Abstract
Colorectal cancer (CRC) is the third most common cancer in the world and is the second leading cause of cancer-related deaths. Several mutations are involved in the development of CRC. The prognostic significance of the KRAS mutation has been discussed in many studies. We aimed to investigate the prognostic significance of the number of KRAS mutations in metastatic CRC (mCRC).
Patients with mutations in the KRAS gene were included in the study. They were divided into 2 groups as single mutation and multiple mutations in the KRAS gene.
For the study, 425 CRC patients were screened. KRAS mutation was positive in 191 patients (45%). One hundred ninety-one patients were included in the study, 171 patients (90%) had single mutations and 20 patients (10%) had multiple mutations. Median progression-free survival was 12.8 months in patients with multiple mutations, while it was 8.8 months in patients with single mutations (P: .05). The median overall survival of patients with multiple mutations was 40.7 months, while it was 22.7 months for patients with single mutations (P = .01)
We found that the presence of multiple mutations in KRAS mutant patients was associated with better overall survival and progression-free survival than a single mutation.
Keywords: colorectal cancer, metastasis, mutations, prognosis, rat sarcoma
1. Introduction
Colorectal cancer (CRC) is the third most common cancer in the world and is the second leading cause of cancer-related deaths.[1] Approximately 20% of CRC patients have metastatic disease at the time of diagnosis.[2] The main goal of treatment in the metastatic phase is to improve the quality of life and prolong survival. Five-year overall survival (OS) is around 14% with conventional chemotherapy agents and biological agents discovered in the last 2 decades.[3]
Several mutations are involved in the development of CRC. Rat sarcoma (RAS) mutations are found in up to 50% of sporadic CRCs and 50% of colonic adenomas larger than 1 cm. They are rarely seen in smaller adenomas.[4,5] The RAS genes encode a group of GTPases that regulate cellular signal transduction by acting as a switch for the transmission of extracellular growth signals to the nucleus.[6] Activating RAS mutations cause the deterioration of RAS protein activity; in this way GTPase activity is decreased, the protein becomes resistant to GTP hydrolysis by GTPase, resulting in a structurally active GTP-bound protein and a continuous growth stimulus.[7,8] Therefore, oncogenic activity occurs. These results suggest that a carcinogen, by mutating the RAS proto-oncogene, activates its oncogenic potential and initiates the tumor formation process. There are 3 members of the RAS gene superfamily: KRAS, HRAS, and NRAS, and approximately 40% of CRCs contain KRAS mutations.[9,10] While KRAS exon 2 mutations constitute the majority of KRAS mutations, exon 3 and exon 4 mutations constitute only 1% to 4% of CRCs.[11] Activating mutations in exons 2 and 3 have similar effects on RAS GTPase activity, whereas exon 4 mutations increase GDP to GTP exchange.[12] KRAS point mutations occur particularly in codon 12 and to a lesser extent in codon 13 and 61.[13] KRAS codon 12 or 13 (exon 2) mutations constitute about 90% of all mutations.[14–22] The prognostic significance of KRAS mutations has been discussed in many studies.[18,23–26] However, its prognostic role in metastatic CRC (mCRC) remains controversial. There is insufficient information in the literature on the prognostic status of mutation of the ras gene and whether it is related to the number of mutations in this gene. Therefore, since there is not enough information in the literature on this subject, in this study we aimed to investigate the prognostic significance of the number of KRAS mutations in mCRC.
2. Materials and methods
2.1. Patients
The data of patients who applied to our center with a diagnosis of mCRC between 2008 and 2019 were reviewed retrospectively. Patients with mutations in the KRAS gene were included in the study. They were divided into 2 groups as single mutation and multiple mutations in the KRAS gene and the data obtained were analyzed.
Inclusion criteria: Patients older than 18 years of age with KRAS mutation analysis, histologically proven CRC and metastatic disease according to the Response Evaluation Criteria in Solid Tumors (RECIST) criteria and who received at least 1 step of oxaliplatin or irinotecan-based chemotherapy regimen. Patients with a second primary tumor, brain metastasis or NRAS and BRAF mutation were excluded from the study.
Age, sex, family history, smoking history, Eastern Cooperative Oncology Group (ECOG) performance status, body mass index, carcinoembryonic antigen (CEA) level, presence of KRAS mutation, tumor localization, histopathological subtype, numbers and localizations of metastatic regions, and systemic treatments they received were recorded.
