary damage in the form of CPDs and the response elicited in the form of γH2AX induction at single-cell resolution. UV irradiation gives rise to 3 distinct populations corresponding to G1, S, and G2/M phases in terms of the levels of primary DNA damage and the response it elicits. Cells were staged in the cell cycle using EdU content in addition to DNA content. (D and E) Bar graphs comparing the mean levels of primary damage in the form of CPDs in control and UV-treated cells and the response in the form of γH2AX. Mean levels of CPDs increase with the increase in DNA content through the cell cycle. Moreover, the S-phase peak observed for γH2AX is missing for the levels of primary damage in the form of CPDs. Cells were irradiated with 10 J/m2 and left to recover for 60 min. Control cells were fixed right after EdU labeling. Error bars are standard errors of the mean. Asterisks mark significant differences with P < 0.01 (t test). Scale bars: 10 μm.
We observed that UV irradiation gives rise to three distinct populations of cells corresponding to G1, S, and G2/M phases on the basis of the levels of CPDs and γH2AX, as reflected in the single cell scatterplots (Fig. 2B and C). Moreover, γH2AX levels are markedly higher in the S-phase cells of the UV-treated population, while such distinction is not observed for the CPD levels (Fig. 2C). In fact, mean levels of CPDs in a UV-treated population increase with the increase in DNA content from G1 to S to G2 phases of the cell cycle, while mean γH2AX levels show a clear peak in S-phase cells (Fig. 2D and E). Furthermore, γH2AX foci do not correspond to regions of high CPD intensity (Fig. 2A).
The above observations on UV-induced primary DNA damage clearly show that it is not the primary DNA damage itself but the response elicited which is cell cycle dependent with a clear peak in the S phase. In particular, UV-induced γH2AX levels in the S phase do not correspond to primary DNA lesions. However, could it represent the DSBs formed from secondary repair intermediates like SSBs during replication? To address this, we investigated the colocalization of the γH2AX foci with a standard marker of DSBs.
UV-induced γH2AX does not also report on the extent of secondary DNA damage in S-phase cells.
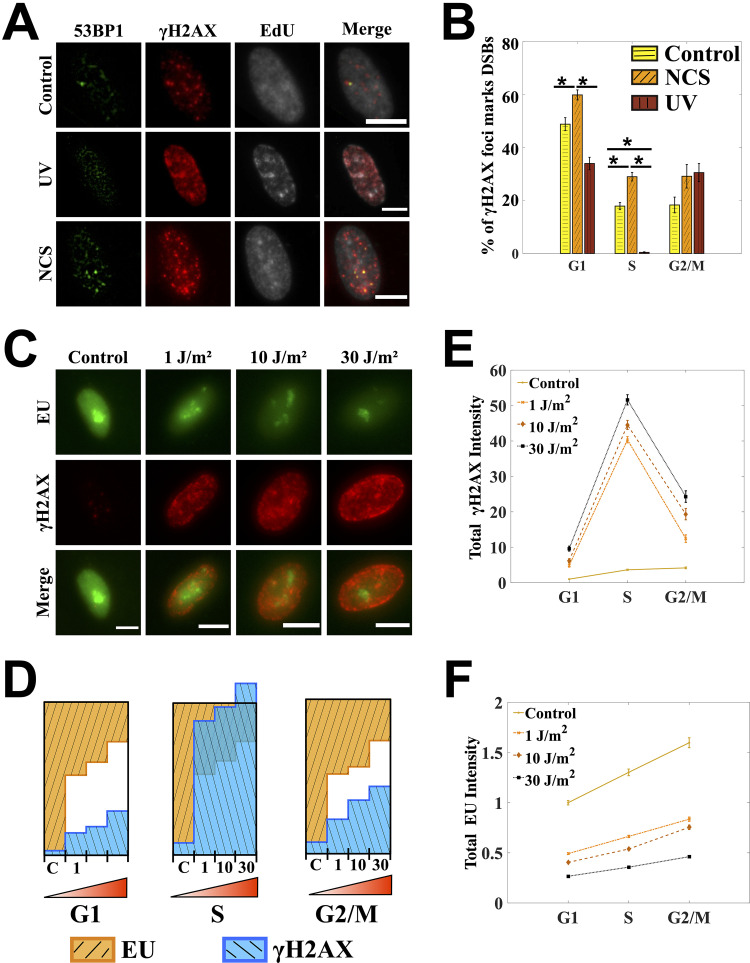
UV-induced DNA adducts can cause replication fork stalling and are repaired by nucleotide excision repair that results in single-strand breaks (SSBs) as repair intermediates. These repair intermediates, if unrepaired, can lead to deleterious DSBs upon DNA replication (13). The protein 53BP1 is a key component of double-strand break repair pathways and, like γH2AX, is known to accumulate at the sites of DSBs within a few minutes of damage (27, 28). It forms distinct foci at the sites of DSBs, where it colocalizes with γH2AX and has been used previously to count the number of DSBs in the cells (17, 29). We used a similar metric whereby if there was more than 50% colocalization between γH2AX and 53BP1 foci at a site, then the site was considered a DSB site (Fig. S2). We used this metric to calculate the percentage of γH2AX foci that corresponded to DSBs in the S-phase cells post-UV.
We found that for the cells treated with UV, the percentage of γH2AX foci marking DSBs was remarkably small in the S-phase cells, unlike the large percentage observed in the cells treated with NCS, a direct double-strand-break-causing agent (Fig. 3A and B). The observation conforms with a previous study showing that only a minority of γH2AX foci after UV irradiation contained double-strand breaks (14). With DSBs out of consideration, we wondered what other kinds of DNA damage could be represented by the UV-induced γH2AX in the S phase of the cell cycle. Rather than going one by one through the possible kinds of secondary damage that UV can cause, we used a proxy that can report fairly accurately on total levels of DNA damage: the comparative levels of total transcriptional activity inside a cell (30, 31).
