Lysine demethylase 6A (KDM6A), also known as UTX, belongs to the KDM6 family of histone H3 lysine 27 (H3K27) demethylases, which also includes UTY and KDM6B (JMJD3). The KDM6A protein contains six tetratricopeptide repeat (TPR) domains and an enzymatic Jumonji C (JmjC) domain that catalyzes the removal of di- and trimethylation on H3K27. KDM6A physically associates with histone H3 lysine 4 monomethyltransferases MLL3 (KMT2C) and MLL4 (KMT2D). Since its identification as an H3K27 demethylase in 2007, studies have reported KDM6A’s critical roles in cell differentiation, development, and cancer.
KEYWORDS: KDM6A/UTX/H3K27 demethylase/enhancer regulation/gene expression, H3K27 demethylase, KDM6A, UTX, cancer, cell differentiation, development, enhancer regulation, gene expression
ABSTRACT
Lysine demethylase 6A (KDM6A), also known as UTX, belongs to the KDM6 family of histone H3 lysine 27 (H3K27) demethylases, which also includes UTY and KDM6B (JMJD3). The KDM6A protein contains six tetratricopeptide repeat (TPR) domains and an enzymatic Jumonji C (JmjC) domain that catalyzes the removal of di- and trimethylation on H3K27. KDM6A physically associates with histone H3 lysine 4 monomethyltransferases MLL3 (KMT2C) and MLL4 (KMT2D). Since its identification as an H3K27 demethylase in 2007, studies have reported KDM6A’s critical roles in cell differentiation, development, and cancer. KDM6A is important for differentiation of embryonic stem cells and development of various tissues. Mutations of KDM6A cause Kabuki syndrome. KDM6A is frequently mutated in cancers and functions as a tumor suppressor. KDM6A is redundant with UTY and functions largely independently of its demethylase activity. It regulates gene expression, likely through the associated transcription factors and MLL3/4 on enhancers. However, KDM6A enzymatic activity is required in certain cellular contexts. Functional redundancy between H3K27 demethylase activities of KDM6A and KDM6B in vivo has yet to be determined. Further understanding of KDM6A functions and working mechanisms will provide more insights into enhancer regulation and may help generate novel therapeutic approaches to treat KDM6A-related diseases.
INTRODUCTION
Chromatin modifications play a major role in regulating gene expression. The methylation of histone at certain lysine residues is one of the best-studied types of chromatin modification. Methylations catalyzed by site-specific histone methyltransferases correlate with gene activation or repression. In 2004, it was discovered that methylation on histone H3 lysine 4 (H3K4) could be reversed by a histone demethylase called lysine-specific demethylase 1 (LSD1 or KDM1A), suggesting that transcriptional regulation by histone methylation is a dynamic and intricate process (1). In 2007, several studies identified a group of histone H3 lysine 27 (H3K27) demethylases consisting of lysine demethylase 6A (KDM6A or UTX), lysine demethylase 6B (KDM6B or JMJD3), and ubiquitously transcribed tetratricopeptide repeat on chromosome Y (UTY) (2–5). KDM6A is an X-linked protein that, similar to KDM6B, contains a catalytic Jumonji C (JmjC) domain which facilitates the removal of the methyl group on di- and trimethylated H3K27 (H3K27me2/3). While UTY is the Y-linked homolog of KDM6A and shares many structural similarities, it does not exert significant demethylase activity (2, 5).
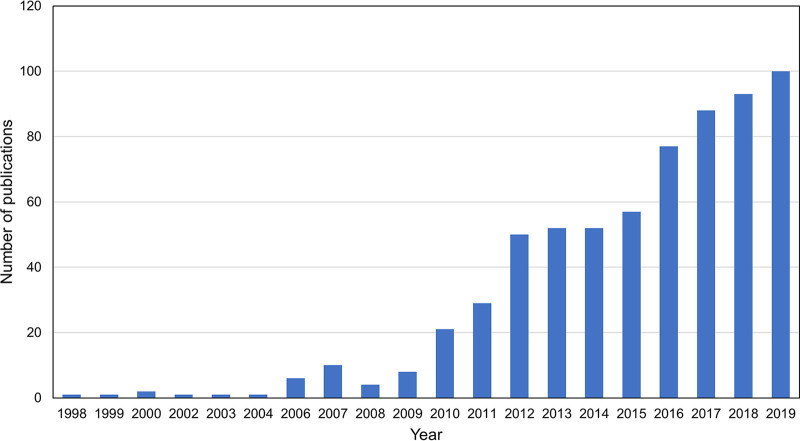
Since its identification as an H3K27 demethylase in 2007, an increasing number of studies are being carried out every year to examine the functions of KDM6A in different biological contexts, with about 100 articles published in 2019 (Fig. 1). Many studies have shown that KDM6A regulates gene expression and enhancer activation independently of its demethylase activity. KDM6A physically and/or functionally interacts with transcription factors (TFs), chromatin-modifying enzymes, including H3K4 methyltransferases MLL3/MLL4 and H3K27 acetyltransferases CBP/p300, and chromatin-remodeling complex SWI/SNF (6–11). KDM6A plays important roles in embryonic stem cell (ESC) differentiation, early embryonic development, and tissue-specific development, including cardiac, mammary, and immune development. KDM6A was also found to be mutated in many types of cancer and to function as a tumor suppressor in mouse cancer models. In this review, we summarize the general features of the KDM6A protein, its interactions with other chromatin regulators, and its roles in enhancer regulation. Current literature on the roles of KDM6A in cell differentiation, embryonic and tissue-specific development, developmental disease Kabuki syndrome (KS), and tumor suppression is discussed. Lastly, we speculate on several future directions to extend the understanding of KDM6A mechanisms and functions.
FIG 1.
Trajectory of publication of KDM6A papers since the identification of the KDM6A gene in 1998. PubMed searches using keywords “kdm6a” and “utx” yielded more than 600 articles published from 1998 to 2019.
THE KDM6A PROTEIN AND ASSOCIATED FACTORS
Basic properties of the KDM6A gene/protein.
The gene encoding KDM6A or UTX is located on the X chromosome in both mice, at XA1.2-1.3, and humans, at Xp11.3. KDM6A escapes X inactivation in both mice and humans (12). In mice, the Kdm6a transcript is 5,918 bp long and contains 29 exons, while the human ortholog is 5,438 bp long and contains 29 exons. In both mice and humans, the KDM6A protein comprises 1,401 amino acids and weighs around 154 kDa, though alternative splicing can produce isoforms with slight variations (Table 1). Sequence alignment between human and mouse KDM6A proteins reveals that they are 97% identical. In developing mouse embryos, KDM6A protein is widely expressed and highly enriched in the heart and neural tissues (8).
TABLE 1.
Basic properties of the KDM6A gene/protein
| Property | Human version | Mouse version |
|---|---|---|
| Gene | KDM6A, UTX | Kdm6a, Utx |
| Chromosome | Xp11.3 | X A1.2-A1.3; X 13.45 cM |
| Ensembl identifier | ENSG00000147050 | ENSMUSG00000037369 |
| Transcript length (bp) | 5,438 | 5,918 |
| No. of exons/no. of introns | 29/28 | 29/28 |
| UniProt identifier | O15550 | O70546 |
| Protein length (amino acids) | 1,401 | 1,401 |
| Protein mol wt (Da) | 154,176.54 | 154,354.83 |
The KDM6A protein contains six tetratricopeptide repeat (TPR) domains at the N terminus and a Jumonji C (JmjC) domain at the C terminus. The catalytic JmjC domain allows KDM6A to demethylate di- or trimethylated lysine specifically at H3K27 without any considerable effect on methylated H3K4, K9, K36, or H4K20 (2, 5). Although KDM6A demethylase activity is not favorable on H3K27me1, it is still capable of demethylating H3K27me1 at high KDM6A dosage (5) or by using histone peptides as substrates (2). Knockdown of KDM6A leads to increased H3K27me3 levels in HeLa cells (3). The 6 TPR domains are dispensable for KDM6A H3K27me2/3 demethylase activity but appear to be important for H3K27me1 demethylation, at least in vitro (5).
KDM6A demethylase activity requires a catalytically active JmjC domain, which has conserved binding sites for cofactors Fe(II) ion and α-ketoglutarate (13). The Fe(II)-binding site is particularly crucial, as point mutations at this binding motif (H1146A and E1148A) can cause a total loss in KDM6A demethylase activity (4, 5). Through hydrogen bonds and hydrophobic interactions, the JmjC domain interacts extensively with residues immediately surrounding the H3K27me2/3 targets, including H3R26, H3A29, H3P30, and H3T32. KDM6A interactions with H3 across multiple regions collectively contribute to its conserved functions in specific binding and demethylation of H3K27me2/3 and not other methylated lysine residues, such as H3K9me3 or H3K36me3 (14).
The JmjC domain-containing KDM6 family.
