ABSTRACT
Bacteria in human milk could directly seed the infant intestinal microbiota, while information about how milk microbiota develops during lactation and how geographic location, gestational hypertensive status, and maternal age influence this process is limited. Here, we collected human milk samples from mothers of term infants at the first day, 2 weeks, and 6 weeks postpartum from 117 longitudinally followed-up mothers (age: 28.7 ± 3.6 y) recruited from three cities in China. We found that milk microbial diversity and richness were the highest in colostrum but gradually decreased over lactation. Microbial composition changed across lactation and exhibited more discrete compositional patterns in 2-week and 6-week milk samples compared with colostrum samples. At phylum level, the abundance of Proteobacteria increased during lactation, while Firmicutes showed the opposite trend. At genus level, Staphylococcus, Streptococcus, Acinetobacter, Pseudomonas, and Lactobacillus were predominant in colostrum samples and showed distinct variations across lactation. Maternal geographic location was significantly associated with the milk microbiota development and the abundance of predominant genus. In addition, milk from mothers with gestational prehypertension had a different and less diverse microbial community at genus level in early lactation times, and contained less Lactobacillus in the 2-week milk samples than those from normotensive mothers. Findings of our study outlined the human milk microbial diversity and community development over lactation, and underscored the importance of maternal geographic locations and gestational hypertensive status on milk microbiota, which might have important implications in the establishment of the infant intestinal microbiota via breastfeeding.
KEYWORDS: Human milk, milk microbiota, maternal factors, gestational blood pressure, lactation
Introduction
Despite previously considered sterile, breast milk is now suggested as a source of bacteria and able to seed the neonatal gut microbiota.1 The early microbial colonization of the infant’s gut plays an essential role in gut maturation, and metabolic and immunologic programming, which has important consequences for early life and long-term health.2 Breastfeeding has a beneficial impact on the establishment of the newborn intestinal microbiome, compared with non-breast-fed infants.3 Breast-fed neonates are likely to harbor more beneficial bacterial populations such as Bifidobacterium and Lactobacillus, and less potentially pathogenic populations such as Clostridium, as compared with formula-fed neonates.3,4 In early life, this beneficial impact of breastfeeding is associated with reduced diarrhea-related gut microbiota dysbiosis, and reduced incidence and severity of infections and metabolic diseases in later life.3–5
Maternal geographic location, diet, age, and stage of lactation are known to influence the nutrients, bioactive compounds, and immunological factors contained in breast milk.6–10 The cytokine content differs in milk depending on the gestational health status, with breast milk from hypertensive mothers having higher levels of pro-inflammatory cytokines than that from normotensive mothers.11 We hypothesized that physiological differences and potential inflammatory triggers that influence human milk composition might also have an impact on bacteria growth. Some studies have investigated the association between maternal geographic location, health status, and breast milk microbiota.12–15 However, cross-sectional design, small sample size, pooled sample collection from different lactation stages, and controversial results are likely to limit the interpretation of the previous findings.
Therefore, we aimed to characterize the human milk microbial diversity and community development during lactation, and examine the associations of maternal geographic location, diet, age, and gestational hypertensive status with milk microbiota in a longitudinally followed-up study.
Results
We collected 117, 113, and 104 milk samples from mothers of term infants at the first day, 2 weeks, and 6 weeks postpartum, respectively (Figure 1). Baseline characteristics and dietary intakes are shown in Table 1. Among all the mothers included in the present study, 34.2% were diagnosed as gestational prehypertension and 27.4% were aged more than 30 years old. After quality filtering and length trimming, 14,968,717 16 S rRNA sequences (total sample n = 334) were analyzed, with an average of 44,816 ± 9,684 sequences per sample.
Figure 1.
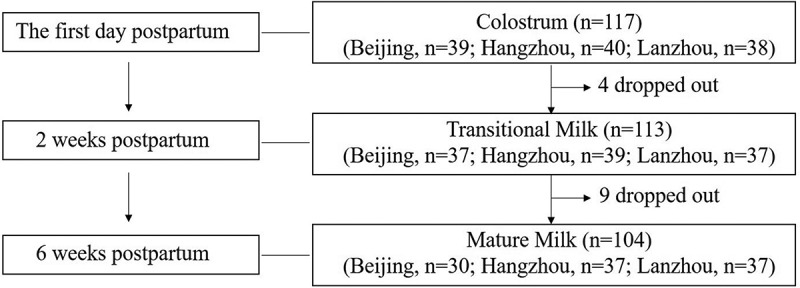
Flow chart of human milk samples collection and measurements during lactation.
Table 1.
Characteristics of maternal demographic and metabolic variables.
