ABSTRACT
Alcohol intake can modify gut microbiota composition, increase gut permeability, and promote liver fibrogenesis. LRP6 is a signal transmembrane protein and a co-receptor for the canonical Wnt signaling pathway. This study compared the curative effect of LRP6-CRISPR on alcohol-related liver injury with that of traditional fecal microbiota transplant (FMT) and investigated the alteration of the gut microbiome following the treatment. A rat model of alcohol-related liver injury was established and injected with lentiviral vectors expressing LRP6-CRISPR or administered with fecal filtrate from healthy rats, with healthy rat served as the control. Liver tissues of rats were examined by HE staining, Sirius staining, and Oil red O staining, respectively. The expression of LRP6 and fibrosis biomarkers were tested by PCR. The fecal sample of rats was collected and examined by 16S rRNA sequencing. Our data indicated that LRP6-CRISPR was more efficient in the prevention of alcohol-related liver injury than FMT. Microbiome analysis showed that alcohol-related liver injury related to gut microbiota dysbiosis, while treatment with LRP6-CRISPR or FMT increased gut microflora diversity and improved gut symbiosis. Further, bacteria specific to the disease stages were identified. Genera Romboutsia, Escherichia-Shigella, Pseudomonas, Turicibacter, and Helicobacter were prevalent in the intestine of rats with alcohol-related liver injury, while the domination of Lactobacillus was found in rats treated with LRP6-CRISPR or FMT. Besides, Lactobacillus and genera belonging to family Lachnospiraceae, Bacteroidales S24-7 group, and Ruminococcaceae were enriched in healthy rats. LRP6-CRISPR and FMT have beneficial effects on the prevention of alcohol-related liver injury, and correspondently, both treatments altered the disrupted gut microflora to a healthy one.
KEYWORDS: LRP6, CRISPR, FMT, gut microbiota, alcohol-related liver injury
Introduction
Alcoholic hepatic fibrosis, a significant type of alcoholic liver disease (ALD), is a lifestyle-associated disease and also a co-factor in many other diseases.[1] Due to the ingested alcohol and its metabolic consequences, alcohol intake can alter the gut microbiota composition and contribute to disease aggravation.[2] Despite its healthy burden, the occurrence, development, detection, and treatment of alcohol-related liver injury remain largely unknown.[3]
Low-density lipoprotein receptor-related protein 6 (LRP6) is a signal transmembrane protein. As a co-receptor for the canonical Wnt pathway, the interaction of LRP6 and Wnt ligand is the first step in the Wnt signal transduction cascade.[4] We have recently demonstrated that RSPO2, which requires LRP6 for activating and amplifying signaling of the Wnt pathway, facilitates HSC activation and arguments liver fibrogenesis by enhancing the canonical Wnt pathway.[5,6] Thus, we hypothesized that the knockout of LRP6 would repress the Wnt signaling activities and subsequently suppress the activation of HSC.
For decades, the gut microbiota is recognized as a major environmental factor influencing liver fibrosis.[7] Qin et al. reported that enteric dysbiosis, especially the translocation of bacteria and their metabolic products across the gut barrier, is involved in the progression of liver fibrosis.[8,9] Alterations of gut microbiota and associated gut dysbiosis may play an essential role in the induction and promotion of liver injury. Thus, the gut microbiota is considered as a potential therapeutic target of a large number of liver diseases due to the existing of gut-liver axis.[10] Lots of randomized-controlled trials show that FMT as a promising treatment in various diseases, such as IBD and Clostridium difficile infections.[11] Although FMT may improve gut symbiosis, its impact on the prevention of alcohol-related liver injury has not been well studied.
This study used clustered regularly interspaced short palindromic repeats (CRISPR) – CRISPR-associated protein 9 (Cas9) system, an efficient genome editing tool, to identify the functions of LRP6 in alcohol-related liver injury. The curative effect of LRP6-CRISPR on alcoholic liver disease was compared with that of traditional fecal microbiota transplant, and the alteration of the gut microbiome following the treatment was investigated.
Results
LRP6-CRISPR attenuated liver fibrogenesis in a rat model of alcoholic liver disease
A rat model of alcohol-related liver injury was established by administrating ethyl alcohol for four weeks (the model). Alcohol-related liver injury rats were then selected for in vivo transduction of lentivirus vector expressing LRP6-CRISPR (the CRISPR group), with healthy rats served as the control. Liver tissues of rats were analyzed by Sirius staining and HE staining, respectively. Sirius-red staining showed decreased liver fibrosis in the CRISPR group compared with the model (Figure 1a). Consistently, HE staining presented similar results. The recovering of vacuoles degeneration was observed in the liver tissues of the CRISPR group (Figure 1b). Oil red O staining indicated the lipid droplets obviously decreased in the liver tissues of the CRISPR group, which might suggest that LRP6-CRISPR relives liver injury induced by ethyl alcohol (Figure 1c). Interestingly, the above experiments presented a similar pattern that the liver tissues of the CRISPR group tended to restore to normal status as the control.
Figure 1.
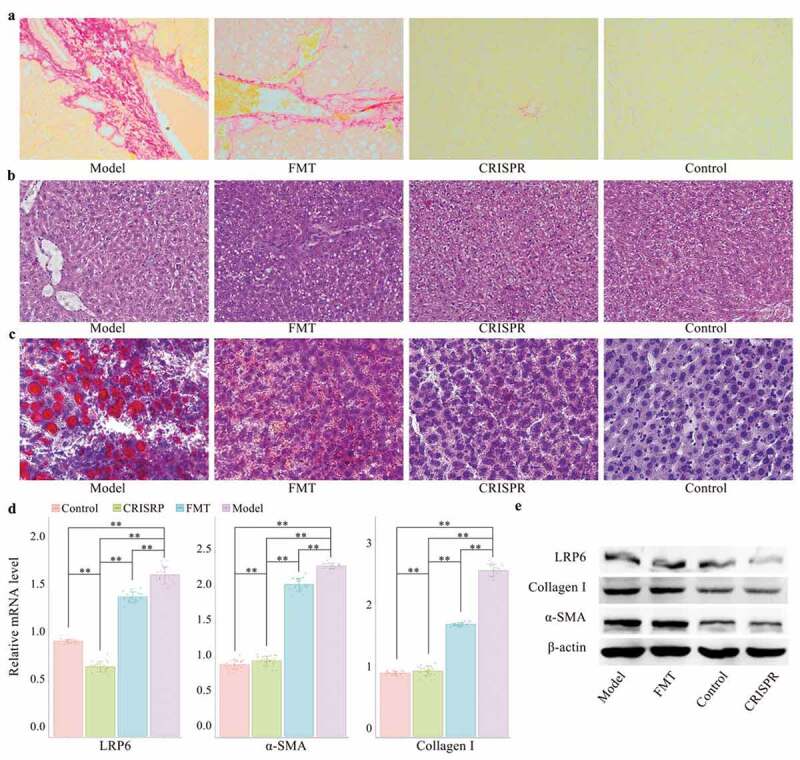
LRP6-CRISPR had a superior efficiency in the prevention of alcohol-related liver injury than FMT in a rat model.
