ABSTRACT
Probiotics are recognized for outcompeting pathogenic bacteria by competitive receptor-mediated colonization and secretion of functional metabolites which are antimicrobial against certain microbes as well as improving host’s gut health and immunity. Recently, we have constructed a bioactive Lactobacillus casei (LC) strain, LC+mcra, by inserting mcra (myosin cross-reactive antigen) gene, which stimulates the conversion of conjugated linoleic acids. In this study, we evaluated the modulation of gut microbiome and protective roles of LC+mcra against pathogenic Salmonella enterica serovar Typhimurium (ST) and enterohemorrhagic E. coli (EHEC) infections in BALB/cJ mice. We observed that LC+mcra colonized efficiently in mice gut intestine and competitively reduced the infection with ST and EHEC in various locations of small and large intestine, specifically cecum, jejunum, and ileum (p < 0.05). Positive modulation of the cecal microbiota, for example, higher relative abundances of Firmicutes, lower relative abundances of Proteobacteria, and increased bacterial species diversity/richness, was detected in ST-challenged mice pretreated with LC+mcra based on 16S metagenomic sequencing. Cytokine gene expression analysis indicated that mice pretreated with LC+mcra associated with attenuated bacterial pathogen-induced gut inflammation. Furthermore, mice fed daily with LC+mcra for one week could protect themselves from the impairments caused by enteric infections with ST or EHEC. These impairments include weight loss, negative hematological changes, intestinal histological alterations, and potential death. This in vivo study suggests that daily consumption of novel conjugated linoleic acids over-producing probiotic effectively improves intestinal microbiota composition and prevents/combats foodborne enteric bacterial infections with pathogenic Salmonella and diarrheagenic E. coli.
KEYWORDS: Probiotic, conjugated linoleic acid, enteric bacterial pathogen, salmonellosis, metagenomics, intestinal microbiota
Introduction
The majority of human gut epithelial surfaces are colonized and safeguarded by a tremendous number of microorganisms including bacteria, viruses, fungi, and protozoans which are known as common gut microflora; each of them is crucial in forming and balancing a complex ecosystem with microbial diversity.1 These large number of microorganisms build up a microbial genetic repertoire approximately 100 times greater than that of the human host.2 Diversity of these microbes, specifically number of diverse bacterial species, is essential for good health and immunity of host.3,4 According to recent reports, human distal gastrointestinal (GI) tract can house more than 1000 distinct bacterial species, and the total number was estimated to be larger than 1014 CFU/gm of fecal material.5 Bacteroidetes, Firmicutes, Proteobacteria, and Actinobacteria are the prevalent bacterial phyla in human gut microbiota and each of these phyla contains dozens of bacterial genus and hundreds of species.6–8
In a gut ecosystem with homeostatic condition, most of the commensal bacteria colonize and survive symbiotically, whereas conditions such as immunodeficiency, malnutrition, and antibiotic-therapy cause dysbiosis and imbalance of commensal bacteria that induce pathogenesis and cause diseases.9,10 Furthermore, broad-spectrum antibiotic therapy or any other detrimental conditions may disturb the gut ecosystem balance long-term or lead to chronically irritated bowels, reducing the number of beneficial bacteria and increasing the number of opportunistic pathogens and their toxic products that further weaken the host defense and/or induce inflammation and damage.11 As a consequence of imbalanced gut microflora, opportunistic pathogens, their produced metabolites, proteins, and/or toxins can take over the gut ecosystem and negatively impact host gut health.
Salmonella and diarrheagenic Escherichia coli generally infect human gut intestine through consumption of contaminated foods and/or drinks.12–14 Once these Gram-negative enteric pathogenic bacteria arrive in host gut, their complex type III secretion systems are activated, enabling them to introduce effector proteins directly into cell cytoplasm. Series of pathogenesis through type III secretion systems induce systematic infections causing acute or chronic inflammation and other serious disorders in the host.15 However, such enteric illness is usually facilitated by compromised gut immunity and dysbiotic gut microbiota which provide those enteric bacterial pathogens with weakened colonization resistance.16 On the other hand, traditional antibiotic therapy has been found to lyse enterohemorrhagic E. coli (EHEC) which further increases the risk for hemolytic-uremic syndrome (HUS), a post infectious sequelae in the patients.17,18 In these situations, procommensal strategies by application of probiotics, prebiotics, and synbiotics can be considered as priority in prevention and treatment of such foodborne bacterial pathogen-induced enteric illness.13,19,20 With a promising scheme, it allows an establishment or recovery of the healthy enteric microbial ecosystem by introducing native, exogenous, or genetically engineered beneficial probiotics without inducing deleterious effects (like antibiotics) on human commensal gut bacteria.16,21
Recently, we constructed and reported the role of a multi-functional Lactobacillus casei probiotic strain overexpressing myosin cross-reactive antigen gene (mcra), named as LC+mcra..22 Several groups of researchers have demonstrated the health-beneficial effects of conjugated linoleic acids, such as anti-carcinogenesis, anti-oxidant, and anti-microbial effects.16,23,24 Similarly, we have also revealed the anti-pathogenic and anti-inflammatory properties of linoleic acids over-producing L. casei (LC+mcra) based on in vitro examination. Here in this study, we aimed to evaluate the protective roles of LC+mcra on modulating/recovering gut intestinal microflora composition and combating/alleviating foodborne enteric bacterial pathogenic infections in vivo based on mice model.
Results
Probiotics preventing ST infection induced physiological abnormalities in mice
The weight of each mouse was monitored every day for the purpose of investigating if probiotics preventive administration could rescue mice from weight loss due to ST/EHEC infection (Figure 1). Within the entire 4-week rearing, a total of 12 mice in control group (no probiotic given), 7 mice in group given wild-type probiotic LC strain, and 1 mouse in group given linoleic acid over-expressed mutant LC+mcra strain were sacrificed due to their health abnormality induced by ST infection. These sacrificed individuals included 8 mice from control and 5 mice from LC treatment found self-death due to ST challenge, but none from LC+mcra treatment, which provided us the ST survival rates as 60% in control group, 75% in LC group, and 100% in LC+mcra group (Figure 2). The death of the mice was generally accompanied with extreme (>20%) weight loss to approximately 8–10 g.
Figure 1.
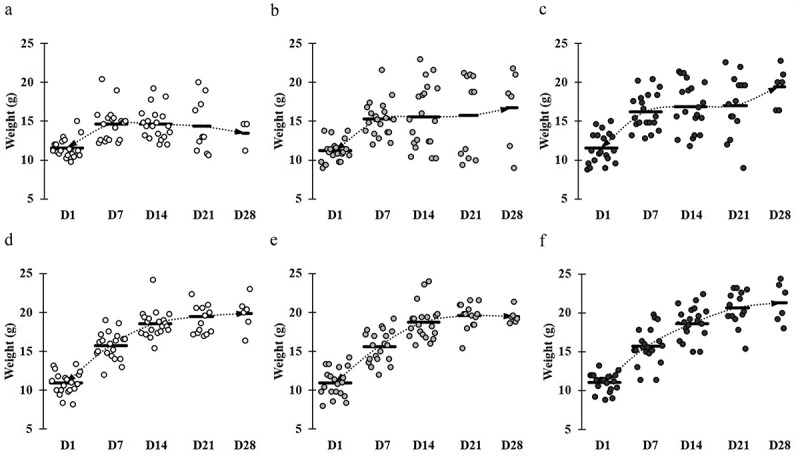
Comparative weight gain and loss in mice across different groups. Mice groups were assigned with the following manner: (a) ST infection, (b) ST infection and LC 1-week pretreatment, (c) ST infection and LC+mcra 1-week pretreatment, (d) EHEC infection, (e) EHEC infection and LC 1-week pretreatment, and (f) EHEC infection and LC+mcra 1-week pretreatment. Each dot indicates individual mouse weight and horizontal bars at each time point indicate averaged weight of mice in accordant group.
Figure 2.
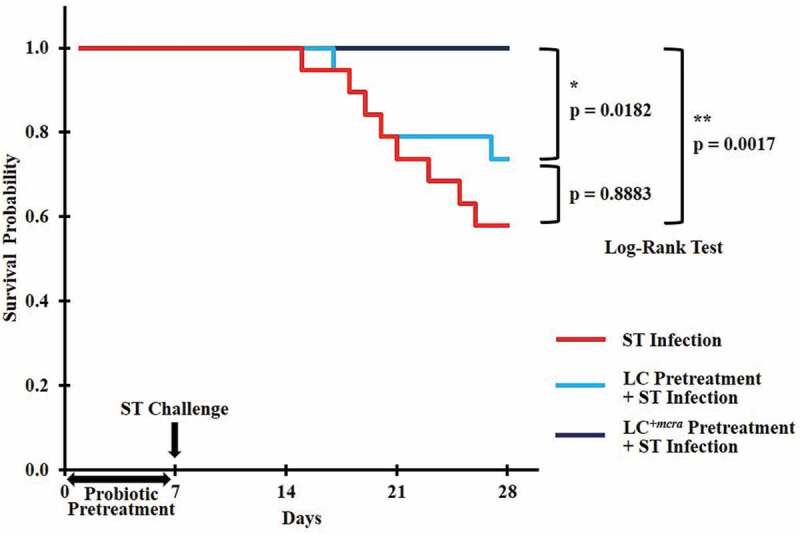
Kaplan-Meier survival curve for ST-infected mice only, or mice pretreated with wild-type probiotic strain LC or bioactive probiotic strain LC+mcra before ST-infection. Survival probability plotted over time and compared at significant level of 0.05 (*) or 0.01 (**) by Log-Rank test.
At the end of week 2, the average weight of mice in control group reached approximately 14–16 g, whereas both groups of mice which were given either LC or LC+mcra gained weight at range of 1–2 g more compared to the control group of mice. Once mice were challenged with ST, the average weight gain trend of mice in control group which was not given probiotic was suspended and remained at 14.65 g during 1st week of post-challenge. Then the weight of those mice decreased to 14.36 g and 13.47 g at 2nd and 3rd post-infection weeks, respectively (Figure 1A). However, the mice which were administered LC+mcra kept continuing to gain average weight. In spite of the negative effect induced by ST infection, mice which were given LC+mcra gained weight at 16.88 g, 17.02 g, and 19.12 g at the 1st, 2nd, and 3rd week of post-infections, respectively (Figure 1C). The wild-type probiotic, LC fed mice exhibited mild effects in maintaining the average body weight during the first two weeks of ST infection and gained approximately 1.5 g weight at the end of 3rd post-infection week (Figure 1B). On the other hand, we failed to observe any negative effects including average weight loss induced by EHEC infection. However, the oral administration of LC+mcra in mice was more effective in stimulating the weight gain by 1.1g and 1.4 g at the 2nd and 3rd post-infection weeks compared with mice administered LC (Figure 1D,E,F).
