Abstract
With obesity rate consistently increasing, a strong relationship between obesity and fatty liver disease has been discovered. More than 90% of bariatric surgery patients also have non-alcoholic fatty liver diseases (NAFLDs). NAFLD and non-alcoholic steatohepatitis (NASH), which are the hepatic manifestations of metabolic syndrome, can lead to liver carcinogenesis. Unfortunately, there is no effective medicine that can be used to treat NASH or liver cancer. Thus, it is critically important to understand the mechanism underlying the development of these diseases. Extensive evidence suggests that microRNA 22 (miR-22) can be a diagnostic marker for liver diseases as well as a treatment target. This review paper focuses on the roles of miR-22 in metabolism, steatosis, and liver carcinogenesis. Literature search is limited based on the publications included in the PubMed database in the recent 10 years.
Keywords: MicroRNA-22(miR-22), Cancer, Liver, Metabolism, Steatosis, Non-alcoholic steatohepatitis, Hepatitis
1. Introduction
MicroRNA 22 (miR-22) is highly conserved across vertebrate species and its expression is ubiquitously expressed in various organs.1–5 The miR-22 gene is located on chromosome 17p13, its cDNA catalyzed by RNA polymerase II is ~1.3 kb. In addition, its transcription start site lacks TATA box.6 Many studies have revealed that miR-22 is implicated in the development of various types of cancer including liver, colon, prostatic, breast cancer, gastric cancers, and many others. In general, miR-22 is considered as a metabolic silencer and a tumor-suppressor. However, its oncogenic effect has been documented as well. Furthermore, miR-22 has many biological functions including inflammatory and immune regulation; arterial smooth muscle cell proliferation and migration regulation; and cardiac and vascular remodeling.7–9 In this review paper, we summarize the role of miR-22 in liver disease development.
2. MiR-22 as a metabolic silencer
MiR-22 is an important regulator of dyslipidemia. It has been shown that miR-22 deficiency prevents high-fat diet (HFD)-induced dyslipidemia by inhibiting the expression of genes (sterol regulatory element binding protein-1 (Srebp-1), CC motif chemokine ligand 2 (Ccl2), interleukin 6 (Il-6), and interferon gamma (Ifng). Thus, miR-22 promotes lipogenesis and inflammation.10
MiR-22 along with miR-34a are up-regulated in the liver of diabetic db/db mice. miR-22 reduces the levels of E1A binding protein p300 (Ep300) as well as transcription factor 7 (Tcf7), and miR-34a decreases the protein level of its target gene Wnt Family Member 1 (Wnt1). Overexpression of miR-22 and miR-34a inhibits Wnt signaling, which leads to increased lipid accumulation in HepG2 cells.11
Fibroblast growth factor 21 (FGF21) is a master metabolic regulator that has a remarkable ability to reverse diabetes and obesity. In addition, FGF21 has regenerative capability and repairs injured tissue. Activation of FGF21 leads to AMPK and ERK1/2 activation. Given the role of FGF21 in metabolism and proliferation, its functions require regulation to avoid metabolism-driven overgrowth, which can be tumorigenic. A recent study has established the relationship between miR-22 and FGF21 and its receptor fibroblast growth factor receptor 1 (FGFR1) expression.12 The levels of miR-22 and FGF21, FGFR1, as well as peroxisome proliferator-activated receptor gamma coactivator 1α (PGC1α) were inversely correlated in human and mouse fatty livers, suggesting that hepatic miR-22 acts as a metabolic silencer. Further mechanistic analysis revealed that miR-22 directly targeted FGFR1. However, miR-22 decreased FGF21 by reducing the occupancy of transcriptional factors peroxisome proliferator-activated receptor α (PPAR α) and PGC1α to their binding motifs. Thus, miR-22 can be considered as a metabolic silencer by inhibiting the expression of FGF21 and its receptor.12 The genes regulated by miR-22 to reduce metabolism are summarized in Table 1.
Table 1.
MiR-22 as a metabolic silencer.
| Regulated genes | Function | Refs. |
|---|---|---|
| Srebp-1, Ccl2, Il-6, Ifng | Lack of miR-22 prohibits fat mass formation and dyslipidemia caused by a high-fat diet | 10 |
| Ep300, Tcf7 | MiR-22 inhibits Wnt signaling leading to increased lipid accumulation in HepG2 cells | 11 |
| FGFR1, FGF21 | Increased hepatic miR-22 and reduced FGF21 are found in hepatic steatosis | 12 |
Abbreviations: miR-22,microRNA 22; Srebp-1, sterol regulatory element binding protein-1; Ccl2, CC motif chemokine ligand 2; Il-6, interleukin 6; Ifng, interferon gamma; Ep300, E1A binding protein p300; Tcf7, transcription factor 7; FGFR1, fibroblast growth factor receptor 1; FGF21, fibroblast growth factor 21.
3. MiR-22 in hepatic steatosis and fibrosis
In consistency with the negative role of miR-22 in regulating hepatic lipid metabolism, miR-22 is increased in various drug-induced steatosis including drugs like valproate, doxycycline, cyclosporin A, and tamoxifen. MiR-22 is a potential biomarker for drug-induced steatosis and can be used to predict the effect of a drug on steatosis development.13 Hepatic miR-22 overexpression also enhances diet and alcohol-induced steatosis. In contrast, reducing miR-22 level up-regulates hepatic FGF21 and FGFR1, leading to AMPK and ERK1/2 activation, which effectively improve alcoholic steatosis in mouse models.12
A combination of serum miR-22 and miR-210, which distinguish F0 fibrosis from any fibrosis, can be noninvasive diagnostic biomarkers to detect the presence of liver fibrosis in children with cystic fibrosis.14
Furthermore, miR-22 levels are inversely correlated with the bone morphogenic protein 7 (BMP7) levels in human livers. BMP7 inhibits the progress of liver cirrhosis by inhibiting the expression of transforming growth factor beta 1 (TGF-β1), blocking the nuclear accumulation of SMAD family member 2/3 (Smad2/3), or increasing the level of Gremlin protein secreted by fibroblasts.15–17 In a carbon tetrachloride-induced cirrhosis mouse models, adeno-associated viruses carrying antisense of miR-22 significantly attenuated the levels of liver fibrosis, portal hypertension, as well as sodium retention caused, possibly by upregulation of BMP7. Thus, increased miR-22 promotes liver cirrhosis through directly targeting BMP7.18 In consistency, microarray screening study showed that “mmu_circ_34116/miR-22-3P/BMP7” signal axis might be involved in the activation of hepatic stellated cells. Furthermore, transfection experiment validated that the expression of alpha-smooth muscle actin (α-SMA) is significantly elevated because of inhibitory expression of mmu_circ_34,116.19
However, miR-22 inhibits galectin-1 and 9, which are implicated in the development of hepatic fibrosis.20,21 Down-regulation of galectin-1 can improve liver fibrosis by reducing α-SMA, desmin, alanine aminotransferase (ALT), aspartate aminotransferase (AST), and total bilirubin.22 Serum galectin-9 levels are positive correlation with liver fibrosis.23 Thus, the role of miR-22 in liver diseases can be complicated. Whether miR-22 via reducing galectins can treat hepatic fibrosis remains to be studied.