To confirm the above findings, liver tissues of rats were further tested by RT-PCR and western blot assays. Aligned with the above findings, the results of the PCR test showed the mRNA level of LRP6 (p < .01), fibrosis biomarker α-SMA (p < .01), and collagen-I (p < .01) in the CRISPR group dropped significantly compared with the model (Figure 1d). No statistical difference was found in the mRNA level of α-SMA and collagen-I between the CRISPR group and the control, whereas the expression of LRP6 was significantly decreased in the CRISPR group compared with the control (p < .01). Consistently, the same results were observed with the western blot assay. Compared with the model, the protein expression of LRP6, α-SMA, and collagen-I was notably down-regulated in the CRISPR group and was close to that of the control (Figure 1e).
LRP6-CRISPR was more efficient in the prevention of alcoholic liver disease than FMT
To validate the curative effect of LRP6-CRISPR on alcoholic liver disease, fibrotic-liver rats were also selected for fecal microbiota transplant. Rats with alcohol-related liver injury were transplanted with fecal filtrate from healthy rats (the FMT group). Liver tissues of rats were analyzed by Sirius staining, HE staining, Oil red O staining, RT-PCR, and western blot assay, respectively. Compared with the CRISPR group, the results of staining experiments indicated FMT has a modest effect in the healing the liver injury caused by ethyl alcohol (Figure 1a, b and c). Consistently, the results of the PCR test confirmed the above findings. Although the mRNA level of fibrosis biomarkers α-SMA and collagen-I was significantly lower in the FMT group than the model (p < .01 for α-SMA and collagen-I), there was still a statistical difference between the FMT group and the control (p < .01 for α-SMA and collagen-I) (Figure 1d). Similar results were also observed with the western blot assay (Figure 1e). The above findings indicated that FMT treatment has little benefit on the relief of alcohol-related liver injury.
Treatment with CRISPR-LRP6 or FMT increased the gut microflora diversity
Fecal samples of rats were collected from the CRISPR group, the FMT group, the model, and the control. 16S rRNA amplicon sequencing was performed on the DNA extracted from the fecal samples. On average 22.29 Mbp clean data per sample was generated. After clustering the operational taxonomic units (OTU) at 97% identity, a total of 1,908 OTUs were recovered from the fecal samples.
Aligned with the previous reports, this study found liver injury correlates to gut microflora dysbiosis.[12,13] Rarefaction curves for Sobs index tended to be smooth, which suggested a reasonable number of samples have been sequenced (Figure 2a). Additionally, the rarefaction curves also indicated the model has lower community richness (Sobs index) than the rests (the CRISPR group, the FMT group, and the control). Interestingly, the rarefaction curve of the CRISPR group was almost overlapped with that of the FMT group, which implied these two groups have similarly observed richness (Sobs index).
Figure 2.
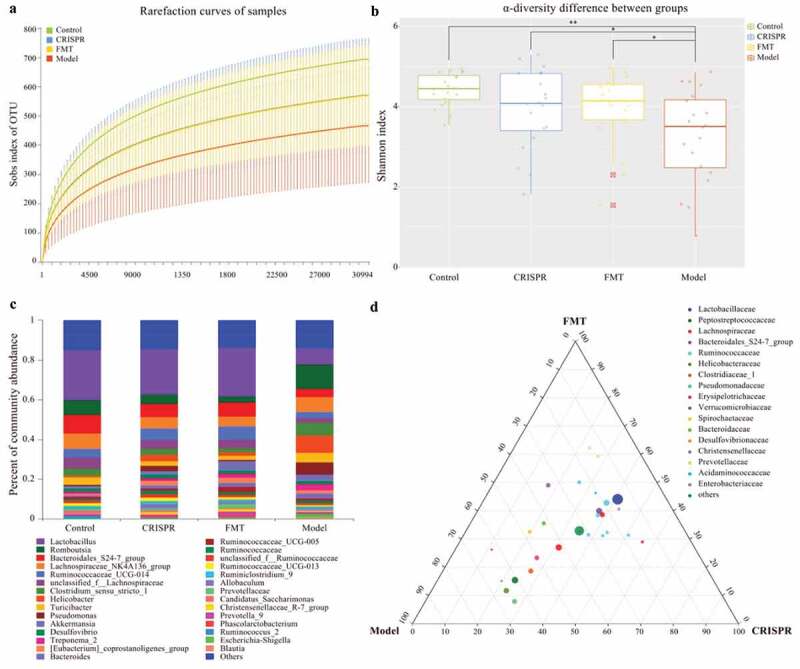
Treatment with CRISPR-LRP6 or FMT improved the gut microflora symbiosis.
The community α-diversity index (Shannon index) of each sample was calculated by removing the outlier values (sample FMT_9). Consistently, results showed the community diversity of the CRISPR group (p = .05), the FMT group (p < .01), and the control (p < .01) were significantly higher than that of the model, and no statistical difference was found between the CRISPR group and the FMT group (p = .20), between the CRISPR group and the control (p = .06), and between the FMT group and the control (p = .26). (Figure 2b). Together, these findings suggested that liver fibrosis was correlated with the gut microbiota dysbiosis and the treatment of CRISPR-LRP6 or FMT increased the gut microflora diversity and tended to be close to that of the control.
Treatment with CRISPR-LRP6 or FMT tended to restore the gut microflora composition
Taxonomic annotations were derived from the SILVA database (version 123), and the community composition of each group (the CRISPR group, the FMT group, model, and the control) was calculated (Figure 2c). Community analysis showed Lactobacillus has the highest abundance in the CRISPR group (22.8%), the FMT group (24.6%), and the control (25%), whereas its abundance was only 8.4% in the model (Table 1). Genus Romboutsia was abundant in the model (12.3%), followed by the control (7.7%), the CRISPR group (4.8%), and the FMT group (3.1%). Bacteroidales S24-7 group (CRISPR: 6.5%, FMT: 7%, control: 9.2%), Lachnospiraceae NK4A136 group (CIRSPR: 5.8%, FMT: 5%, control: 7.6%), Ruminococcaceae UCG-014 (CRISPR: 5.8%, FMT: 6.6%, control: 4.2%), and unclassified Lachnospiraceae (CRISPR: 4.4%, FMT: 4.4%, control: 5.9%) were the dominant genus in the CRISPR group, the FMT group, and the control. On the other hand, Helicobacter (8.9%), Lachnospiraceae NK4A136 group (7.7%), Pseudomonas (6.4%), and Clostridium sensu stricto_1 (6.1%) predominated in the model. In brief, the taxonomic analysis indicated the CRISPR group and the FMT group share a similar pattern of microbial community composition with the control, which was obviously differed from that of the model.
Table 1.
Community composition.