Reduction on colonization of ST and EHEC in probiotics fed mice
Either LC or LC+mcra was orally administered to mice in order to examine their colonization ability in mice gut and evaluate their preventive role in altering enteric pathogenic bacterial colonization and infection in gastrointestinal tract of mice using BALB/cJ mice model. According to the colonization data collected from two individual mice trials, both LC and LC+mcra were able to colonize well in gut of BALB/cJ mice but the genetically modified probiotic strain, LC+mcra, could colonize in the mice gut more aggressively compared to the wild-type LC strain. Further, both LC and LC+mcra significantly reduced the colonization and infection of both enteric bacterial pathogens, ST and EHEC in BALB/cJ mice. We found that mice fed with LC+mcra could defend ST infection remarkably and recover fully within a week of challenge. Specifically, mice highly colonized with LC+mcra strain showed significantly reduced cecal colonization with ST (approximately 1 log CFU/g) compared to the group of mice which were given wild-type LC strain at all three time points (14, 21, and 28 d) (Figure 3A).
Figure 3.
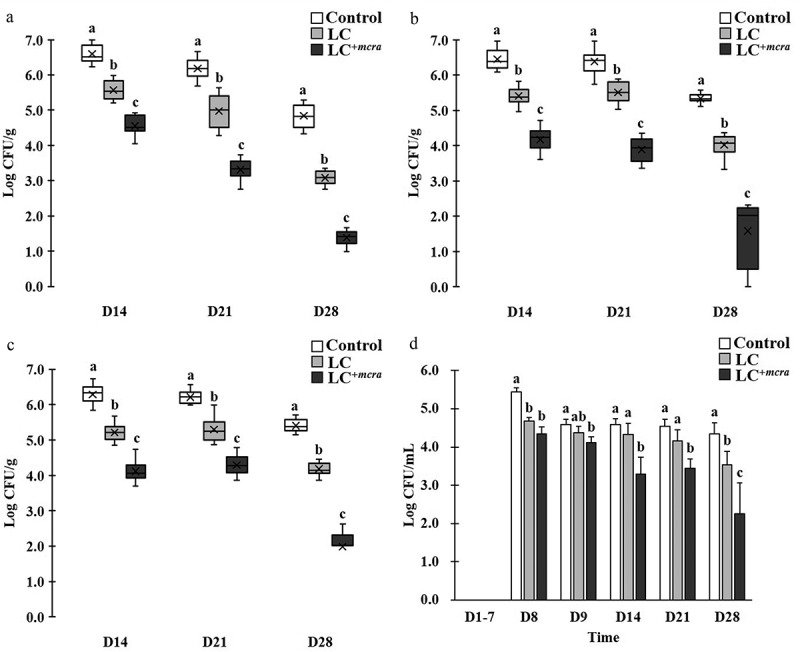
Effect of LC+mcra on reducing colonization of ST in mice gut intestine. The bacterial numbers of ST at 14, 21, and 28 days in cecum (a), jejunum (b), ileum (c), and feces (d) from ST-infected mice with no probiotic treatment, LC, or LC+mcra 1-week pretreatment were investigated in triplicate. Different letters (‘a’ through ‘c’) at individual time point (day 14, 21, or 28) are significantly different (p < 0.05) in the numbers of ST among control, LC pretreatment, and LC+mcra pretreatment.
To compare the colonization of ST in jejunum, we observed that LC or LC+mcra pre-administered mice were colonized with lower number of ST at range of 1.0 to 2.3 log CFU ST per gram jejunum fluids at 1st week post-infection, 0.9 and 2.5 log CFU/g at 2nd week post-infection, and 1.3 and 3.7 log CFU/g at 3rd week post-infection (Figure 3B). Similarly, LC and LC+mcra pre-administered mice were colonized with ST in lower rate of 1.7 and 2.2 log CFU per gram ileum fluids at 1st week post-infection, 0.9 and 1.9 log CFU/g on 2nd week post-infection, and 1.2 and 3.4 log CFU/g on 3rd week post-infection to the control mice (Figure 3C).
The significant reduction on ST gut intestinal colonization was also observed in form of decreased ST fecal shedding (Figure 3D). On the 8th day after mice were challenged with ST, both groups of mice administered with either wild-type probiotic LC or genetically modified probiotic LC+mcra strain were colonized with reduced number (0.8 to 1.1 log CFU/mL) ST in feces but the differences became unsubstantial at the 9th day. However, notable major effectiveness of LC+mcra started to exhibit in mice after 1st week post-infection, at which 1.3 log CFU/mL less ST was recovered from mice feces. In the subsequent two weeks, ST fecal shedding from LC+mcra fed mice were observed to be continuously reduced by 1.1 and 2.1 log CFU/mL.
On the other hand, mice which were pretreated with LC barely reduced the EHEC colonization in jejunum and ileum, whereas mice pretreated with LC+mcra showed significant influence in EHEC colonization resistance (Figure 4). Specifically, LC+mcra fed mice were capable of significantly reducing the colonization of EHEC at 2.3, 1.6, and 0.9 log CFU/g in cecum, 1.6, 1.8, and 2.7 log CFU/g in jejunum, and 2.8, 1.8 and 2.1 log CFU/g in ileum at the 1st, 2nd, and 3rd week post-challenge (Figure 4A,B,C). Meanwhile, consequential decreased EHEC fecal shedding was detected in LC+mcra fed mice as well. However, only insignificant reductions (0.1 to 0.5 CFU EHEC less per mL feces) were found during the first two days after EHEC challenge on EHEC-free mice (the 8th and 9th day). The LC+mcra administration substantially lowered 0.9, 1.9, and 2.2 CFU/mL EHEC fecal shedding at the 1st, 2nd, and 3rd post-infection weeks in comparison with control (Figure 4D).
Figure 4.
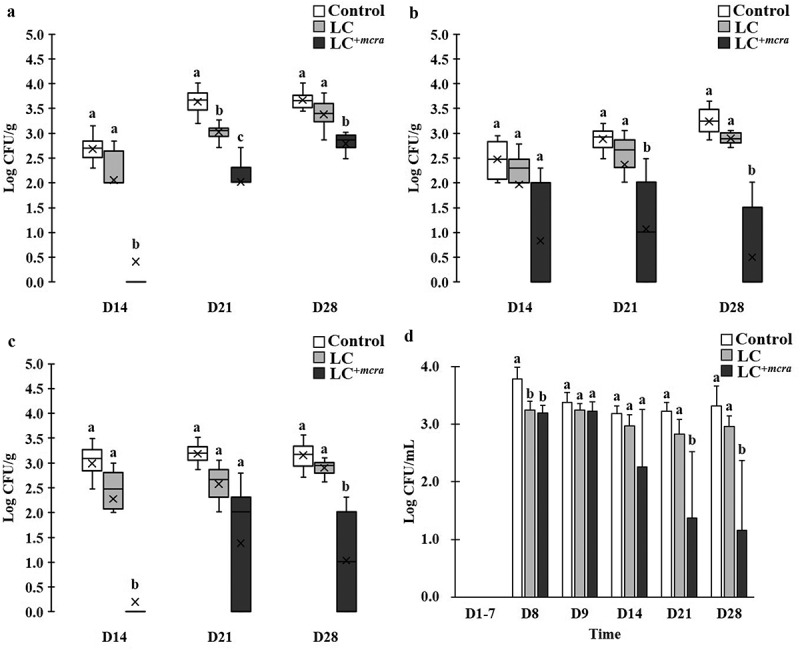
Effect of LC+mcra on reducing colonization of EHEC in mice gut intestine. The bacterial numbers of EHEC at 14, 21, and 28 days in cecum (a), jejunum (b), ileum (c), and feces (d) from EHEC-infected mice with no probiotic treatment, LC, or LC+mcra pretreatment were investigated in triplicate. Different letters (‘a’ through ‘c’) at individual time point (day 14, 21, or 28) are significantly different (p < 0.05) in the numbers of EHEC among control, LC pretreatment, and LC+mcra pretreatment.
Efficient colonization of LC+mcra in mice gut
In order to examine the correlation between probiotic colonization and reduction on intestinal bacterial pathogens, we also compared the colonization level of both LC and LC+mcra in different portion of mice gut (Figure 5). The one-week daily oral administration led to high and stable cecal colonization level of LC+mcra above 106 CFU/g throughout 3 weeks afterwards, which were significantly higher than wild-type LC (Figure 5A). A similar trend was found in mice jejunum (Figure 5B), whereas, LC+mcra only exhibited numerical higher ileum colonization than wild-type LC (Figure 5C).
Figure 5.
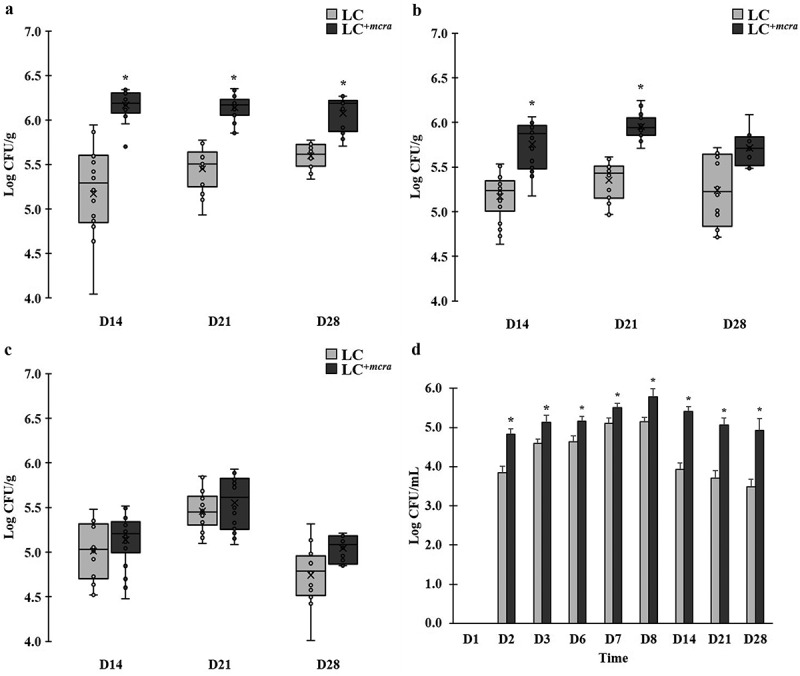
Comparison on colonization levels of LC and LC+mcra in mice gut intestine. The bacterial numbers of specific L. casei at 14, 21, and 28 days in cecum (A), jejunum (B), ileum (C), and feces (D) from mice daily administered with LC or LC+mcra for one week were investigated in triplicate. Asterisk (*) at day 14, 21, or 28 are significantly different (p < 0.05) in the numbers of gut colonized or fecal shedding wild-type LC and LC+mcra.
