ABSTRACT
The intestinal microbiota may be involved, through metabolic gut–brain interactions, in a variety of neurological conditions. In this addendum, we summarize the findings of our recent study investigating the potentially modulatory influence of the microbiome in a transgenic ALS mouse model, and the possible application to human disease. We found that transgenic mice show evidence of dysbiosis, even at the pre-symptomatic stage, and have a more severe disease course under germ-free conditions or after receiving broad-spectrum antibiotics. We demonstrated that Akkermansia muciniphila ameliorated the disease in mice and that this may be due to the production of nicotinamide. We then conducted a preliminary study in human ALS and identified functionally similar alterations within the metagenome. Furthermore, we found that patients with ALS had lower systemic and CSF levels of nicotinamide, suggesting that the changes observed in the mouse model may be relevant to human disease.
KEYWORDS: Amyotrophic lateral sclerosis, als, motor neuron disease, sod1, microbiome, Akkermansia, nicotinamide
Introduction
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease causing relatively selective degeneration of upper and lower motor neurons.1 Host microbiome interactions can influence diseases in general, including neurological conditions.2–7 For example, distinct microbiome and metabolomic profiles are observed in autism spectrum disorder,8 as well as in Alzheimer’s disease.9 Preliminary interventions, such as fecal microbiota transplantation or administration of probiotics may modulate brain activity10 and influence neurological disorders such as multiple sclerosis,11 Alzheimer’s disease,12 Parkinson’s disease,13 and epilepsy.14 Generation of low molecular weight metabolites by the gut microbiome is one postulated mechanism; these compounds are capable of permeating the blood-brain barrier and influence neuronal function.2,3,15,16 Whether the gut microbiome influences ALS is controversial,17–20 leading us to further investigate the functional role of the microbiome in an ALS mouse model and in human disease.21
Lessons learned from the SOD1 transgenic ALS mouse model (Figure)
We used the G93A SOD1 transgenic (SOD1-Tg) ALS mouse model (Figure 1).22 In the first stage of our study, we used broad-spectrum antibiotics to deplete the microbiome of these mice. Microbiome-depleted SOD1-Tg mice had faster motor deterioration compared to SOD1-Tg mice with an intact microbiome. Comparison of the microbiome of SOD1-Tg mice to wild-type (WT) littermates showed SOD1-Tg mice had a distinctly different microbiome composition, even before the onset of motor impairment; thus, suggesting alterations in the gut microbiome of SOD1-Tg mice were not secondary to motor dysfunction. When we examined the effects of the specific strains of commensal microbes on ALS progression, we identified a few commensals that adversely affected the disease, such as Parabacteroides distasonis and Ruminococcus torques. One commensal – Akkermansia muciniphila23 – stood out as having a favorable effect on the motor phenotype in SOD1-Tg mice. We then sought to determine the possible mechanism by which A. muciniphila exerts its benefit on SOD1-Tg mice. The hypothesis underpinning this stage of the study was that A. muciniphila produces or modulates metabolites, which enter the nervous system of SOD1-Tg mice and exert a protective effect. Using a combination of untargeted metabolomic profiling and metagenomics, we narrowed our search down to several metabolites. Of those, two metabolites – nicotinamide (NAM) and phenol sulfate – were further tested in mice. A. muciniphila is able to effectively increase NAM levels in the CSF of SOD1-Tg mice, we investigated whether continuous NAM supplementation could alter the disease course. We found that there was a significant improvement in motor function in the NAM-treated mice, with a non-significant trend toward improved survival. Phenol sulfate administration did not change the disease course.
Figure 1.
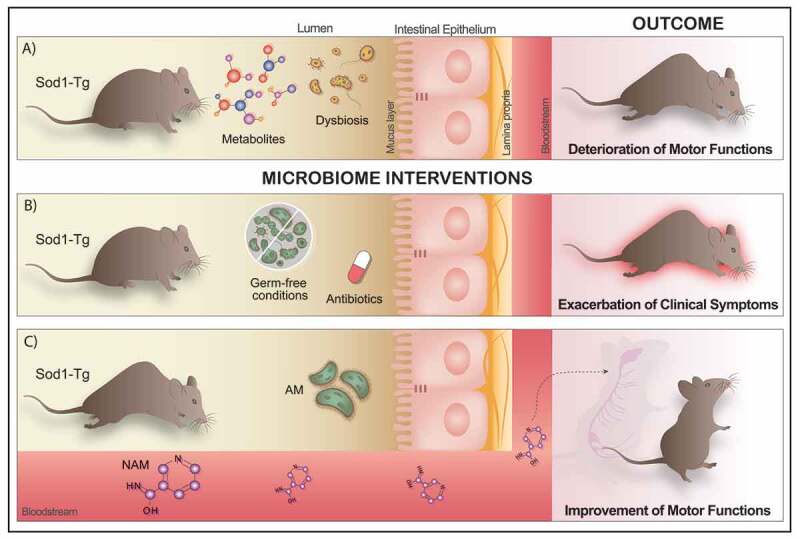
The role of the gut microbiome and its metabolites in an ALS mouse model. (a) SOD-1 transgenic (SOD1-Tg) mouse model of ALS present a distinctly different microbiome composition, even before the onset of any motor impairment and altered metabolites configuration, leading to the deterioration of motor functions after 140 d. (b) Depletion of the gut microbiome by administration of broad-spectrum antibiotics in the drinking water or using germ-free SOD1-Tg mice induces a rapid exacerbation of the clinical symptoms of the disease. (c) Oral administration of Akkermansia muciniphila (AM), a specific commensal bacterium, has beneficial effects on the ALS clinical outcomes, slowing down its progression in SOD1-Tg mice. On the other hand, AM is able to produce metabolites which in turn reach the nervous system through the bloodstream and therefore could impact the course of the disease. Thus, systemic administration of nicotinamide (NAM) via osmotic pumps significantly improved motor functions in the SOD1-Tg treated mice.