| Genera | CRISPR | Control | FMT | Model |
|---|---|---|---|---|
| Lactobacillus | 0.228 | 0.250 | 0.246 | 0.084 |
| Romboutsia | 0.048 | 0.077 | 0.031 | 0.123 |
| Bacteroidales_S24-7_group | 0.065 | 0.092 | 0.070 | 0.040 |
| Lachnospiraceae_NK4A136_group | 0.058 | 0.078 | 0.050 | 0.077 |
| Ruminococcaceae_UCG-014 | 0.059 | 0.042 | 0.066 | 0.029 |
| unclassified_f__Lachnospiraceae | 0.044 | 0.059 | 0.044 | 0.026 |
| Clostridium_sensu_stricto_1 | 0.031 | 0.035 | 0.021 | 0.061 |
| Helicobacter | 0.031 | 0.008 | 0.016 | 0.089 |
| Turicibacter | 0.025 | 0.038 | 0.022 | 0.047 |
| Pseudomonas | 0.027 | 0.006 | 0.008 | 0.064 |
| Akkermansia | 0.015 | 0.010 | 0.044 | 0.030 |
| Desulfovibrio | 0.019 | 0.018 | 0.018 | 0.018 |
| Treponema_2 | 0.012 | 0.011 | 0.020 | 0.029 |
| [Eubacterium]_coprostanoligenes_group | 0.025 | 0.007 | 0.025 | 0.016 |
| Bacteroides | 0.013 | 0.006 | 0.021 | 0.025 |
| Ruminococcaceae_UCG-005 | 0.014 | 0.012 | 0.026 | 0.012 |
| norank_f__Ruminococcaceae | 0.017 | 0.009 | 0.014 | 0.014 |
| unclassified_f__Ruminococcaceae | 0.017 | 0.014 | 0.012 | 0.009 |
| Ruminococcaceae_UCG-013 | 0.019 | 0.014 | 0.012 | 0.007 |
| Ruminiclostridium_9 | 0.015 | 0.011 | 0.011 | 0.009 |
| Allobaculum | 0.023 | 0.002 | 0.012 | 0.006 |
| _unclassified_f__Prevotellaceae | 0.007 | 0.008 | 0.018 | 0.004 |
| Candidatus_Saccharimonas | 0.009 | 0.015 | 0.008 | 0.004 |
| Christensenellaceae_R-7_group | 0.013 | 0.004 | 0.012 | 0.005 |
| Prevotella_9 | 0.007 | 0.005 | 0.015 | 0.004 |
| Phascolarctobacterium | 0.008 | 0.001 | 0.011 | 0.005 |
| Ruminococcus_2 | 0.003 | 0.015 | 0.003 | 0.002 |
| Escherichia-Shigella | 0.004 | 0.001 | 0.003 | 0.014 |
| Blautia | 0.002 | 0.003 | 0.004 | 0.010 |
| Others | 0.144 | 0.149 | 0.136 | 0.138 |
The relative abundance at the genus level (between the CRISPR group, the FMT group, and the model) was compared by ternary analysis. The plot of the ternary analysis showed that most of the genus was distributed along the median of CRISPR and FMT, which indicated the CRISPR group and the FMT group shared a similar microbiota composition and was differed from that of the model (Figure 2d). Analysis results showed Lactobacillus (CRISPR: 40.9%, FMT: 44.1%, model: 15%), and genus belong to family Ruminococcaceae, such as Ruminococcaceae UCG-014 (CRISPR: 38.1%, FMT: 42.8%, model: 19.1%), Ruminococcaceae UCG-013 (CRISPR: 50.6%, FMT: 31.4%, model: 18.1%), unclassified Ruminococcaceae (CRISPR: 43.6%, FMT: 32.3%, model: 24.1%), and Ruminiclostridium (CRISPR: 42.8%, FMT: 31.1%, model: 24.1%) were abundant in the CRISPR group and the FMT group. In comparison, Romboutsia (CRISPR: 23.7%, FMT: 15.4%, model: 60.8%), Helicobacter (CRISPR: 22.9%, FMT: 11.7%, model: 65.3%), and Pseudomonas (CRISPR: 27.4%, FMT: 7.9%, model: 64.7%) were predominated in the model.
Taken together, the above findings suggested the treatment of LRP6-CRISPR or FMT tends to restore the disrupted gut microflora of alcohol-related liver injury rats to a normal status.
Treatment with CRISPR-LRP6 or FMT improved gut symbiosis
Principal co-ordinates analysis (PCoA) was applied to visualize the overall community structure of gut microflora among all fecal samples on the species level (Figure 3a). The first two components accounted for 30.2% (PC1: 18.4%, PC2: 11.8%) of the total variation. A total of four groups (represented by an ellipse) were identified, which stands for the CRISPR group, the FMT group, the model, and the control, respectively. The control ellipse was obviously apart from the model ellipse, which meant they have different community structure of gut microbiota. Notably, the CRISPR group and the FMT group were overlapped and intersected with the model and the control. This result implied the CRISPR group and the FMT group share a similar microbial community structure, which represents an intermediate state transiting from the model to the control.
Figure 3.
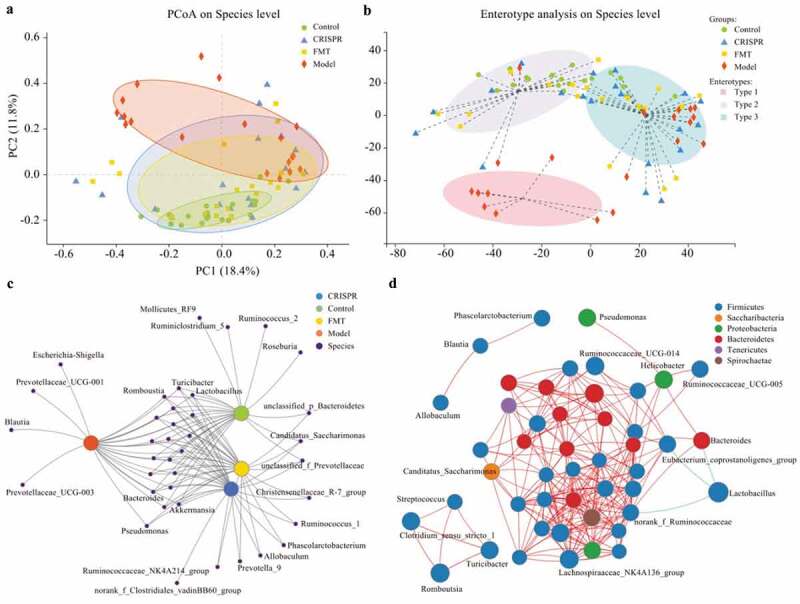
The gut microflora of the CRISPR group and FMT group tended to restore to an equilibrium status similar to the control.
By clustering the fecal samples with similar dominant microbiota structure into one class, the enterotype analysis presented three distinct classes (enterotype), which were type 1, type 2, and type 3, respectively (Figure 3b). Among which, type 1 purely included samples from the model, whereas type 2 almost excluded samples from the model, and type 3 consisted of samples from all groups. Samples from the CRISPR group and the FMT group were distributed evenly in type 2 and type 3.
In summary, the above findings indicated that the alcohol-related liver injury is related to a disrupted gut microflora with low diversity. Further, the treatment of LRP6-CRISPR or FMT improved the gut symbiosis and tended to restore the gut microflora to an equilibrium status similar to the control.
Correlation between samples and gut microflora were identified
For a comprehensive understanding of the relationship between samples and gut microbiota species, network analysis was performed on the abundance data. Co-occurrence network analysis demonstrated the distribution of gut microbiota species between samples and revealed the similarities and differences between groups. Results showed the CRISPR group and the FMT group shared most of the species, and they shared more species with the control than the model (Figure 3c). Co-occurrence network analysis also showed genus Escherichia-Shigella, Prevotellaceae UCG-001, Prevotellaceae UCG-003, and Blautia were related to the model only, whereas genus Fuminococcus_2, Ruminiclostridium_5, Mollicutes_RF9, and Roseburia were only identified in the control.
The plot of correlation network analysis on the genus level illustrated the correlations of the gut microbiota species in the samples. The size of the nodes represented the species abundance in the samples, and the color of the nodes indicated the phylum of the species. A red line indicated a positive correlation between the connected nodes, whereas a green line indicated a negative correlation. Results showed that most of the species belonged to phylum Firmicutes, followed by Bacteroides and Proteobacteria (Figure 3d). Correlation analysis showed that Lactobacillus is negatively correlated with genus norank_f_Ruminococcaceae, Bacteroides, and Eubacterium_coprostanoligenes group.
