Abstract
Objectives:
To investigate the association between dietary fiber density (grams of fiber consumed per 100 kcal) with the gut-muscle axis in older adult men.
Design:
Cross-sectional study.
Setting:
Osteoporotic Fractures in Men (MrOS) cohort participants at Visit 4 (2014–16).
Participants:
Older adult men (average age, 85y) from the MrOS study.
Measurements:
Men who were in the highest tertiles for dietary fiber density and the percentage of whole body lean mass were defined as T3T3 (n=42), whereas men who were in the lowest and intermediate tertiles for these variables were defined as T1T1 (n=32), T1T3 (n=24), and T3T1 (n=13), respectively. Additionally, measures of physical function, including the short physical performance battery (SPPB) score and grip strength were higher in T3T3 when compared with T1T1. Gut bacterial abundance was quantified with use of 16S v4 rRNA sequencing, and the bacterial functional potential was derived from the 16S data with PICRUSt. Chao1, ACE, Shannon, Simpson, and Fisher indices were used as measures of α-diversity. Weighted and unweighted Unifrac, and Bray-Curtis were used as measures of β-diversity. Age, physical activity score, smoking, and number of medications-adjusted DESeq2 models were used to identify bacteria and functions that were different when comparing T3T3 with T1T1, but that were not also different when comparing T3T3 with T1T3 or T3T1.
Results:
α-diversity was not different, but significant differences for β-diversity (unweighted UniFrac, Bray-Curtis) were identified when comparing T3T3 with T1T1. Known butyrate-producing bacteria, including Ruminococcus, Lachnospira, and Clostridia, and gene counts for butyrate production (KEGG IDs: K01034, K01035) were higher in T3T3, when compared with T1T1.
Conclusion:
These data suggest that a high-fiber diet may positively impact butyrate-producing genera and gene counts, which collectively may be involved in mechanisms related to the percentage of whole body lean mass and physical functioning in older adult men. Future studies aimed at testing the causative role of this hypothesis are of interest.
Keywords: Older adults, dietary fiber, microbiome, lean mass, physical function, muscle strength
Introduction
A role for the gut microbiome on the maintenance of whole body lean mass, skeletal muscle mass, and physical function (defined as the gut-muscle axis) has been proposed by several independent research groups (1–5). In support of this, muscle mass and physical function are reduced in germ-free and in antibiotic treated mice (6–10). In humans, higher levels of Prevotellaceae were found in young elite athletes who had a high percentage of whole body lean mass (%WBLM), when compared with age- and sex-matched controls (11). Similarly, Prevotellaceae were elevated in older adults who had a high %WBLM and good physical function, when compared with older adults who had lower values for these variables (12). Although these studies suggest a role for the gut microbiome on the maintenance of lean mass, skeletal muscle mass, and physical functioning, the involved mechanisms are less clear.
One gut microbiome-related pathway that affects the gut-muscle axis involves the short chain fatty acids (SCFAs), acetate, propionate, and butyrate. SCFAs are produced via fiber fermentation by colonic bacteria, and increasing dietary fiber intake results in proportional increases in fecal levels of acetate, propionate, and butyrate (13). SCFAs positively affect the gut-muscle axis. Muscle mass and physical function are increased in young germ-free mice fed a combination of acetate, propionate, and butyrate (7), and %WBLM and the muscle mass/body weight ratio are increased in response to dietary butyrate supplementation in aged mice (14). In young adult humans, fecal SCFAs increased in association with exercise training-induced improvements for %WBLM and endurance exercise capacity (15), and cross-sectionally, fecal levels of SCFAs were increased in the elite athletes of Clarke et al (11), who had higher levels of WBLM when compared with controls.
In older adults, a role for dietary fiber on the gut-muscle axis is supported based on the findings that a low dietary fiber density (< 0.5g of dietary fiber per 100 kcal) is associated with an increased risk for future functional limitation when compared with higher fiber density intakes (> 0.84g/100 kcal) (16), and dietary fiber intake is positively associated with muscle strength (17) and physical functioning (18). More directly, grip strength was increased in older adults in response to 13-weeks of dietary supplementation with the fermentable fiber, inulin (19).
However, despite fiber’s effect on bacterial SCFA production and its association with better physical function, what is notably absent from these studies in older adults is identification of the associated alterations in gut microbiome composition and SCFA-related functions. Accordingly, in the present study we hypothesized that SCFA-producing bacteria and gene counts would be higher in older adult men from the MrOS study who had a high dietary fiber density, a higher proportion of WBLM, and better physical function, when compared with men who had lower values for these variables.
Methods
Subject data
Data were obtained from the Osteoporotic Fractures in Men (MrOS) study (20), which enrolled 5,994 community-dwelling, older adult (> 65y) men from 2000–2002. Study design, recruitment, and eligibility criteria for MrOS have been previously reported (21, 22). Subject demographics, including age, body mass index (BMI), number of medications used by each participant, smoking status (never, former, or current smokers), and values for physical functioning, including the SPPB and grip strength were obtained from study Visit 4, which occurred from May 2014 to May 2016 (23). Dietary intakes were estimated using the Block 98.2 MrOS food frequency questionnaire (FFQ) (NutritionQuest, Berkeley, CA) (24), self-reported physical activity was assessed with the Physical Activity Scale for the Elderly (PASE) (25), and body composition (lean and fat mass, bone mineral content [BMC]) was measured by dual-energy x-ray absorptiometry (DXA) (26). Whole body lean and fat mass and BMC were divided by body weight and multiplied by 100 to yield %WBLM, whole body fat mass (%WBFM), and %BMC. Statistical significance was calculated in R with use of the Welch two sample t-test, with the exception for the comparison between never with former smokers, which was compared using the Pearson chi-squared test.
Microbiome data processing
Stool samples were obtained from 599 men during MrOS study Visit 4, which were sent for microbiome analysis to the Alkek Center for Metagenomics and Microbiome Research at Baylor College of Medicine (Houston, TX). Stool sample collection and 16S rRNA sequencing for these subjects was previously described (27, 28). Briefly, genomic bacterial DNA was extracted from fecal samples, and the 16S v4 rDNA hypervariable region was amplified using the MiSeq platform. Read pairs were demultiplexed, and overlapping reads were merged using USEARCH v9.0.2132. The 599 samples were then run through the standard UPARSE 16S sequence workflow with 99% clustering and a minimum cluster size of 2 to create operational taxonomic units (OTU). The dataset was additionally filtered for rare taxa by removing OTUs that were not present in at least 20% of the samples, thereby resulting in the identification of 495 OTUs. Greengenes 16S rRNA Gene Database version 13.8 (greengenes.lbl.gov) was used for taxonomy prediction. The 495 OTUs classified to 10 phyla, 19 classes, 24 orders, 39 families, and 58 genera.