4. MiR-22 and viral hepatitis
Serum miR-22 and miR-1275 are up-regulated in hepatitis B virus (HBV) patients. The level of those miRNAs are positively correlated with the serum γ-glutamyl transpeptidase levels.24 In consistency, serum level of miR-22 and miR-122 are increased in chronic HBV patients.25,26 Additionally, their expression levels are positively associated with hepatitis B surface antigen (HBsAg) levels and ALT levels. Similarly, elevated circulating miR-22 is found in human immunodeficiency virus (HIV)/hepatitis C virus (HCV) patients and is involved in the etiology of liver injury in HIV patients.27 Further, elevated circulating miR-22 and miR-122 indicates viral replication and liver injury in HBV patients.26
5. Long non-coding RNA (lncRNA) MIR22HG as a tumor suppressor for hepatocellular carcinoma (HCC)
Based on genome-wide lncRNA expression profiles in HCC tissues and paired adjacent non-tumor tissues, lncRNA NR_028502.1 located in 17p13.3, a chromosomal region that is frequently deleted or hypermethylated in liver cancer, is down-regulated in HCC.28,29 NR_028502.1 is annotated as the human miR-22 host gene (MIR22HG).30 Moreover, reduced MIR22HG is related to tumor progression and poor prognosis of patients with HCC. MIR22HG overexpression inhibits proliferation, invasion, and metastasis in HCC cells. In part, LncRNA MIR22HG acts a tumor suppressor for HCC through deriving miR-22–3p to target high mobility group box 1 (HMGB1), thereby inactivating HMGB1 downstream pathways.30
6. MiR-22 as a tumor suppressor
MiR-22 expression levels were analyzed in different types of cancer using information available from the TCGA Data Portal. The studies that have normal specimen number greater than 15 were included in the analysis. The data showed that in comparison with normal specimens, miR-22 levels were differentially expressed based on cancer types. In comparison with normal specimens, its level was reduced in HCC, breast invasive carcinoma, and lung squamous cell carcinoma (Fig. 1A and B).
Fig. 1. MiR-22 expression level in different cancers.
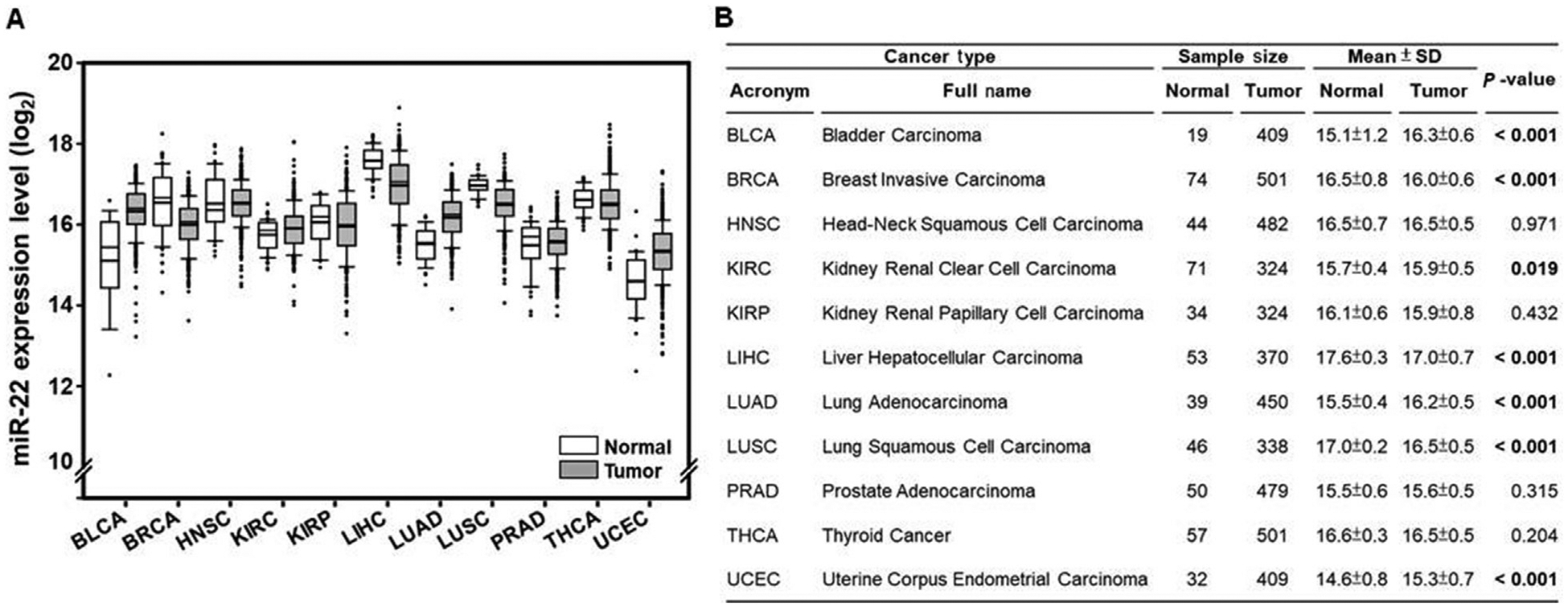
The miR-22 expression level (log2) was analyzed using TCGA Data Portal and data are shown as box plot (white box, normal specimens; gravy box: cancer specimens).
