ABSTRACT
Human milk contains a diverse community of bacteria. The growing appreciation of commensal microbes and increasing availability of high-throughput technology has set the stage for a theory-driven approach to the study of milk microbiota, and translation of this knowledge to improve maternal and child health. We recently profiled the milk microbiota of healthy Canadian mothers and applied theory-driven causal modeling, finding that mode of breast milk feeding (nursing directly at the breast vs. pumping and feeding breast milk from a bottle) was significantly associated with milk microbiota composition. This observation could reflect an increased exposure to pumps and/or a decreased exposure to the infant mouth. Either way, it provides evidence for the retrograde mechanism of milk inoculation. Here, we discuss the implications of this research and related controversies, and raise new questions about the origins and function of milk bacteria.
KEYWORDS: Breastfeeding, milk microbiota, pump, retrograde inoculation, entero-mammary pathway, CHILD cohort
Introduction
The fact that human milk contains bacteria, even when collected aseptically, has been known since the 1920s.1 Yet, milk bacteria are often perceived in an unfavorable light, considered as contaminants introduced during handling, processing, and storage.2 In the 21st century, medical microbiology has undergone a paradigm shift from the exclusive focus on pathogenic bacteria and infectious diseases to appreciating the role of nonpathogenic bacteria and microbial communities in both infectious and noncommunicable diseases, as well as physiologic homeostasis and health.3 This shift in perspective along with the availability of next-generation sequencing has resulted in an exponential increase in the study of host-associated microbial communities (microbiota).4 The culture-independent approach to study milk microbiota is relatively new and poses unique challenges compared to other host-associated microbial niches with higher biomass, such as the gastrointestinal tract. Nevertheless, these methods offer new opportunities to study and understand the microbes present in human milk, identify their sources, mechanistically dissect their role in maternal and infant health, and help inform new microbiome-targeted strategies for health promotion and disease prevention.
Research over the past decade has demonstrated a diverse community of bacterial species in human milk.5 The initial pioneering studies focused primarily on describing the milk microbiota rather than understanding the maternal, infant, milk, and breastfeeding factors that influence its diversity and composition. To address this question, we recently profiled the milk microbiota in 393 healthy Canadian mother-infant dyads in the CHILD birth cohort.6 In accordance with previous studies, we showed that milk microbiota taxonomic composition was highly variable between individuals. By applying multiple statistical methods and theory-driven causal modeling, we provided evidence that mode of breast milk feeding (nursing directly at the breast vs. pumping and feeding breast milk from a bottle) was significantly associated with milk microbiota composition, more than any other factor examined. These findings have improved our understanding of milk bacteria, their origins, and their potential impact on infant health, and raised many new questions for further investigation (Box 1).
Box 1.
Unanswered questions about milk microbiota.
|
Origins of milk bacteria
It was a long-standing belief that milk bacteria were exogenously derived contaminants. Culture-based studies measuring the bacterial load throughout the course of a feeding showed a gradual decline over the first 15 ml of expressed milk,2 providing evidence that the milk closer to the outside environment contains more bacteria. More recently, studies have shown that bacteria consumed orally by lactating women or animals can later be isolated from their milk,7-9 indicating that bacteria may also reach the mammary gland through an ‘internal’ pathway. These two hypotheses regarding the origins of milk microbiota, initially suggested by Fernandez et al.,10 are commonly referred to as the “retrograde transfer” of external bacteria, and the “entero-mammary pathway” for translocation of internal bacteria.10
The proposed entero-mammary pathway involves immune cell-mediated bacterial translocation from the mother’s gastrointestinal tract into the mammary gland, where some of these bacteria are able to colonize the available niche.10 This translocation is thought to occur more frequently during the late stages of pregnancy due to altered tight junction regulation in the intestinal tract, providing a mechanism for the widely held hypothesis that maternal gut microbiota is vertically transferred to the infant via breast milk. Supportive evidence for the entero-mammary pathway includes the presence of a microbial community in colostrum collected even before the first infant feeding,11 vertical transmission of pathogenic bacteria and viruses through breastfeeding,12 and the presence of orally administered probiotic strains in the milk of lactating mothers in both human and animal studies.7-9 Although most research has focused on maternal gut bacteria as the origin of these milk bacteria, it is conceivable that bacterial translocations from the mother’s oral cavity could also contribute bacteria to the mammary gland. The similarity between maternal oral and milk microbiota is supportive of this “oro-mammary translocation” hypothesis.13,14