The fecal shedding number of administered LC were observed to raise after 1st day consumption (Figure 5D). Specifically, LC+mcra fecal shedding colonies gradually increased from 4.8 log CFU/mL, reached 5.8 log CFU/mL at the next day of final daily administration, and slightly decreased to around 5 log CFU/mL after 3 weeks. Whereas, fecal shedding colonies of wild-type LC were observed significantly lower (by 0.4-1.5 log CFU/mL) than LC+mcra. They reached 5.1 log CFU/mL as peak at the next day of final daily administration and ended up with lower than 3.5 log CFU/mL after 3 weeks.
Mice hematology
The hematological changes in mice with ST infection with or without pretreated with probiotic strains at various time points were summarized in Table 3. When compared with control group mice with placebo, ST challenge resulted in dramatic increase of red blood cells (RBC) but decrease of white blood cells (WBC) and platelets (PLT). Both pretreatment of LC or LC+mcra alleviated the increment of RBC and loss of WBC/PLT in mice during Salmonellosis. LC+mcra treatment on mice could further help the mice maintain their normal levels of RBC, WBC, and PLT.
Table 3.
Hematological changes and comparison of mice with and without Salmonella infection.
| Red Blood Cells (106/mm3) |
White Blood Cells (103/mm3) |
Platelets (105/mm3) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day | Control | Infection1 | LC2 | LC+mcra 3 | Control | Infection | LC | LC+mcra | Control | Infection | LC | LC+mcra |
| 14 | 10.15 ± 0.14*b | 13.03 ± 4.20a | 10.84 ± 0.55b | 10.60 ± 0.46b | 3.48 ± 0.35a | 2.95 ± 0.80b | 3.12 ± 0.61a | 3.55 ± 0.42a | 7 ± 3a | 2 ± 2c | 5 ± 2b | 7 ± 3a |
| 21 | 10.34 ± 0.21b | 14.89 ± 3.87a | 10.72 ± 0.65b | 10.51 ± 0.33b | 3.65 ± 0.25a | 2.96 ± 0.40b | 3.13 ± 0.69a | 3.54 ± 0.38a | 8 ± 1a | 2 ± 1c | 5 ± 2b | 8 ± 3a |
| 28 | 10.29 ± 0.13c | 15.64 ± 4.37a | 11.84 ± 2.14b | 10.71 ± 0.53c | 3.59 ± 0.22a | 2.90 ± 0.49b | 3.26 ± 0.30a | 3.51 ± 0.29a | 7 ± 2a | 2 ± 1c | 4 ± 2b | 8 ± 2a |
1Mice with no probiotic protection infected with Salmonella Typhimurium
2Mice pre-administered with wild-type Lactobacillus casei followed by S. Typhimurium infection
3Mice pre-administered with mutant L. casei followed by S. Typhimurium infection
*Means with different letters (a-c) for each type of blood cells (red blood cell, white blood cell, and platelets) in different groups (control, infection, infection following LC pretreatment, and infection following LC+mcra pretreatment) at individual time point (day 14, 21, and 28) are significantly different at p < 0.05
To further evaluate the WBC composition in blood collected from the mice challenged with ST with or without pretreated with probiotic strains, we investigated neutrophils, lymphocytes, monocytes, eosinophils, and basophils counts in different time points, which is summarized in Table 4. The numbers of neutrophils and lymphocytes in blood collected from mice challenged with ST were found to be notably reduced, whereas the monocytes, eosinophils, and basophils levels in mice with salmonellosis were detected to be significantly higher. The pretreatment with either probiotic strain, wild-type LC or mutant LC+mcra, in ST-infected mice assisted in maintenance of their normal WBC composition (including all five cell types studied) at similar levels compared with control group.
Table 4.
White blood cell counts in mice with and without Salmonella infection.
| White Blood Cells | Group | Day |
||
|---|---|---|---|---|
| 14 | 21 | 28 | ||
| Neutrophils (K/µL) |
Control | 1.31 ± 0.40*a | 1.34 ± 0.40a | 1.50 ± 0.21a |
| Infection1 | 0.78 ± 0.36b | 0.41 ± 0.22b | 0.27 ± 0.14b | |
| LC2 | 1.02 ± 0.30ab | 1.07 ± 0.37ab | 1.08 ± 0.48ab | |
| LC+mcra 3 | 1.27 ± 0.56a | 1.16 ± 0.11ab | 1.20 ± 0.23ab | |
| Lymphocytes (K/µL) |
Control | 2.19 ± 0.22a | 2.50 ± 0.13a | 2.44 ± 0.41a |
| Infection | 1.18 ± 0.25b | 0.92 ± 0.15b | 0.73 ± 0.18b | |
| LC | 2.05 ± 0.78a | 2.03 ± 0.13ab | 1.82 ± 0.26a | |
| LC+mcra | 2.37 ± 0.37a | 2.53 ± 0.15a | 2.46 ± 0.52a | |
| Monocytes (K/µL) |
Control | 0.04 ± 0.01b | 0.04 ± 0.02b | 0.04 ± 0.02c |
| Infection | 0.16 ± 0.04a | 0.14 ± 0.06a | 0.19 ± 0.04a | |
| LC | 0.05 ± 0.02b | 0.06 ± 0.03b | 0.08 ± 0.03b | |
| LC+mcra | 0.05 ± 0.02b | 0.05 ± 0.02b | 0.05 ± 0.01c | |
| Eosinophils (K/µL) |
Control | 0.06 ± 0.04b | 0.07 ± 0.03c | 0.07 ± 0.03b |
| Infection | 0.16 ± 0.03a | 0.16 ± 0.04a | 0.17 ± 0.05a | |
| LC | 0.09 ± 0.04b | 0.11 ± 0.01b | 0.09 ± 0.03b | |
| LC+mcra | 0.07 ± 0.03b | 0.06 ± 0.03c | 0.06 ± 0.04b | |
| Basophils (K/µL) |
Control | 0.01 ± 0.01c | 0.01 ± 0.01c | 0.01 ± 0.01c |
| Infection | 0.03 ± 0.01a | 0.03 ± 0.02a | 0.04 ± 0.02a | |
| LC | 0.02 ± 0.01b | 0.02 ± 0.01b | 0.02 ± 0.01b | |
| LC+mcra | 0.01 ± 0.01c | 0.01 ± 0.01c | 0.01 ± 0.01c | |
1Mice with no probiotic protection infected with S. Typhimurium
2Mice pre-administered with wild-type L. casei followed by S. Typhimurium infection
3Mice pre-administered with mutant L. casei followed by S. Typhimurium infection
*Means with different letters (a-c) for each type of WBC (neutrophils, lymphocytes, monocytes, eosinophils, and basophils) in different groups (control, infection, infection following LC pretreatment, and infection following LC+mcra pretreatment) at individual time point (day 14, 21, and 28) are significantly different at p < 0.05
Histopathological changes in mouse cecum
The histological examination of mouse cecal sections is shown in Figure 6. Tissue of cecum collected from the control group mice (Figure 6A) and ST-challenged mice with pre-administration of LC+mcra (Figure 6C) exhibited normal intestinal villi, microvilli, and goblet cells. In comparison, salmonellosis induced variable levels of histological alterations and abnormalities consisting of severe goblet cell depletion, villi/microvilli elimination, and inflammatory infiltrations between circular folds in cecum sections from ST infected mice which were not pretreated with probiotics (Figure 6D,E,F). However, the tissue of cecum collected from the mice administered with LC showed symptoms of salmonellosis, but the induced histopathological changes were mild, such as slight goblet cell reduction and slight changes of villi/microvilli (Figure 6B).
Figure 6.
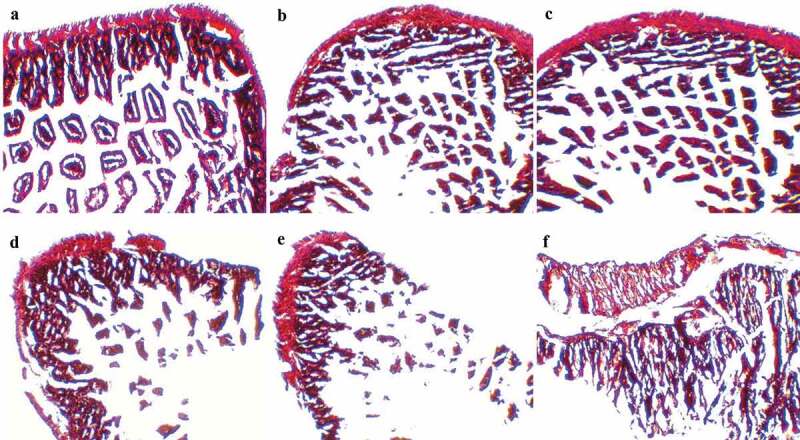
Mice cecum histopathology. Representative H&E-stained cecum sections from experimental groups were showed in panels (a) control mice, (b) intestinal villi and microvilli reduction in ST-infected mice with 1-week LC pretreatment, (c) normal intestinal histology in ST-infected mice with 1-week LC+mcra pretreatment, (d) moderate depletion of goblet cells and villi/microvilli in ST-infected mice, (e) massive elimination of goblet cells and villi/microvilli in ST-infected mice, (f) intestinal inflammation and infiltration at circular folds in ST-infected mice. All images were captured under 100 × .
Regulation on expression of intestinal inflammatory cytokine genes
The regulation of cecal inflammatory cytokine gene expressions during 3-week ST infection as well as 1-week probiotic pre-administration is displayed in Figure 7. Specifically, ST infection induced up-expression of 4 pro-inflammatory cytokines IL-1β, IL-6, INF-γ, TNF-α genes and 1 anti-inflammatory cytokine IL-10 gene in mice cecal tissue cells. The up-regulation levels ranged from 2.2 to 7.8 log folds with the highest values for INF-γ gene and the highest expression at two weeks after ST challenge (Day 21). Another anti-inflammatory cytokine TGF-β gene was found down-regulated in ST infected mice cecum by 1.5 to 2.9 log folds. The expression of intestinal inflammation-related cytokine genes in LC+mcra pretreated mice were manipulated at a positive manner. For example, all 4 pro-inflammatory cytokine genes provoked by ST were suppressed significantly by 1.3 to 5.3 log folds through three weeks after challenging compared to mice with no probiotic protection; expression of anti-inflammatory cytokines IL-10 and TGF-β genes were stimulated notably in comparison with either control or ST infection with no probiotic prevention.
