ABSTRACT
Antimicrobial resistance is one of the largest threats to global health and imposes substantial burdens in terms of morbidity, mortality, and economic costs. The gut is a key conduit for the genesis and spread of antimicrobial resistance in enteric bacterial pathogens. Distinct bacterial species that cause enteric disease can exist as invasive enteropathogens that immediately evoke gastrointestinal distress, or pathobionts that can arise from established bacterial commensals to inflict dysbiosis and disease. Furthermore, various environmental reservoirs and stressors facilitate the evolution and transmission of resistance. In this review, we present a comprehensive discussion on circulating resistance profiles and gene mobilization strategies of the most problematic species of enteric bacterial pathogens. Importantly, we present emerging approaches toward surveillance of pathogens and their resistance elements as well as promising treatment strategies that can circumvent common resistance mechanisms.
KEYWORDS: Antimicrobial resistance, enteric pathogen, pathobiont, mobile genetic element, high-throughput sequencing, whole-genome sequencing, metagenomics
Introduction
Antimicrobial resistance (AMR) is one of the most formidable threats to global human health. A recent report by the Centers for Disease Control and Prevention (CDC) declared that humankind has entered the dreaded “post-antibiotic era,” wherein we face infections resistant to every available treatment option.1 The transmission and spread of multi-drug resistant organisms (MDROs) is facilitated by astronomical increases in human travel and trade within the last few decades.2 Without adequate intervention, global death rates attributable to AMR are projected to surpass that of cancer and reach 10 million deaths per year by 2050.3 Nearly all of the most concerning pathogenic species associated with AMR spend some portion of their lifecycle within the mammalian gut.1,2
The human gastrointestinal (GI) tract contains a highly structured community poised with the potential to grow and transmit MDROs, with the gut microbiome containing an estimated 1014 microorganisms.5 This multi-species ‘organ’ exists in open contact with external factors such as antibiotics and outside organisms that facilitate AMR development and spread. A considerable number of these factors are a product of the past century of human history, during which antimicrobial therapies were discovered and put into wide-scale use for treatment of human infections, animal husbandry, and agriculture.6 AMR is particularly challenging in the realm of hospital-acquired infection, where vulnerable populations are easily colonized by MDROs.1 Even before modern history, life in microbial communities has allowed bacteria to evolve with xenobiotic stressors, natural products or antibiotics, produced by competing microorganisms, resulting in an innate, if not “ancient,” resistome.7
This review focuses on AMR in enteric bacteria, as they are a significant cause of human infection and the human gut serves as a major conduit for the genesis and environmental spread of MDROs. Here we highlight major bacterial species that cause enteric disease, the AMR mechanisms they employ, and the various modes of AMR mobilization. It is important to note that while this review focuses exclusively on bacterial pathogens, there are extensive enteric morbidities and drug resistance associated with viral, protozoal, and fungal microorganisms, which have been reviewed elsewhere.8–11 In the context of enteric bacterial pathogens, we additionally suggest future avenues for prevention and treatment of enteric MDROs. In recent years, genomics- and metagenomics-based methods are increasingly being employed to survey circulating antibiotic resistance genes (ARGs) and predict diverse ARG mobilization strategies.12 A number of alternative solutions to standard antibiotics, in the form of vaccines, alternative antimicrobial targets, or probiotic cocktails, may provide some hope for the mitigation of AMR in the context of enteric disease.13
Part I. Bacterial enemies, foreign and domestic
Within this review, the major enteropathogenic bacterial species are bifurcated into two groups: (1) invasive enteropathogens, which originate from an outside environmental reservoir, and (2) pathobionts, which originate from commensal gut species (Table 1). There are, of course, well-documented deviations from these delineations. For example, we categorize Clostridioides difficile as an invasive enteropathogen, yet it can colonize some hosts asymptomatically.46 Conversely, we classify Escherichia coli as a pathobiont, although some pathotypes of E. coli (discussed further below) are obligate invasive enteropathogens.4 The “switch” between these two categories can often be achieved rapidly through horizontal gene transfer (HGT) of genetic elements such as pathogenicity islands,47 which is discussed in Part II. It is important to note that across these pathogens, there are a range of ad hoc clinical diagnostic standards for determining susceptibility,48 and these practices are often absent or underutilized in the case of anaerobic pathogens such as Clostridioides or Bacteroides species.49
Table 1.
Emerging and notable resistance mechanisms among enteric bacterial pathogens.
| Species | Mechanism | Resistance Conferred | Reference |
|---|---|---|---|
| Invasive Enteropathogens | |||
| Campylobacter species | Enhanced CmeABC multidrug efflux pump | Macrolides, fluoroquinolones, tetracyclines | 14,15 |
| QRDR target site mutations in gyrA | Fluoroquinolones | 16 | |
| Ribosomal target site modification | Macrolides | 17 | |
| ermB-mediated methylation of 23S rRNA | MLSB family, linezolid | 14,18,19 | |
| MDR genomic islands | Aminoglycosides, macrolides, fluoroquinolones | 14,20 | |
| Shigella species | QRDR target site mutations and plasmid-mediated qnrS | Fluoroquinolones | 21 |
| Overexpression of acrA efflux pump gene and plasmid-mediated enhanced efflux through QepAB and OqxAB | Fluoroquinolones | 21–23 | |
| IncF plasmid carriage of mphA or erm genes | Macrolides | 24 | |
| ESBLs including CTX-M, AmpC, and OXA | β-lactams | 21 | |
| Salmonella enterica | Chromosomal QRDR mutations and plasmid-mediated qnr variants | Fluoroquinolones | 25 |
| Plasmid-mediated oqxAB and qepA | Fluoroquinolones, aminoglycosides, and β-lactams | 25 | |
| Salmonella genomic island 1 encoding ACSSuT | Ampicillin, florfenicol, florfenicol, streptomycin, chloramphenicol, spectinomycin, antifolates, and tetracycline | 26 | |
| Plasmid-mediated mobile colistin resistance genes | Colistin | 27 | |
| Plasmid-mediated blaCMY genes | Cephalosporins | 25 | |
| Vibrio cholerae | vex RND family efflux pumps | Erythromycin, novobiocin, penicillins, and polymyxin B | 28 |
| SXT elements | Antifolates, streptomycin, nalidixic acid, tetracycline, and others | 29 | |
| QRDR target site mutations | Fluoroquinolones | 30 | |
| Clostridioides difficile | Tn5398, Tn6194, Tn6215, or Tn916 transfer of ermB or cfr genes | MLSB family and linezolid | 31 |
| Metabolic pathway alterations such as DNA repair and iron uptake | Metronidazole | 32 | |
| Changes in peptidoglycan biosynthesis pathway, likely MurG | Vancomycin | 31 | |
| Mutations in rpoB | Fidaxomicin, rifamycin | 33,34 | |
| Pathobionts | |||
| BFG species | Insertion sequence activation of genes | Carbapenem, β-lactam, metronidazole, and macrolides | 35 |
| Conjugative transposons activated by two-component regulatory system, most notably CTnDOT | Tetracyclines and erythromycin | 36 | |
| bmeABC efflux pump | β-lactams, carbapenems, cephems, metronidazole, quinolones, etc. | 37 | |
| erm gene-mediated methylation of 23S rRNA | MLSB family | 36 | |
| Enterococcus faecalis and Enterococcus faecium | van operon | Glycopeptides | 38 |
| Target site mutations in LiaFSR, gpdD, cls, YycFG | Daptomycin | 39 | |
| Target mutation or methylation of 23S rRNA | Linezolid | 40 | |
| Escherichia coli | AmpC β-lactamase | Cephalosporins | 41 |
| Carbapenemases including NDM, KPC, and OXA | Carbapenems | 42 | |
| Upregulation of acrAB, QRDR point mutations | Fluoroquinolones | 43 | |
| Plasmid-mediated mobile colistin resistance genes (mcr1-9) | Colistin | 44,45 | |
QRDR, quinolone resistance determining region; MLSB, macrolides, lincosamides, streptogramins B; MDR, multi-drug resistance; ESBL, extended-spectrum β-lactamase; ACSSuT, resistance to ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, tetracycline; RND, resistance-nodulation-division; NDM, New Delhi metallo-β-lactamase; KPC, Klebsiella pneumoniae carbapenemase; OXA, oxacillinase.