FIG 3.
UV-induced γH2AX does not also report on the extent of secondary DNA damage in the S-phase cells. (A) Percentages of γH2AX foci marking DNA double-strand breaks (DSBs) were scored by quantifying colocalization between γH2AX and 53BP1 foci using a colocalization metric defined in Fig. S2 in the supplemental material. The cells shown here are in the S phase as marked by the presence of EdU. Images are contrast adjusted to show the foci clearly. (B) Percentage of γH2AX foci which are DSBs for control, UV-, and NCS-treated cells. For UV treatment, only a small fraction of γH2AX foci in the S-phase cells corresponds to DSBs. (C) Global transcription is measured in UV-treated cells by quantifying 5-ethynyl-uridine incorporation. Images were processed with the same contrast adjustments to aid the comparison. (D) Bar graphs for γH2AX and EU are normalized across the four dosages. Resultant bar graphs for EU are inverted over those for γH2AX such that the total height of the two bars corresponding to control population in every cell cycle phase is 1. A clear gap is observed between γH2AX and EU bars for G1 and G2/M phases after UV treatment. The γH2AX bars in the S phase do not just overlap with the EU bars but go past the unit box, showing a disproportionate increase in γH2AX. (E) Mean levels of γH2AX increase with the increase in DNA damage as achieved by increasing the UV dosage in all phases of the cell cycle. For all the dosages, there is a sharp γH2AX peak in the S phase. (F) Increase in DNA damage always leads to decrease in EU as seen across the population treated with different dosages of UV. But within a population there is no dip in EU levels corresponding to the γH2AX peak, showing that γH2AX in the S phase after UV does not reflect on the total extent of DNA damage in those cells. Cells were treated with 10 J/m2 and 1.6 μg/ml NCS for 2 min. Cells were left to recover for 30 min in the presence of EU. Line graphs are showing relative levels with respect to the mean value of G1 phase in control cells. Error bars are standard errors of the mean. Asterisks mark significant differences with P < 0.01 (t test). Scale bars: 10 μm.
UV-induced DNA damage repair depends on the transcriptional status of the damaged DNA: an actively transcribed strand could be repaired via transcription-coupled NER while the rest of the DNA could be repaired via global genomic NER (32). This implies that transcription is a direct target of damage repaired by NER processes and that the total transcriptional activity inside a cell can potentially report on the extent of DNA damage inside a cell. Moreover, inhibition of global transcription is one of the first few responses to UV irradiation (33, 34). In fact, there have been studies reporting inhibition of global transcription as a proxy for DNA damage, especially from UV exposure (30, 31). We used a similar measure and quantified the levels of global transcription by labeling the nascent RNA with 5-ethynyl-uridine (EU) that can later be detected via click chemistry (35).
We observed that an increase in DNA damage by increasing the UV dosage always led to a decrease in global transcription, irrespective of the cell cycle phase (Fig. 3F). In fact the fold decrease in global transcription across the population was almost the same in all the three phases of the cell cycle (see the bar graphs for EU in Fig. 3D). But unlike the G1 and G2/M phases of the cell cycle, the increase in γH2AX levels in the S-phase cells was disproportionately high and was not reflected in the extent of transcriptional inhibition across the population (Fig. 3D). Despite having the highest levels of γH2AX within a population, the S-phase cells did not show a corresponding dip in the global transcription, as one would expect if γH2AX levels in the S phase were indeed reporting on the extent of DNA damage in those cells (Fig. 3E and F).
Taken together, the above observations suggest that all the γH2AX in the S-phase cells after UV irradiation cannot be due to DNA damage. To further investigate the nature of γH2AX in the S-phase cells after UV irradiation, we looked at its localization with respect to replication forks in those cells.
γH2AX colocalizes with the sites of active replication upon UV irradiation.
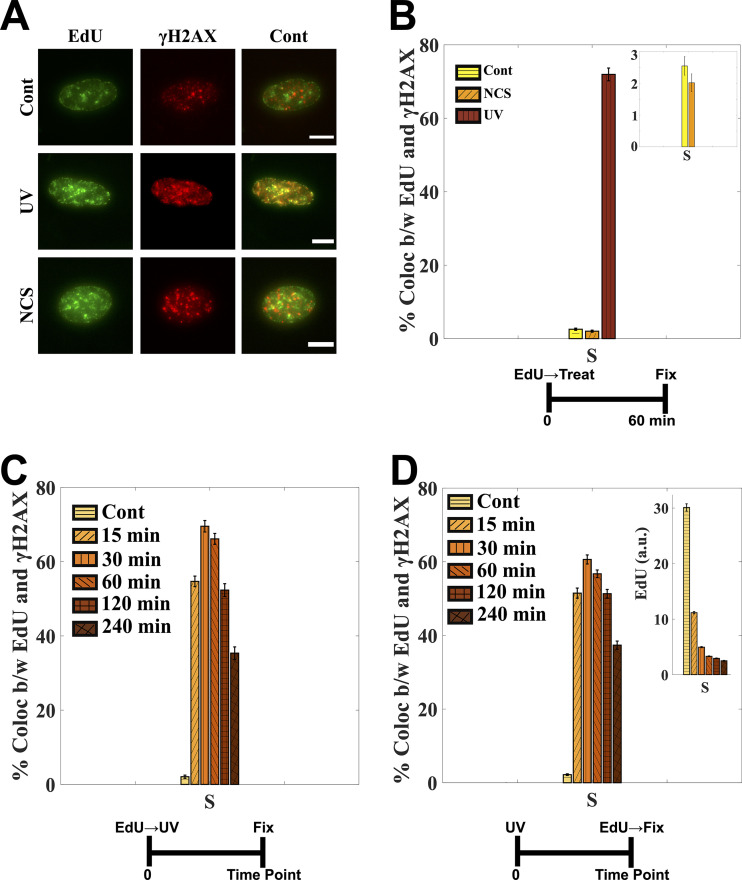
The spatial resolution built into the imaging-based cell cycle staging helped us look at the localization of γH2AX in the nucleus after genotoxic treatments. We observed that γH2AX upon UV irradiation colocalized with the sites of active replication, unlike for NCS treatment, where it colocalized specifically to the sites of DSBs. The percentage of γH2AX foci colocalizing with the sites of active replication, as marked by EdU incorporation, was more than 60% for UV-irradiated cells while less than 3% for control and NCS-treated cells (Fig. 4A and B). This accumulation of γH2AX at the sites of replication began within a few minutes of exposure and peaked at around 30 min post-UV irradiation (Fig. 4C). Although the colocalization between EdU and γH2AX foci in UV-treated populations was largely intact even at higher resolution (Fig. S3), we should emphasize that this might not mean a direct molecular interaction between γH2AX and replication sites but rather might mean that γH2AX is induced in stretches of chromatin proximate to replication forks.