Species that significantly differed between groups were recognized
This study tried to identify the species that significantly differ between groups. The difference of gut microbiota abundance between multiple groups (the CRISPR group, the FMT group, the model, and the control) was analyzed with Kruskal-Wallis H test. Significant difference analysis results showed genera Lactobacillus (p < .01), Bacteroidales S24-7 group (p < .01), and Ruminococcaceae UCG-014 (p = .01) were significantly enriched in the CRISPR group, the FMT group and the control, whereas Romboutsia (p < .01), Turicibacter (p = .03), Pseudomonas (p < .01), and Escherichia-Shigella (p = .01) were dominant in the model (Figure 4a).
Figure 4.
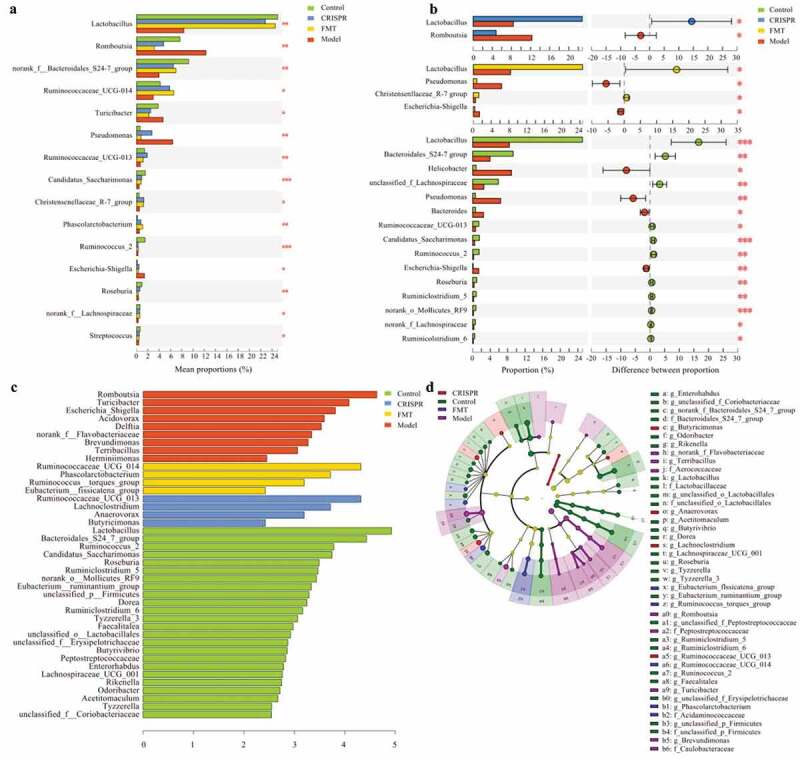
Species that significantly differ between the CRISPR group, the FMT group, the model, and the control were identified.
The difference of gut microbiota abundance between the model and the remain groups was further calculated (Figure 4b). Consistently, results showed that Lactobacillus (p < .05) was enriched in the CRISPR group, whereas Romboutsia (p < .05) was abundant in the model (the CRISPR group vs. the model). Similarly, Lactobacillus (p < .05) and Christensenllaceae R-7 group (p < .05) were enriched in the FMT group, where Escherichia-Shigella (p < .05) and Pseudomonas (p < .05) predominated in the model (the FMT group vs. the model). Further, a noteworthy finding was the enrichment of species Enterococcus faecalis (p < .05) in the model, which was previously reported related to hepatocyte death and alcoholic liver disease .14 In addition, Lactobacillus (p < .01), Bacteroidales S24-7 group (p < .01), unclassified_f_Lachnospiraceae (p < .01), Ruminococcaceae UCG-013 (p < .05) were prevalent in the control, whereas genus Helicobacter (p < .05), Pseudomonas (p < .01), Bacteroides (p < .05), Escherichia-Shigella (p < .01) were enriched in the model (the control vs. the model).
The gut microbial abundance data were further analyzed by linear discriminant analysis to identify bacteria that were specific for the groups. LEfSe (Linear discriminant analysis Effect Size) results were consistent with the analysis of the significant differences. Overall, the abundance of 41 genera was significantly different between groups (Figure 4c, d). Nine genera predominated the model, including Romboutsia (LDA = 4.63), Turicibacter (LDA = 4.08), and Escherichia_Shigella (LDA = .81), which implied these bacteria positively responded to the liver injury induced by ethyl alcohol. Twenty-four genus were abundant in the control, including Lactobacillus (LDA = 4.92), Bacteroidales S24-7 group (LDA = 4.43), and Ruminococcus_2 (LDA = 3.78), which suggested these bacteria might have a protective effect on the liver injury. Besides, Ruminococcaceae UCG_013 (LDA = 3.82), Lachnoclostridium (LDA = 3.23), Anaerovorax (LDA = 2.59), and Butricimonas (LDA = 2.53) were enriched in the CRISPR group, and Ruminococcaceae UCG_014 (LDA = 4.1), Phascolarctobacterium (LDA = 3.71), Ruminococcus torques group (LDA = 3.19), and Eubacterium fissicatena group (LDA = 2.42) were enriched in the FMT group, which implied these bacteria have a positive response to the treatment.
Discussion
Alcoholic liver fibrosis remains a significant global public health problem.[3] Our previous studies established an exciting possibility of intervention in the liver fibrosis by regulating Wnt signaling.[5,15] In this study, we used an animal model that simulated the drinking pattern of alcoholics and attempted to investigate the therapeutic effects of CIRSRP or FMT on alcohol-related liver injury. This study found that the treatment of LRP6-CRISPR or FMT can relieve liver injury in alcohol-related liver injury rats. Further, histological analysis showed that the curative effect of LRP6-CRISPR is superior to that of the FMT. Meanwhile, gut microbiome analysis revealed the treatment of LRP6-CRISPR or FMT restore the disrupted gut microflora caused by alcohol ingestion and its metabolic consequences. Results of 16S gene amplicon sequencing showed rats treated with LRP6-CRISPR or FMT share similar gut microbiota composition, which stands for a transition status from the gut dysbiosis of alcohol-related liver injury rats to an equilibrium status similar as the healthy rats. Besides, species that specific to the disease status were also identified. Our findings indicated that the improvement of gut symbiosis might reflect the therapeutic effect of LRP6-CRISPR or FMT.
Our study used the CRISPR-Cas9 system to knockout the LRP6 gene in rats with alcohol-related liver injury and subsequently suppressed the activities of the Wnt pathway. The CRISPR-Cas9 system is an RNA-guided gene-editing tool for cells and animals at the genomic DNA level.[16,17] This system requires two components, Cas9, a DNA endonuclease for cleaving the target strand, and a single-guide RNA (sgRNA), which accurately guides Cas9 to the location in the genome.[18] The double-strands break inactivates target gene loci and leads to loss of function. A rat model with alcohol-related liver injury was established. Various staining experiments presented significantly improved histological changes in rats treated with LRP6-CRISPR. PCR and WB assays showed decreased expression of fibrosis biomarkers α-SMA and collagen-I. These results confirmed that LRP6-CRISPR effectively relieves liver injury induced by ethyl alcohol.