Professional pathogens: the invasive enteropathogens
Invasive enteropathogens do not typically occupy the human microbiome as commensal species, and upon pathogenesis they can inflict acute intestinal distress including gastroenteritis, inflammation, and diarrhea. If not treated properly, extensive morbidities such as dehydration, bacteremia, shock, and even death may ensue.50 Diarrheal disease accounts for over 1.6 million deaths worldwide and is one of the top five causes of mortality for children under five.51 Many of these diseases are endemic to specific regions, but increased globalization has accelerated international transmission of MDROs.2 Furthermore, common reservoirs of infection include water sources, food, and animals (Figure 1a).2 Invasive enteropathogens employ diverse mechanisms of AMR, which exacerbate the associated burdens on human health and the economy.1
Figure 1.

Major bacterial enteropathogens, antibiotic resistance reservoirs, and pathogenesis in the human gut. (a) The major enteric bacterial species and common reservoirs for proliferation and resistance exchange. (b) A close-up view of the human gut, representing various pathobiont species and pathogenic tendencies. C. diff: Clostridioides difficile, VRE: Vancomycin-resistant enterococcus, BFG: Bacteroides fragilis group. *Indicates E. coli can assume multiple pathogenic manifestations within the gut, as described by Kaper and coauthors.4 Image made with BioRender.
Campylobacter species
Campylobacter species, including C. jejuni and C. coli, are a dominant cause of gastroenteritis and diarrhea, with rates of campylobacteriosis increasing worldwide.52 Acquisition of Campylobacter infection is often foodborne and linked to fecal contamination of water sources (Figure 1a); multiple animal reservoirs, most especially poultry,53 can host Campylobacter species. Campylobacteriosis is typically self-limiting, with empiric use of antibiotics such as fluoroquinolones in settings of acute disease.50 AMR in Campylobacter is highly prevalent in the United States, with over 400,000 cases of drug-resistant campylobacteriosis of the 1.5 million estimated total cases of infection.1,14 Resistance to both azithromycin, the drug of choice, and ciprofloxacin, a key second-line option, have appeared in multiple forms.50,54 Fluoroquinolone resistance can arise through point mutations in the quinolone-resistance-determining region (QRDR) of the fluoroquinolone target gyrA;16 this mechanism of resistance is conserved across many enteropathogens (Table 1). Within Campylobacter, QRDR mutations are synergistic with recently described, “enhanced” versions of the resistance-nodulation-division (RND) multi-drug resistance (MDR)-conferring efflux pump CmeABC.15 This efflux pump system has emerged with mutated regulatory regions that increase transcription of cmeABC and increase resistance to macrolides and fluoroquinolones (Table 1). It is likely that this efflux pump operon is controlled by multiple regulators, some of which may be drug-activated.15,55 Finally, AMR in Campylobacter species may be linked to the organism’s natural competency, allowing it to sample the community for transferable resistance.56 These observed MDR profiles and increasing rates of drug-resistant campylobacteriosis pose Campylobacter species as a serious global health threat.
Shigella species
Shigella bacteria are another major source of food poisoning and diarrheal disease.1,54 In 2016, shigellosis was the second leading cause of diarrheal death worldwide at over 200,000 deaths per year, ranking second only to rotavirus.51 The genus Shigella contains bacteria closely related to E. coli and is comprised of four major pathogenic species: S. dysenteriae, S. flexneri, S. boydii, and S. sonnei.21 Shigella can be transmitted from person-to-person, or through contaminated food sources and water (Figure 1a), while some AMR Shigella outbreaks are associated with international travel and sexual transmission.57 Shigella infections were once highly responsive to cheaper antibiotics such as β-lactams and antifolates, but rising resistance rates have shifted the treatments of choice toward macrolides or fluoroquinolones, with ceftriaxone as an alternative treatment option.50 MDR Shigella can arise through plasmid-borne or integron-mobilized elements encoding multiple types of resistance.21,54 A commonly observed MDR phenotype includes resistance to ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracyclines (ACSSuT).21,24,58 Epidemics driven by MDR Shigella have risen worldwide within the last decade and requires significant intervention efforts to prevent further disease.
