ABSTRACT
Clostridioides difficile is the leading cause of health-care-associated infection throughout the developed world and contributes significantly to patient morbidity and mortality. Typically, antibiotics are used for the primary treatment of C. difficile infections (CDIs), but they are not universally effective for all ribotypes and can result in antibiotic resistance and recurrent infection, while also disrupting the microbiota. Novel targeted therapeutics are urgently needed to combat CDI. Bacteriophage-derived endolysins are required to disrupt the bacterial cell wall of their target bacteria and are possible alternatives to antibiotics. These lytic proteins could potentially replace or augment antibiotics in CDI treatment. We discuss candidate therapeutic lysins derived from phages/prophages of C. difficile and their potential as antimicrobials against CDI. Additionally, we review the antibacterial potential of some recently identified homologues of C. difficile endolysins. Finally, the challenges of endolysins are considered with respect to the development of novel lysin-based therapies.
KEYWORDS: Clostridioides difficile, endolysin; bacteriophage, antimicrobials, clostridioides difficile infections, antibiotics, novel therapy
Introduction
Clostridioides difficile (formerly Clostridium difficile) is a Gram-positive, spore-forming obligate anaerobic bacillus, and the causative agent of healthcare-associated (HA) infectious diarrhea.1 In recent decades C. difficile associated infection has been associated with high morbidity and mortality, in particular in Europe, the USA, Canada, and Australia.2 Some reports recently also describe C. difficile infection (CDI) in Asia.3,4 There are nearly 462,100 CDI cases annually in the United States and at least 12,800 fatalities.5,6 In addition to loss of life, the treatment and management costs of CDI infection are significant, with an estimated annual cost of 800 USD million in the USA and €3,000 million in Europe.7 Although CDI cases in the United Kingdom decreased from 55,498 to 12,275 between 2007 and 2018,8 enormous effort has been put into CDI management strategies.9
CDI is mediated by up to three toxins; toxin A (enterotoxin), toxin B (cytotoxin) and the less common binary CDT toxin.10 These toxins damage the intestinal epithelium cell layer and activate the host inflammatory response that contributes to the disease pathology. The clinical symptoms range from mild to severe diarrhea and, to some instances, extend to potentially life-threatening conditions such as pseudomembranous colitis and toxic megacolon.11
The most common treatment strategy in CDI is to administer antibiotics such as metronidazole, vancomycin, and fidaxomicin. However, recurrence of infection and treatment failure can occur.12 Recent reports suggest there is reduced susceptibility and resistance of C. difficile toward these antibiotics.13,14 This has driven the exploration of alternative therapies to treat infections caused by this bacterium. Several treatment alternatives have gained traction and are at various stages of development; these include new antibiotics, probiotics, fecal microbiota transplantation (FMT), antimicrobial peptides, bacteriocins and phage therapy.15–18 The accessibility of phage genome databases and increased sequencing of phage have kindled interest in the application of phage encoded enzymes, especially endolysins, as alternative therapeutic agents.
Much has been written on the success of endolysins as targeted antimicrobials.19–22 A number of reviews have focused on the advantages, specificity and safety of endolysin therapy.23–28 In this work, we aim to review the existing literature on C. difficile endolysins and discuss their potency and their potential use as antimicrobials in the treatment of CDI.
Antibiotic resistance in Clostridioides difficile
Generally, CDI is initiated following disruption of the normal intestinal microbiota by antibiotics that allow C. difficile, either native or acquired, to proliferate.29 The recommended antibiotics for primary and recurrent CDI are metronidazole, vancomycin and fidaxomicin.1,30 Other antibiotics commonly used for bacterial infections such as cephalosporins, ampicillin, clindamycin, amoxicillin and fluoroquinolones are also associated with a higher risk for CDI.1,31 The evolution of new ribotypes is often associated with acquisition of resistance as a result of inappropriate use of antibiotics.14 Many of the most common C. difficile epidemic ribotypes are associated with multidrug resistance.14,31 However, C. difficile is a spore-forming bacteria that can survive antimicrobial therapy and following germination relapse of CDI can routinely occur.
Several mechanisms of antimicrobial resistance have been identified in C. difficile. These include chromosomal resistance-associated genes, mobile genetic elements (MGEs), alterations in the antibiotic targets of antibiotics and/or in metabolic pathways, and biofilm formation. Examples of chromosomal resistance genes include those encoding β-lactamase-like proteins and penicillin-binding proteins (PBPs) that mediate resistance to the β-lactam antibiotics such as penicillin and cephalosporins.14 The C. difficile genome contains a wide range of mobile elements. MGE-like transposons facilitate the spread of antibiotic resistance genes by the process of conjugation, transduction, and/or transformation among C. difficile and/or between C. difficile and other bacterial species.14 Resistance to antibiotics of the macrolide-lincosamide-streptogramin B (MLSB) family, tetracycline and chloramphenicol in C. difficile is thought to be associated with different transposon families.31 Alterations in the antibiotic targets and/or in metabolic pathways is another important route of resistance development in C. difficile, and this mechanism mediates resistance to rifamycin, fluoroquinolones, metronidazole and vancomycin.14,31 Biofilms help pathogenic bacteria to survive unfavorable environmental stresses, including antibiotics.32 Biofilm formation is potentially involved in metronidazole and vancomycin resistance in C. difficile.14,31
Bacteriophage endolysins
Bacteriophages or phages are viruses that infect and kill bacteria. Bacteriophages can adopt either of two life cycles, lytic and lysogenic. Both virulent and temperate phages may enter the lytic cycle, whereas only temperate phages utilize the lysogenic cycle.33 In most cases, the lytic cycle concludes with cell lysis that leads to cell death (Figure 1a). In single stranded DNA/RNA phages, the genome encodes a lysis effector which inhibits peptidoglycan (PG) biosynthesis from within the bacterium. On the other hand, double stranded DNA (dsDNA) phages utilize phage-encoded muralytic enzymes called endolysins (or lysins) that lead to cell envelope disruption at the final stage of phage reproduction. There are three different lysis mechanisms in dsDNA phages. The most studied and best understood mechanism is canonical lysis, where endolysins require the help of a second phage-encoded protein called a holin to act on the PG layer.34 Holin proteins accumulate and oligomerize in the cytoplasmic membrane (CM) in a time-controlled manner, and trigger depolarization and the formation of holes in the CM. This allows diffusion of endolysin to the membrane, facilitating the destruction of the PG layer (Figure 1b). The second mechanism requires a special class of holins, called pinholins, which form small, heptameric channels that help to depolarize the membrane. In the third mechanism, the lysis of the outer membrane of Gram-negative hosts is facilitated by spanin proteins.35 Spanins form a complex with outer membrane (OM) lipoprotein (o-spanin) and an integral cytoplasmic membrane protein (i-spanin) and disrupt the OM by enzymatic degradation, pore formation and inner membrane-outer membrane fusion.36–38
Figure 1.
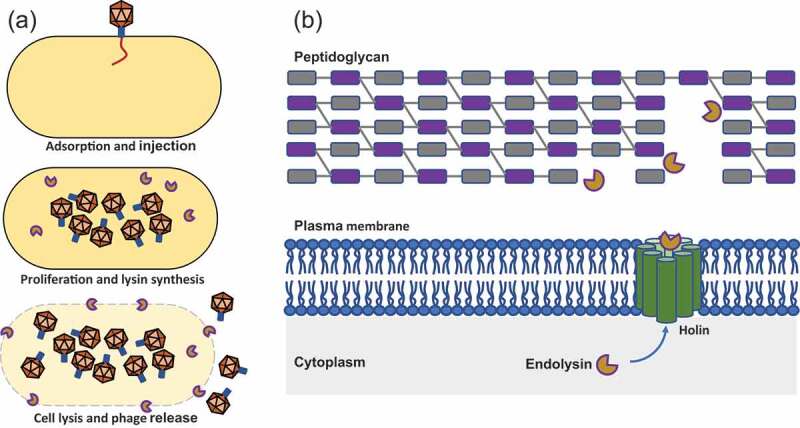
(a) Bacteriophage lytic cycle; (b) mode of action of endolysin against gram-positive cell walls of bacteria.