| All (n = 117) | Beijing (n = 39) | Hangzhou (n = 40) | Lanzhou (n = 38) | p value† | |
|---|---|---|---|---|---|
| Baseline variables | |||||
| Age (years) | 28.7 ± 3.6 | 29.5 ± 2.2‡ | 29.4 ± 4.3 | 27.1 ± 3.6 | 0.005 |
| Height (cm) | 162 ± 4.5 | 164.1 ± 3.3‡ | 160.2 ± 5.3 | 162.1 ± 3.8 | <0.001 |
| Weight (kg) | 68.2 ± 11.4 | 70.7 ± 8.6 | 64.8 ± 14.3 | 69.3 ± 9.7 | 0.06 |
| BMI (kg/m2) | 25.9 ± 4.1 | 26.2 ± 2.9 | 26.4 ± 5.3 | 26.4 ± 3.5 | 0.41 |
| SBP (mm Hg) | 115.3 ± 12.5 | 115.1 ± 7.2 | 116.9 ± 17.1 | 114.0 ± 11.0 | 0.05 |
| DBP (mm Hg) | 73.2 ± 9.5 | 70.1 ± 7.6 | 74.5 ± 11.2 | 74.9 ± 8.7 | 0.60 |
| Baseline dietary intakes | |||||
| Total energy (kcal) | 1734.2 ± 255.2 | 1759.8 ± 306.9 | 1657.4 ± 178.2 | 1788.8 ± 252.8 | 0.06 |
| Fat (g) | 54.8 ± 11.2 | 60.5 ± 13.8‡ | 50.7 ± 8.0 | 53.3 ± 8.5 | <0.001 |
| Carbohydrate (g) | 237.1 ± 45.9 | 224.9 ± 46.7§ | 233.1 ± 44.7 | 254.0 ± 42.4 | 0.02 |
| Protein (g) | 67.2 ± 17.2 | 75.6 ± 21.5‡§ | 60.5 ± 10.5 | 65.7 ± 14.6 | <0.001 |
| Grains (g) | 267.4 ± 53.7 | 249.3 ± 55.9‡ | 277.6 ± 54.8 | 275.4 ± 46.5 | 0.03 |
| Vegetables (g) | 105.3 ± 80.5 | 94.4 ± 63.3§ | 67.3 ± 42.1 | 156.5 ± 99.8¶ | <0.001 |
| Fruits (g) | 107.3 ± 124.6 | 136.1 ± 140.3‡ | 34.6 ± 66.8 | 154.3 ± 122.1¶ | <0.001 |
| Soybean products (g) | 15.3 ± 22.5 | 23.5 ± 29.9‡ | 3.5 ± 7.9 | 19.3 ± 19.0¶ | <0.001 |
| Dairy products (g) | 97.7 ± 114.5 | 67.5 ± 117.3‡ | 53.1 ± 74.9 | 175.5 ± 108.4¶ | <0.001 |
| Eggs and meat (g) | 180.1 ± 76.7 | 212.5 ± 88.8‡§ | 164.1 ± 66.6 | 132.9 ± 48.2 | <0.001 |
| Oils (g) | 28.9 ± 6.2 | 27.9 ± 5.7 | 30.9 ± 6.2 | 27.7 ± 6.2 | 0.04 |
| Dropout rate in 2 weeks | 3.4% | 5.1% | 2.5% | 2.6% | 0.99 |
| Dropout rate in 6 weeks | 11.1% | 23.1% | 7.5% | 2.6% | 0.50 |
*Plus-minus values are means ± SD.
†p values for differences among the three geographic locations were calculated by analysis of variance. When the difference among the regions was significant (p < 0.05), all pairwise comparisons tested for significance with the use of Turkey’s Studentized range test.
‡The value for the consumption in Beijing is significantly different from the value in Hangzhou (p < 0.05).
§ The value for the consumption in Beijing is significantly different from the value in Lanzhou (p < 0.05).
¶ The value for the consumption in Lanzhou is significantly different from the value in Hangzhou (p < 0.05).
BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure.
Human milk microbial diversity and community across lactation time
Human milk microbial diversity (estimated by Shannon index) and richness (estimated by Chao index) were the highest in colostrum, and then decreased across lactation time (both p for time <0.001) (Figure 2(a,b)). The overall microbial community in human milk also differed across the three time points at both phylum and genus levels (both PERMANOVA p for time <0.001) (Figure 2(c,d)). Clustering of colostrum showed a different bacterial composition in comparison with transitional and mature milk samples. At phylum level, Proteobacteria and Firmicutes were dominant in milk samples throughout the lactation period (Figure 3(a)). Firmicutes was dominant in the colostrum samples (52.6%), and then declined to 32.6% in transitional milk samples, and to 31.2% in the mature milk samples (p for time <0.001). In contrast, the relative abundance of Proteobacteria increased from 28.6% in colostrum samples to 52.7% in transitional milk samples, and then to 58.6% in mature milk samples (p for time <0.001). At genus level, Staphylococcus, Streptococcus, Lactobacillus, Acinetobacter, and Pseudomonas were identified as “predominant colonizers” with relative abundances being more than 5% in colostrum samples, and then showed significantly distinct fluctuation across lactation time (Figure 3(b)).
Figure 2.
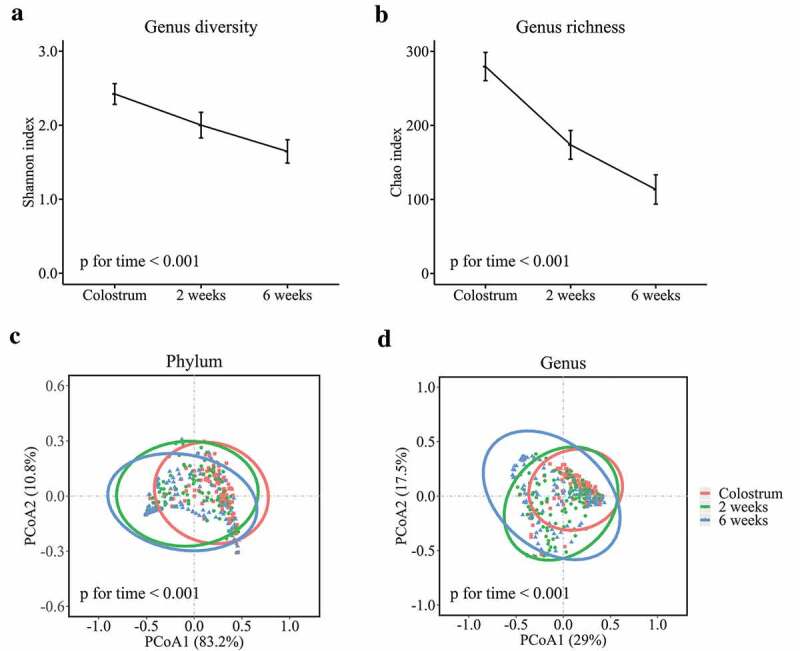
Human milk microbial α diversity at genus level and overall microbial composition during lactation. Microbial diversity calculated by Shannon index (a) and richness calculated by Chao index (b) during lactation. PCoA score plots based on Bray–Curtis distance at phylum (c) and genus (d) levels.