FIG 4.
γH2AX colocalizes with the sites of active replication upon UV irradiation. (A) Cells were labeled with EdU prior to UV or NCS treatment. Images were contrast adjusted to visualize the foci clearly. (B) More than 70% of EdU foci colocalize with γH2AX foci. The colocalization is less than 3% for control and NCS-treated cells (inset). See Fig. S3 in the supplemental material for high-resolution images visualizing EdU and γH2AX foci colocalization. (C) γH2AX starts to accumulate at sites of active replication within a few minutes of UV exposure, as reflected in the percentage of EdU foci colocalizing with γH2AX foci. The colocalization peaks at around 30 min, after which it starts to go down. (D) Cells were labeled with EdU post-UV, right before fixation. As seen from more-or-less intact colocalization between EdU and γH2AX for the same time points as in panel C. γH2AX accumulation does not inhibit EdU incorporation altogether but slows it down (inset). The control populations in all the experiments were fixed right after EdU labeling. See also Fig. S4. Cells were irradiated with 10 J/m2. Error bars are standard errors of the mean. Scale bars: 10 μm.
To investigate how this localization of γH2AX at the replication forks affects the replication itself, we labeled the cells with EdU post-UV but immediately before fixation—unlike the previous experiments where the cells were labeled with EdU right before UV to mark the S-phase cells and replication forks at the time of UV irradiation. We observed that EdU was still incorporated at the sites of γH2AX albeit at lower rates, as inferred from the intact colocalization between EdU and γH2AX and the lower levels of EdU in those cells (Fig. 4D and Fig. S4). This slowing down of replication could be because of translesion synthesis of DNA (TLS) on leading and lagging strands (36), or it could also hint at ongoing NER at those sites. To evaluate these possibilities, we stained the cells for a subunit of replication protein A (RPA) called RPA2 (Fig. S5A). RPA binds with great affinity to single-stranded DNA (ssDNA) in eukaryotic cells during DNA replication and repair. The association of RPA with ssDNA during DNA damage caused by bulky lesions stabilizes the ssDNA, recruits DDR factors to the sites of damage, and activates the S-phase checkpoint (36–38). As expected, we observed that the colocalization of RPA2 with γH2AX increased in the S-phase cells of UV-treated population (Fig. S5A and B). We also observed a significant increase in RPA2 levels in the S-phase cells of the UV-treated population (Fig. S5C) in line with previous reports (37, 38). However, the colocalization between RPA2 and EdU foci remained intact between control and UV-treated populations (Fig. S5D). The above observations suggest that the repair of UV-induced DNA lesions might lead to increased levels of ssDNA as repair intermediates and that these might be responsible for slowing down of replication forks, as also reported previously (39, 40).
We next enquired about the implications of the above observations on the S-phase cells in UV-treated populations. In particular, we asked if the S-phase cells completed the replication of their genomes. We show below that most of the S-phase cells at the time of UV irradiation did complete the replication of their genomes and were then arrested in the G2 phase of the cell cycle.
Secondary DSBs induced by UV at later time points do not peak in the S phase.