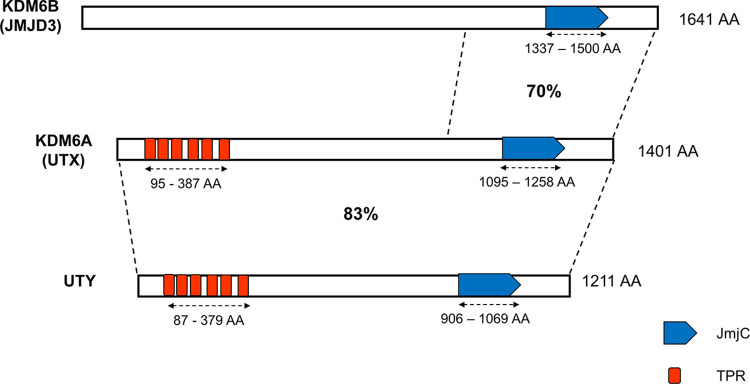
KDM6A is a member of the KDM6 family of H3K27 demethylases, which also contains KDM6B (JMJD3) and UTY (KDM6C) (Fig. 2). The KDM6B protein contains 1,643 and 1,641 amino acids in humans and mice, respectively, and has 70% protein sequence identity with KDM6A in the C-terminal region (5). KDM6B has broad expression during mouse embryonic development, with particularly high expression levels in neural tissues (15). Like KDM6A, KDM6B is capable of H3K27 demethylation. UTY is located on the Y chromosome, contains 1,347 and 1,211 amino acids in humans and mice, respectively, and has 83% protein sequence identity with KDM6A (5). During embryonic development, UTY has expression patterns similar to those of KDM6A, with an enrichment in the neural tube (16). Interestingly, in male embryonic stem cells (ESCs), KDM6A, but not its demethylase activity, positively controls UTY expression. However, in male embryos, UTY expression is not affected by the deletion of Kdm6a (KDM6A knockout [KO]) (7). Unlike KDM6A and KDM6B, UTY shows little to no H3K27 demethylase activity in vivo and in vitro, which can possibly be attributed to some variants of critical amino acids as well as unidentified structural changes of the JmjC domain (2, 5, 16).
FIG 2.
Schematic representation of the mouse KDM6 family of H3K27 demethylases, including KDM6A, KDM6B, and UTY. The catalytic JmjC domains and the TPR domains are indicated. KDM6A shares 83% protein sequence identity with its Y-linked homolog UTY. The C-terminal domain (amino acids [aa] 1174 to 1636) of KDM6B shares 70% protein sequence identity with KDM6A’s C-terminal domain (aa 931 to 1394).
KDM6A is associated with the H3K4 methyltransferase MLL3/4 complex.
KDM6A physically associates with the Set1-like H3K4 methyltransferases and epigenomic writers MLL3 (KMT2C) and MLL4 (KMT2D) in a protein complex that also contains PTIP, PA1, WDR5, RbBP5, ASH2L, DPY30, and NCOA6 and is in the nucleus in mammalian cells (4, 6, 17). KDM6A does not interact with other Set1-like H3K4 methyltransferase complexes, such as Set1A/B or MLL1/2, indicating that KDM6A is a unique subunit of the MLL3/4 complex (6). KDM6A protein stability also depends on the associated MLL3/4 proteins (18). More recent studies found that the TPR domains were necessary to facilitate KDM6A’s association with the MLL3/4 complex. In particular, either the deletion of TPR domains or point mutations (G137V and D336G) in the TPR domains significantly reduce KDM6A’s physical interactions with the MLL3/4 complex (19, 20).
KDM6A REGULATES ENHANCER ACTIVATION LARGELY INDEPENDENTLY OF ITS ENZYMATIC ACTIVITY
KDM6A function is largely independent of its enzymatic activity.
While initial studies identified KDM6A as an H3K27me2/3 demethylase in vitro, follow-up findings have revealed that the roles of KDM6A in differentiation and normal development are independent of its enzymatic activity. In ESCs, KDM6A is required for differentiation along the mesoderm and ectoderm lineages. However, ESCs containing catalytically inactive KDM6A show normal differentiation (7). Meanwhile, catalytically inactive KDM6A is able to rescue the phenotype in KDM6A KO cells (21). In vivo, KDM6A demethylase activity appears to be dispensable for normal development, as homozygous KDM6A enzymatically dead knock-in (KI) female mice were found to be viable and fertile (22). In other organisms, such as Caenorhabditis elegans, KDM6A protein, but not its enzymatic activity, was found to play a critical role in posterior and gonadal development (23). KDM6A function as a tumor suppressor is also independent of its demethylase activity (11, 24).
KDM6A regulates enhancer activation through the MLL3/4 complex.
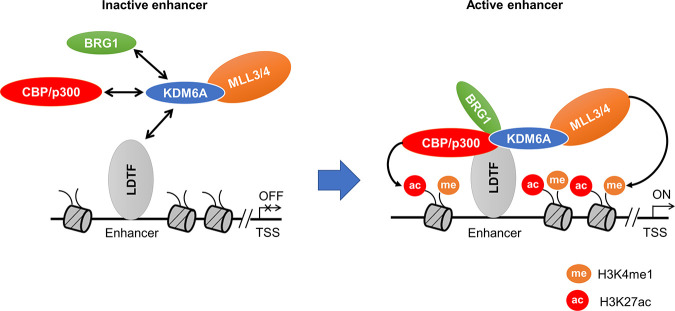
Since KDM6A is a unique subunit of the MLL3/4 complex that is critical for enhancer activation (18, 25), studies have been done to elucidate the potential role of KDM6A in enhancer regulation. It was shown that KDM6A, as a component of the MLL3/4 complex, induced MLL4/p300-regulated enhancer activation and controls the synergy between H3K4me1 and H3K27ac (10). KDM6A recruits the MLL4 complex to target genes by establishing interaction between MLL4 and the retinoic acid receptor (RAR)/retinoid X receptor (RXR) heterodimer. Moreover, KDM6A oversees the cross talk between MLL4 and p300 to synergistically facilitate MLL4-stimulated p300 recruitment and H3K27 acetylation. This establishes a feed-forward regulatory loop involving KDM6A, MLL4, and p300 that gives rise to a cooperative enhancer landscape. On enhancers regulated by this loop, deletion of KDM6A reduces H3K4me1, H3K27ac, and target gene expression but does not affect H3K27me3, suggesting that KDM6A regulates enhancer activation independently of its demethylase activity (Fig. 3) (10). Another study demonstrates that KDM6A loss is associated with repression of some superenhancers (SEs) and activation of others, as marked by H3K27ac and H3K4me1. These effects correlate with the changes in MLL4 occupancy and H3K4me1 signals over SEs in KDM6A KO cells (24).
FIG 3.
The role of KDM6A in facilitating enhancer activation. KDM6A is recruited to the enhancer region by lineage-determining transcription factor (LDTF). KDM6A then facilitates the recruitment of the associated H3K4 methyltransferase MLL3/4 and other chromatin modifiers, including H3K27 acetyltransferase CBP/p300 and the SWI/SNF-dependent chromatin remodeler BRG1. This process promotes histone methylation, acetylation, and chromatin opening, which subsequently activates enhancers and turns on gene transcription.
KDM6A interacts with histone acetyltransferase and chromatin-remodeling complex.
In addition to the MLL3/4 complex, KDM6A has also been reported to physically associate with the H3K27 acetyltransferase CBP and the enzymatic BRG1 (SMARCA4) subunit of the SWI/SNF chromatin-remodeling complex. In Drosophila, KDM6A was found to physically associate with CBP through its TPR domains. KDM6A colocalizes with CBP on many genomic regions marked by H3K27ac. Knockdown of KDM6A in Drosophila S2 cells moderately reduces the global H3K27ac level, suggesting that KDM6A regulates CBP-mediated H3K27ac (9). KDM6A was also found to physically associate with CBP in the breast cancer cell line MCF-7 as well as in vitro (26). Using coimmunoprecipitation followed by Western blotting analysis, Miller et al. provided the initial evidence that KDM6A physically associates with BRG1 in T cells (27). Another study found that KDM6A promotes the interaction between BRG1 and TBX5, a T-box family transcription factor, and plays an enzymatically independent role in the recruitment of the BRG1-containing SWI/SNF complex to the enhancers of target genes (8). A recent study by Gozdecka et al. confirmed the physical association between KDM6A and BRG1 in mouse myeloid cells. They further showed that KDM6A loss in hematopoietic stem and progenitor cells (HSPCs) causes simultaneous downregulation of SWI/SNF-dependent chromatin accessibility and H3K27ac in certain pathways (such as the GATA transcriptional program) and upregulation in other pathways (such as the ETS program) (Fig. 3) (11).
KDM6A IN DIFFERENTIATION, DEVELOPMENT, AND REGENERATION
ESC maintenance and differentiation.
(i) KDM6A is dispensable for ESC maintenance. Several studies have examined the role of KDM6A in the maintenance and differentiation of ESCs. It was consistently reported that in male ESCs, KDM6A KO had no effect on ESC proliferation, survival, or self-renewal. Expression of key ESC identity markers, including Nanog, Oct4, Sox2, and alkaline phosphatase, a marker for undifferentiated ESCs, is unaffected by KDM6A loss (7, 8, 21). UTY expression is also diminished in male KDM6A KO ESCs, thereby eliminating the potential compensatory function by UTY in the absence of KDM6A (7). These results indicate that KDM6A and UTY are dispensable for maintaining ESC identity.