Figure 3.
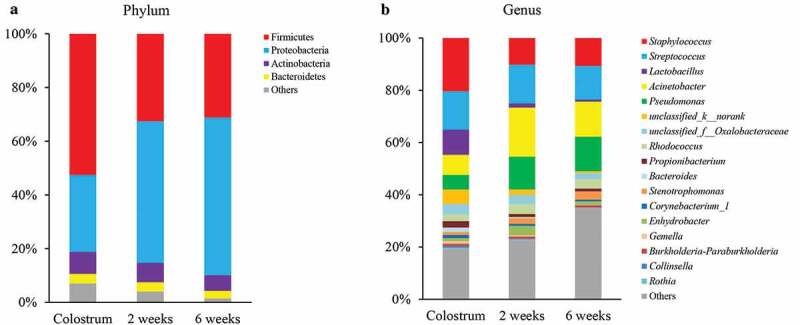
Human milk microbial composition during lactation. (a) at phylum level and (b) at genus level.
Milk microbial diversity and richness, and maternal factors
Milk microbial diversity and richness across the lactation time were affected by maternal geographic location (p for interaction <0.001 for diversity and 0.02 for richness). The milk microbial diversity and richness were significantly different among the three geographic locations in colostrum and mature milk samples (Figure 4(a)). Although gestational hypertensive status was not associated with milk microbial diversity and richness change during lactation time (p for interaction = 0.63 for diversity and 0.91 for richness), lower diversity and richness were observed in milk samples from mothers with gestational prehypertension at each time pointcompared with those from normotensive mothers (Figure 4(b)). We did not find any association between maternal age and milk microbial diversity and richness change over the lactation period (p for interaction = 0.35 for diversity and 0.79 for richness). The milk microbial diversity and richness also did not differ between samples from mothers aged ≤30 and >30 at each lactation time point (Figure 4(c)).
Figure 4.
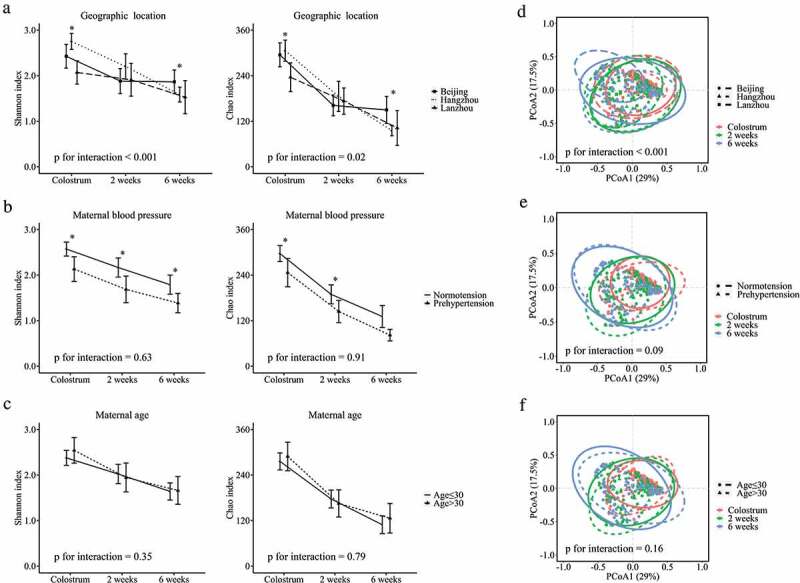
Influence of maternal prenatal factors on human milk microbial α diversity and overall microbial composition at genus level during lactation. (a) geographic location, (b) maternal blood pressure, and (c) maternal age on microbial diversity and richness. (d) geographic location, (e) maternal blood pressure, and (f) maternal age on overall microbial composition at genus level. * p < .05.
Milk microbial community and maternal factors
The association between maternal factors and overall milk microbial community at genus level through lactation is shown in Figure 4(d–f). The maternal geographic location was significantly associated with overall milk microbial community development across lactation time (PERMANOVA p for interaction <0.001, Figure 4(d)). At each lactation time, different overall milk microbial communities were observed in milk samples from the three cities (PERMANOVA p < .001, = 0.003, and <0.001 at the first day, 2 weeks, and 6 weeks postpartum, respectively, Figure S1A-C). The “predominant colonizers” Staphylococcus, Streptococcus, Lactobacillus, and Acinetobacter development were also influenced by maternal geographic location (all p for interaction <0.05) (Figure 5(a–d)). The major difference of these “predominant colonizers” among three cities was observed in mature milk samples.
Figure 5.
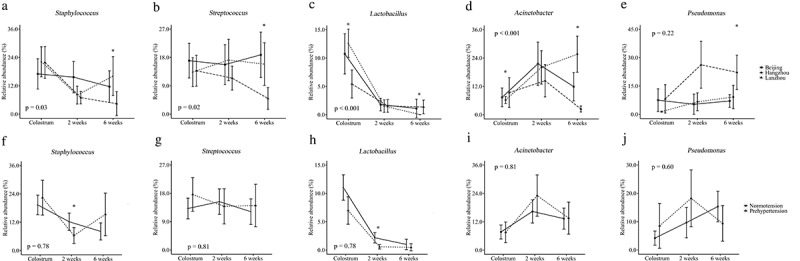
Influence of geographic location and gestational hypertensive status on “predominant colonizers” over lactation at genus level. Geographic location on Staphylococcus (a), Streptococcus (b), Lactobacillus (c), Acinetobacter (d), and Pseudomonas (e). Gestational hypertensive status on Staphylococcus (f), Streptococcus (g), Lactobacillus (h), Acinetobacter (i), and Pseudomonas (j). The p values at the lower or higher left are for the interaction term of time and maternal factor. * p < .05 after FDR correction.
