ABSTRACT
The intestinal mucosal barrier, which is composed of epithelial cells and mucus layers secreted by goblet cells and contains commensal bacteria, constitutes the first line of defense against pathogenic gut microbiota. However, homeostasis between the microbiota and mucus layer is easily disrupted by certain factors, resulting in alteration of the gut microbiota and entry of pathogens to the intestinal mucosal barrier. In this review, we describe the structures and functions of the mucus layer, expound several crucial influencing factors, including diet styles, medications and host genetics, and discuss how pathogenic microorganisms interact with the mucus layer and commensal microbiota, with the understanding that unraveling their complex interactions under homeostatic and dysbiosis conditions in the colon would help reveal some underlying pathogenic mechanisms and thus develop new strategies to prevent pathogenic microbiological colonization.
KEYWORDS: Mucus layer, mucin, commensalism, pathogen, colonization resistance
Introduction
The human colon harbors a diverse community of commensal microorganisms to promote the degradation of some indigestible residues and to resist colonization by pathogenic species under healthy conditions.1 Over the past decade, several studies have focused on the composition of the gut microbiota under healthy and diseased conditions of the host.2 Today, more studies have been carried out to explore the interactions between the microbiota and the host.
The first line of defense against pathogenic microorganisms is the mucosal barrier, which is made up of the epithelium and a protecting overlying host-secreted mucus layer and contains commensal microorganisms.3 Colonic epithelial cells are composed of five differentiated cell types, including enterocytes, enteroendocrine cells, tuft cells, goblet cells, and microfold (M) cells, which are responsible for absorption, hormone secretion, the taste-chemosensory response, mucus production and antigen sampling, respectively.4,5 The mucus secreted by goblet cells continuously replenishes the mucus layer to lubricate and protect the epithelial cells. In recent years, increasing attention has been paid to the role of the mucus barrier owing to changes in dietary habits and lifestyles, such as the reduced consumption of fiber polysaccharides and overuse or abuse of antibiotics.6–8 Such changes in the composition of the colon microbiota may eventually cause damage to the mucus barrier, compromising the resistance to colonization by incoming pathogenic bacteria.9,10
In this review, we describe the structures of the colon mucus layer to gain a better understanding about the mechanisms of the first-line defense against pathogenic gut microbiota. In addition to the interactions between the gut microbiota and the host under homeostatic conditions, we also explore the interactions between the gut microbiota and the host under dysbiosis conditions such as drug usage, diet styles and host genetics. Once the gut environment is disrupted, pathogens have opportunities to invade the host, which will be the focal point of the present review article.
The structures of the colon mucus layer
The mucus layer of the colon contains two layers. The thickness of the outer layer is 100 μm as measured in mice and is approximately twice that of the inner layer.11 The inner layer is stratified into several thin sheets which are densely and firmly attached to the epithelial cells and impervious to bacteria.12 The outer layer, which is the habitat of the commensal bacteria, is less dense and unattached. The protective effect of the colon mucus layer becomes weaker as a result of the increasing permeability of the inner layer, increasing bacterial colonization and encroachment on the epithelial cells once the luminal microbiota migrate from the disrupted outer layer to an interior position. (Figure 1)
Figure 1.
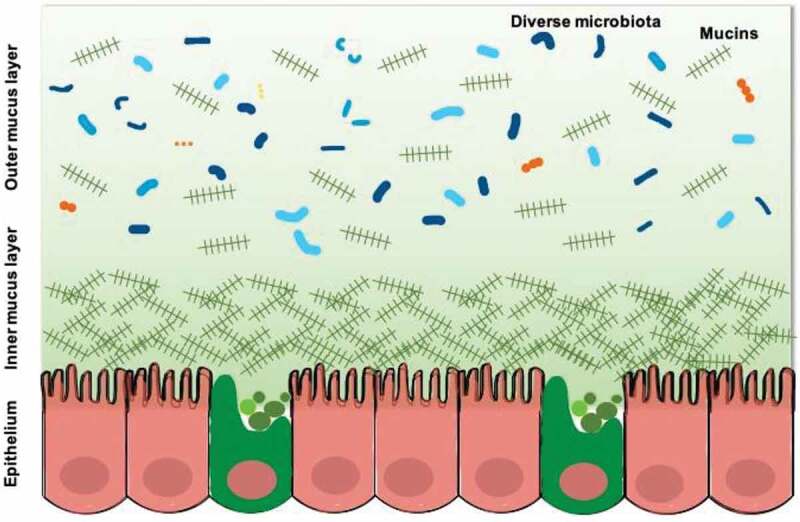
The structures of the mucus layer in the colon.
The mucus layer is mainly composed of mucins that are complex agglomerates of heavily O-glycosylated proteins. Mucins are named in order of their discoveries rather their functions and structures. However, mucins can be functionally classified in two major categories: transmembrane mucins and gel-forming mucins. Transmembrane mucins are usually produced by enterocytes to cover their surface and regulate the local milieu. Gel-forming mucins are secreted by goblet cells and constitute the mucus layer. (Table 1) The MUC2 mucin is the most abundant secretory mucin synthesized and secreted by goblet cells in the colon. The MUC2 monomer forms dimeric COOH-termini and trimeric NH2-termini, which together establish large polymeric netlike structures that make up the stratified inner mucus layer.11,13
Table 1.
