Abstract
The Dominican Republic is one of the two countries on the Hispaniola island, which is part of the Antilles. Hispaniola was affected by the European colonization and massive deportation of African slaves since the XVI century and these events heavily shaped the genetic composition of the present-day population. To shed light about the effect of the European rules, we analyzed 92 single nucleotide polymorphisms on the Y chromosome in 182 Dominican individuals from three different locations. The Dominican Y haplogroup composition was characterized by an excess of northern African/European lineages (59%), followed by the African clades (38%), whereas the Native-American lineages were rare (3%). The comparison with the mitochondrial DNA variability, dominated by African clades, revealed a sex-biased admixture pattern, in line with the colonial society dominated by European men. When other Caribbean and non-Caribbean former colonies were also considered, we noted a difference between territories under a Spanish rule (like the Dominican Republic) and British/French rule, with the former characterized by an excess of European Y lineages reflecting the more permissive Iberian legislation about mixed people and slavery. Finally, we analyzed the distribution in Africa of the Dominican lineages with a putative African origin, mainly focusing on central and western Africa, which were the main sources of African slaves. We found that most (83%) of the African lineages observed in Santo Domingo have a central African ancestry, suggesting that most of the slaves were deported from regions.
Keywords: Y-SNPs, Antilles, Dominican Republic, trans-Atlantic slave trade, African ancestry, central Africa
Significance
Despite the importance of the Dominican Republic in the colonial history of the Caribbeans, little is known about its Y haplogroup composition. Analyzing informative Y markers in Dominican individuals and comparing them with European and African samples, we observed a majority of central African Y lineages in the Dominican sub-Saharan component, suggesting that most of the deported African slaves came from central Africa. We also found a disproportion of European paternal lineages compared with the maternal ones, because of a sex-biased admixture due to the social dominant role of European men. Finally, comparing our results with data from other Greater Antilles, we also observed differences between islands in the paternal ancestry proportion, possibly related to the regulations about mixed unions adopted by the colonists.
Introduction
The Antilles is a group of islands forming an arch between the Florida peninsula and coasts of Venezuela. It is bordered by the Caribbean Sea to the west and south, the Gulf of Mexico to the northwest and the Atlantic Ocean to the east and north. The archipelago is usually divided in Lesser Antilles to the southeast and Greater Antilles to the northwest, composed of the large islands of Cuba, Hispaniola, Puerto Rico, Jamaica, and the Cayman Islands.
According to the archeological evidence, the first human settlements by Native-American people in the Greater Antilles date back to 8,000–5,000 years ago (Rouse 1992). Between 5,000 and 2,000 years ago, other migration waves from the American continent arrived in this group of islands, probably from the Orinoco river basin and through the Lesser Antilles (Bukhari et al. 2017; Schroeder et al. 2018; Fernandes et al. 2020; Nägele et al. 2020). These human groups were probably the ancestors of Tainos, the people inhabiting the Greater Antilles when the European colonists arrived in the XV century. The exact population size of the Tainos at the arrival of the European colonists is hard to estimate: numbers ranging from 100,000 to 1,000,000 have been proposed (Anderson-Córdova 1990, 2017; Guitar 2002), although a recent study based on ancient DNA led to an estimate of 10,000–20,000 individuals (Fernandes et al. 2020). The contact with the Europeans led to a rapid collapse of the Taino population due to three main concurrent reasons: epidemic of infectious diseases from the Old World, wars against colonists or other Caribbean populations and enslavement by Europeans (Rouse 1992; Bukhari et al. 2017); despite the dramatic impact of the European colonization on the Tainos, footprints of their genetic legacy can be still recognized in modern Caribbean populations (Moreno-Estrada et al. 2013; Benn Torres et al. 2015, 2019; Bukhari et al. 2017; Schroeder et al. 2018; Mendisco et al. 2019; Fernandes et al. 2020; Nieves-Colón et al. 2020). The Tainos were exploited to work in the plantations of cotton, coffee, tobacco, and sugarcane, at the base of the economy of the European colonies: with the collapse of the Native population, the introduction of African slaves to the colonies sharply increased (Curtin 1969; Adhikari et al. 2017).
Nowadays, because of its history of migrations and colonization, the population of the Antilles is a mixture of people from Europe, America, and Africa, similarly to other American peoples (Adhikari et al. 2017). Genetic data from autosomes revealed that the proportion of European, American, and African ancestry greatly varies among different American regions, with the Antilles showing a higher degree of African ancestry compared with other American groups because of the massive deportation of African slaves by European colonists (Adhikari et al. 2017).
The analysis of the uniparental markers of the human genomes, that is the mitochondrial DNA (mtDNA) transmitted along the maternal lineage and the male-specific region of the Y chromosome (MSY) with a paternal inheritance, revealed different patterns reflecting the political organization of the European colonies in the Americas, that were ruled by European men with a dominant social role (Adhikari et al. 2017). In general, the maternal lineages show a high proportion of Native-American (30–60%) or African (20–40%) haplogroups, whereas the European lineages are less frequent (∼10%) (Moreno-Estrada et al. 2013; Vilar et al. 2014; Bukhari et al. 2017; Schroeder et al. 2018; Nieves-Colón et al. 2020). On the paternal side, most of the Y haplogroups are of European origin (>70%) or of African origin (20%), whereas the Native-American lineages are very rare or absent (Simms et al. 2012, 2013; Vilar et al. 2014; Benn Torres et al. 2015; Mendisco et al. 2019). Among the African Y lineages, the most frequent is E1b1a-M2, followed by haplogroup B, consistent with the Y haplogroup distribution in western and central Africa (Cruciani et al. 2002; Batini et al. 2011; de Filippo et al. 2011; D’Atanasio et al. 2018), where most of the African slaves came from (Curtin 1969).
In the Greater Antilles, Hispaniola is the second largest island of the archipelago and represents an interesting case, being divided into two political entities: Haiti to the west and the Dominican Republic to the east. The first European colonies in the Antilles were established in the Dominican part of this island, that was ruled by Spanish until 1697, when Haiti passed under the French rule (Coupeau 2008). After this point, the histories of the two halves of Hispaniola partly diverged and the present-day Dominican Republic was characterized by economic decline and political instability due to frequent pirate raids and contrasts with French colonists, who eventually managed to take the control of some Dominican regions (Knight 1997). The first decade of the XVIII century was characterized by struggles due to the unstable French rule, until the Spanish colonists re-established their power. In the meantime, a violent revolution broke up in Haiti in 1791, eventually leading to the independence of the former French colony. After another decade of disorders, the Dominican part of Hispaniola was declared independent in 1821, but it passed under the Haitian control shortly after, briefly reuniting the Hispaniola island under the same rule until the Dominican war of independence in 1844 (Guitar 2007).