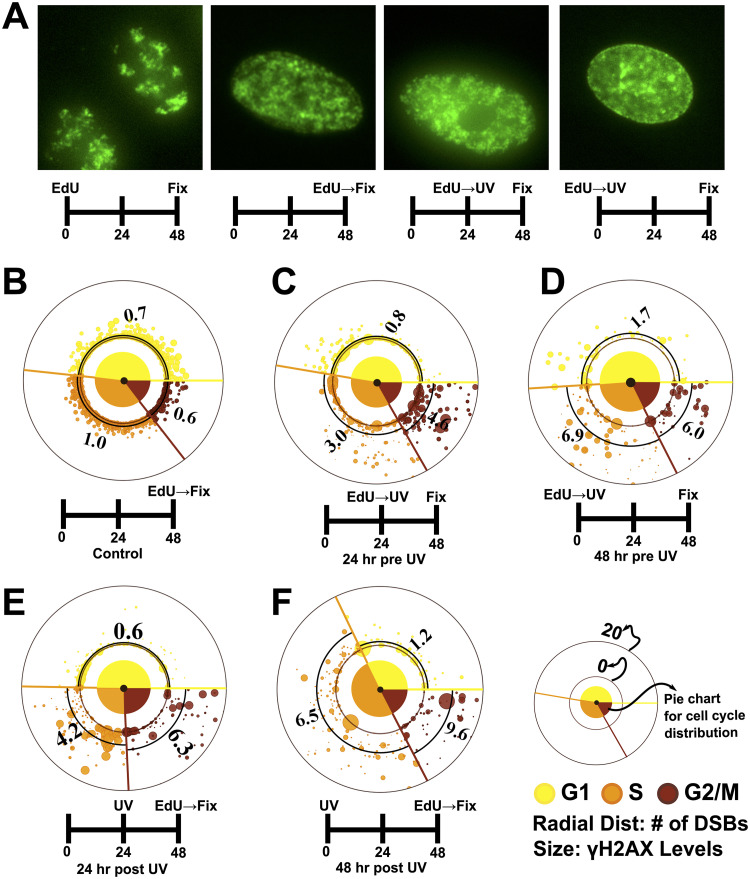
To investigate how the elevated levels of γH2AX affect the S-phase cells at later time points, we labeled the cells with EdU prior to UV and let them recover for 24 to 48 h. We observed that most of the S-phase cells at the time of UV irradiation completed the replication of their DNA and stayed arrested in G2 phase even after 48 h, as reflected by their respective EdU staining patterns and cell cycle distributions (Fig. 5A to D and Fig. S6). This is also evident from the observation that the doubling time of 16.3 h obtained for A549 cells—when calculated with the assumption that there is no cell death or division after UV irradiation within the time periods (24 to 48 h) of observation—agree well with those reported previously (∼18 h) under similar growth conditions (41, 42) (Table S1 and the accompanying calculations in the supplemental material). These observations suggest that most of the EdU-positive cells observed at 24 to 48 h in the populations labeled with EdU right before UV irradiation (24/48 h pre-UV) are exactly those cells which were in the S phase at the time of UV irradiation, as most of the cells in the population failed to divide or undergo mitosis. Also, the DNA contents of most of the EdU-positive cells from such populations were close to the levels of G2-phase cells (Fig. S6C and D), which again suggests that these cells have completed the replication of their genomes. We next quantified the number of DSBs in such populations (24/48 h pre-UV) and observed that the S-phase cells at the time of UV irradiation did not develop as many DSBs at 24 to 48 h post-UV (Fig. 5C and D) as expected from the sharp S-phase peak in γH2AX levels observed within just a few hours of UV exposure (Fig. 1D). Although the cells in G1 phase (in 24/48 h pre-UV) at the time of UV irradiation showed very few DSBs at later time points (Fig. 5C and D), those cells which had exited the G1 arrest by then and had started replicating their DNA (24/48 h post-UV) developed as many DSBs as the ones which had been in the S phase at the time of UV irradiation (Fig. 5E and F, Fig. S7, and Fig. S8B and C). In fact, all three phases seemed to have similar numbers of preapoptotic cells, as apparent from the number of cells showing very bright homogeneous pan-nuclear γH2AX staining with no DSBs (Fig. 5C to F and Fig. S8A). Homogeneous, pan-nuclear γH2AX staining is considered a marker of preapoptotic cells, as reported previously (3, 14, 43).
FIG 5.
Secondary DSBs induced by UV at later time points do not peak in the S phase. (A) EdU staining pattern for control cells post-48 h is strikingly different from that of UV-treated cells, which is very similar to that observed for cells freshly labeled with EdU, showing that UV-treated cells have not gone through a mitosis and are arrested (see also Fig. S6). Images are contrast adjusted to aid the visualization of EdU staining patterns. (B to F) Torus plots comparing the number of DSBs and cell cycle distributions under various treatments. DSBs were quantified as described for Fig. 3. The numbers on the plots are average DSBs in the respective cell cycle phases, as also marked by thick black arcs. The large spots close to the origin represent preapoptotic cells (high levels of γH2AX with no DSBs [see Fig. S8A]). (C and D) The populations labeled with EdU right before UV irradiation have cell cycle distributions similar to that of control cells (B) after 24 and 48 h, indicating that most of the cells have been arrested and have not gone through a mitosis even at 48 h post-UV. Also, unlike the S-phase peak observed in γH2AX levels measured a few hours post-UV treatment, the numbers of DSBs 24 to 48 h post-UV do not peak in the cells which were in the S phase at the time of UV irradiation. (E and F) Cells labeled with EdU post-UV, right before fixation. The S-phase cells here are those cells which have now exited the G1 arrest 24 to 48 h post-UV irradiation. These S-phase cells, on the average, show similar numbers of DSBs as the cells which were in the S phase at the time of UV (C and D), showing that UV causes similar levels of damage to all the cells in a population irrespective of their cell cycle phases (see also Fig. S7 and S8). At least 250 cells were analyzed for all the experiments. Cells were irradiated with 10 J/m2 UV.
Taken together, the above observations suggest that the UV irradiation does not selectively cause higher levels of damage in the S-phase cells; rather, it has equitable effects on all the cells in all the phases of the cell cycle.
DISCUSSION
UV radiation is a common environmental mutagen, making it important to study UV-induced DNA damage and the responses elicited. UV is known to induce replication stress in dividing cells by actuating the dimerization of adjacent pyrimidines that leads to distortions in DNA structures which, if unrepaired, can eventually lead to DSBs, the most lethal of all DNA lesions (44). Most kinds of DNA lesions lead to the phosphorylation of a histone variant H2AX at serine 139 by the master transducers of DNA signals ATM and ATR—which is why γH2AX is considered a definitive marker of DNA damage. UV exposure, in particular, leads to a sharp peak of γH2AX in S-phase cells, which are actively replicating their DNA. This has led to the idea that S-phase cells in a population are most vulnerable to genotoxic insults. In this report, we systematically study the hypothesis by investigating the nature of γH2AX in the S-phase cells after DNA damage.