Expression of mucins in the colon.
| Mucin | Produced by cell type | Function | Reference |
|---|---|---|---|
| Transmembrane mucins | |||
| MUC1 | enterocyte | intracellular signaling; local milieu sensing and regulation at the surface of the enterocytes | 13 |
| MUC3 | enterocyte | 14, 15 | |
| MUC4 | goblet cell; enterocyte | 16 | |
| MUC12 | enterocyte | 17 | |
| MUC13 | enterocyte | 18 | |
| MUC15 | enterocyte | 19 | |
| MUC17 | enterocyte | 20 | |
| MUC20 | enterocyte | 21 | |
| Secreted mucins | |||
| MUC2 | goblet cell; Paneth cell | the skeleton of the mucus layer | 22 |
| MUC5B | goblet cell | 22 | |
Glycoproteins contain different glycans, which provide nutrients and attachment sites for microorganisms and secrete antibodies targeting specific microbial antigens to prevent pathogens from persistently remaining on the intestinal epithelial surfaces.24,25 In addition, O-glycosylated proteins built in the protein core are rich in the amino acids serine, proline and threonine; these structures are called PTS domains or variable number tandem repeats (VNTRs).26 These PTS domains provide sites for the attachment of polysaccharides and bond with them through a series of glycosyltransferase enzymes. Four types of polysaccharide core structures, which are attached by various glycans, are composed of three polysaccharides (galactose, N-acetyl-galactosamine and N-acetyl-glucosamine).27,28 Different glycan chains will be attached to the polysaccharide core structure, and the terminal monosaccharide is usually fucose or sialic acid. Eighty percent of the mucin biomass is from the high polysaccharide content, which simultaneously provides an important nutritional resource for the mucus layer and an attachment site for the gut microbiota.29
Commensal microorganisms and mucus layers
Homeostasis between commensal microorganisms and the mucus layers
The properties of the mucus layer play a crucial role in protecting intestinal homeostasis. In addition to mucins, the mucus layers also contain other substances such as TFF3, RELMβ, and Fcgbp secreted by goblet cells; antimicrobial peptides such as β defensin and lysozymes secreted by Paneth cells; and secretory IgA secreted by enterocytes, all of which contribute to the physical, biochemical and immunological protection of the mucus layer. The question arises of how the complex protective system defends against pathogenic microbiota without attacking the commensal microorganisms. This defense is mainly attributed to the immune system in that the epithelial cells have the ability to distinguish commensal microbiota from pathogenic microbiota by pattern recognition receptors (PRRs), such as cell surface Toll-like receptors (TLRs) and cytoplasmic nucleotide-binding oligomerization domain (NOD) proteins.30–32
Four major phyla account for most of the intestinal microbiota, among which Firmicutes and Bacteroidetes make up more than 90% of the total intestinal microbes, while Proteobacteria and Actinobacteria constitute the rest. However, it is difficult to study the mucus bacteria by simply interpreting a fecal taxonomic analysis. Fecal samples, or even luminal samples, cannot fully represent the mucus populations.33 Several studies also found significant heterogeneity between intestinal luminal, mucosal and fecal microbiota.34–36 Hence, it is necessary to establish innovative methods such as laser capture microdissection and in vitro continuous culture systems to further identify the intestinal bacterial populations growing on mucin surfaces.33,37
Commensal microorganisms rely on absorbing undigested food polysaccharides and endogenic glycans of the host as energy sources to feed themselves. They also rely on binding sites to create niches for themselves. Simple dietary sugars are absorbed in the small intestine, while undigested plant polysaccharides and host glycans are utilized in the colon. To depolymerize these polysaccharides and glycans, the host-associated bacteria produce thousands of carbohydrate-active enzymes (CAZymes), which encompass four different groups of enzymes: glycoside hydrolases, polysaccharide lyases, carbohydrate esterases and glycosyltransferases.4 Hence, the gut microbiome, especially anaerobic bacteria such as Bacteroides, mobilizes its machinery to metabolize complex carbohydrates, leading to the production of diverse metabolites such as short-chain fatty acids (SCFAs), bile acids and other organic acids.
Because of its high polysaccharide content, the mucus layer also serves as a nutrient source for a distinct subset of gut microbiota species. In addition to these direct metabolic impacts, some nutritional generalists could in turn regulate the synthesis and secretion of mucins. Laura et al. found that Bacteroides thetaiotaomicron could exert a positive effect on the thickness and composition of the mucus layer by increasing the differentiation of goblet cells and modulating the expression of mucin-related genes.38 Furthermore, microbiota, especially those colonized in the mucus layer, have proved to possess powerful immunomodulatory properties. Akkermansia muciniphila, a representative mucus-degrading bacterium, exhibits potential anti–inflammatory responses, maintains intestinal integrity and regulates the resident gut microbiota under healthy conditions.39,40 Bacteroides fragilis stimulates interleukin 10 (IL-10) secretion and differentiation of Treg cells to protect the host against colitis by expressing polysaccharide A (PSA).41,42 Faecalibacterium prausnitzii could secrete microbial anti–inflammatory molecules (MAM) to inhibit NF-κB signaling and gut inflammation.43
Dysbiosis between commensal microorganisms and the mucus layers
The dynamic intestinal balance is easily disrupted under conditions such as unhealthy life styles, consumption of drugs, or variants in the host genetics, which would likely result in the loss of beneficial and protective bacterial species, reduced microbial diversity, or even the invasion of pathogens. Here, we discuss some factors affecting the mucus layer. (Figure 2)
Figure 2.