Given the large differences in maternal diet among geographic locations, we assessed the possible influence of maternal dietary intakes on the milk microbiota in colostrum samples. We found no statistically significant association between dietary factors and human milk microbiota composition at genus level (Table S1). We also did not find significant correlations between dietary factors and the five “predominant colonizers” after FDR correction (Table S2).
Although the gestational hypertensive status was not statistically significantly associated with the development of breast milk microbial community over lactation time (PERMANOVA p for interaction = 0.09) (Figure 4(e)), separation of milk microbial communities was observed in colostrum and transitional milk samples from normotensive mothers and mothers with gestational prehypertension (PERMANOVA p = .06 for colostrum and 0.02 for transitional milk) (Figure S1D and E). Additionally, we found that the abundance of Lactobacillus was lower in colostrum (p = .09) and transitional (p = .004) milk samples from mothers with gestational prehypertension compared to normotensive mothers (Figure 5(h)). However, maternal age seemed to have no statistically significant influence on milk microbial community development nor early “predominant colonizers” during lactation (Figure 4(f), Figure S1G-I, and Figure S2).
Discussion
Our longitudinal study provided a new insight into human milk microbiota development throughout the lactation period, and emphasized the importance of maternal geographic location and gestational hypertensive status on human milk microbial diversity and community.
Similar to the nutrients and bioactive components in breast milk, the human milk microbial diversity and richness gradually decreased through lactation.16–18 This trend was also in line with previous cross-sectional studies indicating that the milk microbial diversity was higher in colostrum samples than that in transitional and mature milk samples.19,20 Higher milk microbial diversity and richness observed in colostrum samples might be due to the fact that colostrum had more diverse nutrients and bioactive properties, which could feed the microbes.21 During lactation, milk microbial diversity and richness were associated with geographic location and gestational hypertensive status, with lower level being observed in samples from mothers with gestational prehypertension relative to those from normotensive mothers. Recent evidence has shown that breast milk microbiota can directly seed the infant gut microbiota, the bacterial diversity and composition changes of which were associated with the proportion of daily breast milk consumption in a dose-dependent manner even after the introduction of solid foods.1 Therefore, breast milk with lower microbial diversity might result in lower gut microbiota diversity in infants through breastfeeding. Low infant gut microbial diversity has been reported to be correlated with the risk of necrotizing enterocolitis in early infancy and asthma risk at 7 y of age.22,23
At the phylum level, Firmicutes and Proteobacteria were dominant in the colostrum samples and followed by Actinobacteria and Bacteroidetes, consistent with previous high-throughput studies performed in Canada and Switzerland.24,25 We further extended previous findings by more frequent sampling at the subsequent 2 weeks and 6 weeks postpartum. In particular, the relative abundance of Proteobacteria increased over the lactation period and accounted for 58.6% in mature milk samples. In contrast, the abundance of Proteobacteria was found in very low abundance compared with Firmicutes in the stools of newborn, infants, or healthy adults.26 This might suggest a selective process by which only specific taxa colonized the neonatal gut. One possible reason might be that some certain bacteria in human milk have little or limited effects on the offspring but are present for the mothers' health.27 However, it should be noted that the abundance of Proteobacteria was quite high in the stools of newborns (16%) when compared with the healthy adults (4.5%).26 The neonatal gut is frequently colonized by facultative anaerobes, mainly Proteobacteria species, in the first week of life due to the oxygen abundance.26,28 It could be speculated that the high abundance of Proteobacteria in colostrum might play as the predominant colonizer in shaping the neonatal gut microbial environment via breastfeeding in early life. These facultative anaerobes could help to make the habitat more suitable for successive colonization of strict anaerobe by consuming oxygen, altering the pH, and producing carbon dioxide and nutrients in neonatal gut.26 Further studies are needed to address how and to what degree human milk microbiota colonize the neonatal gut and the mechanism of the selective colonization process in the future.
At genus level, Staphylococcus was particularly high in proportion in the colostrum samples that was likewise consistent with previous studies,25,29 and followed by a sharp decrease thereafter, which was in a manner similar to what had been reported in the infant intestine.30 High proportion of Staphylococcus was found in the intestine of newborns, but their number started to decrease after the first week of life.30 Staphylococcus and Lactobacillus have been reported to be more abundant in the intestine of breast-fed neonates when compared with formula-fed ones,30 suggesting that breast milk feeding appears to be in favor of colonization of the “predominant colonizers” in the gut of the neonates. In contrast, the abundance of Streptococcus remained stable during lactation. These “predominant colonizers” in the human milk are mostly lactose utilizers that could produce lactate. Lactate could be fermented by Lactobacillus, Propionibacteria, and Veillonella to produce propionate, while the accumulation of lactate is likely to confer unfavorable effects.31 The “predominant colonizers” in human milk might play an essential role in the establishment and development of a balanced trophic chain in the neonatal gut via breastfeeding.32 The trophic functions might contribute to a healthy colonization in neonatal gut and might further prevent disorders such as ulcerative colitis resulting from intestinal lactate accumulation.31,33 A recent study has shown that the bacteria from mother’s breast milk and skin were most prominent in their infants’ gut in the first month of life, nearly accounting for 38% of the gut bacteria in primarily breast-fed infants.1 The early “predominant colonizers” in human milk appear to have an ecological advantage over those arriving later and might improve the intestinal microenvironment through breastfeeding, paving the way for the succesivecolonization of other microorganisms in the neonates’ gut.