(ii) KDM6A is necessary for differentiation of ESCs. While KDM6A is dispensable for ESC maintenance, ESC differentiation toward mesoderm and ectoderm requires KDM6A protein but not its demethylase activity. Meanwhile, KDM6A appears to be largely dispensable for endoderm differentiation in mouse ESCs (7, 21). Using an embryoid body (EB) differentiation model, Wang et al. demonstrated that KDM6A was important for mesoderm differentiation of ESCs. They found that KDM6A KO EBs could not differentiate into mesoderm lineage, as the cells retained the expression of Nanog and Oct4 during the later stage of differentiation and could not induce the key mesoderm markers Brachyury (or T) and Wnt3. KDM6A was found to be enriched at the promoter of Brachyury and to regulate Brachyury expression through the Wnt/β-catenin signaling pathway independently of its demethylase activity. Ectopic UTY expression in KDM6A KO cells could rescue Wnt/β-catenin signaling-induced Brachyury expression, suggesting a compensatory function of UTY (7). Morales Torres et al. also confirmed the role of KDM6A, but not its enzymatic activity, in the induction of mesoderm markers during both retinoic acid (RA)-induced monolayer differentiation and spontaneous EB differentiation of ESCs. They further showed that KDM6A KO impaired the induction of key ectoderm markers, such as Sox1, Pax6, Msi1, and Otx2, under both RA-induced and EB differentiation conditions (21). Mechanistically, KDM6A is recruited to the promoters of the marker genes of ectoderm and mesoderm to activate the differentiation programs (21). A recent study with human ESCs showed that KDM6A interacts with 53BP1 to regulate a subset of genes during ESC differentiation into human neural progenitor cells (hNPCs). KDM6A depletion in hNPCs downregulates many neurogenic genes, including SOX4, FOXG1, and SIX3, at day 22 of monolayer differentiation (28).
Early embryonic development.
KDM6A is widely expressed in embryos and is critical for early embryonic development of mice. Homozygous KDM6A KO female embryos (Kdm6a−/−) show more severe defects than KDM6A KO males carrying an intact UTY allele (Kdm6a−/Y). Female homozygous KDM6A KO (Kdm6a−/−) embryos die around embryonic day 12.5 (E12.5) to E13.5 (7, 29, 30). These embryos show severe midgestational embryonic defects around E9.5 to E10.5, including small, abnormal, truncated posterior and cardiovascular and neural tube closure defects, while male KDM6A KO (Kdm6a−/Y) and female heterozygous KO (Kdm6a+/−) embryos appear normal at this stage (7, 16, 29, 30).
The loss of one Kdm6a allele in male mice causes less severe embryonic defects than the deletion of both Kdm6a alleles in females. Several studies found that the majority of KDM6A KO males (Kdm6a−/Y) showed perinatal lethality and only about 20 to 25% could survive through adulthood (7, 16, 29). The surviving males were significantly smaller and less viable than wild-type (WT) males (16, 29). Additionally, male embryos lacking both KDM6A and UTY alleles phenocopy homozygous KDM6A KO females (Kdm6a−/−), indicating that UTY can partially compensate for the loss of KDM6A during embryonic development (16). Interestingly, female heterozygous KDM6A KO (Kdm6a+/−) mice can survive through adulthood and appear normal, whereas KDM6A KO males (Kdm6a−/Y) die around birth, suggesting that one copy of KDM6A would suffice to maintain normal development and that UTY cannot compensate for the loss of KDM6A in postnatal development (7, 16).
Tissue development.
Beside its role in early embryonic development, KDM6A also plays a tissue-specific role in cardiac, mammary, and immune development (Table 2).
TABLE 2.
Summary of mouse models used to study the functions of KDM6A in the development of different tissues
| Tissue | Mouse model | Reference | Observed phenotype |
|---|---|---|---|
| Heart | Kdm6aflox; EIIa-Cre | 8 | Defective cardiac looping; chamber formation around E8.5–E10.5 |
| Kdm6aflox; Sox2-Cre | 16 | ||
| Kdm6aflox; Vasa-Cre | |||
| Kdm6aflox; Prm1-Cre | 7 | ||
| Kdm6aflox; Pgk1-Cre | 30 | ||
| Mammary tissue | Kdm6aflox; MMTV-Cre | 31 | Aberrant luminal cell layout and misplaced alveoli in adult female mice. Unable to nourish offspring. |
| Kdm6a enzyme-dead KI (H1146A/E1148A) | No effect on mammary tissues | ||
| Kdm6aflox; Prm1-Cre | 7 | KO females display anemia at E10.5. | |
| Kdm6aflox; Cre-ERT2 | 30 | KO females display myelodysplasia and splenic erythropoiesis. | |
| Hematopoiesis and T cells | Kdm6aflox; CD4-Cre | 32 | Increased CD4-SP thymocytes and decreased CD4+ T cells in the spleen |
| 34 | Reduced iNKT cells in the liver | ||
| Kdm6aflox; Lck-Cre | 33 | Reduced CD4+ and CD8+ T cells and Tfh cells | |
| Kdm6aflox; Vav-Cre | 35 | Reduced iNKT cells in the liver, thymus, and spleen | |
| Neural tissues | Kdm6aflox; nestin-Cre | 37 | Anxiety-like behaviors, impaired cognitive ability, aberrant hippocampal synaptic transmission and plasticity |
| 38 | Decreased NSC differentiation | ||
| Retina | Kdm6aflox; Dkk3-Cre | 39 | Reduced PKCα-positive bipolar cells in retinas at P10 |
| Muscle | Kdm6aflox; Pax7-CreER | 22 | Impaired muscle regeneration upon injury |
| Kdm6a enzyme-dead KI (H1146A/E1148A) |
(i) Cardiac development. KDM6A, but not its enzymatic activity, is important for embryonic development of cardiac tissues. Female homozygous KDM6A KO embryos show a lack of cardiac looping and defective chamber formation. UTY compensates for KDM6A loss in cardiac development of male embryos (8, 16, 30). The requirements for KDM6A in cardiac development appear to be stage dependent. KDM6A KO embryos show a normal induction of cardiac TFs GATA4, Nkx2.5, SRF, and Tbx5 in the myocardial lineage at E8.5. However, female homozygous KDM6A KO E9 embryos show lower overall levels of Nkx2.5 and Tbx5 than WT embryos (8). Consistent with the observations in vivo, a majority of cardiac lineage-committed embryoid bodies derived from KDM6A KO ESCs failed to develop spontaneous cardiac-like contractions in culture and could not induce expression of many cardiomyocyte genes, such as ANF, α-CA, MLC2v, and Myh6 (8). Mechanistically, KDM6A is recruited to enhancers that contain response elements for key cardiac TFs GATA4, Nkx2.5, SRF, and Tbx5 and acts as a coactivator for these TFs. KDM6A also promotes the interaction between Tbx5 and BRG1 to facilitate the recruitment of BRG1 to enhancers of cardiac genes (8).
(ii) Mammary development. KDM6A controls the development of mammary luminal cell lineage and the expression of luminal cell-specific genes independent of its enzymatic activity. It was shown that the loss of KDM6A in mouse mammary luminal epithelium leads to a disorganized layout of luminal and basal cells, a decrease in luminal cell populations, an increase in basal cell populations, and misplaced alveoli from the main ducts during mammary development. As a consequence, these mice are unable to nourish offspring despite retaining milk production (31). Interestingly, a significant portion of luminal cells show basal cell-like characteristics in KDM6A KO mice. The expression of genes encoding luminal fate-determining TFs, such as STAT3, SOX9, and GATA3, is reduced, whereas basal signature genes, such as Krt5, Krt14, and Acta2, are induced upon KDM6A deletion in luminal cells (31). Mechanistically, more than half of genes enriched in luminal cells are bound by KDM6A at their promoters and enhancers. KDM6A colocalizes with mammary lineage-determining TFs, including NFIB and STAT5, on enhancers of luminal cell-specific genes, such as Sox9 and Krt8, to regulate their expression, suggesting that KDM6A is a transcription coactivator important for maintaining the luminal lineage identity during mammary development (31). The mammary development defects were not observed in enzymatically dead KDM6A KI mice and the global H3K27me3 was unchanged in KDM6A KO mammary luminal cells, indicating that the function of KDM6A in mammary epithelial development is independent of its demethylase activity (31).
(iii) Immune development. Early work has shown that female homozygous KDM6A KO mouse embryos experience anemia around E10.5, suggesting that KDM6A is important for hematopoiesis (7). Another study utilized a tamoxifen-inducible KDM6A KO mouse model to demonstrate that depletion of KDM6A in female mice causes weight loss, anemic bones, myelodysplasia, and splenic erythropoiesis. In contrast, KDM6A depletion appears to have little effect on male mice, suggesting that UTY can compensate for the role of KDM6A in mouse hematopoiesis. Primary HPSCs isolated from KDM6A-deficient female mice also show a defect in cell migration (30). More recent studies have focused on the critical role of KDM6A in the development of T cells, including CD4 T cells and natural killer T (NKT) cells. Using a CD4-Cre-mediated KDM6A conditional knockout mouse model, Manna et al. revealed that the loss of KDM6A results in an increased amount of CD4 singly positive (SP) thymocytes in the thymus and a reduced amount of CD4+ T cells in the spleen while having no effect on cell proliferation, suggesting that KDM6A is important for the late-stage development of CD4+ T cells (32). KDM6A KO downregulates the expression of thymic egress gene S1pr1 in CD4 SP thymocytes, leading to the accumulation of CD4 SP thymocytes and CD4+ lymphopenia. These defects caused by KDM6A KO could not be rescued by UTY, suggesting a requirement for KDM6A demethylase activity (32). Another group used a T cell-specific Lck promoter-driven Cre model to delete KDM6A in mice. The resulting KDM6A KO mice showed moderate decreases in CD4+ and CD8+ T cells in the spleens. They also showed that KDM6A could support the development of CD4+ T follicular helper (Tfh) cells and that KDM6A was required for clearance of chronic viral infection by regulating the expression of Tfh-related genes, such as Il6ra (33).