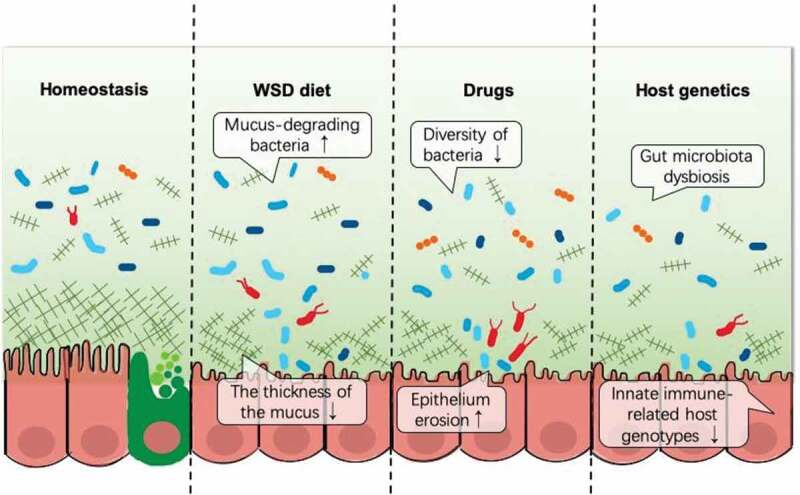
Dysbiosis between commensal microorganisms and the mucus layers.
Diet
Diet has a very significant influence on the gut microbiota and may change the mucus layer directly or indirectly. Different food components could shape the composition, diversity, and richness of the intestinal microbiota. Known to be high in fat and sugar and low in dietary fiber, a western-style diet (WSD) may reduce the diversity of the gut microbiota, cause the disappearance of beneficial species, and impair the mucus layer, especially in the colon. Schroeder et al. found that a reduction in gut microbiota diversity paralleled or even preceded the increase in mucus penetrability under a WSD, and the microbiota transferred from chow-fed mice could reverse the WSD-induced impairment of the mucus physiology.9 Another study reported a similar finding that a WSD and a fructose diet induced a sharp loss in the mucus thickness and defensin expression in the colon.44 Desai et al. further discovered that dietary fiber deprivation promoted the erosion of the mucus by commensals and allowed deeper access to the epithelial cells by some pathogens.10 All of these findings proved that dysbiosis in the gut microbiota may be responsible for mucus defects.
The mechanisms of how diets modulate gut microbial ecology need further consideration. Long-term dietary patterns can shape the gut microbiota, particularly protein and animal fat for Bacteroides versus carbohydrates for Prevotella.45 The ratio of Prevotella/Bacteroides has been used as an indicator of differences in industrialized and nonindustrialized human populations, due to their different diets, e.g., the amount of red meat and dietary fiber.46 For people who have a tendency to eat a more non-Westernized diet, the bacterial taxa in their gut would be enriched in Prevotella to adapt for fiber and carbohydrate fermentation. The main exogenous nutrient source for colonic microorganisms is fiber. A long-term reduction in fiber intake may lead to the permanent extinction of some important microbial species.6 Some other commensal microbial taxa such as Akkermansia muciniphila would instead switch to binding glycans of the mucus layer with the increased expression of the mucus-degrading CAZymes to absorb mucin as a nutrient,47 Bacteroides thetaiotaomicron would shift from fermentable polysaccharides binding mucus glycans as a food source.48 However, some probiotics such as Bifidobacterium strains can repair the mucus defects by strengthening epithelial function and promoting the mucus growth.9 The food source shapes the composition and function of gut microbiota, thereby affecting the interactions between diet and the mucus layer.
Drugs
Conventional antibiotics indiscriminately kill or suppress the growth of both pathogenic and commensal microbiota because of their general bacteriostatic and bactericidal effects. The use of these antibiotics decreases the diversity of the microbiota and compromises resistance to colonization by potential pathogens. These pathogens that colonize in the gut would utilize the mucus as their nutrient source and establish themselves by suppressing commensal microbiota. Carlstedt-Duke et al. found that intestinal mucins broke down in healthy volunteers who were administered with different oral antibiotics, especially bacitracin, clindamycin and vancomycin.49 In addition, several studies demonstrated that a metronidazole treatment reduced the intestinal expression of MUC2 and diminished the thickness of the mucus layer, thereby enhancing its susceptibility to gut pathogens.50,51
Different kinds of antibiotics have different impacts on the gut microbiota. One study chose several different classes of antibiotics to observe the alterations of the microbiota and assess their resistance to C. difficile colonization on a murine model. The results suggested that the diverse antibiotic perturbations varied in their susceptibility to C. difficile colonization due to the diverse microbiome alterations. Individuals who were treated with ampicillin showed the highest amount of C. difficile colonization.52 Antibiotics can directly perturb the taxonomic, genomic and functional features of the microbiota and the species-species interactions rapidly or persistently. A recent work in a mouse model demonstrated that antibiotics could also alter the availability of mucosal carbohydrates by inducing a spike in the abundance of host-derived free sialic acid in the gut, which can be utilized by opportunistic pathogens to enhance their growth and resistance.53 It is worth mentioning that clones carrying resistant genes could be detected even several years after treatment.54 These evidence warrant remarkable prudence in the administration of antibiotics during medical treatments.