KRAS mutation analysis was evaluated with the “real time PCR” method using DNA obtained from paraffin blocks prepared from cancer tissue. With this method, codon 12/13 mutations in exon 2, codon 59/61 in exon 3 and codon 117/146 mutations in exon 4 were detected. Tissue samples used for KRAS mutation analysis were obtained from the primary tumor site by colonoscopy in 153 patients (80%), and by tru-cut biopsy from liver metastasis in 38 patients (20%).
The first-line treatment of patients was one of the following regimens: modified FOLFOX6 or FOLFIRI. The FOLFOX combination included oxaliplatin 85 mg/m2 in 250 mL of 5% dextrose, concurrent with folinic acid (FA) 400 mg/m2/d administered as a 2-hour intravenous infusion, followed by a 10-minute bolus of 5-fluorouracil (5-FU) 400 mg/m2, and a 46-hour continuous infusion of 5- FU 2400 mg/m2. The FOLFIRI combination regimen comprised irinotecan 180 mg/m2, followed by FA 400 mg/m2 in a 2-hour infusion, and then a 10-minute bolus of 5-FU 400 mg/m2, and a 46-hour continuous infusion of 5-FU 2400 mg/m2. For patients receiving anti-VEGF treatment, bevacizumab was administrated either with FOLFOX or FOLFIRI regimens at a dose of 5 mg/kg. The cycles were repeated every 2 weeks.
Tumor response was evaluated every 12 weeks by computed tomography or PET/CT according to RECIST version 1.1 criteria. According to RECIST 1.1 criteria, complete response is disappearance of all target lesions and reduction in the short axis measurement of all pathologic lymph nodes to ≤10 mm, partial response (PR) is defined as ≥30% decrease in tumor size, progressive disease is defined as ≥20% increase in tumor size, and for stable disease neither PR nor progressive disease criteria are met. Objective response rate is defined as the sum of complete response and PR.
2.2. Factors evaluated in univariate and multivariate analysis
Based on previous studies, 15 variables were selected that may have an impact on OS. Variables were divided into 2 categories: age (<65 years or ≥65 years), sex (male or female), ECOG performance status (0–1 or 2), body mass index (<25 kg/m2 or ≥25 kg/m2), tumor localization (right or left), histological subtype (adenocarcinoma or mucinous carcinoma), differentiation (good-medium or bad), KRAS mutation number (single or multiple), CEA level (<5 mg/dl or ≥5 mg/dl), metastasis status (synchronous or metachronous), metastatic region number (1 or ≥2), liver metastasis (present or absent), lung metastasis (present or absent), peritoneal metastasis (present or absent), chemotherapy regimen used in first step treatment (oxaliplatin-based or irinotecan-based), and the use of bevasizumab in the first step (yes or no). Factors with P ≤ .1 in univariate analysis were evaluated by multivariate analysis.
2.3. Statistical analyses
The period from metastatic diagnosis to death was evaluated as OS. The date of first-line treatment and the duration of disease progression or death for any reason were calculated as progression-free survival (PFS).
To evaluate the findings of the study, SPSS 22.0 (Statistical Package for the Social Sciences) computer program was used for statistical analysis. Differences of properties between 2 groups were analyzed with Pearson chi-square and Fisher exact test. Survival analysis was analyzed with the Kaplan-Meier method using the Log-rank test. Survival times were within the 95% CI (confidence interval) range. Cox regression test was used for multivariate analysis. P < .05 was considered statistically significant.
This study was approved by our Institutional Ethics Committee and was conducted according to the principles of the Declaration of Helsinki. (Decision Date: 27.02.2020, Decision No: E1-20-284).