The geographic location is known to influence the infant gut microbiota composition, which is associated with health status in the later life.34–36 In the present study, we found that geographic location also had an impact on milk microbiota diversity and taxonomic composition. The difference was present in the colostrum samples and maintained in 2-week and 6-week milk samples, indicating that the impact of geographic location had a long-term effect on milk microbial community development. Microbial community found in our milk samples were different from those in samples collected in America or European countries, also suggesting that maternal geographic location was able to influence the milk microbial community.19,29 Additionally, long-term maternal dietary choices and probiotic supplements have been suggested to influence human milk microbiota.19,37 One randomized controlled trial showed maternal dietary supplementation of certain probiotic bacteria (L. fermentum and L. salivarius) was able to modify the milk microbiota and could be used as an effective alternative to antibiotics for the treatment of infectious mastitis during lactation.37 In our study, although maternal diet showed significant differences among the three cities, we did not observe the association between dietary intake and milk microbiota. It should be noted that our 1-day 24 h record might not be able to capture the long-term dietary intake of the mothers and we only collected dietary intake information before lactation. Further studies with more detailed dietary intake information, especially during lactation, are needed to explore whether and to what degree the maternal diet could influence the human milk microbiota development.
Gestational hypertensive status was another maternal prenatal factor that could influence the milk microbiota in our study. Notably, the milk microbial diversity and relative abundance of Lactobacillus were lower in colostrum and transitional milk samples from mothers with gestational prehypertension compared to those from normotensive mothers. Lactobacillus has been considered a beneficial population in human milk and has been regarded as a hallmark of the gut microbiota in healthy breast-fed infants as well as Bifidobacteria.31 Infants fed with breast milk lower in Lactobacillus might establish a gut microbial community lower in Lactobacillus, which was associated with a higher risk of gastrointestinal infections.38,39 In contrast, administration of Lactobacillus strain from human milk to 6-month infants could lead to reductions of 27%, 46%, and 30% in the risk of upper respiratory tract infections, gastrointestinal infections, and total number of infections, respectively.40 Prehypertension is not simply a transitional stage between normotensive and hypertensive status in terms of blood pressure levels but rather a state in which gut dysbiosis has already occurred.41 Entero-mammary translocation of the maternal gut microbiota has been proposed to be one of the pathways to explain the origin of the milk microbiota.42 Considering the source of bacteria in milk, it therefore can be speculated that prehypertension that led to the dysbiosis of the mother’s gut microbiota could adversely affect the milk microbiota. Milk microbiota from mothers with prehypertension during pregnancy might act as a potentially unfavorable inoculum for the gut microbiota in infants via breastfeeding.41,43-45 Given that the bacteria in human milk are among the first entering the infant gut, the aberrant milk microbial composition might result inan abnormal assembly of the gut microbiome in infant via breastfeeding and might lead to unfavorable health consequence in infants’ early and later life.
The main strengths of our study include large sample size and longitudinally followed-up study design. In contrast to previous studies, we collected human milk samples from different lactating stages, allowing us to observe a more complete picture of changes in milk microbiota during lactation. Nevertheless, several limitations of this study should be noted. Negative control was not used to measure potential reagent or collection contamination.We cannot fully exclude the possible contribution from skin-associated bacteria and other contaminants. However, it provides us a representative picture of the microbiota that infants ingest during breastfeeding, which might be more biologically relevant to infant health in early life. Other limitations include the lack of information regarding maternal factors such as probiotic intakes, delivery mode, and breastfeeding practices. Breastfeeding practices have been reported to be a determinant of milk microbiota composition, probably due to the exposure of breast to the infant oral cavity which could retrogradely impact the milk microbiota.15 Finally, 16S rRNA gene sequencing has limited capacity to guarantee species identity, further metagenomic studies are required to identify taxa down to species and assess the corresponding function.
In summary, our study outlined the human milk microbiota development over lactation, and underscored the importance of maternal geographic location and gestational hypertensive status on milk microbiota development which might have important relevance in the establishment of the infant gut microbiota via breastfeeding. Further studies are needed to address how human milk microbiota influences infant gut development as well as long-term health.
Subjects and methods
Subjects
Pregnant women with full-term delivery gestation (from 37 to 42 weeks of gestation) were recruited from Women’s Hospital, School of Medicine, Zhejiang University in Hangzhou City (coastal city in southeast China), the Second People’s Hospital of Gansu Province in Lanzhou City (inland city in northwest China), and Chinese PLA General Hospital (inland city in the northeast China). Key inclusion criteria included age range between 20 and 40 years old, body mass index before pregnancy <24 (cutoff point according to the Chinese criteria for overweight), no gestational diabetes, no blood lipid abnormalities before the 20th gestational week, and gestational systolic blood pressure <130 mm Hg and diastolic blood pressure <90 mm Hg before the 20th gestational week. Gestational prehypertension was defined as normotensive before the 20th gestational week but prehypertensive (systolic blood pressure ≥130 mm Hg or diastolic blood pressure ≥80 mm Hg) after 20th gestational week.46 Participants were excluded if they were multiparous mothers or suffering from mastitis or took antibiotics or apparent change in the living environment and dietary habits during lactation. Dietary intakes were calculated by a 1-d 24 h dietary record prior to colostrum samples collection day. All women had self-selected diets without being given any dietary recommendations before sample collection. The study protocol was approved by the Ethics Committee of the College of Biosystem Engineering and Food Science, Zhejiang University, China. Each woman provided written informed consent.
Human milk collection and measurements
Human milk samples were collected on the first day (colostrum), 2 weeks (transitional milk), and 6 weeks (mature milk) postpartum between 10 am and 11 am before the baby feeding on each collection day. All the breast milk samples were collected manually wearing sterile gloves, and aliquoted into sterile tubes with a volume of 2 ml to 5 ml, and then stored at −20°C. After being transported to the laboratory all the human milk samples were then stored at −80°C until analysis. Gestational blood pressure was measured twice (separated by 10–15 minutes) after 5 minutes of rest with the use of OMRON HEM-7112 device (OMRON Corp., Tokyo, Japan) which records blood pressure using an oscillometric technique. Each measurement was obtained on the right arm of participants in a seated position, using an appropriate-sized cuff at the level of the heart. Traditional blood lipid fractions and blood glucose were measured by automatic biochemical analyzer (Siemens Corp., Erlangen, Germany).
