ABSTRACT
There is increasing evidence for the role of gut microbial composition in the pathogenesis of nonalcoholic fatty liver disease (NAFLD). Nonalcoholic steatohepatitis (NASH) is the most serious form of NAFLD where inflammation causes liver damage that can progress to cirrhosis. We have characterized the gut microbiome composition in UK patients with biopsy-proven NASH (n = 65) and compared it to that in healthy controls (n = 76). We report a 7% lower Shannon alpha diversity in NASH patients without cirrhosis (n = 40) compared to controls (p = 2.7x 10−4) and a 14% drop in NASH patients with cirrhosis (n = 25, p = 5.0x 10−4). Beta diversity (Unweighted UniFrac distance) was also significantly reduced in both NASH (p = 5.6x 10−25) and NASH-cirrhosis (p = 8.1x 10−7) groups. The genus most strongly associated with NASH in this study was Collinsella (0.29% abundance in controls, 3.45% in NASH without cirrhosis (False Discovery Rate (FDR) p = .008), and 4.38% in NASH with cirrhosis (FDR p = .02)). This genus, which has been linked previously to obesity and atherosclerosis, was also positively correlated with fasting levels of triglycerides (p = .01) and total cholesterol (p = 1.2x 10−4) and negatively correlated with high-density lipoprotein cholesterol (p = 2.8x 10−6) suggesting that some of the pathways present in this microbial genus may influence lipid metabolism in the host. In patients, we also found decreased abundance of some of the Ruminococcaceae which are known to produce high levels of short-chain fatty acids which can lower inflammation. This may thus contribute to pathology associated with NASH.
KEYWORDS: Fatty liver, nonalcoholic steatohepatitis, cirrhosis, microbiome, Collinsella
Introduction
Nonalcoholic fatty liver disease (NAFLD) is the most common liver disease worldwide1–3 and includes a spectrum of pathologies ranging from simple steatosis through nonalcoholic steatohepatitis (NASH), characterized by the presence of inflammation, to cirrhosis4 which is responsible for 2% of all deaths worldwide5 Around one-third of those with early NASH will progress to advanced fibrosis and cirrhosis within 5–10 years6 but a lack of early symptoms means that patients accumulate significant, sometimes irreversible liver damage by the time they are diagnosed. Therefore, understanding of contributing factors with potential as interventional treatment targets, is urgently sought.
There is a growing body of evidence linking the gut-liver axis to the development of NAFLD. NAFLD has been associated with an increase in intestinal permeability7 and the subsequent infiltration of Gram-negative bacteria could induce liver inflammation via TLR4 signaling, enabling the release of TNF-α and the activation of hepatic stellate cells8 As the liver is linked to the intestine through the portal circulation, gut-derived products including both nutrients from the diet and microbial components will arrive first at the liver.9 Pathogenic interactions between the liver and microbiota range from straightforward effects, such as increased intestinal permeability leading to passage of Gram-negative bacteria into the portal circulation and subsequent inflammation10 to more complex interactions with metabolites produced by the microbiota, under the influence of modifiable factors such as diet. Since the establishment of a gut microbiome characteristic of obesity11 similar links between the intestinal microbiota and the development of NASH have been investigated.
Small-scale studies of the microbiome in NASH have been somewhat contradictory: a study in 16 NASH patients identified a number of associated genera (Parabacteroides and Allisonella), and a longitudinal study has correlated a reduction of intrahepatic triglycerides with a reduction in Firmicutes and increase in Bacteroidetes12 This has been confirmed in a separate study using real-time PCR for 16S rRNA which also associated a drop in Bacteroidetes (in which Prevotella was used as a marker for the entire phylum) with NASH.13 However, an increase in Bacteroidetes abundance was subsequently described in NASH patients.14
Lower alpha diversity has consistently been found to be correlated with obesity and insulin resistance15 higher visceral fat16 and a number of inflammatory conditions.17,18 Since inflammatory processes drive the pathology and progression of NAFLD and are linked to features of the metabolic syndrome including insulin resistance and visceral fat, we hypothesized that patients with NASH will display distinct changes in their microbiota and lower alpha diversity after adjustment for possible confounders such as BMI.
We describe significant differences in the microbiome in a substantial, well-phenotyped cohort of 65 biopsy-proven NASH patients compared to a control cohort of 76 participants, and identify a metabolic potential mechanism by evaluating diet and serum lipids.
Results
Clinical characteristics
The descriptive characteristics of NASH patients and controls are presented in Table 1. The healthy control group was slightly older on average compared to the NASH group and was predominantly females. The control group had a significantly lower median BMI compared to the NASH group; therefore, all analyses were adjusted for age, sex and BMI. All participants in the control group were of White ethnicity; 87% (33/40 NASH and 24/25 NASH-cirrhosis) of the patients were of White ethnicity. No significant differences in demographics were observed between NASH and NASH-cirrhosis groups. Within the NASH group, 1 patient had no fibrosis, 8 patients had fibrosis grade F1, 8 had grade F2, and 23 had grade F3.
Table 1.
Characteristics of the study population.
| Characteristic | Healthy (n = 76) |
NASH (n = 40) |
NASH-cirrhosis (n = 25) |
|---|---|---|---|
| Sex (female/male) | 67/9 | 20/20 | 15/10 |
| Mean age (mean±SEM) | 66.3 ± 1.06 | 60.3 ± 1.6a | 64.0 ± 2.0 |
| Mean BMI (kg/m2, mean±SEM) | 26.5 ± 0.5 | 34.9 ± 1.1a | 34.0 ± 1.1b |
| Type 2 Diabetes | 0 | 22a | 19b |
| Hypertension | 1 | 19a | 14b |
| Shannon diversity (mean±SEM) | 5.83 ± 0.1 | 5.32 ± 0.1a | 5.18 ± 0.2b |
| Observed OTUs (mean±SEM) | 405.3 ± 10.9 | 334.8 ± 9.5a | 313.7 ± 16.9b |
NASH: nonalcoholic steatohepatitis; SEM: standard error of the mean; OTUs: operational taxonomic unit.ap < .05 Healthy vs NASH, bp < .05, Healthy vs NASH-cirrhosis, cp < .05 NASH vs NASH-cirrhosis, ANOVA with a priori contrasts or Kruskal–Wallace followed by Mann–Whitney U test as appropriate based on distribution.