Figure 7.
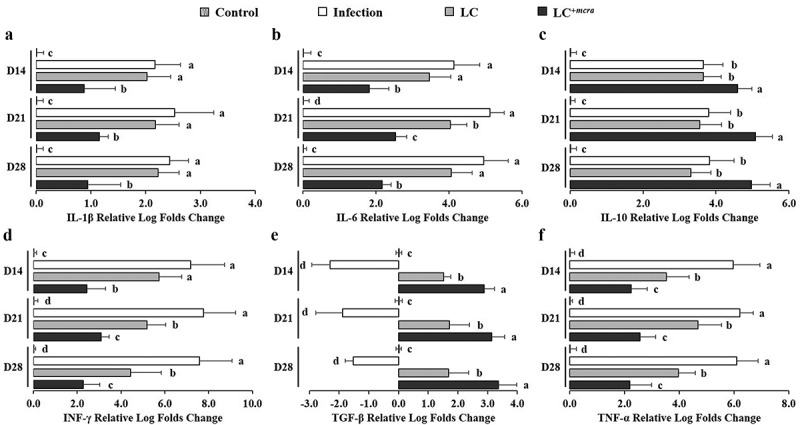
Differential expression levels of mice cecal cytokine genes. The relative log fold changes in expression of IL-1β (a), IL-6 (b), IL-10 (c), INF-γ (d), TGF-β (e), and TNF-α (f) genes from cecum tissue cells collected from control mice, mice under ST infection, mice pretreated with wild-type LC and challenged with ST, or mice pretreated with LC+mcra and challenged with ST were examined in triplicate. Different letters (‘a’ through ‘d’) at individual time point (day 14, 21, or 28) are significantly different (p < 0.05) among groups of control, infection, and infection following probiotic pretreatments.
Modulation of murine gut microbiota due to probiotic pretreatment and salmonellosis
To compare the gut microbial composition in various groups of mice including pretreated with probiotic strains and/or challenged with ST, we randomly selected their cecal contents (5 mice from each group) for 16S metagenomic sequencing and taxonomic classification. According to the taxonomic profile at the phylum level (Figure 8A), Firmicutes were the predominant phylum (63.51%), followed by Bacteroidetes (29.37%), in fecal samples collected from the control group mice which were given placebo (primary control). The relative abundance of Proteobacteria was 0.92% with individual variation from 0.54% to 1.35%. Significant difference in gut microbial community at phylum level was observed in ST infected mice (Figure 8B), specifically the group of mice which were not pretreated with probiotic strains; though the predominant phylum was still Firmicutes (61.26%), the relative abundance of Bacteroidetes was notably decreased to 17.81% and the relative abundance of Proteobacteria boosted to 14.86% (Figure 8B). One-week daily administration of probiotic (LC or LC+mcra) positively shaped the phylum level gut microbiota in the group of mice which were challenged with ST (Figure 8C,D). More specifically, in comparison with ST infected mice which were not pretreated with probiotic (secondary control), the microbial composition of probiotic pretreated mice after challenged with ST were dominated with Firmicutes and they were raised by 5.67 and 13.34% in the groups of mice which were pretreated with LC and LC+mcra, respectively. Whereas in the same pretreated groups of mice, the relative abundances of cecal Proteobacteria were reduced by 13.17 and 14.17%, respectively (Figure 8C,D).
Figure 8.
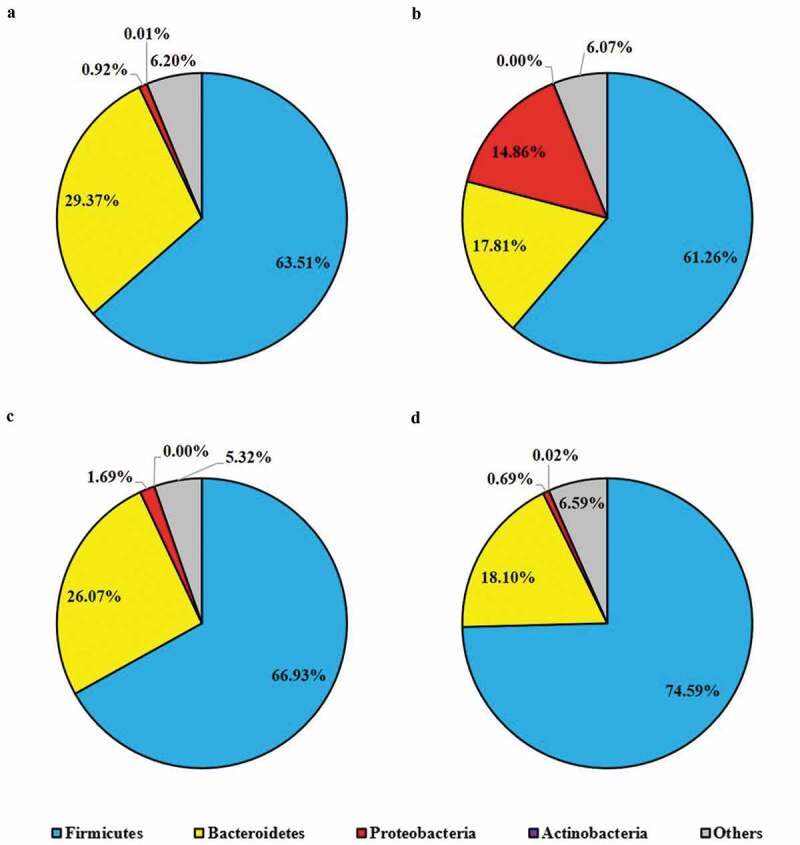
Mice cecal microbial community phylum-level structure. Bacterial distributions at phylum level in cecal contents from individual pooled dataset were depicted in terms of (a) control mice providing placebo and without ST infection, (b) mice infected with ST, (c) mice daily administered with LC for one week followed by ST challenge, and (d) mice daily administered with LC+mcra for one week followed by ST challenge.
At genus level, Bacteroides (18.50%) was identified being the highest abundant in cecal contents of primary control group of mice, followed by Ruminococcus (7.17%), Blautia (7.02%), Johnsonella (4.39%), and Lactobacillus (1.80%) (Figure 9). The relative abundances of Salmonella and Enterobacter were observed less than 0.01% of the total gut bacterial composition in the cecal content of the group of mice which were not challenged ST or challenged with ST but pretreated with probiotic strains (Figure 9). Whereas, the gut microbiota genus in ST-infected mice (without pretreatment with probiotic strains) exhibited significantly higher abundances of Salmonella (5.27%) and Enterobacter (3.72%), but lower abundances of Bacteroides (9.98%), Blautia (5.16%), Johnsonella (3.34%), and Lactobacillus (0.17%) (Figure 9). Other gut microbial genus-level noticeable differences between ST-infected mice and control included reduced abundance of Anaerobranca, Anaeroplasma, and Butyrivibrio but increased abundance of Akkermansia, Desulfobacter, Enterococcus, Klebsiella, Leptolyngbya, Natronincola, Staphylococcus, and Tolumonas, Trabulsiella (Figure 9). Compared with secondary control, probiotic pretreatments notably increased the relative abundances of Bacteroides, Blautia, Escherichia, Johnsonella, and Lactobacillus as well as lowered Salmonella, Enterobacter, Klebsiella, Tolumonas, and Trabulsiella. Particularly, the mice which were pretreated with LC+mcra were partly protected from Salmonella colonization and the relative abundance of Enterobacter was as low as to the control group. Significant escalation of their relative abundances of Bifidobacterium (0.12%), Blautia (8.43%), and Lactobacillus (9.18%) was also detected (Figure 9).
Figure 9.
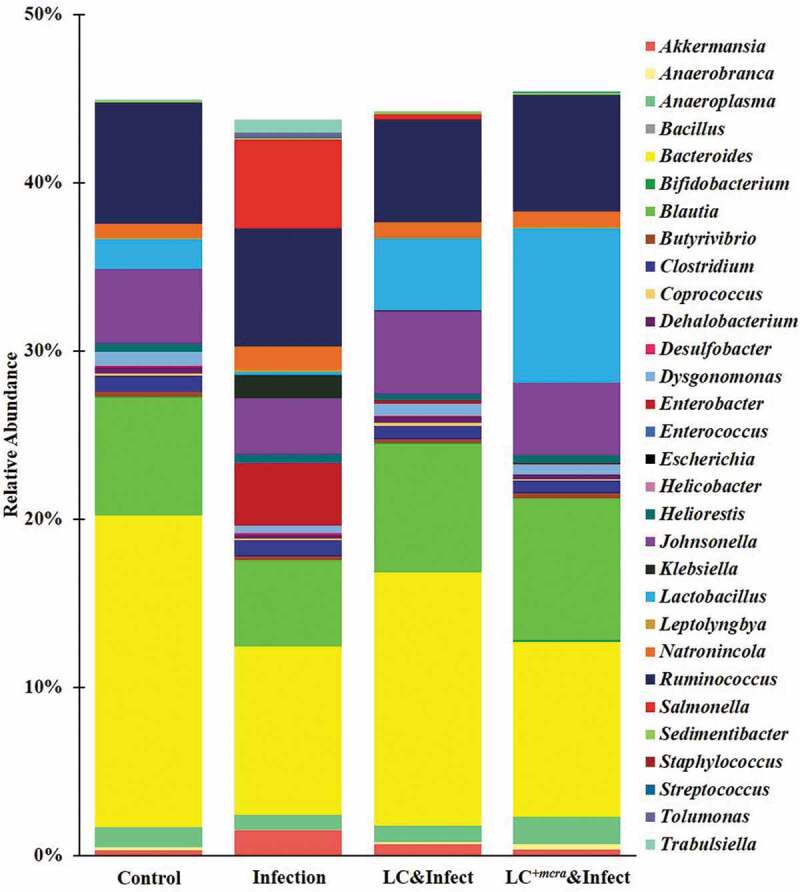
Mice cecal microbiota composition at genus level. Bacterial genus-level community composition in cecal contents from consolidated pool of dataset was compared among control mice providing placebo and without ST infection, mice infected with ST, mice daily administered with LC for one week followed by ST challenge, and mice daily administered with LC+mcra for one week followed by ST challenge. Overall 30 bacterial genera were targeted based on their relative abundances and importance in gut microbiome. The total relative abundances of all targeted 30 genera varied from 43 to 46% in different mice groups.