There are many unanswered questions regarding the biological plausibility of entero-mammary or oro-mammary pathways. For example, does bacterial translocation occur through colonization of immune cells15 or other mechanisms16? Do bacteria transferred to the mammary gland remain viable? Will those bacteria be released or remain within immune cells upon reaching the mammary gland? Do specific bacteria get selected from the gut or oral microbiota for translocation, and how does this occur? Is the translocation directed specifically to the mammary gland? Is this process specifically stimulated or upregulated during pregnancy and postpartum? If so, how is this process regulated? Do these bacteria permanently colonize the mammary gland and remain viable regardless of lactation? Despite these uncertainties, we reasoned that factors affecting the maternal microbiota should be associated with milk microbiota composition if there is a sub-community originating from maternal gut (or mouth). Indeed, we found that maternal characteristics such as body mass index (BMI), atopy, and smoking were associated with milk microbiota composition. However, it is equally plausible that these factors influence the mother’s skin microbiota or the infant’s oral microbiota, which could serve as external sources of milk bacteria – a theory supported by other studies demonstrating high similarity between mothers’ milk, areola and infant oral microbial compositions.13,14
The proposed “retrograde transfer” of external bacteria into the mammary gland is another relatively new area of research. In addition to the areola skin and the infant oral cavity, exogenously derived bacteria could also originate from breast pumps. This mechanism is less appreciated because historically the majority of milk microbiota studies were concerned with intramammary cow’s milk with the primary goal of understanding predisposition to mastitis, which is the most costly disease of dairy animals.17 However, specially in human studies, the focus is now shifting to the offspring and the milk that s/he receives, whether that milk is pumped or provided directly from the breast. Reverse flow of milk from the infant mouth back into the breast occurs during breastfeeding,18 and probably pumping as well, allowing for infant mouth- or pump-associated bacteria to enter the milk duct. Our finding that feeding mode (nursing directly from the breast vs. using a pump) was the most consistent factor associated with milk microbiota composition suggests the exogenously derived bacteria have a stronger role in milk inoculation than the entero-mammary pathway. Increased risk of lactational mastitis in women who pump is also consistent with this proposition.19 Further research is needed to confirm whether bacteria actually enter the mammary gland during nursing and/or pumping, or simply inoculate milk as it is expressed.
Based on our research and the existing literature, we propose a slightly modified nomenclature for the possible origins of milk bacteria: the “oro/entero-mammary pathway” (including translocation of both maternal oral and maternal gut bacteria) and the acquisition of “exogenously derived” bacteria (including maternal skin, infant oral, and pump-associated bacteria) (Figure 1). The extent to which each mechanism influences the milk microbiota load and composition requires further investigation. Additionally, the complex inter-related dispersal routes between different potential sources of milk microbiota should be acknowledged and explored (Figure 2).
Figure 1.
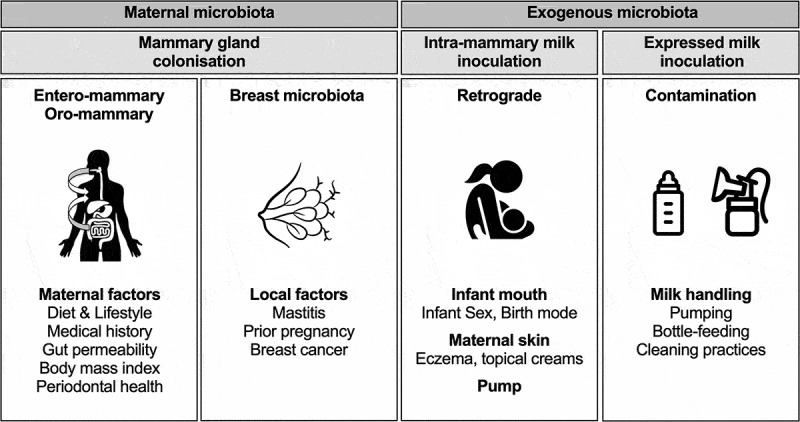
Origins of milk microbiota revisited. Two pathways for the origins of milk microbiota have been hypothesized: the entero-mammary pathway and retrograde translocation.10 We propose that translocation also occurs from the maternal oral cavity (oro-mammary pathway). Further, our study suggests that direct breastfeeding at the breast vs. pump expression could influence the milk microbiota composition6 potentially indicating the pump apparatus as an additional source of retrograde inoculation and/or contamination of the expressed milk. Resident breast microbiota could be another source of mammary gland colonization. Depending on the origin of the bacteria, different factors could be associated with the composition of the milk microbiota.
Figure 2.
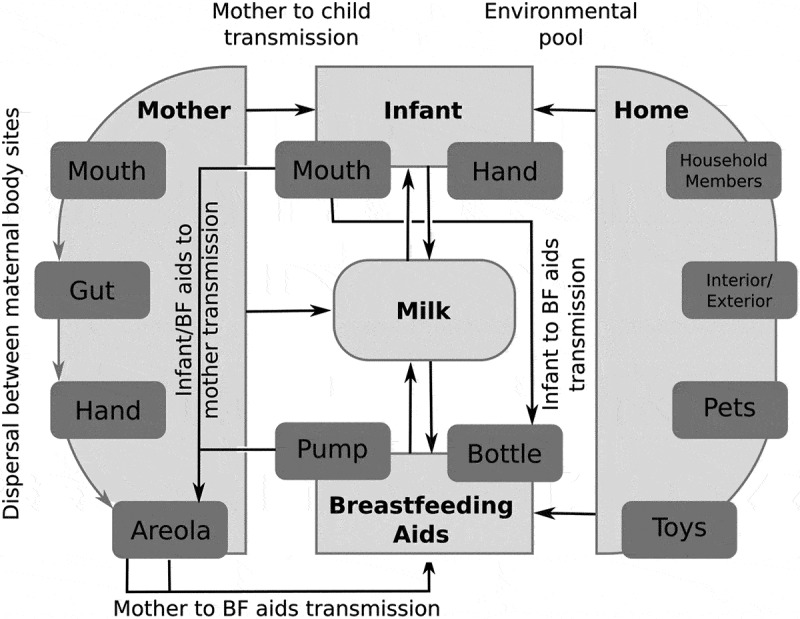
Different dispersal routes of bacteria between maternal body sites, infant, and breastfeeding aids. Bacterial dispersal can occur between different maternal body sites and between infant and breastfeeding aids within the context of the environmental pool of species. This complex dispersal network results in bacterial transmission from 1) mother to infant, 2) infant/breastfeeding aids to mother, and 3) mother to breastfeeding aids. Arrows indicate routes of bacterial transmission. BF, Breastfeeding.