A recent murine longitudinal study by another group – also on the SOD1-Tg mouse model – confirmed that alternations within the microbiome are evident at the pre-symptomatic stage, being found as early as age 37 d – 27 d before motor deterioration.24 In contrast to our study, this study also found changes within the immune landscape, occurring in parallel with dysbiosis; these changes were accompanied by epigenetic alterations, leading the authors to hypothesize that there is interplay between the microbiome, immune system and epigenetic shifts.
As SOD1 mutations are found in only a fraction of ALS patients,25 it will be important to investigate whether similar changes occur in other genetic ALS models and in the absence of identifiable mutations in human disease.26
Potential relevance of our murine findings to human disease
The final stage of our study was to explore whether alterations observed in SOD1-Tg mice, were also present in human disease. We compared stool samples from patients with ALS and healthy household-matched controls. When we examined the global bacterial gene content, there was no clear difference between the abundance of specific bacterial species. However, we found that patients with ALS had a functional decrease in key bacterial genes that were involved in NAM metabolism. Interestingly, we found that several of the genes mapping to the A. muciniphila genome were significantly less abundant in patients with ALS.
Our findings in mice suggest that A. muciniphila may be beneficial in ALS, mediated via changes in the NAM metabolic pathway. To test whether similar changes in metabolic derangements occur in human disease, we obtained serum and CSF samples from patients with ALS and compared them to healthy controls. NAM levels were markedly decreased in human ALS sera and CSF, compared to controls; significant alterations of serum metabolites of NAM were also observed between these groups. NAM levels in patients with ALS were related to levels of the microbiome gene encoding the rate-limiting step of NAM biosynthesis. Interestingly, serum NAM levels tended to be lower in people with more advanced disease.
Taken together, the preliminary human data supports the possibility that microbiome-derived metabolites (such as NAM) may ultimately reach the CNS where they can modify the disease process. Although the murine data provides evidence of cause and effect, the human studies were purely observational and do not support causality.
Our murine study established a possible causal relationship between the microbiome and ALS progression. In order to assess causality in human disease, we believe there are a number of key questions that should be addressed, namely: How may the ALS disease process itself affect the microbiome? Do known ALS risk factors – both genetic and environmental – affect the microbiome? Do changes in the microbiome precede neurodegeneration in human disease? In this regard, it is important to consider that secondary factors such as nutrition, physical activity, and autonomic changes, occurring during the course of the disease, could have an impact on the microbiome and metabolic features as well.
Finally, interventional studies are planned by our group as well as others,27,28 which will hopefully provide evidence of causality, and more importantly therapeutic benefits.
How may ALS affect the microbiome?
The human form of ALS is a complex condition, and there are many ways in which the disease process itself could impact on the microbiome.29,30
Addressing these potentially confounding effects in human disease will be a major challenge to establishing causality.
Bulbar dysfunction affecting food and liquid consumption
ALS causes progressive degeneration of upper and lower motor neurons (UMNs and LMNs, respectively);1 the degeneration includes UMNs within the corticobulbar tract and LMNs within lower cranial nerve motor nuclei. Dysfunction of either of these neuronal populations results in weakness of the muscles involved in mastication, tongue mouth, cheek movement, and protection of the airway during swallowing. Thus, people with ALS tend to eat and drink at a slower rate, often reducing the quantity and changing the quality of the food and liquids they consume. All these changes may impact the microbiome.31 Poor nutrition results in accelerated weight loss, including loss of muscle mass which worsens the prognosis.17 Furthermore, when corticobulbar dysfunction is prominent, patients will often complain that they become highly intolerant to spicy food,32 as this may often trigger laryngospasm.
The bacterial microbiome of the mouth is likely affected by bulbar dysfunction in ALS, as oral hygiene is a challenge in this scenario.33 Oral stasis in ALS would be expected to influence the oral microbiome and, in turn, the intestinal microbiome.34,35 Potential future avenues of research should include a comparison of the microbiome in bulbar versus non-bulbar patients, to ascertain how bulbar dysfunction may affect both the oral and the intestinal microbiome.