KDM6A was also found to be important for invariant NKT (iNKT) cell development in mice. Using a similar CD4-Cre-mediated conditional knockout model, Northrup et al. showed that KDM6A loss leads to a significant reduction of iNKT cells in the liver, in addition to impaired CD4 T cell development (34). Specifically, KDM6A loss in thymocytes blocks the cells from undergoing thymic iNKT maturation. UTY could not support iNKT development in male KDM6A KO mice, indicating that the function of KDM6A is dependent on its demethylase activity (34). Using a Vav-Cre-mediated conditional knockout model, Beyaz et al. demonstrated that KDM6A was required for iNKT cell development in the liver, thymus, and spleen. They further showed that KDM6A was required for the expression of lineage-specific developmental genes, such as Cxcr3, Il2rb, S100a6, and Tbx21, in iNKT cells (35). Mechanistically, KDM6A is recruited to the promoters of many iNKT-specific genes and associates with iNKT lineage-determining TFs JunB and PLZF to regulate the expression of their target genes. The loss of KDM6A could hinder the superenhancer accessibility on iNKT cell identity genes, such as Il2rb and Tbx21, correlating with an increase of H3K27me3 signals on these superenhancers. Enzymatically inactive KDM6A fails to rescue the expression of these cell identity genes in KDM6A KO iNKT cells, suggesting that KDM6A has a demethylase-dependent function in the development of iNKT cells (35). In addition to T cell development, KDM6A has also been implicated in development of B cells (36).
(iv) Development of other tissues. KDM6A also plays a role in the development of other tissues. Two studies used nestin-Cre to conditionally knock out KDM6A in neural stem cells (NSCs) in mice. In one study, KDM6A KO mice show anxiety-like behaviors and impaired spatial learning and memory. The hippocampal regions of these mice displayed aberrant synaptic formation, transmission, and plasticity and impaired dendritic development, suggesting that KDM6A is important for neural development and cognitive behaviors (37). In another study, the loss of KDM6A increased NSC proliferation and downregulated a subset of genes related to NSC differentiation (38). A recent study also suggests that KDM6A is important for the differentiation of a subset of retinal cells. In particular, retina-specific knockout of KDM6A using Dkk3-Cre mice leads to a reduction in protein kinase Cα (PKCα)-positive rod “on” bipolar cells in postnatal day 10 mouse retinas while having no effect on retinal progenitor cell proliferation (39). Using a CRISPR-Cas9 KO system in C2C12 myoblasts to screen a group of candidate lysine demethylases, a group reported that KDM6A was required for osteoblast differentiation in culture (40). However, the in vivo relevance of this study needs to be verified through animal models.
Regeneration.
KDM6A plays a significant role in satellite cell (SC)-mediated muscle fiber regeneration. Using an SC-specific KDM6A KO mouse model, Faralli et al. (22) found that SC-specific KDM6A KO male and female mice experience impaired muscle regeneration after cardiotoxin-induced injury (Table 2). These mice showed a reduction in myofiber density, an increase in necrotic tissues and inflammatory cell infiltration, and a delayed recovery time. Interestingly, female mice with homozygous whole-body enzyme-dead KDM6A KI also showed impaired muscle regeneration after cardiotoxin treatment, whereas heterozygous SC-specific KDM6A KO females showed normal muscle regeneration (Table 2). These results indicate that KDM6A demethylase activity is important for SC-mediated muscle regeneration (22).
Mechanistically, KDM6A mediates the transition between proliferating and differentiating muscle progenitors during regeneration. KDM6A was shown to be dispensable for maintaining SC proliferation and the expression of SC identity genes, including MyoD, Myf5, and Pax7. However, KDM6A and its enzymatic activity are required to initiate differentiation of muscle progenitor cells, as the key TF MYOG could not be induced in either KDM6A KO or KI cells. During differentiation, KDM6A loss reduces the expression of muscle regulatory gene Myog and increases H3K27me3 marks on myotube-specific enhancers. Additionally, differentiating myoblasts isolated from homozygous KDM6A KI mice show an enrichment of H3K27me3 signals at the promoters of myogenic genes, such as Myog, Tnnc2, and Ckm, suggesting that KDM6A regulates the terminal differentiation stage of muscle progenitor cells through a demethylase-dependent function (22).
KDM6A MUTATIONS IN DEVELOPMENTAL DISEASES AND CANCER
Kabuki syndrome.
Kabuki syndrome (KS) is a rare human congenital craniofacial disorder manifesting as abnormal facial features, skeletal deformities, and heart and cognitive defects. Sixty percent to 89% of KS cases are caused by KMT2D (MLL4) mutations, whereas a smaller number of patients (6 to 14%) carry KDM6A mutations (41–44). There are no distinct phenotypic differences between KMT2D and KDM6A mutant KS patients (42). However, some studies reported that KMT2D KS patients appear to have more prominent facial abnormalities, while KDM6A KS patients show more growth defects (41, 45).
A recent study found that KDM6A was critical for craniofacial development in mice and for neural crest (NC) cell survival after the cell migration stage in embryos. NC-specific loss of KDM6A causes human-like Kabuki syndrome in mice (19). That study showed that about 40% of NC-specific KDM6A KO female (Kdm6a−/−) pups died before weaning, while the male KO (Kdm6a−/Y) mice could survive at least until weaning age. All NC-specific male KDM6A KO (Kdm6a−/Y) and surviving female KO (Kdm6a−/−) mice showed growth retardation and aberrant craniofacial features, such as frontonasal hypoplasia, increased facial angle, and reduced palpebral fissures, similar to the human KS phenotype. NC-specific KO (Kdm6a−/Y) males showed milder defects than females (Kdm6a−/−), suggesting that KDM6A loss could be partially compensated by UTY. Mechanistically, KDM6A regulates expression of craniofacial NC genes. In particular, KDM6A KO causes a downregulation of the NC stem cell signaling pathways, including PCP/Wnt/β-catenin and Notch (19).
Interestingly, whole-body homozygous enzyme-dead KI female mice have normal body weight and facial development, suggesting that craniofacial development does not require KDM6A demethylase activity. However, females with one KI and one KO allele show human-like KS features similar to those in NC-specific KO males. These findings suggest that at least one WT KDM6A allele or two enzyme-dead KDM6A alleles are required for normal cranial NC development in mice (19).
KDM6A mutations are frequently present in cancers.
An early study reported that KDM6A mutations are present in many tumor types, including multiple myeloma, esophageal squamous cell carcinoma, renal cell carcinoma, glioblastoma, and breast and colorectal cancers. Among 58 multiple-myeloma patient samples tested, about 10% had KDM6A mutations (46). A more comprehensive study of 4,742 tumor samples from 21 cancer types demonstrated that KDM6A is highly mutated across multiple cancer types, especially in bladder cancer (47). Urothelial bladder carcinoma is the most common type of bladder cancer and one of the most frequent cancers in men in developed regions of the world. Ler et al. used Sanger sequencing to analyze 176 urothelial bladder carcinoma patient samples and reported that KDM6A mutations were found in 45% of nonmuscle invasive urothelial bladder carcinoma, 28% of muscle-invasive tumors, and 28% of tumors of unknown stages. Further combining these findings with additional published data, they showed that KDM6A mutations were present in 29% of urothelial bladder carcinoma samples. The types of KDM6A mutations are widespread across the entire coding region of the gene and include frameshifting (insertion/deletion), nonsense, splice site, and missense mutations (48).
KDM6A mutations are also frequent in pancreatic cancer, one of the most common and lethal cancers in the world. According to public databases of pancreatic cancer genomes, the KDM6A gene is mutated in about 10.7% to 21.6% of pancreatic cancer patient samples (24). Mutations of KDM6A are also present in other cancers, including chronic myelomonocytic leukemia, metastatic castration-resistant prostate cancer, subgroups of medulloblastoma, and acute myeloid leukemia (AML) (46, 49–52).
KDM6A is a tumor suppressor.
