ABSTRACT
Urinary tract infection (UTI) is a common complication in kidney transplant recipients and can lead to significant morbidity and mortality. Recent evidence supports a role for the gut as a source for UTIs but little is known about the relationship between gut commensal bacteria and UTI development. We hypothesized that the abundance of gut commensal bacteria is associated with a lower risk of developing bacteriuria and UTIs. We performed gut microbiome profiling using 16S rRNA gene sequencing of the V4-V5 hypervariable region on 510 fecal specimens in 168 kidney transplant recipients. Fifty-one kidney transplant recipients (30%) developed Enterobacteriaceae bacteriuria within the first 6 months after transplantation (Enterobacteriaceae Bacteriuria Group) and 117 did not (No Enterobacteriaceae Bacteriuria Group). The relative abundances of Faecalibacterium and Romboutsia were significantly higher in the fecal specimens from the No Enterobacteriaceae Bacteriuria Group than those from the Enterobacteriaceae Bacteriuria Group (Adjusted P value<.01). The combined relative abundance of Faecalibacterium and Romboutsia was inversely correlated with the relative abundance of Enterobacteriaceae (r = −0.13, P = .003). In a multivariable Cox Regression, a top tercile cutoff of the combined relative abundance of Faecalibacterium and Romboutsia of ≥13.7% was independently associated with a decreased risk for Enterobacteriaceae bacteriuria (hazard ratio 0.3, P = .02) and Enterobacteriaceae UTI (hazard ratio 0.4, P = .09). In conclusion, we identify bacterial taxa associated with decreased risk for Enterobacteriaceae bacteriuria and Enterobacteriaceae UTI in kidney transplant recipients, which supports future studies on modulating the gut microbiota as a novel treatment for preventing UTIs.
KEYWORDS: Microbiota, bacteriuria, urinary tract infection, Enterobacteriaceae, Faecalibacterium, Romboutsia, Lactobacillus
Introduction
Kidney transplant recipients have increased survival and improved quality of life compared to patients with end-stage renal disease who receive renal replacement therapies.1 However, the immunosuppressive medications used to prevent rejection of the transplanted kidney leads to an immunocompromised state and frequent infectious complications.2 Urinary tract infection (UTI) is the most common infection in kidney transplant recipients,2,3 affecting approximately 20% in the first 3 months after transplantation with Enterobacteriaceae being the most common cause of post-transplant UTI.4 In severe cases, UTI can lead to urosepsis, allograft damage, and mortality in this immunosuppressed population.5-7 Asymptomatic bacteriuria, a positive urine culture without associated symptoms, is also very common and has been associated with development of pyelonephritis and allograft damage.6 Many kidney transplant recipients unfortunately develop recurrent UTIs, which further increase the risk for allograft failure and mortality.8 Prevention of UTI is thus an important unmet need in kidney transplant recipients.
Recent studies support a role for the gut microbiota in the pathogenesis of UTI.9-11 An elegant review by Flores-Mireles et al. reports the first step in the development of UTI as contamination of the peri-urethral space with gut bacteria.12 In a study of non-transplant patients, Paalanne and colleagues found that the gut abundance of E. coli was higher in children with E. coli UTI than in children without E. coli UTI10, suggesting a role of the gut microbiota in UTI development. In a study of 168 kidney transplant recipients at our center, we found that the gut abundance of Escherichia was associated with future development of Escherichia bacteriuria and UTI.9 We further established at the strain level that the E. coli in the urine was genetically most similar to the E. coli in the fecal specimens from the same subject, supporting the concept that the gut is a primary source of UTI.9 Targeting the gut microbiota to modify the risk for UTI development is particularly attractive as currently known clinical risk factors for post-transplant UTI such as gender and age4 are not modifiable. Indeed, recent case reports have reported that fecal microbial transplantation (FMT) is associated with decreased UTI recurrence.13,14 No study, however, has evaluated the relationship between commensal gut bacteria and UTI development and such a study would help to identify important bacterial taxa that could be beneficial for modifying the risk for UTI development.
In the current study, we evaluated the microbial profiles previously characterized in our study of 168 kidney transplant recipients to assess the relationship between the relative abundance of commensal bacterial taxa and the development of Enterobacteriaceae bacteriuria and UTI. We report that the combined relative abundance of Faecalibacterium and Romboutsia is inversely associated with the relative abundance of Enterobacteriaceae and is associated with a decreased risk for Enterobacteriaceae bacteriuria and UTI.
Results
Characteristics of the transplant cohort
One hundred sixty-eight kidney transplant recipients provided 510 fecal specimens within the first 3 months after kidney transplantation. Fifty-one (30%) kidney transplant recipients developed Enterobacteriaceae bacteriuria within the first 6 months after transplantation (Enterobacteriaceae Bacteriuria Group) and 117 did not (No Enterobacteriaceae Bacteriuria Group). The Enterobacteriaceae bacteriuria cases included: 126 E. coli bacteriuria episodes from 36 kidney transplant recipients; 46 Klebsiella species bacteriuria episodes from 20 kidney transplant recipients; 9 Enterobacter cloacae bacteriuria episodes from 2 kidney transplant recipients; and 3 Citrobacter freundii bacteriuria episodes from 3 kidney transplant recipients; and 1 Raoultella ornithinolytica bacteriuria episode from 1 kidney transplant recipient. The median time to the development of first Enterobacteriaceae bacteriuria was 28 days with an interquartile range of 9 to 83. Among the 51 kidney transplant recipients who developed Enterobacteriaceae bacteriuria, 37 (73%) had more than one Enterobacteriaceae bacteriuria within 6 months post-transplantation, and 33 (65%) developed Enterobacteriaceae UTI with 9 (18%) who had more than one Enterobacteriaceae UTI within 6 months post-transplantation. The median time to the development of first Enterobacteriaceae UTI was 45 days with an interquartile range of 13 to 89.