Endolysins are ‘enzybiotics’, a promising class of antibiotics derived from enzymes. Endolysins possess high specificity against the genus or species infected by the phage from which they were derived, which is believed to be one of their key advantages over classical wide-spectrum antibiotics. Thus, these lytic enzymes, specific for undesired pathogenic bacteria, rarely lyse non-target bacteria, including commensals of our microbiota or ‘good’ bacteria in foods (e.g. starter cultures) or those in the environment.39,40 The White house National Action Plan for Combating Antibiotic-resistant Bacteria has listed “phage-derived lysins to kill specific bacteria while preserving the microbiota” among the non-traditional therapeutics of note.41
It is possible to expand the lytic spectrum of an endolysin by the exchange or addition of certain domains beyond the native endolysin’s serovar, species, or even genus specificity.42 Endolysins can also act synergistically when used in combination with each other (i.e. two endolysins with different cleavage specificities) or with other antimicrobial agents.20,43 Other advantages of endolysins over traditional antibiotics are rapid host killing, low chance of resistance development, the potential to kill multi-drug-resistant bacteria, synergism with different antibacterial agents, and the ability to actively work in biofilms as well as on mucosal surfaces.44–49 Nowadays, the emergence of multidrug-resistant pathogens has revitalized the interest in alternative therapies. Due to the unique properties and advantages of endolysins over bacteriophages, endolysins are highly ranked alternatives in eradicating drug-resistant pathogens. Different properties such as specificity and host range,39,50–52 mode of action,53–55resistance development,56,57 stability,58,59 and pharmacokinetics60,61 between bacteriophage and endolysins are summarized in Table 1.
Table 1.
Comparison of bacteriophages and endolysins as antimicrobials.
| Selected properties | Bacteriophage | Endolysin |
|---|---|---|
| Specificity and host range | Generally specific to one bacterial species (or strains within a species). Limited impact on microbiota composition. | Narrow or broad depending on the chemical structure of the targeted macromolecule. Limited impact on microbiota composition. |
| Mode of action | Bacteriolytic activity depends on the titer, multiplicity of infection (MOI), burst size and propagation rate. | Bacteriolytic activity depends on concentration and minimum inhibitory concentration (MIC). |
| Resistance development | Resistance developed by mutation, receptor modification, passive adaptation, restriction-modification, CRISPR-Cas, pseudolysogeny. | Bacteria are less likely to develop resistance to endolysins. |
| Stability | Stability properties dependent on structural protein composition. | Endolysins have a short half-life, but effectively work in short duration due to the rapid mode-of-action. |
| Antibiofilm activity | Relatively effective with limited penetration capacity. | Effective against biofilms with higher penetration capacity. |
| Inflammatory response | Reticuloendothelial system (RES) clearance and immunogenic. | Immunogenic, induction of antibody production. |
| Pharmacokinetics | Not properly defined, self-replicating and can be cleared by immune system. | Evaluated in some endolysins; chemical structure affects penetration, plasma protein binding, and proteolysis degradation. |
| Combined therapy | Synergistic effect possible such as: phage cocktails, phage-protein and antibiotic–phage–protein combination. | Synergistic effect between two endolysins with different catalytic specificities or between an endolysin and an antibiotic. |
Basic structure and enzymatic activity of endolysins
Generally, endolysins have a conserved biological function directed at lysing infected bacterial cells. However, constant evolutionary pressure between bacteriophage and host bacteria has resulted in significant biochemical and structural variations among endolysins.62 Endolysins against Gram-positive and Gram-negative organisms are categorized differently due to the composition of the cell walls of their targets. Gram-positive endolysins are modular in structure with one or two N-terminal enzymatically active domains (EADs) and one or more C-terminal cell-wall binding domains (CBDs), these domains are usually connected by a short linker region.62,63 EADs contain the catalytic mechanism of endolysins, disrupting specific bonds within the bacterial peptidoglycan, whereas CBDs bind to constituents of cell walls to promote EAD localization to its target site and enhance catalytic efficiency. However, sometimes greater lytic activity has been observed in truncated endolysins containing only EADs.20,64 Gram-negative endolysins usually have single globular catalytic domain and lack a CBD.65,66 There have been some reports for Gram-negative endolysins that indicate a modular organization with a CBD at the N-terminus and EAD at the C-terminus which is inverse architecture typical to most Gram-positive endolysins.67,68
PG structure is highly conserved and consists of a polysaccharide of alternating N-acetylmuramic acid (MurNAc) and N-acetylglucosamine (GlcNAc) residues, linked by a β1-4 glycosidic bond (Figure 2). The D-lactoyl group of each MurNAc is linked with a short peptide stem, which is different between bacterial species.69 The tetrapeptide stem found in C. difficile is L-Ala-D-Glu-A2pm-D-Ala (A2pm: 2,6-diaminopimelic acid).70 Endolysins can recognize and digest a specific chemical bond within PG and are classified accordingly: (i) N-acetylmuramoyl-L-alanine amidases cleave the amide bond between N-acetylmuramoyl residues and L-alanine (the first amino acid of the peptide stem), (ii) L-alanoyl-D-glutamate endopeptidases target the bond between L-alanine and D-glutamate, (iii) interpeptide bridge endopeptidases digest the cross-link between peptide stems, (iv) D-glutamyl-m-DAP endopeptidase target bonds between D-glutamate and m-diaminopimelic acid, (v) N-acetyl-D-glucosaminidases hydrolyze the N-acetylglucosaminyl-β-1,4-N-acetylmuramine bond, (vi) N-acetyl-D-muramidases and lytic transglycosylases cleave the N-acetylmuramoyl−1,4-N-acetylglucosamine bond.
Figure 2.
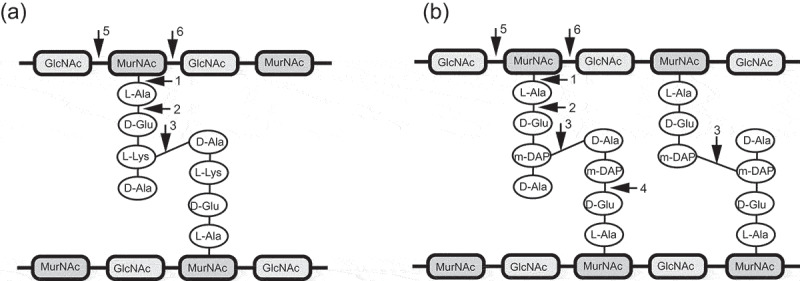
Schemetic presentation of bacterial peptidoglycan and generalized cut sites of peptidoglycan hydrolases. (a) A3α type peptidoglycan of Staphylococcus aureus; (b) A1γ type featuring peptidoglycan of C. difficile. The bonds potentially attacked by endolysins of different enzymatic specificities are indicated by numbers: 1) N-acetylmuramoyl-L-alanine amidases; 2) L-alanoyl-D-glutamate endopeptidases; 3) interpeptide bridge endopeptidases; 4) D-glutamyl-m-DAP endopeptidase; 5) N-acetyl-D-glucosaminidases; 6) N-acetyl-D-muramidases and lytic transglycosylases.
The chemotype of C. difficile PG is A1γ, where a meso-diaminopimelic acid (meso-A2pm) residue at position 3 of the peptide is directly cross-linked to a D-alanine at position 4 of the neighboring peptide. A similar structure is present in Bacillus, Listeria and some Gram-negative species.40,70,71 However, C. difficile also have a very high percentage of 3–3 peptide cross-links between two meso-diaminopimelic acid (meso-A2pm) residues69 (Figure 2b). Endolysin activity against these peptide cross-links has not yet been verified in C. difficile.
Clostridioides difficile endolysins
The use of phage therapy in CDI is limited due to the temperate nature of C. difficile bacteriophages (reported to date). In most cases low titers of lysogenic phages have been recovered after induction with mitomycin C. These phages can easily reintegrate into the host genome following the removal of inducers.72 In the case of reported lytic phages, these may also have access the lysogenic life cycle.73,74 Efforts have focused on alternative options and the exploitation of C. difficile phage endolysins have sparked interest as therapeutic alternatives for CDI. Through published articles and online database searches, we identified sequences of putative endolysins from phage/prophage of C. difficile. The majority of these endolysins are amidases and hydrolases and are summarized in Table 2. The catalytic domains include amidase 3 domains, amidase 2 domains, glucosaminidases and NLPC_P60 domains. The sequence homology among the catalytic amidase domains was analyzed. A BLAST search for the sequence of the catalytic amidase domain of CD27L against the amidase containing endolysins obtained from C. difficile phage or prophage sequences was performed. After curation of the sequence cluster to remove duplicates, the sequences of five amidases (CD27L, phyCD, phiCD38, phiCD119 and CD11) were aligned and analyzed using ESPRIPT90 for sequence conservation. Although the sequence identity is low, several conserved residues were found. The conserved four amino acid residues (His 9, Glu 26, His 84, and Glu 144) are coordinating the zinc ion and responsible for catalytic activity (Figure 3a). The three-dimensional structures of major C. difficile endolysins (CD27L, CDG, phiCD211, phiCDHM11 and phiCDMMP01) have been predicted by homology modeling using SWISS MODEL, an online tool (Figure 3b). The potential templates for target endolysins were identified based on the sequence coverage and percentage of identity between the target and template sequence, except for CD27L whose EAD and CBD three-dimensional structure are already available (PDB code 3QAY and 4CU5). Due to the presence of different catalytic groups, the folding patterns are different in amidases, glucosaminidases and NLPC_P60 containing C. difficile endolysins.