Diversity metrics
A significant decrease in alpha diversity, indicated by Shannon index and operational taxonomic units (OTUs), was observed in both NASH and NASH-cirrhosis groups compared to controls (all p < .05; Table 1; Figure 1). A significant decrease in beta diversity (Unweighted UniFrac), indicative of inter-individual diversity, was also observed between control and NASH groups, although it was slightly higher in the NASH-cirrhosis group compared to the NASH group (Figure 1).
Figure 1.
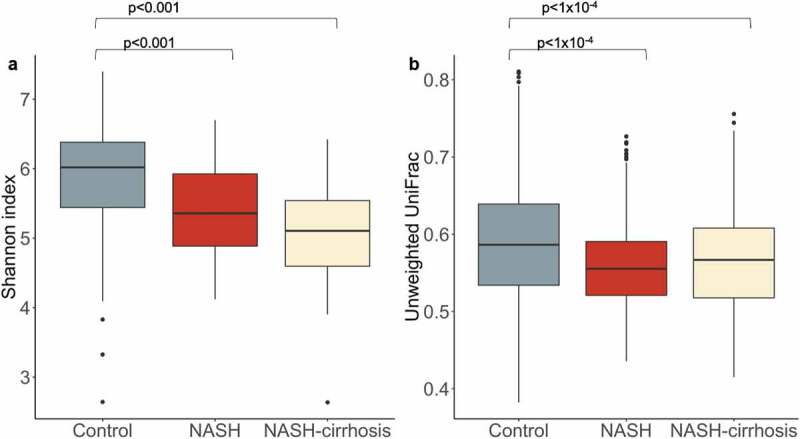
Diversity metrics in control, NASH, and NASH-cirrhosis groups. A: Alpha diversity measured using the Shannon index, B: Beta diversity measured using Unweighted UniFrac distances between all samples in each group. Diversity indices were calculated using the OTU table rarefied to 7000 sequences per sample 50 times and taking the average.
OTUs significantly associated with NASH
Of the 124 OTUs used in the analysis, 27 were significantly associated with NASH after adjustment for covariates (age, gender, and BMI) and false discovery rate (FDR), and 21 of these remained significant in the NASH-cirrhosis group. Several OTUs were significantly associated with a higher likelihood of NASH: most significantly Collinsella, which was also significantly associated with the cirrhosis group (Figures 2 and 3). These OTUs remained associated when patients with white ethnicity only were analyzed (data not shown). The genera most significantly associated with the control group relative to NASH were Clostridium sensu stricto 1, an unidentified genus belonging to the order Mollicutes RF9, Alistipes, and several Ruminococcaceae (Figures 2 and 3).
Figure 2.
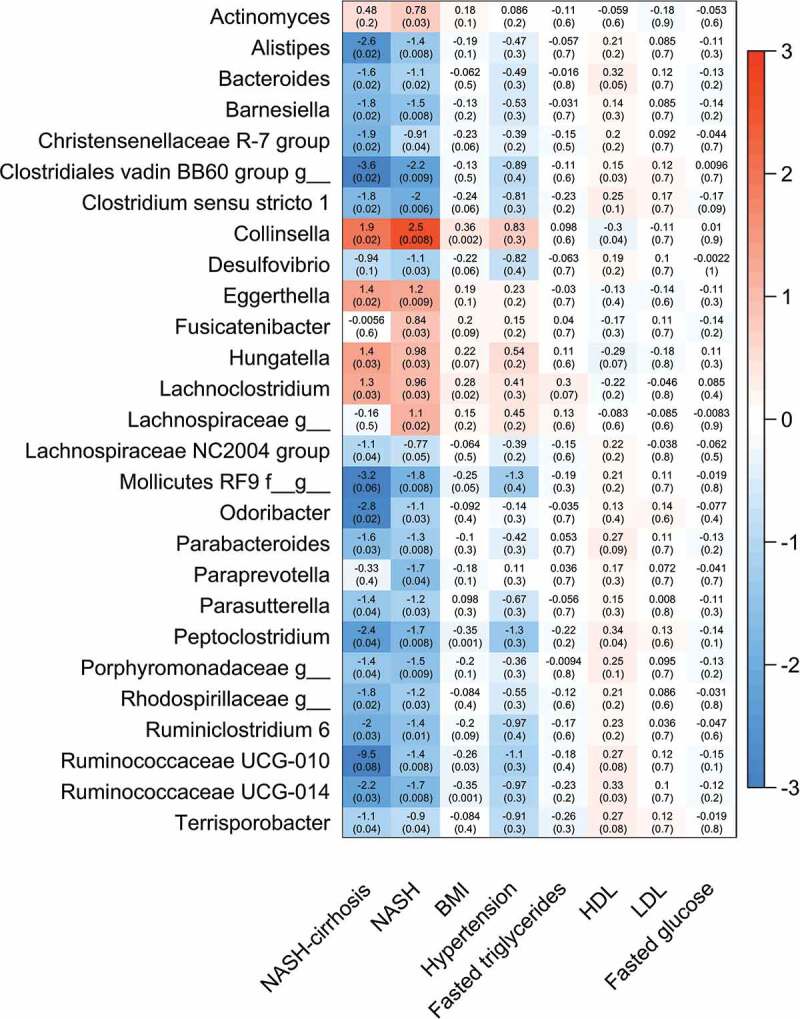
Heat map showing all OTUs (clustered at genus level) significantly associated with NASH or the control group and association with presence of NASH, NASH-cirrhosis, BMI, hypertension, total fasted triglycerides (TG), high-density lipoprotein (HDL), low-density lipoprotein (LDL) and fasted glucose. Values are beta coefficients from linear models adjusted for BMI, age and gender (associations between genera and BMI are adjusted for age and gender only). P values shown in parentheses are adjusted for FDR.
Figure 3.