Determinants of milk microbiota composition
Microbial community composition is regulated by the spatio-temporal dynamics of the microenvironment (niche). Relevant niche characteristics include nutrient resources or lack thereof, pH, salinity, predation, antibiotics, hydrodynamics, and physicochemical conditions.20 In classic microbial ecology, the impact of these factors is assessed on the community dynamic and composition; however, these variables are more challenging to measure and less commonly studied for human microbiota. Rather, the focus of human microbiota studies is often to identify host and/or environmental factors that influence the microbiota composition. We extensively studied the influence of maternal, infant, early life, breastfeeding, and other milk factors on milk microbiota composition using redundancy analysis6 and identified several factors including mode of breastfeeding, lactation stage, and maternal BMI to be associated with the overall composition of the milk microbiota community, albeit with very low redundancy values (each accounting for <2% of the variation in the milk microbiota) (Figure 3a, adapted from Moossavi et al.6). The cumulative association of all the factors assessed in our study in a multivariable analysis explained approximately 25% of the total variation in the milk microbiota composition (Figure 3b).
Figure 3.
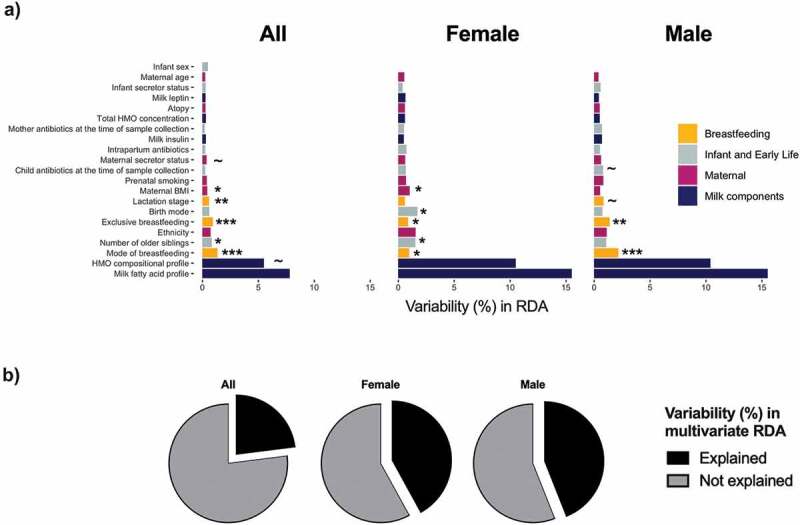
Redundancy analysis of associations of maternal and infant factors with overall milk microbiota composition among 393 dyads in the CHILD cohort. a) Univariate analysis for all infants and stratified by infant sex. b) Cumulative amount explained by all the factors in multivariate analysis. Redundancy values (R2) indicate the % variation explained by each individual factor or each multivariable model. The unexplained variability was calculated by subtracting redundancy value from 100%. BMI, body mass index; HMO, human milk oligosaccharide. P value ~ <0.10, * <0.05, ** <0.01, *** <0.001. Figures are adapted from results originally reported in Moossavi et al.6.
It is notable that host factors explain only a fraction of the microbiota composition in our study and others,21 suggesting that the factors shaping these microbial communities are not well captured by the usual characteristics of interest, such as mode of delivery and antibiotic use. Although we have examined a few components of the milk niche (human milk oligosaccharides [HMOs], fatty acids, and hormones), more ecologically informed parameters (such as Carbon and Nitrogen availability and pH) might be of more direct relevance for future studies. It will also be important to explore the impact of host genetics and immunity. Finally, understanding biotic interactions among bacteria and between bacteria and other microorganisms including fungi and viruses will also be fundamental in understanding the determinants of the milk microbiota composition.