On the genetic side, the two countries of the Hispaniola island show differences in the maternal and paternal genetic composition, possibly due to their different history. Considering the mtDNA diversity, the Dominican Republic shows up to 22% of Native-American lineages, whereas they are virtually absent from Haiti (Bukhari et al. 2017). Considering the Y lineages too, Haiti and the Dominican Republic seem to show a slightly different pattern, but a detailed description of the Y haplogroup composition of the Dominican population is still lacking (Bryc et al. 2010; Simms et al. 2012, 2013; Ono et al. 2019); nonetheless, the analysis of the Dominican Y chromosome variability is of crucial importance to fully understand the effect of the European colonization and the trans-Atlantic slave trade on the genetic diversity of the Antilles, considering the important role of Hispaniola in the colonial Caribbeans and its peculiar history.
To fill this gap in our knowledge, we analyzed 92 Y-SNPs (single nucleotide polymorphisms) in 182 Dominican individuals from three different locations of the Dominican Republic. We observed a proportion of European, African, and Native-American Y haplogroups in line with the other populations from the Antilles, although some differences are present. To assess the western African or central African ancestry of the Dominican samples with a supposedly African Y haplogroup, we then focus on the ethnic and geographic distribution of these clades by analyzing the Y-SNPs in a wider set of ≈5,000 males from our lab collection and extracting the data from relevant populations of the Phase 3 of the 1000 Genomes Project (Poznik et al. 2016).
Materials and Methods
The Sample
Our analysis has been focused on 182 males from three locations in the Dominican Republic: Santo Domingo (N = 50), San Juan de la Maguana (N = 76), and Las Galeras (N = 56). We selected these three locations because of their different population size and geographic location in the Dominican Republic. Santo Domingo, located on the Dominican southern coast, is the capital of the Dominican Republic and it was the first Spanish colony in America. San Juan de la Maguana is one of the oldest cities of the Dominican Republic, in the western part of the country. Finally, Las Galeras is a small village in the east coast of the Samanà Peninsula (fig. 1).
Fig. 1.
Geographic localization of the three sampling places in the Dominican Republic.
Blood or saliva samples have been collected on a voluntary basis more than 20 years ago, after obtaining an appropriate informed consent and according to all the ethical guidelines in effect at the time. DNA extraction was performed in our lab at the Sapienza University of Rome following standard protocols. A subset of the individuals have already been included in previous studies on the mtDNA and Y genetic diversity (Bandelt et al. 2001; Torroni et al. 2001; Salas et al. 2004; D’Atanasio et al. 2018).
We used the Kruskal–Wallis test (a rank-based nonparametric statistical test) (Kruskal and Wallis 1952) to compare the absolute frequencies of Y haplogroups in the three groups.
The haplogroup composition of the Dominican populations was compared with the haplogroup distribution in 695 European, African, and Admixed individuals from the Phase 3 of the 1000 Genomes Project (Poznik et al. 2016) and in about 5,000 African and European subjects from our lab collection, collected over the last decades following all the ethical procedures and already published in our previous studies (Cruciani et al. 2002, 2004, 2007; Trombetta et al. 2015; D’Atanasio et al. 2018).
The study was approved by Policlinico Umberto I Ethical Committee, Sapienza Università di Roma (reference number 1016/10).
Y Haplogroup Analysis
The Y haplogroup affiliation of each subject was assessed by analyzing a set of 92 Y-haplogroup defining SNPs (supplementary table 1, Supplementary Material online). A subset of markers was already analyzed for many populations in previous studies (Cruciani et al. 2002, 2004, 2007, 2010; Scozzari et al. 2014; Trombetta et al. 2015; D’Atanasio et al. 2018), whereas the data for most of the Dominicans and for a subset of people from Western Africa are presented here for the first time. The markers have been selected on the basis on their position in the Y phylogeny and their geographic distribution in Africa, considering the data from the literature when available (Cruciani et al. 2007; Karafet et al. 2008; Scozzari et al. 2014; Van Oven et al. 2014; Trombetta et al. 2015; Poznik et al. 2016; D’Atanasio et al. 2018) and have been analyzed using a hierarchical approach. The DNA fragment around each SNP has been amplified by PCR and then genotyped by Sanger sequencing, DHPLC, or RFLP (supplementary table 1, Supplementary Material online). The frequency data for these SNPs have also been extracted from African, European, and Admixed populations of the Phase 3 of the 1000 Genomes Project (Poznik et al. 2016) (supplementary table 2, Supplementary Material online). We followed the PhyloTree Y for the haplogroup nomenclature, reporting the last derived marker after the lineage (Van Oven et al. 2014).
Y-STR Analysis
For seven Dominican individuals for which the putative place of origin could not be defined based solely on their Y-SNP haplogroup affiliation, we analyzed the 27 Y-STR loci of the Yfiler Plus multiplex (supplementary table 3, Supplementary Material online). The genotyping was performed on 1 ng of genomic DNA, according to the manufacturer’s protocol, and the amplified fragments have been visualized with GeneMapper IDX v.1.4, after an electrophoretic run on a 3500 XL Genetic Analyzer. The Y-STR profiles of the Dominican samples have been then compared with available Yfiler Plus data from Sardinian and from northern, western–central, and eastern African males belonging to a subset of individuals reported in supplementary table 2, Supplementary Material online and previously published (Rapone et al. 2016; Iacovacci et al. 2017; D’Atanasio et al. 2019; Della Rocca et al. 2020). We reconstructed the relationships among samples in a dendrogram using a UPGMA approach with the total number of differences in repeat number as measure of distance between haplotypes (Manhattan distance), as implemented by the software PAST v.4.0 (Hammer et al. 2001).
Admixture Analysis
We estimated the relative genetic contribution of central and western Africans in our Dominican sample using the following formula [modified from Relethford {2012}]:
where i is the ith haplogroup considered for the estimate, PWi and PCi are the frequencies of a specific Y haplogroup in western Africa and central Africa, respectively, whereas PDi is the frequency of the same haplogroup among the chromosomes of African ancestry in the Dominican sample. MCi is the proportion of African lineages observed in the Dominican Republic with a central African ancestry whereas the proportion of admixture from western Africa (MWi) can be estimated as:
Using this approach, we first estimated MWi and MCi for each haplogroup being present in the Dominican Republic, central Africa, and western Africa; then, we estimated the total admixture values MW and MC and their standard error using the formulas for several loci from (Chakraborty 1986).
Results
Major Y Haplogroup Composition of the Dominican Populations
By analyzing a set of 39 basal Y-SNPs we observed a total of 13 main haplogroups among our 182 Dominican samples (table 1).
Table 1.