3. Results
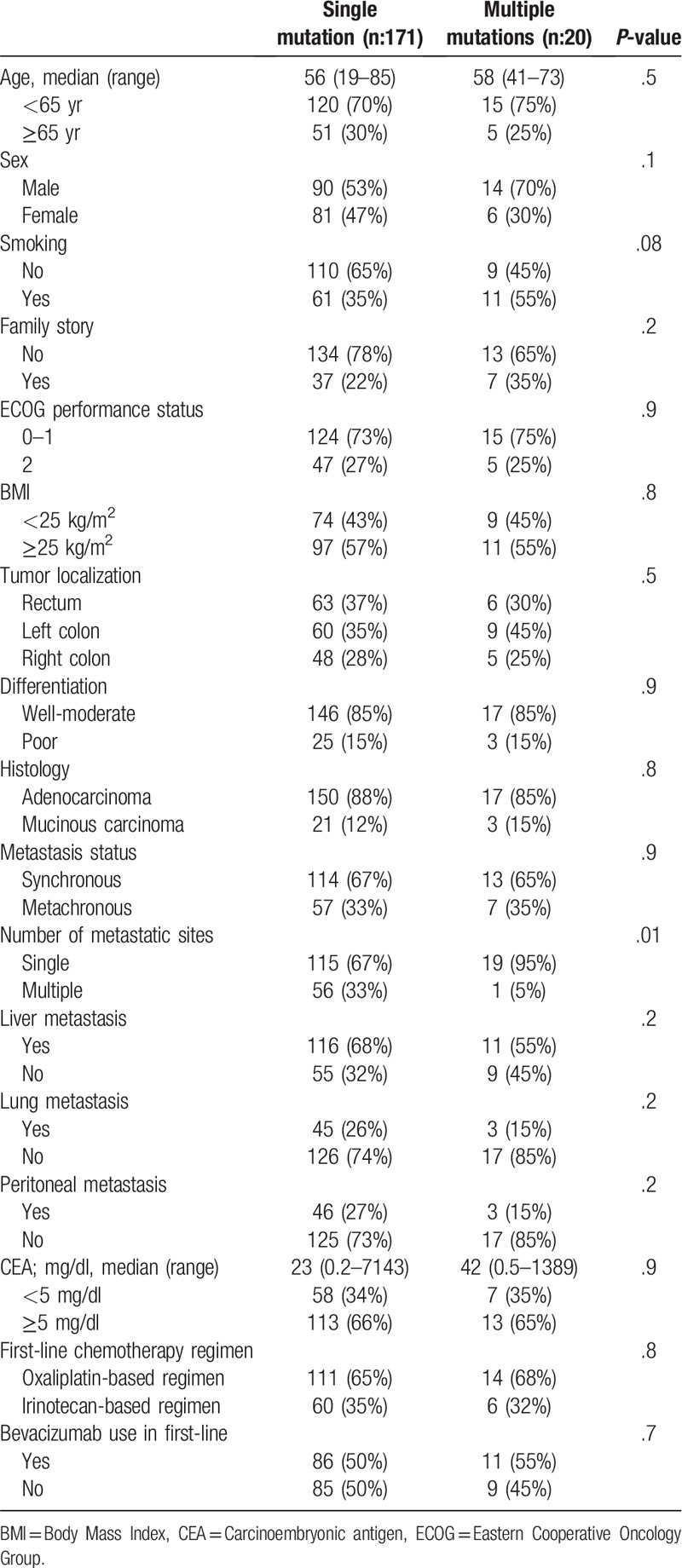
For the study, 425 CRC patients were screened. KRAS mutation was positive in 191 patients (45%). One hundred ninety-one patients were included in the study, 171 patients (90%) had single mutations and 20 patients (10%) had multiple mutations. The median age of all patients was 56 years (range; 19–85) and 54% were male. When we divided patients with KRAS gene mutations into single mutation and multiple mutations, there was no difference in terms of basal features except for the number of metastatic regions. Multiple metastases in patients with single mutations were higher than those with multiple mutations (33% vs 5%, respectively, P = .01). Baseline patient characteristics are given in Table 1. There were exon 2 mutations in 158 (92%), exon 3 mutations in 6 (4%) and exon 4 mutations in 7 (4%) of 171 patients with a single mutation. There were codon 12 mutations in 76% of patients with exon 2 mutations. While 16 (80%) of 20 patients with multiple mutations had more than 1 mutation in exon 2, 3 patients (15%) had exon 2 and 3 comutations, and 1 patient (5%) had exon 2 and 4 comutations. While 19 of 20 patients with multiple mutations had 2 mutations, 1 patient had 3 mutations and all had codon 12 mutations. The 3 most common amino acid mutations in the single mutation group were G12D (n: 51,30%), G12V (n: 38,22%), and G13D (n: 33,19%), and in the multiple mutation group (since there were multiple mutations, the total number exceeds 100%) were G12V (n: 8,40%), G12D (n: 8,40%) and G12C (n: 7,35%). In comparisons of amino acid mutations, OS was 22.8 months in patients with a G13D mutation, 30.6 months in patients with a G12V mutation, 23.3 months in patients with a G12D mutation, and 28.4 months in patients with a G12C mutation. There was no significant difference in terms of OS in amino acid mutation comparisons (G13D vs G12V P = .7, G13D vs G12D P = .8, G13D vs G12C P = .8, G12 V vs G12D P = .4, G12 V vs G12C P = .7, G12D vs G12C P = .7).