Infections and antibiotic exposure
Infections are common in people with ALS, pneumonia being the most frequent, due to a combination of frequent aspiration, difficulty clearing secretions, and incomplete lung expansion.36 Gastrostomy tubes may also become infected37 and immobility may lead to pressure sores and skin infections.38 As such, people with ALS have more frequent exposure to antibiotics, which would impact the microbiome.39 Episodes of infection are a marker of poor prognosis in ALS, and pneumonia is often the cause of death.40–42 Infections could worsen the disease by a multitude of mechanisms, such as exacerbation of respiratory failure or promoting a hypercatabolic state. Nevertheless, it is possible that the effects of antibiotics on the microbiome in ALS could worsen prognosis. Future research directions could include evaluation of the microbiome before and after antibiotic therapy in ALS patients, and an attempt to re-institute the pre-antibiotic microbiome after antibiotic therapy in these patients.
Food supplements and replacements
When dysphagia progresses, most people with ALS opt for gastrostomy feeding.32 There are various liquid formulae available and it is unclear how these affect the microbiome. At earlier stages of the disease, many patients with ALS choose to take a variety of food supplements, vitamins, and minerals, despite lack of evidence of efficacy.43,44 Some of these may also impact the microbiome. Given the plethora of substances that people with ALS ingest in the absence of medical advice, it will be challenging to untangle this subject. That being said, it would be important to assess the relative effects of the various nutritional substitutes – those prescribed by nutritional consultants – on the microbiome.
Drugs
Riluzole and Edaravone are the only FDA-approved disease-modifying medications in ALS. Riluzole is absorbed via the GI tract, while Edaravone is given intravenously, but could still potentially affect the microbiome through the circulation. In addition, a variety of symptomatic treatments are frequency employed, which could all potentially affect the microbiome.32
GI function
Although ALS does not usually cause symptoms related to the autonomic nervous system, constipation is a frequent complaint in people with ALS.32 Indeed, gastrointestinal motor dysfunction – namely delayed gastric emptying and slowed colonic transit – are likely to be an under-recognized feature of ALS.45 Altered dietary habits, dehydration, and the effect of drugs may also contribute. Drugs with anticholinergic effects are often taken to decrease airway secretions and sialorrhea – these drugs affect GI motility, which could have a knock-on effect on the microbiome.
In addition, structural alterations have been demonstrated at the cellular level within the intestine of SOD1-Tg mice.9 The authors found increased permeability and leakiness, occurring together with decreased expression of junctional proteins. These changes were accompanied by accumulation of abnormal Paneth cells, which regulate innate immune responses, and shape the microbiome. They suggest these changes lead to systemic immune-dysregulation, thought to be an important pathogenic process in ALS.46
Other motor dysfunction
A general lack of movement can contribute to poor GI motility and constipation.32 Weakness of abdominal muscles can impair the process of fecal evacuation and contribute to constipation. As such, laxatives suppositories and enemas are frequently used, which can also alter the microbiome.47
Could known risk factors for ALS affect the microbiome?
Our mouse study provided evidence that the microbiome can influence the disease phenotype, but does not give any insight as to whether the microbiome could influence the development of ALS, per se.
In recent years there has been an explosion of genetic research providing insights into the pathogenesis of ALS at the molecular level. Some people with ALS have a family history and have a known pathogenic mutation (such as C9Orf72 or SOD1).2 More importantly, many people without a family history of ALS are found to have genetic alterations which are either know to or suspected of causing ALS.48,49 As such, a concerted effort is being made to delineate the variety of genetic factors that could potentially contribute to the disease.
Nevertheless, there are a number of epidemiological factors that seem to be associated with ALS that have established links with the microbiome. A number of vascular risk factors seem to be protective against the development of ALS;50 these include high BMI,51 type II diabetes,50 and dyslipidemia.52,53 Conversely, cigarette smoking increases the risk of ALS.54 All of these modifying factors have important interactions with the microbiome,55–60 which could be one mechanism via which these factors produce their effect, whether protective or conferring risk. Furthermore, people with high BMI or diabetes consume more non-calorific artificial sweeteners, which in turn produce changes within the microbiome.61,62 It is not clear how the genetic and environmental factors interplay; it has been suggested, for example, that a genetic pre-disposition to athleticism may predispose to ALS, in other words, the same genetic variants that allow high performance in athletics could predispose to ALS.1,63,64 It is also possible that some genetic ALS risk factors could be influencing the microbiome in a way that predisposes to ALS.
A large case–control study in Sweden found that patients with ALS were more likely to have received antibiotic prescriptions in the 8 y prior to diagnosis. The authors speculated that differences could be due to an altered microbiome.65 Unfortunately, no genetic data was presented in this study, as this could indicate whether dysbiosis could be triggering ALS onset in the genetically susceptible, or play a more central etiological role.