We compared the demographical and transplant characteristics between the Enterobacteriaceae Bacteriuria Group and the No Enterobacteriaceae Bacteriuria Group (Table 1). Female gender was significantly more common in the Enterobacteriaceae Bacteriuria Group than in the No Enterobacteriaceae Bacteriuria Group (73% vs 33%, respectively, P < .001, Fisher’s exact test) and cefazolin preoperative prophylaxis was significantly less common in the Enterobacteriaceae Bacteriuria Group than in the No Enterobacteriaceae Bacteriuria Group (73% vs 87%, respectively, P = .03, Fisher’s exact test). Age, African-American race, history of diabetes mellitus, cause of end-stage renal disease, panel reactive antibody status, deceased donor transplantation, delayed graft function, trimethoprim-sulfamethoxazole prophylaxis, anti-thymocyte globulin induction therapy, and prednisone maintenance were not significantly different between the two groups (P > .05, Wilcoxon rank sum test or Fisher’s exact test).
Table 1.
Clinical Characteristics in the No Enterobacteriaceae Bacteriuria Group and the Enterobacteriaceae Bacteriuria Group. P values were calculated using the Fisher’s exact test for dichotomous values and using the Wilcoxon rank sum test for continuous variables. ESRD, end-stage renal disease; DM, diabetes mellitus; HTN, hypertension; PRA panel reactive antibody; PCP, Pneumocystis jiroveci.
| Enterobacteriaceae | No Enterobacteriaceae | ||
|---|---|---|---|
| Bacteriuria Group | Bacteriuria Group | ||
| (N = 51) | (N = 117) | ||
| Characteristic |
N (%) or median |
N (%) or median |
P value |
| Age, Years | 57 | 53 | 0.29 |
| Female Gender | 37 (73%) | 39 (33%) | 3.7 x 10−6 |
| African American Race | 12 (24%) | 32 (27%) | 0.70 |
| History of Diabetes Mellitus | 18 (35%) | 31 (26%) | 0.27 |
| Cause of ESRD – DM | 18 (35%) | 30 (26%) | 0.26 |
| Cause of ESRD – HTN | 7 (14%) | 20 (17%) | 0.65 |
| PRA ≥ 80% | 5 (10%) | 8 (7%) | 0.54 |
| Decreased Donor Transplantation | 17 (33%) | 32 (27%) | 0.46 |
| Delayed Graft Function | 10 (20%) | 18 (15%) | 0.51 |
| Cefazolin Preoperative Abx | 37 (73%) | 102 (87%) | 0.03 |
| Trimethoprim/Sulfamethoxazole PCP Prophylaxis | 50 (98%) | 109 (93%) | 0.28 |
| Anti-thymocyte Globulin Induction | 39 (76%) | 89 (76%) | 0.99 |
| Prednisone Maintenance | 17 (33%) | 28 (24%) | 0.26 |
Gut microbial composition associated with Enterobacteriaceae bacteriuria
Microbiome profiling was previously performed on each of the 510 fecal specimens from the 168 kidney transplant recipients using 16S rRNA gene sequencing of the V4-V5 hypervariable region.9 The median number of bacterial sequences was 15,841 with an interquartile range of 11,262 to 22,510.
Because we wanted to identify the most common bacterial taxa whose relative abundances were significantly associated with Enterobacteriaceae bacteriuria, we compared the relative abundances of the top 10 most common genera in the cohort between the 153 fecal specimens from the 51 patients in the Enterobacteriaceae Bacteriuria Group and the 357 fecal specimens from the 117 patients in the No Enterobacteriaceae Bacteriuria Group (Figure 1a). The relative abundances of Faecalibacterium and Romboutsia were significantly higher in the No Enterobacteriaceae Bacteriuria Group than in the Enterobacteriaceae Bacteriuria Group and the relative abundance of Lactobacillus was significantly lower in the No Enterobacteriaceae Bacteriuria Group than in the Enterobacteriaceae Bacteriuria Group (Adjusted P < .01, Wilcoxon rank sum test, Bonferroni adjustment for multiple hypothesis testing) (Table 2). The relative abundance of Faecalibacterium, the relative abundance of Romboutsia, the relative abundance of Lactobacillus, and the combined relative abundance of Faecalibacterium and Romboutsia are shown over time (Figure 1b-1e). The combined relative abundance of Faecalibacterium and Romboutsia was significantly higher in the No Enterobacteriaceae Bacteriuria Group than in the Enterobacteriaceae Bacteriuria Group (median 4.0% vs. 0.7%, respectively, P < .001, Wilcoxon rank sum test). We also evaluated the subset of 33 kidney transplant recipients with Enterobacteriaceae UTI among the Enterobacteriaceae Bacteriuria Group. The combined relative abundance of Faecalibacterium and Romboutsia was significantly higher in the No Enterobacteriaceae Bacteriuria Group than in the Enterobacteriaceae UTI Group (median 4.0% vs. 1.1%, respectively, P value < .001, Wilcoxon rank sum test).
Figure 1.
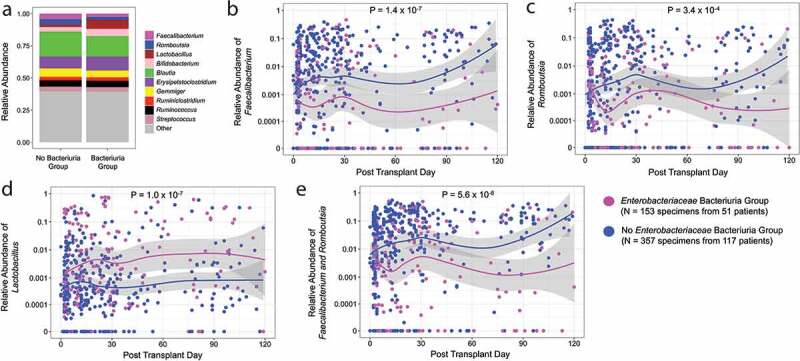
Relative abundances of the most common Genera by Enterobacteriaceae Bacteriuria Group Status. Panel a. The mean relative abundances of the 10 most common genera are represented on the y axis by color and the Enterobacteriaceae Bacteriuria Group status is on the x axis. The Enterobacteriaceae Bacteriuria Group consisted of 153 fecal specimens from 51 patients and the No Enterobacteriaceae Bacteriuria Group consisted of 357 fecal specimens from 117 patients. Panels b – e. The 510 fecal specimens are shown in each graph and each point represents a fecal specimen and the point’s color represents Enterobacteriaceae Bacteriuria Group status. The relative abundance of genera is on the y axis (log10 scale) and the post-transplant day is on the x axis. The line represents a locally estimated scatterplot smoothing curve with 95% confidence intervals in the shaded area. P values were calculated using the Wilcoxon rank sum test. Panel b. Relative abundance of Faecalibacterium is shown with 131 fecal specimens having a zero relative abundance. Panel c. Relative abundance of Romboutsia is shown with 129 specimens having a zero relative abundance. Panel d. Relative abundance of Lactobacillus is shown with 61 specimens having a zero relative abundance. Panel e. Combined relative abundance of Faecalibacterium and Romboutsia with 60 specimens having a zero relative abundance.