92 subjects were removed from further analysis, including: 62 subjects who used antibiotics or probiotics within the 1-month period that preceded stool collection, 8 current smokers, and 9 and 13 participants who did not have values for lean mass and dietary fiber intake, respectively.
Filtering based on reported vs. estimated energy intake
Accurate dietary reporting is important for minimizing the identification of spurious associations for studies that compare associations between diet with the gut microbiome. For example, subjects that self-reported consuming < 800 or > 5000 kcal were excluded from analysis when comparing diet quality with gut microbiome composition (29). These cut-points are seemingly arbitrary, and to attempt to provide greater accuracy, filtering was performed based on comparison of estimated daily energy intake against subjects’ FFQ-reported intake. Estimated energy intake (EEI) was calculated based on age, physical activity (PA) score, weight, and height, where EEI = 662 – (9.53 × age [y]) + PA × (15.91 × weight [kg] + 539.6 × height [m])[30]. To determine a reasonable estimation for the PA coefficient, we considered Visit 4 data for the average number of steps per day that was recorded with the SenseWear® Pro3 Armband (Body Media, Inc., Pittsburgh, PA) (27). Steps/day data was available for 446 of the 507 remaining subjects, and the average number steps per day was 4,604 ± 2978. Less than 5,000 steps per day has been defined as sedentary (31), and accordingly, a value of 1 was assigned to the PA coefficient. The EEI was then calculated for each participant, and subjects that reported a daily energy intake that was < 33% or > 33% of their EEI were removed from further analysis. Based on this criterion, data for 251 subjects were removed, with remaining data for 256 subjects.
Delineation of the dietary fiber density-percentage of whole body lean mass groups
To account for the possibility that subjects that reported a high dietary fiber intake may also have a relatively high energy intake, dietary fiber was divided by total kcal, then multiplied by 100/100, to obtain dietary fiber density (g fiber/100 kcal). Subjects who were in the highest tertile for dietary fiber density and in the highest tertile for the percentage of %WBLM were identified as T3T3 (n=42). Subjects who were in the lowest tertiles for these variables were classified as T1T1 (n=32). Subjects who were in the highest tertile for dietary fiber density but in the lowest tertile for %WBLM were classified as T3T1 (n=13), and as T1T3 for subjects that were in the lowest tertile for dietary fiber density but in the highest tertile for %WBLM (n=24). A summary of this approach, starting from the 599 older adult men that provided stool samples is shown in Figure 1.
Figure 1.
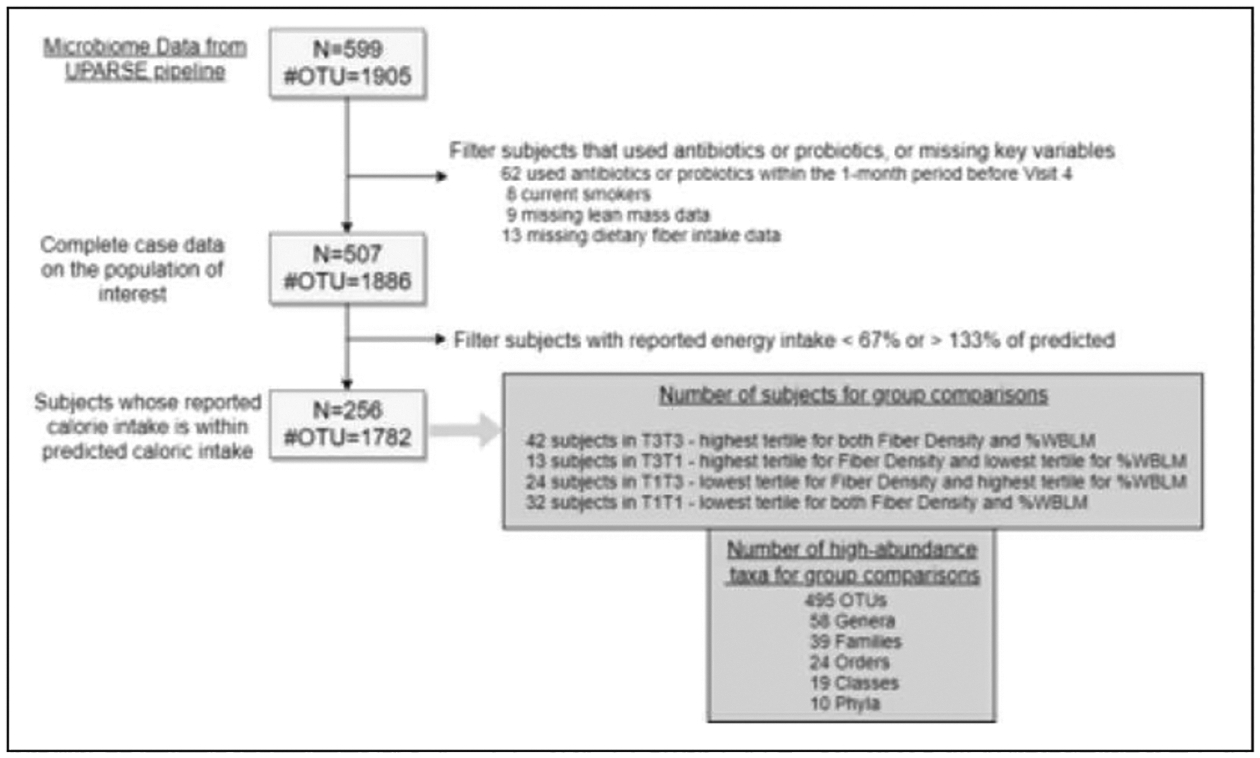
Analytic sample flow diagram
α- and β-diversity measures
α-diversity indices (abundance-coverage estimator (ACE), Chao1, Fisher, Shannon, Simpson) were calculated for each sample based on unfiltered OTU counts. The ACE, Chao1, and Fisher indices were used to evaluate between-group differences in richness, whereas the Simpson and Shannon indices were used as measures of evenness. Differences for α-diversity were evaluated for all 4 groups (T3T3, T1T1, T1T3, T3T1) with use of Kruskal-Wallis testing, and in 2-group pairings (T3T3 vs. T1T1, T3T3 vs. T1T3, T3T3 vs. T3T1) with the Tukey-Kramer-Nemenyi all-pairs test, which includes the Tukey-distance approximation. To evaluate β-diversity, binary (unweighted UniFrac) and abundance-based (Bray-Curtis, weighted UniFrac) measures were used in conjunction with principal coordinates analysis with the phyloseq v 1.28 R package (32). Between-group differences for β-diversity were evaluated using analysis of similarities (ANOSIM)[33]. Statistical significance for α- and β-diversity was considered at a level of 0.05.