An early study showed that reintroduction of WT KDM6A into KDM6A-deficient esophageal squamous carcinoma cells results in slower cell growth, suggesting a role for KDM6A as a tumor suppressor (46). Later studies have reported a role for KDM6A as a tumor suppressor in different cancer types (Table 3). In one study, the loss of KDM6A in a bladder carcinoma cell line increased the cell proliferation rate by about 25%. Analysis of urothelial bladder carcinoma samples revealed that KDM6A loss activates EZH2-dependent transcriptional repression. EZH2 is a histone methyltransferase that, in contrast to KDM6A, adds a methyl group to H3K27 to downregulate gene transcription. Further analysis of KDM6A-deficient urothelial bladder carcinoma cells showed that H3K27me3 signals are enriched at the promoter regions of EZH2 target genes, including PIP5K1B and GHR. This enrichment could be reversed by ectopic KDM6A. These findings suggest that KDM6A functions as a tumor suppressor in urothelial bladder carcinomas by antagonizing EZH2-mediated transcriptional repression (48). Kaneko and Li showed that urothelium-specific KDM6A KO increases bladder cancer risk in female mice. These mice have a significantly lower survival rate upon bladder cancer induction than do female WT mice. The loss of KDM6A also reduces the expression of the tumor suppressor genes Cdkn1a and Perp. Ectopic expression of WT and enzyme-dead KDM6A could suppress cell proliferation and colony formation of a KDM6A-null bladder cancer cell line, suggesting that the tumor suppression function of KDM6A is demethylase independent (53). Another study also used a urothelium-specific KDM6A KO mouse model and reported that KDM6A-deficient mice can develop bladder cancer and have an increased number of CD44-positive bladder stem cells. The loss of KDM6A in these mice also upregulates proinflammatory cytokines such as Cxcl1, Ccl2, and Il6 (54).
TABLE 3.
Summary of mouse models used to study the tumor-suppressive role of KDM6A in different cancer types
| Cancer type | Mouse model | Reference | Observed phenotype |
|---|---|---|---|
| Bladder | Kdm6aflox; Upk2-Cre | 53 | Increased tumor burden and reduced survival rate |
| 54 | Increased cancer development and CD44-positive stem cell proliferation | ||
| Lung | Kdm6aflox; KrasG12D; Ad-Cre | 55 | Increased cancer progression and tumor burden. Shortened life span. |
| Pancreas | Kdm6aflox; KrasG12D; Pdx1-Cre | 24 | More aggressive tumor development. Shortened life span. |
| Kdm6aflox; KrasG12D; Ptf1a-Cre | |||
| Kdm6aflox; KrasG12D; Pdx1-Cre | 20 | ||
| Blood | Kdm6aflox; Mx1-Cre | 11 | Higher chance to develop AML in female KO mice. Increased HSPC self-renewal and decreased HSPC differentiation. |
| Kdm6aflox; CD19-Cre | 36 | Accelerated Eμ-Myc-induced B cell lymphomagenesis | |
| Kdm6aflox; CreER | 57 | Accelerated chromic myelomonocytic leukemia and shortened life span |
Another study has identified KDM6A as a tumor suppressor in a mouse model of metastatic lung cancer. Using the oncogenic KrasG12D-induced lung cancer mouse model, Wu et al. found that deletion of KDM6A promotes lung cancer progression and increases tumor burden (55). KDM6A loss also shortens the life span of KrasG12D mice from 30 weeks to 17 weeks. Tumors extracted from KDM6A KO KrasG12D adult mice are significantly larger and have a higher cell proliferation rate than those from control mice. KDM6A loss promotes primary lung cancer cell growth, which could be rescued by ectopic expression of KDM6A. Notably, KDM6A KO in KrasG12D lung tumors upregulates the expression of H3K27 methyltransferase EZH2 at both the mRNA and protein levels. KDM6A loss also downregulates the expression of tumor suppressors and cell cycle inhibitors CDKN2A and CDKN2B, which may explain the increase in cell proliferation in KDM6A KO tumors (55).
Three studies have identified KDM6A as a tumor suppressor in pancreatic cancer. By CRISPR-Cas9-mediated deletion of KDM6A in two human pancreatic ductal adenocarcinoma cell lines, Watanabe et al. showed that the loss of KDM6A promotes cell proliferation, colony formation, and migration, whereas overexpression of KDM6A could suppress these traits. They further revealed that KDM6A depletion decreases the expression of tumor suppressor genes, including CDKN1A. Interestingly, H3K27ac is downregulated at the enhancers of CDKN1A in KDM6A KO cells (56). Using a KrasG12D-induced cancer mouse model, Andricovich et al. showed that pancreas-specific KO of KDM6A causes squamous-like, metastatic pancreatic cancer in females. These mice show aggressive tumor development and shortened survival time. Interestingly, the induction of squamous-like, metastatic pancreatic cancer in male mice requires the loss of both KDM6A and the homologous UTY (24). Mechanistically, KDM6A loss deregulates MLL4 genomic binding and activates the superenhancers (SEs) controlling the expression of oncogenes Myc, ΔNp63, and Runx3. Ectopic expression of the WT, enzyme-dead KDM6A, or UTY in human KDM6A KO pancreatic tumor cell lines downregulates Myc and ΔNp63 expression and inhibits cell proliferation, indicating that the tumor suppressor role of KDM6A in pancreatic cancer is demethylase activity independent (24). A recent study confirmed the role of KDM6A as a tumor suppressor in pancreatic cancer using the KrasG12D mouse model. Mechanistically, KDM6A is recruited by HNF1A to regulate the enhancer landscape of acinar cells and activate the epithelial gene expression program that indirectly suppresses oncogenic pathways. HNF1A is a homeodomain transcription factor that had been thought to be a pancreatic tumor suppressor. It was also shown that HNF1A loss partially phenocopies KDM6A KO in KrasG12D-induced pancreatic cancer (20).
KDM6A also plays a role in suppressing myeloid leukemogenesis independently of its demethylase activity. Using a hematopoietic stem and progenitor cell (HSPC)-specific KDM6A deletion mouse model, Gozdecka et al. showed that female homozygous KDM6A KO (Kdm6a−/−) mice have a high chance (63%) to develop AML (11). These mice show an increase in HSPC self-renewal and myeloid expansion and a decrease in differentiation. In contrast, female heterozygous KO mice (Kdm6a+/−) and male KDM6A KO mice (Kdm6a−/Y) do not manifest AML. Ectopic expression of KDM6A, enzyme-dead KDM6A, or UTY in a KDM6A/UTY-deficient AML cell line could suppress cell proliferation, suggesting that the function of KDM6A in AML suppression does not require its demethylase activity. KDM6A loss in HSPCs leads to a change in SWI/SNF-dependent chromatin accessibility, indirectly facilitating the oncogenic ETS transcriptional program while simultaneously impairing the tumor-suppressive GATA program (11). In addition to AML, KDM6A has also been identified as a tumor suppressor in other types of blood cancer, including B cell lymphoma, chronic myelomonocytic leukemia, multiple myeloma, and T cell acute lymphoblastic leukemia (T-ALL) (36, 57–60).
Together, these studies demonstrate that KDM6A functions as a tumor suppressor independently of its demethylase activity in a variety of cancer types by regulating different transcriptional programs, especially those involved in oncogenesis and tumor suppression.
Pro-oncogenic role of KDM6A.
Even though many studies have reported the role of KDM6A as a tumor suppressor, some studies have provided evidence to suggest that KDM6A could play a pro-oncogenic role. Despite KDM6A being previously described as a tumor suppressor in T-ALL (59, 60), a 2016 study reported that KDM6A functions as a pro-oncogenic coactivator in a subgroup of T-ALL that expresses the oncogenic TF TAL1. KDM6A physically associates with TAL1 and is recruited by TAL1 to its target genes. Depletion of KDM6A in several TAL1-positive cell lines leads to an increase in apoptosis, whereas overexpression of KDM6A promotes cell growth (61). Another study suggests that KDM6A plays a role in promoting estrogen receptor (ER)-positive breast cancer cell proliferation and migration (26). The loss of KDM6A in a human breast cancer cell line, MCF-7, causes a significant decrease in estrogen-induced cell proliferation. Depletion of KDM6A also suppresses, whereas overexpression of KDM6A enhances, cell migration. Mechanistically, KDM6A colocalizes with ERα on a subset of ERα target genes and regulates their expression in MCF-7 cells. These genes include GREB1, TFF1, MYB, and CXCR4, which are known to be involved in breast cancer development. KDM6A also recruits the H3K27 acetyltransferase CBP to ERα target genes to regulate H3K27ac signals (26). Interestingly, while KDM6A controls the estrogen-induced expression of the oncogene CXCR4, ectopic expression of enzyme-dead KDM6A could not rescue CXCR4 expression in KDM6A KO MCF-7 cells, suggesting that KDM6A plays a demethylase-dependent role in facilitating the ERα-mediated transcriptional program to promote breast cancer cell migration (26). Whether KDM6A facilitates or impedes ER-positive breast cancer progression in vivo remains to be determined.
FUTURE DIRECTIONS
Many studies in the past decade have demonstrated that KDM6A functions in normal development and tumor suppression are largely independent of its enzymatic activity. For example, mouse embryonic midgestational development and development of certain tissues do not require a catalytically active KDM6A (7, 8, 16, 31). As a tumor suppressor in pancreatic cancer and AML, KDM6A also plays a demethylase activity-independent role (11, 24). It remains unclear how KDM6A regulates these processes without its enzymatic function. One hypothesis is that KDM6A regulates gene expression and enhancer activation through its interaction with MLL3/4, since MLL3/4 is known to be critical for enhancer activation and KDM6A protein stability depends on MLL3/4 (Fig. 3) (18, 25). Supporting this hypothesis, it has recently been shown that G137V and D336G mutations in the TPR domains of KDM6A impair its physical interaction with MLL3/4 and destabilize the KDM6A protein. G137V mutation also impairs the ability of KDM6A to suppress colony formation in a soft-agar assay, suggesting that KDM6A functions as a tumor suppressor through its interaction with MLL3/4 (62). The mechanism behind KDM6A genomic targeting also requires further investigation. It has been hinted that certain TFs could interact with KDM6A, which, in turn, recruits MLL3/4 to activate enhancers (20, 24).