Table 2.
Comparison of the Most Abundant Genera between the Enterobacteriaceae Bacteriuria Group and the No Enterobactericaeae Bacteriuria Group. For each of the top 10 genera, the relative abundance of 153 fecal specimens from 51 patients in the Enterobacteriaceae Bacteriuria Group was compared to the relative abundance of 357 fecal specimens from the 117 patients in the No Enterobacteriaceae Bacteriuria Group using a Wilcoxon rank sum test. The adjusted P values were calculated using a Bonferonni correction. In bold are the genera that were significantly different between the groups (Adjusted P < .01).
| No Enterobacteriaceae | Enterobacteriaceae | |||
|---|---|---|---|---|
| Bacteriuria Group | Bacteriuria Group | |||
| (357 specimens, | (153 specimens, | |||
| 117 subjects) | 51 subjects) | |||
| Median Gut Relative | Median Gut Relative | |||
| Genus | Abundance (%) | Abundance (%) | P value | Adjusted P value |
| Lactobacillus | 0.04% | 0.21% | 1.0 x 10–7 | 1.0 x 10−6 |
| Faecalibacterium | 0.80% | 0.03% | 1.4 x 10–7 | 1.4 x 10−6 |
| Romboutsia | 0.39% | 0.06% | 3.4 x 10–4 | 0.003 |
| Blautia | 17.07% | 13.25% | 0.001 | 0.013 |
| Ruminococcus | 2.45% | 1.59% | 0.011 | 0.108 |
| Streptococcus | 0.64% | 1.28% | 0.016 | 0.162 |
| Ruminiclostridium | 1.45% | 0.97% | 0.038 | 0.376 |
| Gemmiger | 1.84% | 0.33% | 0.062 | 0.621 |
| Bifidobacterium | 0.27% | 0.27% | 0.538 | 0.999 |
| Erysipelatoclostridium | 5.45% | 4.68% | 0.699 | 0.999 |
Because female gender was strongly associated with Enterobacteriaceae bacteriuria, we evaluated whether there were differences in the relative abundances of Faecalibacterium, Romboutsia, Lactobacillus, and the combined relative abundance of Faecalibacterium and Romboutsia between female patients and male patients. The relative abundance of Faecalibacterium, Romboutsia, and the combined relative abundance of Faecalibacterium and Romboutsia were significantly lower in the fecal specimens from female patients than in those from male patients (P = .003, P = .03, P = .004, Wilcoxon rank sum test, respectively) and the relative abundance of Lactobacillus was significantly higher in the fecal specimens from female patients in those from male patients (P < .001) (Supplementary Figure 1). We also evaluated whether the relative abundances of these taxa changed after diagnosis of first Enterobacteriaceae bacteriuria or Enterobacteriaceae UTI. The relative abundance of Faecalibacterium, Romboutsia, Lactobacillus, and the combined relative abundance of Faecalibacterium and Romboutsia did not significantly change from the closest specimen prior to diagnosis of first Enterobacteriaceae bacteriuria to the first specimen after Enterobacteriaceae bacteriuria diagnosis (N = 24 patients, P > .05, Wilcoxon signed-rank test) (Supplementary Figure 2) or from the closest specimen prior to diagnosis of first Enterobacteriaceae UTI to the first specimen after Enterobacteriaceae UTI diagnosis (N = 15 patients, P > .05) (Supplementary Figure 3).
The relative abundances of Faecalibacterium, Romboutsia, and Lactobacillus are correlated with relative abundance of Enterobacteriaceae and gut microbial diversity
We next evaluated the relationship between the relative abundance of Faecalibacterium and Romboutsia and the relative abundance of Enterobacteriaceae. The relative abundance of Faecalibacterium (r = −0.09, P = .04, Pearson’s correlation), the relative abundance of Romboutsia (r = −0.10, P = .02), and the combined relative abundance of Faecalibacterium and Romboutsia (r = −0.13, P = .003) were inversely correlated to the relative abundance of Enterobacteriaceae. The relative abundance of Lactobacillus was not correlated with the relative abundance of Enterobacteriaceae (r = 0.05, P = .29). The relative abundance of Lactobacillus was inversely correlated with the combined relative abundance of Faecalibacterium and Romboutsia (r = −0.15, P = 4.5 x 10−4). Taken together with our findings that the gut abundance of uropathogens (e.g. E. coli) is associated with future development of bacteriuria,9 our data suggest that the inverse relationship between the relative abundances of Faecalibacterium and Romboutsia and relative abundance of Enterobacteriaceae could be a factor associated with bacteriuria and UTI development.
We further evaluated whether the relative abundances of Faecalibacterium and Romboutsia were associated with microbial diversity as measured by Shannon diversity index. The relative abundance of Faecalibacterium (r = 0.16, P = 4.0 x 10−4), the relative abundance of Romboutsia (r = 0.08, P = .07), and the combined relative abundance of Faecalibacterium and Romboutsia (r = 0.16, P = 3.0 x 10−4) were positively correlated with the Shannon diversity index and the relative abundance of Lactobacillus was negatively correlated with the Shannon diversity index (r = −0.24, P = 2.8 x 10−8). The positive correlation between the relative abundances of the two genera and the relative abundance of Enterobacteriaceae supports Faecalibacterium and Romboutsia as markers of gut microbial diversity.