Frequency of the 13 Main Y Haplogroups in the Three Sampling Locations of the Dominican Republic
| Haplogroups | Dominicans from Santo Domingo | Dominicans from Las Galeras | Dominicans from San Juan de la Maguana | Total |
|---|---|---|---|---|
| B-M60 | 0.020 | 0.036 | 0.000 | 0.016 |
| E-M33 | 0.020 | 0.018 | 0.000 | 0.011 |
| E1b1a-M2 | 0.220 | 0.393 | 0.382 | 0.341 |
| E1b1b-M215 | 0.020 | 0.036 | 0.053 | 0.038 |
| E-M75 | 0.040 | 0.000 | 0.000 | 0.011 |
| G-M201 | 0.000 | 0.018 | 0.026 | 0.016 |
| I-M170 | 0.040 | 0.054 | 0.079 | 0.060 |
| J2-M172 | 0.200 | 0.143 | 0.132 | 0.154 |
| R1a-M448 | 0.000 | 0.000 | 0.013 | 0.005 |
| R1b-M269 | 0.400 | 0.232 | 0.303 | 0.308 |
| T-M70 | 0.040 | 0.000 | 0.000 | 0.011 |
| Q-M242 | 0.000 | 0.071 | 0.013 | 0.027 |
| N | 50 | 56 | 76 | 182 |
The three Dominican locations do not show significant differences in their Y haplogroup composition as a whole (Kruskal–Wallis test, P > 0.05), so they will be analyzed as a single group hereafter.
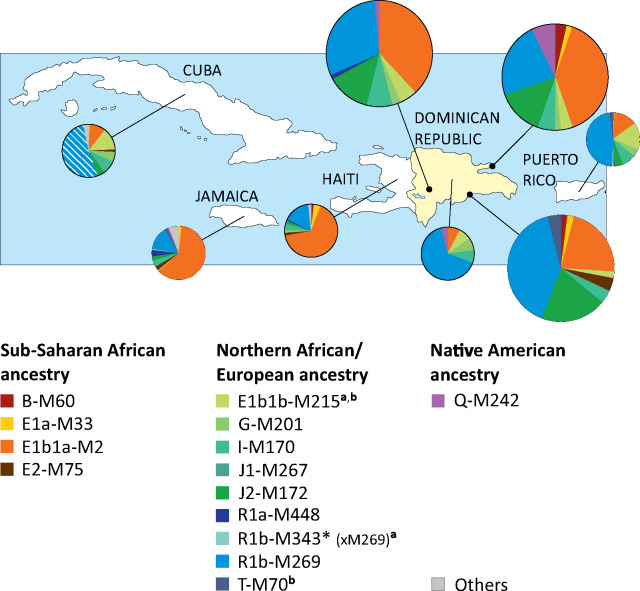
Considering the worldwide ethno-geographic distribution of the Y haplogroups observed in our Dominican samples, we were able to recognize three different ancestry of the paternal lineages: sub-Saharan African (four haplogroups: B-M60, E1a-M33, E1b1a-M2, and E2-M75), northern African or European (six haplogroups: G-M201, I-M170, J1-M267, J2-M172, R1a-M448, and R1b-M269), and Native-American ancestry (one haplogroup: Q-M242). Two of the main lineages (E1b1b-M215 and T-M70) could not be assigned at this level of resolution since they are common in both the Mediterranean and the sub-Saharan region. Because of its history and the geographic barrier represented by the Sahara desert, northern Africa is genetically more similar to southern Europe and Near East than to sub-Saharan Africa (Henn et al. 2012; D’Atanasio et al. 2018) and, for the aim of this study, it will be considered as a northern African/European region. The distribution of the main Y lineages in the Dominican Republic and the other Greater Antilles islands are summarized in figure 2.
Fig. 2.
Y haplogroup distribution of main haplogroups in the Greater Antilles. The larger pies represent the three Dominican populations here analyzed. Data from the literature (smaller pies) for the Dominican Republic and the other islands of the Greater Antilles are also shown (Mendizabal et al. 2008; Bryc et al. 2010; Simms et al. 2012; Vilar et al. 2014; Poznik et al. 2016). The Cuban samples from (Mendizabal et al. 2008) classified as R1-M173* have not been tested for M269 and have been represented with a striped fill. The group “Others” includes all the samples with the ancestral state at the markers here reported and whose ancestry is unknown. aChromosomes from the literature have been assigned to a northern African/European ancestry based on the reported ancestry in the publication and/or the geographic distribution of the derived downstream markers, when available. bThese haplogroups have been assigned to a northern African/European ancestry in our Dominican samples based on successive analyses (see “Major Y Haplogroup Dissection”).
Among the four sub-Saharan African main lineages, the most common is E1b1a-M2, that alone reaches frequencies ranging between 22% and 39% in the three Dominican groups here analyzed. The remaining Dominican samples mostly belong to six European lineages, mainly represented by the western European haplogroup R1b-M269, that was observed with frequencies ranging from 23% to 40%. The Native American component is represented by the Q-M242 haplogroup that was rarely observed (highest frequency: 7%). These observations are in line with previous findings from other Caribbean islands and central and southern American populations (Mendizabal et al. 2008; Bryc et al. 2010; Simms et al. 2012; Vilar et al. 2014; Benn Torres et al. 2015; Poznik et al. 2016; Ono et al. 2019) (fig. 2).
Major Y Haplogroup Dissection
To further shed light on the continental origin of our E1b1b-M215 and T-M70 chromosomes and to determine the exact place of origin of the 13 main haplogroups found in our Dominican samples, we analyzed a set of 53 additional informative markers, selected on the basis of their phylogenetic position and focusing on those able to discriminate among different African regions (supplementary table 2, Supplementary Material online).
The E1b1b-M215 haplogroup is a common lineage spread across three continents: Africa, Asia, and Europe (Chiaroni et al. 2009; Jobling and Tyler-Smith 2017). We analyzed 18 additional Y-SNPs to unambiguously infer the ancestry of our 7 Dominican E1b1b-M215 chromosomes (supplementary table 2, Supplementary Material online). One individual belongs to the Mediterranean E1b1b-M34 (Cruciani et al. 2004; Trombetta et al. 2015). Two samples show the derived state at the V2009 variant, downstream to the V68 SNP and parallel to the E1b1b-M78 clade. The E1b1b-V2009 is quite rare and observed in southern Italy, Sardinia, northern Africa, and among Fulbe people and it probably originated in Europe (Trombetta et al. 2015), suggesting a Mediterranean ancestry of the E1b1b-V2009 Dominican chromosomes. To address this issue, we produced Y-STRs profiles of these individuals and reconstructed their relations with published samples from Sardinia, northern Africa and sub-Saharan Africa using an UPGMA approach, which confirmed the northern African/European ancestry of our Dominican individuals (Rapone et al. 2016; Iacovacci et al. 2017; D’Atanasio et al. 2019). Within E1b1b-M215, one additional sample shows the derived allele at the marker BY1311 (at position hg19: 17082060), internal to the Mediterranean E1b1b-M78/V3536 sub-clade (D’Atanasio et al. 2018). The last three chromosomes belong to the northern African E1b1b-M81 haplogroup (Arredi et al. 2004; Solé-Morata et al. 2017; D’Atanasio et al. 2019).