Interestingly in our study, milk microbiota diversity was influenced by infant sex and we observed sex-specific differences in the strength and significance of associations between microbiota and other factors. For example, maternal BMI was only significantly associated with milk microbiota in milk produced for daughters while the variability explained by mode of breastfeeding was higher for sons (Figure 3a). In addition, the combined contribution of all the above-mentioned factors in multivariable analysis accounted for larger proportion of variation in sex-stratified compared to non-stratified analysis, primarily driven by a higher contribution from HMOs and milk fatty acids after stratification (Figure 3b). Although not widely studied in modern humans, the Trivers-Willard hypothesis suggests that maternal investment in milk composition differs by offspring sex.22 For example, sex-dependent variations have been observed for milk calcium, cortisol, and protein in Rhesus macaques and Macropus eugenii.23-26 Fetal sex is also suggested to affect milk production volume in dairy cattle,27 and we have found sex differences in the duration of breastfeeding (possibly reflecting milk supply) in two independent birth cohorts in Canada and the United Kingdom.28 Whether sex differences in the milk microbiota could also be explained in light of the evolutionary biology or cultural practices needs further investigation.
Pumping, milk microbiota and associated controversies
Although our study was not specifically designed to assess the effect of pumping, we compared milk from mothers who sometimes used a breast pump to express milk for their infant (at least once in the 2 weeks prior to sample collection) to those who did not feed pumped milk to their infant. Although we did not document nursing frequency, it is reasonable to assume it was lower among mothers who pumped; thus, one could argue that we actually (or also) assessed the impact of less vs. more direct exposure to the infant oral cavity. In other words, our results could reflect an increased exposure to pumps and/or a decreased exposure to the infant mouth. Either way, we observed that the bacterial diversity and composition were slightly different between these two scenarios, providing some evidence for the role of retrograde (exogenous) milk inoculation.
Although some bacterial groups containing environmental opportunistic pathogens such as Stenotrophomonas, Pseudomonadaceae, and Enterobacteriaceae were enriched in the milk of mothers who pumped, our observational study could not assess the causal role of pumping, nor the clinical significance of infant exposure to these milk bacteria. It is relevant to note that while they are potential pathogens, Stenotrophomonas and Pseudomonadaceae are common environmental bacteria, frequently detected in drinking water where they do not cause harm in healthy individuals.29 Similarly, species belonging to the Enterobacteriaceae family are common members of the gastrointestinal microbiota and most strains are harmless to the host.30 Overall, in the absence of strain-level information, absolute quantification, and evidence for colonization of these bacteria in the infant gut or upper respiratory tract, it is not possible to confirm the clinical significance of these milk bacteria. Studies designed specifically to address these questions are warranted, given the substantial and increasing use of breast pumps worldwide,31 and evidence that the bacterial content of pump-expressed milk varies considerably depending on the setting, handling, and cleaning of the pump equipment.32 We are aware of one such study (clinicaltrials.gov, NCT03123874)33 and eagerly await the results.
We did not assess the type or other qualitative aspects of pumps used in our study. There is currently limited information about how the mechanics of individual pumps can impact milk bacteriology. In a recent systematic review, there was no difference in milk contamination with known pathogens between manual vs. large electric pumps.34 However, multiple different products exist within each of these broad categories, and new technologies are emerging as the popularity of pumping increases (e.g. hands-free suction-based manual pumps and small wearable electric pumps). In addition, the qualitative aspects of pumps and pump accessories (such as breast shield size and materials used in tubing and bottles) could be important for the potential bacterial transfer and thus requires further investigation.
Storage temperature and duration are important factors to consider, as they can variably influence the integrity and composition of nutrients, bioactives and bacteria in pumped milk.35 Research in the dairy industry and human milk has shown that suboptimal storage could lead to shifts in the milk microbiota composition or viability, and dominance of spoilage bacteria.35 Current guidelines for human milk storage in the context of infant feeding recommend refrigeration for up to 4 days and storage at −18°C or colder for up to 6–12 months.36 In our study we did not observe any associations between milk microbiota composition and refrigeration time prior to processing; however, the duration was generally short and the range was small in our study (mean: 18 h, range: 10 min – 27 h). Further studies are needed to understand the impact of human milk storage conditions on microbiota composition in both research and real-world settings, especially with regards to potential opportunistic pathogens.
Our paper drew considerable media attention to the fact that pumping may impact milk microbiota composition. Some headlines inaccurately stated that pumped milk “causes diseases” or provides “nasty bacteria” to infants, raising understandable concerns from mothers who pump. These unsubstantiated claims oversimplify a complex and emotionally charged issue. It must be acknowledged that, for a variety of reasons, pumping may be the only way for some mothers to provide their own breast milk to their infant, and this is still beneficial compared to formula feeding. The well-established benefits of breast(milk)feeding for both maternal and infant health37 occur through multiple different and still incompletely understood mechanisms, many of which involve other non-microbial components of milk that remain intact upon pumping and storage (e.g. HMOs). Still, our research38,39 and other studies40 do suggest that pumped milk is not equivalent to nursing at the breast, so it is important to continue studying the impact of pumping on milk composition in order to understand and optimize this process. Finally, it is also important to undertake knowledge translation efforts with the recognition that supporting breastfeeding is a societal responsibility and not solely the duty of individual mothers.41
Functional importance of milk microbiota
The functional importance of the milk microbiota is poorly understood. In the dairy industry, the focus of milk microbiota research is heavily geared toward udder health and prevention of mastitis.17 In humans, it is often argued that breast milk not only provides nutrients to the infant, but also serves as a source of prebiotic oligosaccharides and probiotic bacteria contributing to the establishment of the infant gut microbiota.42 This probiotic hypothesis is largely based on comparative analyses identifying a few shared taxa between maternal milk and infant stool that are absent in the infant oral cavity.43,44 However, computational estimations suggest that only 20% of taxa in the infant gut microbiota are shared with breast milk.45 The infant oral microbiota is another possible source of seeding bacteria for the infant gut.46,47 It is suggested that oral microbiota could reach distal segments of the GI tract directly through swallowing saliva, or indirectly through retrograde inoculation of breast milk.44 There is also a theoretical possibility of bacterial exchange in the opposite direction, with milk bacteria seeding the oral cavity.44 Moreover, aspirations during breastfeeding could deliver bacteria to the infant respiratory system.48 Thus, through multiple possible mechanisms and origins, milk bacteria may colonize and/or functionally influence the infant gut, oral cavity and respiratory tract. We did not assess the infant microbiota in our milk microbiota study,6 but these data are available for the CHILD cohort and we are currently undertaking an integrated analysis of milk, stool and nasal microbiota.