Table 1.
Baseline characteristics.
There was no statistical difference in terms of chemotherapy regimens and bevacizumab usage rates in patients with single and multiple mutations in the first step treatment. 65% of patients with a single mutation and 68% of those with multiple mutations received an oxaliplatin-based chemotherapy regimen in the first step. Bevacizumab usage rates were 50% and 55%, respectively.
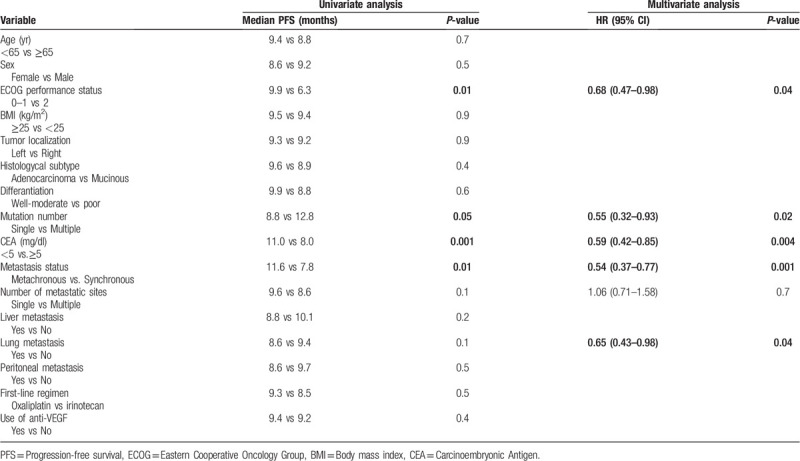
The median follow-up period of our study was 21.0 months (3–95 months). There was no difference in objective response between patients with single and multiple mutations (51% vs 48%, P = .7, respectively). The median PFS of all patients was 9.3 months (95% CI 8.1–10.5). The median PFS was significantly longer for patients with multiple mutations than for patients with a single mutation. Median PFS was 12.8 months for patients with multiple mutations, while it was 8.8 months for patients with single mutations (P = .05). Survival curves are given in Figure 1. In the univariate analysis for PFS, we detected 5 more variables with P ≤ .1. In multivariate analysis for PFS, multiple mutations were found to be an independent prognostic factor associated with good prognosis (HR: 0.55, 95% CI 0.32–0.93, P = .02). Apart from the number of mutations, ECOG performance score of 0 to 1, CEA < 5 mg/dl, metachronous metastasis and absence of lung metastasis were independent positive prognostic factors. Univariate and multivariate analysis results for PFS are given in Table 2.
Figure 1.