Salmonella enterica
Another enteropathogenic species within the Enterobacteriaceae family is Salmonella enterica. The serovars of S. enterica are divided into typhoidal and non-typhoidal Salmonella (NTS), and encompass a group of bacteria that occupy digestive tracts of both animals and humans (Figure 1a).59 Although borne from the same species, the clinical manifestations and the associated immune responses are distinct among serovars. The GI distress associated with typhoid fever is due to the typhoid toxin and the damage it inflicts upon the GI epithelium. Typhoid fever is almost always treated with antibiotics.25,59 This disease is more common to developing countries, while NTS is common to both developed and developing countries. NTS infections present with gastroenteritis and diarrhea.59 NTS is usually self-limiting, and antibiotics are typically avoided since they may induce prolonged shedding of infectious NTS after treatment.59 Inappropriate antibiotic use is a key driver of AMR in Salmonella, and resistant infections often worsen clinical outcomes.1,25,60 Salmonella is notorious for its genomic islands, including Salmonella genomic island 1 (SGI1) carrying the ACSSuT region, encoding MDR (Table 1).26 More recently, some NTS serovars have evolved a novel genomic island encoding streptomycin and azithromycin resistance, which is concerning given that azithromycin is a second-line agent.61 Since Salmonella continues to be a major source of enteric infection, it will likely continue to present severe human health burdens without serious interventions.
Vibrio cholerae
Vibrio cholerae is the causative agent of cholera, a diarrheal disease attributed to upwards of 120,000 deaths per year.62 As with many diarrheal diseases, antibiotics are only required in the case of severe infections. First-line therapy typically includes doxycycline, while azithromycin, ciprofloxacin, and ceftriaxone are alternative therapies.50 Vibrio achieves extensive antibiotic resistance through its natural competency, allowing it to take up mobile genetic elements (MGEs) including plasmids, integrons, conjugative transposons, and SXT elements (Table 1).30,63,64 SXT elements, named for early observations of their conferred resistance to sulfamethoxazole and trimethoprim, are a type of integrative and conjugative element (ICE) that can confer additional resistance to agents such as streptomycin, nalidixic acid, and tetracycline.29 Similar to other diarrheal pathogens, V. cholerae acquires QRDR mutations that result in fluoroquinolone resistance, and can acquire diverse classes of efflux pumps conferring resistance to agents such as erythromycin, penicillins, novobiocin, and polymyxin B (Table 1).28
Cholera outbreaks have recently arisen in many developing countries, with the largest recorded cholera outbreak occurring in Yemen.65 The continual and largely preventable cholera epidemics have been considered “the world’s worst humanitarian crisis” by the United Nations.65 Due to the aggressively large impact on global human health, diverse resistance mechanisms against frontline agents, and the potential for further spread of resistant infections, V. cholerae is one of the highest priority enteric bacterial pathogens.
Clostridioides (Clostridium) difficile
Clostridioides difficile is a leading source of hospital-acquired enteric infection, causing nearly a quarter million infections and over 12,000 deaths per year in the United States alone.1 Although we have classified C. difficile as an invasive enteropathogen, it can asymptomatically colonize the human gut.13 C. difficile infection (CDI) likely occurs because of a loss of host colonization resistance, due to risk factors and co-morbidities such as antibiotic exposure or chemotherapy.66, 45 C. difficile is transmitted and ingested as a metabolically inactive spore. The metabolic environment of a dysbiotic GI tract is thought to facilitate germination of spores, leading to the development of CDI.67 The capacity to exist in various metabolic states, including dormant spores and withinbiofilms, is thought to contribute to its innate resistance to a number of antibiotics and sterilizing agents.31 In a clinical setting, some of the most virulent C. difficile ribotypes are also the most phenotypically drug-resistant. The spectrum of virulence in C. difficile is enhanced by a mobile genome, where 11% of the core genome of C. difficile is made up of MGEs.68,69 These MGEs are primarily represented by conjugative transposons, which are known to harbor MDR (Table 1).31 Transposon-independent resistance to vancomycin, rifampin and others has also been documented.68 Fidaxomicin is a more recently approved treatment option and resistance to date is rare. However, resistance has already been observed through point mutations in rpoB, the β subunit of the RNA polymerase target.33,34 Given that susceptibility testing for anaerobes such as C. difficile is not standard practice in the clinic, the extent of AMR in circulating C. difficile strains may be underestimated.
Breaking bad: the pathobionts
Pathogenic strains evolving from commensal species are known as “pathobionts.”70 Pathobionts are increasingly recognized as a significant source of infection and a key reservoir of AMR. This heightened prominence of pathobionts in today’s society can be largely attributed to advances in modern medicine over the last century. Although the human lifespan is longer than ever, as infectious disease has become less of a threat, extensive antibiotic use within people and the environment (Figure 1b) as well as a substantial rise in vulnerable populations has accommodated the rise of the pathobionts.6
Bacteroides fragilis group (BFG) species
The Bacteroides and Parabacteroides species within the BFG group include some of the most well-characterized commensal GI species, but are also the most commonly isolated organisms in anaerobic extraintestinal infections and increasingly reported to harbor AMR.36 Although the most ubiquitous resistance elements in BFG confer resistance to classes such as tetracyclines and macrolides, resistance to clinically useful agents such as β-lactams, carbapenems, and metronidazole is emerging in the United States and Europe.35,71 Resistance to all three treatment options can be achieved through “activation” of otherwise silent ARGs by insertion sequences.35 Conjugative transposons, most notably CTnDOT, have been well described among BFG species and commonly confer resistance elements against tetracyclines and erythromycin (Table 1).36 Clindamycin resistance has also steadily risen among BFG and is associated with acquisition of erythromycin resistance methylase (erm) genes that originate in gram-positive species.36 AMR only further exacerbates the morbidity and mortality rates associated with anaerobic infection, and further research into resistance mechanisms and prevention measures are desperately needed. Notably, susceptibility testing of anaerobes is not routinely performed in the clinic despite these emerging issues. Future efforts to tailor antibiotic stewardship should include emphasis on novel diagnostics for resistance in BFG and other anaerobes.49
Enterococci
E. faecalis and E. faecium are dominant causes of gram-positive nosocomial infections worldwide.72 Although Enterococci are well-established commensal species in the GI tract, Enterococci also cause extraintestinal infections, including endocarditis and sepsis (Figure 1b).72 Recent epidemics of vancomycin-resistant enterococci (VRE) have caused distinct clinical challenges in finding effective antibiotic regimens. After antibiotic use, enterococcal overgrowth in the GI tract creates an important reservoir for AMR development.72 Vancomycin resistance is acquired through the presence of vanA and vanB operons, which encode inducible synthesis of cell-wall modifications that reduce interactions between vancomycin and the cell wall.73 Additionally, these resistance loci often exist in a plasmidic element, or are associated with a transposable element, increasing the epidemiological threat of vancomycin resistance.74 Daptomycin and linezolid, representing newer antibiotic agents, are key treatment options for VRE infection, but resistance mechanisms are apparent for both agents (Table 1).39,40 VRE is clearly a large public health threat, especially in nosocomial settings, and preventative strategies should be pursued to mitigate the rise of drug resistance in enterococci.