The overall cecal bacterial species diversity was observed minimal in the cecum content of the group of mice which were challenged with ST but not pretreated with probiotic strains but the cecal bacterial species diversity was promoted by LC+mcra pretreatment and protection (Figure 10). Specifically, compared with primary control, the mice group infected with ST exhibited significantly reduced gut intestinal microbial diversity at species level which was indicated by various alpha-diversity indexes including Chao-1, Fisher-alpha, Margalef’s richness, and Simpson (numerically higher), and Shannon (Figure 10). However, the one-week daily pre-administration with LC+mcra, but not with wild-type LC, before ST challenging caused a notably increased bacterial species diversity in cecum compared with secondary control group and even higher in comparison with primary control group.
Figure 10.
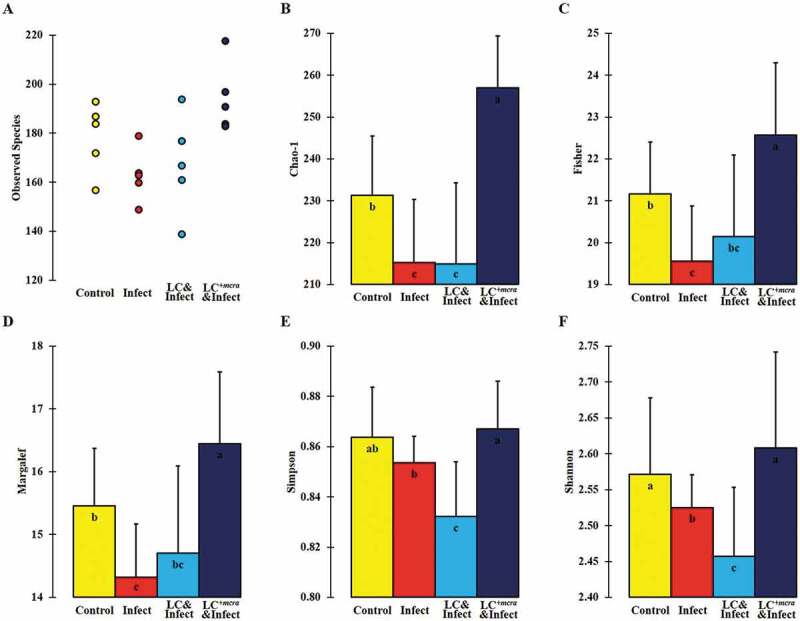
Bacterial diversity at species level in murine cecum. The assessment of alpha-diversity including Observed number of taxa species (A), Chao-1 (B), Fisher’s alpha (C), Margalef’s richness (D), Simpson index (E), and Shannon index (F) was determined and analyzed among control mice providing placebo and without ST infection, mice infected with ST, mice daily administered with LC for one week followed by ST challenge, and mice daily administered with LC+mcra for one week followed by ST challenge. Standard deviations among individual group members were provided. Different letters (‘a’ through ‘c’) are significantly different (p < 0.05) among groups of control, infection, and infection following probiotic pretreatments.
Discussion
The probiotic strain LC+mcra with 7-fold upregulation in its expression level of mcra gene coding linoleate isomerase has been found with prominently significant 21-fold higher rate in total linoleic acids production per bacterial cell.22 In a previous study, we revealed in vitro that LC+mcra could competitively exclude the growth and adhesive activity of both ST and EHEC22 and at meanwhile, suppress their vital virulence gene factors. Moreover, though effectiveness of probiotics in combatting enteric bacterial pathogens is still controversial, several researchers have suggested that secondary metabolites of probiotics such as CLA might enhance their overall in vivo health-beneficial functions.16,25–27 Here in the current study, we systematically and in-depth investigated the double effects of both Lactobacillus and CLA on murine gut health. According to our results, 1-week consecutive consumption of LC+mcra through oral administration efficiently prevented/eased the following Salmonella infection. Although probiotic administration through water might generate variance of bio-availability in mice gut, it is worth mentioning that early-staged oral probiotic gavage possesses high risk in potential induced injury in 3-week-old mouse esophagus. As the balance of bacterial fecal shedding with beneficial and detrimental ratio serves as a key indicator of the gut intestinal health.28 correspondingly we observed in our study the reduced colonization of ST/EHEC in both fecal content and intestinal fluids. Though in this study, we as well as some previous researchers, observed that EHEC were not able to cause enough infection and death in mice, but the Salmonella colonization in mice was claimed to be restricted by bioactive probiotics or functional fatty acids oral supplements in vivo,27,29–31 in which the virulence gene factors of Salmonella were suggested to be manipulated.32,33
On the other hand, probiotic itself was addressed to be capable of reducing intestinal pathogens through physical repellence and colonization resistance.34–38 Interestingly, our in vivo findings in which we observed that either wild-type or genetically engineered L. casei remarkably diminished ST/EHEC colonization in cecum, jejunum, and ileum also agreed with previous reports based on in vitro test.22 Further, LC+mcra displayed more intensive reductions of colonization of ST/EHEC and/or their severity of infection considering the extraneous strengthening effects implemented by its over-promoted CLA production.16 In fact, CLA has been documented and linked with antimicrobial active against several enteric bacterial pathogens including Salmonella though the specific mechanism are still under study.25,39 Most importantly, the in vivo examination based on BALB/cJ mice justified the protective roles of LC+mcra on combating enteric bacterial pathogens, following and matching with previous in vitro outcomes relied on various pathogenic bacterial strains.22,40,41
In most cases, Salmonella infections are associated with diarrhea, weight loss, dramatic alterations in composition of blood cells, as well as death.14,42–44 Accordantly we detected 105–107 CFU/g intestinal colonization of ST induced salmonellosis and caused around 8% weight loss, 52% higher level of RBC, 19% and 71% lower levels of WBC (especially neutrophils and lymphocytes) and PLT, and severe cecal inflammation in the survival mice. The physical, hematological, and gut intestinal abnormalities mentioned above in our in vivo examination contributed in the 40% death rate of mice challenged with enteric bacterial pathogen ST. However, probiotics secreting different types of functional fatty acids initiate attenuation in over-reactive gut inflammation through anti-inflammatory activities,16,22,24 which correlates with the LC+mcra (CLA) mediated relative up-regulation of murine intestinal anti-inflammatory cytokine genes from mice under salmonellosis found in our study. Therefore, apart from the direct colonization competition and repellence, daily administration of probiotics, especially LC+mcra, also prevented regular salmonellosis symptoms and maintained the overall physical and gut health condition of mice through mediating immuno-modulation. In future study, several other tissues including kidney, liver, and lung could be examined to further investigate whether LC+mcra pretreatment prevented ST systemic infection.
To address concerns from the host’s point of view, the maintenance of intact and operative gut intestine physiological condition is crucial in both metabolism and symbiotic intestinal microbiota composition.5,45–47 In our study, LC+mcra and its byproduct CLA prevented ST-induced elimination of goblet cells, villi, and microvilli as well as the inflammatory infiltrations between circular folds in cecum, which maintained the overall functions in terms of intestinal nutrients absorption and profoundly raised the survival rate (0 death) in mice. As a matter of fact, CLA has also been previously tested in colitis and inflammatory bovine disease recovery and showed significant gut health benefits,48,49 but the specific mechanism was not explained. Here, our findings based on CLA are in support of these researches and suggest a protective mechanism from both bacterial colonization and host histology sides.
A balanced gut microbial ecosystem serves as the crucial defense against colonization and infection with enteric pathogens.16,50,51 Salmonella infection could have negatively impacted the gut intestinal microbial composition by diminishing the abundances of Firmicutes including Lactobacillus and Bifidobacterium, and simultaneously favoring the dominance of Proteobacteria inducing follow-up opportunistic infections.52–54 In our study, we observed the increasing abundances of Salmonella and Enterobacter with overall reduced bacterial species diversity following ST challenge in mice, whereas LC+mcra pre-administration successfully prevented the negative shifting of gut microbiota which was induced by ST infection. As a matter of fact, several researchers also observed that CLA-containing diets were able to alter the fatty acids metabolism and maintaining homeostatic gut microflora.55,56 The healthier gut microbial distribution as well as their diversity in specific genus and species are influenced by the consumption of CLA-producing probiotic. Particularly at genus level, the higher abundances of Lactobacillus and Bifidobacterium positively influence the specific gut immuno homeostatic programming and signaling pathways which contributes in defending and excluding endogenous or exogenous pathogens. Furthermore, Blautia usually participates in nutrient absorption and digestion, which contributes in maintaining host’s gut intestinal nutritional status even the balance of gut is disrupted by pathogenic bacterial infection.57,58 On the other hand, reduced abundance of opportunistic pathogenic bacterial colonization like Enterobacter and Klebsiella minimized the risk of gut microbiota dysbiosis and further systematic infection. A balanced intestinal microbiota strengthen the first-line gut intestinal defense system against multiple pathogenic bacterial infections, which may possess a strong correlation and be the explanation of reduced bacterial pathogen colonization and inflammation in mice gut. A follow-up research on accumulative quantity of CLA in murine gut intestine may help in revealing dynamic interactions among the complex microbial ecosystem including established probiotic strains, their metabolic CLA, endogenous gut commensal flora, and exogenous bacterial pathogens.
Based on previous research, EHEC oral challenge on mice with distinct genetic configuration and gut microbial composition can result in various levels of colonization, morbidity, and mortality.59 Specifically, EHEC dose as low as 102 CFU led to intense cecal colonization and death in germ-free mice60,61 whereas for conventional mice model like BALB/c, considerably higher dose of EHEC was requisite to cause diseases.62,63 In some cases, infectious dose of EHEC less than 1010–1011 CFU/mouse failed to even introduce cecal colonization and infection,64,65 which parallel with our findings. Based on the current study, BALB/cJ mice orogastrically challenged with 107 CFU EHEC were colonized at a range from 102 to 104 CFU/g of EHEC in intestinal fluid collected from cecum, jejunum, and ileum but failed to produce any visible physiological abnormalities or mortality in mice. This finding could be explained by the relative resistance in BALB/c mice toward EHEC through shorter shedding duration and producing higher serum/fecal levels of O157-specific IgA.59,64 On the other hand, LC+mcra, as we observed in vitro22 and predicted for in vivo, stood out in reducing the colonization level of EHEC as well as preventing from kidney histological abnormalities and weight loss in BALB/cJ mice. Further research dependent on germ-free or compromised commensal flora mouse model might be substantial in revealing how LC+mcra involved in defending host from EHEC pathogenesis and post-infectious complications.