Taxonomic differential abundance and functional potential analysis
Differential abundance analysis for T3T3 vs. T1T1, T3T3 vs. T1T3, and T3T3 vs. T3T1 was implemented using the DESeq2 v 1.24 R package (34). The DESeq function uses the count data to fit negative binomial generalized linear models and incorporates shrinkage of dispersion estimates and independent filtering for weakly differentially abundant taxa. Models were fitted on complete case data using geometric means as a variance stabilizing transformation and statistical tests to compare fiber density and lean mass groups using Wald tests. Age, PASE score, smoking (former or never), and number of medications were included in the model as categorical covariates. The gut bacterial functional potential in older adults was derived from OTU tables with use of PICRUSt (35), as contained within Microbiomeanalyst.ca (36). Between-group differences (T3T3 vs. T1T1, T3T3 vs. T1T3, and T3T3 vs. T3T1) for KEGG IDs were determined with use of DESeq2 with the same approach that was used for the taxonomic abundance data.
Statistical significance for between-group differences in taxonomy and functions was determined using alpha ≤ 0.05 and the false discovery rate (FDR) using q ≤ 0.30. A FDR of 0.30 indicates that discoveries are likely to be true 7 out of 10 times, which has been suggested to be reasonable in the setting of exploratory analysis (37). Taxa (or functions) that were significantly different when comparing T3T3 with T1T1 and that were not different when comparing T3T3 with T1T3 or T3T1 were considered as bacteria (or functions) that differentiated older men that had higher values for dietary fiber density and %WBLM from older men that had lower values for these variables.
Results
Subject characteristics when comparing T3T3 with T1T1 are shown in Table 1. The two groups were not significantly different in terms of age, number of medications used, and the percentage of former smokers. BMI was significantly lower in T3T3, which had a more favorable body composition, including higher values for %WBLM and %BMC, and a lower %WBFM, when compared with T1T1. T3T3 were more physically active, as indicated by a higher PASE score, and had better physical function, including higher values for the SPPB and grip strength. In terms of dietary factors, although T3T3 consumed less energy, they consumed more dietary fiber, thereby resulting in a higher dietary fiber density. Subject characteristic comparisons for T3T3 with T1T3, and T3T3 with T3T1 are shown in Supplementary Tables 1 and 2, respectively.
Table 1.
Subject characteristics
| T3T3 | T1T1 | p-value | |
|---|---|---|---|
| n | 42 | 32 | |
| Age | 84.4 ± 4.29 | 84.6 ± 4.37 | 0.85 |
| Number of medications | 8.50 ± 4.24 | 9.31 ± 4.93 | 0.46 |
| Former smoker (%) | 42.9 | 50.0 | 0.32 |
| BMI | 23.2 ± 2.50 | 29.3 ± 3.31 | 5.1E-12*** |
| Body composition | |||
| %WBLM | 75.9 ± 4.24 | 63.2 ± 2.19 | < 2.2E-16*** |
| %WBFM | 20.3 ± 4.33 | 33.4 ± 2.18 | < 2.2E-16*** |
| %BMC | 3.73 ± 0.49 | 3.47 ± 0.53 | 0.03* |
| Physical activity, junction | |||
| PASE score | 132.9 ± 57.0 | 97.9 ± 53.2 | 0.008** |
| SPPB | 10.8 ± 1.58 | 8.03 ± 3.49 | 1.6E-04*** |
| Grip strength (N) | 33.5 ± 7.7 | 29.4 ± 6.53 | 0.02* |
| Dietary factors | |||
| Energy intake (kcal) | 1664 ± 343 | 1885±319 | 0.005** |
| Dietary fiber (g/day) | 24.7 ± 5.88 | 12.7 ± 2.98 | < 2.2E-16*** |
| Fiber density (grams of fiber /100 calories) | 1.50 ± 0.25 | 0.68 ± 0.13 | < 2.2E-16*** |
Subject characteristics for older men that were in the highest tertiles for dietary fiber density and the percentage of whole body lean mass (T3T3) were compared against subjects that were in the lowest tertiles (T1T1) for these variables. Values shown represent means ± SD or %, as indicated.
= p ≤ 0.05,
= p ≤ 0.01,
= p ≤ 0.001.
Diversity, taxonomy, and functions
None of the five measures of α-diversity were significantly different when comparing all four groups, or in two-group comparisons for T3T3 with T1T1, T1T3, or T3T1 (Supplementary Figure 1). In terms of β-diversity, group separation was observed when comparing T3T3 with T1T1 with unweighted UniFrac (ANOSIM R=0.09, p=0.006), and Bray-Curtis (ANOSIM R=0.05, p=0.05; Figure 2), whereas weighted UniFrac was close to statistical significance (p=0.10; Supplementary Figure 2).
Figure 2.