The gut microbiota is essential in the maintenance of the balance between health as well as disease and may be considered as a virtual organ of human.[19] Specifically, numerous studies outlined that gut microbiota plays a significant role in the onset and progression of alcoholic liver disease.[2,20] Studies showed that fecal microbiota transplantation improves gut symbiosis.[21] Thus, this study also performed FMT on rats with alcohol-related liver injury to reestablish the balance of intestinal flora disrupted by alcohol ingestion and its metabolic consequences. Rats with alcohol-induced liver fibrosis were administrated with fecal filtrate from healthy rats. Staining experiments demonstrated slightly improved histological changes in the liver tissues of rats with FMT treatment. Results of PCR and WB assays were aligned with those of the staining experiments. These findings suggested FMT has a modest effect in the healing the liver injury caused by ethyl alcohol compared with the LRP6-CRISPR.
A growing number of studies confirmed that gut microbiota involves in the occurrence and development of liver disease.[10,22] The gastrointestinal tract communicates extensively with the liver through the portal vein and biliary tract, which refer to the gut-liver axis. The liver affects the intestine by secreting bile acids and bioactive mediators into the biliary tract while metabolizing products in the intestine is transported to the liver through the portal vein.[23,24] An intestinal barrier, formed by enterocytes tightly bound to adjacent cells, restricts the movement of gut microorganisms and microorganism-derived molecule products from the intestine to the liver while allowing the transport of nutrients across the barrier.[25,26] Infection, high fat diet, and alcohol intake may induce gut dysbiosis and increase intestinal barrier permeability. Upon malfunction of the intestinal barrier, microorganisms and their molecule products can translocate to the liver through the portal vein and promote hepatic injury.[27,28]
Hence, this study examined the gut microbiome of rats with 16S rRNA sequencing. Aligned with the previous reports, our study found alcohol-related liver injury related to gut microbiota dysbiosis.[2] Results of 16S rRNA analysis showed that gut microbiota richness and diversity of rats with alcohol-related liver injury decreased significantly compared with the healthy rats. The gut microflora composition of rats with alcohol-related liver injury also changed. Compared with the healthy rats, the abundance of Lactobacillus and genera belonging to family Bacteroidales S24-7 group, Lachnospiraceae, and Ruminococcaceae in fibrotic-liver rats dropped significantly, while the abundance of Romboutsia, Escherichia-Shigella, Pseudomonas, Helicobacter increased. These findings were following the previous reports that the intestinal dysbiosis in alcohol-related liver injury can be characterized by enrichment of Enterobacteriaceae and reduction of Bacteroidetes and Lactobacillus.[29,30]
Interestingly, the analysis of gut microbiome confirmed the treatment of LRP6-CRISPR or FMT partially restore the gut microbiota composition and improve the gut symbiosis. Under the previous studies, our data indicated that FMT increases the gut microflora diversity.[31,32] Both the gut microbiota richness and diversity in rats receiving fecal microbiota transplantation increased compared with the alcohol-related liver injury rats. Similar results were observed in rats treated with LRP6-CRISPR. Further analysis showed that rats treated with LRP6-CRISPR or FMT shared a similar microbiota composition structure. The ternary analysis revealed the CRISPR group and the FMT group have a similar microbiota composition and was differed from that of the rats with alcohol-related liver injury. Also, PCoA analysis indicated the composition structure of the CRISPR group and the FMT group are overlapped and intersected with the model and the control. Besides, compared with the rats with alcohol-related liver injury, the abundance of Lactobacillus increased in both the CRISPR group and the FMT group. In brief, these findings suggested the CRISPR group had similar alpha-diversity and microbiota composition as the FMT group, which stands for a transition state from the disrupted gut microflora to a healthy one.
Our study further identified the species that specific to the disease stages (rats treated with LRP6-CRISPR or FMT represented a relieved liver fibrosis state). Genera Romboutsia, Pseudomonas, Helicobacter, and Escherichia-Shigella were dominant in the rats with alcohol-related liver injury. Pseudomonas and Escherichia-Shigella were reported reduced in mucosa-associated microbiota of colorectal cancer tissues compared to adjacent tissues.[33] Bashir M, et al. found a decreased relative abundance of Pseudomonas and Escherichia-Shigella after oral vitamin D3 supplementation in the colon.[34] Evidence showed that Helicobacter might be involved in the pathogenesis of alcohol-related liver injury and worsen the course of alcoholic liver cirrhosis.[35–37]
Further, species Enterococcus faecalis was enriched in the alcohol-related liver injury rats. Enterococcus faecalis was recently confirmed as important bacteria in patients with alcoholic liver disease.[14] The products of Enterococcus faecalis, exotoxin and cytolysin, will result in liver injury with poor prognosis. These reports were consistent with our findings.
Further, Lactobacillus was prevalent in rats treated with LRP6-CRISPR or FMT. Genus Lactobacillus is part of the healthy human gastrointestinal flora and considered as probiotics that have been used in food, biotechnology, and therapeutic application.[38,39] A study showed Lactobacillus accumulates in the gut during liver injury and produces IL-22, which helps to restore the damaged gut barrier.[40] Other reports show the domination of gut microbiota by Lactobacillus restore the gut barrier integrity.[41,42] Consistent with the above reports, our findings suggested Lactobacillus may be an essential marker to represent the liver-injury healing stage.
Furthermore, genera belonging to family Bacteroidales S24-7 group, Lachnospiraceae, and Ruminococcaceae were abundant in healthy rats. Family Lachnospiraceae and Ruminococcaceae are beneficial commensals that participate in the production of short-chain fatty acid (SCFA) in the human intestine.[43] SCFA, including acetate, propionate, and butyrate, have a crucial role in energy homeostasis and metabolism and beneficially modulating liver tissue function.[44] Besides, genera belonging to Lachnospiraceae and Ruminococcaceae are reported having the ability to strengthen the integrity of the gut barrier.[45] A decreased abundance of Lachnospiraceae and Ruminococcaceae was related to colon inflammation, cirrhosis, Hepatic encephalopathy, and liver failure.[46–48] Regarding our data, the domination of family Lachnospiraceae and Ruminococcaceae may stand for healthy microbiota composition.
Conclusion
Our study tried to establish a link between the traditional molecular biology and the microbiome and provide a new perspective for the intervention and treatment of that alcohol-related liver injury. In conclusion, treatment of LRP6-CRISPR presented a superior curative effect on alcohol-related liver injury than FMT, while both treatments altered the disrupted gut microflora to a healthy one.
Material and Methods
Study design
This study compared the curative effect of LRP6-CRISPR on the alcoholic liver disease with that of traditional fecal microbiota transplant and investigated the alteration of the gut microbiome following the treatment (Figure 5). Alcohol-related liver injury rats were injected with lentivirus vectors expressing LRP6-CRISPR or administered with fecal filtrate from healthy rats, with healthy rats served as the control. Liver tissues of rats were examined by HE staining, Sirius staining, and Oil red O staining, respectively. The expression of LRP6 and fibrosis biomarkers were tested by PCR. Fecal samples of rats (rats with liver fibrosis, rats injected of LRP6-CRISPR, rats administered with fecal filtrate, and the control) were collected and examined by 16S rRNA sequencing.
Figure 5.
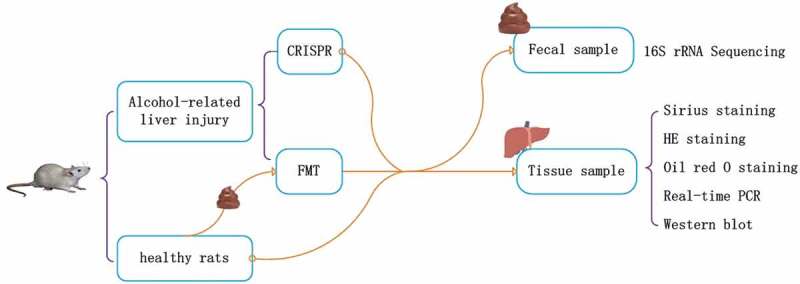
Study design.