There has been a major focus on Bifidobacterium as the most abundant genus in the infant gut and the best-studied bacterium in terms of HMO utilization.49 Assuming Bifidobacterium spp. are evolutionarily selected to be transferred to the infant,50,51 the evidence is scarce that this transfer has evolved to be exclusively from the mother (vertical transfer) or to occur via breast milk. Acknowledging the limits of detection by current methods, Bifidobacterium spp. were detected at low frequency by culture and sequencing in previous milk microbiota studies,52,53 as well as our study where only 40% of milk samples contained amplicon sequencing variants from this genus.53 While vertical transfer of obligate symbionts occurs in some holobionts (e.g. Buchnera aphidicola in aphids), in other cases they can be horizontally transferred from the environment (e.g. Vibrio fischeri in squid) or from other members of the host population (e.g. core Gram negative bacteria in honey bees).54 Similarly in humans, horizontal transfer of bifidobacteria and other bacteria from the environment and other household members likely contributes to the development of the infant gut microbiota,55 regardless of breastfeeding. A fresh outlook based on principles of holobiont ecology56 is required to better understand the functional importance of milk in vertical and horizontal bacterial transmission.
Conclusion and future perspective
Milk microbiota is a hot research topic in the field of developmental origins of health and disease. Building on earlier studies, our recent findings shed new light on the composition and determinants of the milk microbiota, and raise new research questions about the origins, function and analysis of milk bacteria (Box 1). So far, milk microbiota studies have mainly been observational cross-sectional studies based on 16S rRNA amplicon sequencing – a method that is susceptible to reagent contamination when analyzing low biomass samples such as milk, and cannot distinguish between live or dead bacteria. Better understanding of the dynamics and function of milk microbiota requires a comprehensive multi-pronged strategy that (1) assesses viability, activity, and function of milk bacteria; (2) examines the temporality of the microbial load and composition throughout lactation; (3) studies other members of the microbial community including viruses, which can conceivably be vertically transferred more readily than the bacteria,50 and fungi, which could have important health implications; (4) evaluates the interaction between milk microbiota with the maternal and infant immune systems; and (5) experimentally establishes the functional significance of milk microbiota. In addition, the origins of milk bacteria should be rigorously assessed using appropriate experimental study designs and suitable animal models that distinguish between intramammary milk and the expressed milk ingested by the offspring.
While milk bacteriology has been studied since the 19th century57 from an infectious disease perspective, the current appreciation of commensal microbes combined with the increasing availability of novel technologies and methodologies has set the stage for a comprehensive and theory-driven approach to the study of milk microbiota, and the translation of this knowledge to improve the health of the mothers and infants.
Acknowledgments
We thank Hooman Derakhshani and Meredith Brockway for constructive comments on our paper. We also thank John Schellenberg for help with creating figures. The Canadian Institutes of Health Research (CIHR) and the Allergy, Genes and Environment Network of Centers of Excellence (AllerGen NCE) provided core support for the CHILD Study. Milk microbiota sequencing was funded by the Heart and Stroke Foundation and Canadian Lung Association, in partnership with the Canadian Respiratory Research Network and AllerGen NCE. Infrastructure at the Gut Microbiome Laboratory was supported by grants from the Canadian Foundation for Innovation. This research was supported, in part, by the Canada Research Chairs program. SM holds a Research Manitoba Doctoral Studentship. These entities had no role in preparation, review, or approval of the manuscript.
Funding Statement
This work was supported in part by the Canada Research Chairs (MBA) and Research Manitoba Doctoral Studentship (SM).
