Many proteins, including DICER1 and hAgo2, are involved in the biogenesis of microRNAs (miRNAs). Whether hAgo2 regulates DICER1 expression is unknown. Exogenously overexpressed hAgo2 suppressed DICER1 expression at the levels of both protein and mRNA, and the reduction in hAgo2 expression enhanced DICER1 expression. Precursor miRNA processing mediated by DICER1 was also modulated by hAgo2. However, hAgo2 protein did not suppress DICER1 promoter activity. Therefore, hAgo2 protein probably regulates DICER1 expression at the posttranscriptional level.
KEYWORDS: knockdown, knockout, posttranscriptional level, luciferase reporter assay, coimmunoprecipitation
ABSTRACT
Many proteins, including DICER1 and hAgo2, are involved in the biogenesis of microRNAs (miRNAs). Whether hAgo2 regulates DICER1 expression is unknown. Exogenously overexpressed hAgo2 suppressed DICER1 expression at the levels of both protein and mRNA, and the reduction in hAgo2 expression enhanced DICER1 expression. Precursor miRNA processing mediated by DICER1 was also modulated by hAgo2. However, hAgo2 protein did not suppress DICER1 promoter activity. Therefore, hAgo2 protein probably regulates DICER1 expression at the posttranscriptional level. Indeed, hAgo2 protein inhibited the reporter assay of the DICER1 mRNA 3′ untranslated region (3′-UTR). Previous reports have demonstrated that miRNAs (e.g., let-7 and miR-103/107) inhibited DICER1 expression posttranscriptionally. However, hAgo2 still suppressed DICER1 expression in the cells depleted of these miRNAs. Moreover, the reporter activities of the DICER1 mRNA 3′-UTR without these miRNA binding sites were still suppressed by hAgo2. Therefore, in addition to an miRNA-dependent pathway, hAgo2 can also modulate DICER1 expression through an miRNA-independent mechanism. Downregulation of DICER1 expression was further proven to be dependent on both hAgo2 and AUF1 proteins. Interactions of hAgo2 and AUF1 proteins were demonstrated by the coimmunoprecipitation assay. As expected, hAgo2 could not suppress the DICER1 mRNA 3′-UTR reporter with a mutation in the potential AUF1-binding site. Thus, downregulation of DICER1 expression through the 3′-UTR requires both hAgo2 and AUF1.
INTRODUCTION
MicroRNAs (miRNAs), a class of noncoding RNAs, play an important role in the negative regulation of gene expression. Biogenesis of the canonical miRNAs (18 to 25 nucleotides [nt]) involves multiple steps. First, miRNA genes are transcribed by RNA polymerase II as long primary transcripts (pri-miRNAs). These pri-miRNAs then are processed in the nucleus by Drosha to generate precursor miRNA (pre-miRNA; 60 to 120 nt long). Exportin 5 (XPO5) exports these pre-miRNAs from the nucleus to the cytoplasm, followed by cytoplasmic processing by DICER1 to generate mature miRNAs (1). The mature miRNAs are loaded into the Argonaute 2 protein. Ago2 associates with other proteins to form an RNA-induced silencing complex (RISC), which plays a crucial role in the repression of translation or degradation of mRNAs. miRNAs modulate gene expression at the posttranscriptional level mostly by binding to the 3′ untranslated region (3′-UTR) of the target mRNA (2). At present, more than 2,600 miRNAs derived from approximately 1,900 pre-miRNAs in the human genome have been identified (http://www.mirbase.org/).
Proteins responsible for the biogenesis of miRNAs (e.g., Drosha, XPO5, and DICER1) are also essential for the generation of mature miRNAs. Not surprisingly, canonical miRNA production was completely abolished in Drosha-deleted cells (1). miRNA processing is regulated in a complex manner, and its true nature is not completely understood. miRNA abundance can be regulated during transcription of the pre-miRNA at any of the biogenesis steps or at the turnover of the mature miRNA. Therefore, the miRNA levels can be modulated by the proteins, influencing miRNA processing, the variations in the miRNA transcript, and the nuclear export efficiency of the precursor miRNA. In addition to these mechanisms, single-nucleotide polymorphisms (SNPs) also have a pronounced effect on the efficiency of the miRNA processing machinery (3). Various routes of miRNA maturation are tightly regulated by signaling cascades that have also been identified (4). For example, extracellular signal-regulated kinase (ERK) suppresses the pre-miRNA export from the nucleus through phosphorylation of XPO5 at T345/S416/S497 (5). Therefore, the biogenesis of miRNAs is not regulated as a linear, unified mechanism.
The biogenesis of miRNAs is under tight control both temporally and spatially (3). Dysregulation of the miRNA levels can initiate pathological consequences, including carcinogenesis and disorders of the immune system. Not surprisingly, DICER1- or hAgo2-null mice are embryonically lethal (6). Much evidence suggests that the development of human cancers is associated with altered miRNA expression (7, 8), and global miRNA expression is reduced in many cancers (9, 10). Moreover, increasing evidence has demonstrated that there is a low expression of DICER1 in many different types of tumors (11).
Through its influence on miRNA biosynthesis, DICER1 affects cell cycle progression, senescence, stem cell maintenance, and tumorigenesis. DICER1-null mice were embryonic lethal due to the depletion of stem cells. Despite the importance of DICER1 protein in cellular homeostasis, the mechanisms that control its expression are not completely understood (12, 13).
The human Argonaute family can be divided into two subfamilies: one is the AGO subfamily, with members including Ago1, Ago2, Ago3, and Ago4, and the other is the PIWI family, including HIWI1, HIWI2, HIWI3, and HIWI4. The PIWI subfamily members are exclusively expressed in germ line cells, while the AGO subfamily members are broadly expressed in most tissues. In the human AGO family, Ago2 has been demonstrated as the only member with catalytic activity, and it plays an essential role within the RISC in the regulation of small RNA-guided gene silencing processes (14). Different from the other members of the AGO family, Ago2 has also been proved to be indispensable in murine embryonic development based on the finding that knockout of the Ago2 gene was lethal (15). Ago2 has also been revealed to be involved in tumorigenesis through miRNA-dependent or miRNA-independent procedures (16).
The AUF1 RNA-binding proteins, a family of four proteins generated by alternative pre-mRNA splicing, control the stability and/or translation of mRNA targets by recognition of AU-rich sequences within mRNA 3′ untranslated regions (17). The downregulation of DICER1 expression by hAgo2 and different miRNAs was demonstrated previously (18, 19). In this study, we demonstrated that hAgo2 protein, in addition to modulating DICER1 expression via an miRNA-dependent pathway, could also modulate DICER1 expression through an AUF1-dependent but miRNA-independent mechanism.