Animals used
Male Sprague-Dawley (SD) rats (8–10 weeks, 250–300 g) were purchased from Shanghai SLAC Laboratory Animal Co Ltd (Shanghai, China). The animals were housed under standard animal laboratory conditions. All individuals involved in animal research received instructions in experimental methods and the care, maintenance, and handling of animals. All institutional and national guidelines for the care and use of laboratory animals were followed. The protocol of the experiments was approved by the Committee on the Ethics of Animal Experiments of Jiaxing College (JUMC2018-014).
The animal model of alcohol-related liver injury
A modified chronic-plus-binge ethanol feeding model which simulates the drinking pattern of alcoholics was adopted to induce robust liver injury.[49] Eighty male Sprague-Dawley (SD) rats (8–10 weeks old, 250–300 g of body weight) were randomly divided into the following two groups: (i) a control group (n = 20), which was supplied with regular laboratory chow and water and (ii) an alcohol-related liver injury group (n = 60), which was administered with a soya oil and 56% vol/vol alcohol solution instead of water, by starting at 3 ml/kg/day in the first week and then 6 ml/kg/day in the following days. A high level of blood ethanol (150–250 mg/dl) was maintained during the days.
sgRNA plasmid
Single-guide RNA (sgRNA) sequences targeting rat LRP6 (Gene ID: 312781) were designed using online software (http://crispr.mit.edu). This study selected three sgRNA target sequences (target 1, 2, and 3) for LRP6-CRISPR (Table 2).
Table 2.
sgRNA sequences.
| Name | Target | sgRNA |
|---|---|---|
| CRISPR-T1 | GCCAGTGCCAAGGCAACCGA | caccGACGAGTACAAGGTGCCCAG |
| aaacCTGGGCACCTTGTACTCGTC | ||
| CRISPR-T2 | GGAGACGCAGTAAGCGAGGT | caccGACCGGCACAGCATCAAGAA |
| aaacTTCTTGATGCTGTGCCGGTC | ||
| CRISPR-T3 | TCGGAGAGAAGGGATGCGCC | caccCGGCTGAAGAGAACCGCCAG |
| aaacCTGGCGGTTCTCTTCAGCCG |
FMT
Rats with alcohol-related liver injury received antibiotics (ampicillin, 1 mg/ml; neomycin, 1 mg/ml; metronidazole, 1 mg/ml; and vancomycin, 0.5 mg/ml) once a day for one week to deplete the gut microbiota. Each rat in the FMT group was administered with 200 µl of fecal filtrate from healthy rats by oral gavage once a day for five days after the antibiotic treatment (rats in other groups were administered with sterile normal saline). At day 10, rats were sacrificed by CO2 exposure, and liver tissues were harvested.
Fecal sample collection
To account for the cage effects, the fecal samples of rats were collected as below: (i) Eighty healthy rats were co-housed (five rats in one cage) for one week to homogenize the gut microflora before the start of the experiment; (ii) Sixty rats were randomly selected for establishing the alcohol-induced liver injury model, with the remain twenty healthy rats served as the control. Rats in the model and control were housed in different cages; (iii) The alcohol-induced liver injury rats were then randomly divided into three groups, which were model group (20 rats without any further treatment), CRISPR group (20 rats receiving LRP6-CRISPR treatment), and FMT group (20 rats receiving FMT treatment), respectively. Each rat was kept in an individually ventilated cage until collecting fecal pellets. Samples collected from the model group represented the baseline microbiota. Fecal pellets were collected, flash-frozen, and stored at −80°C for later analysis.
DNA extraction and 16S rRNA gene amplicon sequencing
Fecal microbial DNA was extracted from 250 mg frozen sample using Power Fecal DNA Kit (QIAamp, Germantown, MD, USA) following the manufacturer’s instructions. The final DNA concentration and purification were determined by NanoDrop 2000 UV–vis spectrophotometer (Thermo Scientific, Wilmington, USA), and DNA quality was checked by 1% agarose gel electrophoresis. The extracted DNA was stored at −20°C for future processing.
The amplicon library was constructed by amplifying the V3/V4region of 16S rRNA gene using the primer 338F_806R (338F: 5`-ACTCCTACGGGAGGCAGCAG- 3`, 806R: 5`-GGACTACHVGGGTWTCTAAT- 3`). PCR is performed by thermocycler PCR system (GeneAmp 9700, ABI, USA) with TransStartFastpfu DNA Polymerase AP221-02 by using the following conditions: 95°C for 3 min followed by 30 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 45 s, and a final extension step at 72°C for 8 min. The PCR products are quantified using QuantiFluor™ -ST (Promega, U.S.) according to the manufacturer’s protocol. Purified DNA samples were sequenced by using Illumina Miseq. Unqualified reads were removed to obtain clean data for further analysis. On average 22.29 Mbp clean data per sample was generated. After clustering operational taxonomic units (OTU) at 97% identity, a total of 1,908 OTUs were recovered from the fecal samples.
16S rRNA sequence analysis
The sequenced 16S reads were analyzed by using the QIIME software package,[50] the R programming language (R version 3.5.1), and the Python programming language (Python version 3.6). Data decontamination was performed using FLASH[51] and Trimmomatic.[52] OTUs were created by clustering the reads at the similarity threshold of 97% using Usearch (vsesion 7.0, http://drive5.com/uparse/). The taxonomy of each OTU representative sequence was analyzed by RDP Classifier (http://rdp.cme.msu.edu/) against the Bacteria and archaea 16S rRNA gene databases Silva (Release128 http://www.arb-silva.de) using confidence threshold of 0.7. Indexes of α-diversity were calculated using mothur (version v.1.35.1 https://www.mothur.org/). A phylogenetic tree was inferred by using FastTree (version 2.1.3 http://www.microbesonline.org/fasttree/). Principal component analysis (PCA) and PCoA analysis (Principal coordinates analysis) were performed by using R and python for analysis and plotting. With R Ade4 and cluster, enterotyping analysis calculated Jensen-Shannon Distance and PAM(Partitioning Around Medoids) according to the relative abundance of the species level first and then do clustering. Differences analysis was performed by using R for significant difference between groups (Kruskal-Wallis H tests) and Galaxy (http://huttenhower.sph.harvard.edu/galaxy/) for LEfSe (Linear discriminant analysis Effect Size).
Statistical analysis
All statistical analyses were performed using R software (version 3.5.1). Data were presented as mean ± SEM. A value of p<.05 was considered to be statistically significant. A two-tailed Student’s t-test was employed to evaluate the differences between groups. For semi-quantitative analysis of histological staging, nonparametric tests (Mann-Whitney U Test or Kruskal-Wallis Rank Sum Test) were used.
Acknowledgments
We thank Anesthesiology Center in North of Zhejiang Province, Jiaxing key laboratory of neurology and pain medicine, Center for gastroenterology and hepatology connecting with Shanghai.