In addition to antibiotics, some commonly used nonantibiotics such as antidiabetics, proton pump inhibitors (PPIs), nonsteroidal anti–inflammatory drugs (NSAID) and atypical antipsychotics (AAPs), have been found to be associated with alterations in the composition of the gut microbiome.55–58 Maier et al. reported that 24% of more than 1000 market drugs inhibited the growth of at least one strain when cocultured with 40 representative gut bacterial strains, resembling antibiotic-like side effects.59 Although there is little evidence on the direct effects of nonantibiotics on the colon mucus layer and mucins, several studies reported that naproxen could reduce the expression of gastric mucins and that metformin could increase the abundance of the mucus-degrading Akkermansia muciniphila and SCFA-producing microbiota in the gut.60,61These findings hint at a close relationship between some nonantibiotics and the gut mucus layer, which may open a new path for studying the impact of medications on the health of the host.
Host genetics
Some recent studies have demonstrated that host genotypes can affect the composition of the gut microbiome and the abundance of some of the gut microbial taxa in both human and mouse models.62,63 The innate host immune system has been demonstrated to be a major determinant in shaping the microbiome by genome-wide mbQTL studies, especially via the innate molecules of pattern recognition receptors (PRRs) including TLRs, C-type lectins, NOD genes and RIG-I-like receptors.64 Knocking out several of these molecules in animal models resulted in gut dysbiosis.65–67 Several studies in human populations also highlighted the importance of these host genotypes, especially in patients with inflammatory bowel disease (IBD). NOD2 is known to be the strongest immunity-related IBD risk gene which could change the gut microbiome composition.68 The interaction of C-type lectin Dectin-1 with commensal fungi was reported to be related to refractory ulcerative colitis.69 Chen et al. compared monozygotic twins and found that the expression of NLRP12 was downregulated in patients with ulcerative colitis.70 NLRP12 could attenuate colitis by maintaining colonic microbial diversity and promoting protective commensal bacterial growth.70 In addition to immune-related genes, genes related to the mucus barrier and sugar digestion also affect the gut microbiome, and variants in these host genetics could lead to gut dysbiosis and even to the occurrence of the disease.64 However, some major challenges hinder the development of using the host genetics to regulate the gut microbiome. It is difficult to eliminate the environmental factors that mask the effect of genetic variants. The complexity and heterogeneity of the data require extensive histological examination and molecular characterization in future cohort studies.
Pathogenic microorganisms and the mucus layers
The mucus layer serves as an important firewall during the invasion of enteric pathogens such as Clostridium difficile, S. Typhimurium and enterotoxigenic E. coli (ETEC). Here, we focus on how pathogenic microorganisms interact with the mucus layer and commensal microbiota and expound the underlying mechanisms applied by pathogens to promote colonization.
Pathogenic adhesion and damage to mucins
As mentioned above, mucins are highly glycosylated proteins which provide attachment sites for the gut microbiota. Bacteria can express specific proteins to bind to mucins directly, such as mucus-binding proteins (MUBs). Cell-surface appendages, such as pili, fimbriae and flagella, also play a key role in the attachment of bacteria to the mucins. In addition, virulent strains bind more tightly and efficiently to the mucus than avirulent strains.71 Numerous studies have demonstrated that specific pathogens colonize the gut by adhering to the mucins; these include Clostridium difficile, Escherichia coli, Listeria monocytogenes, Salmonellae enterica serotype Typhimurium and Vibrio cholerae.72 In addition, some microbiota could be embedded in self-produced polymeric matrices, known as biofilms, whereby they adhere to other microbial populations and mucosal surfaces for the enhancement of antimicrobial resistance, virulence and other functions.73
Adherence to the mucus layer is a requisite step before pathogenic colonization. The pathogens subsequently disrupt the colon mucus and epithelial tight junctions. Some pathogens have the ability to break colonic mucin structures such as peptide bonds, glycosidic linkages and/or epithelial tight junctions.4 StcE and SslE, both belonging to a metalloprotease of pathogenic E. coli, cleave the mucins to make the epithelial cells accessible.74,75 EHEC also generates mucinases to degrade the mucus and create a nutrient-poor environment near the epithelium where the pathogens most likely colonize.76 Additionally, Shigella utilizes fimbria to attach to the mucins to deliver a subset of effectors via a type III secretion system (T3SS) into the colonic epithelium to alter cellular and immune functions and promote infection.77
Mucin and commensal microbiota in pathogenicity
To colonize the mucus layer, pathogenic bacteria have to compete with the commensal microbiota to harvest nutrients for their expansion. One method is to exploit the oligosaccharides released from the mucins by saccharolytic members of the microbiota. For instance, Bacteroides thetaiotaomicron, widely used as a model of Bacteroides to investigate syntrophic links, has sialidase activity to harvest sialic acid but lacks the catabolic pathway for its utilization. Therefore, the sialic acid released by B. thetaiotaomicron can be catabolized by C. difficile and S. typhimurium to promote pathogen growth and expansion.53 B. thetaiotaomicron can also cleave fucose from host glycans via multiple enzymes, and free fucose can be used as another carbon source for S. typhimurium.53 Fucose is also a signaling molecule to regulate the expression of EHEC’s virulence repertoire.78 EHEC aims to achieve a particular niche by closely adhering to the intestinal enterocytes and competes with the commensal E. coli for nutrients. EHEC can also use specific sugars that commensal E. coli cannot utilize, such as galactose, mannose, hexuranates and ribose.79
There is no doubt that the metabolites of commensal microbiota, such as SCFAs and organic acids, are ubiquitous and abundant in the gut. These metabolites also have diverse impacts on pathogenic colonization. SCFAs such as butyrate protect mice against some pathogens by reinforcing the defense of epithelial cells and suppressing pathogenic virulence gene expression.79,80 Jacobson et al. reported that propionate could directly limit the growth of the pathogen S. typhimurium by disrupting intracellular pH homeostasis to restrain pathogenic colonization.81 The acidification of the SCFAs further promotes mineral solubility and absorption by the colon.82 Ions such as zinc possess anti-microbial ability to avoid gut infections.83,84
However, microbial metabolites also have been demonstrated to enhance pathogen growth under some specific conditions. Zumbrun et al. found that the elevation of the butyrate level in the gut paradoxically enhanced the cell-killing capacity of the Shiga toxin by feeding mice a high fiber diet (HFD).85 Ferreyra et al. illustrated that Clostridium difficile obtained a notable advantage through metabolizing the organic acid succinate, especially after a selective killing of succinate consumers with an antibiotic treatment.86 All of these phenomena seem to illustrate the complexity and variety of the gut microbiota functions under different circumstances, though more in-depth studies are required to explore the underlying mechanisms.