Breast milk microbial DNA extraction, 16S rRNA gene sequencing, and bioinformatics
Before analysis, the human milk samples were thawed and centrifuged at 14,000 rpm for 15 mins at 4°C to separate cells and fat from whey. Thereafter, total DNA was isolated from the pellets by using the MP FastDNA® Spin Kit (MP Biomedicals, Solon, OH, USA) according to manufacturer’s protocols. The final DNA concentration and purification were determined by NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, USA), and DNA quality was checked by 1% agarose gel electrophoresis. The V3-V4 hypervariable regions of the bacteria 16S rRNA gene were amplified with barcode index primers 338F (5'-ACTCCTACGGGAGGCAGCAG-3') and 806R (5'-GGACTACHVGGGTWTCTAAT-3'). The purified amplicons were pooled in equimolar concentration and further paired-end sequencing was performed using an Illumina Miseq instrument (Illumina, San Diego, California, USA) as previous published paper.47 The raw reads were deposited into the NCBI Sequence Read Archive database (Accession Number: PRJNA495111).
Overlapping paired-end reads from the original DNA fragments were merged using FLASH.48 Reads were assigned to each sample according to the unique barcode of each sample and quality-filtered using QIIME. The operational taxonomic unit was clustered with 97% similarity cutoff using UPARSE Pipeline, and chimeric sequences were identified and removed using UCHIME and Silva (SSU123) 16S rRNA database.49,50 Taxonomic ranks were assigned to each sequence using Ribosomal Database Project. In order to calculate α diversity at the genus level, two metrics were applied with Shannon index and Chao index to estimate microbial community diversity and richness, respectively.
Statistical analysis
Linear mixed regression was performed to assess the changing trends of Shannon index and Chao index over lactation time. Principal coordinate analysis (PCoA) based on Bray–Curtis distance and permutational multivariate analysis of variance (PERMANOVA) were performed to compare the overall microbiota composition across lactation time. In the analysis of individual gut microbial taxon through lactation time, data were arcsine square root transformed and analyzed by the linear mixed model.51 When examining the influence of maternal prenatal factors (including geographic location, gestational hypertensive status, and maternal age) on the changing trend of α diversity, overall microbial composition, and individual gut microbial taxa, we included the maternal factors and its interaction term with time in the model. The interaction term provides a test of the hypothesis that the changing trend of α diversity, overall microbial composition, and individual gut microbial taxa over time does not differ by the maternal factor. At each lactation time, whether microbial α diversity or individual genera was influenced by maternal prenatal factors was assessed using Mann–Whitney U test (for hypertensive status and age) or Kruskal–Wallis test (for maternal geographic location), while the influence of maternal prenatal factors on overall microbial composition was examined by PERMANOVA. We also assessed the association between dietary intake and overall microbial composition in colostrum using PERMANOVA and assessed the correlation between dietary intake and individual genera using Spearman’s rank or Pearson’s correlation coefficients depending on the distribution of data. Benjamini-Hochberg’s false discovery rate (FDR) method was used to correct multiple comparisons for all analyses of individual gut microbial taxa. All the statistical analyses were performed in R version 3.5. The main R packages used were ”ape” and ”vegan” for PCoA and PERMANOVA, and “lmerTest” for linear mixed regression.
Supplementary Material
Author contributions
The authors’ contributions were as follows—YW, JJ, ML, and DL: conceived and designed the research; YW, JJ, ML, WT, RZ, JL, JY, and DL: conducted the research; YW, WT, and RZ: analyzed the data; YW: wrote the first draft of the manuscript; JJ, ML, WT, RZ, JL, JY, FW, and DL revised the manuscript; DL: had full access to all data and had final responsibility for the decision to submit for publication; and all authors: interpreted the data and revised the manuscript for important intellectual content and approved the final draft.
Funding Statement
This work was supported by the China Postdoctoral Science Foundation [2019T12058]; National Basic Research Program of China (973 Program) [2015CB663604].