RESULTS
hAgo2 modulates DICER1 expression and precursor miRNA processing.
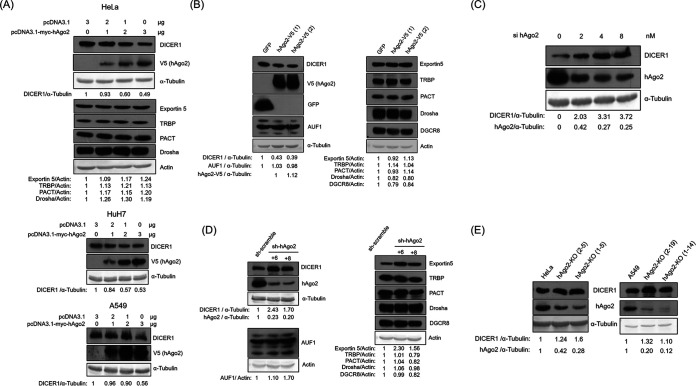
To determine whether hAgo2 can modulate the expression of other proteins involved in the miRNA pathway, the plasmid expressing hAgo2 was transfected into HeLa cells. The expression of DICER1 but not of other proteins involved in the miRNA biogenesis pathway (e.g., Drosha, exportin 5, PACT, and TRBP) was suppressed by transiently expressed hAgo2 (Fig. 1A). The reduction in DICER1 expression by hAgo2 was not cell type specific, since DICER1 expression was also downregulated by transiently expressed hAgo2 in HuH7 and A549 cells (Fig. 1A). Downregulation of DICER1 by hAgo2 was also seen in HeLa cells using stably expressed hAgo2 (Fig. 1B). Therefore, exogenously overexpressed hAgo2 (transiently or stably expressed) suppressed DICER1 expression but not the expression of other proteins involved in the miRNA biogenesis pathway. Furthermore, knockdown of hAgo2 by siRNA (Fig. 1C) or short hairpin RNA (shRNA) (Fig. 1D) enhanced DICER1 expression but not the expression of the other proteins involved in the miRNA biogenesis pathway. Cas9/sgRNA technology was also used to knock out the hAgo2 expression in HeLa and A549 cells. Again, reduction of hAgo2 expression by sgRNA enhanced DICER1 expression in HeLa cells (Fig. 1E, left) and in A549 cells (Fig. 1E, right).
FIG 1.
hAgo2 downregulated DICER1 expression. (A to D) Western blot of various proteins involved in the miRNA pathway, using samples from HeLa cells, HuH7, or A549 cells, transiently transfected with different amounts of either vector or the plasmid expressing hAgo2, as indicated (A), stably transfected with GFP or hAgo2 (two different infection experiments, 1 and 2) (B), transfected with different amounts of siRNA targeting hAgo2, as indicated (C), or stably transfected with shRNA targeting hAgo2 (two different shRNA clones, +6 and +8) (D). (E) Western blot of DICER1 and hAgo2 proteins using samples from HeLa cells (left) or A549 cells (right) transfected with sgRNA targeting hAgo2 (two different clones [2-5 and 1-5] were from HeLa cells, while the other two [2-19 and 1-14] were from A549 cells).
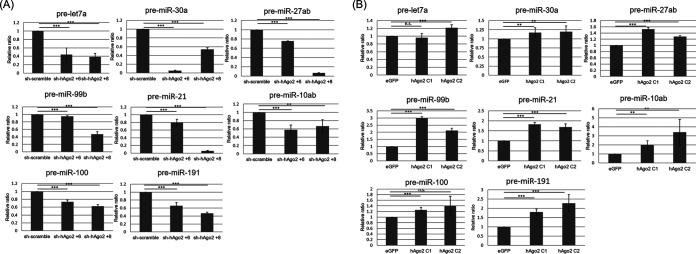
Precursor miRNA processing is mediated by DICER1 to generate mature miRNAs (1). If hAgo2 can regulate DICER1 expression, the precursor miRNA processing will also be modulated by hAgo2. To determine whether the processing of precursor miRNAs was affected by hAgo2, real-time reverse transcription-PCR (RT-PCR) was performed to analyze the amount of eight different pre-miRNAs abundantly expressed in HeLa cells (http://guanlab.ccmb.med.umich.edu/mirmine/index.html). Indeed, the amount of these eight precursor miRNAs was reduced when hAgo2 was knocked down by shRNAs (Fig. 2A), while the amount of these precursor miRNAs increased when hAgo2 was overexpressed (Fig. 2B).
FIG 2.
Real-time RT-PCR to detect the relative amounts of eight different precursor miRNAs using RNA samples from HeLa cells with shRNA targeting hAgo2 (two different clones, +6 and +8) (A) or overexpression of hAgo2 (two different clones, C1 and C2) (B). Results in panels A and B are the means (±SD) from three independent experiments.
hAgo2 affects DICER1 expression through regulating its 3′-UTR.
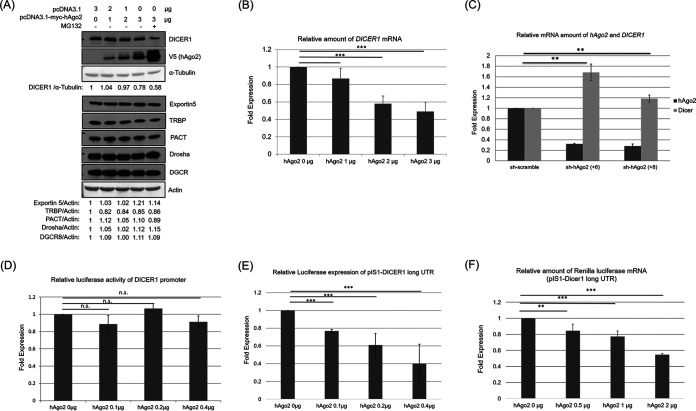
To determine whether hAgo2 facilitates the degradation of DICER1 protein, proteasome inhibitor MG132 was used (Fig. 3A). MG132 can inhibit 20S proteasome activity by its covalently binding to the active site of this proteasome complex (20). MG132 treatment stabilized the exogenously expressed hAgo2, but it did not reverse the suppression of DICER1 by hAgo2. This result suggested that hAgo2 did not facilitate the degradation of DICER1 protein. To determine whether hAgo2 affected DICER1 expression at the mRNA level, real-time RT-PCR was performed (Fig. 3B). Indeed, overexpressed hAgo2 suppressed DICER1 expression at the mRNA level in a dose-dependent manner. On the other hand, the amount of DICER1 mRNA increased when hAgo2 was knocked down by shRNAs (Fig. 3C). A luciferase assay was then performed to see if hAgo2 exerted its suppression on the DICER1 promoter (21). However, hAgo2 protein did not suppress the DICER1 promoter activity (Fig. 3D). Therefore, hAgo2 protein probably regulates DICER1 expression at the posttranscriptional level. Regulation of DICER1 expression at its 3′-UTR by miRNAs and/or AUF1 has been reported previously (18, 19, 22). To determine whether hAgo2 suppressed DICER1 expression at its 3′-UTR, the renilla luciferase gene was placed on the upstream region of the DICER1 mRNA 3′-UTR, and the reporter activity was reduced by hAgo2 (Fig. 3E). The mRNA level of this reporter was also reduced by hAgo2 dose dependently (Fig. 3F). Thus, hAgo2 downregulated DICER1 expression through DICER1’s mRNA 3′-UTR.