Haplogroup T-M70 is observed at low frequency in Europe, Africa, and Near East, where probably it originated (Underhill et al. 2001; King et al. 2007; Nogueiro et al. 2010; Mendez et al. 2011; Zhong et al. 2011; Marques et al. 2016) (supplementary table 2, Supplementary Material online). The Y-STR analysis of these chromosomes allowed us to classify them in the northern African/Europe group, since they form a cluster with northern Africans individuals.
After the molecular dissection, all our E1b1b-M215 and T-M70 samples were unambiguously assigned to a northern African/European ancestry (fig. 2).
Regarding the sub-Saharan lineages, we selected and analyzed five additional markers defining internal lineages of the B-M60 haplogroup: all B chromosomes in our Dominican samples belong to the B2b-M109 subclade, that is mainly observed in central Africa and is very rare in western Africa (supplementary table 2, Supplementary Material online) (Cruciani et al. 2002; Wood et al. 2005; Batini et al. 2011; Barbieri et al. 2012).
The E1b1a-M2 Dominican subjects belong to 15 different internal lineages (supplementary table 2, Supplementary Material online). The most frequent E1b1a-M2 subclade is E1b1a-U174, which is observed at a frequency as high as 21% in our Dominican sample. This clade is common in central Africa and it is frequently observed in eastern African and southern Africa, whereas it is quite rare in western Africa: its geographic distribution reflect the Bantu expansion, to which this lineage has been associated (de Filippo et al. 2011; Poznik et al. 2016). The other E1b1a-M2 lineages are not frequently observed among our Dominican groups, including the E1b1a-M2/Z15939 clade, which is found at high frequencies (30–50%) among western Africans and Fulbe people (supplementary table 2, Supplementary Material online) (D’Atanasio et al. 2018).
We analyzed two SNPs internal to the E1a-M33 clade, namely Z932 and M44. We found no individuals belonging to the E1a-M44 subclade, which is often observed among Fulbe people (supplementary table 2, Supplementary Material online). Our Dominican samples were found to be E1a-M33* or E1a-Z932, with the former rarely observed and the latter commonly found in both central and western Africa (supplementary table 2, Supplementary Material online).
Regarding the African E2-M75 lineage, we analyzed two internal clades defined by the M41 and M85 Y-SNPs: both Dominican samples show the ancestral allele at these markers and belong to the paragroup E2-M75*, which has been observed in both western and central Africa (supplementary table 2, Supplementary Material online).
In total, we observed about 59% of Dominican samples with northern African/European ancestry (11 different haplogroups), about 38% of sub-Saharan ancestry (19 haplogroups) and less than 3% of Native-American ancestry (a single haplogroup).
Ancestry Inferences for the African Component of the Dominican Y Chromosome Diversity
Taking into account the haplogroup distribution of the sub-Saharan lineages, we tried to assign to a specific sub-Saharan region our Dominican samples based on their haplogroup affiliation. To this aim, we subdivided the sub-Saharan Africa in four macroregions: western Africa, central Africa, eastern Africa, and southern Africa (supplementary fig. 1, Supplementary Material online). Our aim was to analyze how the trans-Atlantic slave trade shaped the composition of the putatively sub-Saharan Y clades: for this reason, we focused only on the western African and central African macroregions, which have been the main source areas of African slaves (Eltis and Richardson 2010), ignoring the lineages with a different sub-Saharan origin (i.e. eastern Africa or southern Africa) (supplementary table 2, Supplementary Material online).
Our sample collection includes also nomadic populations, namely Fulbe, Tuareg, and Bantu. The former occupies a wide area of the Sahelian belt, stretching from Senegal to Sudan. Although their origins are still matter of debate, the first records of their presence place them in western Africa, so they have been considered of western African origin. Tuareg are Sahelian nomadic people; however, their northern African origin is well known (Pereira et al. 2010; Ottoni et al. 2011), so they can be grouped with northern Africans and we did not include them in this analysis. Bantu people are widespread in sub-Saharan Africa as a consequence of the Bantu expansion, that begun no more than 5,000 years ago and has been linked with the spread of agriculture and iron technology. The Bantu’s place of origin has been located in central Africa (between Cameroon and Nigeria), where their expansion started towards eastern and southern Africa (Beleza et al. 2005; de Filippo et al. 2011; Ansari Pour et al. 2013; Campbell et al. 2014). Taking into account the Bantu influx on the sub-Saharan people, we classified the Y lineages related to the Bantu expansion as central African.
To assign each Dominican sample to western or central Africa, we considered the distribution of their Y haplogroup in the African continent, assigning the sample to one of the two areas if the frequency difference was at least an order of magnitude. Doubtful cases have been further dissected analyzing their Y-STR profiles with the UPGMA approach (supplementary table 3, Supplementary Material online). This Y-STR analysis allowed us to define as western African the E1a-M33* individual, whereas it was not possible to unambiguously assign the E1a-Z932 and the E2-M75 individuals.
Using this frequency approach, we were able to assign 47 Dominican individuals (≈26% of our sample) to central Africa, 14 (≈7%) to western Africa and only other 8 individuals have not been assigned (table 2).
Table 2.
Y Haplogroup Frequencies of the Sub-Saharan Haplogroups in the Dominican Republic Compared with Their Frequency in Western and Central Africa and Ancestry of the Same Y Lineages Inferred from Their Frequencies
| Haplogroups | Dominican Republic | Central Africa | Western Africa | Ancestry |
|---|---|---|---|---|
| B2a-M109 | 0.02 | 0.07 | 0.00 | Central Africa |
| E1a-M33* | 0.01 | 0.00 | 0.00 | Western Africa |
| E1a-Z932 | 0.01 | 0.02 | 0.06 | Unknown |
| E1b1a-CTS10066 | 0.01 | 0.00 | 0.04 | Western Africa |
| E1b1a-M58 | 0.01 | 0.00 | 0.00 | Unknown |
| E1b1a-M10 | 0.01 | 0.02 | 0.00 | Central Africa |
| E1b1a-V2003* | 0.01 | 0.00 | 0.02 | Western Africa |
| E1b1a-V3224* | 0.01 | 0.00 | 0.00 | Western Africa |
| E1b1a-U209* | 0.02 | 0.02 | 0.03 | Unknown |
| E1b1a-M4215 | 0.01 | 0.04 | 0.00 | Central Africa |
| E1b1a-V3862* | 0.01 | 0.02 | 0.00 | Central Africa |
| E1b1a-V2580 | 0.04 | 0.01 | 0.00 | Central Africa |
| E1b1a-U290 | 0.02 | 0.10 | 0.04 | Central Africa |
| E1b1a-V7937* | 0.01 | 0.00 | 0.00 | Western Africa |
| E1b1a-M191* | 0.02 | 0.00 | 0.00 | Western Africa |
| E1b1a-M4313 | 0.01 | 0.01 | 0.00 | Central Africa |
| E1b1a-U174 | 0.14 | 0.21 | 0.03 | Central Africa |
| E1b1a-Z15939 | 0.03 | 0.00 | 0.35 | Western Africa |
| E2-M75* | 0.01 | 0.01 | 0.01 | Unknown |
Our data suggest a disproportion of central African ancestry compared with the western African one. Two haplogroups were particularly informative about the sub-Saharan origin of our Dominican samples, namely E1b1a-U174 and E1b1a-Z15939. These two lineages show a clear differentiation between central and western Africa. E1b1a-U174 is frequent in central Africa, whereas it is much less common in western Africa: its distribution in the African continent has been linked to the Bantu expansion. The E1b1a-Z15939 lineage show a different distribution, being mainly observed in western Africa, whereas in central Africa it is virtually observed only among Fulbe people (supplementary table 2, Supplementary Material online) (D’Atanasio et al. 2018).