Conclusion
The intestinal mucosal barrier physically protects the gut epithelial cells against pathogenic microbiota. Mucins are the main structural components of the mucus layer, emphasizing their essential biological status. The integrity of the mucus layers depends on a number of genetic and environmental factors. These factors shape the gut microbiota and change the mucus layer directly or indirectly. Once the homeostasis between the mucus layer and commensal microbiota is disrupted, pathogenic microbiota would have opportunities to attach to and encroach upon the mucus layer, leading to enteric diseases.
A deep understanding of the mechanisms underlying the associations between the mucus layer, commensal microbiota and pathogenic microbiota can help develop new preventive strategies and therapies. The prudent use of certain drugs, especially antibiotics, is advised due to their side effects on the microbiome. Moreover, some healthy diet styles should be advocated to promote a stronger intestinal ecosystem. Dietary fiber, which is considered a key ancestral prebiotic, can help preserve gut ecology and regulate macronutrients and host physiology.46 Currently, polymicrobial biofilms, mostly consisting of pathogenic species, have been observed in oral and intestinal infections, IBD and colorectal cancer.87,88 These biofilms appear to be an early warning signal pronouncing degenerative changes in the gut mucosa. However, there are technological challenges inherent in in situ microbiome research. Clinically reflective models of intestinal biofilms, such as organoids, are urgently needed to grasp the functional dynamics of this complex community.37,89 Although more recent research has started to illustrate specific interactions of gut microbiota with mucins, epithelial cells or immune cells, there is still a long way to go before we can determine their relationships.
Funding Statement
This research project was supported by the National Natural Science Foundation of China [81700463 and 81871734].
References
- 1.Sassone-Corsi M, Raffatellu M.. No vacancy: how beneficial microbes cooperate with immunity to provide colonization resistance to pathogens. J Immunol. 2015;194:4081–4087. doi: 10.4049/jimmunol.1403169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin JJ, Li RQ, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–U70. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 4.Martens EC, Neumann M, Desai MS. Interactions of commensal and pathogenic microorganisms with the intestinal mucosal barrier. Nat Rev Microbiol. 2018;16:457–470. doi: 10.1038/s41579-018-0036-x. [DOI] [PubMed] [Google Scholar]
- 5.Gerbe F, Legraverend C, Jay P. The intestinal epithelium tuft cells: specification and function. Cell Mol Life Sci. 2012;69:2907–2917. doi: 10.1007/s00018-012-0984-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529:212–U08. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonnenburg ED, Sonnenburg JL. Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab. 2014;20:779–786. doi: 10.1016/j.cmet.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 9.Schroeder BO, Birchenough GMH, Stahlman M, Arike L, Johansson MEV, Hansson GC, Bäckhed F. Bifidobacteria or fiber protects against diet-induced microbiota-mediated colonic mucus deterioration. Cell Host Microbe. 2018;23:27–40 e7. doi: 10.1016/j.chom.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. 2016;167:1339–1353. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johansson MEV, Larsson JMH, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A. 2011;108:4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansson MEV, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brayman M, Thathiah A, Carson DD. MUC1: a multifunctional cell surface component of reproductive tissue epithelia. Reprod Biol Endocrinol. 2004;2:4. doi: 10.1186/1477-7827-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gum JR, Hicks JW, Swallow DM, Lagace RL, Byrd JC, Lamport DT, Siddiki B, Kim YS. Molecular cloning of cDNAs derived from a novel human intestinal mucin gene. Biochem Biophys Res Commun. 1990;171:407–415. doi: 10.1016/0006-291X(90)91408-K. [DOI] [PubMed] [Google Scholar]
- 15.Pratt WS, Crawley S, Hicks J, Ho J, Nash M, Kim YS, Gum JR, Swallow DM. Multiple transcripts of MUC3: evidence for two genes, MUC3A and MUC3B. Biochem Biophys Res Commun. 2000;275:916–923. doi: 10.1006/bbrc.2000.3406. [DOI] [PubMed] [Google Scholar]
- 16.Audie JP, Janin A, Porchet N, Copin MC, Gosselin B, Aubert JP. Expression of human mucin genes in respiratory, digestive, and reproductive tracts ascertained by in situ hybridization. J Histochem Cytochem. 1993;41:1479–1485. doi: 10.1177/41.10.8245407. [DOI] [PubMed] [Google Scholar]
- 17.Moehle C, Ackermann N, Langmann T, Aslanidis C, Kel A, Kel-Margoulis O, Schmitz-Madry A, Zahn A, Stremmel W, Schmitz G, et al. Aberrant intestinal expression and allelic variants of mucin genes associated with inflammatory bowel disease. J Mol Med (Berl). 2006;84:1055–1066. doi: 10.1007/s00109-006-0100-2. [DOI] [PubMed] [Google Scholar]
- 18.Williams SJ, Wreschner DH, Tran M, Eyre HJ, Sutherland GR, McGuckin MA. MUC13, a novel human cell surface mucin expressed by epithelial and hemopoietic cells. J Biol Chem. 2001;276:18327–18336. doi: 10.1074/jbc.M008850200. [DOI] [PubMed] [Google Scholar]