FIG 3.
hAgo2 downregulates DICER1 expression by affecting the 3′-UTR of its mRNA. (A) Western blot using protein samples from HeLa cells transiently transfected with expression plasmids with or without the MG132 treatment, as indicated. (B) Real-time RT-PCR to detect the DICER1 mRNA amount using RNA samples from HeLa cells transiently transfected with expression plasmids for hAgo2. (C) Real-time RT-PCR to detect the relative mRNA amount of hAgo2 and DICER1 using RNA samples from HeLa cells with shRNA targeting hAgo2 (two different clones, +6 and +8). (D) HeLa cells were transfected with the following plasmids: pGL3-DICER1-prom or pGL3-Basic, 0.1 μg; pRL-TK, 0.01 μg; pcDNA3-myc-hAgo2 (or pcDNA3 empty vector), different doses of 0.1, 0.2, and 0.4 μg. Cells were harvested, and luciferase activity was measured 48 h after transfection. (E) HeLa cells were transfected with the following plasmids: pIS1-DICER1-long UTR or pIS1, 0.1 μg; PIS0, 0.01 μg; pcDNA3-myc-hAgo2 (or pcDNA3 empty vector), different doses of 0.1, 0.2, and 0.4 μg. The cells were harvested, and luciferase activity was measured 48 h after transfection. (F) HeLa cells were transfected with the following plasmids: pIS1-DICER1-long UTR and pcDNA3-myc-hAgo2 (or pcDNA3 empty vector) with different doses of 0, 0.5, 1, and 2 μg. The cells were harvested, and the luciferase mRNA level was determined 48 h after transfection using real-time RT-PCR. Results in panels B, C, D, E, and F are the means (±SD) from three independent experiments.
Downregulation of DICER1 expression by hAgo2 is independent of miRNAs.
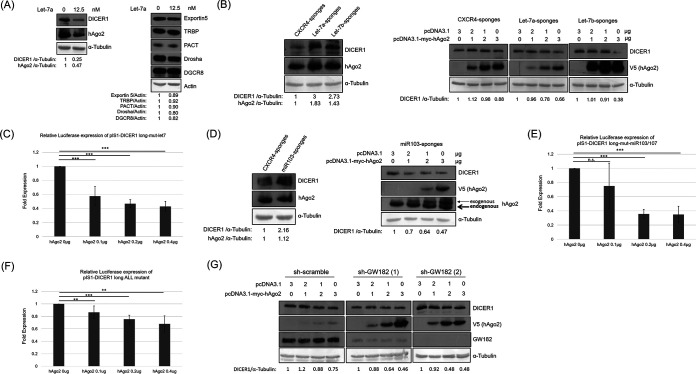
It has been reported that miRNA let-7 suppressed DICER1 expression through its 3′-UTR (19). To verify this, synthetic let-7a was transfected into HeLa cells. Exogenous let-7a suppressed the expression of DICER1 and hAgo2 but not the other proteins involved in the miRNA biogenesis pathway (Fig. 4A). To determine whether the downregulation of DICER1 expression by hAgo2 occurs through let-7, sponges were used to remove let-7a and let-7b while a nontargeting miRNA sponge based on a CXCR4 sequence devoid of any miRNA seed motif was used as a control (Fig. 4B) (23). As expected, the removal of let-7a or let-7b upregulated the expression of both DICER1 and hAgo2 (Fig. 4B, left). Moreover, the exogenously expressed hAgo2 still suppressed DICER1 expression dose dependently in the cells with let-7a sponge or let-7b sponge (Fig. 4B, right). Not surprisingly, the hAgo2 protein still suppressed the reporter activity of the DICER1 mRNA 3′-UTR with a mutation in the let-7 binding site (Fig. 4C). It has been reported that miR103/miR107 suppressed DICER1 expression through its 3′-UTR (18). To determine whether the downregulation of DICER1 expression by hAgo2 occurs through miR103/miR107, sponges for miR103/miR107 were used (Fig. 4D). As expected, removal of miR103/miR107 upregulated DICER1 expression (Fig. 4D, left), while hAgo2 still suppressed DICER1 expression in these cells (Fig. 4D, right). Furthermore, hAgo2 protein suppressed the reporter activity of the DICER1 mRNA 3′-UTR with a mutation at the miR103/miR107 binding site (Fig. 4E) and even without the binding sites of let-7 and miR103/miR107 (Fig. 4F). The GW182 protein is required to form a RISC (24). Moreover, disruption of the GW bodies impairs RNA interference (25). To determine whether the downregulation of DICER1 by hAgo2 is through the formation of a RISC, GW182 knockdown cells were established. In these GW182 knockdown HeLa cells, transiently expressed hAgo2 still downregulated DICER1 expression (Fig. 4G). Together, these results suggested that downregulation of DICER1 expression by hAgo2 is an event independent of specific miRNA functions, e.g., let-7 and miR103/miR107.
FIG 4.
Downregulation of DICER1 by hAgo2 is not through miRNAs. (A) Western blot of various proteins involved in the miRNA pathway using samples from HeLa cells transfected with miRNA Let-7a. (B, left) Western blot of DICER1 and hAgo2 proteins using samples from HeLa cells stably transfected with control sponge (labeled as CXCR4) or sponges to remove Let-7a or Let-7b. (Right) Western blotting of DICER1 protein using samples from HeLa cells stably transfected with different sponges and transiently transfected with the plasmid expressing hAgo2. (C) Renilla luciferase activity was measured using samples from HeLa cells with different amounts of pIS1-DICER1 long-mut-let7 (pIS0 empty firefly luciferase plasmid as control). (D, left) Western blotting of DICER1 and hAgo2 proteins using samples from HeLa cells stably transfected with sponges to remove miR103/107. (Right) Western blotting of DICER1 protein using samples from HeLa cells stably transfected with miR103/107 sponges and transiently transfected with the plasmid expressing hAgo2. (E) Luciferase activity was measured using samples from HeLa cells with pIS1-DICER1 long-mut-miR103/107. (F) Renilla luciferase activity was measured using samples from HeLa cells with the pIS1-DICER1 long ALL mutant. (G) Western blotting of DICER1 protein using samples from HeLa cells stably transfected with shRNA targeting GW182 (two different shRNA clones) and transiently transfected with expressing plasmids for hAgo2. Results in panels C, E, and F are the means (±SD) from three independent experiments.