In conclusion, the current study has demonstrated a substantial influence of CLA over-producing probiotic strain, LC+mcra exerted on Salmonella and pathogenic E. coli infections in conventional mice. Specifically, mice orally given LC+mcra daily for one week minimized EHEC colonization and protected themselves from ST-facilitated serious salmonellosis which was observed by notably reduced fecal shedding and intestinal colonization of ST, amelioration on acute inflammation, and prevention on hematological and histological abnormalities. In depth metagenomic analysis revealed that LC+mcra pretreatment modulated mice cecal bacterial community with increased diversity which are predominated with comparative higher Firmicutes and lower Proteobacteria. The outstanding protective roles of LC+mcra against ST and EHEC infection plus its profound effectiveness over wild-type LC may provide a promising option for prophylaxis on pathogenic Salmonella and diarrheagenic E. coli infections and reduce enteric bacterial infections.
Methods
Bacterial strain and growth conditions
Lactobacillus casei (LC, ATCC 334) and our laboratory generated linoleic acid over-expressed L. casei, LC+mcra22,40 were used as probiotics while Salmonella enterica serovar Typhimurium (ST, ATCC 14028) and enterohemorrhagic Escherichia coli EDL933 (EHEC, ATCC700927) were chosen as enteric bacterial pathogens in this study. Lactobacillus strains LC and LC+mcra were grown on regular MRS agar and MRS agar containing 100 mg/mL 5-fluorouracil separately at 37°C for 24 h in the presence of 5% CO2 (Forma™ Scientific CO2 water jacketed incubator, Thermo Fisher Scientific Inc., Waltham, MA, USA).22 ST and EHEC were grown on LB agar (EMD Chemicals Inc., Gibbstown, NJ, USA) for 18 h at 37°C under aerobic conditions (Thermo Scientific MAXQ 4450, Thermo Fisher Scientific Inc., Waltham, MA, USA).
Mouse model and animal experiments
The 3-week-old BALB/cJ Mice (approximately 8–10 g) were purchased from The Jackson Laboratory (Bar Harbor, ME USA) and reared in static micro-isolating cages with cellulose Bio-Performance bedding and huts as environmental enrichment. Teklad standard rodent diet and regular tap water were provided for mice feeding and drinking, respectively. A total of 90 mice (45 males and 45 females) were used for each trial. Following a completely randomized method, 90 mice were randomly assigned to 9 groups (designated A1 to C3) resulting in 10 mice per group; two cages were assigned to each group with a total of 5 mice per cage. Mice cages were changed weekly, and each individual mouse was weighed and monitored with health examinations daily. At the end of the second, third, and fourth week, 3, 3, and 4 mice from each group respectively, were randomly selected and euthanized with CO2 inhalation in euthanasia chamber for organ samples collection.
Feeding probiotic to BALB/cJ mice and challenging with ST and EHEC
Overnight culture of LC or LC+mcra in MRS broth were diluted in fresh 5 mL MRS broth at 1:50 and allowed for 3 h further growth. The bacterial cells in exponential phase were harvested following centrifugation at 3,000 × g for 15 min, PBS washing, and resuspension in 1.0 mL PBS. A final concentration of 1011 CFU/mL was adjusted with PBS and used to feed mice. The design of in vivo mouse trial was summarized in Table 1. Probiotic (either 109 CFU/mL LC or LC+mcra) cells were maintained in water bottle filled with regular tap water for group B and C and fed to mice from Day 1 to Day 7. Control mice, group A, was fed with regular tap water only.
Table 1.
Mice groups, numbers per group, and their treatment/infection.
| Probiotic Treatment (daily during 1st week) |
Pathogen challenge (beginning of 2nd week) |
||||||
|---|---|---|---|---|---|---|---|
| Group (#) | Mice (n) | PBS | LC1 | LC+mcra 2 | PBS | ST3 | EHEC4 |
| A1 | 10 | + | - | - | + | - | - |
| B1 | 10 | - | + | - | + | - | - |
| C1 | 10 | - | - | + | + | - | - |
| A2 | 10 | + | - | - | - | + | - |
| B2 | 10 | - | + | - | - | + | - |
| C2 | 10 | - | - | + | - | + | - |
| A3 | 10 | + | - | - | - | - | + |
| B3 | 10 | - | + | - | - | - | + |
| C3 | 10 | - | - | + | - | - | + |
1Wild-type L. casei; 2Mutant L. casei; 3S. Typhimurium; 4 Enterohemorrhagic E. coli
Overnight culture of ST and EHEC bacterial cells in LB broth were diluted in fresh 5 mL LB broth at 1:50 and allowed for 3–4 h further growth at 37°C. The exponential phase bacterial cells were harvested and washed by centrifugation at 3,000 × g for 15 min and resuspended in 1.0 mL of PBS. A final concentration of bacterial cells was adjusted to 108 CFU/mL in PBS. On Day 7, an aliquot of 100 µL ST or EHEC suspension containing approximately 107 CFU was fed to mice in groups 2 or 3 respectively, with oral gavage, and the mice were reared thereafter for another 3 weeks. Mice in group 1 was orogastrically fed with 100 µL PBS and served as control.
Sample collection and processing
In order to estimate the bacterial fecal shedding, fecal samples were collected from each mouse in sterile Whirl-Pak bags using sterile spoons at Day 1, 2, 3, 6, 7, 8, 9, 14, 21, and 28 for PBS serial dilution and plating on specific agar plates.66 In order to investigate the bacterial colonization in mice gut, intestine, ileum, jejunum, and cecum from each euthanized mouse were separated and harvested. Then the ileum, jejunum, and cecal fluids were serial diluted with PBS, followed by plating on specific agar plates. Specifically, MRS agar for LC, MRS agar containing 100 mg/mL 5-fluorouracil for LC+mcra, XLT-4 agar for ST, and MacConkey agar for EHEC were used, respectively.
Mice cecum was kept in RNA Later for further RNA extraction, cDNA reverse transcription, and inflammation-related gene expression level analysis. For hematological analysis, the blood samples from each mouse was collected from heart in VACUETTER® Heparin tubes (Greiner Bio-One, Monroe, NC, USA) and further analyzed with a ProCyte Dx® Hematology Analyzer (IDEXX, Westbrook, ME, USA) according to the manufacturer’s instructions.
RNA extraction, cDNA synthesis, and quantitative RT-PCR
Extraction of mice intestinal RNA was carried out using TRIzol® Reagent (Life Technologies Co., Carlsbad, CA, USA) following previous methods.67 The cDNA synthesis was performed according to the manufacture’s instruction of qScript cDNA SuperMix. The PCR reaction mixture containing 10 µL PerfeCTa SYBR Green Fast Mix (Quanta Biosciences, Beverly, MA, USA), 2 µL of each 100 nM primer (Table 2),68,69 2 µL of cDNA (10 ng), and 4 µL of RNase-free water was amplified using an Eco Real-Time PCR system with 30 s denaturation at 95°C, followed by 40 cycles of 95°C for 5 s, 55°C for 15 s, and 72°C for 10 s. All the relative transcription levels of target genes were estimated by comparative fold change. The CT values of genes were normalized to the housekeeping gene, and the relative expression levels of target genes were calculated by the comparative method.70 Quantitative RT-PCR was carried out in triplicate.
Table 2.
Primers used for qRT-PCR analysis of mice intestinal cytokine gene expressions.
| Gene | Primer | Sequence (5ʹ-3ʹ) | Function |
|---|---|---|---|
| 18S rRNA | Sense | TTAGAGTGTTCAAAGCAGGCCCGA | Housekeeping gene |
| Antisense | TCTTGGCAAATGCTTTCGCTCTGG | ||
| IL-1β | Sense | TTTGAAGTTGACGGACCCC | Inflammatory cytokine gene |
| Antisense | TGTGCTGCTGCGAGATTTG | ||
| IL-6 | Sense | ACACATGTTCTCTGGGAAATCGTGG | Pro-/Anti-inflammatory cytokine gene |
| Antisense | TCTGCAAGTGCATCATCGTTGTTCA | ||
| IL-10 | Sense | GCTCTTACTGACTGGCATGAG | Anti-inflammatory cytokine gene |
| Antisense | CGCAGCTCTAGGAGCATGTG | ||
| IFN-γ | Sense | ATTGCGGGGTTGTATCTGGG | Pro-inflammatory cytokine gene |
| Antisense | GGGTCACTGCAGCTCTGAAT | ||
| TGF-β | Sense | GCGTGCTAATGGTGGACCGCA | Anti-inflammatory cytokine gene |
| Antisense | CGGGCACTGCTTCCCGAATGT | ||
| TNF-α | Sense | GGAACACGTCGTGGGATAATG | Inflammatory cytokine gene |
| Antisense | GGCAGACTTTGGATGCTTGTT |
Histopathology analysis
Intestinal tissue samples were taken from mice after euthanization and were stored in neutral buffered formalin (4% formaldehyde; pH 7.4) at 4°C for further processing. Once the samples were removed from fixative, they were dehydrated with increasing concentrations of ethanol, cleared in xylene, and embedded in paraffin. Microtome (LEICA RM2065, Leica Biosystems, Buffalo Grove, IL, USA) was used to harvest 5 μm thick paraffin sections followed by heat fixing at 37°C overnight. Then the slices were stained with hematoxylin and eosin and mounted with DPX mounting medium 13512 (Electron Microscopy Sciences, Hatfield, PA, USA). Histological observations were performed under a light microscope (BA210E, Motic Asia, Hong Kong, China).
Metagenomic sequencing and analysis
Mice cecal contents were harvested during the 4th week (Day 22–28) and 5 samples from each group of control (PBS only), ST infection, LC pretreatment followed by ST infection, or LC+mcra pretreatment followed by ST infection were randomly selected for metagenomics analysis. Microbial genomic DNA extraction was carried out using QIAamp Fast DNA Stool Kit (QIAGEN, Valencia, CA, USA) following instructions from the manufacturer. The variable V3 and V4 regions of microbial 16S rRNA gene were targeted for phylogenetic classifications. DNA libraries were prepared for equimolar-pooling using Nextera DNA Library Preparation Kit and Nextera Index Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Paired-end sequencing (2 × 300 bp) was conducted on Illumina MiSeq using MiSeq v3 600-cycle kit (Illumina, San Diego, CA, USA). Sequence data was processed through MiSeq Reporter – BaseSpace for FASTQ Workflow generation followed by taxonomic classification based on Greengenes database (http://greengenes.lbl.gov/). Demultiplexing was performed using only perfect index recognition (mismatch = 0) followed by removing PhiX reads. 16S sequence length below 1250 bp or with more than 50 wobble bases was filtered, and all entries classified with no genus or species were also filtered. The relative abundances and alpha/beta-diversity indices were calculated using ‘vegan’, ‘phyloseq’, and ‘metagenomeseq’ R package and plotted in Excel.