Potential future avenues of human research
Observational studies
A longitudinal study in pre-symptomatic ALS gene carriers would be a critical step to understanding the temporal relationship between microbiome alterations, and disease course. It is of particular importance to determine whether dysbiosis is occurring prior to symptom onset in human disease, as neurophysiological and neuropathological changes precede ALS symptom onset.1,66 We have initiated such a study in pre-symptomatic C9Orf72 gene carriers. Longitudinal studies in the broader ALS population, starting at diagnostic suspicion, would also be valuable in identifying changes occurring earlier in the disease.
Therapeutic interventions
Studying the microbiome in ALS may uncover metabolic changes affecting the disease process. Our study established that A. muciniphila increases NAM levels in the CSF of SOD1-Tg mice, and treating these mice with NAM improved motor function.21 Although it is tempting to extrapolate these changes to other progressive neurological conditions, current evidence in both humans and mice suggests otherwise; for example, Akkermansia species may be enriched in Alzheimer’s disease,9,67 Parkinson’s disease,68 and multiple sclerosis.69,70
In addition to therapeutic fecal microbiota transplantation trials in ALS, our findings also support metabolite-based treatment strategies, such as NAM supplementation.
Conclusion
The microbiome may be altered in ALS mice and can influence disease progression in this model. Some of these changes seem to be relevant to human disease. The mechanisms underlying these changes could involve an interplay between the microbiome, structural changes within the gut, gut-derived metabolites, and the immune system. We believe that interventional human studies are now warranted. Whether the microbiome could have an etiological role in ALS is an important avenue for further research.
Acknowledgments
We thank the members of the Elinav lab, and members of the DKFZ cancer-microbiome division for discussions, and apologize to authors whose work was not included due to space constraints. E.E. is supported by: Y. and R. Ungar; the Abisch Frenkel Foundation for the Promotion of Life Sciences; the Gurwin Family Fund for Scientific Research; the Leona M. and Harry B. Helmsley Charitable Trust; the Crown Endowment Fund for Immunological Research; the Else Kroener Fresenius Foundation; the estate of J. Gitlitz; the estate of L. Hershkovich; the Benoziyo Endowment Fund for the Advancement of Science; the Adelis Foundation; J. L. and V. Schwartz; A. and G. Markovitz; A. and C. Adelson; the French National Center for Scientific Research (CNRS); D. L. Schwarz; The V. R. Schwartz Research Fellow Chair; L. Steinberg; J. N. Halpern; A. Edelheit, and by grants funded by the European Research Council; the Israel Science Foundation; the Helmholtz Foundation E.E. is the Sir Marc & Lady Tania Feldmann Professorial Chair in Immunology, Weizmann Institute of Science, Israel, Director, Microbiome Research Division, DKFZ, Heidelberg, Germany, is a senior fellow, Canadian Institute of Advanced Research (CIFAR) and an international scholar, The Bill & Melinda Gates Foundation and Howard Hughes Medical Institute (HHMI).
Competing interests
E.E. is a paid consultant at DayTwo and BiomX. None of this work is related to, shared with or licensed to these or any other commercial entity. None of the other authors have competing interests related to this work.
References
- 1.Turner MR, Hardiman O, Benatar M, Brooks BR, Chio A, de Carvalho M, Ince PG, Lin C, Miller RG, Mitsumoto H, et al. Controversies and priorities in amyotrophic lateral sclerosis. Lancet Neurol. 2013. March;12(3):310–322. doi: 10.1016/S1474-4422(13)70036-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, Challis C, Schretter CE, Rocha S, Gradinaru V, et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of parkinson’s disease. Cell. 2016. December;167(6):1469–1480.e12. doi: 10.1016/j.cell.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharon G, Cruz NJ, Kang D-W, Gandal MJ, Wang B, Kim Y-M, Zink EM, Casey CP, Taylor BC, Lane CJ, et al. Human gut microbiota from autism spectrum disorder promote behavioral symptoms in mice. Cell. 2019. May;177(6):1600–1618.e17. doi: 10.1016/j.cell.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Theije CGM, Wopereis H, Ramadan M, van Eijndthoven T, Lambert J, Knol J, Garssen J, Kraneveld AD, Oozeer R.. Altered gut microbiota and activity in a murine model of autism spectrum disorders. Brain Behav Immun. 2014. March;37:197–206. doi: 10.1016/j.bbi.