Escherichia coli
Like other commensals, one of the most well-defined benefits of Escherichia coli to the human host is its ability to inhibit colonization by exogenous gut pathogens.75 In addition to the basal level of AMR that E. coli may harbor as a ‘commensal,’76 certain E. coli strains are highly pathogenic and form at least six pathotypes capable of causing both GI and extra-intestinal disease.4 The specific course of antibiotic therapy for E. coli infection is often guided by pathotype and/or strain type.50 For example, in the case of Shiga toxin-producing E. coli, antibiotics are frequently avoided as they can exacerbate associated morbidities. In addition, among Enterobacteriaceae bacteria such as Escherichia, Salmonella, Shigella, Enterobacter, and Klebsiella, AMR elements of many classes are shared between these genera.44,77-79 Carbapenem-resistant Enterobacteriaceae (CRE) have been listed as an “urgent threat” by the CDC, while ESBL-producing Enterobacteriaceae have been categorized as a “serious threat”.1 The supposed agents of “last resort” include carbapenems and colistin, but as expected, resistance has arisen for these antibiotics over the last decade. Various β-lactamases and carbapenemases are increasingly documented among E. coli isolates (Table 1).41,42 mcr is an emerging ARG against colistin in Enterobacteriaceae, and at least nine mcr homologs (mcr-1-9) have been identified (Table 1).45 This gene encodes a phosphatidylethanolamine transferase that transfers a phosphatidylethanolamine molecule to lipid A within the gram-negative cell membrane, thus making the bacteria less susceptible to the action of colistin.45 Although E. coli is a proven commensal occupant of the human GI tract and a genetic workhorse in laboratory settings, it is responsible for highly-concerning MDR profiles worldwide and can represent a true “superbug.” Since resistance to virtually every clinically utilized antibiotic can be rapidly disseminated among pathogenic Enterobacteriaceae, it is critical to pursue preventative and alternative measures to treat these infections.
Part II. Questionable arms deals: the evolution and acquisition of drug resistance
Commensal gut microbes and pathogens constantly evolve under the pressures of antibiotic exposure, whether in the gut or in an environmental reservoir. Depending on the bacteria’s ancestral lineage and the extent of allelopathy occurring in the microbe’s previous habitat, some bacteria are more intrinsically antibiotic-resistant.7,80 Here in Part II, we distill the diverse landscape of bacterial AMR mechanisms into a spectrum of antibiotic susceptibility, whereby a number of diverse pathways can lead to less susceptible organisms (Figure 2).
Figure 2.
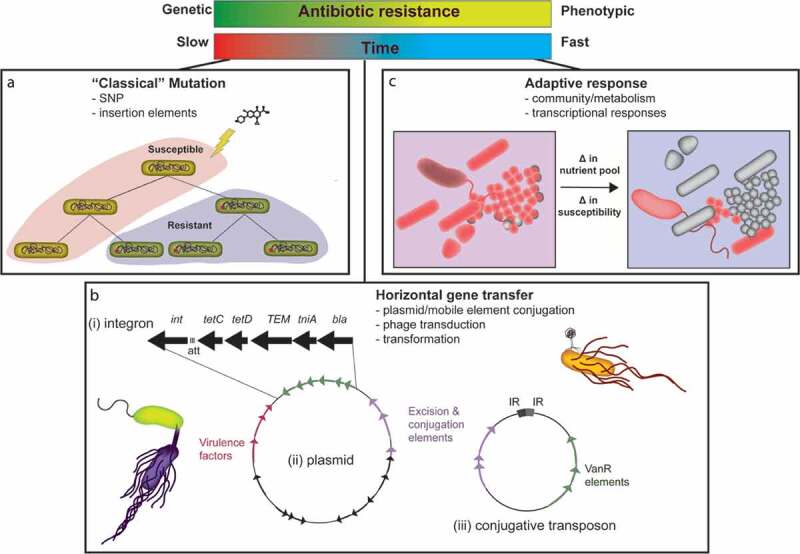
Manifestations of drug resistance. Bacteria become less susceptible to antibiotics through multiple mechanisms, from genetic acquisition of resistance to phenotypic responses to antibiotics. These mechanisms fall into three categories: classic mutation (a), horizontal gene transfer (b), and adaptive response (c). (a) Mutation via genome replication or intragenomic rearrangement can result in the acquisition of resistance. This process requires a series of bacterial generations to cause selection in the population for the inheritance of the resistance mutation. (b) Horizontal transfer of genetic material through multiple mechanisms in enteric pathogens, mainly bacterial conjugation, results in the more rapid acquisition of resistance elements. Mobile genetic elements such as integrons (i), plasmids (ii), or transposons (iii) can be transferred by conjugation. (c) An enteric pathogen’s susceptibility to antibiotics can be determined or altered based on the microbial community. In this example, a change in nutrient pools within a polymicrobial community can prompt a change in the inherent susceptibility of certain organisms to a given antibiotic. Often these susceptibility changes are mediated by changes in bacterial metabolism.
At one end of the spectrum are “classical” resistance mechanisms, including spontaneous mutations (Figure 2a) that modify target enzymes, alter transcription of select genes, or bypass antibiotic activity. In addition to nonsynonymous point mutations, these spontaneous mutations can also come in the form of insertion elements.81 Such evolved changes are inherited by the daughter cells through vertical transfer in subsequent generations.
Moving along the spectrum (Figure 2b), HGT is a phenomenon encouraged by the density of microbial communities both in the gut and in environmental reservoirs. We discuss movement of genetic elements in invasive enteropathogens and pathobionts through distinct examples of pathogen evolution (Figure 2b). HGT is positioned somewhere in the middle of the spectrum whereby bacteria acquire and shed genetic traits encoding resistance, often resulting in AMR acquisition more rapidly than spontaneous mutations.47,82
At the very end of the spectrum, bacteria are primed to respond to antibiotics through phenotypic variation, fluxes in bacterial metabolic state, programmed responses to the antibiotic/environment, or a systems-level effect of the bacterial community in the environment (Figure 2c).83 While these phenotypic states may not result in outright resistance, they provoke tolerance in the bacterial population and allow time for the organisms to acquire extensive AMR through other mechanisms.84 Genetic mobilization and adaptive strategies that result in clinically significant AMR are further described below.