- 19.Pallesen LT, Berglund L, Rasmussen LK, Petersen TE, Rasmussen JT. Isolation and characterization of MUC15, a novel cell membrane-associated mucin. Eur J Biochem. 2002;269:2755–2763. doi: 10.1046/j.1432-1033.2002.02949.x. [DOI] [PubMed] [Google Scholar]
- 20.Gum JR Jr., Crawley SC, Hicks JW, Szymkowski DE, Kim YS. MUC17, a novel membrane-tethered mucin. Biochem Biophys Res Commun. 2002;291:466–475. doi: 10.1006/bbrc.2002.6475. [DOI] [PubMed] [Google Scholar]
- 21.Dhanisha SS, Guruvayoorappan C, Drishya S, Abeesh P. Mucins:Structural diversity, biosynthesis, its role in pathogenesis and as possible therapeutic targets. Crit Rev Oncol Hematol. 2018;122:98–122. doi: 10.1016/j.critrevonc.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 22.Johansson MEV, Hansson GC. Immunological aspects of intestinal mucus and mucins. Nat Rev Immunol. 2016;16:639–649. doi: 10.1038/nri.2016.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ambort D, Johansson MEV, Gustafsson JK, Nilsson HE, Ermund A, Johansson BR, Koeck PJB, Hebert H, Hansson GC. Calcium and pH-dependent packing and release of the gel-forming MUC2 mucin. Proc Natl Acad Sci U S A. 2012;109:5645–5650. doi: 10.1073/pnas.1120269109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antoni L, Nuding S, Wehkamp J, Stange EF. Intestinal barrier in inflammatory bowel disease. World J Gastroenterol. 2014;20:1165–1179. doi: 10.3748/wjg.v20.i5.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGuckin MA, Linden SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nat Rev Microbiol. 2011;9:265–278. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- 26.Johansson ME, Sjovall H, Hansson GC. The gastrointestinal mucus system in health and disease. Nat Rev Gastroenterol Hepatol. 2013;10:352–361. doi: 10.1038/nrgastro.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larsson JM, Karlsson H, Sjovall H, Hansson GC. A complex, but uniform O-glycosylation of the human MUC2 mucin from colonic biopsies analyzed by nanoLC/MSn. Glycobiology. 2009;19::756–766. doi: 10.1093/glycob/cwp048. [DOI] [PubMed] [Google Scholar]
- 28.Juge N. Microbial adhesins to gastrointestinal mucus. Trends Microbiol. 2012;20:30–39. doi: 10.1016/j.tim.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Kim YS, Ho SB. Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr Gastroenterol Rep. 2010;12:319–330. doi: 10.1007/s11894-010-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukata M, Abreu MT. Pathogen recognition receptors, cancer and inflammation in the gut. Curr Opin Pharmacol. 2009;9:680–687. doi: 10.1016/j.coph.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim YS, Ho SB. Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr Gastroenterol Rep. 2010;12:319–330. doi: 10.1007/s11894-010-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moncada DM, Kammanadiminti SJ, Chadee K. Mucin and Toll-like receptors in host defense against intestinal parasites. Trends Parasitol. 2003;19:305–311. doi: 10.1016/S1471-4922(03)00122-3. [DOI] [PubMed] [Google Scholar]
- 33.Lavelle A, Lennon G, O’Sullivan O, Docherty N, Balfe A, Maguire A, Mulcahy HE, Doherty G, O’Donoghue D, Hyland J, et al. Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut. 2015;64:1553–1561. doi: 10.1136/gutjnl-2014-307873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zoetendal EG, von Wright A, Vilpponen-Salmela T, Ben-Amor K, Akkermans ADL, de Vos WM. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol. 2002;68:3401–3407. doi: 10.1128/aem.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmed N, Hong P-Y, Croix JA, Greenberg E, Gaskins HR, Mackie RI. Pyrosequencing-based analysis of the mucosal microbiota in healthy individuals reveals ubiquitous bacterial groups and micro-heterogeneity. PLoS One. 2011:6. doi: 10.1371/journal.pone.0025042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z, Geng J, Tang X, Fan H, Xu J, Wen X, Ma Z, Shi P. Spatial heterogeneity and co-occurrence patterns of human mucosal-associated intestinal microbiota. Isme J. 2014;8:881–893. doi: 10.1038/ismej.2013.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macfarlane S, Woodmansey EJ, Macfarlane GT. Colonization of mucin by human intestinal bacteria and establishment of biofilm communities in a two-stage continuous culture system. Appl Environ Microbiol. 2005;71:7483–7492. doi: 10.1128/aem.71.11.7483-7492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wrzosek L, Miquel S, Noordine ML, Bouet S, Chevalier-Curt MJ, Robert V, Philippe C, Bridonneau C, Cherbuy C, Robbe-Masselot C, et al. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. 2013;11. doi: 10.1186/1741-7007-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Derrien M, Belzer C, de Vos WM. Akkermansia muciniphila and its role in regulating host functions. Microb Pathog. 2017;106:171–181. doi: 10.1016/j.micpath.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 40.Geerlings S, Kostopoulos I, de Vos W, Belzer C. Akkermansia muciniphila in the human gastrointestinal tract: when, where, and how? Microorganisms. 2018;6:75. doi: 10.3390/microorganisms6030075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 42.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hornef MW, Pabst O. Real friends: faecalibacterium prausnitziisupports mucosal immune homeostasis. Gut. 2016;65:365–367. doi: 10.1136/gutjnl-2015-310027. [DOI] [PubMed] [Google Scholar]