Table 2.
The list of endolysins of C. difficile phages/prophages.
| Phages/Prophages | Putative endolysin (Name of ORF) | Domain and features | References |
|---|---|---|---|
| phiCD119 | ORF35 | MurNAc-LAA/Amidase_3 | 75 |
| phiC2 | phiC2p38 & phiC2p22 |
MurNAc-LAA/Amidase_3 & NLPC_P60/pfam00877* | 76,77 |
| phiCD27 | gp34 | MurNAc-LAA/Amidase_3 | 78 |
| phiCD6356 | phiCD6356_28 | MurNAc-LAA/Amidase_3 | 79 |
| phiCD38-2 | gp23 | MurNAc-LAA/Amidase_3 | 80 |
| phiMMP02 | gp34 | MurNAc-LAA/Amidase_3 | 72 |
| phiMMP04 | gp26 & gp16 | MurNAc-LAA/Amidase_3 & NLPC_P60/pfam00877* | 72 |
| phiCDMH1 | gp34 & gp24 | MurNAc-LAA/Amidase_3 & NLPC_P60/pfam00877* | 81 |
| phiCDHM13 | gp25 & gp15 | MurNAc-LAA/Amidase_3 & NLPC_P60/pfam00877* | 82 |
| phiCDHM14 | gp25 & gp15 | MurNAc-LAA/Amidase_3 & NLPC_P60/pfam00877* | 82 |
| phiCDHM19 | gp40 & gp26 | MurNAc-LAA/Amidase_3 & NLPC_P60/pfam00877* | 82 |
| phiCD211 | PHICD211_20039 & PHICD211_20040 |
MurNAc-LAA/Amidase_3 & Glucosaminidase/pfam01832* | 83 |
| phiCDIF1296T | CDIF1296T_phi042 & CDIF1296T_phi043 |
MurNAc-LAA/Amidase_3 & Glucosaminidase/pfam01832* | 84 |
| phiCD24-1 | PHICD2401_20030 | MurNAc-LAA/Amidase_3 | 83 |
| phiCD111 | PHICD111_20024 | MurNAc-LAA/Amidase_3 | 83 |
| phiCD146 | PHICD146_20023 | MurNAc-LAA/Amidase_3 | 83 |
| phiMMP01 | PHIMMP01_20036 & PHIMMP01_20024 |
MurNAc-LAA/Amidase_3 & Glucosaminidase and NLPC_P60* | 72,83 |
| phiMMP03 | PHIMMP03_20039 & PHIMMP03_20024 | MurNAc-LAA/Amidase_3 & Glucosaminidase and NLPC_P60* | 72,83 |
| phiCD481-1 | PHICD48101_20027 | CwlA/Amidase_2 | 85 |
| phiCD505 | PHICD505_20034 | MurNAc-LAA/Amidase_3 | 85 |
| phiCD506 | PHICD506_20027 | MurNAc-LAA/Amidase_3 | 85 |
| phiCDHM11 | phiCDHM11_gp25 & phiCDHM11_gp15 |
MurNAc-LAA/Amidase_3 & NLPC_P60/pfam00877* | 86 |
| phiCDKM9 | CDHM9_32 | MurNAc-LAA/Amidase_3 | 87 |
| phiCDKM15 | CDKM15_37 | MurNAc-LAA/Amidase_3 | 87 |
| phiSemix9P1 | Semix9P1_phi34 | MurNAc-LAA/Amidase_3 | 88 |
| C. difficile 630 | YP_001088405 & WP_009895119.1 |
MurNAc-LAA/Amidase_3 | 20,89 |
| C. difficile DA00211 | EQH20562.1 | CwlA/Amidase_2 | 89 |
*Present study.
Figure 3.
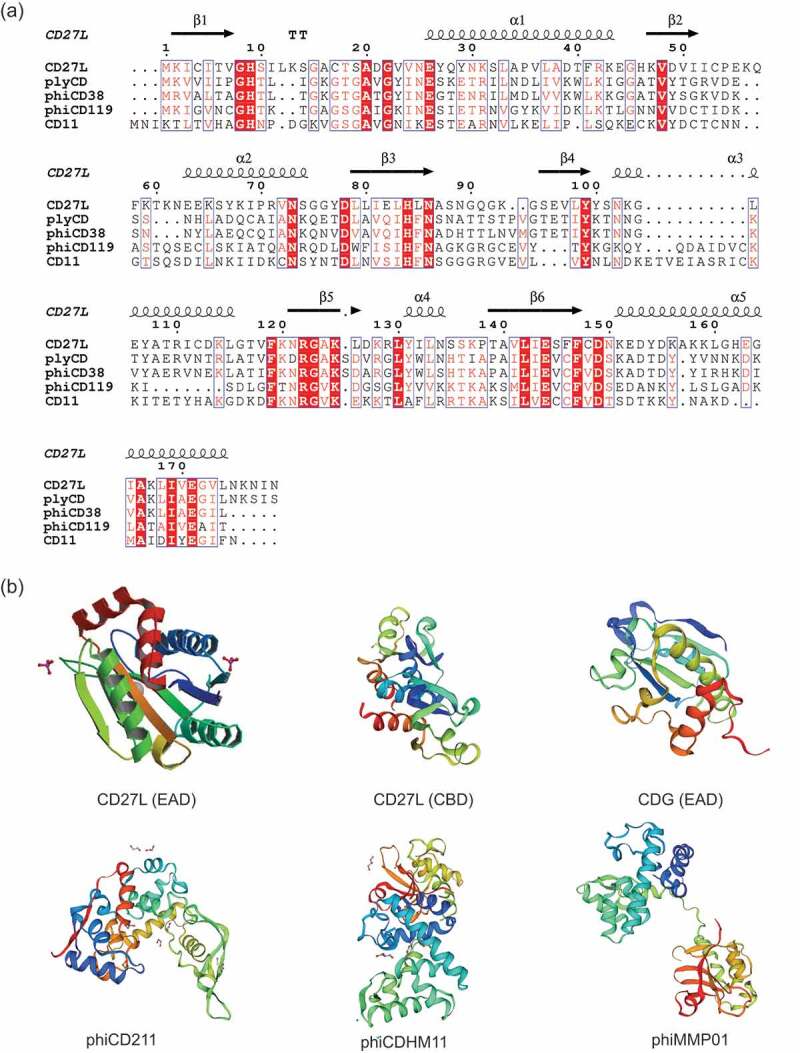
Sequence and structural analysis of C. difficile endolysins. (a) Sequence of the amidase containing catalytic domain of CD27L aligned to other amidase containing endolysins of C. difficile. Multiple-sequence alignment was performed by Clustal Omega and visualized by ESPript using CD27L endolysin structure (PDB code 3QAY) as the query. The position of amino acid residues based on CD27L amidase domain is shown. Secondary structure of CD27L1–179 is displayed, with arrows indicating beta strands and ribbons indicating alpha helices. Conserved residues can be visualized as white text on a red background, while amino acids with similar properties written in red text; (b) protein 3D structure of enzymatically active domains (EAD) and cell-wall binding domains (CBD). Homology modeling was performed by Swiss-model server (https://swissmodel.expasy.org/). The predicted model of CDG, phiCD211, phiCDHM11 and phiCDMMP01 were generated using protein data bank (PDB) templates 2L47, 5WQW, 4HPE and 4FDY, respectively.
CD27L
The endolysin CD27L is derived from C. difficile bacteriophage φCD27, a temperate bacteriophage belonging to the Myoviridae family with a genome length of 50,930 bp.78 This was the first C. difficile endolysin to be cloned and recombinantly expressed. The CD27L endolysin is 270 amino acids with an estimated molecular weight of ~30 kDa. CD27L is a modular endolysin that contains one EAD and one CBD. It has no transmembrane regions or signal peptide regions. The catalytic domain of CD27L belongs to the N-acetylmuramoyl-L-alanine amidase class (amidase 3). This lysin is active against a panel of 30 different C. difficile strains, including the hypervirulent ribotype 027, while a selection of the commensal bacteria of the GI tract are insensitive.78 Endolysin delivery to the gut environment is a challenge, and one approach is to use a genetically modified lactic acid bacterium. Crude protein extracts from Lactococcus lactis expressing CD27L have been shown to successfully lyse target cells to a similar degree to that observed for E. coli expressed lysin.78 It was found that CD27L lyses cells over a broad pH range, indicating it should remain active in the GI tract environment. Furthermore, the CD27L lysin catalytic domain, CD27L1-179, is more effective and exhibits a higher degree of specificity than the full-length endolysin.64 However, CD27L activity has not been tested an in vivo study.