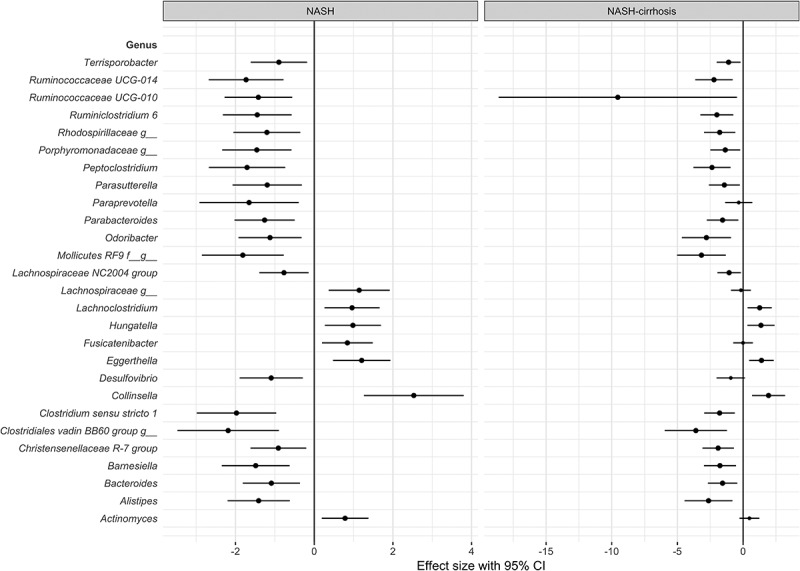
Forest plot of effect sizes with 95% confidence intervals showing association of all significant (FDR adjusted p < .05) OTUs with NASH and NASH-cirrhosis compared to control cohort. Smaller dots on the NASH-cirrhosis arm indicate a loss of statistical significance.
Figure 4 displays the relative abundance of each OTU significantly associated with presence of NASH. Collinsella showed a 12-fold increase in relative abundance in the NASH group, increasing from 0.29% to 3.44%, and to 4.38% in NASH-cirrhosis patients. Conversely, the genera Allistipes, Peptoclostridium, Paraprevotella, and two Ruminococcaceae showed statistically significant reductions in the both NASH groups relative to controls.
Figure 4.
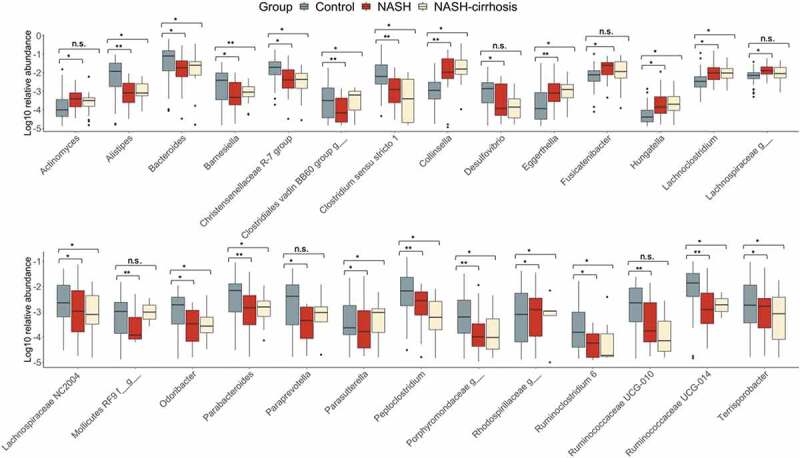
Relative abundance of all OTUs significantly associated (FDR p < .05) with the NASH group. *p < .05, **p < .01.
A number of genera had significantly reduced abundance in NASH-only participants. However, of these, Actinomyces, Fusicatenibacter, an unknown Lachnospiracae, an unknown Mollicutes RF9, paraprevotella, and Ruminococcaceae UCG-010 were not significantly altered in the cirrhosis group compared to controls. Aside from Mollicutes RF9, which had a similar relative abundance in the NASH-cirrhosis group to in controls, the remaining five genera all displayed similar changes in abundance to NASH patients without cirrhosis, but this did not meet the FDR-adjusted significance threshold.
Microbial associations with dietary components
We then assessed the link between nutrient intake and the abundance of taxa to investigate the potential impact of diet on microbes which could contribute to the development of NASH/NASH-cirrhosis. Figure 5 shows a heatmap summarizing Spearman correlations between nutrient intake calculated from a Food Frequency Questionnaire (FFQ) and the bacterial genera found to be associated with presence of NASH. The genera that showed significant positive associations with fiber (NSP) were Eggerthella (r = 0.646; p = .047) and Ruminococcaceae UCG-014 (r = 0.853; p = .003), whereas Collinsella (r = –0.785; p = .015) showed a negative correlation. Collinsella showed a positive correlation (r = 0.647; p = .051) with total fat although it did not reach statistical significance, whereas Alistipes and Eggerthella were significantly correlated with fat intake (r = −0.517; p = .040 and r = −0.717; p = .033, respectively). The correlation between Collinsella and sugar was r = 0.512 (p = .062) and the correlation with total carbohydrates was r = 0.326 (p = .822).
Figure 5.
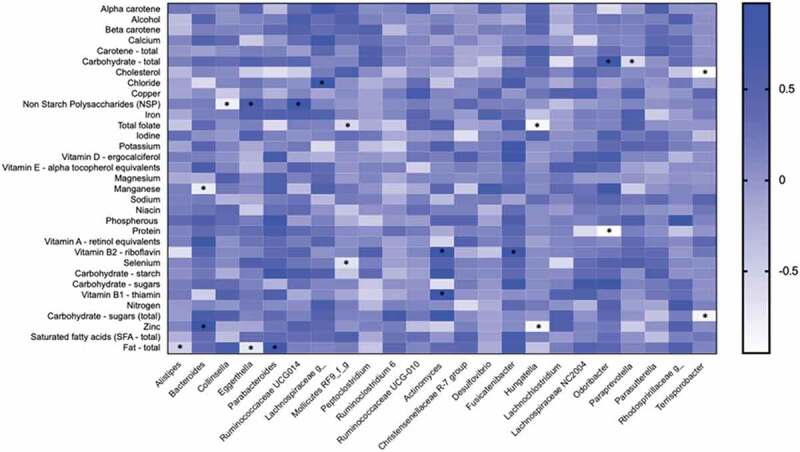
Heatmap depicting the correlation between the abundance of genera significantly associated with NASH and NASH-cirrhosis and dietary components. The intensity of the color represents the degree of association between bacterial abundances and nutrients as measured by Spearman’s correlations. The asterisk symbols indicate the associations that are significant after adjusting for FDR.