We first show that not all genotoxic treatments lead to a γH2AX peak in the S phase of the cell cycle; UV treatment causes γH2AX to peak in the S phase but NCS, a DSB-causing agent, does not. We also show that the UV-induced S-phase peak in γH2AX levels is not reflected in the levels of primary (in terms of CPDs) or secondary DNA damage (in terms of DSBs) caused by UV. This demonstrates that it is the damage response, and not DNA damage itself, that peaks in the S phase post-UV irradiation. We also find that the increase in γH2AX after UV irradiation in the S phase within a population does not square with the decrease in global transcription—a known measure for the extent of DNA damage, especially of that induced by UV exposure, inside a cell (31). We further observed that γH2AX colocalized with the sites of active replication and that those sites continued incorporating EdU even 4 h past UV irradiation, although at lower speeds. Furthermore, the enhanced colocalization of RPA and γH2AX in a UV-treated population possibly hints at underlying mechanisms with which cells deal with stalled replication forks from bulky DNA lesions, via either translesion synthesis or repriming events downstream of those lesions in both leading and lagging strands of DNA (39, 40). Such events are reported to produce large stretches of ssDNA at replication forks which initiate DNA damage signaling by recruiting DDR proteins (39, 40). These observations suggest that γH2AX in the S-phase cells post-UV exposure mark damage response rather than the extent of total damage in those cells. The inference is further strengthened by the observation that, unlike the sharp S-phase peak observed in γH2AX levels measured a few hours post-UV treatment, the numbers of DSBs induced at 24- to 48-h time points post-UV increase with the increase in DNA content as cells move from G1 to S to G2 phases of the cell cycle, similar to what is observed in cells treated with NCS. Furthermore, the G1-phase cells which exit the G1 arrest and start replicating their DNA (as marked by EdU) at 24- to 48-h time points develop as many DSBs as those cells which were in the S phase at the time of UV irradiation. Taken together, our results suggest two things: (i) vulnerability to the exogenous sources of DNA damage depends more on the DNA content than on the cell cycle stages, and (ii) the γH2AX foci in the S phase after UV irradiation are not the sites of DNA damage—they colocalize with the active replication forks at the time of UV irradiation. This also suggests that the sites of active replication post-UV irradiation do not correspond to primary DNA lesions, which is in congruence with a recent study on the yeast Saccharomyces cerevisiae system showing that UV-induced DNA lesions are spatially and temporally separated from the active replication forks (45).
Although these results have been shown just in the context of UV-induced replication stress, they might also hold true for replication stress induced by other genotoxic agents, such as camptothecin, 4NQO, or cisplatin, where similar observations regarding γH2AX have been reported (8–10, 46). This poses new questions as to the roles of γH2AX in the maintenance of genome stability, which hitherto have been confined to the mediation of DNA repair inside a cell. It also highlights the need to look at chromatin modifications such as γH2AX with the broader view involving the gamut of processes that chromatin modifications control. Although previous experiments with H2AX knockout cells have shown that γH2AX does not affect the viability of cells upon UV irradiation (47), in the future it will be interesting to investigate the possible roles of γH2AX in maintaining the stability of replication sites after UV irradiation. It would shed light on the possible roles and importance of γH2AX in fork stability in the mammalian system analogous to that seen recently in the yeast Schizosaccharomyces pombe (48).
MATERIALS AND METHODS
Cell culture and treatments.
A549 human cells were used for all the experiments. Cells were grown in Dulbecco's modified Eagle medium (DMEM)–F-12 (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco) without antibiotic. Cells were tested negative for any bacterial contamination, including mycoplasma. Cells were passaged every ∼4 days and were allowed to grow for at least 24 h before starting any experiments.
Cells were labeled with 10 μM 5-ethynyl-2’-uridine (EdU) for 15 min before or after UV as appropriate, to mark true S-phase cells.
For UV treatment, an Analytik Jena UVP cross-linker CL-1000 was used. The instrument was set to the appropriate dosage in joules per square meter before irradiation. The cells in plates were washed twice with phosphate-buffered saline (PBS) before irradiating them with 254 nm UV light. The irradiation was done in the absence of any liquid medium in the plates.
Neocarzinostatin was used at a 1.6 μg/ml concentration. The cells were treated with the chemical-containing medium for 2 min, after which the cells were washed twice with PBS and fresh medium was added to the plates. Unless mentioned otherwise, control populations were always fixed right after EdU labeling. Cells were left to recover after all treatments for 60 min unless mentioned otherwise.
For all the experiments, including long time point (24 and 48 h) experiments, the cells were plated at the same time and treated exactly the same except for their respective experimental conditions.
Immunofluorescence and click chemistry.
Cells were fixed with 4% paraformaldehyde (PFA) in PBS for 15 min followed by permeabilization with 0.3% Triton X-100 in PBS for 10 min. For immunofluorescence (IF) combined with click chemistry, permeabilization was followed by click chemistry detection of EU or EdU followed by the usual immunofluorescence, as described below.
For immunofluorescence-based detection of protein levels, cells were blocked for nonspecific binding by using Block, a 5% solution of bovine serum albumin (BSA; Sigma A2153) in PBS, for 30 min at room temperature (RT) followed by labeling with the primary antibodies in Block for 60 min at RT. The cells were then washed twice with PBS and labeled with the secondary antibodies in Block for 60 min at RT.
The protocol for click chemistry was adopted from a previous method (49). In brief, cells were equilibrated with Tris-buffered saline (TBS) buffer before the addition of click reaction cocktail (CRC) to label the cells with azide dyes. A 1-ml solution of CRC was prepared as follows: (i) 730 μl of 100 mM Tris (pH ∼8.5) plus (ii) 15 μl of 100 mM CuSO4 plus 41 μl of 50 mM THPTA (tris-hydroxypropyltriazolylmethylamine) (Sigma 762342) ligand plus 1 μl of 6 mM azide dye plus (iii) 100 μl of 10% glucose in 10% glycerol plus 4 μl of >20,000 U/ml stock of catalase (Sigma C3515) plus 8 μl of 500 U/ml glucose oxidase (Sigma G2133) plus (iv) 100 μl of 1 M sodium ascorbate.