Statistical analysis
All data were analyzed by the SPSS software. Comparison among multiple mice groups were performed with the one-way analysis of variance followed by Tukey’s and Bonferroni’s tests. Survival rate among multiple mice groups were analyzed with Log-Rank test. For all tests, significant differences were considered on the basis of p values below a significant level of 0.05.
Acknowledgments
The authors would like to thank Dr. Vinod Nagarajan, Zabdiel Alvarado Martinez, Arpita Aditya, for their assistance during animal experiments. We also thank Dr. Rachel Dennis and Jasmine Mengers for their guide on histopathological studies.
Funding Statement
This study was funded by Tier-1 (2945060) grants awarded to Debabrata Biswas.
Contributors
MP conceived the study, designed and carried out the experiments, analyzed the data, and wrote the manuscript. ZT contributed to in vivo experiments and manuscript draft. PP and CBe assisted in experimental preparation, sample processing, and microbiological experiments. CBi participated in reviewing and editing the manuscript. JM provided sequencing resources and helped in metagenomics. DB supervised the research and finalized the manuscript. All authors approved the final manuscript.
Ethics approval and consent to participate
Mice in vivo experiments were performed in ABSL2 facilities in Department of Animal and Avian Sciences, University of Maryland in accordant with protocol #R-NOV-17-55 approved by the Institutional Animal Care and Use Committee (IACUC). The best effort was made for minimizing the suffer of animals. To ensure animal welfare, mice were monitored and recorded for physical appearance and body weight on a daily basis during experimental period.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Tlaskalová-Hogenová H, Tpánková R, Kozáková H, Hudcovic T, Vannucci L, Tuková L, Rossmann P, Hrní T, Kverka M, Zákostelská Z, et al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol Immunol. 2011;8:110–120. doi: 10.1038/cmi.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu L, Chen X, Skogerbø G, Zhang P, Chen R, He S, Huang DW.. The human microbiome: A hot spot of microbial horizontal gene transfer. Genomics. 2012;100:165–270. doi: 10.1016/j.ygeno.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 3.Fujimura KE, Slusher NAN, Cabana MD. Role of the gut microbiota in defining human health. Expert Rev Anti. 2010;8:435–454. Internet. http://www.expert-reviews.com/doi/abs/10.1586/eri.10.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simon JC, Marchesi JR, Mougel C, Selosse MA. Host-microbiota interactions: from holobiont theory to analysis. Microbiome. 2019;7:1–5. doi: 10.1186/s40168-019-0619-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hillman ET, Lu H, Yao T, Nakatsu CH. Microbial Ecology along the Gastrointestinal Tract. Microbes Environ. 2017;32:300–313. doi: 10.1264/jsme2.ME17017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003;361:512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 7.Vedantam G, Hecht DW. Antibiotics and anaerobes of gut origin. Curr Opin Microbiol. 2003;6:457–461. [DOI] [PubMed] [Google Scholar]
- 8.Shreiner AB, Kao JY, Young VB. The gut microbiome in health and in disease. Curr Opin Gastroenterol. 2015;31:69–75. doi: 10.1097/MOG.0000000000000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev. 2007;20:593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segal LN, Blaser MJ. A brave new world: the lung microbiota in an era of change. Ann Am Thorac Soc Vol 11, Supplement 1, pp S21–S27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beaugerie L, Petit JC. Antibiotic-associated diarrhoea. Best Pract Res Clin Gastroenterol. 2004;18:337–352. doi: 10.1016/j.bpg.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Peng M, Salaheen S, Buchanan RL, Biswas D. Alterations of Salmonella enterica Serovar Typhimurium Antibiotic Resistance under Environmental Pressure. Appl Environ Microbiol. 2018;84:e01173–18. Internet [accessed 2018 September20]. http://www.ncbi.nlm.nih.gov/pubmed/30054356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng M, Salaheen S, Biswas D. Animal health: antibiotic global issues. In: Alfen NKV editor. Encyclopedia of Agriculture and Food Systems, Elsevier Inc. Press. 2014;346–357. [Google Scholar]
- 14.Peng M, Salaheen S, Almario JA, Tesfaye B, Buchanan R, Biswas D. Prevalence and antibiotic resistance pattern of Salmonella serovars in integrated crop-livestock farms and their products sold in local markets. Environ Microbiol. 2016;18:1654–1665. doi: 10.1111/1462-2920.13265. [DOI] [PubMed] [Google Scholar]
- 15.Coburn B, Sekirov I, Finlay BB. Type III secretion systems and disease. Clin Microbiol Rev. 2007;20:535–549. doi: 10.1128/CMR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng M, Biswas D. Short Chain and Polyunsaturated Fatty Acids in Host Gut Health and Foodborne Bacterial Pathogen Inhibition. Crit Rev Food Sci Nutr. 2017;57:3987–4002. Internet http://www.tandfonline.com/doi/full/10.1080/10408398.2016.1203286. [DOI] [PubMed] [Google Scholar]
- 17.Panos GZ, Betsi GI, Falagas ME. Systematic review: are antibiotics detrimental or beneficial for the treatment of patients with Escherichia coli O157: h7infection? Aliment Pharmacol Ther. 2006;24:731–742. doi: 10.1111/apt.2006.24.issue-5. [DOI] [PubMed] [Google Scholar]
- 18.Goldwater PN, Bettelheim KA. Treatment of enterohemorrhagic Escherichia coli (EHEC) infection and hemolytic uremic syndrome (HUS). BMC Med. 2012;10:1–8. doi: 10.1186/1741-7015-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salaheen S, Peng M, Biswas D. Replacement of conventional antimicrobials and preservatives in food production to improve consumer safety and enhance health benefits. In: Rai VR and Bai JA editors. Microbial Food Safety and Preservation Techniques. CRC press. 2014;305–328. [Google Scholar]
- 20.Peng M, Patel P, Nagarajan V, Bernhardt C, Carrion M, Biswas D. Feasible options to control colonization of enteric pathogens with designed synbiotics. In: Watson R and Preedy V. Dietary Interventions in Gastrointestinal Diseases. Elsevier Inc. Press. 2019;135–149. [Google Scholar]
- 21.Peng M, Aryal U, Cooper B, Biswas D. Metabolites produced during the growth of probiotics in cocoa supplementation and the limited role of cocoa in host-enteric bacterial pathogen interactions. Food Control. 2015;53:124–133. Internet. doi: 10.1016/j.foodcont.2015.01.014. [DOI] [Google Scholar]
- 22.Peng M, Tabashsum Z, Patel P, Bernhardt C, Biswas D. Linoleic Acids Overproducing Lactobacillus casei Limits Growth, Survival, and Virulence of Salmonella Typhimurium and Enterohaemorrhagic Escherichia coli. Front Microbiol. 2018;9:2663. Internet [accessed 2018 December17]. https://www.frontiersin.org/article/10.3389/fmicb.2018.02663/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Shea EF, Cotter PD, Stanton C, Ross RP, Hill C. Production of bioactive substances by intestinal bacteria as a basis for explaining probiotic mechanisms: bacteriocins and conjugated linoleic acid. Int J Food Microbiol. 2012;152:189–205. doi: 10.1016/j.ijfoodmicro.2011.05.025. [DOI] [PubMed] [Google Scholar]
- 24.Yang B, Chen H, Stanton C, Ross RP, Zhang H, Chen YQ, Chen W. Review of the roles of conjugated linoleic acid in health and disease. J Funct Foods. 2015;15:314–325. doi: 10.1016/j.jff.2015.03.050. [DOI] [Google Scholar]
- 25.Byeon JI, Song HS, Oh TW, Kim YS, Choi BD, Kim HC, Kim JO, Shim KH, Ha YL. Growth inhibition of foodborne and pathogenic bacteria by conjugated linoleic acid. J Agric Food Chem. 2009;57:3164–3172. Internet. http://www.scopus.com/inward/record.url?eid=2-s2.0-65349162210&partnerID=40&md5=9610d56bb99211a5bae2b01378ad8ecd. [DOI] [PubMed] [Google Scholar]
- 26.Garner CD, Antonopoulos DA, Wagner B, Duhamel GE, Keresztes I, Ross DA, Young VB, Altier C. Perturbation of the small intestine microbial ecology by streptomycin alters pathology in a Salmonella enterica serovar typhimurium murine model of infection. Infect Immun. 2009;77:2691–2702. doi: 10.1128/IAI.01570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snel J, Born L, van der Meer R. Dietary fish oil impairs induction of gamma-interferon and delayed-type hypersensitivity during a systemic Salmonella enteritidis infection in rats. APMIS. 2010;118:578–584. doi: 10.1111/j.1600-0463.2010.02655.x. [DOI] [PubMed] [Google Scholar]
- 28.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willamil J, Creus E, Francisco Pérez J, Mateu E, Martín-Orúe SM. Effect of a microencapsulated feed additive of lactic and formic acid on the prevalence of Salmonella in pigs arriving at the abattoir. Arch Anim Nutr. 2011;65:431–444. doi: 10.1080/1745039X.2011.623047. [DOI] [PubMed] [Google Scholar]