Serum lipids
Based on our finding that the abundance of Collinsella is 12-fold higher in NASH patients and correlation with fat intake, we explored the association between Collinsella and circulating lipid sub-types to investigate any changes in fat metabolism that may arise which impact on disease pathways (Table 2). Fasting serum levels of triglycerides (TG), and saturated fatty acids were significantly higher in the NASH and NASH-cirrhosis group than in controls (Supplementary Table 1) whereas levels of low-density lipoprotein cholesterol (LDL-C) and high-density lipoprotein cholesterol (HDL-C) were lower in patients. Correlation between these lipids and presence or absence of NASH/NASH-cirrhosis was then assessed (Supplementary Table 2). HDL-C was the most strongly associated lipid with NAFLD showing a strong negative correlation (Pearson’s r = −0.75 p = 1.9x 10−17), TG and LDL-C and fatty acids were also correlated with presence of disease.
Table 2.
Correlation between relative abundance of Collinsella and serum lipids in the total study population.
| Metabolite | Correlation with Collinsella relative abundance (Pearson’s r and p value) |
|---|---|
| Triglycerides (mmol/L) | 0.22 (0.01) |
| Total cholesterol (mmol/L) | 0.328 (1.2x 10−4) |
| VLDL-C (mmol/L) | 0.14 (0.11) |
| HDL-C (mmol/L) | −0.39 (2.8x 10−6) |
| LDL-C (mmol/L) | −0.16 (0.071) |
| Saturated fatty acids (mmol/L) | 0.09 (0.32) |
| Remnant cholesterol (mmol/L) | 0.0021 (0.98) |
VLDL-C: very low-density cholesterol; HDL-C: high-density lipoprotein cholesterol; LDL-C: low-density lipoprotein cholesterol.
We then tested whether any of these markers were also correlated with the abundance of the genus Collinsella (Table 2) and found that HDL-C was inversely associated with its abundance (r = −0.39, p = 2.8x 10−6). A significant positive correlation was seen between the relative abundance of Collinsella and both serum TG (r = 0.21, p = .049) and total cholesterol (r = 0.328, p = 1.2x 10−4).
Discussion
Links between the microbiome and the development of NAFLD are now well established,19,20 but the details of changes in specific genera are lacking. We aimed to compare a substantial cohort of biopsy-proven NASH and NASH-cirrhosis patients, with a well-characterized control cohort sourced from the TwinsUK study.
The genus most strongly associated with NASH in this study, Collinsella, has been previously linked to obesity and atherosclerosis in human studies,21–23 showing a fourfold increase in abundance in obesity and 3.5-fold in type 2 diabetes.24 It has also been found to be highly abundant in untreated patients with hypercholesterolemia compared to controls and patients treated with statins.25 NMR spectroscopy in a mouse model has shown a significant correlation between members of the Coriobacteriaceae family, which includes Collinsella, and hepatic triglycerides.26 Collinsella has been linked to pro-inflammatory dysbiosis in type 2 diabetes24 and with circulating insulin21 suggestive of a mechanism for promotion of NAFLD pathology.
Insulin resistance and dyslipidemia are characteristics of NAFLD.27,28 Consistent with this, we show significant increases in total triglycerides, VLDL-C and saturated fatty acids, and concomitant reduction in HDL-C in both NASH and NASH-cirrhosis groups. We also report that the abundance of Collinsella is negatively correlated with serum HDL-C levels and positively correlated with TG, which suggests that some of the pathways present in this microbial genus may influence lipid metabolism in the host. This is consistent with previous work in healthy adults correlating Collinsella abundance with total cholesterol29 and in agreement with animal studies.26,30 We observed a significant inverse correlation between the relative abundance of Collinsella and dietary fiber (Figure 5) as previously reported in obese women.21 Dietary fiber and the gut-derived metabolites that are associated with its breakdown (i.e. short-chain fatty acids) have been implicated in the etiology of metabolic disease including NAFLD.31 Increasing dietary fiber intake could be considered a strategy for modulating the composition of the gut microbiome32 reducing dysbiosis with the potential to reduce pathological features.
Collinsella spp. have been shown to metabolize bile acids to oxo-bile acid intermediates. Production of these secondary bile acids may increase intestinal permeability and contribute to the development of NAFLD.33,34 Administration of the antibiotic rifaximin to decrease 7α-dehydroxylating bacteria (including Collinsella) has been linked to a drop in secondary fecal bile acid concentrations.35 This provides a convincing mechanism by which Collinsella could mediate the progression of NAFLD, but further work is required to establish this link and determine the specific species or group of species that could be the cause.
We identified an OTU belonging to the putative order Mollicutes RF9 significantly associated with the control cohort. Members of the Mollicutes class have previously been linked to obesity36, but their functional roles are unclear.37 Recent work38 has correlated abundance of Mollicutes RF39 with circulating indolepropionic acid, a potent antioxidant produced by the microbiota and associated with a lower risk of type II diabetes39,40 thus providing a likely mechanism protective for NAFLD.