We further analyzed the relationships between miR-22 levels and HCC clinical features. The data showed that the level of miR-22 was inversely associated with the depth of HCC invasion. T3 and T4 cancers had lower miR-22 level compared with T1 and T2 (Fig. 2A and E). In addition, HCC patients at stage III or IV had lower miR-22 level than those at stages I and II (Fig. 2B and E). Furthermore, miR-22 expression level was positively correlated with overall survival and disease-free survival (Fig. 2C–E). Thus, miR-22 can be considered as tumor suppressor for HCC.
Fig. 2. The associations between miR-22 levels and HCC clinical features.
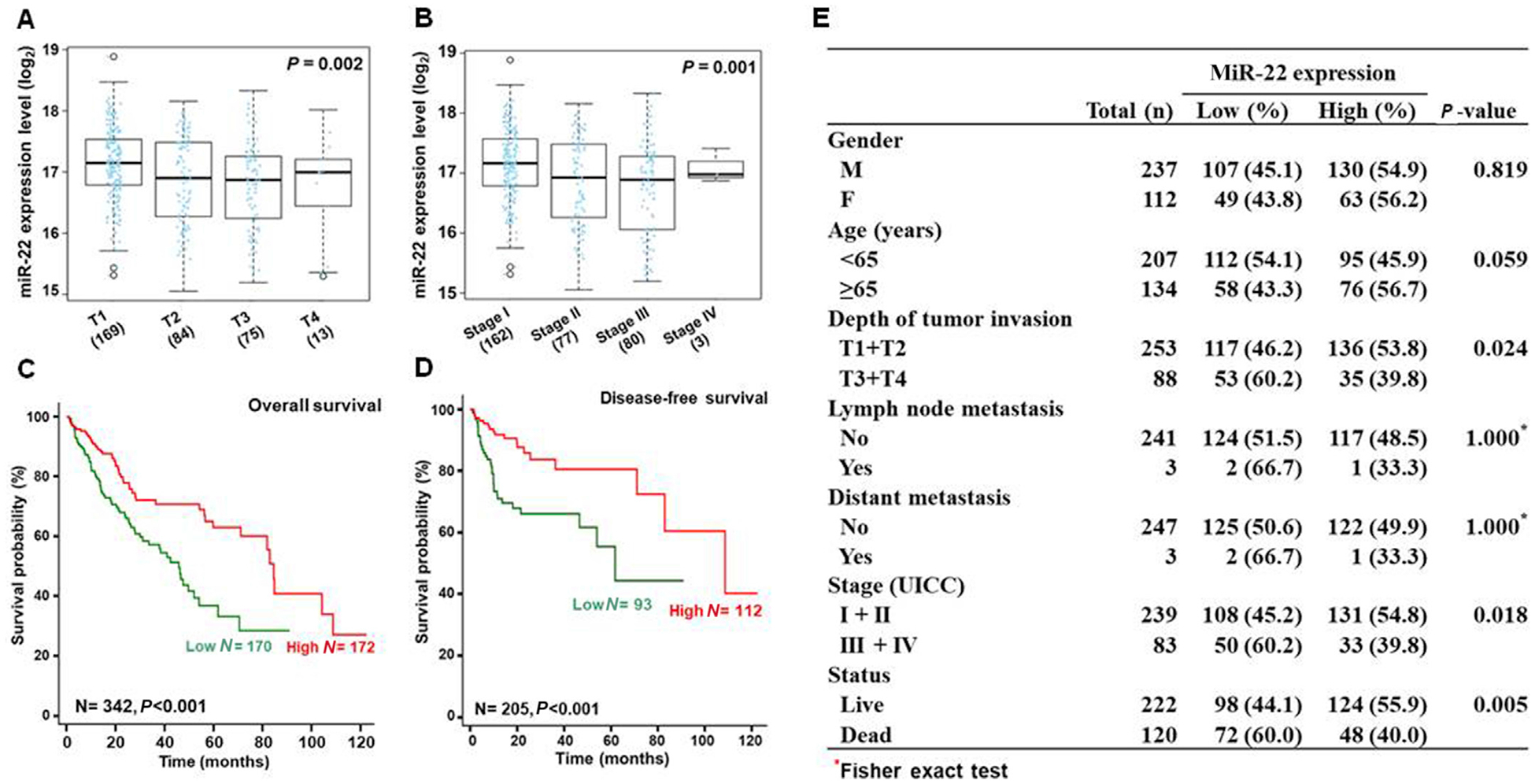
The correlation between miR-22 expression level (log2) and (A) the depth of tumor invasion and (B) tumor stages. Kaplan-Meier curves showed the relationships between miR-22 levels and (C) overall survival and (D) disease-free survival. P-values were calculated by the log-rank test. Clinical features and P values are summarized in (E).
It has been shown that low expression of miR-22 is associated with poor prognosis in hepatoma patients.31 In addition, reduced hepatic or serum miR-22 is shown in HBV-associated HCC patients. However, no significant difference of serum miR-22 levels was found between benign liver disease and non-HBV-related HCC patients. In consistency with our data analysis, miR-22 levels were negatively correlated with tumor size, lymph node metastasis, TNM stage, pathological type, differentiation grade, liver cirrhosis, serum alpha-fetoprotein (AFP) and HBV DNA copy number.32 Moreover, another study also shows that serum miR-22 and miR-199a-3p in combined with AFP have a high accuracy in early detection of HCC patients with chronic hepatitis C (Table 2).33
Table 2.
MiR-22 as a liver cancer diagnostic marker.
| Diagnostic indicator | Disease | Refs. |
|---|---|---|
| Reduced expression of miR-22 | Poor prognosis in hepatoma in patients | 31 |
| Reduced hepatic or serum miR-22 | HBV-associated HCC patients. | 32 |
| Serum miR-22 and miR-199a-3p in combination with AFP | Early phase of HCC in patients with chronic HCV | 33 |
Abbreviations: miR, microRNA; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; AFP, alpha-fetoprotein.