- 30.Sunkara LT, Achanta M, Schreiber NB, Bommineni YR, Dai G, Jiang W, Lamont S, Lillehoj HS, Beker A, Teeter RG, et al. Butyrate enhances disease resistance of chickens by inducing antimicrobial host defense peptide gene expression. PLoS One. 2011;6:e27225. doi: 10.1371/journal.pone.0027225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sunkara LT, Jiang W, Zhang G. Modulation of Antimicrobial Host Defense Peptide Gene Expression by Free Fatty Acids. PLoS One. 2012;7:e49558. doi: 10.1371/journal.pone.0049558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hung CC, Garner CD, Slauch JM, Dwyer ZW, Lawhon SD, Frye JG, Mcclelland M, Ahmer BMM, Altier C. The intestinal fatty acid propionate inhibits Salmonella invasion through the post-translational control of HilD. Mol Microbiol. 2013;87:1045–1060. doi: 10.1111/mmi.12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun Y, O’Riordan MXD. Regulation of bacterial pathogenesis by intestinal short-chain fatty acids. Adv Appl Microbiol. 2013;85:92–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKenney PT, Pamer EG. From Hype to Hope: the Gut Microbiota in Enteric Infectious Disease. Cell. 2015;163:1326–1332. doi: 10.1016/j.cell.2015.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amalaradjou MAR, Bhunia AK. Modern Approaches in Probiotics Research to Control Foodborne Pathogens. Adv Food Nutr Res. 2012;67:185–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng M, Reichmann G, Biswas D. Lactobacillus casei and its byproducts alter the virulence factors of foodborne bacterial pathogens. J Funct Foods. 2015;15:418–428. Internet. doi: 10.1016/j.jff.2015.03.055. [DOI] [Google Scholar]
- 38.Peng M, Bitsko E, Biswas D. Functional properties of peanut fractions on the growth of probiotics and foodborne bacterial pathogens. J Food Sci. 2015;80:M635–41. doi: 10.1111/1750-3841.12785. [DOI] [PubMed] [Google Scholar]
- 39.Meraz-Torres LS, Hernandez-Sanchez H. Conjugated Linoleic Acid in Dairy Products: A Review. Am J Food Technol. 2012;7:176–179. doi: 10.3923/ajft.2012.176.179. [DOI] [Google Scholar]
- 40.Tabashsum Z, Peng M, Salaheen S, Comis C, Biswas D. Competitive elimination and virulence property alteration of Campylobacter jejuni by genetically engineered Lactobacillus casei. Food Control. 2018;85:283–291. doi: 10.1016/j.foodcont.2017.10.010. [DOI] [Google Scholar]
- 41.Tabashsum Z, Peng M, Kahan E, Rahaman SO, Biswas D. Effect of Conjugated Linoleic Acid Overproducing Lactobacillus with Berry Pomace Phenolic Extracts on Campylobacter jejuni Pathogenesis. Food Funct. 2018;10:296–303. Internet [accessed 2018 December17]. http://pubs.rsc.org/en/Content/ArticleLanding/2018/FO/C8FO01863D. [DOI] [PubMed] [Google Scholar]
- 42.Oracz G, Feleszko W, Golicka D, Maksymiuk J, Klonowska A, Szajewska H. Rapid Diagnosis of Acute Salmonella Gastrointestinal Infection. Clin Infect Dis. 2003;36:112–115. doi: 10.1086/cid.2003.36.issue-1. [DOI] [PubMed] [Google Scholar]
- 43.Barba-Vidal E, Buttow Roll VF, Garcia Manzanilla E, Torrente C, Moreno Muñoz JA, Pérez JF, Martín-Orúe SM. Blood parameters as biomarkers in a Salmonella spp. disease model of weaning piglets. PLoS One. 2017;12:e0186781. doi: 10.1371/journal.pone.0186781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santos RL, Tsolis RM, Bäumler AJ, Adams LG. Hematologic and serum biochemical changes in Salmonella ser Typhimurium-infected calves. Am J Vet Res. 2002. doi: 10.2460/ajvr.2002.63.1145. [DOI] [PubMed] [Google Scholar]
- 45.Ohland CL, MacNaughton WK. Probiotic bacteria and intestinal epithelial barrier function. AJP Gastrointest Liver Physiol. 2010;298:G807–19. doi: 10.1152/ajpgi.00243.2009. [DOI] [PubMed] [Google Scholar]
- 46.Rugge M, Pennelli G, Pilozzi E, Fassan M, Ingravallo G, Russo VM, Di Mario F. Gastritis: the histology report. Dig Liver Dis. 2011;43:S373–84. doi: 10.1016/S1590-8658(11)60593-8. [DOI] [PubMed] [Google Scholar]
- 47.Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J. 2017;474:1823–1836. doi: 10.1042/BCJ20160510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Viladomiu M, Hontecillas R, Yuan L, Lu P, Bassaganya-Riera J. Nutritional protective mechanisms against gut inflammation. J Nutr Biochem. 2013;24:929–939. doi: 10.1016/j.jnutbio.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bassaganya-Riera J, Viladomiu M, Pedragosa M, de Simone C, Carbo A, Shaykhutdinov R, Jobin C, Arthur JC, Corl BA, Vogel H, et al. Probiotic bacteria produce conjugated linoleic acid locally in the gut that targets macrophage PPAR γ to suppress colitis. PLoS One. 2012;7:e31238. doi: 10.1371/journal.pone.0031238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ubeda C, Djukovic A, Isaac S. Roles of the intestinal microbiota in pathogen protection. Clin Transl Immunol. 2017;6:e128. doi: 10.1038/cti.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iacob S, Iacob DG, Luminos LM. Intestinal Microbiota as a Host Defense Mechanism to Infectious Threats. Front Microbiol. 2019;9:1–9. Internet [accessed 2019 February26]. https://www.frontiersin.org/article/10.3389/fmicb.2018.03328/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, Salzman N. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun. 2008;76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Videnska P, Sisak F, Havlickova H, Faldynova M, Rychlik I. Influence of Salmonella enterica serovar Enteritidis infection on the composition of chicken cecal microbiota. BMC Vet Res. 2013;9:1–8. doi: 10.1186/1746-6148-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Argüello H, Estellé J, Zaldívar-López S, Jiménez-Marín Á, Carvajal A, López-Bascón MA, Crispie F, O’Sullivan O, Cotter PD, Priego-Capote F, et al. Early Salmonella Typhimurium infection in pigs disrupts Microbiome composition and functionality principally at the ileum mucosa. Sci Rep. 2018;8:1–12. doi: 10.1038/s41598-018-26083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kho ZY, Lal SK. The human gut microbiome - A potential controller of wellness and disease. Front Microbiol. 2018;9:1–23. doi: 10.3389/fmicb.2018.01835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marques TM, Wall R, O’Sullivan O, Fitzgerald GF, Shanahan F, Quigley EM, Cotter PD, Cryan JF, Dinan TG, Ross RP, et al. Dietary trans-10, cis-12-conjugated linoleic acid alters fatty acid metabolism and microbiota composition in mice. Br J Nutr. 2015;113:1–11. doi: 10.1017/S0007114514004206. [DOI] [PubMed] [Google Scholar]
- 57.O’Neill I, Schofield Z, Hall LJ. Exploring the role of the microbiota member Bifidobacterium in modulating immune-linked diseases. Emerg Top Life Sci. 2017;1:333–349. doi: 10.1042/ETLS20170058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Reddy DN. Role of the normal gut microbiota. World J Gastroenterol. 2015;21:8787–8803. doi: 10.3748/wjg.v21.i10.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mohawk KL, O’Brien AD. Mouse Models of Escherichia coli O157: h7Infection and Shiga Toxin Injection. J Biomed Biotechnol. 2011;2011:1–17. Internet http://www.hindawi.com/journals/bmri/2011/258185/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Isogai E, Isogai H, Takeshi K, Nishikawa T. Protective effect of Japanese green tea extract on gnotobiotic mice infected with an Escherichia coli O157: h7strain. Microbiol Immunol. 1998;42:125–128. doi: 10.1111/j.1348-0421.1998.tb02260.x. [DOI] [PubMed] [Google Scholar]
- 61.Eaton KA, Friedman DI, Francis GJ, Tyler JS, Young VB, Haeger J, Abu-Ali G, Whittam TS. Pathogenesis of renal disease due to enterohemorrhagic Escherichia coli in germ-free mice. Infect Immun. 2008;76:3054–3063. doi: 10.1128/IAI.01626-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karpman D, Council H, Svensson M, Scheutz F, Aim P, Svanborg C. The role of lipopolysaccharide and Shiga-like toxin in a mouse model of Escherichia coli O157: h7infection. J Infect Dis. 1997;175:611–620. doi: 10.1086/516490. [DOI] [PubMed] [Google Scholar]
- 63.Brando RJF, Miliwebsky E, Bentancor L, Deza N, Baschkier A, Ramos MV, Fernández GC, Meiss R, Rivas M, Palermo MS. Renal damage and death in weaned mice after oral infection with Shiga toxin 2-producing Escherichia coli strains. Clin Exp Immunol. 2008;153:297–306. doi: 10.1111/j.1365-2249.2008.03698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Conlan JW, Perry MB. Susceptibility of Three Strains of Conventional Adult Mice to Intestinal Colonization by an Isolate of _Escherichia coli_ O157:H7. Can J Microbiol. 1998;44:800–805. [DOI] [PubMed] [Google Scholar]
- 65.Nagano K, Taguchi K, Hara T, Yokoyama SI, Kawada K, Mori H. Adhesion and colonization of enterohemorrhagic Escherichia coli O157: h7in cecum of mice. Microbiol Immunol. 2003;47:125–132. [DOI] [PubMed] [Google Scholar]
- 66.Salaheen S, Jaiswal E, Joo J, Peng M, Ho R, OConnor D, Adlerz K, Aranda-Espinoza JH, Biswas D. Bioactive extracts from berry byproducts on the pathogenicity of Salmonella Typhimurium. Int J Food Microbiol. 2016;237:128–135. Internet. doi: 10.1016/j.ijfoodmicro.2016.08.027. [DOI] [PubMed] [Google Scholar]
- 67.Peng M, Zhao X, Biswas D. Polyphenols and tri-terpenoids from Olea europaea L. in alleviation of enteric pathogen infections through limiting bacterial virulence and attenuating inflammation. J Funct Foods. 2017;36:132–143. Internet. doi: 10.1016/j.jff.2017.06.059. [DOI] [Google Scholar]
- 68.Dann SM, Le C, Choudhury BK, Liu H, Saldarriaga O, Hanson EM, Cong Y, Eckmann L. Attenuation of Intestinal Inflammation in Interleukin-10-Deficient Mice Infected with Citrobacter rodentium. Infect Immun. 2014;81:1949–1958. doi: 10.1128/IAI.00066-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H-L, Lu L, Wang X-S, Qin L-Y, Wang P, Qiu S-P, Wu H, Huang F, Zhang -B-B, Shi H-L, et al. Alteration of Gut Microbiota and Inflammatory Cytokine/Chemokine Profiles in 5-Fluorouracil Induced Intestinal Mucositis. Front Cell Infect Microbiol. 2017;7:1–14. doi: 10.3389/fcimb.2017.00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-delta-delta-CT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]