Several genera belonging to the order Clostridiales were also associated with the control group in this study. Of these, we found that Ruminococcaceae showed significant positive correlations with fiber intake. Several Ruminococcaceae have previously been inversely associated with cirrhosis in small studies including patients with a variety of etiologies,41,42 and with NAFLD.43,44 Ruminococcaceae are mainly responsible for fermenting fiber and other plant components of the diet such as inulin and cellulose, producing short-chain fatty acids (SCFA) which can both be utilized by the host for energy45 and display anti-inflammatory properties in the gut.46
We report a lower alpha diversity in NASH patients, further reduced in those with cirrhosis, similar to previous work comparing NAFLD patients without advanced fibrosis to those with cirrhosis.47 We also demonstrate parallel changes in beta diversity, with a decline in our 40 NASH patients (of which 23 had advanced fibrosis) compared to controls but with no further reduction in cirrhotics. This indicates that the microbiome in the general population is both more diverse and more stable than that of both NASH and NASH-cirrhosis patients, with inter-individual variation increasing in cirrhosis. In agreement with earlier publications, we found a notable decrease in Bacteroides abundance in both the NASH group and NASH-cirrhosis groups. Unlike a previous report in pediatric NAFLD,48 we did not see a significant decrease in Oscillspira; increases in Blautia and Ruminococcus were observed but did not achieve statistical significance. Increases in Blautia and Ruminococcus were similarly observed in a cirrhosis group in the aforementioned Caussy et al. study47 but not in their cohort without advanced fibrosis. This suggests that some of these changes may reflect different stages of severity of liver disease and highlights the importance of in-depth hepatic phenotyping.
We acknowledge several limitations in our study. The control group is predominantly female whereas both NASH and cirrhosis groups are more evenly gender balanced. The effect of gender on the microbiome is well established49,50 and may therefore have some effect on our results, but this has been adjusted for in all statistical comparisons. There are also acknowledged issues with using 16S reads as a proxy indicator for abundance51 compared to absolute quantification with genus or species-specific PCR primers or a metagenomic sequencing approach. The study presented here is also purely observational; a germ-free animal model would be required to definitively verify the links we propose between Collinsella abundance, NASH, and lipid metabolism. Furthermore, the associations between nutrients and the gut microbiome would require further evaluations to explore their interdependence in the etiology of NASH and NASH-cirrhosis. The diet-gut microbiome associations presented are based on statistical associations from cross-sectional observations, and do not prove that causal relationships exist. Further explorations into causal impacts would require dietary interventions. In order to untangle the effect of nutrients on host-microbiome crosstalk, future studies should also consider looking at fecal metabolites such as SCFAs. Fecal bile acid data were also not available for this study; this would be required to study the metabolic effects of Collinsella in more detail, as well as the gut-liver axis in general.52 Thus, our study could be improved by looking at bacterial metabolic responses to diet which may be more informative in assessing the effect of certain nutrients on health, compared to the gut microbiota itself.
Overall, we have determined the relationship between gut microbiome composition and diversity and disease in the largest biopsy-proven NASH cohort to date. We confirm the findings from previous studies14,47 showing a reduced alpha diversity in NASH patients and further decrease in cirrhosis patients. We also report, for the first time, that the genus Collinsella is significantly increased in NASH patients with and without cirrhosis. This association with pathology is suggested to be linked with effects on host lipid metabolism and diet.
Materials and methods
Study participants
The 65 adult patients included in this study were prospectively recruited from hepatology clinics at Nottingham University Hospitals NHS Trust. Patients gave informed consent to participate in research studies in accordance with the 1975 Declaration of Helsinki. The studies were approved by NHS Regional Ethics Committees (REC reference 12/WM/0288 and GM010201). NASH was diagnosed on the basis of the following criteria: appropriate exclusion of other causes of liver disease including alcohol, drugs, autoimmune or viral hepatitis, or cholestatic or metabolic/genetic liver disease; a weekly ethanol consumption of <140 g in women and <210 g in men and a liver biopsy showing steatohepatitis with or without cirrhosis. The Fatty Liver Inhibition of Progression (FLIP) Consortium algorithm was used as a diagnostic indicator where histological diagnosis of NASH requires presence of steatosis, ballooning, and lobular inflammation, and those without all three features are identified as having fatty liver while those lacking steatosis (<5% hepatocytes with fat accumulation) are not considered as having NASH53 Fibrosis was scored following the CRN grading system54 by a single pathologist. Patients were stratified into two groups: NASH (40 patients without cirrhosis) and NASH with cirrhosis (25 patients). All patients in the NASH-cirrhosis group had compensated cirrhosis at the time of sample collection.
Seventy-six healthy controls were included from the UK Adult Twin Registry (TwinsUK), a large cohort of volunteer adult twins from the United Kingdom hosted by the Department of Twin Research and Genetic Epidemiology at St. Thomas’ Hospital, King’s College London.55This cohort contained 13 twin pairs (9 monozygotic pairs and 4 dizygotic pairs); all others had a twin who was not included.
Sequencing
Fecal samples were provided at study visits and immediately frozen at −80°C. DNA was extracted by homogenizing in lysis buffer (500mM NaCl, 50mM Tris-HCl, 50mM EDTA, 4% SDS) with bead beating (MagNA Lyser, Roche) followed by extraction using the QIAmp DNA Stool Mini Kit (Qiagen; cat 51504). DNA was diluted to 20ng/µl for 16S rRNA amplification and sequencing at the Genetic Laboratory, Erasmus Medical Center, Rotterdam, the Netherlands. Water negative controls were included from extraction, through PCR to sequencing and select samples were sequenced in duplicate for quality control.
The V4 region of the 16S ribosomal RNA gene was amplified using universal primers 355F (CCAGACTCCTACGGGAGGCAGC) and 806R (GGACTACHVGGGTWTCTAAT). Amplified DNA was sequenced on the MiSeq platform (Illumina). Read filtering and clustering was carried out using the MYcrobiota pipeline.56 Briefly, chimeric sequences were filtered using the VSEARCH algorithm within Mothur, and reads were clustered into operational taxonomic units (OTUs) using closed-reference clustering against the SILVA database v132 based on a 97% similarity. Diversity metrics (Shannon index observed OTUs and Unweighted UniFrac) were calculated by rarefying the OTU table down to 7000 sequences per sample 50 times and taking the average. These analyses were carried out in QIIME 2 (v2018.11).