While many findings have shown the enzymatic activity-independent role of KDM6A in normal development, several studies have shown that KDM6A demethylase activity is important in certain cellular contexts, such as T cell development and muscle regeneration (22, 35). Future work is needed to validate the tissue-specific role of KDM6A demethylase activity in vivo by utilizing enzyme-dead KDM6A KI mice. Unlike mammals, Drosophila has only one KDM6 ortholog, dUtx. It was reported that dUtx demethylase activity was important for embryonic and larval development, as enzyme-dead dUtx Drosophila flies die during the larval stage. These observations in Drosophila suggest that there could be some redundancy between the enzymatic activities of KDM6A and other KDM6 family members, in particular KDM6B, in mammals (63). One approach to study this redundancy is to generate mice harboring both demethylase-inactive KDM6A and KDM6B. It also remains unclear whether KDM6A enzymatic activity can target nonhistone substrates other than the known target H3K27me2/3.
The rapid progression and development of new tools and technology have vastly accelerated the studies of protein structure and function, which can be utilized to facilitate a more in-depth understanding of the KDM6A protein. Glycogen synthase kinase J4 (GSK-J4) is a potent small-molecule inhibitor for KDM6A. It can effectively inhibit KDM6A enzymatic activity, with a half-maximum inhibitory concentration (IC50) of 6.6 μM. However, this inhibitor is not specific to KDM6A, as it can also potently inhibit KDM6B (IC50 = 8.6 μM) and other demethylases (64). The development of a highly specific small-molecule inhibitor of KDM6A would facilitate in-depth studies to distinguish the functions of KDM6A protein and its enzymatic activity. Advanced live-cell imaging has been one of the most rapidly developing technologies in the past decade and could be employed as a powerful tool to determine KDM6A cellular localization and its dynamic interactions with TFs, MLL3/4, or other chromatin regulators during enhancer activation, gene expression, and cell differentiation. Recently, cryo-electron microscopy (cryo-EM) has been used to study the structure of the MLL3 catalytic module consisting of the catalytic SET domain of MLL3 and associated regulatory factors ASH2L, RBBP5, WDR5, and DPY30 (65). However, the catalytic C-terminal SET domain of MLL3 does not associate with KDM6A. Future studies using cryo-EM are needed to determine how KDM6A interacts with full-length MLL3/4 in the intact holocomplex.
As KDM6A is a known tumor suppressor in many cancer types, many attempts have been made to develop potential therapeutics to counteract cancer growth in KDM6A-mutated tumors. For example, KDM6A-deficient urothelial bladder carcinoma cells were found to be more sensitive to EZH2 inhibitors, such as GSK343 and GSK126. These inhibitors could effectively suppress in vivo onset and growth of KDM6A-deficient tumors in mice (48). Consistent with the study, another EZH2 inhibitor, JQEZ5, suppresses the growth of KDM6A-deficient lung tumors in a KrasG12D mouse model (55). In pancreatic cancer, KDM6A-deficient cell lines are sensitive to the bromodomain and extraterminal domain inhibitors (24). These findings suggest promising strategies for developing treatments of KDM6A-mutated cancers, which should be further explored. Finally, mutation of KDM6A is one of the main causes of Kabuki syndrome (KS). Since this rare genetic disorder currently has no cure, targeted gene therapy can be a potential treatment to correct the KDM6A mutations in KS patients.
KDM6A has also been proposed to be an oxygen sensor (66). It was shown that KDM6A protein and the isolated KDM6A enzymatic domain display low oxygen affinities in vitro. Hypoxia appears to moderately increase cellular H3K27me3 levels in a KDM6A-dependent manner in several cell types. Using myogenesis as a model system, the authors showed that either hypoxia treatment or KDM6A depletion blocks cell differentiation. These results suggest that hypoxia inhibits KDM6A-mediated removal of H3K27me3 in cells (66). However, it is unclear whether hypoxia directly inhibits KDM6A enzymatic activity. Future work is needed to establish the functional relevance of KDM6A as an oxygen sensor in vivo.
ACKNOWLEDGMENTS
We apologize to colleagues whose work on KDM6A cannot be cited in this review due to space limitation. We thank Ji-Eun Lee, Kaitlin McKernan, Zhizhong Ren, and Guojia Xie for proofreading.
This work was supported by a grant from the Intramural Research Program of NIDDK, NIH, to K.G.
REFERENCES
- 1.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 2.Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, Roberts TM, Chang HY, Shi Y. 2007. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 3.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 4.Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R. 2007. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 5.Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. 2007. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci U S A 104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, Ge K. 2007. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem 282:20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang C, Lee JE, Cho YW, Xiao Y, Jin Q, Liu C, Ge K. 2012. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci U S A 109:15324–15329. doi: 10.1073/pnas.1204166109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S, Lee JW, Lee SK. 2012. UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev Cell 22:25–37. doi: 10.1016/j.devcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tie F, Banerjee R, Conrad PA, Scacheri PC, Harte PJ. 2012. Histone demethylase UTX and chromatin remodeler BRM bind directly to CBP and modulate acetylation of histone H3 lysine 27. Mol Cell Biol 32:2323–2334. doi: 10.1128/MCB.06392-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang SP, Tang Z, Chen CW, Shimada M, Koche RP, Wang LH, Nakadai T, Chramiec A, Krivtsov AV, Armstrong SA, Roeder RG. 2017. A UTX-MLL4-p300 transcriptional regulatory network coordinately shapes active enhancer landscapes for eliciting transcription. Mol Cell 67:308–321.e6. doi: 10.1016/j.molcel.2017.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gozdecka M, Meduri E, Mazan M, Tzelepis K, Dudek M, Knights AJ, Pardo M, Yu L, Choudhary JS, Metzakopian E, Iyer V, Yun H, Park N, Varela I, Bautista R, Collord G, Dovey O, Garyfallos DA, De Braekeleer E, Kondo S, Cooper J, Göttgens B, Bullinger L, Northcott PA, Adams D, Vassiliou GS, Huntly BJP. 2018. UTX-mediated enhancer and chromatin remodeling suppresses myeloid leukemogenesis through noncatalytic inverse regulation of ETS and GATA programs. Nat Genet 50:883–894. doi: 10.1038/s41588-018-0114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenfield A, Carrel L, Pennisi D, Philippe C, Quaderi N, Siggers P, Steiner K, Tam PP, Monaco AP, Willard HF, Koopman P. 1998. The UTX gene escapes X inactivation in mice and humans. Hum Mol Genet 7:737–742. doi: 10.1093/hmg/7.4.737. [DOI] [PubMed] [Google Scholar]