Antibiotic administration is associated with relative abundances of Faecalibacterium, Romboutsia, and Lactobacillus
Given that antibiotics alter the gut microbiota,15,16 we evaluated the relationship between antibiotic administration and the relative abundances of Faecalibacterium, Romboutsia, and Lactobacillus. In the cohort, 89 kidney transplant recipients (53%) received additional antibiotics aside from surgical antibiotic prophylaxis and Pneumocystis jiroveci prophylaxis within the first 3 months after transplantation (Abx Group) and 79 did not (No Abx Group). These additional antibiotics were used for: presumed or documented infection in 50 patients (56%) with the most common documented infection being bacteriuria or UTI; antibiotic prophylaxis in 38 patients (43%) with the most common indication being ureteral stent removal; and gastric motility in 1 patient (1%). Among the 89 patients who had antibiotic exposure, 69 (78%) had a stool specimen before first antibiotic exposure. The relative abundance of Faecalibacterium was significantly higher in the 235 fecal specimens from the No Abx Group than in the 275 fecal specimens from the Abx Group (median 1.5% vs. 0.1%, P < .001, Wilcoxon rank sum test); the relative abundance of Romboutsia was significantly higher in the 235 fecal specimens from the No Abx Group than in the 275 fecal specimens from the Abx Group (median 1.3% vs. 0.1%, P < .001, Wilcoxon rank sum test); the relative abundance of Lactobacillus was significantly lower in the 235 fecal specimens from the No Abx Group than in the 275 specimens from the Abx Group (median 0.04% vs. 0.1%, P < .001, Wilcoxon rank sum test); the combined relative abundance of Faecalibacterium and Romboutsia was significantly higher in the 235 fecal specimens from the No Abx Group than in the 275 fecal specimens from the Abx Group (median 6.9% vs. 0.8%, P < .001, Wilcoxon rank sum test). The relative abundance of Faecalibacterium, the relative abundance of Romboutsia, the relative abundance of Lactobacillus, and the combined relative abundance of Faecalibacterium and Romboutsia are shown over time (Supplemental Figure 4). Our data reveals that antibiotic administration is associated with lower relative abundance of Faecalibacterium and Romboutsia and higher relative abundance of Lactobacillus.
We further analyzed the relationship between antibiotic administration and the relative abundances of Faecalibacterium and Romboutsia by antibiotic subgroup. The most common antibiotics used in the cohort were: beta-lactams (Beta-lactam Group, N = 56 patients) and fluoroquinolones (Fluoroquinolone Group, N = 51 patients). The relative abundance of Faecalibacterium, the relative abundance of Romboutsia, and the combined relative abundance of Faecalibacterium and Romboutsia were all significantly higher in the 235 fecal specimens from the No Abx Group than in the 177 fecal specimens from the Beta-lactam Group and the relative abundance of Lactobacillus was significantly lower in the 235 fecal specimens from the No Abx Group than in the 177 fecal specimens from the Beta-lactam Group (P < .001, respectively, Wilcoxon rank sum test). The relative abundance of Faecalibacterium, the relative abundance of Romboutsia, and the combined relative abundance of Faecalibacterium and Romboutsia were all significantly higher in the 235 fecal specimens from the No Abx Group than in the 154 fecal specimens from the Fluoroquinolone Group (P < .001, respectively, Wilcoxon rank sum test) and the relative abundance of Lactobacillus was significantly lower in the 235 fecal specimens from the No Abx Group than in the 154 fecal specimens from the Flouroquinolone Group (P = .005, Wilcoxon rank sum test).
The combined relative abundance of Faecalibacterium and Romboutsia is associated with a decreased risk for Enterobacteriaceae bacteriuria and UTI
We next evaluated whether the relative abundances of the identified genera are associated with a decreased risk for future development of Enterobacteriaceae bacteriuria and UTI. We utilized a time-dependent Cox regression to evaluate for the future risk of both Enterobacteriaceae bacteriuria and UTI. As a cutoff for relative abundance, we utilized the top tercile cutoff where at least a third of the kidney transplant recipients reached the threshold during the first 3 months after transplantation.
With a top tercile cutoff of 7% abundance, a high relative abundance of Faecalibacterium was significantly associated with a lower risk of development of Enterobacteriaceae bacteriuria (Hazard ratio [HR] 0.3, 95% confidence interval [CI] 0.1–0.7, P = .01). With a top tercile cutoff of 7% abundance, a high relative abundance of Romboutsia was not significantly associated with a lower risk of development of Enterobacteriacaeae bacteriuria (HR 0.8, 95% CI 0.4–1.5, P = .43). With a top tercile cutoff of 1.6%, a high relative abundance of Lactobacillus was significantly associated with an increased risk of development of Enterobacteriaceae bacteriuria (HR 3.0, 95% CI 1.7–5.4, P < .001). With a top tercile cutoff of 13.7%, a high combined relative abundance of Faecalibacterium and Romboutsia was significantly associated with a lower risk of future development of Enterobacteriaceae bacteriuria (HR 0.2, 95% CI 0.1–0.7, P = .008).
With a top tercile cutoff of 7% abundance, a high relative abundance of Faecalibacterium was not significantly associated with a lower risk of future development of Enterobacteriaceae UTI (HR 0.6, 95% CI 0.2–1.5, P = .23). With a top tercile cutoff of 7% abundance, a high relative abundance of Romboutsia was not significantly associated with a lower risk of development of Enterobacteriacaeae UTI (HR 0.8, 95% CI 0.3–1.8, P = .53). With a top tercile cutoff of 1.6%, a high relative abundance of Lactobacillus was significantly associated with an increased risk of development of Enterobacteriaceae UTI (HR 3.0, 95% CI 1.5–6.0, P = .002). However, with a top tercile cutoff of 13.7%, a high combined relative abundance of Faecalibacterium and Romboutsia was significantly associated with a lower risk of future development of Enterobacteriaceae UTI (HR 0.3, 95% CI 0.1–0.9, P = .04).
In order to account for significant risk factors such as gender and cefazolin antibiotic prophylaxis, we performed a multivariable cox regression analysis for the development of Enterobacteriaceae bacteriuria and UTI with the combined relative abundance of Faecalibacterium and Romboutsia as a time-dependent covariate and first antibiotic administration as a time dependent covariate (Table 3). In multivariate analysis, the combined relative abundance of Faecalibacterium and Romboutsia of greater than 13.7% was significantly associated with future development of Enterobacteriaceae bacteriuria (HR 0.3, 95% CI 0.1–0.8, P = .02) (Table 3A) and future development of Enterobacteriaceae UTI (HR 0.4, 95% CI 0.1–1.2, P = .09) (Table 3B). We also evaluated the relative abundance of Lactobacillus as a time-dependent covariate and first antibiotic administration as a time dependent covariate. In multivariate analysis, the relative abundance of Lactobacillus of greater than 1.6% was significantly associated with future development of Enterobacteriaceae bacteriuria (HR 2.4, 95% CI 1.3–4.2, P = .003) and future development of Enterobacteriaceae UTI (HR 2.3, 95% CI 1.1–4.7, P = .02) (Supplemental Table 1).