Lipid measurements
Serum lipids were quantified using a high-throughput1 H NMR platform (Nightingale Health, Helsinki, Finland) using 500Mhz and 600MhH protein nuclear magnetic resonance spectroscopy as described previously.57,58
Diet-microbiome analysis
Self-reported dietary intake was assessed by food frequency questionnaires following the EPIC-Norfolk guidelines.59 Individuals answered questions on intake of 131 food groups and nutrient intakes (weight/day) were derived from the UK Nutrient Database using the FETA software (version 2).59 A total of 33 nutrients were estimated from this calculation. Associations between relative abundances of bacterial taxa shown to be significantly associated with NASH or the control group, and nutrient intakes (adjusted for total energy (kcal)) were examined by Spearman correlation.
Statistical analysis
OTUs with a relative abundance of <0.1% in every sample were removed, and relative OTU abundances were inverse normal transformed before further analysis. Associations between NASH and NASH-cirrhosis groups and OTU abundance at genus level were predicted using a general linear model adjusted for age, gender and BMI. OTUs were assumed to be significantly associated with NASH or cirrhosis with a p value <.05 after adjusting for FDR. All statistical analyses were carried out in R v3.5.2.
Supplementary Material
Acknowledgments
We are grateful to Melanie Lingaya for technical assistance including DNA extraction. We thank the clinical fellows (Naaventhan Palaniyappan and Robert Scott) and nurse team at the NIHR Nottingham Biomedical Research Centre for assisting with recruitment. Sample collection was funded by the National Institute for Health Research (NIHR) Nottingham Biomedical Research Centre (reference no: BRC-1215-20003). The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health. The team at the Genetic Laboratory, Erasmus Medical Center, Rotterdam, are acknowledged for 16S amplification and sequencing. We thank Philip Kaye (Department of Histopathology, Nottingham University Hospitals NHS Trust) for scoring all patient biopsies.
Funding Statement
This work was funded by the NIHR Nottingham Biomedical Research Centre [BRC-1215-20003, flexifund award], the Nottingham Molecular Pathology Node [MRC/EPSRC grant MR/N005953/1], the MRC AimHy study [MR/M016560/1] and a CDRF project grant. TwinsUK receives funding from the Wellcome Trust and the EU Seventh Framework Programme. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Disclosure statement
AMV is a consultant for Zoe Global Ltd.
Contributions
JIG, AMV, GPA: Obtained funding, developed concept and designed the study. JIG: study co-ordination. SA, EA, AV, JIG, AMV: Analysed and interpreted the data and drafted the manuscript. All authors reviewed and revised the final manuscript.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Browning JD, Cohen JC, Hobbs HH.. Patatin-like phospholipase domain-containing 3 and the pathogenesis and progression of pediatric nonalcoholic fatty liver disease. Hepatology. 2010;52:1189–1192. doi: 10.1002/hep.23946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, Charlton M, Sanyal AJ. The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the american association for the study of liver diseases, American college of gastroenterology, and the American gastroenterological association. Hepatology. 2012;55:2005–2023. doi: 10.1002/hep.25762. [DOI] [PubMed] [Google Scholar]
- 3.Kelly T, Yang W, Chen C-S, Reynolds K, He J. Global burden of obesity in 2005 and projections to 2030. Int J Obes. 2008;32:1431–1437. doi: 10.1038/ijo.2008.102. [DOI] [PubMed] [Google Scholar]
- 4.Higuera-de la Tijera F, Servín-Caamaño AI. Pathophysiological mechanisms involved in non-alcoholic steatohepatitis and novel potential therapeutic targets. World J Hepatol. 2015;7:1297–1301. doi: 10.4254/wjh.v7.i10.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murray CJL, Vos T, Lozano R, Naghavi M, Flaxman AD, Michaud C, Ezzati M, Shibuya K, Salomon JA, Abdalla S, et al. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: a systematic analysis for the global burden of disease study 2010. Lancet Lond Engl. 2012;380:2197–2223. doi: 10.1016/S0140-6736(12)61689-4. [DOI] [PubMed] [Google Scholar]
- 6.Calzadilla Bertot L, Adams LA. The natural course of non-alcoholic fatty liver disease. International Journal Of Molecular Sciences. 2016;17:774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miele L, Valenza V, La Torre G, Montalto M, Cammarota G, Ricci R, Mascianà R, Forgione A, Gabrieli ML, Perotti G, et al. Increased intestinal permeability and tight junction alterations in nonalcoholic fatty liver disease. Hepatology. 2009;49:1877–1887. doi: 10.1002/hep.22848. [DOI] [PubMed] [Google Scholar]
- 8.Csak T, Ganz M, Pespisa J, Kodys K, Dolganiuc A, Szabo G. Fatty acid and endotoxin activate inflammasomes in mouse hepatocytes that release danger signals to stimulate immune cells. Hepatology. 2011;54:133–144. doi: 10.1002/hep.v54.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandl K, Kumar V, Eckmann L. Gut-liver axis at the frontier of host-microbial interactions. Am J Physiol-Gastrointest Liver Physiol. 2017;312:G413–9. doi: 10.1152/ajpgi.00361.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moreira APB, Texeira TFS, Ferreira AB, Peluzio MDCG, Alfenas RDCG. Influence of a high-fat diet on gut microbiota, intestinal permeability and metabolic endotoxaemia. Br J Nutr. 2012;108:801–809. [DOI] [PubMed] [Google Scholar]
- 11.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 12.Wong VW-S, Tse C-H, Lam TT-Y, Wong GL-H, Chim AM-L, Chu WC-W, Yeung DK-W, Law PT-W, Kwan H-S, Yu J, et al. Molecular characterization of the fecal microbiota in patients with nonalcoholic steatohepatitis – a longitudinal study. PLoS One. 2013;8:e62885. doi: 10.1371/journal.pone.0062885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mouzaki M, Comelli EM, Arendt BM, Bonengel J, Fung SK, Fischer SE, McGilvray ID, Allard JP. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology. 2013;58:120–127. doi: 10.1002/hep.26319. [DOI] [PubMed] [Google Scholar]