Disclosure of potential conflicts of interest
The authors declare no conflicts of interest.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Abbreviations
- PCoA
principal coordinate analysis
- PERMANOVA
permutational multivariate analysis of variance
- FDR
false discovery rate
References
- 1.Pannaraj PS, Li F, Cerini C, Bender JM, Yang S, Rollie A, Adisetiyo H, Zabih S, Lincez PJ, Bittinger K, et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 2017;171(7):647–654. doi: 10.1001/jamapediatrics.2017.0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maynard CL, Elson CO, Hatton RD, Weaver CT.. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489(7415):231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jost T, Lacroix C, Braegger C, Chassard C. Impact of human milk bacteria and oligosaccharides on neonatal gut microbiota establishment and gut health. Nutr Rev. 2015;73(7):426–437. doi: 10.1093/nutrit/nuu016. [DOI] [PubMed] [Google Scholar]
- 4.Hosea BH, Cicalo MC, Holland CD, Field CJ. The immunological components of human milk. Adv Food Nutr Res. 2008;54:45–80. doi: 10.1016/S1043-4526(07)00002-2. [DOI] [PubMed] [Google Scholar]
- 5.Ho NT, Li F, Lee-Sarwar KA, Tun HM, Brown BP, Pannaraj PS, Bender JM, Azad MB, Thompson AL, Weiss ST, et al. Meta-analysis of effects of exclusive breastfeeding on infant gut microbiota across populations. Nat Commun. 2018;9(1):4169. doi: 10.1038/s41467-018-06473-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nunes M, Da SC, Bosa VL, Bernardi JR, Werlang I, Goldani MZ. Could a remarkable decrease in leptin and insulin levels from colostrum to mature milk contribute to early growth catch-up of SGA infants? BMC Pregnancy Childbirth. 2017;17(1):410. doi: 10.1186/s12884-017-1593-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Innis SM. Human milk: maternal dietary lipids and infant development. Proc Nutr Soc. 2007;66(3):397–404. doi: 10.1017/S0029665107005666. [DOI] [PubMed] [Google Scholar]
- 8.Mehta R, Petrova A. Biologically active breast milk proteins in association with very preterm delivery and stage of lactation. J Perinatol. 2011;31(1):58–62. doi: 10.1038/jp.2010.68. [DOI] [PubMed] [Google Scholar]
- 9.Koenig A, de Albuquerque DE, Barbosa SF, Vaz FA. Immunologic factors in human milk: the effects of gestational age and pasteurization. J Hum Lact. 2005;21(4):439–443. doi: 10.1177/0890334405280652. [DOI] [PubMed] [Google Scholar]
- 10.Ruan C, Liu X, Man H, Ma X, Lu G, Duan G, DeFrancesco CA, Connor WE. Milk composition in women from five different regions of China: the great diversity of milk fatty acids. J Nutr. 1995;125(12):2993–2998. doi: 10.1093/jn/125.12.2993. [DOI] [PubMed] [Google Scholar]
- 11.Zambruni M, Villalobos A, Somasunderam A, Westergaard S, Nigalye M, Turin CG, Zegarra J, Bellomo S, Mercado E, Ochoa TJ, et al. Maternal and pregnancy-related factors affecting human milk cytokines among Peruvian mothers bearing low-birth-weight neonates. J Reprod Immunol. 2017;120:20–26. doi: 10.1016/j.jri.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khodayar-Pardo P, Mira-Pascual L, Collado MC, Martinez-Costa C. Impact of lactation stage, gestational age and mode of delivery on breast milk microbiota. J Perinatol. 2014;34(8):599–605. doi: 10.1038/jp.2014.47. [DOI] [PubMed] [Google Scholar]
- 13.Kumar H, Du Toit E, Kulkarni A, Aakko J, Linderborg KM, Zhang Y, Nicol MP, Isolauri E, Yang B, Collado MC, et al. Distinct patterns in human milk microbiota and fatty acid profiles across specific geographic locations. Front Microbiol. 2016;7:1619. doi: 10.3389/fmicb.2016.01619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dave V, Street K, Francis S, Bradman A, Riley L, Eskenazi B, Holland N. Bacterial microbiome of breast milk and child saliva from low-income Mexican-American women and children. Pediatr Res. 2016;79(6):846–854. doi: 10.1038/pr.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moossavi S, Sepehri S, Robertson B, Bode L, Goruk S, Field CJ, Lix LM, de Souza RJ, Becker AB, Mandhane PJ, et al. Composition and variation of the human milk microbiota are influenced by maternal and early-life factors. Cell Host Microbe. 2019;25(2):324–335. doi: 10.1016/j.chom.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 16.Jiang J, Xiao H, Wu K, Yu Z, Ren Y, Zhao Y, Li K, Li J, Li D. Retinol and alpha-tocopherol in human milk and their relationship with dietary intake during lactation. Food Funct. 2016;7(4):1985–1991. doi: 10.1039/c5fo01293g. [DOI] [PubMed] [Google Scholar]
- 17.Xu G, Davis JC, Goonatilleke E, Smilowitz JT, German JB, Lebrilla CB. Absolute quantitation of human milk oligosaccharides reveals phenotypic variations during lactation. J Nutr. 2017;147(1):117–124. doi: 10.3945/jn.116.238279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao Y, Weber D, Xu W, Durbin-Johnson BP, Phinney BS, Lonnerdal B. Absolute quantification of human milk caseins and the whey/casein ratio during the first year of lactation. J Proteome Res. 2017;16(11):4113–4121. doi: 10.1021/acs.jproteome.7b00486. [DOI] [PubMed] [Google Scholar]
- 19.Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr. 2012;96(3):544–551. doi: 10.3945/ajcn.112.037382. [DOI] [PubMed] [Google Scholar]
- 20.Murphy K, Curley D, O’Callaghan TF, O’Shea CA, Dempsey EM, O’Toole PW, Ross RP, Ryan CA, Stanton C. The composition of human milk and infant faecal microbiota over the first three months of life: a pilot study. Sci Rep. 2017;7:40597. doi: 10.1038/srep40597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tripathi V. Bioactive compounds of colostrum and its application. Food Rev Int. 2006;3(22):225–244. doi: 10.1080/87559120600694606. [DOI] [Google Scholar]
- 22.Warner BB, Deych E, Zhou Y, Hall-Moore C, Weinstock GM, Sodergren E, Shaikh N, Hoffmann JA, Linneman LA, Hamvas A, et al. Gut bacteria dysbiosis and necrotising enterocolitis in very low birthweight infants: a prospective case-control study. Lancet. 2016;387(10031):1928–1936. doi: 10.1016/S0140-6736(16)00081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abrahamsson TR, Jakobsson HE, Andersson AF, Bjorksten B, Engstrand L, Jenmalm MC. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy. 2014;44(6):842–850. doi: 10.1111/cea.12253. [DOI] [PubMed] [Google Scholar]