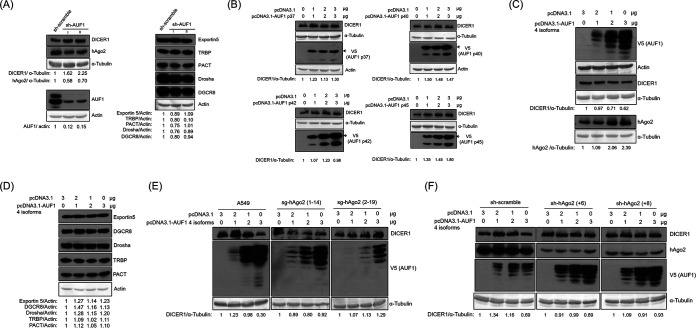
Downregulation of DICER1 by AUF1(s) requires hAgo2.
There are four isoforms of AUF1 proteins (26). It has been reported that AUF1(s) suppressed miRNA production by reducing DICER1 expression (22). To verify this, all four AUF1 isoforms were knocked down with shRNAs (Fig. 5A). As expected, the reduction in AUF1 expression by shRNAs upregulated DICER1 expression (Fig. 5A, left) but not the expression of the other proteins (exportin 5, TRBP, PACT, Drosha, and DGCR8) involved in the miRNA biogenesis pathway (Fig. 5A, right). Each AUF1 isoform (p37, p40, p42, or p45) was transiently transfected into HeLa cells individually. None of the AUF1 isoforms alone suppressed DICER1 expression (Fig. 5B). When the four AUF1 isoforms were cotransfected together, DICER1 expression was downregulated (Fig. 5C). In contrast, transient overexpression of all four AUF1 isoforms did not affect the expression of those proteins involved in the miRNA biogenesis pathway (Fig. 5D). It has been reported that cooperation of AUF1 and hAgo2 proteins can suppress the expression of many genes through their 3′-UTRs (27). To determine whether the suppression of DICER1 by AUF1 is dependent on hAgo2, cells with reduced hAgo2 expression were used. A549 cells with reduced hAgo2 expression by Cas9/single guide RNA (sgRNA) were transiently transfected with the plasmids expressing the four isoforms of AUF1, and DICER1 expression was not suppressed (Fig. 5E). In HeLa cells with shRNAs to knock down hAgo2 expression, overexpression of the four isoforms of AUF1 did not suppress the level of DICER1 expression (Fig. 5F). These results demonstrated that downregulation of DICER1 by AUF1(s) requires the presence of hAgo2.
FIG 5.
Downregulation of DICER1 by AUF1(s) requires hAgo2. (A) Western blot of various proteins involved in the miRNA pathway using samples from HeLa cells stably transfected with shRNA targeting AUF1 (two different shRNA clones, I and II). (B) Western blotting of DICER1 protein using samples from HeLa cells transiently transfected with the expression plasmids for one of the four different isoforms of AUF1 (p37, p40, p42, or p45). The expected proteins were marked by arrows, while the smaller proteins were probably the degraded products. (C) Western blot of DICER1 and hAgo2 proteins using samples from HeLa cells transiently transfected with different amounts of either vector or the plasmids expressing four different forms of AUF1, as indicated. (D) Western blot of various proteins involved in the miRNA pathway using samples from HeLa cells transiently transfected with the expression plasmids for the four isoforms of AUF1. (E) Western blot of DICER1 protein using samples from A549 cells stably transfected with sgRNA targeting hAgo2 (two different clones [2-19 and 1-14] from A549 cells) and transiently transfected with expression plasmids for four different forms of AUF1. (F) Western blotting of DICER1 protein using samples from HeLa cells stably transfected with shRNA targeting hAgo2 (two different shRNA clones, +6 and +8) and transiently transfected with expression plasmids for four different forms of AUF1.
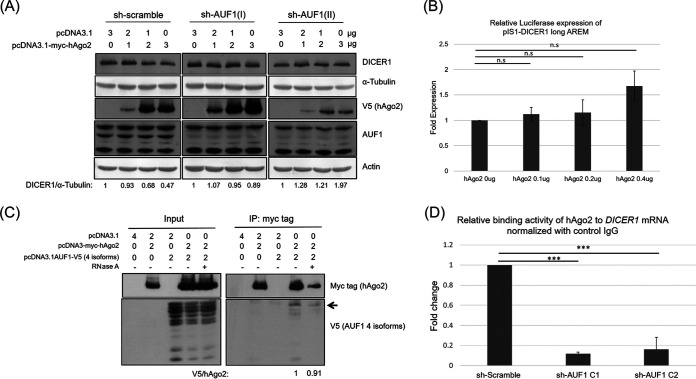
Suppression of DICER1 expression requires both hAgo2 and AUF1.
It has been reported that both AUF1 and Ago2 could control the decay of selected target mRNAs (27). AUF1 could also promote let-7b loading on Ago2 (28). To determine whether the suppression of DICER1 expression by hAgo2 is dependent on AUF1, HeLa cells with shRNAs to knock down AUF1 expression were used. In this case, overexpressed hAgo2 suppressed DICER1 expression in control cells but not in the AUF1 knockdown cells (Fig. 6A). Therefore, downregulation of DICER1 by hAgo2 requires the presence of AUF1(s). Cooperation of hAgo2 and AUF1(s) suppressed the expression of many genes through their 3′-UTR (27). There is a potential AUF1 binding site (AU-rich segment, nt 2068 to 2080 of 3′-UTR, TAATATTTATAAA) at the 3′-UTR of DICER1 mRNA, but it has not been confirmed as an AUF1 binding site. The hAgo2 protein did not reduce the reporter activity of the DICER1 mRNA 3′-UTR when the potential AUF1 binding site was mutated (from TAATATTTATAAA to CCGGCGGCCGCCG) (Fig. 6B). If suppression of DICER1 expression requires both hAgo2 and AUF1(s), protein-protein interactions between these two proteins are expected, and this was confirmed by the coimmunoprecipitation in HEK293T cells (Fig. 6C). Although p45 is not the most abundant isoform among the four exogenously expressed AUF1s, it was the most abundantly precipitated by hAgo2 (Fig. 6C). To determine whether hAgo2 interacts with AUF1 through mRNA, samples were treated with RNase A before coimmunoprecipitation. Under this condition, AUF1 is still precipitated by hAgo2 protein (binding ratio of AUF1 to hAgo2 is changed from 1 to 0.91) (Fig. 6C). Therefore, interaction of hAgo2 and AUF1 is not mRNA dependent. Furthermore, to determine whether AUF1 affects the binding of hAgo2 to DICER1 mRNA, real-time RT-PCR to detect the DICER1 mRNA level was conducted after immunoprecipitation using anti-hAgo2. As expected, hAgo2 could not bind to DICER1 mRNA in the cells that lack AUF1 (Fig. 6D).