In both cases, our Dominican sample show frequencies similar to central Africa for these two clades, further suggesting a predominant central Africa ancestry. To quantify the relative contribution of the two sub-Saharan regions in the Dominican people, we estimated its admixture considering the frequencies of the Dominican Y clades also observed in central and western Africa. The admixture analysis for the two sub-Saharan regions resulted in 83% (±0.1%) central African contribution and a 17% (±0.1%) contribution of western Africa with respect to the African haplogroups in our Dominican sample. These values approximately correspond to a 32% central African contribution and a 6% western African contribution after taking into account that the sub-Saharan component in our Dominican sample is about 38%. However, our results should be interpreted with the due cautions due to the difference in sample size and extent of sampling areas between our western African and central African groups.
Discussion
The African Ancestry of the Dominican Y Lineages
The trans-Atlantic slave trade, started by the Spanish colonists in the XV century and lasted until the XIX century, led to the deportation in America of about 10 million Africans to be exploited in the European colonies (Curtin 1969; Adhikari et al. 2017). It has been estimated from available archival records that approximately 5 million slaves were deported in the Caribbean region, with approximately 1 million only in the Hispaniola island (Voyages: The Trans-Atlantic Slave Trade Database, www.slavevoyages.org). The large majority of the African slaves were embarked in the harbors along the western–central Atlantic coasts from Senegal to Angola (Campbell et al. 2014), however little is known about the exact place of origin of the captives. Usually, the trans-Atlantic slave trade is divided into two main phases: the first one started just after the European colonization and involved mainly western African people from the Senegambia, accounting for less than 20% of all the deported slaves; the second peaked in the XVIII century and accounted for the large majority of the trade, deporting people from the Bight of Benin and Biafra (Eltis and Richardson 2010). Our data from the Y chromosome are in line with this scenario, observing a minority of western African lineages in the Dominican sample, whereas most of their Y clades seem to come from central Africa.
Interestingly, our results are also in line with data from mtDNA (Bandelt et al. 2001; Salas et al. 2004; Bukhari et al. 2017) and from genome-wide studies, that showed a larger amount of haplotypes of putative central African origin in the Caribbean populations (Moreno-Estrada et al. 2013; Gouveia et al. 2020); these findings suggest that both enslaved women and men in the Caribbean came mainly from the same African regions.
The exact place of origin within the broad central African region is not easy to state and our resolution level does not allow us to resolve this. However, some clues points to an origin from the coastal regions rather than the Sahelian area. Considering the Y haplogroup composition in our Dominican sample, we can note that the clades frequently observed in the Sahel are usually rare or absent. A good example is represented by some lineages internal to the E1b1a-M2 haplogroup, such as E1b1a-M10 and E1b1a-V5280, which are observed mainly in the Sahelian groups (D’Atanasio et al. 2018). Although the E1b1a-M2 is the most frequent African haplogroup in the Dominican Republic and in the whole Caribbean archipelago, these lineages are absent or very rare (supplementary table 2, Supplementary Material online). A stronger clue is represented by the American distribution of the R1b-V88 haplogroup, which is very frequent in the lake Chad basin and sharply decreases moving west and south (Cruciani et al. 2010; D’Atanasio et al. 2018). This haplogroup has not been observed in our Dominican sample and has been rarely reported from other Caribbean locations (Mendizabal et al. 2008; Bryc et al. 2010; Simms et al. 2012; Vilar et al. 2014; Benn Torres et al. 2015; Poznik et al. 2016; Mendisco et al. 2019), in line with a coastal origin of slaves rather than Sahelian. However, it is worth noting that this lineage was observed in an African slave dating back to the XVII century from the Caribbean island of Saint Martin (Schroeder et al. 2015), suggesting that slaves could come from other regions of the African continent, albeit at a lesser extent compared with western and central Africa (Heinz et al. 2015; Muzzio et al. 2018; Ongaro et al. 2019).
Sex-Biased Admixture in the Dominican Republic
The collapse of the Native populations and massive deportation of Africans heavily shaped the genetic features of the American people, with different degree of admixture in different areas depending on both the population size and social factors. Among the Iberian colonists, there was a marked predominance of males, leading to an early admixture between Iberian men and Native women. The interethnic admixture has continued to be common over time in the Iberian colonies, because of their legislation and patriarchal structure of the society. Although Europeans were at the top of the society and interethnic marriage was illegal, exceptions were allowed (e.g. change of ethnicity affiliation) and there were no consequences for European men for having children with native or African women, with a high rate of illegitimacy. Other elements of social fluidity were the fact that, according to the Iberian legislation, Native Americans were not officially slaves, African slaves could be freed under some circumstances and the mixed individuals were recognized with a distinct status (Wade 2009; Snyder 2015; Adhikari et al. 2017). All these aspects led to a higher level of admixture in the Iberian colonies compared with the British ones, where the slavery legislation and rules against interethnic relations were stricter and the social barriers were stronger (Skidmore 1972; Adhikari et al. 2017).
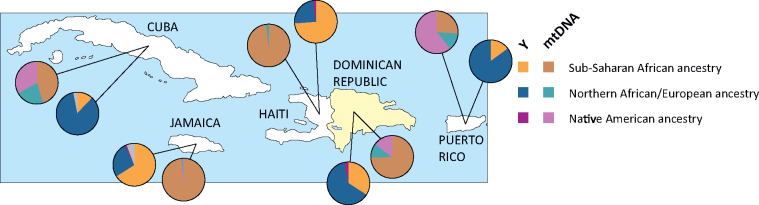
This sex-biased admixture is clearly shown by the differences between the Y and mtDNA haplogroup composition, with a relatively high proportion of Native Americans and African maternal lineages compared with the paternal lineages, which show a high frequency of European haplogroups (figs. 2 and 3) (Simms et al. 2012, 2013; Moreno-Estrada et al. 2013; Vilar et al. 2014; Bukhari et al. 2017; Schroeder et al. 2018; Benn Torres et al. 2019; Mendisco et al. 2019; Nieves-Colón et al. 2020).