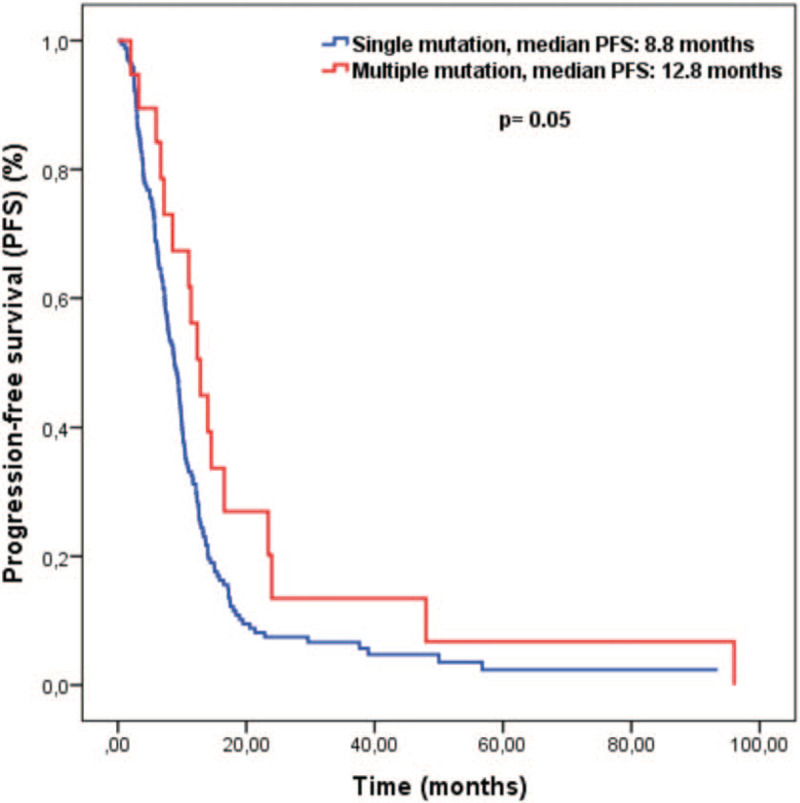
Kaplan-Meier curves for progression-free survival.
Table 2.
Univariate and multivariate analysis for progression-free survival.
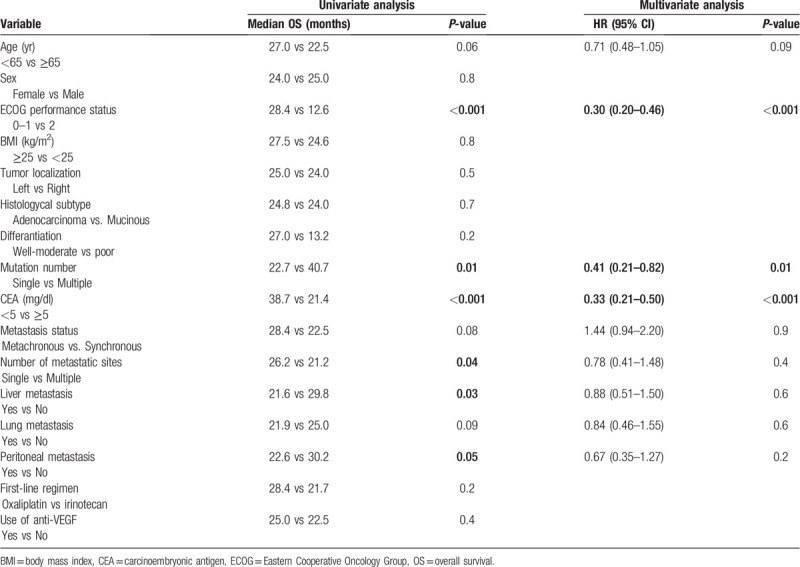
The median OS of all patients was 24.7 months (95% CI 21.5–27.8). Median OS was significantly longer in patients with multiple mutations than patients with a single mutation. The median OS in patients with multiple mutations was 40.7 months, while it was 22.7 months for patients with single mutations (P = .01). Survival curves are given in Figure 2. In the univariate analysis for OS, we found 8 more variables with P ≤ .1. In the multivariate analysis for OS, it was observed that multiple mutations were independent prognostic factors associated with a good prognosis (HR: 0.41, 95% CI 0.21–0.82, P = .01). Except for the number of mutations, ECOG performance score of 0 to 1 and CEA < 5 mg/dl were found to be positive prognostic factors independent of OS. Single and multivariate analysis results for OS are given in Table 3.
Figure 2.