Major horizontal gene transfer mechanisms: pathogens sample their reservoir
Bacterial HGT occurs by conjugation, transformation, and phage transduction (Figure 2b). Conjugation is the most well-studied mechanism of HGT and likely the major contributor to AMR in enteric pathogens.47 Conjugation requires secretion machinery, either encoded on a plasmid or within the chromosome, to inject genetic material directly into a neighboring cell.47 The types of genetic material that undergo conjugal transfer are expanding and evolving. Conjugative plasmids, conjugative transposons, and integrative and conjugative elements (ICEs), often integrons, represent defined genetic units that can be transferred by conjugation.85 While these terms imply distinct DNA entities transferred by a single defined mechanism, the mechanisms of bacterial genetic mobility exhibit further complexity in that MGEs may contain other MGEs. (Figure 2b, panels i,iii).85 Conjugative plasmids contain elements mediating plasmid replication and transfer, and can also encode additional adaptive elements such as ARGs or virulence factors (Figure 2b, panel ii).86 Integrons contain an integrase gene and an att recombination site.87 Conjugative transposons contain sequence elements to excise, transfer, and integrate the entire ‘jumping’ fragment into a new genetic location.88 These elements can be as large as 65 kb, in the case of CTnDOT, a frequently transferred conjugative transposon carrying AMR in Bacteriodes.36 Given that enteric pathogens are under immense pressure to adapt to the GI environment and associated antibiotic exposures, new combinations of MGEs continue to emerge, as discussed below.
Rather than acquire DNA through conjugation, bacterial species can also acquire exogenous DNA through transformation, which is the direct uptake of DNA from the surrounding environment that has been excreted or released from lysed cells. It is well-established that bacterial competence is heavily dependent on the environment, which often conditions an organism’s competency.89 In the case of enteric pathogens, genera such as Campylobacter and Vibrio possess competence systems and can take up exogenous DNA that may confer resistance.89,90 While there is no evidence of transformation occurring in vivo, many of these pathogens are capable and poised to take up exogenous DNA in their environmental reservoirs.
A final and underappreciated mechanism of HGT is phage transduction, which has an emerging role in shaping the fitness and resistance of enteric pathogens. Phages are DNA or RNA viruses encapsulated by a protein coat or capsid, and replicate within a bacterial cell, possibly incorporating their genomes into the bacterial chromosome.91 There is controversial evidence of phage transduction of resistance in vivo and, additionally, multiple lines of evidence indicating that phages from multiple environments are circulating ARGs.91–93 Compelling and recent in vivo studies involving head-to-head competition of two E. coli strains in a mouse gut demonstrated that phage-mediated genetic exchange is responsible for adaptation of an invading E. coli species, underlining the likelihood of phage transduction as a mechanism of AMR spread.94 The proportion of mobilized AMR due to phage transduction in human enteric disease remains an open question.
The mobilome of enteric pathogens: the ubiquity of mobility
Three concerning phenomena regarding MGEs are their mobility between environments and humans, their mobility across surprisingly unrelated bacterial taxa, and the potential for permanent integration of MGEs into the genome.2 These movements are captured in a growing appreciation for the “mobilome”, the group of all genetic elements that move within a chromosome, between chromosomes, and often, through plasmids. AMR acquisition via sampling of this mobilome often occurs in environmental niches that are external to the human host (Figure 1a). Campylobacter is found in multiple animal hosts and environmental niches, and thus has the opportunity to sample multiple ARG reservoirs. Remarkably, C. jejuni and C. coli can be isolated from livestock with phenotypic resistance profiles identical to those found in the clinic, especially in the case of fluoroquinolones.16,95 These ARGs appear to cluster on genomic islands or on circulating plasmids, indicating the frequent exchange of genetic material across environments or bacteria.95,96 In a subsample of E. coli isolates from both humans and poultry, a novel incompatibility group of plasmids has been discovered carrying a blaCMY mobile element.97 In addition, multiple classes of integrons carrying AMR have been found in both commensal E. coli and S. enterica from ruminants.98,99 Future efforts should better characterize the spatiotemporal dynamics of these and other environmental MGEs as it relates to both human disease and sources of AMR in our agricultural and waste practices.
An alarming feature of HGT is that it can occur between a range of bacteria across species and even phylogenies, especially via conjugative elements. Perhaps the most appreciated exchange of resistance occurs within the Enterobacteriaceae. Within this group of bacteria, containing Salmonella, Shigella, and Escherichia, as many as 28 different plasmid types are circulating.77 Findings from a mouse model have recapitulated this interspecies transfer: in the mouse gut, a conjugative plasmid could move between S. enterica and a commensal E. coli species.100 Sequence-based analyses of ARGs found in certain pathogens indicates that elements may have been acquired from distant bacterial genera. For example, macrolide resistance elements, including erm genes, were identified in a number of animal-derived isolates of Campylobacter. Their closest gene homologs were found to originate from gram-positive genera.101 As previously described, enterococcal species can exchange van operons, encoding vancomycin resistance, via conjugative plasmids.74,102 Most evidence indicates that this conjugation occurs frequently within the genus, but there is in vitro evidence that Enterococcus spp. can conjugate with distantly related gut commensals such as Lactococcus spp. or Bifidobacterium spp.103 In the gram-positive pathogens, there is strong evidence that a Tn5397 conjugative transposon, encoding resistance to tetracycline, can move between C. difficile and E. faecalis.104 The extensive interspecies exchange of genetic information highlights the importance of systems-level approaches to identify patterns of ARG movement across diverse bacterial species.
The worrying outcome of the movement of such plasmids is not only the acquisition of resistance, but the potential for permanent deposition of mobile resistance elements into the pathogen’s chromosome. Perhaps one of the most notorious examples of this comes from ESBL plasmids, a family of conjugative plasmids responsible for MDR against multiple classes of β-lactams.105 β-lactamases of plasmid origin now appear to be chromosomally encoded in Salmonella106 and E. coli isolates (Figure 2b).42,78 In the gram-positive enteric pathogens, some C. difficile isolates have evidence of a ‘cryptic plasmid’ within their genome, an extrachromosomal piece of DNA that may be the result of a recombination event between a phage and plasmid.69,107 This genomic mobility within bacterial populations underlines the need to broaden our perspective on the factors that encourage the transition of AMR elements from the mobilome to the genome.