The order of reagent addition was important. Reagents ii and iii were prepared separately in two 1.5-ml tubes. The tube containing THPTA, CuSO4, and azide dye was left to sit for at least 5 min before Tris was added to the tube, after which the contents of the other tube were added to the tube containing THPTA. Finally, sodium ascorbate was added.
Once the CRC was prepared, the TBS in plates was replaced with CRC. The labeling was performed for 15 min, after which the cells were washed with PBS and then with 100 mM EDTA in PBS for 5 min to remove excess Cu ions from the cells, after which the cells were treated for the usual immunofluorescence. Last, the cells were labeled with 1 μg/ml Hoechst 33342 in PBS for 10 min.
For measuring global transcription, cells were labeled with 5-ethynyl-uridine (EU) immediately after UV irradiation for 30 min. EU gets incorporated into actively transcribing RNAs and marks the translational activity inside the cells that can later be detected using click chemistry. EU (CLK-N002-10) and EdU (CLK-N001-100) were procured from Jena Bioscience. The stock solutions were made in PBS.
For CPD staining, cells were denatured right after permeabilization with 2 N HCl at 37°C for 15 min. Cells were then washed once with PBS and then neutralization was performed with 0.2 M Borax buffer with pH ∼8.5 for 5 min at RT. Click chemistry and IF followed as described above. DNA denaturation required longer Hoechst staining achieved by adding Hoechst directly to the secondary antibody solutions.
Antibodies used.
Antibodies used were as follows: anti-γH2AX from Merck (05-636) and Cell Signaling Technologies (9718) at 1:500 dilution, anti-53BP1 from Abcam (ab175933) at 1:1,000 dilution, anti-RPA2/32 from Abcam (ab2175) at 1:500 dilution, and anti-pATM from Abcam (ab36810) at 1:250 dilution. Anti-CPD antibodies from Cosmo Bio (TDM-2 clone, CAC-NM-DND-001) were used at a 1:250 dilution. Highly cross-adsorbed Alexa Fluor-labeled secondary antibodies raised in goat from Invitrogen were always used at a 1:500 dilution.
Fluorophores.
The fluorophores were kept constant across the experiments. Click chemistry was always performed with azide Alexa Fluor 488. Mouse antibodies (which included anti-γH2AX) were always detected with Alexa Fluor 546 and rabbit antibodies (which included anti-53BP1) with Alexa Fluor 594. DNA was stained with a minor groove binder Hoechst 33342.
Microscopy.
Imaging was always performed with freshly prepared 0.1% solution of p-phenylene diamine (PPD) in PBS as a mounting medium to reduce photobleaching of Hoechst 33342.
Images were acquired at 14-bit resolution using the fully automated Olympus IX83 microscope on a Retiga 6000 (QImaging) camera. Images were taken using a 40×, 0.75-numerical-aperture (NA) air objective. Four planes, 2 μm apart, were taken for every field. For higher-resolution images, 25 planes at 300 nm apart were taken for every field using a 60×, 1.42-NA oil objective. Filter sets from Olympus and Chroma Technology Corp. (49309 ET, Orange no. 2 FISH and 49310 ET, Red no. 2 FISH) were used to prevent any bleed through among the fluorophores used.
Data and image analyses.
Cells were first staged in the cell cycle based on their DNA content using an automated Matlab routine developed earlier (8). Briefly, the images from all the channels were first average projected and the resulting images were used for further analyses. The average-projected image from the DAPI channel was used to segment out nuclei. The mask thus obtained was used to quantify DNA content, EdU content, and protein contents. Once staged, the cells were later redistributed cross the cell cycle stages based on their EdU intensities to identify true S-phase cells. For experiments where it was not possible to combine EdU-based staging of cells, cells were staged based just on their DNA contents (Fig. 3C to F). All the analyses included at least 1,000 cells in total for all the three cell cycle phases, unless specified otherwise.
The focus-counting module was adopted from our previous study (50). Briefly, the maximum-projected images were convoluted with a Laplacian of Gaussian (LOG) filter with parameters set to enhance the foci of size typically observed in our experiments. The resulting image was then used to obtain a binary mask using an automated thresholding algorithm. The binary mask thus obtained was used to count the foci as well as to quantify colocalization between images from two different channels.
Deconvolution was performed using ImageJ (National Institutes of Health). Synthetic 3D center-aligned PSF images of size 255 × 255 × 25 (25 planes, each of 255 by 255 pixels) were generated using the Richards-Wolf algorithm available on PSF Generator plug-in from www.epfl.ch with appropriate parameters for wavelengths and refractive indices. The resultant PSF image was used to deconvolve the experimental image stack employing Richardson-Lucy algorithm using DeconvolutionLab2 plug-in from www.epfl.ch with iterations set to 500.
Statistically significant differences were identified using a t test with unequal variance. The analyses were performed in Matlab using the function ttest2 with the parameter “Vartype” set to “unequal.”
All images and data were analyzed using Matlab. For graphs, Matlab and Python 3 were used. All codes and programs used in the study are available upon request.
Supplementary Material
ACKNOWLEDGMENTS
A.M. and S.D. thank Tamal Das for access to the UV cross-linker. We also thank the two anonymous reviewers for their constructive feedback that helped us convey our results more effectively.
A.M., S.D., and S.R. conceived and designed the experiments. S.D. performed and analyzed the experiments. S.D., S.R., and A.M. interpreted the data. S.D. and A.M. wrote the manuscript. A.M. monitored the project and associated grants.
We declare no conflicts of interest.