Table 3.
Multivariable Cox Regression for Enterobacteriaceae Bacteriuria and Enterobacteriaceae UTI. Univariate Cox regression analysis was performed for each of the characteristics and the development of Enterobacteriaceae bacteriuria or Enterobacteriaceae UTI. The combined relative abundance of Faecalibacterium and Romboutsia (cutoff of 13.8%) was analyzed as a time-dependent covariate and first antibiotic administration was analyzed as a time-dependent covariate. For characteristics that were significantly associated with either Enterobacteriaceae bacteriuria or Enterobacteriaceae UTI (P < .10), a multivariable Cox Regression was performed with the significantly associated characteristics. Table 3A. Multivariable Cox Regression for Enterobacteriaceae bacteriuria. Table 3B. Multivariable Cox Regression for Enterobacteriaceae UTI. ESRD, end stage renal disease; DM, diabetes mellitus; HTN, hypertension; PRA panel reactive antibody; PCP, Pneumocystis jiroveci.
| Table 3A | ||||||||
| Risk Factors for Enterobacteriaceae UTI |
Univariate Analysis |
|
Multivariate Analysis |
|
||||
| Characteristic | HR (95% CI) | P value | HR (95% CI) | P value | ||||
| Age, Years | 1.0 (1.0–1.0) | 0.54 | ||||||
| Female Gender | 4.1 (2.2–7.6) | 6.8 x 10−6 | 4.4 (2.3–8.2) | 3.9 x 10−6 | ||||
| African American Race | 0.9 (0.4–1.6) | 0.65 | ||||||
| History of Diabetes Mellitus | 1.4 (0.8–2.4) | 0.29 | ||||||
| Cause of ESRD – DM | 1.4 (0.8–2.5) | 0.29 | ||||||
| Cause of ESRD – HTN | 0.9 (0.4–2.1) | 0.82 | ||||||
| PRA ≥ 80% | 1.1 (0.4–2.8) | 0.81 | ||||||
| Decreased Donor Transplantation | 1.2 (0.7–2.2) | 0.49 | ||||||
| Delayed Graft Function | 1.2 (0.6–2.5) | 0.53 | ||||||
| Cefazolin Preoperative Abx | 0.5 (0.2–0.9) | 0.01 | 0.5 (0.2–0.9) | 0.02 | ||||
| Trimethoprim/Sulfamethoxazole PCP Prophylaxis | 3.2 (0.4–23.2) | 0.25 | ||||||
| Anti-thymocyte Globulin Induction | 1.1 (0.6–2.1) | 0.82 | ||||||
| Prednisone Maintenance | 1.3 (0.7–2.4) | 0.34 | ||||||
| First Antibiotic Administration | 2.1 (1.1–3.8) | 0.02 | 1.4 (0.7–2.6) | 0.33 | ||||
| Relative Abundance of Faecalibacterium & Romboutsia |
0.2 (0.1–0.7) |
0.008 |
0.3 (0.1–0.8) |
0.02 |
||||
| Table 3B | ||||||||
| Risk Factors for Enterobacteriaceae UTI |
Univariate Analysis |
|
Multivariate Analysis |
|
||||
| Characteristic | HR (95% CI) | P value | HR (95% CI) | P value | ||||
| Age, Years | 1.0 (1.0–1.0) | 0.31 | ||||||
| Female Gender | 4.4 (2.0–9.7) | 2.8 x 10–4 | 4.4 (2.0–9.9) | 3.2 x 10–4 | ||||
| African American Race | 0.8 (0.3–1.8) | 0.55 | ||||||
| History of Diabetes Mellitus | 1.3 (0.7–2.7) | 0.44 | ||||||
| Cause of ESRD – DM | 1.2 (0.6–2.5) | 0.60 | ||||||
| Cause of ESRD – HTN | 0.5 (0.2–1.9) | 0.33 | ||||||
| PRA ≥ 80% | 0.7 (0.2–3.0) | 0.63 | ||||||
| Decreased Donor Transplantation | 1.1 (0.5–2.2) | 0.88 | ||||||
| Delayed Graft Function | 0.9 (0.3–2.3) | 0.80 | ||||||
| Cefazolin Preoperative Abx | 0.5 (0.2–1.1) | 0.07 | 0.5 (0.2–1.2) | 0.12 | ||||
| Trimethoprim/Sulfamethoxazole PCP Prophylaxis | – – | – – | ||||||
| Anti-thymocyte Globulin Induction | 1.0 (0.5–2.3) | 0.95 | ||||||
| Prednisone Maintenance | 1.0 (0.5–2.1) | 0.99 | ||||||
| First Antibiotic Administration | 2.2 (1.1–4.7) | 0.03 | 1.5 (0.7–3.3) | 0.29 | ||||
| Relative Abundance of Faecalibacterium & Romboutsia | 0.3 (0.1–0.9) | 0.04 | 0.4 (0.1–1.2) | 0.09 | ||||
Discussion
In this study, we identified that high relative abundances of two taxa, Faecalibacterium and Romboutsia, are associated with a decreased risk for Enterobacteriaceae bacteriuria and UTI in kidney transplant recipients. We further report an inverse relationship of the relative abundances of these two taxa with the relative abundance of Enterobacteriaceae. These data provide further support for a growing notion that gut commensal organisms are associated with lower risk of infectious complications, which is well-established for Clostridioides difficile disease.
Recent studies have shown that the relative abundance of pathogenic bacteria is associated with UTI development. Thanert and colleagues evaluated 14 non-transplant patients with recurrent UTIs or non-recurrent UTIs and found that the gut was one of three different sources for recurrence of UTIs.11 In a study that we performed at our center, we observed that the gut abundance of Escherichia was associated with future development of Escherichia bacteriuria and UTI.9 These studies investigated the relationship between the relative abundance of pathogenic bacteria and UTI development. The current study is the first, to the best of our knowledge, to investigate whether high relative abundance of commensal organisms is associated with lower rates of UTI development.