- 13.Klose RJ, Kallin EM, Zhang Y. 2006. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 14.Sengoku T, Yokoyama S. 2011. Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A. Genes Dev 25:2266–2277. doi: 10.1101/gad.172296.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burgold T, Voituron N, Caganova M, Tripathi PP, Menuet C, Tusi BK, Spreafico F, Bévengut M, Gestreau C, Buontempo S, Simeone A, Kruidenier L, Natoli G, Casola S, Hilaire G, Testa G. 2012. The H3K27 demethylase JMJD3 is required for maintenance of the embryonic respiratory neuronal network, neonatal breathing, and survival. Cell Rep 2:1244–1258. doi: 10.1016/j.celrep.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 16.Shpargel KB, Sengoku T, Yokoyama S, Magnuson T. 2012. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet 8:e1002964. doi: 10.1371/journal.pgen.1002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel SR, Kim D, Levitan I, Dressler GR. 2007. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev Cell 13:580–592. doi: 10.1016/j.devcel.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J-E, Wang C, Xu S, Cho Y-W, Wang L, Feng X, Baldridge A, Sartorelli V, Zhuang L, Peng W, Ge K. 2013. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife 2:e01503. doi: 10.7554/eLife.01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shpargel KB, Starmer J, Wang C, Ge K, Magnuson T. 2017. UTX-guided neural crest function underlies craniofacial features of Kabuki syndrome. Proc Natl Acad Sci U S A 114:E9046–E9055. doi: 10.1073/pnas.1705011114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalisz M, Bernardo E, Beucher A, Maestro MA, Del Pozo N, Millán I, Haeberle L, Schlensog M, Safi SA, Knoefel WT, Grau V, de Vas M, Shpargel KB, Vaquero E, Magnuson T, Ortega S, Esposito I, Real FX, Ferrer J. 2020. HNF1A recruits KDM6A to activate differentiated acinar cell programs that suppress pancreatic cancer. EMBO J 39:e102808. doi: 10.15252/embj.2019102808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morales Torres C, Laugesen A, Helin K. 2013. Utx is required for proper induction of ectoderm and mesoderm during differentiation of embryonic stem cells. PLoS One 8:e60020. doi: 10.1371/journal.pone.0060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faralli H, Wang C, Nakka K, Benyoucef A, Sebastian S, Zhuang L, Chu A, Palii CG, Liu C, Camellato B, Brand M, Ge K, Dilworth FJ. 2016. UTX demethylase activity is required for satellite cell-mediated muscle regeneration. J Clin Invest 126:1555–1565. doi: 10.1172/JCI83239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vandamme J, Lettier G, Sidoli S, Di Schiavi E, Nørregaard Jensen O, Salcini AE. 2012. The C. elegans H3K27 demethylase UTX-1 is essential for normal development, independent of its enzymatic activity. PLoS Genet 8:e1002647. doi: 10.1371/journal.pgen.1002647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andricovich J, Perkail S, Kai Y, Casasanta N, Peng W, Tzatsos A. 2018. Loss of KDM6A activates super-enhancers to induce gender-specific squamous-like pancreatic cancer and confers sensitivity to BET inhibitors. Cancer Cell 33:512–526.e8. doi: 10.1016/j.ccell.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Froimchuk E, Jang Y, Ge K. 2017. Histone H3 lysine 4 methyltransferase KMT2D. Gene 627:337–342. doi: 10.1016/j.gene.2017.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie G, Liu X, Zhang Y, Li W, Liu S, Chen Z, Xu B, Yang J, He L, Zhang Z, Jin T, Yi X, Sun L, Shang Y, Liang J. 2017. UTX promotes hormonally responsive breast carcinogenesis through feed-forward transcription regulation with estrogen receptor. Oncogene 36:5497–5511. doi: 10.1038/onc.2017.157. [DOI] [PubMed] [Google Scholar]
- 27.Miller SA, Mohn SE, Weinmann AS. 2010. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell 40:594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X, Xu B, Mulvey B, Evans M, Jordan S, Wang YD, Pagala V, Peng J, Fan Y, Patel A, Peng JC. 2019. Differentiation of human pluripotent stem cells into neurons or cortical organoids requires transcriptional co-regulation by UTX and 53BP1. Nat Neurosci 22:362–373. doi: 10.1038/s41593-018-0328-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welstead GG, Creyghton MP, Bilodeau S, Cheng AW, Markoulaki S, Young RA, Jaenisch R. 2012. X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proc Natl Acad Sci U S A 109:13004–13009. doi: 10.1073/pnas.1210787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thieme S, Gyárfás T, Richter C, Özhan G, Fu J, Alexopoulou D, Muders MH, Michalk I, Jakob C, Dahl A, Klink B, Bandola J, Bachmann M, Schröck E, Buchholz F, Stewart AF, Weidinger G, Anastassiadis K, Brenner S. 2013. The histone demethylase UTX regulates stem cell migration and hematopoiesis. Blood 121:2462–2473. doi: 10.1182/blood-2012-08-452003. [DOI] [PubMed] [Google Scholar]
- 31.Yoo KH, Oh S, Kang K, Wang C, Robinson GW, Ge K, Hennighausen L. 2016. Histone demethylase KDM6A controls the mammary luminal lineage through enzyme-independent mechanisms. Mol Cell Biol 36:2108–2120. doi: 10.1128/MCB.00089-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manna S, Kim JK, Baugé C, Cam M, Zhao Y, Shetty J, Vacchio MS, Castro E, Tran B, Tessarollo L, Bosselut R. 2015. Histone H3 lysine 27 demethylases Jmjd3 and Utx are required for T-cell differentiation. Nat Commun 6:8152. doi: 10.1038/ncomms9152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cook KD, Shpargel KB, Starmer J, Whitfield-Larry F, Conley B, Allard DE, Rager JE, Fry RC, Davenport ML, Magnuson T, Whitmire JK, Su MA. 2015. T follicular helper cell-dependent clearance of a persistent virus infection requires T cell expression of the histone demethylase UTX. Immunity 43:703–714. doi: 10.1016/j.immuni.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Northrup D, Yagi R, Cui K, Proctor WR, Wang C, Placek K, Pohl LR, Wang R, Ge K, Zhu J, Zhao K. 2017. Histone demethylases UTX and JMJD3 are required for NKT cell development in mice. Cell Biosci 7:25. doi: 10.1186/s13578-017-0152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beyaz S, Kim JH, Pinello L, Xifaras ME, Hu Y, Huang J, Kerenyi MA, Das PP, Barnitz RA, Herault A, Dogum R, Haining WN, Yilmaz ÖH, Passegue E, Yuan GC, Orkin SH, Winau F. 2017. The histone demethylase UTX regulates the lineage-specific epigenetic program of invariant natural killer T cells. Nat Immunol 18:184–195. doi: 10.1038/ni.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Zhang Y, Zheng L, Liu M, Chen CD, Jiang H. 2018. UTX is an escape from X-inactivation tumor-suppressor in B cell lymphoma. Nat Commun 9:2720. doi: 10.1038/s41467-018-05084-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang GB, Zeng YQ, Liu PP, Mi TW, Zhang SF, Dai SK, Tang QY, Yang L, Xu YJ, Yan HL, Du HZ, Teng ZQ, Zhou FQ, Liu CM. 2017. The histone H3K27 demethylase UTX regulates synaptic plasticity and cognitive behaviors in mice. Front Mol Neurosci 10:267. doi: 10.3389/fnmol.2017.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lei X, Jiao J. 2018. UTX affects neural stem cell proliferation and differentiation through PTEN signaling. Stem Cell Rep 10:1193–1207. doi: 10.1016/j.stemcr.2018.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Umutoni D, Iwagawa T, Baba Y, Tsuhako A, Honda H, Aihara M, Watanabe S. 2020. H3K27me3 demethylase UTX regulates the differentiation of a subset of bipolar cells in the mouse retina. Genes Cells 25:402–412. doi: 10.1111/gtc.12767. [DOI] [PubMed] [Google Scholar]
- 40.Munehira Y, Yang Z, Gozani O. 2017. Systematic analysis of known and candidate lysine demethylases in the regulation of myoblast differentiation. J Mol Biol 429:2055–2065. doi: 10.1016/j.jmb.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyake N, Koshimizu E, Okamoto N, Mizuno S, Ogata T, Nagai T, Kosho T, Ohashi H, Kato M, Sasaki G, Mabe H, Watanabe Y, Yoshino M, Matsuishi T, Takanashi J, Shotelersuk V, Tekin M, Ochi N, Kubota M, Ito N, Ihara K, Hara T, Tonoki H, Ohta T, Saito K, Matsuo M, Urano M, Enokizono T, Sato A, Tanaka H, Ogawa A, Fujita T, Hiraki Y, Kitanaka S, Matsubara Y, Makita T, Taguri M, Nakashima M, Tsurusaki Y, Saitsu H, Yoshiura K, Matsumoto N, Niikawa N. 2013. MLL2 and KDM6A mutations in patients with Kabuki syndrome. Am J Med Genet A 161a:2234–2243. doi: 10.1002/ajmg.a.36072. [DOI] [PubMed] [Google Scholar]
- 42.Bögershausen N, Gatinois V, Riehmer V, Kayserili H, Becker J, Thoenes M, Simsek-Kiper P, Barat-Houari M, Elcioglu NH, Wieczorek D, Tinschert S, Sarrabay G, Strom TM, Fabre A, Baynam G, Sanchez E, Nürnberg G, Altunoglu U, Capri Y, Isidor B, Lacombe D, Corsini C, Cormier-Daire V, Sanlaville D, Giuliano F, Le Quan Sang KH, Kayirangwa H, Nürnberg P, Meitinger T, Boduroglu K, Zoll B, Lyonnet S, Tzschach A, Verloes A, Di Donato N, Touitou I, Netzer C, Li Y, Geneviève D, Yigit G, Wollnik B. 2016. Mutation update for Kabuki syndrome genes KMT2D and KDM6A and further delineation of X-linked Kabuki syndrome subtype 2. Hum Mutat 37:847–864. doi: 10.1002/humu.23026. [DOI] [PubMed] [Google Scholar]
- 43.Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, Lee C, Turner EH, Smith JD, Rieder MJ, Yoshiura K, Matsumoto N, Ohta T, Niikawa N, Nickerson DA, Bamshad MJ, Shendure J. 2010. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet 42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adam MP, Banka S, Bjornsson HT, Bodamer O, Chudley AE, Harris J, Kawame H, Lanpher BC, Lindsley AW, Merla G, Miyake N, Okamoto N, Stumpel CT, Niikawa N, Kabuki Syndrome Medical Advisory Board . 2019. Kabuki syndrome: international consensus diagnostic criteria. J Med Genet 56:89–95. doi: 10.1136/jmedgenet-2018-105625. [DOI] [PubMed] [Google Scholar]