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 5.De Angelis M, Piccolo M, Vannini L, Siragusa S, De Giacomo A, Serrazzanetti DI, Cristofori F, Guerzoni ME, Gobbetti M, Francavilla R. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS One. 2013. October;8(10):e76993. doi: 10.1371/journal.pone.0076993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mazzini L, Mogna L, De Marchi F, Amoruso A, Pane M, Aloisio I, Cionci NB, Gaggìa F, Lucenti A, Bersano E, et al. Potential role of gut microbiota in ALS pathogenesis and possible novel therapeutic strategies. J Clin Gastroenterol. 2018;52(Suppl 1):S68–S70. Proceedings from the 9th Probiotics, Prebiotics and New Foods, Nutraceuticals and Botanicals for Nutrition & Human and Microbiota Health Meeting, held in Rome, Italy from September 10 to 12, 2017. doi: 10.1097/MCG.0000000000001042. [DOI] [PubMed] [Google Scholar]
- 7.Erber AC, Cetin H, Berry D, Schernhammer ES. The role of gut microbiota, butyrate and proton pump inhibitors in amyotrophic lateral sclerosis: a systematic review. Int J Neurosci. 2019. December:1–9. DOI: 10.1080/00207454.2019.1702549. [DOI] [PubMed] [Google Scholar]
- 8.De Angelis M, Francavilla R, Piccolo M, De Giacomo A, Gobbetti M. Autism spectrum disorders and intestinal microbiota. Gut Microbes. 2015;6(3):207–213. doi: 10.1080/19490976.2015.1035855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogt NM, Kerby RL, Dill-McFarland KA, Harding SJ, Merluzzi AP, Johnson SC, Carlsson CM, Asthana S, Zetterberg H, Blennow K, et al. Gut microbiome alterations in Alzheimer’s disease. Sci Rep. 2017. October;7(1):13537. doi: 10.1038/s41598-017-13601-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tillisch K, Labus J, Kilpatrick L, Jiang Z, Stains J, Ebrat B, Guyonnet D, Legrain-Raspaud S, Trotin B, Naliboff B, et al. Consumption of fermented milk product with probiotic modulates brain activity. Gastroenterology. 2013. June;144(7):1394–401, 1401.e1. doi: 10.1053/j.gastro.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kouchaki E, Tamtaji OR, Salami M, Bahmani F, Daneshvar Kakhaki R, Akbari E, Tajabadi-Ebrahimi M, Jafari P, Asemi Z. Clinical and metabolic response to probiotic supplementation in patients with multiple sclerosis: A randomized, double-blind, placebo-controlled trial. Clin Nutr. 2017;36(5):1245–1249. doi: 10.1016/j.clnu.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 12.Akbari E, Asemi Z, Daneshvar Kakhaki R, Bahmani F, Kouchaki E, Tamtaji OR, Hamidi GA, Salami M. Effect of probiotic supplementation on cognitive function and metabolic status in Alzheimer’s disease: a randomized, double-blind and controlled trial. Front Aging Neurosci. 2016. November;8:256. doi: 10.3389/fnagi.2016.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamtaji OR, Taghizadeh M, Daneshvar Kakhaki R, Kouchaki E, Bahmani F, Borzabadi S, Oryan S, Mafi A, Asemi Z. Clinical and metabolic response to probiotic administration in people with Parkinson’s disease: A randomized, double-blind, placebo-controlled trial. Clin Nutr. 2019;38(3):1031–1035. doi: 10.1016/j.clnu.2018.05.018. [DOI] [PubMed] [Google Scholar]
- 14.Lum GR, Olson CA, Hsiao EY. Emerging roles for the intestinal microbiome in epilepsy. Neurobiol Dis. 2020. February;135:104576. doi: 10.1016/j.nbd.2019.104576. [DOI] [PubMed] [Google Scholar]
- 15.Blacher E, Levy M, Tatirovsky E, Elinav E. Microbiome-modulated metabolites at the interface of host immunity. J Immunol. 2017. January;198(2):572–580. doi: 10.4049/jimmunol.1601247. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Kasper LH. The role of microbiome in central nervous system disorders. Brain Behav Immun. 2014. May;38:1–12. doi: 10.1016/j.bbi.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu S, Yi J, Zhang Y-G, Zhou J, Sun J. Leaky intestine and impaired microbiome in an amyotrophic lateral sclerosis mouse model. Physiol Rep. 2015;3(4):Apr. doi: 10.14814/phy2.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y-G, Wu S, Yi J, Xia Y, Jin D, Zhou J, Sun J. Target intestinal microbiota to alleviate disease progression in amyotrophic lateral sclerosis. Clin Ther. 2017. February;39(2):322–336. doi: 10.1016/j.clinthera.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang X, Wang X, Yang S, Meng F, Wang X, Wei H, Chen T. Evaluation of the microbial diversity in amyotrophic lateral sclerosis using high-throughput sequencing. Front Microbiol. 2016. September;7:1479. doi: 10.3389/fmicb.2016.01479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brenner D, Hiergeist A, Adis C, Mayer B, Gessner A, Ludolph AC, Weishaupt JH. The fecal microbiome of ALS patients. Neurobiol Aging. 2018;61:132–137. doi: 10.1016/j.neurobiolaging.2017.09.023. [DOI] [PubMed] [Google Scholar]