References
- 1.Dudgeon LS, Jewesbury RC.. The bacteriology of human milk. J Hyg (Lond). 1924;23:64–10. doi: 10.1017/s0022172400008470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.West PA, Hewitt JH, Murphy OM. The influence of methods of collection and storage on the bacteriology of human milk. J Appl Bacteriol. 1979;46:269–277. doi: 10.1111/j.1365-2672.1979.tb00820.x. [DOI] [PubMed] [Google Scholar]
- 3.Vayssier-Taussat M, Albina E, Citti C, Cosson JF, Jacques MA, Lebrun MH, Le Loir Y, Ogliastro M, Petit M-A, Roumagnac P, et al. Shifting the paradigm from pathogens to pathobiome: new concepts in the light of meta-omics. Front Cell Infect Microbiol. 2014;4:29. doi: 10.3389/fcimb.2014.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cani PD. Human gut microbiome: hopes, threats and promises. Gut. 2018;67:1716–1725. doi: 10.1136/gutjnl-2018-316723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fitzstevens JL, Smith KC, Hagadorn JI, Caimano MJ, Matson AP, Brownell EA. Systematic review of the human milk microbiota. Nutr Clin Pract. 2016;32:354–364. doi: 10.1177/0884533616670150. [DOI] [PubMed] [Google Scholar]
- 6.Moossavi S, Sepehri S, Robertson B, Bode L, Goruk S, Field CJ, Lix LM, de Souza RJ, Becker AB, Mandhane PJ, et al. Composition and variation of the human milk microbiome is influenced by maternal and early life factors. Cell Host Microbe. 2019;25:324–335. doi: 10.1016/j.chom.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 7.Treven P, Mrak V, Bogovic Matijasic B, Horvat S, Rogelj I. Administration of probiotics lactobacillus rhamnosus GG and lactobacillus gasseri K7 during pregnancy and lactation changes mouse mesenteric lymph nodes and mammary gland microbiota. J Dairy Sci. 2015;98:2114–2128. doi: 10.3168/jds.2014-8519. [DOI] [PubMed] [Google Scholar]
- 8.Abrahamsson TR, Sinkiewicz G, Jakobsson T, Fredrikson M, Björkstén B. Probiotic lactobacilli in breast milk and infant stool in relation to oral intake during the first year of life. J Pediatr Gastroenterol Nutr. 2009;49:349–354. doi: 10.1097/MPG.0b013e31818f091b. [DOI] [PubMed] [Google Scholar]
- 9.de Andres J, Jimenez E, Chico-Calero I, Fresno M, Fernandez L, Rodriguez JM. Physiological translocation of lactic acid bacteria during pregnancy contributes to the composition of the milk microbiota in mice. Nutrients. 2018;10:14. doi: 10.3390/nu10010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez L, Langa S, Martin V, Maldonado A, Jimenez E, Martin R, Rodriguez JM. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res. 2013;69:1–10. doi: 10.1016/j.phrs.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Damaceno QS, Souza JP, Nicoli JR, Paula RL, Assis GB, Figueiredo HC, Azevedo V, Martins FS. Evaluation of potential probiotics isolated from human milk and colostrum. Probiotics Antimicrob Proteins. 2017;9:371–379. doi: 10.1007/s12602-017-9270-1. [DOI] [PubMed] [Google Scholar]
- 12.Lawrence RM, Lawrence RA. Breast milk and infection. Clin Perinatol. 2004;31:501–528. doi: 10.1016/j.clp.2004.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drell T, Stsepetova J, Simm J, Rull K, Aleksejeva A, Antson A, Tillmann V, Metsis M, Sepp E, Salumets A, et al. The influence of different maternal microbial communities on the development of infant gut and oral microbiota. Sci Rep. 2017;7:9940. doi: 10.1038/s41598-017-09278-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams JE, Carrothers JM, Lackey KA, Beatty NF, Brooker SL, Peterson HK, Steinkamp KM, York MA, Shafii B, Price WJ, et al. Strong multivariate relations exist among milk, oral, and fecal microbiomes in mother-infant dyads during the first six months postpartum. J Nutr. 2019;149:902–914. doi: 10.1093/jn/nxy299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fung TC, Bessman NJ, Hepworth MR, Kumar N, Shibata N, Kobuley D, Wang K, Ziegler CGK, Goc J, Shima T, et al. Lymphoid-tissue-resident commensal bacteria promote members of the IL-10 cytokine family to establish mutualism. Immunity. 2016;44:634–646. doi: 10.1016/j.immuni.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Damgaard C, Magnussen K, Enevold C, Nilsson M, Tolker-Nielsen T, Holmstrup P, Nielsen CH. Viable bacteria associated with red blood cells and plasma in freshly drawn blood donations. PLoS One. 2015;10:e0120826. doi: 10.1371/journal.pone.0120826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derakhshani H, Fehr KB, Sepehri S, Francoz D, De Buck J, Barkema HW, Plaizier JC, Khafipour E. Invited review: microbiota of the bovine udder: contributing factors and potential implications for udder health and mastitis susceptibility. J Dairy Sci. 2018;101:10605–10625. doi: 10.3168/jds.2018-14860. [DOI] [PubMed] [Google Scholar]
- 18.Gardner H, Kent JC, Lai CT, Mitoulas LR, Cregan MD, Hartmann PE, Geddes DT. Milk ejection patterns: an intra-individual comparison of breastfeeding and pumping. BMC Pregnancy Childbirth. 2015;15:156. doi: 10.1186/s12884-015-0583-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johns HM, Forster DA, Amir LH, McLachlan HL. Prevalence and outcomes of breast milk expressing in women with healthy term infants: a systematic review. BMC Pregnancy Childbirth. 2013;13:212. doi: 10.1186/1471-2393-13-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saleem M. Microbiome community ecology: fundamentals and applications. Springer; 2015. [Google Scholar]