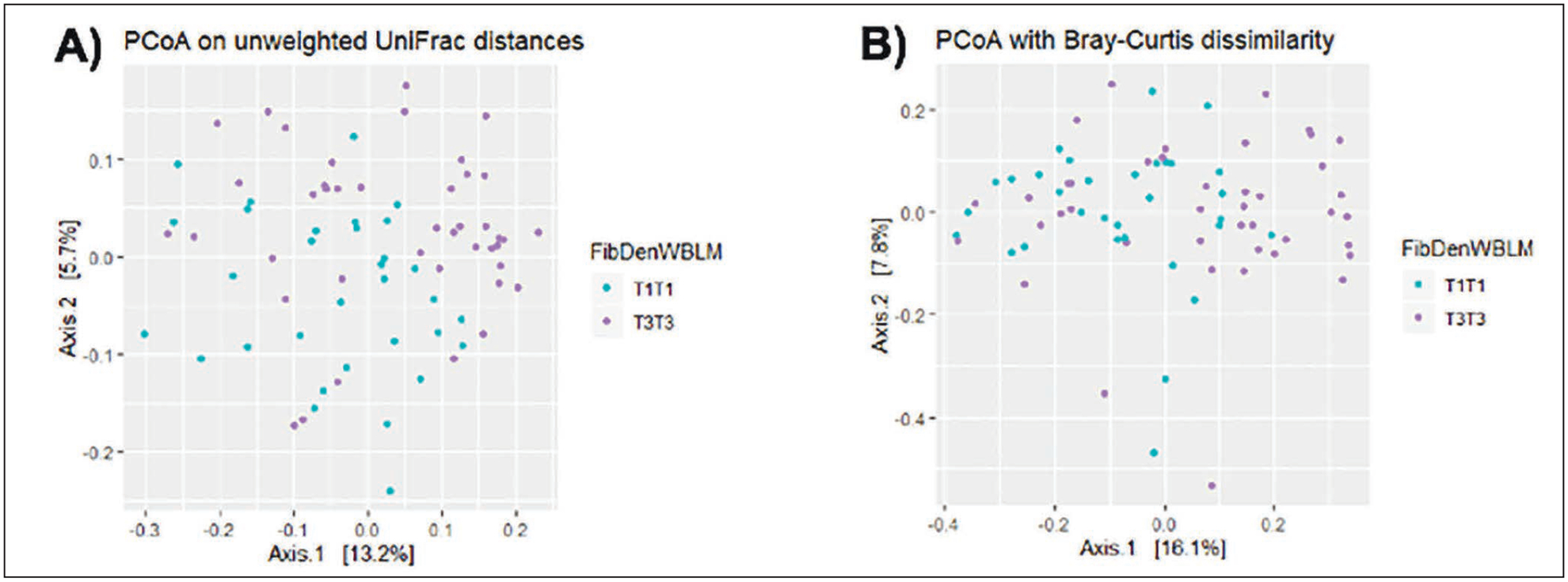
β-diversity measures when comparing T3T3 with T1T1.
Significant differences for group separation were not identified when all 4 groups were compared with unweighted or weighted UniFrac, or with Bray-Curtis, or in two-group comparisons for T3T3 with T1T3 or T3T1 (Supplementary Figure 2).
Seventy-nine OTUs were significantly different when comparing T3T3 with T1T1, and also were not different when comparing T3T3 with T1T3 or T3T1 (data not shown). Bacteria at the phyla, class, order, family, and genus taxonomic levels that were significantly different in terms of percent relative abundance when comparing T3T3 with T1T1, and that were not different when comparing T3T3 with T1T3 or T3T1 are shown in Table 2. Bacteria that were more abundant in T3T3 when compared with T1T1 include phyla-level Tenericutes and Lentisphaerae, class-level RF3 and [Lentisphaeria], order-level ML615J-28 and Victivallales, family-level Victivallaceae, Synergistaceae, and Dehalobacteriaceae, and genus-level Lachnobacterium, Clostridium, SMB53, Ruminococcus, Odoribacter, and Lachnospira. Bacteria that were lower in T3T3 when compared with T1T1 include class-level Betaproteobacteria, order-level Burkholderiales and Turicibacterales, family-level Porphyromonadaceae, Streptococcaceae, and Alcaligenaceae, and genus-level Coprobacillus and Parabacteroides. Genus-level relative abundance for each of the four groups is visually represented in Figure 3.
Table 2.
Bacteria that are significantly different when comparing T3T3 with T1T1 and that are not also different when comparing T3T3 with T1T3 or T3T1
| T3T3 (% ± SD) | T1T1 (% ± SD) | Log2FC (± SE) | p-value | FDR | |
|---|---|---|---|---|---|
| Phyla | |||||
| Tenericutes | 1.11 ± 2.11 | 0.37 ± 1.03 | −2.06 ± 0.79 | 0.01 | 0.05 |
| Lentisphaerae | 0.08 ± 0.18 | 0.01 ± 0.03 | −3.21 ± 1.26 | 0.01 | 0.05 |
| Class | |||||
| RF3 | 0.23 ± 0.39 | 0.06 ± 0.19 | −2.95 ± 1.04 | 0.004 | 0.08 |
| [Lentisphaeria] | 0.08 ± 0.18 | 0.00 ± 0.01 | −3.19 ± 1.29 | 0.01 | 0.12 |
| Betaproteobacteria | 0.99 ± 0.84 | 1.71 ± 2.13 | 1.05 ± 0.47 | 0.02 | 0.14 |
| Order | |||||
| ML615J-28 | 0.23 ± 0.39 | 0.06 ± 0.19 | −2.70 ± 1.03 | 0.01 | 0.18 |
| Victivallales | 0.08 ± 0.18 | 0.01 ± 0.04 | −3.03 ± 1.29 | 0.02 | 0.18 |
| Burkholderiales | 0.99 ± 0.84 | 1.71 ± 2.13 | 1.06 ± 0.48 | 0.03 | 0.18 |
| Turicibacterales | 0.03 ± 0.06 | 0.05 ± 0.26 | 1.65 ± 0.81 | 0.04 | 0.23 |
| Family | |||||
| Porphyromonadaceae | 3.34 ± 3.24 | 4.89 ± 3.02 | 1.13 ± 0.35 | 0.001 | 0.02 |
| Victivallaceae | 0.10 ± 0.21 | 0.01 ± 0.04 | −3.33 ± 1.27 | 0.01 | 0.08 |
| Streptococcaceae | 0.24 ± 0.51 | 0.43 ± 1.10 | 1.19 ± 0.55 | 0.03 | 0.16 |
| Alcaligenaceae | 1.09 ± 0.97 | 1.80 ± 2.27 | 1.11 ± 0.50 | 0.03 | 0.16 |
| Synergistaceae | 0.02 ± 0.05 | 0.00 ± 0.01 | −2.49 ± 1.17 | 0.03 | 0.16 |
| Dehalobacteriaceae | 0.03 ± 0.07 | 0.01 ± 0.01 | −1.45 ± 0.60 | 0.04 | 0.17 |
| Genus | |||||
| Lachnobacterium | 0.29 ± 0.62 | 0.04 ± 0.10 | −3.12 ± 0.74 | 2.4E-05 | 0.0003 |
| Clostridium | 1.80 ± 3.01 | 0.38 ± 0.36 | −1.39 ± 0.46 | 0.002 | 0.02 |
| SMB53 | 0.30 ± 0.96 | 0.06 ± 0.13 | −1.73 ± 0.60 | 0.004 | 0.03 |
| Ruminococcus | 8.30 ± 7.11 | 4.59 ± 4.07 | −0.78 ± 0.29 | 0.01 | 0.05 |
| Coprobacillus | 0.01 ± 0.01 | 0.02 ± 0.05 | 1.50 ± 0.65 | 0.02 | 0.10 |
| Parabacteroides | 4.47 ± 4.06 | 6.05 ± 3.44 | 0.83 ± 0.36 | 0.02 | 0.10 |
| Dehalobacterium | 0.04 ± 0.09 | 0.01 ± 0.02 | −1.30 ± 0.62 | 0.04 | 0.14 |
| Odoribacter | 0.64 ± 0.96 | 0.49 ± 0.93 | −1.16 ± 0.56 | 0.04 | 0.14 |
| Lachnospira | 2.42 ± 2.13 | 1.44 ± 1.77 | −0.75 ± 0.37 | 0.04 | 0.14 |
Figure 3.