FIG 6.
Both hAgo2 and AUF1(s) are required to downregulate DICER1 expression. (A) Western blotting of DICER1 protein using samples from HeLa cells stably transfected with shRNA targeting AUF1 (two different shRNA clones, I and II) and transiently transfected with an expression plasmid for hAgo2. (B) Luciferase activity was measured using samples from HeLa cells with pIS1-DICER1 long AREM. (C) Coimmunoprecipitation of hAgo2 and AUF1 proteins. Forty-eight hours after transfection of HEK293T cells with different plasmids, as indicated, cell lysates from these cells were directly analyzed by Western blotting (input) for expression control or immunoprecipitated with the anti-Myc antibody prior to Western blotting (bottom). AUF1 p45 protein was marked by an arrow. Results in panel B are the means (±SD) from three independent experiments.
Suppression of DICER1 expression requires almost the entire hAgo2.
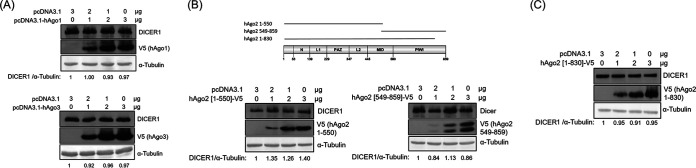
There are four members in the Ago family (29). To determine whether other members of the Ago family, in addition to hAgo2, could suppress DICER1 expression, plasmids expressing hAgo1 or hAgo3 were separately transiently transfected into HeLa cells (Fig. 7A). Neither hAgo1 (Fig. 7A, upper) nor hAgo3 (Fig. 7A, lower) suppressed DICER1 expression. Thus, neither hAgo1 nor hAgo3 downregulated DICER1 expression. To identify the region(s) within hAgo2 responsible for the downregulation of DICER1 expression, plasmids expressing the N terminus (amino acids [aa] 1 to 550) or the C terminus (aa 549 to 849) of hAgo2 were transiently transfected into HeLa cells individually (Fig. 7B). Neither hAgo2 fragment suppressed the DICER1 expression (Fig. 7B). Other hAgo2 C-terminal deletions (aa 1 to 530, aa 1 to 610, and aa 1 to 814) were transiently transfected into HeLa cells individually, and none of them suppressed DICER1 expression either (data not shown). Moreover, hAgo2 did not suppress DICER1 expression even when only the C-terminal 29 aa was deleted (aa 1 to 830 of hAgo2) (Fig. 7C). Therefore, almost the entire hAgo2 protein is required to suppress DICER1 expression.
FIG 7.
Western blot of DICER1 protein using samples from HeLa cells transiently transfected with an expression plasmid for hAgo1 (A, upper), hAgo3 (A, bottom), the N-terminal 550 aa of hAgo2 (B, left), the C terminus of hAgo2 (B, right), or hAgo2 (1 to 830 aa) (C).
DISCUSSION
The importance of miRNAs (and/or expression of DICER1 and hAgo2) in human cancers is well established (18). For example, the widespread downregulation of miRNAs is often observed in many human cancers. Indeed, global repression of miRNA biogenesis by suppression of the key components of miRNA processing machinery, such as Drosha, DICER1, and exportin 5, promotes cellular transformation and tumorigenesis (30, 31). It was also reported that c-Myc broadly repressed the transcription of miRNA genes. The results of this study indicated that hAgo2 can modulate DICER1 expression and the processing of its precursor miRNA (Fig. 1 and 2), which should be important in the regulation of miRNA biogenesis and even in cell physiology regulated by miRNA expression, e.g., development (32) and allergy (33).
Exogenously overexpressed hAgo2 (transiently expressed and stably expressed) (Fig. 1A and B) suppressed DICER1 expression in a dose-dependent manner. Furthermore, reduction of hAgo2 by exogenous siRNA increased DICER1 expression in a dose-dependent manner (Fig. 1C). However, reduction of hAgo2 (knockdown by shRNA or knockout by sgRNA) (Fig. 1D and E) increased DICER1 expression but not dose dependently. The lack of dose dependence could be due to experimental variations caused by clonal selection.
In agreement with previous reports, removing let-7 or miR103/107 with sponges upregulated DICER1 expression (Fig. 4B and D). Interestingly, hAgo2 expression was also increased when let-7 was removed (Fig. 4B). Inhibition of hAgo2 expression by let-7 (Fig. 4A) should be caused indirectly through lin-41. It has been reported that let-7 downregulated the expression of lin-41, which stimulated hAgo2 expression (34).
Exogenously overexpressed hAgo2 did not enhance AUF1 expression (Fig. 1B), and knockdown of hAgo2 did not reduce AUF1 expression (Fig. 1D), suggesting that hAgo2 did not regulate AUF1 expression. On the other hand, exogenous transient expression of AUF1 increases hAgo2 levels (Fig. 5C), while knockdown of AUF1 reduces hAgo2 levels (Fig. 5A), indicating that AUF1 regulated the expression of hAgo2. AUF1(s) may regulate hAgo2 expression indirectly through an undefined protein, because it is well known that AUF1(s) can downregulate the expression of many genes through their 3′-UTRs (35). Therefore, AUF1 may downregulate the expression of one specific gene through its 3′-UTR, whose expression then suppresses hAgo2 expression.
The suppression of DICER1 expression by hAgo2 (Fig. 1) and the regulation of hAgo2 expression by AUF1 (Fig. 5A and C) imply that downregulation of DICER1 by AUF1 is through hAgo2. This hypothesis is supported by the finding that downregulation of DICER1 by AUF1 is dependent on hAgo2 (Fig. 5E and F). However, downregulation of DICER1 by hAgo2 is also AUF1 dependent (Fig. 6A). Therefore, the cooperation of hAgo2 and AUF1 is required for the downregulation of DICER1 expression, which is not surprising, since the decay of selected target mRNAs has been reported previously to be controlled by the combinatorial binding of mRNA by AUF1 and hAgo2 proteins (27). Coimmunoprecipitation of hAgo2 and AUF1(s) has been demonstrated previously (36). Our results also showed that RNase A treatment did not disrupt the binding of hAgo2 to AUF1 (Fig. 6C). Our results further demonstrated that hAgo2 could not interact with DICER1 mRNA in the cells depleted of AUF1 (Fig. 6D), and hAgo2 did not reduce the reporter activity of the DICER1 mRNA 3′-UTR when the potential AUF1 binding site was mutated (Fig. 6B), indicating hAgo2 interacts with DICER1 mRNA through AUF1.