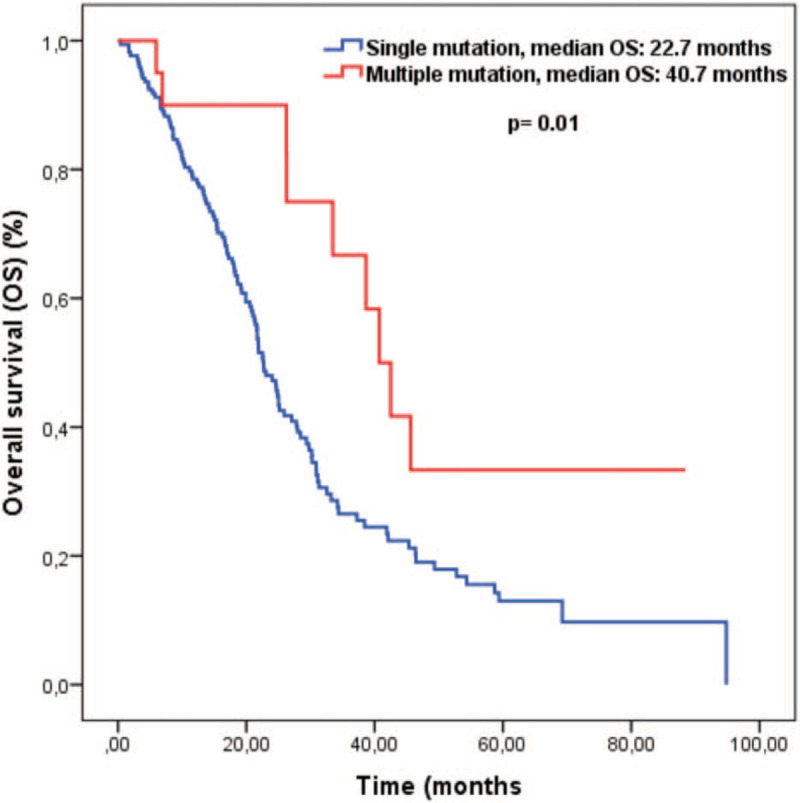
Kaplan-Meier curves for overall survival.
Table 3.
Univariate and multivariate analysis for overall survival.
4. Discussion
To our knowledge, this is the first study that has investigated the prognostic significance of the number of KRAS mutations in patients with mCRC. In this study, we found the incidence of KRAS mutations to be 45% and this result was similar to other studies in Turkey.[27,28] A single KRAS mutation was determined in 90% of patients, of which 92% had a mutation in exon 2, 4% in exon 3 and 4% in exon 4. 10% of patients had multiple KRAS mutations. 76% of exon 2 mutations were codon 12 mutations. Eighty percent of patients with multiple mutations had these mutations in exon 2. Ergun et al showed that 47% of patients with mCRC had a KRAS mutation, of which 92% had a mutation in exon 2, 5% in exon 3, 5% in exon 4, and 12% in multiple exons or codon locations.[27] In another study with 353 Chinese CRC patients, a KRAS mutation was detected in 52.7% of patients and about 80.1% of KRAS mutant patients had mutations in exon 2, 4.3% in exon 3 and 15.5% in exon 4.[29] In the same study, among exon 2 mutations, most of the patients had single mutations in codon 12.[29] Different results between studies may be related to ethnic and environmental factors.
Some of the previous studies have shown that the presence of a KRAS mutation is associated with poor prognosis while others showed no effect on prognosis.[20,30] In the mentioned studies, patients with and without KRAS mutation were compared. In this study, we examined the relationship between the number of mutations and prognosis in KRAS mutant mCRC patients. The median PFS and OS values were statistically higher in patients with multiple mutations (8.8 vs 12.8 months, respectively) than those with a single mutation (22.7 vs 40.7 months, respectively). In analysis for PFS and OS, the presence of multiple mutations was evaluated as an independent prognostic factor associated with good cancer prognosis. To the best of our knowledge, in the literature, we could not find any study investigating the relationship between mutation count and survival in patients with KRAS mutant CRC.
The multicenter RASCAL study[17] investigated the prognostic significance of codon 12 and codon 13 mutations of exon 2, and found that only codon 12 mutations were independently associated with an increased risk of recurrence and death. Another study showed that KRAS exon 3 mutations may provide less aggressive biological behavior, because an exon 3 mutation was found to be associated with a lower TNM stage and a smaller/less invasive tumor.[29] In this study, KRAS mutant patients were evaluated among themselves. In 80% of patients with multiple mutations, in addition to the codon 12 mutation, there were different mutations in exon 2. Twenty percent of our patients with multiple mutations had exon 3 or exon 4 mutations. The presence of more than 1 mutation may suppress the poor prognostic effect of a single mutation. In addition, exon 3 and exon 4 mutations may be less aggressive than exon 2 mutations.
In a meta-analysis of 5 large studies, G12C and G13D mutations were reported to be associated with the worse OS.[18] In another study, no difference was found between G12V, G12D, and G13D mutations in terms of PFS and OS.[27] We cannot currently explain whether the number of multiple mutations is associated with good prognosis because spot mutations silence or activate each other, or if some specific amino acid mutations (such as G13D) are related to a worse prognosis.