7. The mechanism by which miR-22 acts as a tumor suppressor
Several mechanisms by which miR-22 acts as a liver cancer suppressor have been uncovered. miR-22 inhibits the development of HCC through directly targeting LncRNA NEAT1 and AKT serine/threonine kinase 2 (AKT2), which are overexpressed in human HCC specimens in comparison with adjacent normal tissue.34 Both NEAT1 and AKT2 are implicated in the development of HCC by increasing proliferation and invasion while inhibiting apoptosis in HCC cells.34–36
MiR-22 can also directly target the heterogeneous nuclear ribonucleoprotein A1 (HNRNPA1), a potential diagnostic marker for HBV-related HCC.37 As an oncogene, HNRNPA1 promotes HBV-related HCC via the EGFR signaling pathway. Additionally, HNRNPA1 is negatively correlated with the overall survival of HCC patients.
MiR-22 is reduced in folate deficiency-conditioned HCC cell lines including SK-Hep1 and Mahlavu.38 MiR-22 overexpression reduces the number of spheres in both liver cancer Sk-Hep1 and Mahlavu (MDA-MB-453) cells, and the opposite is observed by inhibiting miR-22. It has been shown that reduced miR-22 causes folate deficiency-induced cancer stem-like phenotypes via increasing histone deacetylase 4 (HDAC4), zinc finger E-box binding homeobox 2 (ZEB2), and octamer-binding transcription factor 4 (OCT4), but decreasing paired related homeobox 1 (PRRX1).38
MiR-22 also silences galectin-1 and 9, which specifically bind to β-galactoside sugars. Galectin-1 is overexpressed in HCC and promotes HCC progression.39–41 The expression of miR-22 is negatively correlated with the expression of galectin-1. The expression of galectin-1 is increased in hepatic stellate cells (HSCs) isolated from HCC tissues. MiR-22 inhibits the HSC-induced T cell apoptosis and cytokine production promoted by HSC-derived galectin-1 in HCC.20 Moreover, elevated galectin-1 and low CD3 expression levels is associated with poor prognosis in HCC patients. Further, Galectin-9 is increased while miR-22 is decreased in human liver cancer tissues and cell lines. MiR-22 inhibits lymphocyte apoptosis and tumor cell proliferation in HCC cells via silencing galectin-9.21
MiR-22 can directly target cell cycle gene expression.42 Cyclin A2 (CCNA2) is a direct miR-22 target gene in both liver and colon cancer cells.42 MiR-22 overexpression as well as chemicals that induce miR-22 expression can reduce CCNA2 protein and increase the number of G0/G1 in human liver cancer Huh7 and colon cancer HCT116 cells.42
Silencing multiple protein deacetylases is another mechanism by which miR-22 has anti-cancer effects. HDAC1 is a novel miR-22 target recently uncovered by our group using colon cancer cells.43 In a miR-22-dependent manner, histone deacetylase (HDAC) inhibitors reduce HDAC1, HDAC4, and sirtuin 1 (SIRT1), which are highly expressed in the liver and colon cancer specimens. Upon miR-22 induction, reduced HDAC1, HDAC4, and SIRT1 occupied the transcriptional regulatory region of the retinoic acid receptor beta (RARβ) and nuclear receptor subfamily 4 group A member 1 (NUR77) genes leads to increased H3K9 acetylation of the RARβ and NUR77 genes. Therefore, miR-22-reduced protein deacetylases simultaneously induce NUR77 and RARβ expression, as well as, their nuclear export converting their transcriptional effect into apoptotic effect.43
Nuclear factor κB (NF-κB) regulates many biological processes including liver tumorigenesis.44 MiR-22 inhibits NF-κB activity through targeting NF-κB coactivator, namely nuclear receptor coactivator 1 (NCOA1).44 The mechanisms by which miR-22 functions as a HCC suppressor is summarized in Table 3 and Fig. 3.
Table 3.
The mechanisms by which miR-22 acts as a liver cancer suppressor.
| Cancer models | Target genes | Function of the target genes | Refs. |
|---|---|---|---|
| Human HCC | NEAT1, AKT2 | Promote proliferation and invasion, inhibit Apoptosis | 34–36 |
| HBV-related HCC | HNRNPA1 | Regulates EGFR signaling | 37 |
| Folate deficiency-conditioned HCC cells | HDAC4, ZEB2, OCT4, PRRX1 | Regulate gene expression | 38 |
| Human HCC | Galectin-1, Galectin-9 | Promote T cell apoptosis and cytokine production | 39–41 |
| Huh7 and HCT116cells | CCNA2 | Regulates cell cycle | 42 |
| HCT116 and DLD-1 cell | HDAC1, HDAC4, SIRTJ, NUR77, RARb | Epigenetic and transcriptional regulation leading to apoptosis of cancer cell | 43 |
| Huh7 cell | NCOA1, NF-kB | Transcriptional regulation | 44 |
Abbreviations: miR-22, microRNA 22; HCC, hepatocellular carcinoma; AKT2, AKT serine/threonine kinase 2; HBV, hepatitis B virus; HNRNPA1, heterogeneous nuclear Ribonucleoprotein A1; HDAC, histone deacetylase; ZEB2, Zinc finger E-box binding homeobox 2; OCT4, octamer-binding transcription factor 4; PRRX1, decreased paired related homeobox 1; CCNA2, cyclin A2; SIRT1, sirtuin 1; NUR77, nuclear receptor subfamily 4 group A member 1; RARβ, retinoic acid receptor beta; NCOA1, nuclear receptor coactivator 1; NF-κB, nuclear factor kappa B.
Fig. 3. The regulation and function of miR-22.