PlyCD
The lysin PlyCD is contained within a prophage in a multidrug resistance C. difficile strain, CD630.20 PlyCD is 262 amino acids with a molecular weight of ~28 kDa. The amino acid sequence identity between PlyCD and CD27L is 33%, with 34.6% identity between the catalytic domains. Both the full-length PlyCD and the recombinantly expressed truncated PlyCD1-174 are active against a number of C. difficile strains. Indeed, PlyCD1-174 has significantly greater lytic activity (>4-log kill) and a broader spectrum of activity while retaining high specificity toward C. difficile versus other commensal bacterial species. Additionally, a combination of PlyCD1-174 with a subinhibitory dose of vancomycin was significantly more successful in vitro against C. difficile compared to PlyCD1-174 or vancomycin alone.20 The functional capability of PlyCD1-174 was further confirmed by an in vivo mouse model study where the PlyCD treated mice showed increased survival and a delay in morbidity and mortality rate as compared to the buffer-treated control animals. Moreover, an ex vivo treatment model of mouse colon infection showed PlyCD1–174 functioned effectively in the presence of intestinal contents and significantly reducing colonizing C. difficile compared to buffer control.20
CDG and CD11
The lysin proteins CDG and CD11 were identified in C. difficile DA00211 and C. difficile 630, respectively.89 Both lysins were expressed recombinantly and were found to be effective against C. difficile cells in a dose-dependent manner, reducing levels by up to four logs within five hours. These lytic proteins were active against clinical isolates of C. difficile, while no activity was observed against Bacillus or Staphylococcal species. The biocatalytic mechanism showed that these enzymes cleave bonds between N-acetylmuramoyl and L-alanine within the cell wall PG.89
LHD
The modular structure of lysin proteins can facilitate the modification of bacteriolytic activity, specificity, solubility, and other physicochemical properties of these proteins in order to design novel antimicrobials. A lysin, LHD, was engineered to have the catalytic domain of a lysin protein from a C. difficile bacteriophage phiC2 fused with the functional domain of a human defensin protein HD5 by a 3-repeating unit linker; (GGGGS)3.91 The reason for choosing the catalytic domain of phiC2 is that phiC2 is present in the majority of human isolates of C. difficile92 and it may have a wide spectrum of lytic activity. On the other hand, human defensin protein HD5 has been documented to inhibit hypervirulent C. difficile strains.93 So, the hybrid would be more active than these individual antimicrobials. It was found that this lysin-human defensin fusion protein was active against several clinical C. difficile strains, including the epidemic 027, 078, 012, and 087 strains that are prevalent in many different regions of world.94 The minimum inhibitory concentration (MIC) of LHD was lower than the MIC of the lysin protein LCD. The fusion protein was also active in a broader pH range (6.0, 7.0, and 8.0). In an in vivo mouse model, the LHD treated group had reduced symptoms mortality from CDI compared to the control buffer treated group. In addition, LHD significantly decreased the C. difficile spore count and toxin production in feces of the infected mice.91
Cell wall hydrolases
Cell wall hydrolases (CWHs) are classified based on their origin as endolysins, exolysins and/or autolysins.95,96 Some of the C. difficile phage/prophages contain putative CWH sequences that may have potent lytic activity as endolysins (Table 2). The major catalytic domains present in C. difficile phage/prophage CWHs are NlpC/P60 and glucosaminidase. The NlpC/P60 domain is described as a superfamily and has a diverse range of catalytic activity including cleaving N-acetylmuramate-L-alanine linkages and the 4–3 linkage between D-Glu and m-DAP residues.97–99 The glucosaminadases hydrolyze the glycosidic bond of the sugar backbone. The putative N-acetylglucosaminidases (EC.3.2.1.96) that are present in C. difficile phages/prophages were also present in the prophage LambdaSa2 of Streptococcus agalactiae and exhibit β-D-N-acetylglucosaminidase activity.100
Bacterial resistance to endolysins
Bacteriophage endolysins have a unique attribute in comparison to intact phages and antibiotics, in that resistance development is an extremely rare event. Generally, antibiotics work by inhibiting essential metabolic pathways of bacteria leading to cell death.101 However, bacteria have found ways to overcome this adverse situation by using alternative metabolic pathways. It is difficult for bacteria to find means of resistance to endolysin as they bind to and degrade highly conserved peptidoglycan targets within the cell wall.102 Any mutations leading to endolysin resistance would be damaging to the integrity of the cell and thus a very rare event.103 Although no attempts have been made to study lysin-resistance development in C. difficile, there are some studies using other bacterial strains that have investigated repeated lysin exposure and revealed resistance did not develop to either native or engineered phage lysins.40,104-106 Some CWHs of C. difficile have more than one catalytic domain and this theoretically lowers the chance of mutation in multiple target sites in bacteria. Similar observations have been made with S. aureus endolysins.107
Safety and current trials
Bacteriophages are naturally an integral part of the human microbiota and thus the release of phage-derived lysins is unlikely to have a harmful effect on human health.108 The safety of phage lysins has been confirmed in a number of animal model systems.20,47,109,110 The impact of lytic proteins on inflammatory responses and/or their toxicity has been evaluated in animal models and it was observed that the administration of some lysin proteins, for example, Cpl-1 and MV-L, triggered an immune response which ultimately resulted in the production of antibodies against this protein.111,112 In another study, low levels of antibodies and/or cytokine production were observed in animals compared with untreated controls following endolysin treatment.113,114 Despite the number of animal trials published, only a few lysins have undergone human clinical trials. SAL200 is the first endolysin-based therapeutic formulation against MRSA. It is derived from Staphylococcus phage SAP-1 that infects Staphylococci, including MRSA and vancomycin-resistant S. aureus (VRSA) strains. Recently, protein SAL200 was evaluated in humans by intravenous infusion as part of a phase 1 clinical trial. Single ascending intravenous doses (0.1 mg/kg to 10 mg/kg) were applied to healthy male volunteers in order to assay pharmacokinetics, pharmacodynamics, and tolerance of SAL200.61 No serious adverse effects as well as recurrence of infection were observed in volunteers except more than three participants noticed mild and temporary effects like fatigue, headaches and myalgia. Another endolysin-based product called Staphefekt SA.100, developed by Dutch biotech company Micreos, has been available in Europe since 2017 for human use. Staphefekt SA.100 is an engineered chimeric endolysin for topical skin application that specifically binds to the cell wall of S. aureus and cleaves the cell membrane via endopeptidase and putative amidase activities.115 In a case study on three human subjects with chronic and recurrent S. aureus-related dermatoses showed that Staphefekt™ improved the clinical symptoms, but they rapidly recurred if the treatment was ceased. Potentially due to the recolonization of S. aureus from the nose and environment. It has also been shown that the long term daily use of Staphefekt did not result in generation of bacterial resistance during chronic and recurrent S. aureus treatment.22 A multi-center, placebo-controlled, double-blinded and randomized superiority trial study (ClinicalTrials.gov, NCT02840955) of Staphefekt showed application on the skin, targeting only S. aureus and leaving skin commensals unharmed, improves S. aureus-related skin infections, such as eczema, acne, and rosacea.116 Staphefekt is registered as a (class 1) medical device in Europe and available as an over-the-counter treatment in the form of a cream or gels. There are several other products at different stages of clinical trials, some with promising results that will pave the way for future endolysin-based therapies.107,117
Challenges of endolysin therapy in CDI
There are several challenges facing the commercialization of endolysins, for instance, large-scale production and formulation, targeted delivery and regulatory framework amongst others. Until now, the use of endolysins as human therapeutics has not been approved in the United States or Europe, except for the endolysin-based product Staphefekt which is marketed in the EU as a ‘medical device’.22 Although the role of phage-based lysins is already established and examined in different animal infection models, the study of C. difficile endolysins is still limited. It may be due to some experimental limitations, particularly the development of targeted enteric delivery of such endolysins. To date, only two studies were performed where C. difficile lysins were examined in animal models for clinical development.20,91 In both studies, the targeted delivery of endolysins to the gut was by direct administration via oral gavage. Lysins are non-replicating protein molecules, with short in-vivo half-lives.33,118 The enteral delivery of phage lytic proteins faces the challenge of maintaining enzyme activity at low pH and in the presence of proteolytic enzymes of the stomach. To avoid such obstacles, the development of novel delivery strategies is important. Nanoparticle-based anticancer drug delivery to eukaryotic cells are becoming more popular as a treatment strategy.119,120 It also offers possibilities to effectively target bacterial cells.121,122 Endolysins could also be encapsulated in polymeric nanoparticles to give protection from the harsh gastric environment. Another proposed innovative approach is to deliver and preserve lysins in the gastrointestinal tract via engineered lactic acid bacteria that actively excrete the endolysin during gut transit.123,124 The C. difficile phage endolysin CD27L has been successfully expressed in Lactococcus lactis MG1363.78 Additionally, the probiotic strain Lactobacillus johnsonii FI9785 was precisely engineered to deliver and secrete an endolysin active against C. perfringens.124
Conclusion
Clostridioides difficile is recognized as the leading cause of nosocomial and community-acquired diarrhea associated with exposure to antibiotics. Antibiotic resistance and a high rate of recurrence limit the usefulness of current antimicrobials used for primary C. difficile infection; alternative solutions are urgently required that work effectively while maintaining the gut microbiota. Phage lytic proteins show great potential in this regard as a replacement for, or as an additional therapy to, the use of traditional antibiotics in CDI. In fact, several endolysins active against C. difficile are currently being investigated in this regard. Major features, such as high catalytic activity, modular structure, and the possibility of engineering, support the development of these novel alternatives to conventional antibiotic therapy. The possibility of resistance development to phage lytic proteins remains a real possibility worthy of further investigation. To date, preclinical and clinical studies demonstrate that endolysins are safe and very effective antimicrobials.