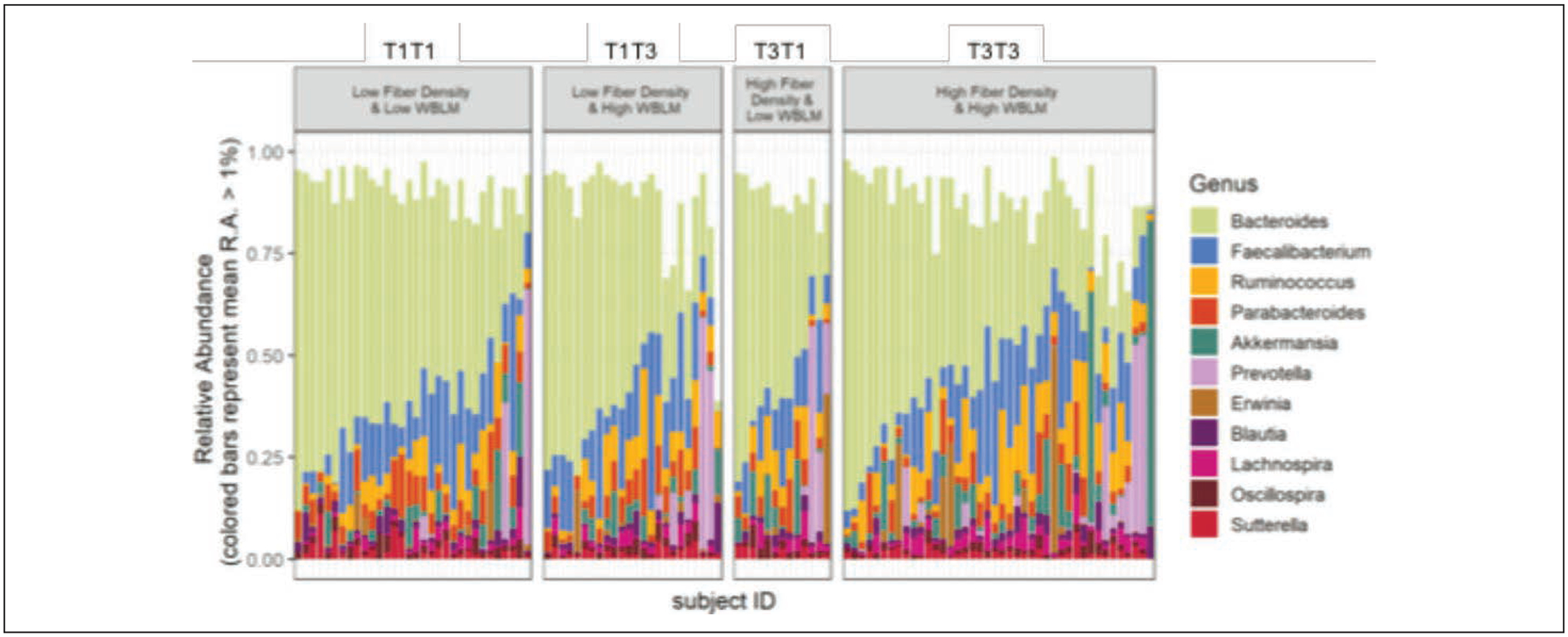
Genus-level relative abundance for T1T1, T1T3, T3T1, T3T3.
Percent relative abundance (± SD) for bacteria that are significantly different at each taxonomic level when comparing T3T3 with T1T1, but that are not different when comparing T3T3 with T1T3 or T3T1 are shown in order of significance with log 2-fold change (Log2FC) ± standard error (SE) values.
Bacteria that were significantly different when comparing their percent relative abundance in T3T3 with T1T1 but that also were different when comparing T3T3 with T1T3 or T3T1 are shown in Supplementary Tables 3 and 4, respectively.
Ninety-one KEGG IDs (functions) were significantly different when comparing T3T3 with T1T1, and were not different when comparing T3T3 with T1T3 or T3T1. Twelve functions related to SCFA production are shown in Table 3, with the remaining 79 functions shown in Supplementary Table 5. Butyrate and acetoacetyl-CoA, and acetyl-CoA producing genes (K01034 and K01035, K00171, respectively), and K03563, which positively regulates acetyl-CoA synthesis[38] were higher in T3T3 when compared with T1T1. Conversely, 3 genes in the (methyl)-malonate semialdehyde pathway for acetyl-CoA or propionyl-CoA production (K03336, K03337, K00140), and 2 genes involved in acetyl-CoA production (K004019, K00132) were higher in T1T1, when compared with T3T3. Additionally, 3 genes for glucoside hydrolases (K04844, K05989, K07406) were higher in T1T1, when compared with T3T3.
Table 3.