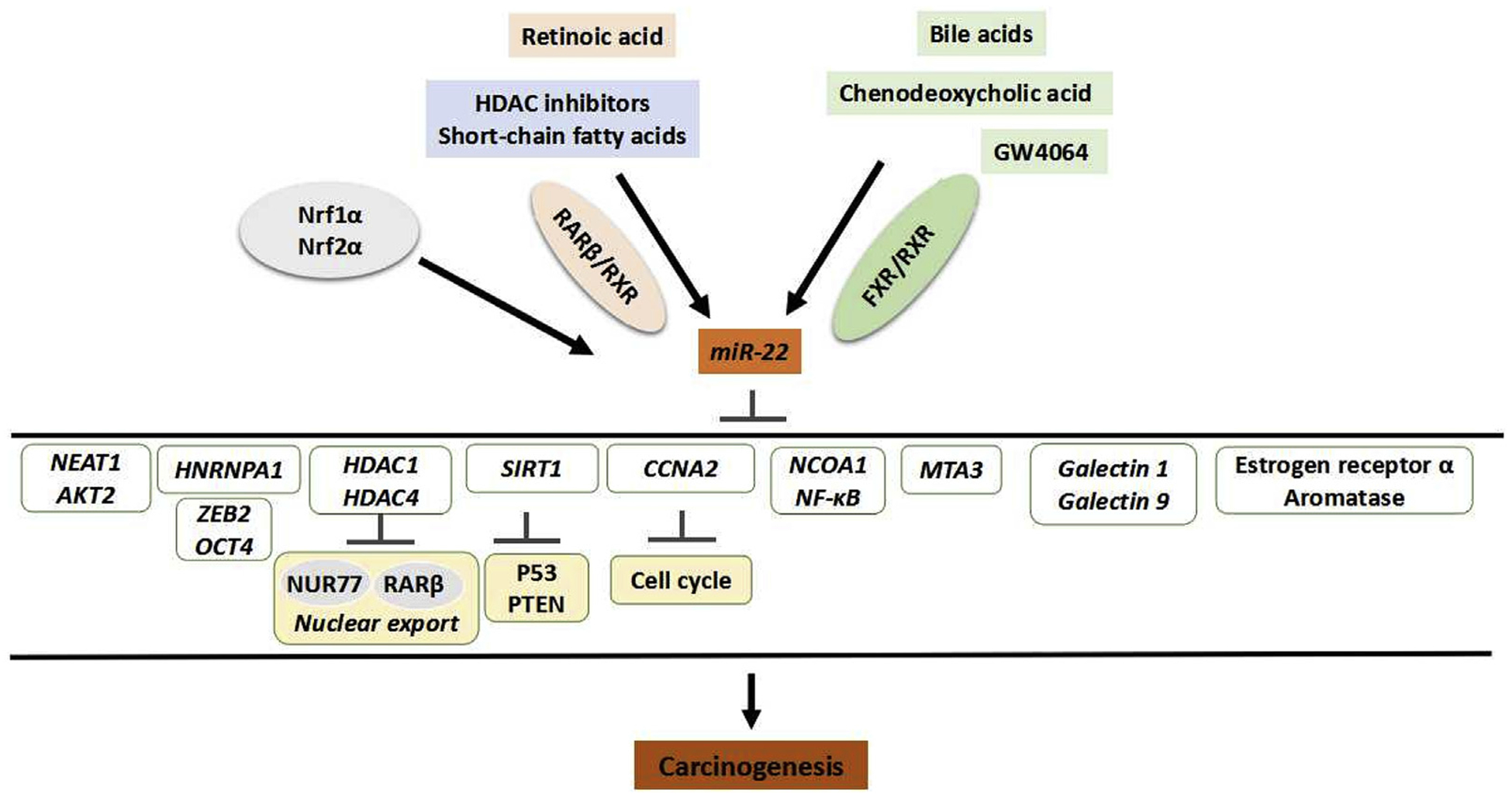
MiR-22 can be induced by ligands for nuclear receptors such as retinoic acid, bile acids, as well as chemicals that have HDAC inhibitory property. Transcription factor Nrf1α and Nuf2α also induce miR-22. All those chemicals and transcriptional factors have known anti-cancer effects. Abbreviations: miR-22,microRNA 22; HDAC, histone deacetylase; Nrf1α, nuclear respiratory factor 1; Nrf2α, NF-E2-related factor 2a; RARβ, retinoic acid receptor beta; FXR, farnesoid X receptor; HNRNPA1, heterogeneous nuclear ribonucleoprotein A1; ZEB2, Zinc finger E-box binding homeobox 2; OCT4, octamer-binding transcription factor 4; NUR77, nuclear receptor subfamily 4 group A member 1; SIRT1, sirtuin 1; PTEN, phosphatase and tensin homolog; CCNA2, cyclin A2; NCOA1, nuclear receptor coactivator 1; NF-κB, nuclear factor kappa B; MTA3, metastasis associated 1 family member 3.