Funding Statement
SIM have received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement number [754535].
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Leffler DA, Lamont JT.. Clostridium difficile infection. N Engl J Med. 2015;372:1539–15. doi: 10.1056/NEJMra1403772. [DOI] [PubMed] [Google Scholar]
- 2.Kuijper EJ, Coignard B, Tull P. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect. 2006;12(Suppl 6):2–18. doi: 10.1111/j.1469-0691.2006.01580.x. [DOI] [PubMed] [Google Scholar]
- 3.Borren NZ, Ghadermarzi S, Hutfless S, Ananthakrishnan AN. The emergence of Clostridium difficile infection in Asia: a systematic review and meta-analysis of incidence and impact. PLoS One. 2017;12(5):e0176797. doi: 10.1371/journal.pone.0176797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.T V R, Kimura T. The epidemiology of Clostridium difficile Infection in Japan: a systematic review. Infect Dis Ther. 2018;7(1):39–70. doi: 10.1007/s40121-018-0186-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guh AY, Mu Y, Winston LG, Johnston H, Olson D, Farley MM, Wilson LE, Holzbauer SM, Phipps EC, Dumyati GK, et al. Trends in U.S. Burden of Clostridioides difficile infection and outcomes. N Engl J Med. 2020;382:1320–1330. doi: 10.1056/NEJMoa1910215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2019. Atlanta (GA): U.S. Department of Health and Human Services , CDC;2019. doi: 10.15620/cdc:82532. [DOI] [Google Scholar]
- 7.Bouza E. Consequences of Clostridium difficile infection: understanding the healthcare burden. Clin Microbiol Infect. 2012;18:5–12. doi: 10.1111/1469-0691.12064. [DOI] [PubMed] [Google Scholar]
- 8.Thelwall S, Hope R. Annual epidemiological commentary: gram-negative bacteraemia, MRSA bacteraemia, MSSA bacteraemia and C. difficile infections, up to and including financial year April 2017 to March 2018. Public Health England. 2018. [Google Scholar]
- 9.Hughes GJ, Nickerson E, Enoch DA, Ahluwalia J, Wilkinson C, Ayers R, Brown NM. Impact of cleaning and other interventions on the reduction of hospital-acquired Clostridium difficile infections in two hospitals in England assessed using a breakpoint model. J Hosp Infect. 2013;84:227–234. doi: 10.1016/j.jhin.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 10.Elliott B, Androga GO, Knight DR, Riley TV. Clostridium difficile infection: evolution, phylogeny and molecular epidemiology. Infect Genet Evol. 2017;49:1–11. doi: 10.1016/j.meegid.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 11.Schäffler H, Breitrück A. Clostridium difficile – from colonization to infection. Front Microbiol. 2018;9:646. doi: 10.3389/fmicb.2018.00646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Surawicz CM, Brandt LJ, Binion DG, Ananthakrishnan AN, Curry SR, Gilligan PH, McFarland LV, Mellow M, Zuckerbraun BS. Guidelines for diagnosis, treatment, and prevention of clostridium difficile infections. Am J Gastroenterol. 2013;108(4):478–498. doi: 10.1038/ajg.2013.4. [DOI] [PubMed] [Google Scholar]
- 13.Kociolek LK, Gerding DN. Breakthroughs in the treatment and prevention of Clostridium difficile infection. Nat Rev Gastroenterol Hepatol. 2016;13(3):150–160. doi: 10.1038/nrgastro.2015.220. [DOI] [PubMed] [Google Scholar]
- 14.Spigaglia P. Recent advances in the understanding of antibiotic resistance in Clostridium difficile infection. Ther Adv Infect Dis. 2016;3(1):23–42. doi: 10.1177/2049936115622891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rea MC, Alemayehu D, Ross RP, Hill C. Gut solutions to a gut problem: bacteriocins, probiotics and bacteriophage for control of Clostridium difficile infection. J Med Microbiol. 2013;62(Pt 9):1369–1378. doi: 10.1099/jmm.0.058933-0. [DOI] [PubMed] [Google Scholar]
- 16.Zucca M, Scutera S, Savoia D. Novel avenues for Clostridium difficile infection drug discovery. Expert Opin Drug Discov. 2013;8(4):459–477. doi: 10.1517/17460441.2013.770466. [DOI] [PubMed] [Google Scholar]
- 17.Nale JY, Redgwell TA, Millard A, Clokie MRJ. Efficacy of an optimised bacteriophage cocktail to clear clostridium difficile in a batch fermentation model. Antibiot (Basel, Switzerland). 2018;7(1):13. doi: 10.3390/antibiotics7010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liubakka A, Vaughn BP. Clostridium difficile infection and fecal microbiota transplant. AACN Adv Crit Care. 2016;27(3):324–337. doi: 10.4037/aacnacc2016703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Briers Y, Walmagh M, Van Puyenbroeck V, Cornelissen A, Cenens W, Aertsen A, Oliveira H, Azeredo J, Verween G, Pirnay J-P, et al. Engineered endolysin-based “Artilysins” to combat multidrug-resistant gram-negative pathogens. MBio. 2014;5(4):e01379–14. doi: 10.1128/mBio.01379-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q, Euler CW, Delaune A, Fischetti VA. Using a novel lysin to help control Clostridium difficile infections. Antimicrob Agents Chemother. 2015;59(12):7447–7457. doi: 10.1128/AAC.01357-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen Y, Barros M, Vennemann T, Gallagher DT, Yin Y, Linden SB, Heselpoth RD, Spencer DJ, Donovan DM, Moult J, et al. A bacteriophage endolysin that eliminates intracellular streptococci. Elife. 2016;5. doi: 10.7554/eLife.13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Totte JEE, van Doorn MB, Pasmans SGMA. Successful treatment of chronic staphylococcus aureus-related dermatoses with the topical endolysin staphefekt SA.100: a report of 3 cases. Case Rep Dermatol. 2017;9(2):19–25. doi: 10.1159/000473872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gondil VS, Harjai K, Chhibber S. Endolysins as emerging alternative therapeutic agents to counter drug-resistant infections. Int J Antimicrob Agents. 2020;55(2):105844. doi: 10.1016/j.ijantimicag.2019.11.001. [DOI] [PubMed] [Google Scholar]
- 24.Gutierrez D, Fernandez L, Rodriguez A, Garcia P. Are phage lytic proteins the secret weapon to kill staphylococcus aureus? MBio. 2018;9(1). doi: 10.1128/mBio.01923-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerstmans H, Criel B, Briers Y. Synthetic biology of modular endolysins. Biotechnol Adv. 2018;36(3):624–640. doi: 10.1016/j.biotechadv.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 26.Gerstmans H, Rodriguez-Rubio L, Lavigne R, Briers Y. From endolysins to Artilysin(R)s: novel enzyme-based approaches to kill drug-resistant bacteria. Biochem Soc Trans. 2016;44(1):123–128. doi: 10.1042/BST20150192. [DOI] [PubMed] [Google Scholar]
- 27.Van Tassell ML, Angela Daum M, Kim J-S, Miller MJ. Creative lysins: listeria and the engineering of antimicrobial enzymes. Curr Opin Biotechnol. 2016;37:88–96. doi: 10.1016/j.copbio.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 28.Briers Y, Lavigne R. Breaking barriers: expansion of the use of endolysins as novel antibacterials against Gram-negative bacteria. Future Microbiol. 2015;10(3):377–390. doi: 10.2217/fmb.15.8. [DOI] [PubMed] [Google Scholar]
- 29.Sun X, Hirota SA. The roles of host and pathogen factors and the innate immune response in the pathogenesis of Clostridium difficile infection. Mol Immunol. 2015;63(2):193–202. doi: 10.1016/j.molimm.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ofosu A. Clostridium difficile infection: a review of current and emerging therapies. Ann Gastroenterol. 2016;29(2):147–154. doi: 10.20524/aog.2016.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spigaglia P, Mastrantonio P, Barbanti F. Antibiotic resistances of Clostridium difficile. Adv Exp Med Biol. 2018;1050:137–159. doi: 10.1007/978-3-319-72799-8_9. [DOI] [PubMed] [Google Scholar]
- 32.Ethapa T, Leuzzi R, Ng YK, Baban ST, Adamo R, Kuehne SA, Scarselli M, Minton NP, Serruto D, Unnikrishnan M, et al. Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. J Bacteriol. 2013;195(3):545–555. doi: 10.1128/JB.01980-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Flaherty S, Ross RP, Coffey A. Bacteriophage and their lysins for elimination of infectious bacteria: review article. FEMS Microbiol Rev. 2009;33(4):801–819. doi: 10.1111/j.1574-6976.2009.00176.x. [DOI] [PubMed] [Google Scholar]