In this context, the Dominican Republic is in line with the observations from other Caribbean and non-Caribbean American regions. All the mtDNA African lineages account for 61% of the maternal haplogroups, with the most frequent mtDNA lineages being the sub-Saharan L1c, L2a, L3b, and L3d, all reaching frequencies higher than 10%. The Native lineages are present at a frequency of 22%, slightly more than the European maternal clades (19%) (Torroni et al. 2001; Bukhari et al. 2017). From a Y haplogroup perspective, the European lineages represent the majority of haplogroups, followed by the African clades and with a very rare presence of Native clades (fig. 1). This genetic pattern reflects the society organization of the Iberian colonies, which the Dominican Republic was part of, characterized by a high degree of interethnic relationships between European men and Native American or African women.
Comparison between the Dominican Republic and Haiti and Other Antilles
Extending the analysis to the whole Antilles archipelago, the overall Y haplogroup pattern shows European and African lineages at high frequencies, with rare or absent Native lineages. The relative frequencies of European and African clades seem to be correlated with the type of colonial rule. In the British and French colonies, the segregation of slaves was stronger than in the Iberian territories, the interethnic marriages prohibited and no rights were recognized to children of parents of different ethnicity. Therefore, in the former British and French territories, the separation between the two main groups (i.e. European colonists and African slaves) was sharper and the admixture reduced. For this reason, the European Y lineages are usually less frequent than the African ones, partly reflecting the higher number of African slaves compared with European rulers in the colonies (Simms et al. 2011; Benn Torres et al. 2015, 2019; Mendisco et al. 2019). An example is represented by the Jamaica island in the Greater Antilles, under the British rule since 1655 and characterized by a higher proportion of Y African lineages compared with the European ones (Simms et al. 2012). Another example is represented by Haiti, which shares the Hispaniola island with the Dominican Republic, although the two countries have experienced different colonial rules and different histories. Differently from the Dominican Republic, Haiti passed under the French rule in 1697. Although the French law recognized the status of the admixed people (defined “coloreds,” from the French gens de couleur) and some rights were allowed to them, the slavery conditions were particularly harsh and brutal, with a high death rate of the African slaves and continuously new deportation from Africa. Because of the harsh conditions of the slaves and on the wave of the French revolution, a slave rebellion broke out in the colony that eventually ended with the independence of Haiti and a massacre of most of the white population (Coupeau 2008). This is reflected by the mtDNA and Y haplogroup composition of the Haitians. Considering the mtDNA haplogroup composition, no Native American lineages are reported and the European clades are rare (<2%), whereas more than 98% of the population show an African haplogroup (fig. 3). The Y haplogroup composition differ from the other American areas: the African lineages account for 70% of the Y lineages, with the sub-Saharan E1b1a-M2 clade reaching a frequency higher than 60%, whereas the European paternal lineages are less common compared with other former colonies (25%), and no Native clades have been reported (fig. 1). In the Dominican part of the island, the haplogroup composition dramatically changes, with a higher proportion of European Y lineages (fig. 2) and African mtDNA clades (Torroni et al. 2001; Bukhari et al. 2017).
Fig. 3.
Comparison between the ancestry of the Y and mtDNA lineages in the Greater Antilles. mtDNA data are from (Vilar et al. 2014; Bukhari et al. 2017; Ono et al. 2019; Zaidi and Makova 2019), Y chromosome data are the same as in figure 1, collapsing our results for the three Dominican groups with Dominican data from the literature.
The difference in the social organization between Iberian and British (and, at lesser extent, French and Dutch) colonies is reflected also by genome-wide analysis (Ongaro et al. 2019) and is also mirrored by the perceived ancestry, characterized by a high proportion of “mixed ancestry” and lower proportion of European, Native American, and African ancestries in the former Iberian colonies from central and southern America (Adhikari et al. 2017). However, in the former British or French colonies the African ancestry is usually the most reported. Interestingly, the Dominican Republic is the only Antillean region with a high proportion of perceived “mixed ancestry,” similar to the values reported in the former Iberian colonies from the continent and differently from the other Antilles, previously under the British, French, and Dutch rule (Adhikari et al. 2017).
Despite the differences in their relative proportion, the Y lineages observed in the Antilles (and in former colonial territories in general) are mainly the same (fig. 1). In particular, the presence of the same sub-Saharan Y haplogroups seems to suggest that the trans-Atlantic slavery routes and slave source regions were largely the same for most of the American colonies (Voyages: The Trans-Atlantic Slave Trade Database, www.slavevoyages.org).
Conclusions
Our analysis of the Y chromosome diversity of the Dominican Republic allow us to shed light on the colonial history of this country and of the Greater Antilles in general. First, our results suggest that most of the African slaves came from central Africa rather than western Africa, with minor inputs from other regions. Although our level of resolution does not allow an analysis of the geographic origins of slaves at a finer scale, the absence or paucity in the Dominican Republic of Y haplogroups frequent in the internal regions of central Africa points to the coastal regions as main slave source areas. Second, we observed an excess of European paternal lineages and Native or African maternal clades due to a sex-biased admixture, in line with the predominant role of European males in the colonial society. Third, the frequency of European or African paternal lineages varies in the Antilles and seems to be correlated with the type of colonial rule: the Iberian legislation was less strict as regards interethnic unions, leading to higher levels of admixture and higher frequencies of European Y haplogroups compared with colonies formerly under the British (or French and Dutch) rule.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Data Availability Statement
The data that support the findings of this study are available in the Supplementary material.
Supplementary Material
Acknowledgments
We are grateful to all the donors for providing biological samples. We also thank Chiara Della Rocca for Y-STR genotyping. This work was supported by the project “Organization of the settlements of the Tainos in the east and northern areas of the Dominican Republic” funded by DGSP—Office VI of MEACI [to A.C.] and by Sapienza University of Rome [grant number RM118164337D9395 to F.C.].