Although there is no data on the prognostic significance of the number of mutations in colon cancer, our findings can suggest differences in tumor biology between single mutation and multiple mutations in terms of tumor aggressiveness and treatment sensitivity that might be interfered with by different point of mutations in the KRAS gene.
This study had some limitations. The number of patients was relatively small, and since a treatment comparison could not be made, we could not analyze the predictive effect of multiple mutations. In order to avoid confounding bias that may be caused by mutations in the NRAS gene, we included only patients with KRAS gene mutations. However, being the first study on this subject and evaluating 15 different variables are the strengths of our study.
In summary, we found that the presence of multiple mutations in KRAS mutant patients was associated with better OS and PFS than the presence of a single mutation. More mechanistic studies with larger patient numbers are needed to support our hypotheses.
Author contributions
Conceptualization: Gokhan Ucar.
Data curation: Gokhan Ucar, Merve Dirikoc.
Methodology: Yakup Ergun, Selin Akturk Esen, Yusuf Acikgoz.
Project administration: Gokhan Ucar, Yakup Ergun, Selin Akturk Esen.
Visualization: Yakup Ergun, Oznur Bal, Dogan Uncu.
Writing – original draft: Selin Akturk Esen.
Writing – review & editing: Selin Akturk Esen, Yusuf Acikgoz, Irfan Esen.
Footnotes
Abbreviations: BMI = body mass index, CEA = carcinoembryonic antigen, CR = complete response, CRC = colorectal cancer, ECOG = Eastern Cooperative Oncology Group, OS = overall survival, PD = progressive disease, PFS = progression free survival, PR = partial response, RAS = rat sarcoma, RECIST = Response Evaluation Criteria in Solid Tumors.
How to cite this article: Ucar G, Ergun Y, Esen SA, Acikgoz Y, Dirikoc M, Esen İ, Bal Ö, Uncu D. Prognostic and predictive value of KRAS mutation number in metastatic colorectal cancer. Medicine. 2020;99:39(e22407).
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
The authors have no conflicts of interests to disclose.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019; 69: 7–34. 2019/01/09. DOI: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- [2].Evaluation of Genomic Applications in P and Prevention Working G. Recommendations from the EGAPP Working Group. Can testing of tumor tissue for mutations in EGFR pathway downstream effector genes in patients with metastatic colorectal cancer improve health outcomes by guiding decisions regarding anti-EGFR therapy? Genet Med 2013;15:517–27. [DOI] [PubMed] [Google Scholar]
- [3].American Cancer Society. Cancer Facts & Figures 2020. Atlanta: American Cancer Society; 2020. Available at: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and- statistics/annual-cancer-facts-and-figures/2020/cancer-facts-and-figures-2020.pdf. [access date April 01, 2020]. [Google Scholar]
- [4].Takayama T, Ohi M, Hayashi T, et al. Analysis of K-ras, APC, and beta-catenin in aberrant crypt foci in sporadic adenoma, cancer, and familial adenomatous polyposis. Gastroenterology 2001;121:599–611. [DOI] [PubMed] [Google Scholar]
- [5].Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N Engl J Med 1988;319:525–32. [DOI] [PubMed] [Google Scholar]
- [6].Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature 1991;349:117–27. [DOI] [PubMed] [Google Scholar]
- [7].Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer 2007;7:295–308. [DOI] [PubMed] [Google Scholar]
- [8].Mor A, Philips MR. Compartmentalized Ras/MAPK signaling. Annu Rev Immunol 2006;24:771–800. [DOI] [PubMed] [Google Scholar]
- [9].Lea IA, Jackson MA, Li X, et al. Genetic pathways and mutation profiles of human cancers: site- and exposure-specific patterns. Carcinogenesis 2007;28:1851–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Rajagopalan H, Bardelli A, Lengauer C, et al. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature 2002;418:934. [DOI] [PubMed] [Google Scholar]