Funding Statement
This work was supported by grants from the Natural Science Foundation of Zhejiang Province (LY17H030012, LY16H030016). Key medical discipline in Jiaxing—Infectious disease(2019-ZC-02). Key medical discipline in Jiaxing –Gastroenterology(2019-ZC-08)
Author contributions
Linghua Yu participated in the design, data analysis, and interpretation, and drafted the manuscript. Linlin Wang, HuixingYi, and Xiaojun Wu participated in the interpretation of experiments and helped to finalize the paper.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed
Data and code availability
16S rRNA sequence data can be accessed from NCBI BioProjectPRJNA543718. Our analysis use standard, open source software. R packages are available on CRAN (cran.r-project.org), Galaxy (usegalaxy.org), and the Python programming language (Python version 3.6).
References
- 1.Crabb DW, Im GY, Szabo G, JL Mellinger, Lucey MR.. Diagnosis and treatment of alcohol-related liver diseases: 2019 practice guidance from the American association for the study of liver diseases. Hepatology. 2020;71(1):306–333. doi: 10.1002/hep.30866. [DOI] [PubMed] [Google Scholar]
- 2.Bajaj JS. Alcohol, liver disease and the gut microbiota. Nat Rev Gastroenterol Hepatol. 2019;16(4):235–246. doi: 10.1038/s41575-018-0099-1. [DOI] [PubMed] [Google Scholar]
- 3.Bataller R, Gao B. Liver fibrosis in alcoholic liver disease. Semin Liver Dis. 2015;35(2):146–156. doi: 10.1055/s-00000069. [DOI] [PubMed] [Google Scholar]
- 4.Go GW. Low-density lipoprotein receptor-related protein 6 (LRP6) is a novel nutritional therapeutic target for hyperlipidemia, non-alcoholic fatty liver disease, and atherosclerosis. Nutrients. 2015;7(6):4453–4464. doi: 10.3390/nu7064453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin X, Yi, H., Wang, L., Wu, W., Wu, X, and Yu, L. RSPOs facilitated HSC activation and promoted hepatic fibrogenesis. Oncotarget. 2016;7(39):63767–63778. doi: 10.18632/oncotarget.11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xinguang Y, Huixing, Y., Xiaowei, W., Xiaojun, W. and Linghua, Y. R-spondin1 arguments hepatic fibrogenesis in vivo and in vitro. J Surg Res. 2015;193(2):598–605. [DOI] [PubMed] [Google Scholar]
- 7.Tilg H, Cani PD, Mayer EA. Gut microbiome and liver diseases. Gut. 2016;65(12):2035–2044. doi: 10.1136/gutjnl-2016-312729. [DOI] [PubMed] [Google Scholar]
- 8.Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513(7516):59–64. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 9.Qin N, Le Chatelier E, Guo J, Prifti E, Li L, Ehrlich SD. Qin et al. reply. Nature. 2015;525(7569):E2–3. doi: 10.1038/nature14852. [DOI] [PubMed] [Google Scholar]
- 10.Federico A, Dallio M, Caprio GG, Ormando VM, Loguercio C. Gut microbiota and the liver. Minerva Gastroenterol Dietol. 2017;63(4):385–398. doi: 10.23736/S1121-421X.17.02375-3. [DOI] [PubMed] [Google Scholar]
- 11.de Groot PF, Frissen MN, de Clercq NC, Nieuwdorp M. Fecal microbiota transplantation in metabolic syndrome: history, present and future. Gut Microbes. 2017;8(3):253–267. doi: 10.1080/19490976.2017.1293224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagymasi K, Bacsárdi A, Egresi A, Berta E, Tulassay Z, Lengyel G. The role of gut microbiota in chronic liver diseases, and treatment possibilities. Orv Hetil. 2018;159(36):1465–1474. doi: 10.1556/650.2018.31178. [DOI] [PubMed] [Google Scholar]
- 13.Yan AW, Schnabl B. Bacterial translocation and changes in the intestinal microbiome associated with alcoholic liver disease. World J Hepatol. 2012;4(4):110–118. doi: 10.4254/wjh.v4.i4.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duan Y, Llorente C, Lang S, Brandl K, Chu H, Jiang L, White RC, Clarke TH, Nguyen K, Torralba M, et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature. 2019;575(7783):505–511. doi: 10.1038/s41586-019-1742-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin X, Yi H, Wu W, Shu J, Wu X, Yu L. R-spondin2 activates hepatic stellate cells and promotes liver fibrosis. Dig Dis Sci. 2014;59(10):2452–2461. doi: 10.1007/s10620-014-3208-1. [DOI] [PubMed] [Google Scholar]
- 16.Shen Y, Cohen JL, Nicoloro SM, Kelly M, Yenilmez B, Henriques F, Tsagkaraki E, Edwards YJK, Hu X, Friedline RH, et al. CRISPR-delivery particles targeting nuclear receptor-interacting protein 1 (Nrip1) in adipose cells to enhance energy expenditure. J Biol Chem. 2018;293(44):17291–17305. doi: 10.1074/jbc.RA118.004554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci USA. 2013;110(34):13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su KC, Tsang M-J, Emans N, Cheeseman IM. CRISPR/Cas9-based gene targeting using synthetic guide RNAs enables robust cell biological analyses. Mol Biol Cell. 2018;29(20):2370–2377. doi: 10.1091/mbc.E18-04-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sgambato D, Miranda, A., Romano, L. and Romano, M. Gut microbiota and gastric disease. Minerva Gastroenterol Dietol. 2017;63(4):345–354. doi: 10.23736/S1121-421X.17.02380-7. [DOI] [PubMed] [Google Scholar]
- 20.Vassallo G, Mirijello A, Ferrulli A, Antonelli M, Landolfi R, Gasbarrini A, Addolorato G. Review article: alcohol and gut microbiota - the possible role of gut microbiota modulation in the treatment of alcoholic liver disease. Aliment Pharmacol Ther. 2015;41(10):917–927. doi: 10.1111/apt.2015.41.issue-10. [DOI] [PubMed] [Google Scholar]
- 21.Suez J, Zmora N, Zilberman-Schapira G, Mor U, Dori-Bachash M, Bashiardes S, Zur M, Regev-Lehavi D, Ben-Zeev Brik R, Federici S, et al. Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell. 2018;174(6):1406–1423.e16. doi: 10.1016/j.cell.2018.08.047. [DOI] [PubMed] [Google Scholar]
- 22.Gkolfakis P, Dimitriadis G, Triantafyllou K. Gut microbiota and non-alcoholic fatty liver disease. Hepatobiliary Pancreat Dis Int. 2015;14(6):572–581. doi: 10.1016/S1499-3872(15)60026-1. [DOI] [PubMed] [Google Scholar]
- 23.Xiao L, Pan G. An important intestinal transporter that regulates the enterohepatic circulation of bile acids and cholesterol homeostasis: the apical sodium-dependent bile acid transporter (SLC10A2/ASBT). Clin Res Hepatol Gastroenterol. 2017;41(5):509–515. doi: 10.1016/j.clinre.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Arab JP, Martin-Mateos RM, Shah VH. Gut-liver axis, cirrhosis and portal hypertension: the chicken and the egg. Hepatol Int. 2018;12(Suppl 1):24–33. doi: 10.1007/s12072-017-9798-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schnabl B, Brenner DA. Interactions between the intestinal microbiome and liver diseases. Gastroenterology. 2014;146(6):1513–1524. doi: 10.1053/j.gastro.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shao T, Zhao C, Li F, Gu Z, Liu L, Zhang L, Wang Y, He L, Liu Y, Liu Q, et al. Intestinal HIF-1alpha deletion exacerbates alcoholic liver disease by inducing intestinal dysbiosis and barrier dysfunction. J Hepatol. 2018;69(4):886–895. doi: 10.1016/j.jhep.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henao-Mejia J, Elinav E, Thaiss CA, Licona-Limon P, Flavell RA. Role of the intestinal microbiome in liver disease. J Autoimmun. 2013;46:66–73. doi: 10.1016/j.jaut.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Guerville M, Boudry G. Gastrointestinal and hepatic mechanisms limiting entry and dissemination of lipopolysaccharide into the systemic circulation. Am J Physiol Gastrointest Liver Physiol. 2016;311(1):G1–g15. doi: 10.1152/ajpgi.00098.2016. [DOI] [PubMed] [Google Scholar]