- 21.Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, Valles-Colomer M, Vandeputte D, et al. Population-level analysis of gut microbiome variation. Science. 2016. April 29;352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 22.Veller C, Haig D, Nowak MA. The trivers-willard hypothesis: sex ratio or investment? Proc Biol Sci. 2016;283:pii:20160126. doi: 10.1098/rspb.2016.0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinde K. First-time macaque mothers bias milk composition in favor of sons. Curr Biol. 2007;17:R958–9. doi: 10.1016/j.cub.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 24.Hinde K, Foster AB, Landis LM, Rendina D, Oftedal OT, Power ML. Daughter dearest: sex-biased calcium in mother’s milk among rhesus macaques. Am J Phys Anthropol. 2013;151:144–150. doi: 10.1002/ajpa.22229. [DOI] [PubMed] [Google Scholar]
- 25.Sullivan EC, Hinde K, Mendoza SP, Capitanio JP. Cortisol concentrations in the milk of rhesus monkey mothers are associated with confident temperament in sons, but not daughters. Dev Psychobiol. 2011;53:96–104. doi: 10.1002/dev.20483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robert KA, Braun S. Milk composition during lactation suggests a mechanism for male biased allocation of maternal resources in the tammar wallaby (Macropus eugenii). PLoS One. 2012;7:e51099. doi: 10.1371/journal.pone.0051099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barreiro R, Regal P, Diaz-Bao M, Vazquez BI, Cepeda A. Effects of bovine pregnancy on the fatty acid composition of milk: the significance for humans needs. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2017;34:608–616. doi: 10.1080/19440049.2016.1277270. [DOI] [PubMed] [Google Scholar]
- 28.Azad MB, Hinde K, Timpson NJ, Moossavi S, Becker AB, Mandhane P, Turvey SE, Moraes TJ, Lefebvre D, Subbarao P, et al. More milk for daughters? Sex differentiated breastfeeding in two birth cohorts. Breastfeeding Med. 2018;13:A-11. [Google Scholar]
- 29.Ashbolt NJ. Microbial contamination of drinking water and human health from community water systems. Curr Environ Health Rep. 2015;2:95–106. doi: 10.1007/s40572-014-0037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Litvak Y, Mon KKZ, Nguyen H, Chanthavixay G, Liou M, Velazquez EM, Kutter L, Alcantara MA, Byndloss MX, Tiffany CR, et al. Commensal enterobacteriaceae protect against salmonella colonization through oxygen competition. Cell Host Microbe. 2019;25:128–39 e5. doi: 10.1016/j.chom.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martucci J. History of medicine: breast Pumping. AMA J Ethics. 2013;15:791–797. doi: 10.1001/virtualmentor.2013.15.9.mhst1-1309. [DOI] [Google Scholar]
- 32.Haiden N, Pimpel B, Assadian O, Binder C, Kreissl A, Repa A, Thanhäuser M, Roberts CD, Berger A. Comparison of bacterial counts in expressed breast milk following standard or strict infection control regimens in neonatal intensive care units: compliance of mothers does matter. J Hosp Infect. 2016;92:226–228. doi: 10.1016/j.jhin.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 33.Reyes SM, Allen DL, Williams J, McGuire MA, McGuire MK, Hay AG, Rasmussen KM. Sources of bacterial contamination in home-expressed human milk. Breastfeeding Med. 2018;13:A-50. [Google Scholar]
- 34.Becker GE, Smith HA, Cooney F. Methods of milk expression for lactating women. Cochrane Database Syst Rev. 2016;9:CD006170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peters MDJ, McArthur A, Munn Z. Safe management of expressed breast milk: a systematic review. Women and Birth. 2016;29:473–481. doi: 10.1016/j.wombi.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 36.Center for Disease Control and Prevention . Proper storage and preparation of breast milk. [accessed 2019 August6]. URL: https://www.cdc.gov/breastfeeding/recommendations/handling_breastmilk.htm,
- 37.Victora CG, Bahl R, Barros AJD, França GVA, Horton S, Krasevec J, Murch S, Sankar MJ, Walker N, Rollins NC. Breastfeeding in the 21st century: epidemiology, mechanisms, and lifelong effect. Lancet. 2016;387:475–490. doi: 10.1016/S0140-6736(15)01024-7. [DOI] [PubMed] [Google Scholar]
- 38.Klopp A, Vehling L, Becker AB, Subbarao P, Mandhane PJ, Turvey SE, Lefebvre DL, Sears MR, CHILD Study Investigators, Azad MB. Modes of infant feeding and the risk of childhood asthma: a prospective birth cohort study. J Pediatr. 2017;190:192–199. doi: 10.1016/j.jpeds.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 39.Azad MB, Vehling L, Chan D, Klopp A, Nickel NC, McGavock JM, Becker AB, Mandhane PJ, Turvey SE, Moraes TJ, et al. Infant feeding and weight gain: separating breast milk from breastfeeding and formula from food. Pediatrics. 2018;142:pii:e20181092. doi: 10.1542/peds.2018-1092. [DOI] [PubMed] [Google Scholar]
- 40.Soto-Ramirez N, Kar S, Zhang H, Karmaus W. Infant feeding patterns and eczema in children in the first 6 years of life. Clin Exp Allergy. 2017;47:1285–1298. doi: 10.1111/cea.12998. [DOI] [PubMed] [Google Scholar]
- 41.Office of the Surgeon General (US); Centers for Disease Control and Prevention (US); Office on Women’s Health (US). The Surgeon General’s Call to Action to Support Breastfeeding. Rockville (MD): Office of the Surgeon General (US) ; 2011. Breastfeeding from the public health perspective. [accessed 2019 Aug 10]. Available from: https://www.ncbi.nlm.nih.gov/books/NBK52684/.