Rising rates of multidrug resistant uropathogens and stagnating development of novel antimicrobials17 have led to an increased need for novel therapies to treat UTIs. Modulation of the gut microbiota via FMT constitute one major new line of research. In a study of 8 non-transplant patients with recurrent C. difficile infections, FMT was associated with a significant decrease in UTI recurrence.14 In a case of a heart and kidney transplant recipient who had recurrent vancomycin-resistant Enterococcus (VRE) bacteremia and UTIs, FMT was performed because of recurrent C. difficile infection and was associated with resolution of the VRE infections.18 In another case, FMT was performed in a kidney transplant recipient who had recurrent multidrug resistant K. pneumoniae UTIs, which was associated with eradication of the multidrug resistant K. pneumoniae and resolution of K. pneumoniae UTIs.13 However, FMT is largely an untargeted approach and most recently has been associated with lethal complications in immunocompromised patients.19 Thus there is a need to identify commensal bacterial taxa that could confer protection against pathogenic bacteria. Our study’s identification of higher relative abundance of Faecalibacterium and Romboutsia as being associated with decreased risk for Enterobacteriaceae UTI may thus help to better personalize the use of FMT for patients with recurrent Enterobacteriaceae UTIs. It could also lead to development of personalized consortia of probiotics for the prevention of UTIs.
Importantly, we also found an inverse correlation between the relative abundances of Faecalibacterium and Romboutsia and the relative abundance of Enterobacteriaceae. In this study, we did not test this relationship in vitro so we do not know the mechanism for this inverse relationship in our cohort. We speculate that one of the mechanisms by which low relative abundances of Faecalibacterium or Romboutsia could inhibit growth of Enterobacteriaceae is through short-chain fatty acid (SCFA) production. An elegant study by the Pamer group found that a mixture of short-chain fatty acids (SCFAs) and acidic pH inhibited the growth of E. coli and K. pneumoniae in vivo and that their growth is inhibited because of intracellular acidification from SCFAs.20 Faecalibacterium has been described as one of the most abundant and important producers of the SCFA butyrate in the intestine.21 Species in the Romboutsia taxa are also predicted to be associated with production of the SCFA acetate.22 It is plausible that even though the relative abundances of Faecalibacterium or Romboutsia are low, these bacteria produce sufficient SCFAs to have protective effects on the growth and relative abundances of Enterobacteriaceae.
In this study, we also found that antibiotic administration was associated with decreased relative abundance of Faecalibacterium and decreased relative abundance of Romboutsia. While antibiotics have been described to reduce various commensal bacterial taxa,15,16 our study highlights the effects of antibiotics on these two taxa that we have identified as associated with future development of Enterobacteriaceae bacteriuria and UTI. Our data suggests that antibiotic administration could lead to the development of infections beyond C. difficile disease. Interestingly, female kidney transplant recipients had lower relative abundance of Faecalibacterium and Romboutsia and higher relative abundance of Lactobacillus than male transplant recipients. Importantly, we controlled for gender in the Cox Regression analysis for development of Enterobacteriaceae bacteriuria and UTI as female gender is independently associated with development of UTI in kidney transplant recipients.4 While female gender was a significant risk factor for Enterobacteriaceae bacteriuria and UTI, the relative abundance of the identified taxa were also independently associated with development of the outcomes.
Interestingly, we also report that the relative abundance of Lactobacillus is associated with an increased hazard ratio for the development of Enterobacteriaceae bacteriuria and UTI. With respect to microbial interactions with Lactobacillus, we have found that the relative abundance of Lactobacillus is correlated with decreased microbial diversity, is negatively correlated with the combined relative abundance of Faecalibacterium and Romboutsia, but not correlated with the relative abundance of Enterobacteriaceae. Vaginal suppositories of Lactobacillus crispatus has been associated with decreased development of UTI 23 and mixed data exists on the use of oral Lactobacillus species for the prevention of UTI.24 It is possible that the gut abundance of Lactobacillus does not directly correlate with vaginal or urinary abundances of Lactobacillus. Furthermore, from our correlational data, Lactobacillus could be a biomarker for decreased microbial diversity manifested by decreased relative abundance of commensal bacterial taxa such as Faecalibacterium and Romboutsia.
There are several limitations to our study. While we had comprehensive evaluation of bacteriuria development given that kidney transplant recipients provided urine specimens at every clinical outpatient visit, we retrospectively reviewed the medical charts to identify UTI symptoms. It is possible that some patients did not have UTI symptoms recorded in the medical record and thus some UTIs could be misclassified as bacteriuria, which could explain discrepancies between the Cox Regression analyses for bacteriuria and UTI. However, given the critical clinical importance of documenting and treating UTIs in kidney transplant recipients, we believe that most symptomatic UTIs were captured. Diet has also been shown to change the gut microbiota25 and we did not evaluate the effect of diet on the gut microbiota in the transplant recipients in this cohort. The use of probiotics such as Lactobacillus in the cohort was not obtained in the transplant population and reflects another limitation of the study.
In conclusion, we identified that high relative abundances of Faecalibacterium and Romboutsia are associated with decreased risk for Enterobacteriaceae bacteriuria and UTI development in kidney transplant recipients. Our data support future studies evaluating the use of gut microbial based therapies for the prevention of recurrent UTIs in the kidney transplant population, which may be further generalizable to non-transplant patients with recurrent UTIs.
Patients and methods
Kidney transplant cohort
One hundred sixty-eight kidney transplant recipients who had kidney transplantations from August 2015 to November 2016 at NewYork-Presbyterian Hospital – Weill Cornell Medical Center were recruited to provide serial fecal specimens in the first 3 months after transplantation. The Weill Cornell Medicine Institutional Review Board approved this study (IRB # 1207012730) and each of the patients provided written informed consent. Kidney transplant recipients at our center undergo routine microscopic urinalysis and conventional urine culture at every clinical visit (approximately twice weekly in the first month, weekly in the second month, every 2 weeks in the third month, and monthly in the fourth, fifth, and sixth month), providing a comprehensive evaluation of bacteriuria. Kidney transplant recipients were considered to have Enterobacteriaceae bacteriuria if they had a positive urine culture (≥10,000 colony-forming units of a species from the Enterobacteriaceae family per mL of urine) during the first 6 months after transplantation and to have Enterobacteriaceae UTI if they had Enterobacteriaceae bacteriuria and symptoms of dysuria, frequency, urgency, or fever26 during the first 6 months after transplantation. Urine specimens were analyzed according to standard of care procedures at NewYork Presbyterian Hospital – Weill Cornell Medical Center clinical microbiology laboratory. Kidney transplant recipients receive surgical pre-operative antibiotic prophylaxis (most commonly cefazolin) and Pneumocystis jiroveci prophylaxis (most commonly trimethoprim-sulfamethoxazole).