- 21.Blacher E, Bashiardes S, Shapiro H, Rothschild D, Mor U, Dori-Bachash M, Kleimeyer C, Moresi C, Harnik Y, Zur M, et al. Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature. 2019. July;572(7770):474–480. doi: 10.1038/s41586-019-1443-5. [DOI] [PubMed] [Google Scholar]
- 22.Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, Alexander DD, Caliendo J, Hentati A, Kwon YW, Deng HX. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994. June;264(5166):1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 23.Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl Environ Microbiol. 2007. December;73(23):7767–7770. doi: 10.1128/AEM.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Figueroa-Romero C, Guo K, Murdock BJ, Paez-Colasante X, Bassis CM, Mikhail KA, Raue KD, Evans MC, Taubman GF, McDermott AJ, et al. Temporal evolution of the microbiome, immune system and epigenome with disease progression in ALS mice. Dis Model Mech. 2019. November;13(2). doi: 10.1242/dmm.040832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rowland LP. Ameliorating amyotrophic lateral sclerosis. N Engl J Med. 2010. March;362(10):953–954. doi: 10.1056/NEJMcibr0912229. [DOI] [PubMed] [Google Scholar]
- 26.Rothstein JD. Of mice and men: reconciling preclinical ALS mouse studies and human clinical trials. Ann Neurol. 2003. April;53(4):423–426. doi: 10.1002/ana.10561. [DOI] [PubMed] [Google Scholar]
- 27.Mandrioli J, Amedei A, Cammarota G, Niccolai E, Zucchi E, D’Amico R, Ricci F, Quaranta G, Spanu T, Masucci L. FETR-ALS study protocol: a randomized clinical trial of fecal microbiota transplantation in amyotrophic lateral sclerosis. Front Neurol. 2019. September;10:1021. doi: 10.3389/fneur.2019.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.A study of protein metabolism, microbiome and investigational probiotic use in patients with ALS - full text view - clinicalTrials.gov. [Online]. [Accessed 21March 2020]. https://clinicaltrials.gov/ct2/show/NCT03324399.
- 29.Rowin J, Xia Y, Jung B, Sun J. Gut inflammation and dysbiosis in human motor neuron disease. Physiol. Rep. 2017. September;5(18):e13443. doi: 10.14814/phy2.13443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhai C-D, Zheng -J-J, An B-C, Huang H-F, Tan Z-C. Intestinal microbiota composition in patients with amyotrophic lateral sclerosis: establishment of bacterial and archaeal communities analyses. Chin Med J (Engl). 2019. August;132(15):1815–1822. doi: 10.1097/CM9.0000000000000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh RK, Chang H-W, Yan D, Lee KM, Ucmak D, Wong K, Abrouk M, Farahnik B, Nakamura M, Zhu TH, et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017. April;15(1):73. doi: 10.1186/s12967-017-1175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson CE, McVey AL, Rudnicki S, Dimachkie MM, Barohn RJ. Symptom management and end-of-life care in amyotrophic lateral sclerosis. Neurol Clin. 2015. November;33(4):889–908. doi: 10.1016/j.ncl.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bergendal B, McAllister A. Orofacial function and monitoring of oral care in amyotrophic lateral sclerosis. Acta Odontol Scand. 2017. April;75(3):179–185. doi: 10.1080/00016357.2016.1276212. [DOI] [PubMed] [Google Scholar]
- 34.Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, Bienenstock J, Cryan JF. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci USA. 2011. September;108(38):16050–16055. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao L, Xu T, Huang G, Jiang S, Gu Y, Chen F. Oral microbiomes: more and more importance in oral cavity and whole body. Protein Cell. 2018. May;9(5):488–500. doi: 10.1007/s13238-018-0548-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sorenson EJ, Stalker AP, Kurland LT, Windebank AJ. Amyotrophic lateral sclerosis in Olmsted County, Minnesota, 1925 to 1998. Neurology. 2002. July;59(2):280–282. doi: 10.1212/WNL.59.2.280. [DOI] [PubMed] [Google Scholar]
- 37.ProGas Study Group . Gastrostomy in patients with amyotrophic lateral sclerosis (ProGas): a prospective cohort study. Lancet Neurol. 2015. July;14(7):702–709. DOI: 10.1016/S1474-4422(15)00104-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen JH, Wu SC, Chen HJ, Kao CH, Tseng CH, Tsai CH. Risk of developing pressure sore in amyotrophic lateral sclerosis patients - a nationwide cohort study. J Eur Acad Dermatol Venereol. 2018. September;32(9):1589–1596. doi: 10.1111/jdv.14911. [DOI] [PubMed] [Google Scholar]