This project was funded by intramural funds at TIFR Hyderabad from the Department of Atomic Energy (DAE), and partially by a DST SERB Early Career Research Award (ECR/2016/000907) to A.M. S.R. was supported by a DST SERB National Post-Doctoral Fellowship (N-PDF) scheme (PDF/2016/002781).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Peterson CL, Laniel M-A. 2004. Histones and histone modifications. Curr Biol 14:R546–51. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. 2007. Chromatin modifications and their function. Cell 128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Meyer B, Voss KO, Tobias F, Jakob B, Durante M, Taucher-Scholz G. 2013. Clustered DNA damage induces pan-nuclear H2AX phosphorylation mediated by ATM and DNA-PK. Nucleic Acids Res 41:6109–6118. doi: 10.1093/nar/gkt304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L, Dai W, Lu L. 2014. Osmotic stress-induced phosphorylation of H2AX by polo-like kinase 3 affects cell cycle progression in human corneal epithelial cells. J Biol Chem 289:29827–29835. doi: 10.1074/jbc.M114.597161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaneko H, Igarashi K, Kataoka K, Miura M. 2005. Heat shock induces phosphorylation of histone H2AX in mammalian cells. Biochem Biophys Res Commun 328:1101–1106. doi: 10.1016/j.bbrc.2005.01.073. [DOI] [PubMed] [Google Scholar]
- 6.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 7.Maréchal A, Zou L. 2013. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol 5:a012716. doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhuppar S, Mazumder A. 2018. Measuring cell cycle-dependent DNA damage responses and p53 regulation on a cell-by-cell basis from image analysis. Cell Cycle 17:1358–1371. doi: 10.1080/15384101.2018.1482136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prendergast ÁM, Cruet-Hennequart S, Shaw G, Barry FP, Carty MP. 2011. Activation of DNA damage response pathways in human mesenchymal stem cells exposed to cisplatin or γ-irradiation. Cell Cycle 10:3768–3777. doi: 10.4161/cc.10.21.17972. [DOI] [PubMed] [Google Scholar]
- 10.Huang X, Traganos F, Darzynkiewicz Z. 2003. DNA damage induced by DNA topoisomerase I- and topoisomerase II- inhibitors detected by histone H2AXphosphorylation in relation to the cell cycle phase and apoptosis. Cell Cycle 2:613–618. doi: 10.4161/cc.2.6.565. [DOI] [PubMed] [Google Scholar]
- 11.Zhao H, Traganos F, Darzynkiewicz Z. 2010. Kinetics of the UV-induced DNA damage response in relation to cell cycle phase. Correlation with DNA replication. Cytometry A 77:285–293. doi: 10.1002/cyto.a.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartek J, Lukas C, Lukas J. 2004. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol 5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 13.Kuzminov A, Dianov GL. 2001. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc Natl Acad Sci U S A 98:8241–8246. doi: 10.1073/pnas.131009198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Feraudy S, Revet I, Bezrookove V, Feeney L, Cleaver JE. 2010. A minority of foci or pan-nuclear apoptotic staining of gammaH2AX in the S phase after UV damage contain DNA double-strand breaks. Proc Natl Acad Sci U S A 107:6870–6875. doi: 10.1073/pnas.1002175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Probst AV, Dunleavy E, Almouzni G. 2009. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol 10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- 16.Bou Kheir T, Lund AH. 2010. Epigenetic dynamics across the cell cycle. Essays Biochem 48:107–120. doi: 10.1042/bse0480107. [DOI] [PubMed] [Google Scholar]
- 17.Karanam K, Kafri R, Loewer A, Lahav G. 2012. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol Cell 47:320–329. doi: 10.1016/j.molcel.2012.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenburger T. 2006. DNA repair and mutagenesis, second edition. ASM Press, Washington, DC. [Google Scholar]
- 19.Hoeijmakers JHJ. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 20.Salic A, Mitchison TJ. 2008. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A 105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stiff T, Walker SA, Cerosaletti K, Goodarzi AA, Petermann E, Concannon P, O'Driscoll M, Jeggo PA. 2006. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J 25:5775–5782. doi: 10.1038/sj.emboj.7601446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernández L, Terradas M, Martín M, Tusell L, Genescà A. 2013. Highly sensitive automated method for DNA damage assessment: gamma-H2AX foci counting and cell cycle sorting. Int J Mol Sci 14:15810–15826. doi: 10.3390/ijms140815810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T. 2007. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol 9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aten JA, Stap J, Krawczyk PM, van Oven CH, Hoebe RA, Essers J, Kanaar R. 2004. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science 303:92–95. doi: 10.1126/science.1088845. [DOI] [PubMed] [Google Scholar]
- 25.Atsumi Y, Minakawa Y, Ono M, Dobashi S, Shinohe K, Shinohara A, Takeda S, Takagi M, Takamatsu N, Nakagama H, Teraoka H, Yoshioka K. i. 2015. ATM and SIRT6/SNF2H mediate transient H2AX stabilization when DSBs form by blocking HUWE1 to allow efficient γH2AX foci formation. Cell Rep 13:2728–2740. doi: 10.1016/j.celrep.2015.11.054. [DOI] [PubMed] [Google Scholar]