It is interesting that the p45 isoform, unlike the other AUF1 isoforms, could be precipitated down more abundantly by hAgo2 protein (Fig. 6C). This result implied that interactions between the p45 isoform of AUF1 and hAgo2 proteins downregulated DICER1 expression. However, none of the AUF1 isoforms alone (p37, p40, p42, or p45) suppressed DICER1 expression (Fig. 5B). Therefore, in addition to the p45 isoform, other AUF1 isoforms may participate in the suppression of DICER1 expression through interactions with the p45 isoform. Further investigations are required to resolve this issue.
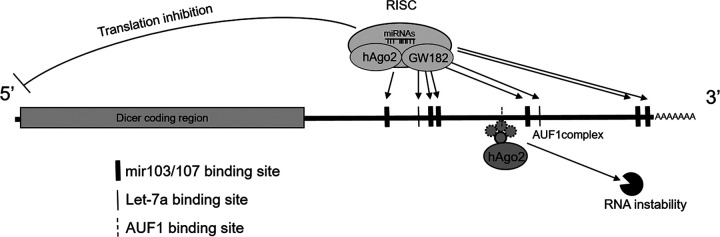
A model is proposed for this study (Fig. 8). DICER1 expression can be regulated at its 3′-UTR through miRNA-dependent and miRNA-independent pathways. In addition to an miRNA (let-7 and miRNA 103/107)-regulated mechanism, hAgo2 and AUF1 can also modulate DICER1 expression at its 3′-UTR through an miRNA-independent pathway.
FIG 8.
Proposed model for this study. DICER1 expression could be downregulated by hAgo2 and different miRNAs (e.g., let-7a or miRNA 103/107). In addition to this mechanism, hAgo2, in collaboration with AUF1s, could also suppress DICER1 expression through an miRNA-independent pathway. In the AUF1 complex, the circle with a solid line represents p45, while the three circles with dotted lines represent p37, p40, and p42.
MATERIALS AND METHODS
Cell culture.
HeLa cells were cultured in RPMI medium 1640 (Gibco) containing 2 g/liter NaHCO3, 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco). A549 cells were cultured in Ham’s F-12 nutrient mixture Kaighn's modification, containing 2.5 g/liter NaHCO3, 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. HuH-7 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 3.7 g/liter NaHCO3, 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. HEK293T cells were cultured in DMEM containing 2 g/liter NaHCO3, 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 1% NEAA (Gibco). All cultured cells were maintained at 37°C with 5% CO2.
Plasmid construction and DNA transfection.
The expression plasmids used in this study were constructed using standard protocols as described in our previous studies (37). The primer sequences used for cloning are available upon request. The plasmid pcDNA3-myc-Ago2 was a gift from G. J. Hannon (38). These expression plasmids were all verified by DNA sequencing. pIS1 DICER1 long UTR (Addgene plasmid number 21649), pIS1 DICER1 long-mut-let-7 (Addgene plasmid number 21651), pIS1 DICER1 long-mut-miR103/107 (Addgene plasmid number 21652), pIS1 DICER1 long-mut-all (Addgene plasmid number 21653), pIS0 (Addgene plasmid number 12178), and pIS1 (Addgene plasmid number 12179) were obtained from David Bartel as a gift (39). Deletion of the AU-rich element (ARE) at DICER1’s 3′-UTR was performed by using a QuikChange Lightning site-directed mutagenesis kit (Agilent, CA) using pIS1 DICER1 long UTR as a template. pGL3-DICER1-Prom (Addgene plasmid number 25851) was a gift from David Fisher (21). The miRNA sponges (23) were purchased from RNAi core facilities (http://rnai.genmed.sinica.edu.tw); 13 repeats of each miRNA-sponge sequence were cloned into pAS7w.mCherry-CMV d2EGFP plasmid (see Table S1 in the supplemental material). The polyethyleneimine (PEI; linear, molecular weight of 25,000) used to transfect DNA into cells was commercially available (Polysciences Inc.).
RNA extraction and real-time RT-PCR.
Total RNAs, extracted from the cells using TRIzol reagent (Invitrogen, Thermo Fisher Scientific) or an miRNeasy minikit (Qiagen, Netherlands) according to the manufacturer’s instructions, were treated with DNase I and converted into cDNAs using oligo(dT) (for the mRNA analysis) or random hexamers (for the precursor miRNA analysis) as the primers. A high-capacity cDNA reverse transcription kit (Applied Biosystems, ThermoFisher Scientific) was used for the reverse transcription. A LabStar SYBR qPCR kit (TAIGEN Bioscience Corporation) was used for the real-time PCR. The primers used for the real-time RT-PCR are listed in Table S1. In these reactions, beta-actin mRNA (for the mRNA analysis) or U6 RNA (for the precursor miRNA analysis [40]) was used as the internal control. All experiments were repeated at least three times, and the real-time RT-PCR procedures followed the MIQE guidelines (41).
Protein expression and Western blotting.
For transient expression, cells (HeLa, HuH7, or A549) were transfected with empty vector or different amounts of expressing plasmids (e.g., hAgo2), as indicated. Forty-eight hours after transfection, proteins from these cells were collected and analyzed with SDS-PAGE, followed by Western blotting. For stable expression, HeLa cells were infected with a lentiviral vector encoding GFP or hAgo2 (duplicate). After puromycin selection, proteins from these cells were collected and analyzed with SDS-PAGE, followed by Western blotting. The Western blotting results are representative of three or more experiments.
Our previous procedures were followed for Western blotting (42). The primary antibodies used for the analyses in this study were antibodies against DICER1, hAgo2, DGCR8, Drosha, XPO5, TRBP, and PACT (Abcam, Cambridge, UK), against Myc tag clone 4A6 (Merck Millipore), against V5 tag (Bio-Rad), against beta-actin and alpha-tubulin (Genetex), and against GFP and GW182 proteins (Santa Cruz Biotechnology). In this assay, beta-actin or alpha-tubulin protein was used as the loading control.
siRNA and shRNA knockdown experiments.
The siRNA against hAgo2 was purchased from Dharmacon company using the sequence published previously (43). The shRNA knockdown (e.g., hAgo2 or AUF1) experiments were conducted using the lentiviral expression system (http://rnai.genmed.sinica.edu.tw) according to the manufacturer’s instructions and our previous procedures (44). Two shRNA clones with different target sequences against one specific cellular gene were used. The shRNA knockdown reagents were obtained from the National RNAi Core Facility located at the Institute of Molecular Biology/Genomic Research Center, Academia Sinica, Taiwan.