- 39.Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology. 2010. November;156(Pt 11):3216–3223. doi: 10.1099/mic.0.040618-0. [DOI] [PubMed] [Google Scholar]
- 40.Chiò A, Logroscino G, Hardiman O, Swingler R, Mitchell D, Beghi E, Traynor BG, Eurals Consortium . Prognostic factors in ALS: A critical review. Amyotroph Lateral Scler. 2009. December;10(5–6):310–323. doi: 10.3109/17482960802566824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corcia P, Pradat P-F, Salachas F, Bruneteau G, le Forestier N, Seilhean D, Hauw -J-J, Meininger V. Causes of death in a post-mortem series of ALS patients. Amyotroph Lateral Scler. 2008;9(1):59–62. doi: 10.1080/17482960701656940. [DOI] [PubMed] [Google Scholar]
- 42.Westeneng H-J, Debray TPA, Visser AE, van Eijk RPA, Rooney JPK, Calvo A, Martin S, McDermott CJ, Thompson AG, Pinto S, et al. Prognosis for patients with amyotrophic lateral sclerosis: development and validation of a personalised prediction model. Lancet Neurol. 2018. March;17(5):423–433. doi: 10.1016/S1474-4422(18)30089-9. [DOI] [PubMed] [Google Scholar]
- 43.Kim S, Chung SE, Lee S, Park J, Choi S, Kim S. Experience of complementary and alternative medicine in patients with amyotrophic lateral sclerosis and their families: A qualitative study. Amyotroph Lateral Scler Frontotemporal Degener. 2016. January;17(3–4):191–197. doi: 10.3109/21678421.2015.1125504. [DOI] [PubMed] [Google Scholar]
- 44.Adams J, Lee M, Peng W. Critical review of complementary and alternative medicine use in amyotrophic lateral sclerosis: prevalence and users’ profile, decision-making, information seeking, and disclosure in the face of a lack of efficacy. Neurodegener Dis. 2018. September;18(4):225–232. doi: 10.1159/000492946. [DOI] [PubMed] [Google Scholar]
- 45.Toepfer M, Folwaczny C, Klauser A, Riepl RL, Müller-Felber W, Pongratz D. Gastrointestinal dysfunction in amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord. 1999. December;1(1):15–19. doi: 10.1080/146608299300079484. [DOI] [PubMed] [Google Scholar]
- 46.Beers DR, Appel SH. Immune dysregulation in amyotrophic lateral sclerosis: mechanisms and emerging therapies. Lancet Neurol. 2019. February;18(2):211–220. doi: 10.1016/S1474-4422(18)30394-6. [DOI] [PubMed] [Google Scholar]
- 47.Tropini C, Moss EL, Merrill BD, Ng KM, Higginbottom SK, Casavant EP, Gonzalez CG, Fremin B, Bouley DM, Elias JE, et al. Transient osmotic perturbation causes long-term alteration to the gut microbiota. Cell. 2018. June;173(7):1742–1754.e17. doi: 10.1016/j.cell.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cooper-Knock J, Robins H, Niedermoser I, Wyles M, Heath PR, Higginbottom A, Walsh T, Kazoka M, Project MinE ALS Sequencing Consortium, Ince P. G., Hautbergue G. M., McDermott C. J., Kirby J., and Shaw P. J.. Targeted genetic screen in amyotrophic lateral sclerosis reveals novel genetic variants with synergistic effect on clinical phenotype. Front Mol Neurosci. 2017. November;10:370. doi: 10.3389/fnmol.2017.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cady J, Allred P, Bali T, Pestronk A, Goate A, Miller TM, Mitra RD, Ravits J, Harms MB, Baloh RH. Amyotrophic lateral sclerosis onset is influenced by the burden of rare variants in known amyotrophic lateral sclerosis genes. Ann Neurol. 2015. January;77(1):100–113. doi: 10.1002/ana.24306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kioumourtzoglou M-A, Rotem RS, Seals RM, Gredal O, Hansen J, Weisskopf MG. Diabetes mellitus, obesity, and diagnosis of amyotrophic lateral sclerosis: a population-based study. JAMA Neurol. 2015. August;72(8):905–911. doi: 10.1001/jamaneurol.2015.0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Reilly ÉJ, Wang H, Weisskopf MG, Fitzgerald KC, Falcone G, McCullough ML, Thun M, Park Y, Kolonel LN, Ascherio A. Premorbid body mass index and risk of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Frontotemporal Degener. 2013. April;14(3):205–211. doi: 10.3109/21678421.2012.735240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dupuis L, Corcia P, Fergani A, Gonzalez De Aguilar JL, Bonnefont-Rousselot D, Bittar R, Seilhean D, Hauw JJ, Lacomblez L, Loeffler JP, et al. Dyslipidemia is a protective factor in amyotrophic lateral sclerosis. Neurology. 2008. March;70(13):1004–1009. doi: 10.1212/01.wnl.0000285080.70324.27. [DOI] [PubMed] [Google Scholar]
- 53.Dupuis L, Pradat P-F, Ludolph AC, Loeffler J-P. Energy metabolism in amyotrophic lateral sclerosis. Lancet Neurol. 2011. January;10(1):75–82. doi: 10.1016/S1474-4422(10)70224-6. [DOI] [PubMed] [Google Scholar]