It is interesting to note that the potential tumor-promoting effect of miR-22 has also been revealed in an animal model. miR-22 inhibits the expression of methionine adenosyltransferase 1A (Mat1a) and methylenetetrahydrofolate reductase (Mthfr) in early preneoplastic livers of rats treated by 2-acetylaminofluorene. The reduced expression of Mat1a and Mthfr genes by miR-22 and miR-29b is a main driver to promote liver carcinogenesis in the studied model.45
8. The mechanisms by which the expression of miR-22 is regulated
Knockout hepatic nuclear respiratory factor 1 (Nrf1α) causes oncogenic activation of NF-E2-related factor 2 (Nrf2) and leads to the development of NASH and hepatoma. Thus, Nrf1α functions as a dominant tumor suppressor. It has been shown that both Nrf1α and Nrf2 regulate miR-22 expression via binding to the antioxidant response element (ARE) site of the miR-22 promoter.46
There are several chemicals that can induce the expression of miR-22 including catalpol, an iridoid glucoside. Catalpol induces miR-22 and reduces cell proliferation, invasion, and migration. Catalpol also increases apoptotic rates and G0/G1 phase of Huh7 and HCLMM2 cells. Catalpol exerts anti-tumor effects through up-regulating miR-22–3p, which reduces the metastasis associated 1 family member 3 (MTA3) expression by directly targeting MTA3.47
MiR-22 is induced in human liver cancer Huh7 cells treated with sodium butyrate.42,48 Sodium butyrate treatment or forced miR-22 overexpression increases the ROS production and reduces SIRT1 expression. Down-regulation of miR-22 counteracts the effects of butyrate in Huh7cells including the induction of apoptosis via ROS production, cytochrome c release, and activation of caspase-3. Furthermore, anti-miR-22 also reverses the inhibition of cell growth and proliferation mediated by sodium butyrate.48
In addition to butyrate, other short-chain fatty acids (SCFAs) that have histone deacetylase (HDAC) inhibitory property such as propionate and valerate as well as synthetic HDAC inhibitor suberanilohydroxamic acid (SAHA) can also induce miR-22 as demonstrated using colon cancer cells.43 In contrast, SCFAs that lack HDAC inhibitory effect such as formate and acetate do not have an effect in inducing miR-22.43 Additionally, retinoic acid (RA), which is produced by butyrate-induced aldehyde dehydrogenase 1 family member A1(ALDH1A1), also induces miR-22. Furthermore, when HDAC inhibitors are used in combination with RA, the induction of miR-22 reaches to a higher level than a single chemical treatment. Such induction is mediated via retinoic acid receptor β (RARβ) binding to a direct repeat 5 (DR5) motif found in the regulatory region of the miR-22.43
Bile acids via its receptor farnesoid X receptor (FXR) induces miR-22 by direct binding to an invert repeat 1 (IR-1) motif located in the regulatory region of the miR-22.42 Both endogenous FXR ligand chenodeoxycholic acid and synthetic FXR ligand GW4064 increase miR-22 in Huh7 liver and HCT116 colon cells.42 In addition, semi-synthetic bile acid, obeticholic acid, which is in clinical trials to treat NASH, also increases miR-22 expression in Huh7 liver cells.12 Furthermore, by inducing FGF21 signaling, miR-22 inhibitors can improve the effect of obeticholic acid in improving insulin sensitivity as demonstrated in Western diet-induced obese mice.12
The liver is a testosterone-responsive organ. Testosterone regulates liver metabolism, inhibits hepatic immune responses and even promotes liver carcinogenesis.49–54 Testosterone treatment of female mice induces hepatic miR-22, miR-690, miR-122, let-7A, miR-30D, and let-7D. An androgen response element (ARE) has been found in the miR-122 promoter, but not in the other five induced miRNAs. Therefore, the mechanism by which testosterone induces miR-22 remains to be uncovered. The induction of miR-22 leads to reduced expression of estrogen receptor α and aromatase, thus resulting in estrogen signal inhibition.55
The chemicals that have anti-cancer effects and can increase the expression level of miR-22 are summarized in Table 4 and Fig. 3. It is interesting to note that most of those miR-22 inducers have metabolic stimulating effects, and yet miR-22 functions as a metabolic silencer.
Table 4.
MiR-22 inducers.
| MiR-22 inducers | Refs. |
|---|---|
| Catalpol | 47 |
| Bile acids, Chenodeoxycholic acid, GW4064, Obeticholic acid | 12,42 |
| Retinoic acid | 43 |
| HDAC inhibitors: butyrate, propionate, valerate, suberanilohydroxamic acid | 42,43,48 |
| Testosterone | 55 |
Abbreviations: miR-22, microRNA 22; HDAC, histone deacetylase.
9. Conclusion
The level of miR-22 rises in hepatic steatosis and declines in liver cancer. Thus, miR-22 inhibition can treat NAFLD, and yet miR-22 inducers or mimics can be useful treating liver cancer. Indeed, the cancer treatment effects of miR-22 inducers includes catalpol, butyrate, retinoic acid, and HDAC inhibitors have been revealed.
Metabolism driven by FGF21 leads to AMPK and ERK1/2 activation thereby supporting growth and cell proliferation. Surprisingly, metabolism enhancers such as bile acids, testosterone, and retinoic acid induce the expression of miR-22, which silences FGF21 and its receptor. The simultaneous induction of miR-22 as well as FGF21 signaling likely maintain FGF21 homeostasis and restrict persistent ERK1/2 activation. In other words, concomitant induction of FGF21 and miR-22 can be a way to maintain FGF21 homeostasis and thus insulin sensitivity. However, the reduction of miR-22 may improve the efficacy of AMPK activators by increasing hepatic FGF21.
The expression level of miR-22 changes in a dynamic way as liver disease progresses. To use miR-22 as a drug target, the status of miR-22 needs to be monitored. miR-22 is also ubiquitously expressed and it has many other important biological functions such as immunity regulation.56,57 In other types of cancer, miR-22 may function differently. Targeted delivery of miR-22 inducer or silencer should be considered to avoid unwanted effects.
Acknowledgements
This study was supported by grants funded by the National Institutes of Health U01CA179582 and R01CA222490, UC Davis Comprehensive Cancer Center seed grant, and UC Davis Global Affairs seed grant. The authors thank Ying Hu, Mindy Huynh, and Xingru Zhu for editing the manuscript.
Footnotes
Declaration of competing interest
The authors declare that there is no conflict of interest.