- 24.Jost T, Lacroix C, Braegger C, Chassard C. Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br J Nutr. 2013;110(7):1253–1262. doi: 10.1017/S0007114513000597. [DOI] [PubMed] [Google Scholar]
- 25.Urbaniak C, Angelini M, Gloor GB, Reid G. Human milk microbiota profiles in relation to birthing method, gestation and infant gender. Microbiome. 2016;4:1. doi: 10.1186/s40168-015-0145-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33(9):496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 27.Heikkila MP, Saris PE. Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J Appl Microbiol. 2003;95(3):471–478. doi: 10.1046/j.1365-2672.2003.02002.x. [DOI] [PubMed] [Google Scholar]
- 28.Guaraldi F, Salvatori G. Effect of breast and formula feeding on gut microbiota shaping in newborns. Front Cell Infect Microbiol. 2012;2:94. doi: 10.3389/fcimb.2012.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boix-Amoros A, Collado MC, Mira A. Relationship between milk microbiota, bacterial load, macronutrients, and human cells during lactation. Front Microbiol. 2016;7:492. doi: 10.3389/fmicb.2016.00492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jost T, Lacroix C, Braegger CP, Chassard C. New insights in gut microbiota establishment in healthy breast fed neonates. PLoS One. 2012;7(8):e44595. doi: 10.1371/journal.pone.0044595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chassard C, de Wouters T, Lacroix C. Probiotics tailored to the infant: a window of opportunity. Curr Opin Biotechnol. 2014;26:141–147. doi: 10.1016/j.copbio.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 32.Duncan SH, Louis P, Flint HJ. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol. 2004;70(10):5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vernia P, Caprilli R, Latella G, Barbetti F, Magliocca FM, Cittadini M. Fecal lactate and ulcerative colitis. Gastroenterology. 1988;95(6):1564–1568. doi: 10.1016/s0016-5085(88)80078-7. [DOI] [PubMed] [Google Scholar]
- 34.Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Acta Paediatr. 2009;98(2):229–238. doi: 10.1111/j.1651-2227.2008.01060.x. [DOI] [PubMed] [Google Scholar]
- 35.Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F, Adams H, van Ree R, Stobberingh EE. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA birth cohort study. Gut. 2007;56(5):661–667. doi: 10.1136/gut.2006.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sjogren YM, Jenmalm MC, Bottcher MF, Bjorksten B, Sverremark-Ekstrom E. Altered early infant gut microbiota in children developing allergy up to 5 years of age. Clin Exp Allergy. 2009;39(4):518–526. doi: 10.1111/j.1365-2222.2008.03156.x. [DOI] [PubMed] [Google Scholar]
- 37.Arroyo R, Martin V, Maldonado A, Jimenez E, Fernandez L, Rodriguez JM. Treatment of infectious mastitis during lactation: antibiotics versus oral administration of Lactobacilli isolated from breast milk. Clin Infect Dis. 2010;50(12):1551–1558. doi: 10.1086/652763. [DOI] [PubMed] [Google Scholar]
- 38.Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. 1999;69(5):1035S–1045S. doi: 10.1093/ajcn/69.5.1035s. [DOI] [PubMed] [Google Scholar]
- 39.Wold AE, Adlerberth I. Breast feeding and the intestinal microflora of the infant–implications for protection against infectious diseases. Adv Exp Med Biol. 2000;478:77–93. doi: 10.1007/0-306-46830-1_7. [DOI] [PubMed] [Google Scholar]
- 40.Maldonado J, Canabate F, Sempere L, Vela F, Sanchez AR, Narbona E, Lopez-Huertas E, Geerlings A, Valero AD, Olivares M, et al. Human milk probiotic Lactobacillus fermentum CECT5716 reduces the incidence of gastrointestinal and upper respiratory tract infections in infants. J Pediatr Gastroenterol Nutr. 2012;54(1):55–61. doi: 10.1097/MPG.0b013e3182333f18. [DOI] [PubMed] [Google Scholar]
- 41.Li J, Zhao F, Wang Y, Chen J, Tao J, Tian G, Wu S, Liu W, Cui Q, Geng B, et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome. 2017;5(1):14. doi: 10.1186/s40168-016-0222-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGuire MK, McGuire MA. Got bacteria? The astounding, yet not-so-surprising, microbiome of human milk. Curr Opin Biotechnol. 2017;44:63–68. doi: 10.1016/j.copbio.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 43.Fraser A, Nelson SM, Macdonald-Wallis C, Sattar N, Lawlor DA. Hypertensive disorders of pregnancy and cardiometabolic health in adolescent offspring. Hypertension. 2013;62(3):614–620. doi: 10.1161/HYPERTENSIONAHA.113.01513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sprockett D, Fukami T, Relman DA. Role of priority effects in the early-life assembly of the gut microbiota. Nat Rev Gastroenterol Hepatol. 2018;15(4):197–205. doi: 10.1038/nrgastro.2017.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gomez-Arango LF, Barrett HL, McIntyre HD, Callaway LK, Morrison M, Dekker NM. Increased systolic and diastolic blood pressure is associated with altered gut microbiota composition and butyrate production in early pregnancy. Hypertension. 2016;68(4):974–981. doi: 10.1161/HYPERTENSIONAHA.116.07910. [DOI] [PubMed] [Google Scholar]
- 46.Sibai BM. Diagnosis and management of gestational hypertension and preeclampsia. Obstet Gynecol. 2003;102(1):181–192. doi: 10.1016/S0029-7844(03)00475-7 [DOI] [PubMed] [Google Scholar]
- 47.Wan Y, Wang F, Yuan J, Li J, Jiang D, Zhang J, Li H, Wang R, Tang J, Huang T, et al. Effects of dietary fat on gut microbiota and faecal metabolites, and their relationship with cardiometabolic risk factors: a 6-month randomised controlled-feeding trial. Gut. 2019;68(8):1417–1429. doi: 10.1136/gutjnl-2018-317609. [DOI] [PubMed] [Google Scholar]
- 48.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 51.de la Cuesta-zuluaga J, Mueller NT, Corrales-Agudelo V, Velasquez-Mejia EP, Carmona JA, Abad JM, Escobar JS. Metformin is associated with higher relative abundance of mucin-degrading akkermansia muciniphila and several short-chain fatty acid-producing microbiota in the gut. Diabetes Care. 2017;40(1):54–62. doi: 10.2337/dc16-1324. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


