Abstract
Motivation
Identification of new molecules promising for treatment of HIV-infection and HIV-associated disorders remains an important task in order to provide safer and more effective therapies. Utilization of prior knowledge by application of computer-aided drug discovery approaches reduces time and financial expenses and increases the chances of positive results in anti-HIV R&D. To provide the scientific community with a tool that allows estimating of potential agents for treatment of HIV-infection and its comorbidities, we have created a freely-available web-resource for prediction of relevant biological activities based on the structural formulae of drug-like molecules.
Results
Over 50 000 experimental records for anti-retroviral agents from ChEMBL database were extracted for creating the training sets. After careful examination, about seven thousand molecules inhibiting five HIV-1 proteins were used to develop regression and classification models with the GUSAR software. The average values of R2 = 0.95 and Q2 = 0.72 in validation procedure demonstrated the reasonable accuracy and predictivity of the obtained (Q)SAR models. Prediction of 81 biological activities associated with the treatment of HIV-associated comorbidities with 92% mean accuracy was realized using the PASS program.
Availability and implementation
Freely available on the web at http://www.way2drug.com/hiv/.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
According to World Health Organization estimates (https://www.who.int/gho/hiv/en/), in 2017 about 36.9 million people live with HIV/AIDS worldwide. Highly active anti-retroviral therapy represented by combination of 2–4 anti-retroviral drugs maximally suppresses the HIV virus, terminates the progression of HIV disease and prevents onward transmission of HIV. However, the existing medicines do not provide a complete cure, exhibit severe adverse effects and cause the appearance of resistant strains (Geronikaki et al., 2016). Therefore, the further efforts on the discovery of new safer and more potent anti-HIV agents are needed (Zhan et al., 2016).
Computational prediction of HIV inhibitors applied at the early stages of anti-retroviral drug discovery decreases the number of compounds to be synthesized and tested. The only web server for estimating of anti-retroviral activity (Qureshi et al., 2018) predicts inhibition of HIV-1 protease, reverse transcriptase and integrase with moderate predictivity (Pearson correlation coefficient ranged from 0.68 to 0.78 for different targets and various endpoints). Since the strong performance of GUSAR in comparison with many other popular (Q)SAR methods has been demonstrated (Filimonov et al., 2009; Zakharov et al., 2014a), one may expect that this approach will provide reasonable accuracy and predictivity of anti-retroviral activity. We decided to additionally provide an estimation of biological activities relevant to the therapy of HIV-associated comorbidities, to estimate the multi-target action of compounds (Anighoro et al., 2014; Croset et al., 2014).
2 Materials and methods
Over 50 000 experimental records for anti-retroviral activities were extracted from the ChEMBL database version 24 (www.ebi.ac.uk/chembl/). After careful curation in accordance with the modern recommendations (Fourches et al., 2016; Tropsha, 2010), we obtained training sets of 1440 HIV-1 protease (PR, catalytic site) inhibitors, 1393 reverse transcriptase (RT, polymerase action) inhibitors, 1436 integrase (IN, strand transfer) inhibitors, 2566 TAT inhibitors and 105 Rev inhibitors (RRE RNA interaction), respectively, for further modeling. All information about curation procedure, modeling methods of GUSAR and PASS software is available in Supplementary Material (Sections S1–S5). Training sets are also available (Section S8).
All five targets were characterized by IC50 as the endpoints of the particular inhibitory activity. On average, pIC50 values varied from 5 to 10 negative log units; thus, this data may be used for development of (Q)SAR models (Zakharov et al., 2014b). To discriminate the active compounds, the cutoff IC50 < 1 μM was applied.
3 Results
For PR, RT, IN and Rev inhibitors QSAR models with reasonable accuracy and predictivity estimated by the leave-many-out cross validation procedure were built using GUSAR (Table 1).
Table 1.
Characteristics of the QSAR models
| HIV-1 target | N | R 2 | Q 2 | RMSE |
|---|---|---|---|---|
| PR | 1440 | 0.966 | 0.810 | 0.64 |
| RT | 1393 | 0.943 | 0.677 | 0.60 |
| IN | 1436 | 0.969 | 0.830 | 0.56 |
| Rev | 105 | 0.933 | 0.554 | 0.34 |
Note: N: number of compounds in the training set; R2: square of regression coefficient; Q2: cross-validated R2; RSME: root-mean-square error.
An example plot of predicted versus observed endpoint values is given in Figure 1 for PR inhibitors.