- 44.Volynets V, Louis S, Pretz D, Lang L, Ostaff MJ, Wehkamp J, Bischoff SC. Intestinal barrier function and the gut microbiome are differentially affected in mice fed a western-style diet or drinking water supplemented with fructose. J Nutr. 2017;147:770–780. doi: 10.3945/jn.116.242859. [DOI] [PubMed] [Google Scholar]
- 45.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Makki K, Deehan EC, Walter J, Backhed F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe. 2018;23:705–715. doi: 10.1016/j.chom.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 47.Deehan EC, Duar RM, Armet AM, Perez-Munoz ME, Jin ML, Walter J. Modulation of the gastrointestinal microbiome with nondigestible fermentable carbohydrates to improve human health. Microbiol Spectr. 2017:5. doi: 10.1128/microbiolspec.BAD-0019-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gorden JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 49.Carlstedt-Duke B, Høverstad T, Lingaas E, Norin KE, Saxerholt H, Steinbakk M, Midtvedt T. Influence of antibiotics on intestinal mucin in healthy subjects. Eur J Clin Microbiol. 1986;5:634–638. doi: 10.1007/bf02013287. [DOI] [PubMed] [Google Scholar]
- 50.Willing BP, Russell SL, Finlay BB. Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat Rev Microbiol. 2011;9:233–243. doi: 10.1038/nrmicro2536. [DOI] [PubMed] [Google Scholar]
- 51.Wlodarska M, Willing B, Keeney KM, Menendez A, Bergstrom KS, Gill N, Russell SL, Vallance BA, Finlay BB. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbatedcitrobacter rodentium-induced colitis. Infect Immun. 2011;79:1536–1545. doi: 10.1128/iai.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schubert AM, Sinani H, Schloss PD. Antibiotic-induced alterations of the murine gut microbiota and subsequent effects on colonization resistance against clostridium difficile. MBio. 2015;6:e00974. doi: 10.1128/mBio.00974-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology. 2010;156:3216–3223. doi: 10.1099/mic.0.040618-0. [DOI] [PubMed] [Google Scholar]
- 55.Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, Prifti E, Vieira-Silva S, Gudmundsdottir V, Krogh Pedersen H, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Imhann F, Bonder MJ, Vich Vila A, Fu J, Mujagic Z, Vork L, Tigchelaar EF, Jankipersadsing SA, Cenit MC, Harmsen HJM, et al. Proton pump inhibitors affect the gut microbiome. Gut. 2016;65:740–748. doi: 10.1136/gutjnl-2015-310376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rogers MAM, Aronoff DM. The influence of non-steroidal anti-inflammatory drugs on the gut microbiome. Clin Microbiol Infect. 2016;22:178 e1- e9. doi: 10.1016/j.cmi.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Flowers SA, Evans SJ, Ward KM, McInnis MG, Ellingrod VL. Interaction between atypical antipsychotics and the gut microbiome in a bipolar disease cohort. Pharmacotherapy. 2017;37:261–267. doi: 10.1002/phar.1890. [DOI] [PubMed] [Google Scholar]
- 59.Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, Brochado AR, Fernandez KC, Dose H, Mori H, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018;555:623–628. doi: 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jaworski T, Sarosiek I, Sostarich S, Roeser K, Connor M, Brotze S, Wallner G, Sarosiek J. Restorative impact of rabeprazole on gastric mucus and mucin production impairment during naproxen administration: its potential clinical significance. Dig Dis Sci. 2005;50:357–365. doi: 10.1007/s10620-005-1611-3. [DOI] [PubMed] [Google Scholar]
- 61.de la Cuesta-zuluaga J, Mueller NT, Corrales-Agudelo V, Velásquez-Mejía EP, Carmona JA, Abad JM, Escobar JS. Metformin is associated with higher relative abundance of mucin-degradingakkermansia muciniphilaand several short-chain fatty acid–producing microbiota in the gut. Diabetes Care. 2017;40:54–62. doi: 10.2337/dc16-1324. [DOI] [PubMed] [Google Scholar]
- 62.Suzuki TA, Phifer-Rixey M, Mack KL, Sheehan MJ, Lin D, Bi K, Nachman MW. Host genetic determinants of the gut microbiota of wild mice. Mol Ecol. 2019;28:3197–3207. doi: 10.1111/mec.15139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goodrich JK, Davenport ER, Waters JL, Clark AG, Ley RE. Cross-species comparisons of host genetic associations with the microbiome. Science. 2016;352:532–535. doi: 10.1126/science.aad9379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kurilshikov A, Wijmenga C, Fu J, Zhernakova A. Host genetics and gut microbiome: challenges and perspectives. Trends Immunol. 2017;38:633–647. doi: 10.1016/j.it.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 65.Pang X, Xiao X, Liu Y, Zhang R, Liu J, Liu Q, Wang P, Cheng G. Mosquito C-type lectins maintain gut microbiome homeostasis. Nat Microbiol. 2016:1. doi: 10.1038/nmicrobiol.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rehman A, Sina C, Gavrilova O, Hasler R, Ott S, Baines JF, Schreiber S, Rosenstiel P. Nod2 is essential for temporal development of intestinal microbial communities. Gut. 2011;60:1354–1362. doi: 10.1136/gut.2010.216259. [DOI] [PubMed] [Google Scholar]