References
- 1.Chen B, Tang H, Liu X, et al. miR-22 as a prognostic factor targets glucose transporter protein type 1 in breast cancer. Canc Lett. 2015;356:410–417. [DOI] [PubMed] [Google Scholar]
- 2.Huang ZP, Wang DZ. miR-22 in cardiac remodeling and disease. Trends Cardiovasc Med. 2014;24:267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng Y, Huang W, Wani M, Yu X, Ashraf M. Ischemic preconditioning potentiates the protective effect of stem cells through secretion of exosomes by targeting Mecp2 via miR-22. PloS One. 2014;9, e88685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi C, Xu X. MicroRNA-22 is down-regulated in hepatitis B virus-related hepatocellular carcinoma. Biomed Pharmacother. 2013;67:375–380. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Li Y, Ding M, Zhang H, Xu X, Tang J. Molecular mechanisms and clinical applications of miR-22 in regulating malignant progression in human cancer (Review). Int J Oncol. 2017;50:345–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bar N, Dikstein R. miR-22 forms a regulatory loop in PTEN/AKT pathway and modulates signaling kinetics. PloS One. 2010;5, e10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang B, Yao Q, Xu D, Zhang JA. MicroRNA-22–3p as a novel regulator and therapeutic target for autoimmune diseases. Int Rev Immunol. 2017;36: 176–181. [DOI] [PubMed] [Google Scholar]
- 8.Huang SC, Wang M, Wu WB, et al. Mir-22–3p inhibits arterial smooth muscle cell proliferation and migration and neointimal hyperplasia by targeting HMGB1 in arteriosclerosis obliterans. Cell Physiol Biochem. 2017;42: 2492–2506. [DOI] [PubMed] [Google Scholar]
- 9.Huang ZP, Wang DZ. miR-22 in smooth muscle cells: a potential therapy for cardiovascular disease. Circulation. 2018;137:1842–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diniz GP, Huang ZP, Liu J, et al. Loss of microRNA-22 prevents high-fat diet induced dyslipidemia and increases energy expenditure without affecting cardiac hypertrophy. Clin Sci. 2017;131:2885–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaur K, Pandey AK, Srivastava S, Srivastava AK, Datta M. Comprehensive miRNome and in silico analyses identify the Wnt signaling pathway to be altered in the diabetic liver. Mol Biosyst. 2011;7:3234–3244. [DOI] [PubMed] [Google Scholar]
- 12.Hu Y, Liu HX, Jena PK, Sheng L, Ali MR, Wan YJ. miR-22 inhibition reduces hepatic steatosis via FGF21 and FGFR1 induction. JHEP Rep. 2020;2(2), 100093 10.1016/j.jhepr.2020.100093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez-Riera M, Conde I, Tolosa L, et al. New microRNA biomarkers for drug-induced steatosis and their potential to predict the contribution of drugs to non-alcoholic fatty liver disease. Front Pharmacol. 2017;8:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cook NL, Pereira TN, Lewindon PJ, Shepherd RW, Ramm GA. Circulating microRNAs as noninvasive diagnostic biomarkers of liver disease in children with cystic fibrosis. J Pediatr Gastroenterol Nutr. 2015;60:247–254. [DOI] [PubMed] [Google Scholar]
- 15.Yang T, Chen SL, Lu XJ, Shen CY, Liu Y, Chen YP. Bone morphogenetic protein 7 suppresses the progression of hepatic fibrosis and regulates the expression of gremlin and transforming growth factor beta1. Mol Med Rep. 2012;6:246–252. [DOI] [PubMed] [Google Scholar]
- 16.Zhong L, Wang X, Wang S, Yang L, Gao H, Yang C. The anti-fibrotic effect of bone morphogenic protein-7(BMP-7) on liver fibrosis. Int J Med Sci. 2013;10: 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou GL, Zuo S, Lu S, et al. Bone morphogenetic protein-7 represses hepatic stellate cell activation and liver fibrosis via regulation of TGF-beta/Smad signaling pathway. World J Gastroenterol. 2019;25:4222–4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji D, Li B, Shao Q, Li F, Li Z, Chen G. MiR-22 suppresses BMP7 in the development of cirrhosis. Cell Physiol Biochem. 2015;36:1026–1036. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Y, Lv X, Qu H, et al. Preliminary screening and functional analysis of circular RNAs associated with hepatic stellate cell activation. Gene. 2018;677: 317–323. [DOI] [PubMed] [Google Scholar]
- 20.You Y, Tan JX, Dai HS, et al. MiRNA-22 inhibits oncogene galectin-1 in hepatocellular carcinoma. Oncotarget. 2016;7:57099–57116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Q, Jiang W, Zhuang C, et al. microRNA-22 downregulation of galectin-9 influences lymphocyte apoptosis and tumor cell proliferation in liver cancer. Oncol Rep. 2015;34:1771–1778. [DOI] [PubMed] [Google Scholar]
- 22.Jiang ZJ, Shen QH, Chen HY, Yang Z, Shuai MQ, Zheng SS. Galectin-1 gene silencing inhibits the activation and proliferation but induces the apoptosis of hepatic stellate cells from mice with liver fibrosis. Int J Mol Med. 2019;43: 103–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujita K, Niki T, Nomura T, et al. Correlation between serum galectin-9 levels and liver fibrosis. J Gastroenterol Hepatol. 2018;33:492–499. [DOI] [PubMed] [Google Scholar]
- 24.Akamatsu S, Hayes CN, Tsuge M, et al. Differences in serum microRNA profiles in hepatitis B and C virus infection. J Infect. 2015;70:273–287. [DOI] [PubMed] [Google Scholar]
- 25.Hayes CN, Akamatsu S, Tsuge M, et al. Hepatitis B virus-specific miRNAs and Argonaute2 play a role in the viral life cycle. PloS One. 2012;7, e47490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arataki K, Hayes CN, Akamatsu S, et al. Circulating microRNA-22 correlates with microRNA-122 and represents viral replication and liver injury in patients with chronic hepatitis B. J Med Virol. 2013;85:789–798. [DOI] [PubMed] [Google Scholar]
- 27.Anadol E, Schierwagen R, Elfimova N, et al. Circulating microRNAs as a marker for liver injury in human immunodeficiency virus patients. Hepatology. 2015;61:46–55. [DOI] [PubMed] [Google Scholar]
- 28.Zheng J, Xiong D, Sun X, et al. Signification of hypermethylated in cancer 1 (HIC1) as tumor suppressor gene in tumor progression. Cancer Microenviron. 2012;5:285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao X, He M, Wan D, et al. The minimum LOH region defined on chromosome 17p13.3 in human hepatocellular carcinoma with gene content analysis. Canc Lett. 2003;190:221–232. [DOI] [PubMed] [Google Scholar]