Adaptive resistance: physiology engenders resistance
Phenotypic tolerance, or decreased susceptibility to antibiotics occurring independently of classical resistance mutations or mobilome-mediated acquisition of ARGs, can occur because of an innate cellular property or regulatory circuit present in the organism. Even in monoclonal in vitro studies, heterogeneity within a bacterial population, generated from stochastic properties such as permeability and gene expression, can lead to antibiotic tolerance, or decreased susceptibility in a subpopulation of the culture.108–110 Phase variation, wherein bacteria reversibly vary phenotypic properties within a population, can also reduce susceptibility.111
The addition of antibiotics or other stressors to a population results in bacterial adaptation which may be mediated through multiple forms of regulation, leading to antibiotic resistance and/or tolerance. For example, in the E. cloacae complex, a response-regulator complex senses change in cation concentrations and activates transcription of lipid A modification enzymes, causing a temporary increase in resistance to colistin.112 In Enterobacteriaceae, transcription of efflux pumps is often controlled by cellular sensing of antibiotics.113 In gram-positive bacteria, inducible macrolide resistance is regulated transcriptionally and post-transcriptionally, often upregulating programs to protect the ribosome or pump out the macrolide.114
The state of the bacterial population itself can prompt antibiotic tolerance and reduced susceptibility. In some distinct multicellular communities, such as bacterial biofilms, cells physically organize and form extracellular protective structures in a manner that promotes resistance for a subpopulation of cells or in the total population.115 In the case of Enterococci, they appear to form microcolonies in the gut which are biofilm-like communities. Within these communities, it is hypothesized that conjugative transfer of plasmids is greatly increased, possibly amplifying the resistance of the community.116 In Campylobacter, natural competency seems to be amplified in biofilms, facilitating the uptake of ARGs.56
More recently, a non-canonical form of altered drug susceptibility called heteroresistance has been characterized, wherein a subpopulation of a monoisolate culture survives and replicates in the presence of certain antibiotics while the remaining population is killed off. The resistance phenotype observed in heteroresistant cells is characteristically transient and reversible after removal of the antibiotic stressor, underlining an intimate link between heteroresistance and cellular physiology.117 In the case of E. cloacae, heteroresistance appears dependent on the presence of phoQ, a broad regulator of a number of cellular processes, including colistin resistance genes.118,119 C. difficile has also been described to have heteroresistance to metronidazole through an unknown mechanism.120 This phenomenon has been studied in other organisms and is not driven by one regulatory mechanism but rather, a number of diverse, stochastic cellular processes that are organism- and antibiotic-specific. Future efforts in continuing to understand, diagnose, and treat heteroresistance are greatly warranted. Furthermore, understanding the effect of the complex environment of the human gut on both innate drug susceptibility and phenotypic tolerance of enteric pathogens will be key to the development of novel therapeutic strategies.
Part III. The way forward: harnessing next-generation methods to monitor AMR and effectively treat infections
Despite our recent foray into the dreaded post-antibiotic era,1 we are also entering a time of unprecedented technological advancement in biomedicine, genetics, and bioinformatics. These tools provide immense promise for improved AMR surveillance and diagnostics, as well as effective alternative strategies to treat and prevent AMR enteric infections. High-throughput sequencing (HTS) of both isolated species and metagenomic samples can be coupled with bioinformatics tools to predict and catalog ARGs as well as associated MGEs.12,121 In addition, we discuss alternative treatment options that circumvent traditional antibiotic therapies, which have encouraged resistance in the past.
Predicting antibiotic resistance
Next-generation sequencing-based methods have increased capacities in the realm of pathogen surveillance and AMR profile characterization. These techniques can be performed on both isolate collections derived from the same species, or metagenomic samples representing mixed microbial communities, such as stool, soil, or wastewater. The promising utility and application of these genomics-based methods for the purposes of both pathogen surveillance and studying AMR has been thoroughly reviewed,12,121 and we present examples of how these methods have already provided incredibly useful insights into AMR surveillance, spread, and persistence in enteric bacteria. Furthermore, the potential for translating genomics techniques to clinical use is also discussed.
Comparative genomics of clinical isolates for MDRO surveillance
In recent years, sequencing-based methods have proven to be an invaluable tool for surveillance of enteric pathogen isolates and their corresponding resistomes. For instance, in the United States, the National Antimicrobial Resistance Monitoring System (NARMS) has used whole-genome sequencing (WGS) for the surveillance of Campylobacter53 and NTS.122 For both studies, hundreds of isolates were analyzed and showed a high degree of correlation between AMR genotype and culture-based resistance phenotypes. The genotype-phenotype correlation of Campylobacter ranged from 68–100% for identified ARGs, and based on one house-keeping gene, species-level resolution of the Campylobacter species could be achieved.53 The genotype-phenotype correlation was over 99% positive for NTS and revealed the first instance of ESBL carriage of Salmonella collected from retail meats in the United States.122
Another example of using comparative genomics to study isolate collections is a recent exploration of linezolid-resistant E. faecium isolates from the United States and Pakistan.123 Forty-nine draft genomes were constructed through Illumina WGS and 52 E. faecium genomes were obtained from public databases. The genetic mechanisms of linezolid resistance were distinct between the two geographic sites, with isolates from the USA having 23S rRNA mutations and Pakistan isolates having acquired ARGs including efflux pump genes and the chloramphenicol-florfenicol resistance (cfr) methyltransferase.123 Furthermore, MGEs associated with transposases and phage were proximal to the efflux gene optrA, suggesting HGT as a potential means of disseminating linezolid resistance. Disparate genetic mechanisms can therefore lead to similar resistance profiles among isolate cohorts. Clearly, WGS can uncover a comprehensive view of the multiple paths that lead to resistance and how they are mobilized.
Metagenomics-based methods to functionally characterize resistomes in the human gut
In addition to isolate collections, WGS-based methods are undoubtedly useful for phylogenetic and resistome analyses of metagenomic samples. In the case of functional metagenomics, HTS analyses of genomes can be coupled with culture-based approaches to infer the functional resistome of a metagenomic sample.12 This is especially useful in the case of novel resistance elements that are previously unannotated and likely not captured through standard bioinformatics-based approaches.