SCFA-related bacterial functions (KEGG IDs) that are significantly different when comparing T3T3 with T1T1 but that are not different when comparing T3T3 with T1T3 or T3T1
| KEGG ID | Function | T3T3 (% ± SD) | T1T1 (% ± SD) | Log2FC (± SE) | p-value | FDR |
|---|---|---|---|---|---|---|
| K00132 | Acetaldehyde dehydrogenase | 3.6E-06 ± 9.7E-06 | 1.2E-05 ± 2.3E-05 | 1.77± 0.46 | 0.0001 | 0.02 |
| K00140 | Malonate or methylmalonate-semialdehyde dehydrogenase | 1.2E-05 ± 2.6E-05 | 2.8E-05 ± 6.1E-05 | 1.86 ± 0.63 | 0.003 | 0.19 |
| K00171 | Pyruvate ferredoxin oxidoreductase δ-subunit | 2.1E-05 ± 3.3E-05 | 6.6E-06 ± 7.2E-06 | −1.92 ± 0.45 | 1.6E-05 | 0.01 |
| K01034 | Butyrate-acetoacetate CoA-transferase | 4.3E-06 ± 1.0E-05 | 2.9E-06 ± 8.5E-05 | −1.98 ± 0.71 | 0.01 | 0.22 |
| K01035 | Butyrate-acetoacetate CoA-transferase | 4.3E-06 ± 1.0E-05 | 2.9E-06 ± 8.5E-05 | −2.45 ± 0.69 | 0.004 | 0.04 |
| K03336 | 3D-(3,5/4)-trihydroxycyclohexane-1,2-dione acylhydrolase | 3.2E-06 ± 9.8E-06 | 1.3E-05 ± 2.4E-05 | 1.93 ± 0.54 | 0.0004 | 0.04 |
| K03337 | 5-deoxy-glucuronate isomerase | 3.2E-06 ± 9.8E-06 | 1.3E-05 ± 2.4E-05 | 1.93 ± 0.54 | 0.0004 | 0.04 |
| K03563 | Carbon storage regulator | 1.2E-04 ± 1.0E-04 | 6.7E-05 ± 6.0E-05 | −0.90 ± 0.34 | 0.01 | 0.29 |
| K04019 | Ethanolamine utilization protein | 7.4E-06 ± 1.2E-05 | 1.8E-05 ± 3.0E-05 | 1.05 ± 0.38 | 0.01 | 0.22 |
| K04844 | Hypothetical glycosyl hydrolase | 2.6E-06 ± 9.9E-06 | 1.1E-05 ± 2.3E-05 | 2.46 ± 0.62 | 0.0001 | 0.02 |
| K05989 | α-L-rhamnosidase | 2.0E-05 ± 1.9E-05 | 3.5E-05 ± 3.9E-05 | 0.84 ± 0.29 | 0.004 | 0.19 |
| K07406 | α-galactosidase | 6.1E-06 ± 1.0E-05 | 2.0E-05 ± 2.8E-05 | 1.76 ± 0.42 | 2.7E-05 | 0.01 |
Percent relative abundance (± SD) for bacterial functions are shown in numerical order based on KEGG IDs with log 2-fold change (Log2FC) ± standard error (SE) values.
Discussion
The primary goals of the present study were to compare gut microbiome composition and SCFA-related functions in older men from the MrOS study who differed in terms of dietary fiber density and %WBLM. Additionally, men who were in the highest tertiles for fiber density and %WBLM had better physical function than men who were in the lowest tertiles for fiber density and %WBLM. Significant differences for β-diversity were identified for men that were in the highest tertiles of dietary fiber density and %WBLM, when compared with men that had lower values for these variables. More specifically, butyrate-producing bacteria, including Ruminococcus, Lachnospira, and Clostridia, and genes related to butyrate and SCFA production were higher in T3T3 when compared with T1T1. Collectively, these data suggest candidate butyrate-producing bacterial genera and fecal bacterial butyrate production as a potential mechanism that may link dietary fiber intake with higher levels of %WBLM and physical function in older adult men.
In support of our findings, muscle mass and physical function are higher in mice fed a high-fiber diet in conjunction with increased intestinal levels of Ruminococcus (10). Higher levels of Ruminococcus and butyrate-producing genes, and lower levels of Porphyromonadaceae and Parabacteroides were identified in less frail older adult humans that had a larger calf circumference (as an indirect measure of muscle mass) and a higher dietary fiber intake, when compared with more frail older adults (39). Parabacteroides are positively associated with fat mass in rats (40), a potentially relevant finding because Parabacteroides were increased in T1T1, a group that had a higher %WBFM than T3T3. Investigating further, Ruminococcus and Lachnospiraceae (the bacterial family corresponding to genus-level Lachnospira) are increased, and Parabacteroides are decreased in response to dietary fiber supplementation in overweight humans (41). Collectively, these data suggest that Ruminococcus, Lachnospiraceae, Porphyromonadaceae, and Parabacteroides may be fiber-responsive bacterial taxa that are involved in mechanisms related to the maintenance of body composition and physical function.
Ruminococcus, Lachnospira, and Clostridium contain butyrate-producing genes (42), a finding that may explain the higher levels of butyrate-producing genes (K01034, K01035; BCoAT) in T3T3, when compared with T1T1. Moreover, although we did not directly quantify fecal SCFAs, BCoAT is positively associated with fecal levels of butyrate (15), evidence that suggests higher fecal butyrate production in T3T3. Similarly, Lachnospira are increased in association with exercise training-induced improvements in %WBLM, and with a corresponding increase for fecal levels of butyrate (15). Beyond butyrate production, three functions for glycoside hydrolases (K07406, K05989, K04844) were lower in T3T3 when compared with T1T1, a finding that suggests higher fecal SCFAs in T3T3. Glycoside hydrolase inhibition allows more sugars to be available for colonic bacterial fermentation to SCFAs, as evidenced by the increase in fecal SCFAs in response to supplementation with acarbose (43), which inhibits intestinal glucoside hydrolases (44). However, in disagreement with this hypothesis, although two KEGG IDs (K00171, K03563) involved with acetyl-CoA production were higher in T3T3, alternatively, two genes involved in the conversion of ethanolamine to acetyl-CoA (K04019, K00132), and three of the five genes involved in the conversion of D-2,3-Diketo-4-deoxy-epi-inositol into acetyl-CoA (or propionyl-CoA) were higher T1T1, evidence that raises doubt about higher levels of acetate or propionate in older adult men who had higher values for fiber density, %WBLM, and physical functioning.