- 34.Young R. Phage lysis: three steps, three choices, one outcome. J Microbiol. 2014;52(3):243–258. doi: 10.1007/s12275-014-4087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berry J, Rajaure M, Pang T, Young R. The spanin complex is essential for lambda lysis. J Bacteriol. 2012;194(20):5667–5674. doi: 10.1128/JB.01245-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young R, Way J, Way S, Yin J, Syvanen M. Transposition mutagenesis of bacteriophage lambda: a new gene affecting cell lysis. J Mol Biol. 1979;132(3):307–322. doi: 10.1016/0022-2836(79)90262-6. [DOI] [PubMed] [Google Scholar]
- 37.Krupovic M, Cvirkaite-Krupovic V, Bamford DH. Identification and functional analysis of the Rz/Rz1-like accessory lysis genes in the membrane-containing bacteriophage PRD1. Mol Microbiol. 2008;68(2):492–503. doi: 10.1111/j.1365-2958.2008.06165.x. [DOI] [PubMed] [Google Scholar]
- 38.Berry J, Savva C, Holzenburg A, Young R. The lambda spanin components Rz and Rz1 undergo tertiary and quaternary rearrangements upon complex formation. Protein Sci. 2010;19(10):1967–1977. doi: 10.1002/pro.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson D, Loomis L, Fischetti VA. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc Natl Acad Sci U S A. 2001;98(7):4107–4112. doi: 10.1073/pnas.061038398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmelcher M, Donovan DM, Loessner MJ. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012;7:1147–1171. doi: 10.2217/fmb.12.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Secretary O of the P . Fact sheet: Obama administration releases national action plan to combat antibiotic-resistant bacteria. Washington (DC): The White House; 2015. [Google Scholar]
- 42.Haddad Kashani H, Schmelcher M, Sabzalipoor H, Seyed Hosseini E, Moniri R. Recombinant endolysins as potential therapeutics against antibiotic-resistant staphylococcus aureus: current status of research and novel delivery strategies. Clin Microbiol Rev. 2018. doi: 10.1128/CMR.00071-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmelcher M, Powell AM, Camp MJ, Pohl CS, Donovan DM. Synergistic streptococcal phage λSA2 and B30 endolysins kill streptococci in cow milk and in a mouse model of mastitis. Appl Microbiol Biotechnol. 2015;99(20):8475–8486. doi: 10.1007/s00253-015-6579-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopez R, Garcia E, Garcia P, Garcia JL. The pneumococcal cell wall degrading enzymes: a modular design to create new lysins? Microb Drug Resist. 1997;3(2):199–211. doi: 10.1089/mdr.1997.3.199. [DOI] [PubMed] [Google Scholar]
- 45.Loessner MJ, Kramer K, Ebel F, Scherer S. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol Microbiol. 2002;44(2):335–349. doi: 10.1046/j.1365-2958.2002.02889.x. [DOI] [PubMed] [Google Scholar]
- 46.Briers Y, Schmelcher M, Loessner MJ, Hendrix J, Engelborghs Y, Volckaert G, Lavigne R. The high-affinity peptidoglycan binding domain of Pseudomonas phage endolysin KZ144. Biochem Biophys Res Commun. 2009;383(2):187–191. doi: 10.1016/j.bbrc.2009.03.161. [DOI] [PubMed] [Google Scholar]
- 47.Yang H, Linden SB, Wang J, Yu J, Nelson DC, Wei H. A chimeolysin with extended-spectrum streptococcal host range found by an induced lysis-based rapid screening method. Sci Rep. 2015;5:17257. doi: 10.1038/srep17257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Domenech M, Garcia E, Moscoso M. In vitro destruction of Streptococcus pneumoniae biofilms with bacterial and phage peptidoglycan hydrolases. Antimicrob Agents Chemother. 2011;55(9):4144–4148. doi: 10.1128/AAC.00492-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuch R, Nelson D, Fischetti VA. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature. 2002;418(6900):884–889. doi: 10.1038/nature01026. [DOI] [PubMed] [Google Scholar]
- 50.Santamaria RI, Bustos P, Sepulveda-Robles O, Lozano L, Rodríguez C, Fernández JL, Juárez S, Kameyama L, Guarneros G, Dávila G, et al. Narrow-host-range bacteriophages that infect Rhizobium etli associate with distinct genomic types. Appl Environ Microbiol. 2014;80(2):446–454. doi: 10.1128/AEM.02256-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Attai H, Rimbey J, Smith GP, Brown PJB. Expression of a peptidoglycan hydrolase from lytic bacteriophages Atu_ph02 and Atu_ph03 triggers lysis of agrobacterium tumefaciens. Appl Environ Microbiol. 2017;83:23. doi: 10.1128/AEM.01498-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Flaherty S, Coffey A, Meaney W, Fitzgerald GF, Ross RP. The recombinant phage lysin LysK has a broad spectrum of lytic activity against clinically relevant staphylococci, including methicillin-resistant Staphylococcus aureus. J Bacteriol. 2005;187(20):7161–7164. doi: 10.1128/JB.187.20.7161-7164.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abedon ST. Phage therapy dosing: the problem(s) with multiplicity of infection (MOI). Bacteriophage. 2016;6(3):e1220348. doi: 10.1080/21597081.2016.1220348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haddad Kashani H, Fahimi H, Dasteh Goli Y, Moniri R, Novel Chimeric A. Endolysin with antibacterial activity against methicillin-resistant staphylococcus aureus. Front Cell Infect Microbiol. 2017;7:290. doi: 10.3389/fcimb.2017.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Love MJ, Bhandari D, Dobson RCJ, Billington C. Potential for bacteriophage endolysins to supplement or replace antibiotics in food production and clinical care. Antibiot (Basel, Switzerland). 2018;7(1). doi: 10.3390/antibiotics7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rahimzadeh G, Gill P, Rezai MS. Endolysins of bacteriophages as an anti-methicillin resistant staphylococcus aureus infection in children: a narrative review. J Pediatr Rev. 2017;6. In Press. doi: 10.5812/jpr.11562. [DOI] [Google Scholar]
- 57.Fernandez L, Gonzalez S, Campelo AB, Martinez B, Rodriguez A, Garcia P. Downregulation of autolysin-encoding genes by phage-derived lytic proteins inhibits biofilm formation in staphylococcus aureus. Antimicrob Agents Chemother. 2017;61:5. doi: 10.1128/AAC.02724-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fischetti VA. Development of phage lysins as novel therapeutics: a historical perspective. Viruses. 2018;10(6):310. doi: 10.3390/v10060310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jonczyk E, Klak M, Miedzybrodzki R, Gorski A. The influence of external factors on bacteriophages – review. Folia Microbiol (Praha). 2011;56(3):191–200. doi: 10.1007/s12223-011-0039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Payne RJH, Jansen VAA. Pharmacokinetic principles of bacteriophage therapy. Clin Pharmacokinet. 2003;42(4):315–325. doi: 10.2165/00003088-200342040-00002. [DOI] [PubMed] [Google Scholar]
- 61.Jun SY, Jang IJ, Yoon S, Jang K, Yu K-S, Cho JY, Seong M-W, Jung GM, Yoon SJ, Kang SH, et al. Pharmacokinetics and tolerance of the phage endolysin-based candidate drug SAL200 after a single intravenous administration among healthy volunteers. Antimicrob Agents Chemother. 2017;61:6. doi: 10.1128/AAC.02629-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loessner MJ. Bacteriophage endolysins – current state of research and applications. Curr Opin Microbiol. 2005;8(4):480–487. doi: 10.1016/j.mib.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 63.Rodriguez-Rubio L, Gutierrez D, Donovan DM, Martinez B, Rodriguez A, Garcia P. Phage lytic proteins: biotechnological applications beyond clinical antimicrobials. Crit Rev Biotechnol. 2016;36(3):542–552. doi: 10.3109/07388551.2014.993587. [DOI] [PubMed] [Google Scholar]