Fecal specimen collections
Patients were instructed to collect fecal specimens using a Fisherbrand toilet specimen collection kit (Fisher Scientific, New Hampton, NH, USA). The fecal specimen was aliquoted and stored at −80°C. Patients were asked to collect fecal specimens at post-transplant week 1, week 2, week 4, week 12, and during episodes of UTIs and diarrhea.
DNA extraction and 16S rRNA gene sequencing
DNA extraction, 16S rRNA gene amplification, and deep sequencing of the 16S rRNA amplicon were previously performed and complete details of the protocol can be found in Magruder et al.9 In brief, DNA was isolated using a bead-beater phenol-chloroform extraction method. The V4-V5 region of the 16S rRNA gene (563 F and 926 R) was amplified with barcodes for multiplexing. The PCR amplicons were quantified and pooled. The Illumina TruSeq Sample Preparation protocol was used to add Illumina barcodes and adaptors (Illumina Inc., San Diego, CA, USA). The products were sequenced on an Illumina MiSeq Instrument (250 base pair by 250 base pair).
Bioinformatics and statistical analyses
Details of the bioinformatics to assign taxonomy are found in Magruder et al. In brief, the 16S rRNA gene paired-end reads were merged and demultiplexed. The UPARSE pipeline27 was used to group sequences into operational taxonomic units by 97% distance-based similarity and remove potential chimeric sequences. Taxonomic assignment was done using nucleotide BLAST28 with NCBI RefSeq29 as the reference training set with a minimum E-value threshold of 10−10. The distribution of categorical values was analyzed using two-tailed Fisher’s exact test and the distribution of continuous variables was compared using the two-tailed Wilcoxon rank sum test. Correlation between two continuous variables was analyzed using a Pearson’s correlation. A Cox Regression Hazard Model was utilized to assess the relationship between the relative abundance of specific taxa and the development of bacteriuria or UTI. In this model, the relative abundance of the specific taxa was a time-ever dependent covariate where it was assumed that the relative abundance was not crossed until the first time the relative abundance value crossed the threshold. A multivariable Cox Regression was performing including significant clinical variables. All analyses were performed in R 3.3.3.
Supplementary Material
Acknowledgments
We thank Dr. Manikkam Suthanthiran for his assistance with study design and data analysis for the manuscript and Dr. Eric Pamer for his assistance with study design, microbiome sequencing, and data analyses for the manuscript.
Funding Statement
This research work was supported, in part, by National Institute of Allergy and Infectious Diseases [K23 AI 124464].
Disclosure statement
L.F.W. receives research support from Accelerate Diagnostics, Inc., Affinity Biosensors, LLC, BioFire Diagnostics, LLC, and Hardy Diagnostics and consulting fees from Roche Molecular System, Inc., and Shionogi; M.J.S. receives consulting fees from Achaogen and Shionogi and has received research support from Accelerate Diagnostics, LLC, BioFire Diagnostics, LLC, Merck, Allergan, and Affinity Biosensors. D.M.D. receives consulting fees from Veloxis Pharmaceutical, Inc. and Allovir, Inc. J.R.L. receives research support from BioFire Diagnostics, LLC. D.M.D. and J.R.L. are inventors on patent US-2020-0048713-A1, entitled “METHODS OF DETECTING CELL-FREE DNA IN BIOLOGICAL SAMPLES.”
Data availability
All sequencing data and the deidentified clinical data will be made available upon publication at accession number phs001879 in the database of Genotypes and Phenotypes (dbGaP). Local institutional review board approval will be necessary to obtain the data.
Supplementary material
Supplemental data for this article can be accessed here
References
- 1.Wolfe RA, Ashby VB, Milford EL, Ojo AO, Ettenger RE, Agodoa LY, Held PJ, Port FK.. Comparison of mortality in all patients on dialysis, patients on dialysis awaiting transplantation, and recipients of a first cadaveric transplant. N Engl J Med. 1999;341(23):1725–12. doi: 10.1056/NEJM199912023412303. [DOI] [PubMed] [Google Scholar]
- 2.Goldman JD, Julian K. Urinary tract infections in solid organ transplant recipients: guidelines from the American society of transplantation infectious diseases community of practice. Clin Transplant. 2019;33(9):e13507. doi: 10.1111/ctr.13507. [DOI] [PubMed] [Google Scholar]
- 3.Alangaden GJ, Thyagarajan R, Gruber SA, Morawski K, Garnick J, El-Amm JM, West MS, Sillix DH, Chandrasekar PH, Haririan A. Infectious complications after kidney transplantation: current epidemiology and associated risk factors. Clin Transplant. 2006;20(4):401–409. doi: 10.1111/j.1399-0012.2006.00519.x. [DOI] [PubMed] [Google Scholar]
- 4.Lee JR, Bang H, Dadhania D, Hartono C, Aull MJ, Satlin M. Independent risk factors for urinary tract infection and for subsequent bacteremia or acute cellular rejection: a single-center report of 1166 kidney allograft recipients. Transplantation 2013; 96:732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abbott KC, Swanson SJ, Richter ER, Bohen EM, Agodoa LY, Peters TG, Barbour G, Lipnick R, Cruess DF. Late urinary tract infection after renal transplantation in the United States. Am J Kidney Dis. 2004;44(2):353–362. doi: 10.1053/j.ajkd.2004.04.040. [DOI] [PubMed] [Google Scholar]
- 6.Ariza-Heredia EJ, Beam EN, Lesnick TG, Cosio FG, Kremers WK, Razonable RR. Impact of urinary tract infection on allograft function after kidney transplantation. Clin Transplant. 2014;28(6):683–690. doi: 10.1111/ctr.12366. [DOI] [PubMed] [Google Scholar]