Genes other than those involved in SCFA production that may have a role in the diet-gut-muscle axis include K00274 and K13479, which were higher in T1T1 when compared with T3T3. K00274 encodes monoamine oxidase, which can produce methylglyoxal from aminoacetone. Methylglyoxal impairs muscle development. Exposure to methylglyoxal reduces myotube formation and induces myotube atrophy in C2C12 muscle cells (45, 46). K13479 encodes xanthine dehydrogenase, which converts xanthine into uric acid. Elevated serum levels of uric acid are associated with reduced muscle mass and physical function in older adults (47, 48). However, whether circulating levels of methylgloxal or uric acid were increased in T1T1 when compared with T3T3 was not evaluated in the present study. Separately, 7 genes (K00068, K02773, K02774, K02775, K02783, K02821, K07816) involved in the production of sugar phosphates were higher in T3T3 when compared with T1T1, but their role in the diet-gut-muscle axis is unknown.
Our study had several limitations. First, women were not enrolled into the MrOS study, and approximately 90% of MrOs participants were white-whether our findings will be similarly identified in older adult women, or are generalizable to other ethnicities is unknown. Second, microbiome data were obtained and analyzed for one time point-beyond identification of associations between the gut microbiome with outcome variables at one study visit, repeated measurements over time test the reproducibility of these associations (49). Accordingly, longitudinal studies may be an important strategy to further elucidate the role of dietary fiber on the gut-muscle axis. Third, although data for 74 subjects were included when comparing T3T3 with T1T1, smaller group sizes for T3T1 (n=13) and T1T3 (n=24) may limit the strength of our findings. Validation of our findings in a larger cohort are of interest. Fourth, use of 16S rDNA to obtain gene counts is a measure of the maximal bacterial functional potential, but may not represent actively expressing genes. Moreover, there is significant overlap for many of the genes (i.e., acetyl CoA, propionyl CoA production) that are derived from the 16S rDNA data, which can make it difficult to predict if a given metabolite is up- or down-regulated. Fifth, the present study was association-based, and causation cannot be determined. Collectively, based on these data, future studies aimed at testing the effect of a high-fiber dietary intervention on the gut-muscle axis in older adults, including direct quantification of fecal SCFAs, are of interest.
Supplementary Material
Funding:
This work was supported by National Institute on Aging (K01AG050700) and Boston Claude D. Pepper Older Americans Independence Center (5P30AG031679) grants to MSL, and a USDA grant (58-1950-4-003) to the NEPS laboratory at the Jean Mayer HNRCA at Tufts University. The Osteoporotic Fractures in Men (MrOS) Study is supported by National Institutes of Health funding. The following institutes provide support: the National Institute on Aging, the National Institute of Arthritis and Musculoskeletal and Skin Diseases, the National Center for Advancing Translational Sciences, and NIH Roadmap for Medical Research under the following grant numbers: U01 AG027810, U01 AG042124, U01 AG042139, U01 AG042140, U01 AG042143, U01 AG042145, U01 AG042168, U01 AR066160, and UL1 TR000128.
Footnotes
Conflict of Interest: None declared.
Ethical Standards: The institutional review board at each of the 6 MrOS study sites approved the study protocol, and written informed consent was obtained from all participants.
References
- 1.Grosicki GJ, Fielding RA, and Lustgarten MS, Gut Microbiota Contribute to Age-Related Changes in Skeletal Muscle Size, Composition, and Function: Biological Basis for a Gut-Muscle Axis. Calcif Tissue Int, 2018. 102(4): p. 433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Sire R, et al. , Skeletal muscle-gut axis: emerging mechanisms of sarcopenia for intestinal and extra intestinal diseases. Minerva Gastroenterol Dietol, 2018. 64(4): p. 351–362. [DOI] [PubMed] [Google Scholar]
- 3.Ni Lochlainn M, Bowyer RCE, and Steves CJ, Dietary Protein and Muscle in Aging People: The Potential Role of the Gut Microbiome. Nutrients, 2018. 10(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Picca A, et al. , Gut Dysbiosis and Muscle Aging: Searching for Novel Targets against Sarcopenia. Mediators Inflamm, 2018. 2018: p. 7026198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ticinesi A, et al. , Gut Microbiota, Muscle Mass and Function in Aging: A Focus on Physical Frailty and Sarcopenia. Nutrients, 2019. 11(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu YJ, et al. , Effect of intestinal microbiota on exercise performance in mice. J Strength Cond Res, 2015. 29(2): p. 552–8. [DOI] [PubMed] [Google Scholar]
- 7.Lahiri S, et al. , The gut microbiota influences skeletal muscle mass and function in mice. Sci Transl Med, 2019. 11(502). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manickam R, et al. , Metronidazole Causes Skeletal Muscle Atrophy and Modulates Muscle Chronometabolism. Int J Mol Sci, 2018. 19(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nay K, et al. , Gut bacteria are critical for optimal muscle function: a potential link with glucose homeostasis. Am J Physiol Endocrinol Metab, 2019. 317(1): p. E158–E171. [DOI] [PubMed] [Google Scholar]
- 10.Okamoto T, et al. , Microbiome potentiates endurance exercise through intestinal acetate production. Am J Physiol Endocrinol Metab, 2019. 316(5): p. E956–E966. [DOI] [PubMed] [Google Scholar]
- 11.Clarke SF, et al. , Exercise and associated dietary extremes impact on gut microbial diversity. Gut, 2014. 63(12): p. 1913–20. [DOI] [PubMed] [Google Scholar]
- 12.Fielding RA, et al. , Muscle strength is increased in mice that are colonized with microbiota from high-functioning older adults. Exp Gerontol, 2019. 127: p. 110722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fredstrom SB, et al. , Apparent fiber digestibility and fecal short-chain fatty acid concentrations with ingestion of two types of dietary fiber. JPEN J Parenter Enteral Nutr, 1994. 18(1): p. 14–9. [DOI] [PubMed] [Google Scholar]
- 14.Walsh ME, et al. , The histone deacetylase inhibitor butyrate improves metabolism and reduces muscle atrophy during aging. Aging Cell, 2015. 14(6): p. 957–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allen JM, et al. , Exercise Alters Gut Microbiota Composition and Function in Lean and Obese Humans. Med Sci Sports Exerc, 2018. 50(4): p. 747–757. [DOI] [PubMed] [Google Scholar]