- 45.Banka S, Lederer D, Benoit V, Jenkins E, Howard E, Bunstone S, Kerr B, McKee S, Lloyd IC, Shears D, Stewart H, White SM, Savarirayan R, Mancini GM, Beysen D, Cohn RD, Grisart B, Maystadt I, Donnai D. 2015. Novel KDM6A (UTX) mutations and a clinical and molecular review of the X-linked Kabuki syndrome (KS2). Clin Genet 87:252–258. doi: 10.1111/cge.12363. [DOI] [PubMed] [Google Scholar]
- 46.van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O'Meara S, Teague J, Butler A, Hinton J, Latimer C, Andrews J, Barthorpe S, Beare D, Buck G, Campbell PJ, Cole J, Forbes S, Jia M, Jones D, Kok CY, Leroy C, Lin M-L, McBride DJ, Maddison M, Maquire S, McLay K, Menzies A, Mironenko T, Mulderrig L, Mudie L, Pleasance E, Shepherd R, Smith R, Stebbings L, Stephens P, Tang G, Tarpey PS, Turner R, Turrell K, Varian J, West S, Widaa S, Wray P, Collins VP, Ichimura K, Law S, Wong J, Yuen ST, Leung SY, Tonon G, DePinho RA, Tai Y-T, Anderson KC, Kahnoski RJ, Massie A, Khoo SK, Teh BT, Stratton MR, Futreal PA. 2009. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet 41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. 2014. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ler LD, Ghosh S, Chai X, Thike AA, Heng HL, Siew EY, Dey S, Koh LK, Lim JQ, Lim WK, Myint SS, Loh JL, Ong P, Sam XX, Huang D, Lim T, Tan PH, Nagarajan S, Cheng CW, Ho H, Ng LG, Yuen J, Lin PH, Chuang CK, Chang YH, Weng WH, Rozen SG, Tan P, Creasy CL, Pang ST, McCabe MT, Poon SL, Teh BT. 2017. Loss of tumor suppressor KDM6A amplifies PRC2-regulated transcriptional repression in bladder cancer and can be targeted through inhibition of EZH2. Sci Transl Med 9:eaai8312. doi: 10.1126/scitranslmed.aai8312. [DOI] [PubMed] [Google Scholar]
- 49.Jankowska AM, Makishima H, Tiu RV, Szpurka H, Huang Y, Traina F, Visconte V, Sugimoto Y, Prince C, O'Keefe C, Hsi ED, List A, Sekeres MA, Rao A, McDevitt MA, Maciejewski JP. 2011. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood 118:3932–3941. doi: 10.1182/blood-2010-10-311019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, Asangani IA, Ateeq B, Chun SY, Siddiqui J, Sam L, Anstett M, Mehra R, Prensner JR, Palanisamy N, Ryslik GA, Vandin F, Raphael BJ, Kunju LP, Rhodes DR, Pienta KJ, Chinnaiyan AM, Tomlins SA. 2012. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, Chalhoub N, Baker SJ, Huether R, Kriwacki R, Curley N, Thiruvenkatam R, Wang J, Wu G, Rusch M, Hong X, Becksfort J, Gupta P, Ma J, Easton J, Vadodaria B, Onar-Thomas A, Lin T, Li S, Pounds S, Paugh S, Zhao D, Kawauchi D, Roussel MF, Finkelstein D, Ellison DW, Lau CC, Bouffet E, Hassall T, Gururangan S, Cohn R, Fulton RS, Fulton LL, Dooling DJ, Ochoa K, Gajjar A, Mardis ER, Wilson RK, Downing JR, Zhang J, Gilbertson RJ. 2012. Novel mutations target distinct subgroups of medulloblastoma. Nature 488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, Leiserson MDM, Miller CA, Welch JS, Walter MJ, Wendl MC, Ley TJ, Wilson RK, Raphael BJ, Ding L. 2013. Mutational landscape and significance across 12 major cancer types. Nature 502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaneko S, Li X. 2018. X chromosome protects against bladder cancer in females via a KDM6A-dependent epigenetic mechanism. Sci Adv 4:eaar5598. doi: 10.1126/sciadv.aar5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kobatake K, Ikeda KI, Nakata Y, Yamasaki N, Ueda T, Kanai A, Sentani K, Sera Y, Hayashi T, Koizumi M, Miyakawa Y, Inaba T, Sotomaru Y, Kaminuma O, Ichinohe T, Honda ZI, Yasui W, Horie S, Black PC, Matsubara A, Honda H. 2020. Kdm6a deficiency activates inflammatory pathways, promotes M2 macrophage polarization, and causes bladder cancer in cooperation with p53 dysfunction. Clin Cancer Res 26:2065–2079. doi: 10.1158/1078-0432.CCR-19-2230. [DOI] [PubMed] [Google Scholar]
- 55.Wu Q, Tian Y, Zhang J, Tong X, Huang H, Li S, Zhao H, Tang Y, Yuan C, Wang K, Fang Z, Gao L, Hu X, Li F, Qin Z, Yao S, Chen T, Chen H, Zhang G, Liu W, Sun Y, Chen L, Wong KK, Ge K, Chen L, Ji H. 2018. In vivo CRISPR screening unveils histone demethylase UTX as an important epigenetic regulator in lung tumorigenesis. Proc Natl Acad Sci U S A 115:E3978–E3986. doi: 10.1073/pnas.1716589115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watanabe S, Shimada S, Akiyama Y, Ishikawa Y, Ogura T, Ogawa K, Ono H, Mitsunori Y, Ban D, Kudo A, Yamaoka S, Tanabe M, Tanaka S. 2019. Loss of KDM6A characterizes a poor prognostic subtype of human pancreatic cancer and potentiates HDAC inhibitor lethality. Int J Cancer 145:192–205. doi: 10.1002/ijc.32072. [DOI] [PubMed] [Google Scholar]
- 57.Zheng L, Xu L, Xu Q, Yu L, Zhao D, Chen P, Wang W, Wang Y, Han G, Chen CD. 2018. Utx loss causes myeloid transformation. Leukemia 32:1458–1465. doi: 10.1038/s41375-018-0011-6. [DOI] [PubMed] [Google Scholar]
- 58.Ezponda T, Dupéré-Richer D, Will CM, Small EC, Varghese N, Patel T, Nabet B, Popovic R, Oyer J, Bulic M, Zheng Y, Huang X, Shah MY, Maji S, Riva A, Occhionorelli M, Tonon G, Kelleher N, Keats J, Licht JD. 2017. UTX/KDM6A loss enhances the malignant phenotype of multiple myeloma and sensitizes cells to EZH2 inhibition. Cell Rep 21:628–640. doi: 10.1016/j.celrep.2017.09.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van der Meulen J, Sanghvi V, Mavrakis K, Durinck K, Fang F, Matthijssens F, Rondou P, Rosen M, Pieters T, Vandenberghe P, Delabesse E, Lammens T, De Moerloose B, Menten B, Van Roy N, Verhasselt B, Poppe B, Benoit Y, Taghon T, Melnick AM, Speleman F, Wendel HG, Van Vlierberghe P. 2015. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood 125:13–21. doi: 10.1182/blood-2014-05-577270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ntziachristos P, Tsirigos A, Welstead GG, Trimarchi T, Bakogianni S, Xu L, Loizou E, Holmfeldt L, Strikoudis A, King B, Mullenders J, Becksfort J, Nedjic J, Paietta E, Tallman MS, Rowe JM, Tonon G, Satoh T, Kruidenier L, Prinjha R, Akira S, Van Vlierberghe P, Ferrando AA, Jaenisch R, Mullighan CG, Aifantis I. 2014. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature 514:513–517. doi: 10.1038/nature13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benyoucef A, Palii CG, Wang C, Porter CJ, Chu A, Dai F, Tremblay V, Rakopoulos P, Singh K, Huang S, Pflumio F, Hébert J, Couture JF, Perkins TJ, Ge K, Dilworth FJ, Brand M. 2016. UTX inhibition as selective epigenetic therapy against TAL1-driven T-cell acute lymphoblastic leukemia. Genes Dev 30:508–521. doi: 10.1101/gad.276790.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kato H, Asamitsu K, Sun W, Kitajima S, Yoshizawa-Sugata N, Okamoto T, Masai H, Poellinger L. 2020. Cancer-derived UTX TPR mutations G137V and D336G impair interaction with MLL3/4 complexes and affect UTX subcellular localization. Oncogene 39:3322–3335. doi: 10.1038/s41388-020-1218-3. [DOI] [PubMed] [Google Scholar]
- 63.Copur Ö, Müller J. 2018. Histone demethylase activity of Utx is essential for viability and regulation of HOX gene expression in Drosophila. Genetics 208:633–637. doi: 10.1534/genetics.117.300421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heinemann B, Nielsen JM, Hudlebusch HR, Lees MJ, Larsen DV, Boesen T, Labelle M, Gerlach L-O, Birk P, Helin K. 2014. Inhibition of demethylases by GSK-J1/J4. Nature 514:E1–E2. doi: 10.1038/nature13688. [DOI] [PubMed] [Google Scholar]
- 65.Xue H, Yao T, Cao M, Zhu G, Li Y, Yuan G, Chen Y, Lei M, Huang J. 2019. Structural basis of nucleosome recognition and modification by MLL methyltransferases. Nature 573:445–449. doi: 10.1038/s41586-019-1528-1. [DOI] [PubMed] [Google Scholar]
- 66.Chakraborty AA, Laukka T, Myllykoski M, Ringel AE, Booker MA, Tolstorukov MY, Meng YJ, Meier SR, Jennings RB, Creech AL, Herbert ZT, McBrayer SK, Olenchock BA, Jaffe JD, Haigis MC, Beroukhim R, Signoretti S, Koivunen P, Kaelin WG Jr.. 2019. Histone demethylase KDM6A directly senses oxygen to control chromatin and cell fate. Science 363:1217–1222. doi: 10.1126/science.aaw1026. [DOI] [PMC free article] [PubMed] [Google Scholar]