CRISPR/Cas9 knockout of hAgo2.
To knock out hAgo2 in A549 and HeLa cells, all-in-one lenti CRISPR/Cas9 plasmid (pAll-Cas9.Ppuro) with an sgRNA-targeted human EIF2C2 gene was used (http://rnai.genmed.sinica.edu.tw). Two sgRNA clones with different target sequences were used. A549 or HeLa cells were cultured and infected with recombinant lentiviral vector with Cas9-sgAgo2. Twenty-four hours after infection, the cells were trypsinized and serially diluted in 96-well plates with 2 μg/ml puromycin-containing DMEM for the selection of stable clones. Every stable cell clone was picked and further amplified in 24-well plates. Proteins from each stable cell clone were collected and analyzed with SDS-PAGE, followed by Western blotting. The hAgo2 protein amount in these stable knockout clones, compared with that of control cells, was reduced, but not completely eliminated (data not shown). For each sgRNA, only one cell cone with the least hAgo2 expression was selected for sequencing to verify the gene mutation after the amplification of DNA fragments around the target site. Two different knockout cell clones (one for each sgRNA) from each cell type then were used for further analysis.
Luciferase assay.
HeLa cells (1 × 105) were transfected with the reporter (pIS1 DICER1 long UTR, pIS1 DICER1 long-mut-let-7, pIS1 DICER1 long-mut-miR103/107, pIS1 DICER1 long-mut-all, or pIS1 DICER1 long UTR AREM), the internal control plasmid (pIS0), and different expressing plasmids (e.g., hAgo2). Cells were harvested 72 h after transfection. The Dual-Luciferase assay system (Promega) was used for the luciferase assays by following the manufacturer’s instructions and our previous procedures (44). In each experiment, triplicate samples were analyzed. The results shown were the averages from three different experiments. Analysis of variance (ANOVA) was used to compare the means among groups. The post hoc Tukey test was used to test the means between two groups.
Coimmunoprecipitation and RNA immunoprecipitation.
The coimmunoprecipitation assay was conducted in HEK293T cells by following our previous procedures (44). In this experiment, expression plasmids for pcDNA3-myc-hAgo2 and/or pcDNA3.1-AUF1 for all four isoforms were used. Protein lysates were collected and treated with or without 3 μg RNase A for 10 min at 25°C. RNA immunoprecipitation was done by following the protocol provided by Abcam (https://docs.abcam.com/pdf/protocols/rip-protocol.pdf), with modifications. In brief, protein lysates were collected and incubated with either control IgG or Ago2 antibody at 4°C overnight. Protein A magnetic beads were added to precipitate the antibody-bound proteins, followed by purification of RNA with TRIzol reagent (Invitrogen, Thermo Fisher Scientific). RNAs were reverse transcribed to cDNA with random hexamer and analyzed by real-time PCR.
Supplementary Material
ACKNOWLEDGMENTS
We thank G. J. Hannon, D. Bartel, and D. Fisher for providing the expression plasmids. The RNAi reagents were obtained from the National RNAi Core Facility, located at the Institute of Molecular Biology/Genomic Research Center, Academia Sinica, which is supported by grants from the NSC National Research Program for Genomic Medicine (NSC 94-3112-B-001-003 and NSC 94-3112-B-001-018-Y).
This work was supported by grants from the Ministry of Science and Technology, Republic of China (MOST 106-2320-B-320-009-MY3), to Shih-Yen Lo and from the Tzu Chi University to Shih-Yen Lo (TCIRP106001-04Y1) and to Hui-Chun Li (TCIRP106001-02Y1).
We have no competing interests to declare.
Authors made the following contributions: designed research, S.-Y.L.; performed research, C.-H.Y., T.-S.K., C.-H.W. and K.-C.S.; analyzed data, H.-C.L. and S.-Y.L.; wrote the paper, H.-C.L. and S.-Y.L.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Kim YK, Kim B, Kim VN. 2016. Re-evaluation of the roles of DROSHA, Exportin 5, and DICER in microRNA biogenesis. Proc Natl Acad Sci U S A 113:E1881–E1889. doi: 10.1073/pnas.1602532113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ha M, Kim VN. 2014. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 3.Slezak-Prochazka I, Durmus S, Kroesen BJ, van den Berg A. 2010. MicroRNAs, macrocontrol: regulation of miRNA processing. RNA 16:1087–1095. doi: 10.1261/rna.1804410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen J, Hung MC. 2015. Signaling-mediated regulation of MicroRNA processing. Cancer Res 75:783–791. doi: 10.1158/0008-5472.CAN-14-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun HL, Cui R, Zhou J, Teng KY, Hsiao YH, Nakanishi K, Fassan M, Luo Z, Shi G, Tili E, Kutay H, Lovat F, Vicentini C, Huang HL, Wang SW, Kim T, Zanesi N, Jeon YJ, Lee TJ, Guh JH, Hung MC, Ghoshal K, Teng CM, Peng Y, Croce CM. 2016. ERK activation globally downregulates miRNAs through phosphorylating exportin-5. Cancer Cell 30:723–736. doi: 10.1016/j.ccell.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. 2003. Dicer is essential for mouse development. Nat Genet 35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 7.Hata A, Lieberman J. 2015. Dysregulation of microRNA biogenesis and gene silencing in cancer. Sci Signal 8:re3. doi: 10.1126/scisignal.2005825. [DOI] [PubMed] [Google Scholar]
- 8.Huang JT, Wang J, Srivastava V, Sen S, Liu SM. 2014. MicroRNA machinery genes as novel biomarkers for cancer. Front Oncol 4:113. doi: 10.3389/fonc.2014.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. 2005. MicroRNA expression profiles classify human cancers. Nature 435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 10.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. 2008. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene 27:1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 11.Swahari V, Nakamura A, Deshmukh M. 2016. The paradox of dicer in cancer. Mol Cell Oncol 3:e1155006. doi: 10.1080/23723556.2016.1155006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hinkal GW, Grelier G, Puisieux A, Moyret-Lalle C. 2011. Complexity in the regulation of Dicer expression: Dicer variant proteins are differentially expressed in epithelial and mesenchymal breast cancer cells and decreased during EMT. Br J Cancer 104:387–388. doi: 10.1038/sj.bjc.6606022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurzynska-Kokorniak A, Koralewska N, Pokornowska M, Urbanowicz A, Tworak A, Mickiewicz A, Figlerowicz M. 2015. The many faces of Dicer: the complexity of the mechanisms regulating Dicer gene expression and enzyme activities. Nucleic Acids Res 43:4365–4380. doi: 10.1093/nar/gkv328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z. 2007. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell 129:1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 15.Morita S, Horii T, Kimura M, Goto Y, Ochiya T, Hatada I. 2007. One Argonaute family member, Eif2c2 (Ago2), is essential for development and appears not to be involved in DNA methylation. Genomics 89:687–696. doi: 10.1016/j.ygeno.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Ye Z, Jin H, Qian Q. 2015. Argonaute 2: a novel rising star in cancer research. J Cancer 6:877–882. doi: 10.7150/jca.11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White EJ, Matsangos AE, Wilson GM. 2017. AUF1 regulation of coding and noncoding RNA. Wiley Interdiscip Rev RNA 8:10.1002/wrna.1393. doi: 10.1002/wrna.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, Enzo E, Guzzardo V, Rondina M, Spruce T, Parenti AR, Daidone MG, Bicciato S, Piccolo S. 2010. A MicroRNA targeting dicer for metastasis control. Cell 141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 19.Tokumaru S, Suzuki M, Yamada H, Nagino M, Takahashi T. 2008. let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis 29:2073–2077. doi: 10.1093/carcin/bgn187. [DOI] [PubMed] [Google Scholar]
- 20.Hayashi M, Saito Y, Kawashima S. 1992. Calpain activation is essential for membrane fusion of erythrocytes in the presence of exogenous Ca2+. Biochem Biophys Res Commun 182:939–946. doi: 10.1016/0006-291x(92)91822-8. [DOI] [PubMed] [Google Scholar]
- 21.Levy C, Khaled M, Robinson KC, Veguilla RA, Chen PH, Yokoyama S, Makino E, Lu J, Larue L, Beermann F, Chin L, Bosenberg M, Song JS, Fisher DE. 2010. Lineage-specific transcriptional regulation of DICER by MITF in melanocytes. Cell 141:994–1005. doi: 10.1016/j.cell.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abdelmohsen K, Tominaga-Yamanaka K, Srikantan S, Yoon JH, Kang MJ, Gorospe M. 2012. RNA-binding protein AUF1 represses Dicer expression. Nucleic Acids Res 40:11531–11544. doi: 10.1093/nar/gks930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ebert MS, Neilson JR, Sharp PA. 2007. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfaff J, Hennig J, Herzog F, Aebersold R, Sattler M, Niessing D, Meister G. 2013. Structural features of Argonaute-GW182 protein interactions. Proc Natl Acad Sci U S A 110:E3770–E3779. doi: 10.1073/pnas.1308510110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jakymiw A, Lian S, Eystathioy T, Li S, Satoh M, Hamel JC, Fritzler MJ, Chan EK. 2005. Disruption of GW bodies impairs mammalian RNA interference. Nat Cell Biol 7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- 26.Wagner BJ, DeMaria CT, Sun Y, Wilson GM, Brewer G. 1998. Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms. Genomics 48:195–202. doi: 10.1006/geno.1997.5142. [DOI] [PubMed] [Google Scholar]
- 27.Wu X, Chesoni S, Rondeau G, Tempesta C, Patel R, Charles S, Daginawala N, Zucconi BE, Kishor A, Xu G, Shi Y, Li ML, Irizarry-Barreto P, Welsh J, Wilson GM, Brewer G. 2013. Combinatorial mRNA binding by AUF1 and Argonaute 2 controls decay of selected target mRNAs. Nucleic Acids Res 41:2644–2658. doi: 10.1093/nar/gks1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoon JH, Jo MH, White EJ, De S, Hafner M, Zucconi BE, Abdelmohsen K, Martindale JL, Yang X, Wood WH III, Shin YM, Song JJ, Tuschl T, Becker KG, Wilson GM, Hohng S, Gorospe M. 2015. AUF1 promotes let-7b loading on Argonaute 2. Genes Dev 29:1599–1604. doi: 10.1101/gad.263749.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hock J, Meister G. 2008. The Argonaute protein family. Genome Biol 9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang LI, Wang C, Liu S, Zhao Y, Liu C, Guo Z. 2016. Prognostic significance of Dicer expression in hepatocellular carcinoma. Oncol Lett 11:3961–3966. doi: 10.3892/ol.2016.4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaksman O, Hetland TE, Trope CG, Reich R, Davidson B. 2012. Argonaute, Dicer, and Drosha are up-regulated along tumor progression in serous ovarian carcinoma. Hum Pathol 43:2062–2069. doi: 10.1016/j.humpath.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 32.Neilson JR, Zheng GX, Burge CB, Sharp PA. 2007. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev 21:578–589. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu TX, Rothenberg ME. 2018. MicroRNA. J Allergy Clin Immunol 141:1202–1207. doi: 10.1016/j.jaci.2017.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rybak A, Fuchs H, Hadian K, Smirnova L, Wulczyn EA, Michel G, Nitsch R, Krappmann D, Wulczyn FG. 2009. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat Cell Biol 11:1411–1420. doi: 10.1038/ncb1987. [DOI] [PubMed] [Google Scholar]
- 35.DeMaria CT, Brewer G. 1996. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J Biol Chem 271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- 36.Chang N, Yi J, Guo G, Liu X, Shang Y, Tong T, Cui Q, Zhan M, Gorospe M, Wang W. 2010. HuR uses AUF1 as a cofactor to promote p16INK4 mRNA decay. Mol Cell Biol 30:3875–3886. doi: 10.1128/MCB.00169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen JS, Li HC, Lin SI, Yang CH, Chien WY, Syu CL, Lo SY. 2015. Cleavage of Dicer protein by I7 protease during vaccinia virus infection. PLoS One 10:e0120390. doi: 10.1371/journal.pone.0120390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 39.Mayr C, Bartel DP. 2009. Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang J, Lee EJ, Gusev Y, Schmittgen TD. 2005. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res 33:5394–5403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 42.Yang CH, Li HC, Shiu YL, Ku TS, Wang CW, Tu YS, Chen HL, Wu CH, Lo SY. 2017. Influenza A virus upregulates PRPF8 gene expression to increase virus production. Arch Virol 162:1223–1235. doi: 10.1007/s00705-016-3210-3. [DOI] [PubMed] [Google Scholar]
- 43.Wilson JA, Zhang C, Huys A, Richardson CD. 2011. Human Ago2 is required for efficient microRNA 122 regulation of hepatitis C virus RNA accumulation and translation. J Virol 85:2342–2350. doi: 10.1128/JVI.02046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang CH, Li HC, Ku TS, Wu PC, Yeh YJ, Cheng JC, Lin TY, Lo SY. 2017. Hepatitis C virus down-regulates SERPINE1/PAI-1 expression to facilitate its replication. J Gen Virol 98:2274–2286. doi: 10.1099/jgv.0.000901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.