- 64.Mayer MJ, Garefalaki V, Spoerl R, Narbad A, Meijers R. Structure-based modification of a Clostridium difficile-targeting endolysin affects activity and host range. J Bacteriol. 2011;193(19):5477–5486. doi: 10.1128/JB.00439-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oliveira H, Vilas Boas D, Mesnage S, Kluskens LD, Lavigne R, Sillankorva S, Secundo F, Azeredo J. Structural and enzymatic characterization of ABgp46, a novel phage endolysin with broad anti-gram-negative bacterial activity. Front Microbiol. 2016;7:208. doi: 10.3389/fmicb.2016.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Callewaert L, Walmagh M, Michiels CW, Lavigne R. Food applications of bacterial cell wall hydrolases. Curr Opin Biotechnol. 2011;22(2):164–171. doi: 10.1016/j.copbio.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 67.Briers Y, Walmagh M, Lavigne R. Use of bacteriophage endolysin EL188 and outer membrane permeabilizers against Pseudomonas aeruginosa. J Appl Microbiol. 2011;110(3):778–785. doi: 10.1111/j.1365-2672.2010.04931.x. [DOI] [PubMed] [Google Scholar]
- 68.Oliveira H, Azeredo J, Lavigne R, Kluskens L. Bacteriophage endolysins as a response to emerging foodborne pathogens. Trends Food Sci Technol. 2012;28:103–115. doi: 10.1016/j.tifs.2012.06.016. [DOI] [Google Scholar]
- 69.Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32(2):149–167.doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 70.Peltier J, Courtin P, El Meouche I, Lemée L, M-P C-C, Pons J-L. Clostridium difficile has an original peptidoglycan structure with a high level of N-acetylglucosamine deacetylation and mainly 3-3 cross-links. J Biol Chem. 2011;286(33):29053–29062. doi: 10.1074/jbc.M111.259150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van Tassell ML, Ibarra-Sánchez LA, Hoepker GP, Miller MJ. Hot topic: antilisterial activity by endolysin PlyP100 in fresh cheese. J Dairy Sci. 2017;100(4):2482–2487. doi: 10.3168/jds.2016-11990. [DOI] [PubMed] [Google Scholar]
- 72.Meessen-Pinard M, Sekulovic O, Fortier L-C. Evidence of in vivo prophage induction during Clostridium difficile infection. Appl Environ Microbiol. 2012;78(21):7662–7670. doi: 10.1128/AEM.02275-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hargreaves KR, Clokie MRJ. Clostridium difficile phages: still difficult? Front Microbiol. 2014:5. doi: 10.3389/fmicb.2014.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nale JY, Spencer J, Hargreaves KR, Trzepiński P, Douce GR, Clokie MRJ. Bacteriophage combinations significantly reduce clostridium difficile growth in vitro and proliferation in vivo. Antimicrob Agents Chemother. 2016;60:968–981. doi: 10.1128/AAC.01774-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Govind R, Fralick JA, Rolfe RD. Genomic organization and molecular characterization of Clostridium difficile bacteriophage PhiCD119. J Bacteriol. 2006;188(7):2568–2577. doi: 10.1128/JB.188.7.2568-2577.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goh S, T V R, Chang BJ. Isolation and characterization of temperate bacteriophages of Clostridium difficile. Appl Environ Microbiol. 2005;71(2):1079–1083. doi: 10.1128/AEM.71.2.1079-1083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goh S, Ong PF, Song KP, T V R, Chang BJ. The complete genome sequence of Clostridium difficile phage phiC2 and comparisons to phiCD119 and inducible prophages of CD630. Microbiology. 2007;153(Pt3):676–685. doi: 10.1099/mic.0.2006/002436-0. [DOI] [PubMed] [Google Scholar]
- 78.Mayer MJ, Narbad A, Gasson MJ. Molecular characterization of a Clostridium difficile bacteriophage and its cloned biologically active endolysin. J Bacteriol. 2008;190(20):6734–6740. doi: 10.1128/JB.00686-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Horgan M, O’Sullivan O, Coffey A, Fitzgerald GF, van Sinderen D, McAuliffe O, Ross RP. Genome analysis of the Clostridium difficile phage PhiCD6356, a temperate phage of the Siphoviridae family. Gene. 2010;462(1–2):34–43. doi: 10.1016/j.gene.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 80.Sekulovic O, Meessen-Pinard M, Fortier L-C. Prophage-stimulated toxin production in Clostridium difficile NAP1/027 lysogens. J Bacteriol. 2011;193(11):2726–2734. doi: 10.1128/JB.00787-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hargreaves KR, Kropinski AM, Clokie MRJ. What does the talking?: quorum sensing signalling genes discovered in a bacteriophage genome. PLoS One. 2014;9(1):e85131. doi: 10.1371/journal.pone.0085131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hargreaves KR, Flores CO, Lawley TD, Clokie MRJ. Abundant and diverse clustered regularly interspaced short palindromic repeat spacers in Clostridium difficile strains and prophages target multiple phage types within this pathogen. MBio. 2014;5(5):e01045–13. doi: 10.1128/mBio.01045-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boudry P, Semenova E, Monot M, Datsenko KA, Lopatina A, Sekulovic O, Ospina-Bedoya M, Fortier L-C, Severinov K, Dupuy B, et al. Function of the CRISPR-Cas system of the human pathogen Clostridium difficile. MBio. 2015;6(5):e01112–15. doi: 10.1128/mBio.01112-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wittmann J, Riedel T, Bunk B, Sproer C, Gronow S, Overmann J. Complete genome sequence of the novel temperate Clostridium difficile phage phiCDIF1296T.GenomeAnnounc.2015;3(4).doi: 10.1128/genomeA.00839-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sekulovic O, Garneau JR, Neron A, Fortier L-C. Characterization of temperate phages infecting Clostridium difficile isolates of human and animal origins. Appl Environ Microbiol. 2014;80(8):2555–2563. doi: 10.1128/AEM.00237-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hargreaves KR, Clokie MRJ, Taxonomic A. Review of Clostridium difficile phages and proposal of a novel genus, “Phimmp04likevirus”. Viruses. 2015;7(5):2534–2541. doi: 10.3390/v7052534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rashid SJ, Barylski J, Hargreaves KR, Millard AA, Vinner GK, Clokie MRJ. Two novel myoviruses from the North of Iraq reveal insights into Clostridium difficile phage diversity and biology. Viruses. 2016;8:11. doi: 10.3390/v8110310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Riedel T, Wittmann J, Bunk B, Schober I, Spröer C, Gronow S, Overmann J. A Clostridioides difficile bacteriophage genome encodes functional binary toxin-associated genes. J Biotechnol. 2017;250:23–28. doi: 10.1016/j.jbiotec.2017.02.017. [DOI] [PubMed] [Google Scholar]
- 89.Mehta KK, Paskaleva EE, Wu X, Grover N, Mundra RV, Chen K, Zhang Y, Yang Z, Feng H, Dordick JS, et al. Newly identified bacteriolytic enzymes that target a wide range of clinical isolates of Clostridium difficile. Biotechnol Bioeng. 2016;113(12):2568–2576. doi: 10.1002/bit.26029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42(W1):W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peng Z, Wang S, Gide M, Zhu D, Lamabadu Warnakulasuriya Patabendige HM, Li C, Cai J, Sun X. A novel bacteriophage lysin-human defensin fusion protein is effective in treatment of clostridioides difficileinfection in mice. Front Microbiol. 2019:9. doi: 10.3389/fmicb.2018.03234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Roy Chowdhury P, Demaere M, Chapman T, Worden P, Charles IG, Darling AE, Djordjevic SP. Comparative genomic analysis of toxin-negative strains of Clostridium difficile from humans and animals with symptoms of gastrointestinal disease. BMC Microbiol. 2016;16(1). doi: 10.1186/s12866-016-0653-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Furci L, Baldan R, Bianchini V, Trovato A, Ossi C, Cichero P, Cirillo DM. New role for human α-defensin 5 in the fight against hypervirulent Clostridium difficile strains. Infect Immun. 2015;83(3):986–995. doi: 10.1128/IAI.02955-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shin JH, Chaves-Olarte E, Warren CA. Clostridium difficile Infection. Microbiol Spectr. 2016;4(3). doi: 10.1128/microbiolspec.EI10-0007-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vermassen A, Leroy S, Talon R, Provot C, Popowska M, Desvaux M. Cell wall hydrolases in bacteria: insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Front Microbiol. 2019;10:331. doi: 10.3389/fmicb.2019.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vollmer W, Joris B, Charlier P, Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev. 2008;32(2):259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 97.Payne KM, Hatfull GF. Mycobacteriophage endolysins: diverse and modular enzymes with multiple catalytic activities. PLoS One. 2012;7(3):e34052. doi: 10.1371/journal.pone.0034052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sheehan MM, Garcia JL, Lopez R, Garcia P. The lytic enzyme of the pneumococcal phage Dp-1: a chimeric lysin of intergeneric origin. Mol Microbiol. 1997;25(4):717–725. doi: 10.1046/j.1365-2958.1997.5101880.x. [DOI] [PubMed] [Google Scholar]