Literature Cited
- Adhikari K, Chacón-Duque JC, Mendoza-Revilla J, Fuentes-Guajardo M, Ruiz-Linares A. 2017. The genetic diversity of the Americas. Annu Rev Genomics Hum Genet. 18(1):277–296. [DOI] [PubMed] [Google Scholar]
- Anderson-Córdova KF. 1990. Hispaniola and Puerto Rico: Indian acculturation and heterogeneity, 1492–1550 [Ph. D. dissertation]. Yale University. Ann Arbor: University Microfilms. [Google Scholar]
- Anderson-Córdova KF. 2017. Surviving Spanish Conquest: Indian fight, flight, and cultural transformation in Hispaniola and Puerto Rico. Tuscaloosa: University of Alabama Press. [Google Scholar]
- Ansari Pour N, Plaster CA, Bradman N. 2013. Evidence from Y-chromosome analysis for a late exclusively eastern expansion of the Bantu-speaking people. Eur J Hum Genet. 21(4):423–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arredi B, et al. 2004. A predominantly Neolithic origin for Y-chromosomal DNA variation in North Africa. Am J Hum Genet. 75(2):338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H-J, et al. 2001. Phylogeography of the human mitochondrial haplogroup L3e: a snapshot of African prehistory and Atlantic slave trade. Ann Hum Genet. 65(6):549–563. [DOI] [PubMed] [Google Scholar]
- Barbieri C, et al. 2012. Contrasting maternal and paternal histories in the linguistic context of Burkina Faso. Mol Biol Evol. 29(4):1213–1223. [DOI] [PubMed] [Google Scholar]
- Batini C, et al. 2011. Signatures of the preagricultural peopling processes in sub-Saharan Africa as revealed by the phylogeography of early Y chromosome lineages. Mol Biol Evol. 28(9):2603–2613. [DOI] [PubMed] [Google Scholar]
- Beleza S, Gusmão L, Amorim A, Carracedo A, Salas A. 2005. The genetic legacy of western Bantu migrations. Hum Genet. 117(4):366–375. [DOI] [PubMed] [Google Scholar]
- Benn Torres J, et al. 2015. Genetic diversity in the lesser Antilles and its implications for the settlement of the Caribbean Basin. PLoS One 10(10):e0139192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benn Torres J, et al. 2019. Analysis of biogeographic ancestry reveals complex genetic histories for indigenous communities of St. Vincent and Trinidad. Am J Phys Anthropol. 169(3):482–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryc K, et al. 2010. Genome-wide patterns of population structure and admixture among Hispanic/Latino populations. Proc Natl Acad Sci. 107(Suppl 2):8954–8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari A, Luis JR, Alfonso-Sanchez MA, Garcia-Bertrand R, Herrera RJ. 2017. Taino and African maternal heritage in the Greater Antilles. Gene 637:33–40. [DOI] [PubMed] [Google Scholar]
- Campbell MC, Hirbo JB, Townsend JP, Tishkoff SA. 2014. The peopling of the African continent and the diaspora into the new world. Curr Opin Genet Dev. 29:120–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R. 1986. Gene admixture in human populations: models and predictions. Am J Phys Anthropol. 29(S7):1–43. [Google Scholar]
- Chiaroni J, Underhill PA, Cavalli-Sforza LL. 2009. Y chromosome diversity, human expansion, drift, and cultural evolution. Proc Natl Acad Sci. 106(48):20174–20179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupeau S. 2008. The history of Haiti. Westport (CT: ): Greenwood Press. [Google Scholar]
- Cruciani F, et al. 2002. A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am J Hum Genet. 70(5):1197–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciani F, et al. 2004. Phylogeographic analysis of haplogroup E3b (E-M215) Y chromosomes reveals multiple migratory events within and out of Africa. Am J Hum Genet. 74(5):1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciani F, et al. 2007. Tracing past human male movements in Northern/Eastern Africa and Western Eurasia: new clues from Y-chromosomal haplogroups E-M78 and J-M12. Mol Biol Evol. 24(6):1300–1311. [DOI] [PubMed] [Google Scholar]
- Cruciani F, et al. 2010. Human Y chromosome haplogroup R-V88: a paternal genetic record of early mid Holocene trans-Saharan connections and the spread of Chadic languages. Eur J Hum Genet. 18(7):800–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin PD. 1969. The Atlantic slave trade: a census. Madison (WI: ): University of Wisconsin Press. [Google Scholar]
- D’Atanasio E, et al. 2018. The peopling of the last Green Sahara revealed by high-coverage resequencing of trans-Saharan patrilineages. Genome Biol. 19(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Atanasio E, et al. 2019. Rapidly mutating Y-STRs in rapidly expanding populations: discrimination power of the Yfiler Plus multiplex in northern Africa. Forensic Sci Int Genet. 38:185–194. [DOI] [PubMed] [Google Scholar]
- de Filippo C, et al. 2011. Y-chromosomal variation in sub-Saharan Africa: insights into the history of Niger-Congo groups. Mol Biol Evol. 28(3):1255–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Rocca C, et al. Forthcoming 2020. Ethnic fragmentation and degree of urbanization strongly affect the discrimination power of Y-STR haplotypes in central Sahel. Forensic Sci Int Genet. doi:10.1016/j.fsigen.2020.102374 [DOI] [PubMed]
- Eltis D, Richardson D. 2010. Atlas of the transatlantic slave trade. New Haven (CT: ): Yale University Press; Available from: https://yalebooks.yale.edu/book/9780300212549/atlas-transatlantic-slave-trade (Accessed March 18, 2020). [Google Scholar]
- Fernandes DM, et al. 2020. A genetic history of the pre-contact Caribbean. bioRxiv doi:10.1101/2020.06.01.126730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia MH, et al. 2020. Origins, admixture dynamics and homogenization of the African gene pool in the Americas. Mol Biol Evol. 37:1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guitar L. 2002. Documenting myth of Taíno extinction. Available from: http://www.kacike.org/GuitarEnglish.pdf (Accessed June 23, 2020).
- Guitar L. 2007. Dominican Republic history by Hispaniola. Available from: https://web.archive.org/web/20070601040005/http://www.hispaniola.com/dominican_republic/info/history.php (Accessed June 24, 2020).