Neonates represent an especially vulnerable patient population, and therefore the characterization of pathogens and AMR within the neonatal gut microbiome is of intense interest. A recent study employed both shotgun sequencing metagenomic and functional metagenomics to characterize the gut microbiome and resistome of preterm infants who had spent their first few months of life in the neonatal intensive care unit.124 Analyses of fecal samples collected longitudinally during the first two years of life revealed that MDROs, including Enterobacteriaceae, are enriched after antibiotic exposure and persistently colonize the gut microbiota of neonates. Sequencing of metagenomic fragments identified as phenotypic resistance determinants through functional metagenomics revealed that the median identity of ARGs to a commonly used AMR database was only 32%.124 This highlights the discrepancy between currently available annotated resistance elements and the plethora of functional, circulating ARGs that are not identifiable by conventional genetic methods.
Intelligent methods to characterize mobilization and activation of AMR elements
As discussed in Part II, a key contribution to the true extent of drug resistance is not only the presence of ARGs, but also the ability to mobilize genetic elements in a manner that either transfers or activates their function. Computational tools to study these phenomena are increasingly being developed and improved; we describe a few examples of available tools in this section.125 Phasefinder is a recently developed tool to identify DNA-inversion events that result in phase variation, and has been used to identify invertible promoters upstream of ARGs in Bacteriodes.111 Site-specific, integrative MGEs can be detected using MGEfinder, which is especially suited for identifying transposable elements and can capture integration sites with apparent effects on AMR in both in vitro adaptive evolution experiments and in clinical isolates.126 HGT (or lateral gene transfer) events can be revealed with tools such as WAAFLE (http://huttenhower.sph.harvard.edu/waafle) or DarkHorse,127 and options are available for both isolates and metagenomes.125 Hi-C is a novel experimental approach that provides resolution on the association of plasmids with specific bacterial species, which is often lost during standard HTS practices.128 Long-read sequencing is also an emerging and increasingly cost-effective strategy to achieve better resolution on plasmid vs. chromosomal elements, and can assist with covering genomic sites that do not always attain adequate coverage such as repeated DNA elements at insertion sequence sites found upstream of the cfiA carbapenemase in BFG.129 These and other emerging technologies will likely prove essential in identifying elements beyond strict ARG sequences that can have a substantial impact on the extent of AMR in genomes or metagenomes.
Harnessing genomics for use in clinical microbiology
Genomics technology could be a beneficial tool for rapid and comprehensive resistance detection in clinical microbiology labs, yet certain hurdles remain before clinical implementation of these technologies. Current workflows can be time and cost-prohibitive, given the demand within a clinical setting. A recent study highlighted that genomics pipelines are highly variable between labs, with discordant results from the same sample depending on the ARG database and/or pipeline used, and further discordance with phenotypic resistance.130 Marrying WGS with phenotypic resistance is additionally complicated by the observation that phenotypic resistance can occur independently of the simple presence or mutation of one chromosomal ARG. Furthermore, functional metagenomics studies have revealed that a substantial fraction of genes that encode phenotypic resistance in clinical samples are not captured in current ARG repositories.124,131 Pinpointing ARGs to the causative pathogen in community samples can also be obscured by the fact that commensal bacteria often harbor innate or acquired ARGs.
Despite these obstacles, interest in the field and continual advances in genomics technology will likely spur this resource into routine clinical use in the near future. ARG repositories such as the Comprehensive Antibiotic Resistance Depository (CARD)132 and Resfinder133 are routinely updated and improved, and are therefore increasingly more reliable for comprehensive and accurate resistance detection. Advances in long-read sequencing technology, which can provide real-time diagnostic information, are enabling rapid, species-level resolution of ARG-harboring pathogens from metagenomics samples.134 A recent review suggests a simplified genomics pipeline for routine clinical use.48 Genomics technology has enormous potential for not only improving resistance detection in the clinic, but also for species identification, tracking virulence, and epidemiology.
Treating the untreatable: alternative strategies for preventing and managing AMR enteric infections
It is commonly accepted that standard antibiotic therapies, once considered “miracle drugs,” are inadequate long-term solutions for drug-resistant infections.1 Resistance has been observed to virtually every clinically utilized agent, and even therapies with novel targets deployed in recent years have been rapidly met with resistance.3 The quandary of rapid resistance foiling the use of even the newest antimicrobial agents demands that alternative solutions be developed to manage the burden of enteric bacterial infections.
Vaccines
Vaccines are a promising strategy to mitigate infections without encouraging the spread of AMR. Alongside antibiotics, vaccines are one of the most life-saving innovations of the 20th century. Vaccines can have long-term protective effects and harness the host immune system to clear pathogens before they can get a foothold on pathogenesis. Vaccines have already eradicated smallpox and nearly abolished polio infection.135 Licensed oral vaccines already exist for the two major enteric bacterial pathogens Salmonella typhi and Vibrio cholerae. Current research efforts toward Shigella, enterotoxigenic E. coli (ETEC), Campylobacter, and S. paratyphi are ongoing.136 Developing countries with endemic sources of enteric infection would stand to receive the most benefits from new vaccines.
Empowering the commensals
An emerging view is that gut dysbiosis, or a shift in the phylogenetic composition of the gut microbiome away from community compositions typically considered “healthy,” contributes to the success of enteric pathogens. This has inspired the development of modern therapeutic strategies to restore healthy microbiomes and therefore “empower” commensal populations in the gut to prevent pathogen colonization and proliferation. The healthy gut microbiota possesses metabolic functions that impair pathogen proliferation through direct nutrient competition or other indirect mechanisms.137,138 The path toward increased use of alternative therapies, such as probiotics and fecal microbiota transfer (FMT), can be forged by a deeper understanding of commensal-pathogen interactions and how AMR traits affect these interactions.139
Various efforts to harness beneficial bacteria to ameliorate enteric disease have been put forth in recent years. Bifidobacterium are a well-established component of the commensal gut microbiome and may provide protection against pathogen colonization early in life.140 B. infantis was given to a group of healthy term infants, resulting in a 90% decrease in ARGs in comparison to control infants.141 However, it is likely that the therapeutic solution to many states of dysbiosis will not be one bacteria, but rather, a community of bacteria. FMT may provide some efficacy in the case of recurrent CDI, as it may restore colonization resistance.142 Yet, a number of recent studies indicate that this procedure may have adverse outcomes due to transfer of possibly pathogenic, drug-resistant organisms.143 Rigorous screening of donor stools to avoid transfer of MDROs as well as efforts toward formulating a more defined “synthetic microbiota” may offer some hope toward avoiding MDRO transfer. In fact, for CDI, a number of synthetic cocktails of bacterial taxa have been discovered that appear to inhibit C. difficile proliferation in mouse models of disease.144,145 Further studies into the constituents of colonization resistance in a healthy human microbiota that provide protection against AMR enteric disease would spur progress toward this clinically promising approach.