- 42.Moossavi S, Miliku K, Sepehri S, Khafipour E, Azad MB. The prebiotic and probiotic properties of human milk: implications for infant immune development and pediatric asthma. Front Pediatr. 2018;6:197. doi: 10.3389/fped.2018.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Asnicar F, Manara S, Zolfo M, Truong DT, Scholz M, Armanini F, Ferretti P, Gorfer V, Pedrotti A, Tett A, et al. Studying vertical microbiome transmission from mothers to infants by strain-level metagenomic profiling. mSystems. 2017;2:e00164–16. doi: 10.1128/mSystems.00164-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biagi E, Quercia S, Aceti A, Beghetti I, Rampelli S, Turroni S, Faldella G, Candela M, Brigidi P, Corvaglia L. The bacterial ecosystem of mother’s milk and infant’s mouth and gut. Front Microbiol. 2017;8:1214. doi: 10.3389/fmicb.2017.01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pannaraj PS, Li F, Cerini C, Bender JM, Yang S, Rollie A, Adisetiyo H, Zabih S, Lincez PJ, Bittinger K, et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 2017;171:647–654. doi: 10.1001/jamapediatrics.2017.0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Costello EK, Carlisle EM, Bik EM, Morowitz MJ, Relman DA. Microbiome assembly across multiple body sites in low-birthweight infants. MBio. 2013;4:e00782–13. doi: 10.1128/mBio.00782-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509:357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biesbroek G, Bosch AA, Wang X, Keijser BJ, Veenhoven RH, Sanders EA, Bogaert D. The impact of breastfeeding on nasopharyngeal microbial communities in infants. Am J Respir Crit Care Med. 2014;190:298–308. doi: 10.1164/rccm.201401-0073OC. [DOI] [PubMed] [Google Scholar]
- 49.Thomson P, Medina DA, Garrido D. Human milk oligosaccharides and infant gut bifidobacteria: molecular strategies for their utilization. Food Microbiol. 2018;75:37–46. doi: 10.1016/j.fm.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 50.Duranti S, Lugli GA, Mancabelli L, Armanini F, Turroni F, James K, Ferretti P, Gorfer V, Ferrario C, Milani C, et al. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome. 2017;5:66. doi: 10.1186/s40168-017-0282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Milani C, Mancabelli L, Lugli GA, Duranti S, Turroni F, Ferrario C, Mangifesta M, Viappiani A, Ferretti P, Gorfer V, et al. Exploring vertical transmission of bifidobacteria from mother to child. Appl Environ Microbiol. 2015;81:7078–7087. doi: 10.1128/AEM.02037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jost T, Lacroix C, Braegger CP, Rochat F, Chassard C. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ Microbiol. 2014;16:2891–2904. doi: 10.1111/1462-2920.12238. [DOI] [PubMed] [Google Scholar]
- 53.Moossavi S, Atakora F, Miliku K, Sepehri S, Robertson B, Duan QL, Becker AB, Mandhane PJ, Turvey SE, Moraes TJ, et al. Integrated analysis of human milk microbiota with oligosaccharides and fatty acids in the CHILD cohort. Front Nutr. 2019;6:58. doi: 10.3389/fnut.2019.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haag KL. Holobionts and their hologenomes: evolution with mixed modes of inheritance. Genet Mol Biol. 2018;41:189–197. doi: 10.1590/1678-4685-GMB-2017-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lane AA, McGuire MK, McGuire MA, Williams JE, Lackey KA, Hagen EH, Kaul A, Gindola D, Gebeyehu D, Flores KE, et al. Household composition and the infant fecal microbiome: the INSPIRE study. Am J Phys Anthropol. 2019;169:526–539. doi: 10.1002/ajpa.23843. [DOI] [PubMed] [Google Scholar]
- 56.Simon JC, Marchesi JR, Mougel C, Selosse MA. Host-microbiota interactions: from holobiont theory to analysis. Microbiome. 2019;7:5. doi: 10.1186/s40168-019-0619-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.[No authors listed] . Bacteria in milk and its products. Science. 1889;14:116–118. doi: 10.1126/science.ns-14.341.116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Center for Disease Control and Prevention . Proper storage and preparation of breast milk. [accessed 2019 August6]. URL: https://www.cdc.gov/breastfeeding/recommendations/handling_breastmilk.htm,