- [11].Edkins S, O’Meara S, Parker A, et al. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol Ther 2006;5:928–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Janakiraman M, Vakiani E, Zeng Z, et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res 2010;70:5901–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bos JL. Ras oncogenes in human cancer: a review. Cancer Res 1989;49:4682–9. [PubMed] [Google Scholar]
- [14].Peeters M, Kafatos G, Taylor A, et al. Prevalence of RAS mutations and individual variation patterns among patients with metastatic colorectal cancer: a pooled analysis of randomised controlled trials. Eur J Cancer 2015;51:704–1713. [DOI] [PubMed] [Google Scholar]
- [15].Colussi D, Brandi G, Bazzoli F, et al. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. Int J Mol Sci 2013;14:16365–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cerottini JP, Caplin S, Saraga E, et al. The type of K-ras mutation determines prognosis in colorectal cancer. Am J Surg 1998;175:198–202. [DOI] [PubMed] [Google Scholar]
- [17].Andreyev HJ, Norman AR, Cunningham D, et al. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J Natl Cancer Inst 1998;90:675–84. [DOI] [PubMed] [Google Scholar]
- [18].Samowitz WS, Curtin K, Schaffer D, et al. Relationship of Ki-ras mutations in colon cancers to tumor location, stage, and survival: a population-based study. Cancer Epidemiol Biomarkers Prev 2000;9:1193–7. [PubMed] [Google Scholar]
- [19].Yoon HH, Tougeron D, Shi Q, et al. KRAS codon 12 and 13 mutations in relation to disease- free survival in BRAF-wild-type stage III colon cancers from an adjuvant chemotherapy trial (N0147 alliance). Clin Cancer Res 2014;20:3033–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Modest DP, Ricard I, Heinemann V, et al. Outcome according to KRAS-, NRAS- and BRAF- mutation as well as KRAS mutation variants: pooled analysis of five randomized trials in metastatic colorectal cancer by the AIO colorectal cancer study group. Ann Oncol 2016;27:1746–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Taieb J, Zaanan A, Le Malicot K, et al. Prognostic effect of BRAF and KRAS mutations in patients with stage iii colon cancer treated with leucovorin, fluorouracil, and oxaliplatin with or without cetuximab: a post hoc analysis of the PETACC-8 trial. JAMA Oncol 2016;2:643–53. [DOI] [PubMed] [Google Scholar]
- [22].Taieb J, Le Malicot K, Shi Q, et al. Prognostic value of BRAF and KRAS mutations in MSI and MSS stage III colon cancer. J Natl Cancer Inst 2017;109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Roth AD, Tejpar S, Delorenzi M, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol 2010;28:466–74. [DOI] [PubMed] [Google Scholar]
- [24].Andreyev HJ, Norman AR, Cunningham D, et al. Kirsten ras mutations in patients with colorectal cancer: the ’RASCAL II’ study. Br J Cancer 2001;85:692–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bazan V, Migliavacca M, Zanna I, et al. Specific codon 13 K-ras mutations are predictive of clinical outcome in colorectal cancer patients, whereas codon 12 K-ras mutations are associated with mucinous histotype. Ann Oncol 2002;13:1438–46. [DOI] [PubMed] [Google Scholar]
- [26].Dinu D, Dobre M, Panaitescu E, et al. Prognostic significance of KRAS gene mutations in colorectal cancer--preliminary study. J Med Life 2014;7:581–7. [PMC free article] [PubMed] [Google Scholar]
- [27].Ergun Y, Acikgoz Y, Bal O, et al. KRAS codon 12 and 13 mutations may guide the selection of irinotecan or oxaliplatin in first-line treatment of metastatic colorectal cancer. Expert Rev Mol Diagn 2019;19:1131–40. [DOI] [PubMed] [Google Scholar]
- [28].Karadayi H, AÇ, Alkac M, et al. Molecular analysis of Kras and Nras mutations in a series of 720 patients for guiding the treatment of metastatic colorectal cancer in Turkey. J Clin Oncol 2016;34:e13115. [Google Scholar]
- [29].Guo F, Gong H, Zhao H, et al. Mutation status and prognostic values of KRAS, NRAS, BRAF and PIK3CA in 353 Chinese colorectal cancer patients. Sci Rep 2018;8:6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Andreatos N, Ronnekleiv-Kelly S, Margonis GA, et al. From bench to bedside: clinical implications of KRAS status in patients with colorectal liver metastasis. Surg Oncol 2016;25:332–8. [DOI] [PubMed] [Google Scholar]