- 7.Pelle G, Vimont S, Levy PP, Hertig A, Ouali N, Chassin C, Arlet G, Rondeau E, Vandewalle A. Acute pyelonephritis represents a risk factor impairing long-term kidney graft function. Am J Transplant. 2007;7(4):899–907. doi: 10.1111/j.1600-6143.2006.01700.x. [DOI] [PubMed] [Google Scholar]
- 8.Britt NS, Hagopian JC, Brennan DC, Pottebaum AA, Santos CAQ, Gharabagi A, Horwedel TA. Effects of recurrent urinary tract infections on graft and patient outcomes after kidney transplantation. Nephrol Dial Transplant. 2017;32(10):1758–1766. doi: 10.1093/ndt/gfx237. [DOI] [PubMed] [Google Scholar]
- 9.Magruder M, Sholi AN, Gong C, Zhang L, Edusei E, Huang J, Albakry S, Satlin MJ, Westblade LF, Crawford C. Gut uropathogen abundance is a risk factor for development of bacteriuria and urinary tract infection. Nat Commun. 2019;10(1):5521. doi: 10.1038/s41467-019-13467-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paalanne N, Husso A, Salo J, Pievilainen O, Tejesvi MV, Koivusaari P, Pirttilä AM, Pokka T, Mattila S, Jyrkäs J, et al. Intestinal microbiome as a risk factor for urinary tract infections in children. Eur J Clin Microbiol Infect Dis. 2018;37(10):1881–1891. doi: 10.1007/s10096-018-3322-7. [DOI] [PubMed] [Google Scholar]
- 11.Thanert R, Reske KA, Hink T, Wallace MA, Wang B, Schwartz DJ, et al. Comparative genomics of antibiotic-resistant uropathogens implicates three routes for recurrence of urinary tract infections. mBio. 2019;10(4):e01977-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13(5):269–284. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hocquart M, Pham T, Kuete E, Tomei E, Lagier JC, Raoult D. Successful fecal microbiota transplantation in a patient suffering from irritable bowel syndrome and recurrent urinary tract infections. Open Forum Infect Dis. 2019;6(10):ofz398. doi: 10.1093/ofid/ofz398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tariq R, Pardi DS, Tosh PK, Walker RC, Razonable RR, Khanna S. Fecal microbiota transplantation for recurrent clostridium difficile infection reduces recurrent urinary tract infection frequency. Clin Infect Dis. 2017;65(10):1745–1747. doi: 10.1093/cid/cix618. [DOI] [PubMed] [Google Scholar]
- 15.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6(11):e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sader HS, Castanheira M, Duncan LR, Flamm RK. Antimicrobial susceptibility of enterobacteriaceae and pseudomonas aeruginosa isolates from United States medical centers stratified by infection type: results from the International Network for Optimal Resistance Monitoring (INFORM) surveillance program, 2015-2016. Diagn Microbiol Infect Dis. 2018;92(1):69–74. doi: 10.1016/j.diagmicrobio.2018.04.012. [DOI] [PubMed] [Google Scholar]
- 18.Stripling J, Kumar R, Baddley JW, Nellore A, Dixon P, Howard D, Ptacek T, Lefkowitz EJ, Tallaj JA, Benjamin WH, et al. Loss of vancomycin-resistant enterococcus fecal dominance in an organ transplant patient with clostridium difficile colitis after fecal microbiota transplant. Open Forum Infect Dis. 2015;2(2):ofv078. doi: 10.1093/ofid/ofv078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeFilipp Z, Bloom PP, Torres Soto M, Mansour MK, Sater MRA, Huntley MH, Turbett S, Chung RT, Chen Y-B, Hohmann EL, et al. Drug-resistant E. coli bacteremia transmitted by fecal microbiota transplant. N Engl J Med. 2019;381(21):2043–2050. doi: 10.1056/NEJMoa1910437. [DOI] [PubMed] [Google Scholar]
- 20.Sorbara MT, Dubin K, Littmann ER, Moody TU, Fontana E, Seok R, Leiner IM, Taur Y, Peled JU, van den Brink MRM. Inhibiting antibiotic-resistant Enterobacteriaceae by microbiota-mediated intracellular acidification. J Exp Med. 2019;216(1):84–98. doi: 10.1084/jem.20181639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miquel S, Martin R, Rossi O, Bermudez-Humaran LG, Chatel JM, Sokol H, Thomas M, Wells JM, Langella P. Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol. 2013;16(3):255–261. doi: 10.1016/j.mib.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Gerritsen J, Hornung B, Renckens B, van Hijum S, Martins Dos Santos VAP, Rijkers GT, Schaap PJ, de Vos WM, Smidt H. Genomic and functional analysis of Romboutsia ilealis CRIB T reveals adaptation to the small intestine. PeerJ. 2017;5:e3698. doi: 10.7717/peerj.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stapleton AE, Au-Yeung M, Hooton TM, Fredricks DN, Roberts PL, Czaja CA, Yarova-Yarovaya Y, Fiedler T, Cox M, Stamm WE. Randomized, placebo-controlled phase 2 trial of a Lactobacillus crispatus probiotic given intravaginally for prevention of recurrent urinary tract infection. Clin Infect Dis. 2011;52(10):1212–1217. doi: 10.1093/cid/cir183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beerepoot MA, Ter Riet G, Nys S, van der Wal WM, de Borgie CA, de Reijke TM. Lactobacilli vs antibiotics to prevent urinary tract infections: a randomized, double-blind, noninferiority trial in postmenopausal women. Arch Intern Med. 2012;172(9):704–712. doi: 10.1001/archinternmed.2012.777. [DOI] [PubMed] [Google Scholar]
- 25.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollyer I, Ison MG. The challenge of urinary tract infections in renal transplant recipients. Transpl Infect Dis. 2018;20(2):e12828. doi: 10.1111/tid.12828. [DOI] [PubMed] [Google Scholar]
- 27.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 28.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 29.Tatusova T, Ciufo S, Fedorov B, O’Neill K, Tolstoy I. RefSeq microbial genomes database: new representation and annotation strategy. Nucleic Acids Res. 2014;42(D1):D553–9. doi: 10.1093/nar/gkt1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data and the deidentified clinical data will be made available upon publication at accession number phs001879 in the database of Genotypes and Phenotypes (dbGaP). Local institutional review board approval will be necessary to obtain the data.