- Hammer O, Harper DAT, Ryan PD. 2001. PAST: paleontological Statistics software package for education and data analysis. Palaeontol Electron. 4:1–9. [Google Scholar]
- Heinz T, et al. 2015. The genomic legacy of the transatlantic slave trade in the Yungas Valley of Bolivia. PLoS One 10(8):e0134129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn BM, et al. 2012. Genomic ancestry of North Africans supports back-to-Africa migrations Schierup, MHE, editor. PLoS Genet. 8(1):e1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovacci G, et al. 2017. Forensic data and microvariant sequence characterization of 27 Y-STR loci analyzed in four Eastern African countries. Forensic Sci Int Genet. 27:123–131. [DOI] [PubMed] [Google Scholar]
- Jobling MA, Tyler-Smith C. 2017. Human Y-chromosome variation in the genome-sequencing era. Nat Rev Genet. 18:485–497. [DOI] [PubMed] [Google Scholar]
- Karafet TM, et al. 2008. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 18(5):830–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King TE, et al. 2007. Africans in Yorkshire? The deepest-rooting clade of the Y phylogeny within an English genealogy. Eur J Hum Genet. 15(3):288–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight FW. 1997. General history of the Caribbean. Paris, France: UNESCO Publishing; and London and Oxford: Macmillan Publishers Ltd. [Google Scholar]
- Kruskal WH, Wallis WA. 1952. Use of ranks in one-criterion variance analysis. J Am Stat Assoc. 47(260):583–621. [Google Scholar]
- Marques SL, Gusmão L, Amorim A, Prata MJ, Alvarez L. 2016. Y chromosome diversity in a linguistic isolate (Mirandese, NE Portugal): Y chromosome diversity in Mirandese speakers. Am J Hum Biol. 28(5):671–680. [DOI] [PubMed] [Google Scholar]
- Mendez FL, et al. 2011. Increased resolution of Y chromosome haplogroup t defines relationships among populations of the Near East, Europe, and Africa. Hum Biol. 83(1):39–53. [DOI] [PubMed] [Google Scholar]
- Mendisco F, et al. 2019. Tracing the genetic legacy in the French Caribbean islands: a study of mitochondrial and Y‐chromosome lineages in the Guadeloupe archipelago. Am J Phys Anthropol. 170:507–518. [DOI] [PubMed] [Google Scholar]
- Mendizabal I, et al. 2008. Genetic origin, admixture, and asymmetry in maternal and paternal human lineages in Cuba. BMC Evol Biol. 8(1):213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Estrada A, et al. 2013. Reconstructing the population genetic history of the Caribbean Tarazona-Santos, E, editor. PLoS Genet. 9(11):e1003925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzzio M, et al. 2018. Population structure in Argentina. PLoS One 13(5):e0196325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nägele K, et al. 2020. Genomic insights into the early peopling of the Caribbean. Science 369:456–460. [DOI] [PubMed] [Google Scholar]
- Nieves-Colón MA, et al. 2020. Ancient DNA reconstructs the genetic legacies of precontact Puerto Rico communities Mulligan, C, editor. Mol Biol Evol. 37(3):611–626. [DOI] [PubMed] [Google Scholar]
- Nogueiro I, Manco L, Gomes V, Amorim A, Gusmão L. 2010. Phylogeographic analysis of paternal lineages in NE Portuguese Jewish communities: paternal lineages in the Portuguese Jews. Am J Phys Anthropol. 141(3):373–381. [DOI] [PubMed] [Google Scholar]
- Ongaro L, et al. 2019. The genomic impact of European colonization of the Americas. Curr Biol. 29(23):3974–3986.e4. [DOI] [PubMed] [Google Scholar]
- Ono T, et al. 2019. Comparative study between Helicobacter pylori and host human genetics in the Dominican Republic. BMC Evol Biol. 19:197. doi:10.1186/s12862-019-1526-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottoni C, et al. 2011. Deep into the roots of the Libyan Tuareg: a genetic survey of their paternal heritage. Am J Phys Anthropol. 145(1):118–124. [DOI] [PubMed] [Google Scholar]
- Pereira L, et al. 2010. Linking the sub-Saharan and West Eurasian gene pools: maternal and paternal heritage of the Tuareg nomads from the African Sahel. Eur J Hum Genet. 18(8):915–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poznik GD, et al. 2016. Punctuated bursts in human male demography inferred from 1,244 worldwide Y-chromosome sequences. Nat Genet. 48(6):593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapone C, et al. 2016. Forensic genetic value of a 27 Y-STR loci multiplex (Yfiler® Plus kit) in an Italian population sample. Forensic Sci Int Genet. 21:e1–5. [DOI] [PubMed] [Google Scholar]
- Relethford JH. 2012. Human population genetics. Hoboken (NJ: ): Blackwell Pub. [Google Scholar]
- Rouse I. 1992. The Tainos. New Haven (CT: ): Yale University Press. [Google Scholar]
- Salas A, et al. 2004. The African Diaspora: mitochondrial DNA and the Atlantic slave trade. Am J Hum Genet. 74(3):454–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder H, et al. 2015. Genome-wide ancestry of 17th-century enslaved Africans from the Caribbean. Proc Natl Acad Sci. 112:3669–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder H, et al. 2018. Origins and genetic legacies of the Caribbean Taino. Proc Natl Acad Sci USA. 115(10):2341–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scozzari R, et al. 2014. An unbiased resource of novel SNP markers provides a new chronology for the human Y chromosome and reveals a deep phylogenetic structure in Africa. Genome Res. 24(3):535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms TM, et al. 2011. Paternal lineages signal distinct genetic contributions from British Loyalists and continental Africans among different Bahamian islands. Am J Phys Anthropol. 146(4):594–608. [DOI] [PubMed] [Google Scholar]
- Simms TM, et al. 2012. Y-chromosomal diversity in Haiti and Jamaica: contrasting levels of sex-biased gene flow. Am J Phys Anthropol. 148(4):618–631. [DOI] [PubMed] [Google Scholar]
- Simms TM, et al. 2013. Y-STR diversity and sex-biased gene flow among Caribbean populations. Gene 516(1):82–92. [DOI] [PubMed] [Google Scholar]
- Skidmore TE. 1972. Toward a comparative analysis of race relations since abolition in Brazil and the United States. J Lat Am Stud. 4(1):1–28. [Google Scholar]
- Snyder TL. 2015. Women, race, and the law in early America. Oxf Res Encycl Am Hist. doi:10.1093/acrefore/9780199329175.013.12 [Google Scholar]
- Solé-Morata N, et al. 2017. Whole Y-chromosome sequences reveal an extremely recent origin of the most common North African paternal lineage E-M183 (M81). Sci Rep. 7(1):15941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, et al. 2001. Do the four clades of the mtDNA haplogroup L2 evolve at different rates? Am J Hum Genet. 69(6):1348–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta B, et al. 2015. Phylogeographic refinement and large scale genotyping of human Y chromosome haplogroup E provide new insights into the dispersal of early pastoralists in the African continent. Genome Biol Evol. 7(7):1940–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, et al. 2001. The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Human Genet. 65(1):43–62. [DOI] [PubMed] [Google Scholar]
- Van Oven M, Van Geystelen A, Kayser M, Decorte R, Larmuseau MHD. 2014. Seeing the wood for the trees: a minimal reference phylogeny for the human Y chromosome. Hum Mutat. 35(2):187–191. [DOI] [PubMed] [Google Scholar]
- Vilar MG, et al. 2014. Genetic diversity in Puerto Rico and its implications for the peopling of the Island and the West Indies. Am J Phys Anthropol. 155(3):352–368. [DOI] [PubMed] [Google Scholar]
- Wade P. 2009. Race and sex in Latin America. London: Pluto Press; Available from: https://www.amazon.it/Race-Latin-America-Peter-Wade/dp/0745329497 (Accessed March 17, 2020). [Google Scholar]
- Wood ET, et al. 2005. Contrasting patterns of Y chromosome and mtDNA variation in Africa: evidence for sex-biased demographic processes. Eur J Hum Genet. 13(7):867–876. [DOI] [PubMed] [Google Scholar]
- Zaidi AA, Makova KD. 2019. Investigating mitonuclear interactions in human admixed populations. Nat Ecol Evol. 3(2):213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, et al. 2011. Extended Y chromosome investigation suggests postglacial migrations of modern humans into East Asia via the Northern Route. Mol Biol Evol. 28(1):717–727. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available in the Supplementary material.