- 99.Boyce JD, Davidson BE, Hillier AJ. Identification of prophage genes expressed in lysogens of the Lactococcus lactis bacteriophage BK5-T. Appl Environ Microbiol. 1995;61(11):4099–4104. doi: 10.1128/AEM.61.11.4099-4104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pritchard DG, Dong S, Kirk MC, Cartee RT, Baker JR. LambdaSa1 and LambdaSa2 prophage lysins of Streptococcus agalactiae. Appl Environ Microbiol. 2007;73(22):7150–7154. doi: 10.1128/AEM.01783-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gerstmans H, Rodríguez-Rubio L, Lavigne R, Briers Y. From endolysins to Artilysin®s: novel enzyme-based approaches to kill drug-resistant bacteria. Biochem Soc Trans. 2016;44(1):123–128. doi: 10.1042/BST20150192. [DOI] [PubMed] [Google Scholar]
- 102.Fischetti VA. Bacteriophage endolysins: A novel anti-infective to control Gram-positive pathogens. Int J Med Microbiol. 2010;300(6):357–362. doi: 10.1016/j.ijmm.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fischetti VA. Using phage lytic enzymes to control pathogenic bacteria. BMC Oral Health. 2006;6(S1). doi: 10.1186/1472-6831-6-S1-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Loeffler JM, Nelson D, Fischetti VA. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science. 2001:80. doi: 10.1126/science.1066869. [DOI] [PubMed] [Google Scholar]
- 105.Gilmer DB, Schmitz JE, Euler CW, Fischetti VA. Novel bacteriophage lysin with broad lytic activity protects against mixed infection by streptococcus pyogenes and methicillin-resistant staphylococcus aureus. Antimicrob Agents Chemother. 2013;57:2743–2750. doi: 10.1128/AAC.02526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pastagia M, Euler C, Chahales P, Fuentes-Duculan J, Krueger JG, Fischetti VA. A novel chimeric lysin shows superiority to mupirocin for skin decolonization of methicillin-resistant and -sensitive staphylococcus aureus strains. Antimicrob Agents Chemother. 2011;55:738–744. doi: 10.1128/AAC.00890-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gutiérrez D, Fernández L, Rodríguez A, García P. Are phage lytic proteins the secret weapon to kill staphylococcus aureus? MBio. 2018:9. doi: 10.1128/mBio.01923-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Navarro F, Muniesa M. Phages in the human body. Front Microbiol. 2017:8. doi: 10.3389/fmicb.2017.00566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jun SY, Jung GM, Yoon SJ, Choi YJ, Koh WS, Moon KS, Kang SH, et al. Preclinical safety evaluation of intravenously administered SAL200 containing the recombinant phage endolysin SAL-1 as a pharmaceutical ingredient. Antimicrob Agents Chemother. 2014;58:2084–2088. doi: 10.1128/AAC.02232-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Peng Z, Wang S, Gide M, Zhu D, Lamabadu Warnakulasuriya Patabendige HM, Li C, Cai J, Sun X. A novel bacteriophage lysin-human defensin fusion protein is effective in treatment of clostridioides difficile infection in mice. Front Microbiol. 2018;9:3234. doi: 10.3389/fmicb.2018.03234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rashel M, Uchiyama J, Ujihara T, Uehara Y, Kuramoto S, Sugihara S, Yagyu K-I, Muraoka A, Sugai M, Hiramatsu K, et al. Efficient elimination of multidrug-resistant staphylococcus aureus by cloned lysin derived from bacteriophage φMR11. J Infect Dis. 2007;196:1237–1247. doi: 10.1086/521305. [DOI] [PubMed] [Google Scholar]
- 112.Entenza JM, Loeffler JM, Grandgirard D, Fischetti VA, Moreillon P. Therapeutic effects of bacteriophage Cpl-1 lysin against Streptococcus pneumoniae endocarditis in rats. Antimicrob Agents Chemother. 2005;49(11):4789–4792. doi: 10.1128/AAC.49.11.4789-4792.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Daniel A, Euler C, Collin M, Chahales P, Gorelick KJ, Fischetti VA. Synergism between a novel chimeric lysin and oxacillin protects against infection by methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2010;54(4):1603–1612. doi: 10.1128/AAC.01625-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Witzenrath M, Schmeck B, Doehn JM, Tschernig T, Zahlten J, Loeffler JM, Zemlin M, Müller H, Gutbier B, Schütte H, et al. Systemic use of the endolysin Cpl-1 rescues mice with fatal pneumococcal pneumonia. Crit Care Med. 2009;37(2):642–649. doi: 10.1097/CCM.0b013e31819586a6. [DOI] [PubMed] [Google Scholar]
- 115.Herpers BL, Badoux P, Pietersma F, Eichenseher F, Loessner MJ. Specific lysis of methicillin susceptible and resistant Staphylococcus aureus by the endolysin Staphefekt SA.100TM. Presented at the 24th European Congress of Clinical Microbiology and Infectious Diseases (ECCMID) 2014, Barcelona, Spain, 10–13 May 2014; abstr. R144. [Google Scholar]
- 116.Totté J, de Wit J, Pardo L, Schuren F, van Doorn M, Pasmans S. Targeted anti-staphylococcal therapy with endolysins in atopic dermatitis and the effect on steroid use, disease severity and the microbiome: study protocol for a randomized controlled trial (MAAS trial). Trials. 2017;18(1). doi: 10.1186/s13063-017-2118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Czaplewski L, Bax R, Clokie M, Dawson M, Fairhead H, Fischetti VA, Foster S, Gilmore BF, Hancock REW, Harper D, et al. Alternatives to antibiotics – a pipeline portfolio review. Lancet Infect Dis. 2016;16(2):239–251. doi: 10.1016/S1473-3099(15)00466-1. [DOI] [PubMed] [Google Scholar]
- 118.Loeffler JM, Djurkovic S, Fischetti VA. Phage lytic enzyme Cpl-1 as a novel antimicrobial for pneumococcal bacteremia. Infect Immun. 2003;71(11):6199–6204. doi: 10.1128/IAI.71.11.6199-6204.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liong M, Lu J, Kovochich M, Xia T, Ruehm SG, Nel AE, Tamanoi F, Zink JI. Multifunctional inorganic nanoparticles for imaging, targeting, and drug delivery. ACS Nano. 2008;2(5):889–896. doi: 10.1021/nn800072t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Steiniger SCJ, Kreuter J, Khalansky AS, Skidan IN, Bobruskin AI, Smirnova ZS, Severin SE, Uhl R, Kock M, Geiger KD, et al. Chemotherapy of glioblastoma in rats using doxorubicin-loaded nanoparticles. Int J Cancer. 2004;109(5):759–767. doi: 10.1002/ijc.20048. [DOI] [PubMed] [Google Scholar]
- 121.Kell AJ, Stewart G, Ryan S, Peytavi R, Boissinot M, Huletsky A, Bergeron MG, Simard B. Vancomycin-modified nanoparticles for efficient targeting and preconcentration of gram-positive and gram-negative bacteria. ACS Nano. 2008;2(9):1777–1788. doi: 10.1021/nn700183g. [DOI] [PubMed] [Google Scholar]
- 122.Choi KH, Lee HJ, Park BJ, Wang KK, Shin EP, Park JC, Kim YK, Oh MK, Kim YR, et al. Photosensitizer and vancomycin-conjugated novel multifunctional magnetic particles as photoinactivation agents for selective killing of pathogenic bacteria. Chem Commun. 2012. doi: 10.1039/c2cc17766h. [DOI] [PubMed] [Google Scholar]
- 123.Gervasi T, Lo Curto R, Minniti E, Narbad A, Mayer MJ. Application of lactobacillus johnsonii expressing phage endolysin for control of Clostridium perfringens. Lett Appl Microbiol. 2014;59(4):355–361. doi: 10.1111/lam.12298. [DOI] [PubMed] [Google Scholar]
- 124.Gervasi T, Horn N, Wegmann U, Dugo G, Narbad A, Mayer MJ. Expression and delivery of an endolysin to combat Clostridium perfringens. Appl Microbiol Biotechnol. 2014;98(6):2495–2505. doi: 10.1007/s00253-013-5128-y. [DOI] [PMC free article] [PubMed] [Google Scholar]