- 14.Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD, Gill SR. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57:601–609. doi: 10.1002/hep.26093. [DOI] [PubMed] [Google Scholar]
- 15.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto J-M, Kennedy S, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 16.Beaumont M, Goodrich JK, Jackson MA, Yet I, Davenport ER, Vieira-Silva S, Debelius J, Pallister T, Mangino M, Raes J, et al. Heritable components of the human fecal microbiome are associated with visceral fat. Genome Biol. 2016;17:189. doi: 10.1186/s13059-016-1052-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EGAbramson SB, et al. Expansion of intestinal prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schnabl B, Brenner DA. Interactions between the intestinal microbiome and liver diseases. Gastroenterology. 2014;146:1513–1524. doi: 10.1053/j.gastro.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Safari Z, Gérard P. The links between the gut microbiome and non-alcoholic fatty liver disease (NAFLD). Cell Mol Life Sci. 2019;76:1541–1558. doi: 10.1007/s00018-019-03011-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomez-Arango LF, Barrett HL, Wilkinson SA, Callaway LK, McIntyre HD, Morrison M, Nitert MD. Low dietary fiber intake increases Collinsella abundance in the gut microbiota of overweight and obese pregnant women. Gut Microbes. 2018;9:189–201. doi: 10.1080/19490976.2017.1406584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karlsson FH, Fåk F, Nookaew I, Tremaroli V, Fagerberg B, Petranovic D, Bäckhed F, Nielsen J. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun. 2012;3:1245. doi: 10.1038/ncomms2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kourosh A, Luna RA, Balderas M, Nance C, Anagnostou A, Devaraj S, Davis CM. Fecal microbiome signatures are different in food-allergic children compared to siblings and healthy children. Pediatr Allergy Immunol Off Publ Eur Soc Pediatr Allergy Immunol. 2018;29:545–554. doi: 10.1111/pai.12904. [DOI] [PubMed] [Google Scholar]
- 24.Candela M, Biagi E, Soverini M, Consolandi C, Quercia S, Severgnini M, Peano C, Turroni S, Rampelli S, Pozzilli P, et al. Modulation of gut microbiota dysbioses in type 2 diabetic patients by macrobiotic Ma-Pi 2 diet. Br J Nutr. 2016;116:80–93. doi: 10.1017/S0007114516001045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan TJ, Ahmed YM, Zamzami MA, Siddiqui AM, Khan I, Baothman OAS, Mehanna MG, Kuerban A, Kaleemuddin M, Yasir M. Atorvastatin treatment modulates the gut microbiota of the hypercholesterolemic patients. Omics J Integr Biol. 2018;22:154–163. doi: 10.1089/omi.2017.0130. [DOI] [PubMed] [Google Scholar]
- 26.Claus SP, Ellero SL, Berger B, Krause L, Bruttin A, Molina J, Paris A, Want EJ, Waziers ID, Cloarec O, et al. Colonization-induced host-gut microbial metabolic interaction. mBio. 2011;2:e00271–10. doi: 10.1128/mBio.00271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanyal AJ, Campbell–Sargent C, Mirshahi F, Rizzo WB, Contos MJ, Sterling RK, Luketic VA, Shiffman ML, Clore JN. Nonalcoholic steatohepatitis: association of insulin resistance and mitochondrial abnormalities. Gastroenterology. 2001;120:1183–1192. doi: 10.1053/gast.2001.23256. [DOI] [PubMed] [Google Scholar]
- 28.Katsiki N, Mikhailidis DP, Mantzoros CS. Non-alcoholic fatty liver disease and dyslipidemia: an update. Metabolism. 2016;65:1109–1123. doi: 10.1016/j.metabol.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Lahti L, Salonen A, Kekkonen RA, Salojärvi J, Jalanka-Tuovinen J, Palva A, Orešič M, de Vos WM. Associations between the human intestinal microbiota, Lactobacillus rhamnosus GG and serum lipids indicated by integrated analysis of high-throughput profiling data. PeerJ. 2013;1:e32. doi: 10.7717/peerj.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martínez I, Wallace G, Zhang C, Legge R, Benson AK, Carr TP, Moriyama EN, Walter J. Diet-induced metabolic improvements in a hamster model of hypercholesterolemia are strongly linked to alterations of the gut microbiota. Appl Environ Microbiol. 2009;75:4175–4184. doi: 10.1128/AEM.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Canfora EE, Meex RCR, Venema K, Blaak EE. Gut microbial metabolites in obesity, NAFLD and T2DM. Nat Rev Endocrinol. 2019;15:261–273. doi: 10.1038/s41574-019-0156-z. [DOI] [PubMed] [Google Scholar]
- 32.Makki K, Deehan EC, Walter J, Bäckhed F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe. 2018;23:705–715. doi: 10.1016/j.chom.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 33.Doden H, Sallam LA, Devendran S, Ly L, Doden G, Daniel SL, Alves JMP, Ridlon JM. Metabolism of oxo-bile acids and characterization of recombinant 12α-hydroxysteroid dehydrogenases from bile acid 7α-dehydroxylating human gut bacteria. Appl Environ Microbiol. 2018;84(10). doi: 10.1128/AEM.00235-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stenman LK, Holma R, Eggert A, Korpela R. A novel mechanism for gut barrier dysfunction by dietary fat: epithelial disruption by hydrophobic bile acids. Am J Physiol-Gastrointest Liver Physiol. 2012;304:G227–34. doi: 10.1152/ajpgi.00267.2012. [DOI] [PubMed] [Google Scholar]