- 16.Tomey KM, et al. , Dietary intake related to prevalent functional limitations in midlife women. Am J Epidemiol, 2008. 167(8): p. 935–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tak YJ, et al. , Association of Handgrip Strength with Dietary Intake in the Korean Population: Findings Based on the Seventh Korea National Health and Nutrition Examination Survey (KNHANES VII-1), 2016. Nutrients, 2018. 10(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu IC, et al. , Association between dietary fiber intake and physical performance in older adults: a nationwide study in Taiwan. PLoS One, 2013. 8(11): p. e80209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buigues C, et al. , Effect of a Prebiotic Formulation on Frailty Syndrome: A Randomized, Double-Blind Clinical Trial. Int J Mol Sci, 2016. 17(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MrOS Online. San Francisco Coordinating Center, California Pacific Medical Center Research institution & University of California, San Francisco. [Google Scholar]
- 21.Blank JB, et al. , Overview of recruitment for the osteoporotic fractures in men study (MrOS). Contemp Clin Trials, 2005. 26(5): p. 557–68. [DOI] [PubMed] [Google Scholar]
- 22.Orwoll E, et al. , Design and baseline characteristics of the osteoporotic fractures in men (MrOS) study--a large observational study of the determinants of fracture in older men. Contemp Clin Trials, 2005. 26(5): p. 569–85. [DOI] [PubMed] [Google Scholar]
- 23.Cawthon PM, et al. , Strong Relation Between Muscle Mass Determined by D3-creatine Dilution, Physical Performance, and Incidence of Falls and Mobility Limitations in a Prospective Cohort of Older Men. J Gerontol A Biol Sci Med Sci, 2019. 74(6): p. 844–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boucher B, et al. , Validity and reliability of the Block98 food-frequency questionnaire in a sample of Canadian women. Public Health Nutr, 2006. 9(1): p. 84–93. [DOI] [PubMed] [Google Scholar]
- 25.Washburn RA, et al. , The Physical Activity Scale for the Elderly (PASE): development and evaluation. J Clin Epidemiol, 1993. 46(2): p. 153–62. [DOI] [PubMed] [Google Scholar]
- 26.Laddu DR, et al. , Trajectories of the relationships of physical activity with body composition changes in older men: the MrOS study. BMC Geriatr, 2017. 17(1): p. 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langsetmo L, et al. , The Association between Objectively Measured Physical Activity and the Gut Microbiome among Older Community Dwelling Men. J Nutr Health Aging, 2019. 23(6): p. 538–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abrahamson M, et al. , Successful collection of stool samples for microbiome analyses from a large community-based population of elderly men. Contemp Clin Trials Commun, 2017. 7: p. 158–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, et al. , Dietary quality and the colonic mucosa-associated gut microbiome in humans. Am J Clin Nutr, 2019. 110(3): p. 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medicine, I.o., Dietary Reference Intakes: The Essential Guide to Nutrient Requirements, ed. Otten JJ, Hellwig JP, and Meyers LD. 2006, Washington, DC: The National Academies Press; 1344. [Google Scholar]
- 31.Tudor-Locke C and Bassett DR Jr., How many steps/day are enough? Preliminary pedometer indices for public health. Sports Med, 2004. 34(1): p. 1–8. [DOI] [PubMed] [Google Scholar]
- 32.McMurdie PJ and Holmes S, phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One, 2013. 8(4): p. e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clarke KR, Non-parametric multivariate analyses of changes in community structure. Australian Journal of Ecology, 1993. 18(1): p. 117–143. [Google Scholar]
- 34.Love MI, Huber W, and Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 2014. 15(12): p. 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langille MG, et al. , Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol, 2013. 31(9): p. 814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dhariwal A, et al. , MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res, 2017. 45(W1): p. W180–W188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lustgarten MS and Fielding RA, Metabolites related to renal function, immune activation, and carbamylation are associated with muscle composition in older adults. Exp Gerontol, 2017. 100: p. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei B, et al. , Global regulatory mutations in csrA and rpoS cause severe central carbon stress in Escherichia coli in the presence of acetate. J Bacteriol, 2000. 182(6): p. 1632–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Claesson MJ, et al. , Gut microbiota composition correlates with diet and health in the elderly. Nature, 2012. 488(7410): p. 178–84. [DOI] [PubMed] [Google Scholar]
- 40.Lecomte V, et al. , Changes in gut microbiota in rats fed a high fat diet correlate with obesity-associated metabolic parameters. PLoS One, 2015. 10(5): p. e0126931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benitez-Paez A, et al. , A Multi-omics Approach to Unraveling the Microbiome-Mediated Effects of Arabinoxylan Oligosaccharides in Overweight Humans. mSystems, 2019. 4(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vital M, Howe AC, and Tiedje JM, Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio, 2014. 5(2): p. e00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith BJ, et al. , Changes in the gut microbiome and fermentation products concurrent with enhanced longevity in acarbose-treated mice. BMC Microbiol, 2019. 19(1): p. 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hermansen K and Thomsen CH, [Acarbose, a glucosidase inhibitor: a new therapeutic principle in diabetes mellitus]. Ugeskr Laeger, 1996. 158(18): p. 2564–8. [PubMed] [Google Scholar]
- 45.Baig MH, et al. , Methylglyoxal and Advanced Glycation End products: Insight of the regulatory machinery affecting the myogenic program and of its modulation by natural compounds. Sci Rep, 2017. 7(1): p. 5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tseng YT, et al. , Protective effects of Liuwei dihuang water extracts on diabetic muscle atrophy. Phytomedicine, 2019. 53: p. 96–106. [DOI] [PubMed] [Google Scholar]
- 47.Beavers KM, et al. , Low relative skeletal muscle mass indicative of sarcopenia is associated with elevations in serum uric acid levels: findings from NHANES III. J Nutr Health Aging, 2009. 13(3): p. 177–82. [DOI] [PubMed] [Google Scholar]
- 48.Huang C, et al. , An inverted J-shaped association of serum uric acid with muscle strength among Japanese adult men: a cross-sectional study. BMC Musculoskelet Disord, 2013. 14: p. 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt TSB, Raes J, and Bork P, The Human Gut Microbiome: From Association to Modulation. Cell, 2018. 172(6): p. 1198–1215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