Fig. 1.
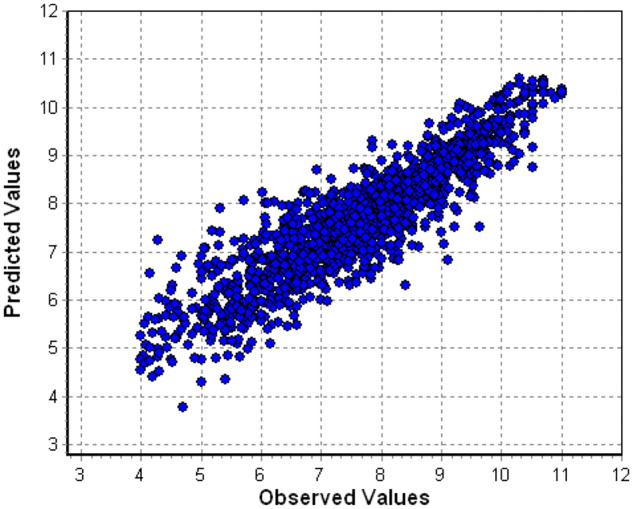
Protease inhibitors: predicted vs. observed pIC50 values
For TAT inhibitors QSAR models with good predictivity could not be built, which may be explained by the high heterogeneity of the experimental conditions under which this endpoint has been measured. However, we were able to obtain classification models with reasonable characteristics: N = 2566; Sensitivity = 0.966; Specificity = 0.810 and G-mean = 0.822. Examples of prediction are given in Supplementary Material (Section S7).
Eighty-one biological activities relevant for treatment of HIV-associated comorbidities may be estimated using classification method implemented in PASS (Filimonov et al., 2018). The list of biological activities is given in Supplementary Material (Section S6).
On the basis of these GUSAR and PASS models, we have developed a freely available web resource to provide predictions for anti-retroviral targets inhibition and HIV-associated comorbidities treatment based on the structural formula of a compound under study.
The user can draw a chemical structure and choose the particular HIV target of interest. Prediction can be obtained for single-component, uncharged, structures with a molecular weight of less than 1250 Da and having at least three carbon atoms.
The user has immediate access to predictions for compounds taken from the Open NCI database (https://cactus.nci.nih.gov/download/nci/) that was developed in the framework of the NCI Developmental Therapeutics Program (DTP). For some compounds samples from the Open NCI database may be available from DTP for testing in bioassays (https://dtp.cancer.gov/organization/dscb/obtaining/default.htm).
Funding
This work was supported by the RFBR-NIH grant No. 17-54-30015-NIH_a.
Conflict of Interest: none declared.
Supplementary Material
References
- Anighoro A. et al. (2014) Polypharmacology: challenges and opportunities in drug discovery. J. Med. Chem., 57, 7874.. [DOI] [PubMed] [Google Scholar]
- Croset S. et al. (2014) The functional therapeutic chemical classification system. Bioinformatics, 30, 876.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filimonov D.A. et al. (2009) QNA based ‘Star Track’ QSAR approach. SAR QSAR Environ. Res., 20, 679.. [DOI] [PubMed] [Google Scholar]
- Filimonov D.A. et al. (2018) Computer-aided prediction of biological activity spectra for chemical compounds: opportunities and limitations. Biomed. Chem. Res. Meth., 1, e00004. [Google Scholar]
- Fourches D. et al. (2016) Trust, but verify II: a practical guide to chemogenomics Data Curation. J. Chem. Inf. Model, 56, 1243.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geronikaki A. et al. (2016) Anti-HIV agents: current status and recent trends. Topics Med. Chem., 29, 37. [Google Scholar]
- Qureshi A. et al. (2018) HIVprotI: an integrated web based platform for prediction and design of HIV proteins inhibitors. J. Cheminform., 10, 12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropsha A. (2010) Best practices for QSAR model development, validation, and exploitation. Mol. Inf., 29, 476.. [DOI] [PubMed] [Google Scholar]
- Zakharov A.V. et al. (2014a) A new approach to radial basis function approximation and its application to QSAR. J. Chem. Inf. Model, 54, 713.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharov A.V. et al. (2014b) QSAR modeling of imbalanced high-throughput screening data in PubChem. J. Chem. Inf. Model, 54, 705.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan P. et al. (2016) Anti-HIV drug discovery and development: current innovations and future trends. J. Med. Chem., 59, 2849.. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