- 35.Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, Takei H, Muto A, Nittono H, Ridlon JM, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58:949–955. doi: 10.1016/j.jhep.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turnbaugh PJ. Microbes and diet-induced obesity: fast, cheap, and out of control. Cell Host Microbe. 2017;21:278–281. doi: 10.1016/j.chom.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menni C, Hernandez MM, Vital M, Mohney RP, Spector TD, Valdes AM. Circulating levels of the anti-oxidant indoleproprionic acid are associated with higher gut microbiome diversity. Gut microbes. 2019;1–8. doi: 10.1080/19490976.2019.1586038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Mello VD, Paananen J, Lindström J, Lankinen MA, Shi L, Kuusisto J, Pihlajamäki J, Auriola S, Lehtonen M, Rolandsson O, et al. Indolepropionic acid and novel lipid metabolites are associated with a lower risk of type 2 diabetes in the finnish diabetes prevention study. Sci Rep. 2017;7:46337. doi: 10.1038/srep46337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valdes AM, Walter J, Segal E, Spector TD. Role of the gut microbiota in nutrition and health. BMJ. 2018;361:k2179. doi: 10.1136/bmj.k2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bajaj JS, Ridlon JM, Hylemon PB, Thacker LR, Heuman DM, Smith S, Sikaroodi M, Gillevet PM. Linkage of gut microbiome with cognition in hepatic encephalopathy. Am J Physiol-Gastrointest Liver Physiol. 2011;302:G168–75. doi: 10.1152/ajpgi.00190.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bajaj JS, Hylemon PB, Ridlon JM, Heuman DM, Daita K, White MB, Monteith P, Noble NA, Sikaroodi M, Gillevet PM. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol-Gastrointest Liver Physiol. 2012;303:G675–85. doi: 10.1152/ajpgi.00152.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang W, Wu N, Wang X, Chi Y, Zhang Y, Qiu X, Hu Y, Li J, Liu Y. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci Rep. 2015;5:8096. doi: 10.1038/srep08096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raman M, Ahmed I, Gillevet PM, Probert CS, Ratcliffe NM, Smith S, Greenwood R, Sikaroodi M, Lam V, Crotty P, et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2013;11(868–875.e3). doi: 10.1016/j.cgh.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 45.Scheppach W, Weiler F. The butyrate story: old wine in new bottles? Curr Opin Clin Nutr Metab Care. 2004;7:563. doi: 10.1097/00075197-200409000-00009. [DOI] [PubMed] [Google Scholar]
- 46.Kles KA, Chang EB. Short-chain fatty acids impact on intestinal adaptation, inflammation, carcinoma, and failure. Gastroenterology. 2006;130:S100–5. doi: 10.1053/j.gastro.2005.11.048. [DOI] [PubMed] [Google Scholar]
- 47.Caussy C, Tripathi A, Humphrey G, Bassirian S, Singh S, Faulkner C, Bettencourt R, Rizo E, Richards L, Xu ZZ, et al. A gut microbiome signature for cirrhosis due to nonalcoholic fatty liver disease. Nat Commun. 2019;10:1406. doi: 10.1038/s41467-019-09455-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chierico FD, Nobili V, Vernocchi P, Russo A, Stefanis CD, Gnani D, Furlanello C, Zandonà A, Paci P, Capuani G, et al. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology. 2017;65:451–464. doi: 10.1002/hep.v65.2. [DOI] [PubMed] [Google Scholar]
- 49.Haro C, Rangel-Zúñiga OA, Alcalá-Díaz JF, Gómez-Delgado F, Pérez-Martínez P, Delgado-Lista J, Quintana-Navarro GM, Landa BB, Navas-Cortés JA, Tena-Sempere M, et al. Intestinal microbiota is influenced by gender and body mass index. PLoS One. 2016;11:e0154090. doi: 10.1371/journal.pone.0154090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, Valles-Colomer M, Vandeputte D, et al. Population-level analysis of gut microbiome variation. Science. 2016;352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 51.Tkacz A, Hortala M, Poole PS. Absolute quantitation of microbiota abundance in environmental samples. Microbiome. 2018;6:110. doi: 10.1186/s40168-018-0491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu H-X, Keane R, Sheng L, Wan Y-JY. Implications of microbiota and bile acid in liver injury and regeneration. J Hepatol. 2015;63:1502–1510. doi: 10.1016/j.jhep.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bedossa P, Poitou C, Veyrie N, Bouillot J-L, Basdevant A, Paradis V, Tordjman J, Clement K. Histopathological algorithm and scoring system for evaluation of liver lesions in morbidly obese patients. Hepatol Baltim Md. 2012;56:1751–1759. doi: 10.1002/hep.25889. [DOI] [PubMed] [Google Scholar]
- 54.Brunt EM, Kleiner DE, Wilson LA, Belt P, Neuschwander-Tetri BA. NASH clinical research network (CRN). Nonalcoholic fatty liver disease (NAFLD) activity score and the histopathologic diagnosis in NAFLD: distinct clinicopathologic meanings. Hepatol Baltim Md. 2011;53:810–820. doi: 10.1002/hep.24127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moayyeri A, Hammond CJ, Valdes AM, Spector TD. Cohort Profile: twinsUK and healthy ageing twin study. Int J Epidemiol. 2013;42:76–85. doi: 10.1093/ije/dyr207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boers SA, Hiltemann SD, Stubbs AP, Jansen R, Hays JP. Development and evaluation of a culture-free microbiota profiling platform (MYcrobiota) for clinical diagnostics. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol. 2018;37:1081–1089. doi: 10.1007/s10096-018-3220-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valdes AM, Ravipati S, Pousinis P, Menni C, Mangino M, Abhishek A, Chapman V, Barrett DA, Doherty M. Omega-6 oxylipins generated by soluble epoxide hydrolase are associated with knee osteoarthritis. J Lipid Res. 2018;59:1763–1770. doi: 10.1194/jlr.P085118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soininen P, Kangas AJ, Würtz P, Tukiainen T, Tynkkynen T, Laatikainen R, Järvelin M-R, Kähönen M, Lehtimäki T, Viikari J, et al. High-throughput serum NMR metabonomics for cost-effective holistic studies on systemic metabolism. Analyst. 2009;134:1781–1785. doi: 10.1039/b910205a. [DOI] [PubMed] [Google Scholar]
- 59.Mulligan AA, Luben RN, Bhaniani A, Parry-Smith DJ, O’Connor L, Khawaja AP, Forouhi NGKhaw K-T. A new tool for converting food frequency questionnaire data into nutrient and food group values: FETA research methods and availability. BMJ Open 2014; 4:e004503. Available from: http://www.srl.cam.ac.uk/epic/nutmethod/FFQ.shtml [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