- 67.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT, et al. Metabolic syndrome and altered gut microbiota in mice lacking toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Philpott DJ, Sorbara MT, Robertson SJ, Croitoru K, Girardin SE. NOD proteins: regulators of inflammation in health and disease. Nat Rev Immunol. 2014;14:9–23. doi: 10.1038/nri3565. [DOI] [PubMed] [Google Scholar]
- 69.Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen L, Wilson JE, Koenigsknecht MJ, Chou WC, Montgomery SA, Truax AD, Brickey WJ, Packey CD, Maharshak N, Matsushima GK, et al. NLRP12 attenuates colon inflammation by maintaining colonic microbial diversity and promoting protective commensal bacterial growth. Nat Immunol. 2017;18:541–551. doi: 10.1038/ni.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vimal D, Khullar M, Gupta S, Ganguly NK. Intestinal mucins: the binding sites for Salmonella typhimurium. Mol Cell Biochem. 2000;204:107–117. doi: 10.1023/A:1007015312036. [DOI] [PubMed] [Google Scholar]
- 72.Sicard J-F, Le Bihan G, Vogeleer P, Jacques M, Harel J. Interactions of intestinal bacteria with components of the intestinal mucus. Front Cell Infect Microbiol. 2017:7. doi: 10.3389/fcimb.2017.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Vos WM. Microbial biofilms and the human intestinal microbiome. NPJ Biofilms Microbiomes. 2015:1. doi: 10.1038/npjbiofilms.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Valeri M, Paccani SR, Kasendra M, Nesta B, Serino L, Pizza M, Soriani M. Pathogenic E-coli exploits ssle mucinase activity to translocate through the mucosal barrier and get access to host cells. PLoS One. 2015;10:e0117486. doi: 10.1371/journal.pone.0117486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grys TE, Siegel MB, Lathem WW, Welch RA. The StcE protease contributes to intimate adherence of enterohemorrhagic Escherichia coli O157: H7to host cells. Infect Immun. 2005;73:1295–1303. doi: 10.1128/IAI.73.3.1295-1303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Curtis MM, Hu ZP, Klimko C, Narayanan S, Deberardinis R, Sperandio V. The gut commensal bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe. 2014;16:759–769. doi: 10.1016/j.chom.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ashida H, Ogawa M, Mimuro H, Kobayashi T, Sanada T, Sasakawa C. Shigella are versatile mucosal pathogens that circumvent the host innate immune system. Curr Opin Immunol. 2011;23:448–455. doi: 10.1016/j.coi.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 78.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Baumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535:85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–U791. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 81.Jacobson A, Lam L, Rajendram M, Tamburini F, Honeycutt J, Pham T, Van Treuren W, Pruss K, Stabler SR, Lugo K, et al. A gut commensal-produced metabolite mediates colonization resistance to salmonella infection. Cell Host Microbe. 2018;24:296–307. doi: 10.1016/j.chom.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baye K, Guyot JP, Mouquet-Rivier C. The unresolved role of dietary fibers on mineral absorption. Crit Rev Food Sci Nutr. 2017;57:949–957. doi: 10.1080/10408398.2014.953030. [DOI] [PubMed] [Google Scholar]
- 83.QS Medeiros PH, Ledwaba SE, Bolick DT, Giallourou N, Yum LK, Costa DV, Oriá RB, Barry EM, Swann JR, Lima AÂ, et al. A murine model of diarrhea, growth impairment and metabolic disturbances with Shigella flexneri infection and the role of zinc deficiency. Gut Microbes. 2019;1–16. doi: 10.1080/19490976.2018.1564430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reed S, Neuman H, Moscovich S, Glahn RP, Koren O, Tako E. Chronic Zinc deficiency alters chick gut microbiota composition and function. Nutrients. 2015;7:9768–9784. doi: 10.3390/nu7125497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zumbrun SD, Melton-Celsa AR, Smith MA, Gilbreath JJ, Merrell DS, O’Brien AD. Dietary choice affects Shiga toxin-producing Escherichia coli (STEC) O157: H7colonization and disease. Proc Natl Acad Sci U S A. 2013;110:E2126–33. doi: 10.1073/pnas.1222014110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ferreyra JA, Wu KJ, Hryckowian AJ, Bouley DM, Weimer BC, Sonnenburg JL. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe. 2014;16:770–777. doi: 10.1016/j.chom.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tytgat HLP, Nobrega FL, van der Oost J, de Vos WM. Bowel biofilms: tipping points between a healthy and compromised gut? Trends Microbiol. 2019;27:17–25. doi: 10.1016/j.tim.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 88.Wake N, Asahi Y, Noiri Y, Hayashi M, Motooka D, Nakamura S, Gotoh K, Miura J, Machi H, Iida T, et al. Temporal dynamics of bacterial microbiota in the human oral cavity determined using an in situ model of dental biofilms. NPJ Biofilms Microbiomes. 2016;2. doi: 10.1038/npjbiofilms.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Røder HL, Sørensen SJ, Burmølle M. Studying bacterial multispecies biofilms: where to start? Trends Microbiol. 2016;24:503–513. doi: 10.1016/j.tim.2016.02.019. [DOI] [PubMed] [Google Scholar]