- 29.Yan AW, E. Fouts D, Brandl J, Stärkel P, Torralba M, Schott E, Tsukamoto H, E. Nelson K, A. Brenner D, Schnabl B, et al. Enteric dysbiosis associated with a mouse model of alcoholic liver disease. Hepatology. 2011;53(1):96–105. doi: 10.1002/hep.24018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tuomisto S, Pessi T, Collin P, Vuento R, Aittoniemi J, Karhunen PJ. Changes in gut bacterial populations and their translocation into liver and ascites in alcoholic liver cirrhotics. BMC Gastroenterol. 2014;14:40. doi: 10.1186/1471-230X-14-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taur Y, Coyte, K., Schluter, J., Robilotti, E., Figueroa, C., Gjonbalaj, M., Littmann, E.R., Ling, L., Miller, L., Gyaltshen, Y. and Fontana, E. Reconstitution of the gut microbiota of antibiotic-treated patients by autologous fecal microbiota transplant. Sci Transl Med. 2018;10(460):eaap9489. doi: 10.1126/scitranslmed.aap9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paramsothy S, Nielsen S, Kamm MA, Deshpande NP, Faith JJ, Clemente JC, Paramsothy R, Walsh AJ, van den Bogaerde J, Samuel D, et al. Specific bacteria and metabolites associated with response to fecal microbiota transplantation in patients with ulcerative colitis. Gastroenterology. 2019;156(5):1440–1454.e2. doi: 10.1053/j.gastro.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Gao Z, Guo B, Gao R, Zhu Q, Qin H. Microbiota disbiosis is associated with colorectal cancer. Front Microbiol. 2015;6:20. doi: 10.3389/fmicb.2015.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bashir M, Prietl B, Tauschmann M, Mautner SI, Kump PK, Treiber G, Wurm P, Gorkiewicz G, Högenauer C, Pieber TR, et al. Effects of high doses of vitamin D3 on mucosa-associated gut microbiome vary between regions of the human gastrointestinal tract. Eur J Nutr. 2016;55(4):1479–1489. doi: 10.1007/s00394-015-0966-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pogorzelska J, Łapińska M, Kalinowska A, Łapiński TW, Flisiak R. Helicobacter pylori infection among patients with liver cirrhosis. Eur J Gastroenterol Hepatol. 2017;29(10):1161–1165. doi: 10.1097/MEG.0000000000000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waluga M, Kukla, M., Żorniak, M., Bacik, A., and Kotulski, R. From the stomach to other organs: helicobacter pylori and the liver. World J Hepatol. 2015;7(18):2136–2146. doi: 10.4254/wjh.v7.i18.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okushin K, Tsutsumi T, Ikeuchi K, Kado A, Enooku K, Fujinaga H, Moriya K, Yotsuyanagi H, Koike K. Helicobacter pylori infection and liver diseases: epidemiology and insights into pathogenesis. World J Gastroenterol. 2018;24(32):3617–3625. doi: 10.3748/wjg.v24.i32.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duar RM, Lin XB, Zheng J, Martino ME, Grenier T, Pérez-Muñoz ME, Leulier F, Gänzle M, Walter J. Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol Rev. 2017;41(Supp_1):S27–s48. doi: 10.1093/femsre/fux030. [DOI] [PubMed] [Google Scholar]
- 39.Goldstein EJ, Tyrrell KL, Citron DM. Lactobacillus species: taxonomic complexity and controversial susceptibilities. Clin Infect Dis. 2015;60(Suppl 2):S98–107. doi: 10.1093/cid/civ072. [DOI] [PubMed] [Google Scholar]
- 40.Nakamoto N, Amiya T, Aoki R, Taniki N, Koda Y, Miyamoto K, Teratani T, Suzuki T, Chiba S, Chu P-S, et al. Commensal lactobacillus controls immune tolerance during acute liver injury in mice. Cell Rep. 2017;21(5):1215–1226. doi: 10.1016/j.celrep.2017.10.022. [DOI] [PubMed] [Google Scholar]
- 41.Hanash AM, Dudakov J, Hua G, O’Connor M, Young L, Singer N, West M, Jenq R, Holland A, Kappel L, et al. Interleukin-22 protects intestinal stem cells from immune-mediated tissue damage and regulates sensitivity to graft versus host disease. Immunity. 2012;37(2):339–350. doi: 10.1016/j.immuni.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leclercq S, Matamoros S, Cani PD, Neyrinck AM, Jamar F, Stärkel P, Windey K, Tremaroli V, Bäckhed F, Verbeke K, et al. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc Natl Acad Sci USA. 2014;111(42):E4485–93. doi: 10.1073/pnas.1415174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bajaj JS. The role of microbiota in hepatic encephalopathy. Gut Microbes. 2014;5(3):397–403. doi: 10.4161/gmic.28684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chambers ES, Morrison DJ, Frost G. Control of appetite and energy intake by SCFA: what are the potential underlying mechanisms? Proc Nutr Soc. 2015;74(3):328–336. doi: 10.1017/S0029665114001657. [DOI] [PubMed] [Google Scholar]
- 45.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux -J-J, Blugeon S, Bridonneau C, Furet J-P, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of crohn disease patients. Proc Natl Acad Sci USA. 2008;105(43):16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei D, Wang Y, Zhu B, Li L. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54(2):562–572. doi: 10.1002/hep.24423. [DOI] [PubMed] [Google Scholar]
- 47.Bajaj JS, Kassam Z, Fagan A, Gavis EA, Liu E, Cox IJ, Kheradman R, Heuman D, Wang J, Gurry T, et al. Fecal microbiota transplant from a rational stool donor improves hepatic encephalopathy: A randomized clinical trial. Hepatology. 2017;66(6):1727–1738. doi: 10.1002/hep.29306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen L, Wilson, J.E., Koenigsknecht, M.J., Chou, W.C., Montgomery, S.A., Truax, A.D., Brickey, W.J., Packey, C.D., Maharshak, N., Matsushima, G.K., and Plevy, S.E. NLRP12 attenuates colon inflammation by maintaining colonic microbial diversity and promoting protective commensal bacterial growth. Nat Immunol. 2017;18(5):541–551. doi: 10.1038/ni.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bertola A, Mathews S, Ki SH, Wang H, Gao B. Mouse model of chronic and binge ethanol feeding (the NIAAA model). Nat Protoc. 2013;8(3):627–637. doi: 10.1038/nprot.2013.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lawley B, Tannock GW. Analysis of 16S rRNA gene amplicon sequences using the QIIME software package. Methods Mol Biol. 2017;1537:153–163. [DOI] [PubMed] [Google Scholar]
- 51.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
16S rRNA sequence data can be accessed from NCBI BioProjectPRJNA543718. Our analysis use standard, open source software. R packages are available on CRAN (cran.r-project.org), Galaxy (usegalaxy.org), and the Python programming language (Python version 3.6).