- 26.Yokoyama H, Mizutani R. 2014. Structural biology of DNA (6–4) photoproducts formed by ultraviolet radiation and interactions with their binding proteins. Int J Mol Sci 15:20321–20338. doi: 10.3390/ijms151120321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bekker-Jensen S, Lukas C, Melander F, Bartek J, Lukas J. 2005. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J Cell Biol 170:201–211. doi: 10.1083/jcb.200503043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson L, Henderson C, Adachi Y. 2001. Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol Cell Biol 21:1719–1729. doi: 10.1128/MCB.21.5.1719-1729.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsouroula K, Furst A, Rogier M, Heyer V, Maglott-Roth A, Ferrand A, Reina-San-Martin B, Soutoglou E. 2016. Temporal and spatial uncoupling of DNA double strand break repair pathways within mammalian heterochromatin. Mol Cell 63:293–305. doi: 10.1016/j.molcel.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Calkins AS, Iglehart JD, Lazaro J-B. 2013. DNA damage-induced inhibition of rRNA synthesis by DNA-PK and PARP-1. Nucleic Acids Res 41:7378–7386. doi: 10.1093/nar/gkt502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia N, Nakazawa Y, Guo C, Shimada M, Sethi M, Takahashi Y, Ueda H, Nagayama Y, Ogi T. 2015. A rapid, comprehensive system for assaying DNA repair activity and cytotoxic effects of DNA-damaging reagents. Nat Protoc 10:12–24. doi: 10.1038/nprot.2014.194. [DOI] [PubMed] [Google Scholar]
- 32.Lans H, Marteijn JA, Schumacher B, Hoeijmakers JHJ, Jansen G, Vermeulen W. 2010. Involvement of global genome repair, transcription coupled repair, and chromatin remodeling in UV DNA damage response changes during development. PLoS Genet 6:e1000941. doi: 10.1371/journal.pgen.1000941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lavigne MD, Konstantopoulos D, Ntakou-Zamplara KZ, Liakos A, Fousteri M. 2017. Global unleashing of transcription elongation waves in response to genotoxic stress restricts somatic mutation rate. Nat Commun 8:1–15. doi: 10.1038/s41467-017-02145-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tufegdžić Vidaković A, Mitter R, Kelly GP, Neumann M, Harreman M, Rodríguez-Martínez M, Herlihy A, Weems JC, Boeing S, Encheva V, Gaul L, Milligan L, Tollervey D, Conaway RC, Conaway JW, Snijders AP, Stewart A, Svejstrup JQ. 2020. Regulation of the RNAPII pool is integral to the DNA damage response. Cell 180:1245–1261.e21. doi: 10.1016/j.cell.2020.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jao CY, Salic A. 2008. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci U S A 105:15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marians KJ. 2018. Lesion bypass and the reactivation of stalled replication forks. Annu Rev Biochem 87:217–238. doi: 10.1146/annurev-biochem-062917-011921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michael WM, Ott R, Fanning E, Newport J. 2000. Activation of the DNA replication checkpoint through RNA synthesis by primase. Science 289:2133–2137. doi: 10.1126/science.289.5487.2133. [DOI] [PubMed] [Google Scholar]
- 38.Zou L, Elledge SJ. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 39.Lopes M, Foiani M, Sogo JM. 2006. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell 21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 40.Kurat CF, Yeeles JTP, Patel H, Early A, Diffley JFX. 2017. Chromatin controls DNA replication origin selection, lagging-strand synthesis, and replication fork rates. Mol Cell 65:117–130. doi: 10.1016/j.molcel.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brower M, Carney DN, Oie HK, Gazdar AF, Minna JD. 1986. Growth of cell lines and clinical specimens of human non-small cell lung cancer in a serum-free defined medium. Cancer Res 46:798–806. [PubMed] [Google Scholar]
- 42.Akram KM, Lomas NJ, Spiteri MA, Forsyth NR. 2013. Club cells inhibit alveolar epithelial wound repair via TRAIL-dependent apoptosis. Eur Respir J 41:683–694. doi: 10.1183/09031936.00213411. [DOI] [PubMed] [Google Scholar]
- 43.Moeglin E, Desplancq D, Conic S, Oulad-Abdelghani M, Stoessel A, Chiper M, Vigneron M, Didier P, Tora L, Weiss E. 2019. Uniform widespread nuclear phosphorylation of histone H2AX is an indicator of lethal DNA replication stress. Cancers (Basel) 11:355. doi: 10.3390/cancers11030355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinha RP, Häder DP. 2002. UV-induced DNA damage and repair: a review. Photochem Photobiol Sci 1:225–236. doi: 10.1039/b201230h. [DOI] [PubMed] [Google Scholar]
- 45.Wong RP, García-Rodríguez N, Zilio N, Hanulová M, Ulrich HD. 2019. Processing of DNA polymerase-blocking lesions during genome replication is spatially and temporally segregated from replication forks. Mol Cell 77:3–16.e4. doi: 10.1016/j.molcel.2019.09.015. [DOI] [PubMed] [Google Scholar]
- 46.Berniak K, Rybak P, Bernas T, Zarębski M, Biela E, Zhao H, Darzynkiewicz Z, Dobrucki JW. 2013. Relationship between DNA damage response, initiated by camptothecin or oxidative stress, and DNA replication, analyzed by quantitative 3D image analysis. Cytometry A 83:913–924. doi: 10.1002/cyto.a.22327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Revet I, Feeney L, Bruguera S, Wilson W, Dong TK, Oh DH, Dankort D, Cleaver JE. 2011. Functional relevance of the histone γH2Ax in the response to DNA damaging agents. Proc Natl Acad Sci U S A 108:8663–8667. doi: 10.1073/pnas.1105866108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mejia-Ramirez E, Limbo O, Langerak P, Russell P. 2015. Critical function of γH2A in S-phase. PLoS Genet 11:e1005517. doi: 10.1371/journal.pgen.1005517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Löschberger A, Niehörster T, Sauer M. 2014. Click chemistry for the conservation of cellular structures and fluorescent proteins: ClickOx. Biotechnol J 9:693–697. doi: 10.1002/biot.201400026. [DOI] [PubMed] [Google Scholar]
- 50.Dhuppar S, Mazumder A. 2020. Investigating cell cycle-dependent gene expression in the context of nuclear architecture at a single allele resolution. J Cell Sci 133:jcs246330. doi: 10.1242/jcs.246330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.