- 54.Calvo A, Canosa A, Bertuzzo D, Cugnasco P, Solero L, Clerico M, De Mercanti S, Bersano E, Cammarosano S, Ilardi A, et al. Influence of cigarette smoking on ALS outcome: a population-based study. J Neurol Neurosurg Psychiatry. 2016. November;87(11):1229–1233. doi: 10.1136/jnnp-2016-313793. [DOI] [PubMed] [Google Scholar]
- 55.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006. December;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 56.Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, Fu H, Xue X, Lu C, Ma J, et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. 2018. March;359(6380):1151–1156. doi: 10.1126/science.aao5774. [DOI] [PubMed] [Google Scholar]
- 57.Koh A, Molinaro A, Ståhlman M, Khan MT, Schmidt C, Mannerås-Holm L, Wu H, Carreras A, Jeong H, Olofsson LE, et al. Microbially produced imidazole propionate impairs insulin signaling through mTORC1. Cell. 2018. November;175(4):947–961.e17. doi: 10.1016/j.cell.2018.09.055. [DOI] [PubMed] [Google Scholar]
- 58.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto J-M, Kennedy S, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013. August;500(7464):541–546. DOI: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 59.Depommier C, Everard A, Druart C, Plovier H, Van Hul M, Vieira-Silva S, Falony G, Raes J, Maiter D, Delzenne NM, et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med. 2019. July;25(7):1096–1103. doi: 10.1038/s41591-019-0495-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shanahan ER, Shah A, Koloski N, Walker MM, Talley NJ, Morrison M, Holtmann GJ. Influence of cigarette smoking on the human duodenal mucosa-associated microbiota. Microbiome. 2018. August;6(1):150. doi: 10.1186/s40168-018-0531-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suez J, Korem T, Zilberman-Schapira G, Segal E, Elinav E. Non-caloric artificial sweeteners and the microbiome: findings and challenges. Gut Microbes. 2015. April;6(2):149–155. doi: 10.1080/19490976.2015.1017700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suez J, Korem T, Zeevi D, Zilberman-Schapira G, Thaiss CA, Maza O, Israeli D, Zmora N, Gilad S, Weinberger A, et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature. 2014. October;514(7521):181–186. doi: 10.1038/nature13793. [DOI] [PubMed] [Google Scholar]
- 63.Gotkine M, Friedlander Y, Hochner H. Triathletes are over-represented in a population of patients with ALS. Amyotroph Lateral Scler Frontotemporal Degener. 2014. December;15(7–8):534–536. doi: 10.3109/21678421.2014.932383. [DOI] [PubMed] [Google Scholar]
- 64.Huisman MHB, Seelen M, de Jong SW, Dorresteijn KRIS, van Doormaal PTC, van der Kooi AJ, de Visser M, Schelhaas HJ, van den Berg LH, Veldink JH. Lifetime physical activity and the risk of amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2013. September;84(9):976–981. doi: 10.1136/jnnp-2012-304724. [DOI] [PubMed] [Google Scholar]
- 65.Sun J, Zhan Y, Mariosa D, Larsson H, Almqvist C, Ingre C, Zagai U, Pawitan Y, Fang F. Antibiotics use and risk of amyotrophic lateral sclerosis in Sweden. Eur J Neurol. 2019. November;26(11):1355–1361. doi: 10.1111/ene.13986. [DOI] [PubMed] [Google Scholar]
- 66.Chiu AY, Zhai P, Dal Canto MC, Peters TM, Kwon YW, Prattis SM, Gurney ME. Age-dependent penetrance of disease in a transgenic mouse model of familial amyotrophic lateral sclerosis. Mol Cell Neurosci. 1995. August;6(4):349–362. doi: 10.1006/mcne.1995.1027. [DOI] [PubMed] [Google Scholar]
- 67.Dodiya HB, Kuntz T, Shaik SM, Baufeld C, Leibowitz J, Zhang X, Gottel N, Zhang X, Butovsky O, Gilbert JA, et al. Sex-specific effects of microbiome perturbations on cerebral Aβ amyloidosis and microglia phenotypes. J Exp Med. 2019. July;216(7):1542–1560. doi: 10.1084/jem.20182386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hill-Burns EM, Debelius JW, Morton JT, Wissemann WT, Lewis MR, Wallen ZD, Peddada SD, Factor SA, Molho E, Zabetian CP, et al. Parkinson’s disease and Parkinson’s disease medications have distinct signatures of the gut microbiome. Mov Disord. 2017. February;32(5):739–749. doi: 10.1002/mds.26942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cekanaviciute E, Yoo BB, Runia TF, Debelius JW, Singh S, Nelson CA, Kanner R, Bencosme Y, Lee YK, Hauser SL, et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc Natl Acad Sci USA. 2017. October;114(40):10713–10718. doi: 10.1073/pnas.1711235114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berer K, Gerdes LA, Cekanaviciute E, Jia X, Xiao L, Xia Z, Liu C, Klotz L, Stauffer U, Baranzini SE, et al. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc Natl Acad Sci USA. 2017. October;114(40):10719–10724. doi: 10.1073/pnas.1711233114. [DOI] [PMC free article] [PubMed] [Google Scholar]