- 30.Zhang DY, Zou XJ, Cao CH, et al. Identification and functional characterization of long non-coding RNA MIR22HG as a tumor suppressor for hepatocellular carcinoma. Theranostics. 2018;8:3751–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Yang Y, Yang T, et al. MicroRNA-22, downregulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumourigenicity. Br J Canc. 2010;103:1215–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiao DD, Yang J, Lei XF, et al. Expression of microRNA-122 and microRNA-22 in HBV-related liver cancer and the correlation with clinical features. Eur Rev Med Pharmacol Sci. 2017;21:742–747. [PubMed] [Google Scholar]
- 33.Zekri AN, Youssef AS, El-Desouky ED, et al. Serum microRNA panels as potential biomarkers for early detection of hepatocellular carcinoma on top of HCV infection. Tumour Biol. 2016;37:12273–12286. [DOI] [PubMed] [Google Scholar]
- 34.Zhou X, Wang X, Zhou Y, Cheng L, Zhang Y. Long noncoding RNA NEAT1 promotes cell proliferation and invasion and suppresses apoptosis in hepatocellular carcinoma by regulating miRNA-22–3p/akt2 in vitro and in vivo. OncoTargets Ther. 2019;12:8991–9004. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Grabinski N, Ewald F, Hofmann BT, et al. Combined targeting of AKT and mTOR synergistically inhibits proliferation of hepatocellular carcinoma cells. Mol Canc. 2012;11:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang L, Sun J, Pan Z, et al. Long non-coding RNA NEAT1 promotes hepatocellular carcinoma cell proliferation through the regulation of miR-129–5p-VCP-Ikap-paB. Am J Physiol Gastrointest Liver Physiol. 2017;313:G150–G156. [DOI] [PubMed] [Google Scholar]
- 37.Ke RS, Zhang K, Lv LZ, et al. Prognostic value and oncogene function of heterogeneous nuclear ribonucleoprotein A1 overexpression in HBV-related hepatocellular carcinoma. Int J Biol Macromol. 2019;129:140–151. [DOI] [PubMed] [Google Scholar]
- 38.Su YH, Huang WC, Huang TH, et al. Folate deficient tumor microenvironment promotes epithelial-to-mesenchymal transition and cancer stem-like phenotypes. Oncotarget. 2016;7:33246–33256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang PF, Li KS, Shen YH, et al. Galectin-1 induces hepatocellular carcinoma EMT and sorafenib resistance by activating FAK/PI3K/AKT signaling. Cell Death Dis. 2016;7, e2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spano D, Russo R, Di Maso V, et al. Galectin-1 and its involvement in hepatocellular carcinoma aggressiveness. Mol Med. 2010;16:102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leung Z, Ko FCF, Tey SK, et al. Galectin-1 promotes hepatocellular carcinoma and the combined therapeutic effect of OTX008 galectin-1 inhibitor and sorafenib in tumor cells. J Exp Clin Canc Res. 2019;38:423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang F, Hu Y, Liu HX, Wan YJ. MiR-22-silenced cyclin A expression in colon and liver cancer cells is regulated by bile acid receptor. J Biol Chem. 2015;290: 6507–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu Y, French SW, Chau T, et al. RARbeta acts as both an upstream regulator and downstream effector of miR-22, which epigenetically regulates NUR77 to induce apoptosis of colon cancer cells. Faseb J. 2019;33:2314–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takata A, Otsuka M, Kojima K, et al. MicroRNA-22 and microRNA-140 suppress NF-kappaB activity by regulating the expression of NF-kappaB coactivators. Biochem Biophys Res Commun. 2011;411:826–831. [DOI] [PubMed] [Google Scholar]
- 45.Koturbash I, Melnyk S, James SJ, Beland FA, Pogribny IP. Role of epigenetic and miR-22 and miR-29b alterations in the downregulation of Mat1a and Mthfr genes in early preneoplastic livers in rats induced by 2-acetylaminofluorene. Mol Carcinog. 2013;52:318–327. [DOI] [PubMed] [Google Scholar]
- 46.Qiu L, Wang M, Hu S, et al. Oncogenic activation of Nrf2, though as a master antioxidant transcription factor, liberated by specific knockout of the full-length Nrf1alpha that acts as a dominant tumor repressor. Cancers. 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao L, Wang Y, Liu Q. Catalpol inhibits cell proliferation, invasion and migration through regulating miR-22–3p/MTA3 signalling in hepatocellular carcinoma. Exp Mol Pathol. 2019;109:51–60. [DOI] [PubMed] [Google Scholar]
- 48.Pant K, Yadav AK, Gupta P, Islam R, Saraya A, Venugopal SK. Butyrate induces ROS-mediated apoptosis by modulating miR-22/SIRT-1 pathway in hepatic cancer cells. Redox Biol. 2017;12:340–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clodfelter KH, Holloway MG, Hodor P, Park SH, Ray WJ, Waxman DJ. Sex-dependent liver gene expression is extensive and largely dependent upon signal transducer and activator of transcription 5b (STAT5b): STAT5b-dependent activation of male genes and repression of female genes revealed by microarray analysis. Mol Endocrinol. 2006;20:1333–1351. [DOI] [PubMed] [Google Scholar]
- 50.Krucken J, Dkhil MA, Braun JV, et al. Testosterone suppresses protective responses of the liver to blood-stage malaria. Infect Immun. 2005;73:436–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wunderlich F, Dkhil MA, Mehnert LI, et al. Testosterone responsiveness of spleen and liver in female lymphotoxin beta receptor-deficient mice resistant to blood-stage malaria. Microb Infect. 2005;7:399–409. [DOI] [PubMed] [Google Scholar]
- 52.Drinkwater NR, Hanigan MH, Kemp CJ. Genetic and epigenetic promotion of murine hepatocarcinogenesis. Prog Clin Biol Res. 1990;331:163–76. [PubMed] [Google Scholar]
- 53.Kemp CJ Drinkwater NR. The androgen receptor and liver tumor development in mice. Prog Clin Biol Res. 1990;331:203–214. [PubMed] [Google Scholar]
- 54.Nagasue N Kohno H. Hepatocellular carcinoma and sex hormones. HPB Surg. 1992;6:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delic D, Grosser C, Dkhil M, Al-Quraishy S, Wunderlich F. Testosterone-induced upregulation of miRNAs in the female mouse liver. Steroids. 2010;75: 998–1004. [DOI] [PubMed] [Google Scholar]
- 56.Ren Q, Huang Y, He Y, Wang W, Zhang X. A white spot syndrome virus microRNA promotes the virus infection by targeting the host STAT. Sci Rep. 2015;5:18384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liang X, Liu Y, Mei S, et al. MicroRNA-22 impairs anti-tumor ability of dendritic cells by targeting p38. PloS One. 2015;10:e0121510. [DOI] [PMC free article] [PubMed] [Google Scholar]