Phage therapy
In addition to transporting genetic material between cells, phages have also exhibited potent antimicrobial activity against clinically important bacterial species.13 Phages can confer direct lytic effects on the target species or serve as payload carriers for antibacterial or antivirulence targets. Phages have high species specificity and have shown superior biofilm penetration to standard antibiotics.146 Although phage therapy is not currently an authorized therapy in Western countries, it has already shown promise as an experimental treatment in compassionate use programs.13 Phage therapy could serve as an ideal treatment for enteric pathogens, as phage treatment is not expected to encourage AMR and its specificity avoids damaging neighboring commensals.
Unusual targets and new combination therapies
Promising new treatment strategies in the preclinical pipeline include inhibitors against entirely new targets such as virulence factors and bacterial metabolic pathways that are absent in humans yet crucial for microbial survival, and targets that are highly implicated in adaptive resistance mechanisms such as persistence, tolerance, and heteroresistance. A recent review highlighted antimicrobial peptides and inhibitors of LpxC, which inhibit the first committed step in the biosynthesis of lipid A, as two inhibitor groups with broad interest in the field. Both inhibitor sets are especially promising for the treatment of gram-negative bacteria.13 Furthermore, antivirulence targets such as those that have well-established roles in persistence are of interest. New inhibitor series targeting the caseinolytic P protease system, an essential quality control process in many bacterial species such as VRE, can cause unchecked activation of the protease and nonspecific protein degradation, leading to cell death. These drugs seem especially effective in killing of stationary phase bacteria and persisters.147
The field has also developed a focus on other antibacterial strategies such as repurposing existing drugs, developing potentiators which enhance the activity of standard antibiotic therapies, or developing immunomodulators that harness the host immune system against the threat of infection.13 As an example, immunogenic activity derived from cholera toxin-conjugated siderophores protected mice from Salmonella infection.148 Furthermore, improved GI localization of metronidazole by conjugation to reutericyclin from Lactobacillus improved outcomes in a hamster model of CDI.148,149 It is important to continue to improve targeted delivery mechanisms to the GI tract such that collateral damage to the human and microbiome are minimized. Finally, combinations of existing antimicrobials have yielded surprisingly effective activity against recalcitrant pathogens such as heteroresistant bacteria. CRE clinical isolates (Enterobacter, Escherichia, and Klebsiella) display resistance to carbapenems but are sensitized in the case of treatment with multiple antibiotics.117 Rather than producing iterative homologs of existing antibiotics that may perpetuate existing AMR mechanisms, these alternative approaches offer promising and potentially life-saving options to mitigate enteric AMR infections.
Concluding remarks
The definition of enteric pathogenesis has undergone considerable restructuring in the last several decades. A new appreciation for the complex relationship between the host microbiome, enteric pathogens, and AMR has shaped this understanding. AMR acquisition in the context of enteric disease appears to be a “systems-level” phenomenon in many cases, and is more complicated than the one gene, one bug explanation of AMR. Fortunately, the continual development of genomics-based approaches and bioinformatic tools is improving our understanding of what constitutes a healthy gut, an enteric infection, and drug resistance.12,48
Enteric bacterial pathogens will continue to exist, evolve, and present medical challenges to the human population. AMR is clearly an ever-present, innate, and ubiquitous feature of these pathogens that we must continue to strive toward understanding in order to formulate effective prevention and treatment strategies.1,2 In the lineup of pathogens discussed herein, we highlighted invasive enteropathogens and pathobionts, both of which cause life-threatening disease and are often recalcitrant to recommended treatment options because of wide-spread AMR. Within these organisms, we defined unique pathogenesis mechanisms, reservoirs of AMR transmission, and gene mobilization strategies. Furthermore, we presented the application and promising potential of next-generation surveillance, treatment, and diagnostic measures.
Although we presented several cutting-edge approaches to understanding and ameliorating the burden of AMR in enteric bacteria, other approaches beyond the scope of this review could have tremendous impact on alleviating the health burdens attributable to enteric disease. This includes changes in global health policies that will increase access to enteric disease treatment and prevention.51,65 Better management of antibiotic exposure in the environment and in the prevention of disease would undoubtedly reduce the spread of AMR worldwide.3
Acknowledgments
We thank Eric C. Keen and Alaric W. D’Souza for thoughtful comments on this manuscript
Funding Statement
This work was supported in part by awards to G.D. through the National Institute of Allergy and Infectious Diseases (NIAID: https://www.niaid.nih.gov/), the Eunice Kennedy Shriver National Institute of Child Health & Human Development (NICHD: https://www.nichd.nih.gov/), and the National Center for Complementary and Integrative Health (NCCIH: https://nccih.nih.gov/) of the National Institutes of Health (NIH) under award numbers [R01AI123394, R01HD092414, and R01AT009741], respectively; the National Institute for Occupational Safety and Health (NIOSH: https://www.cdc.gov/niosh/index.htm) of the US Centers for Disease Control and Prevention (CDC) under award number [R01OH011578]; and the Congressionally Directed Medical Research Program (CDMRP: https://cdmrp.army.mil/prmrp/default) of the US Department of Defense (DOD) under award number [W81XWH1810225]. This work was additionally supported by an award to M.J.W. through the National Cancer Institute (NCI: https://cancer.gov) of the NIH under award number [T32 CA113275-12] and to S.R.S.F. through the National Institute of Child Health and Human Development (NICHD: https://www.nichd.nih.gov) of the NIH under award number [T32 HD004010]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
References
- 1.U.S. Department of Health and Human Services, Centers for Disease Control and Prevention . Antibiotic resistance threats in the United States, 2019. Atlanta (GA): CDC; 2019. [Google Scholar]
- 2.Holmes AH, Moore LS, Sundsfjord A, Steinbakk M, Regmi S, Karkey A, Guerin PJ, Piddock LJ.. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet. 2016;387(10014):176–21. doi: 10.1016/S0140-6736(15)00473-0. [DOI] [PubMed] [Google Scholar]
- 3.O’Neill J. Tackling drug-resistant infections globally: final report and recommendations. London: The Review on Antimicrobial Resistance; 2016. http://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf. [Google Scholar]
- 4.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2(2):123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 5.Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J. 2017;474(11):1823–1836. doi: 10.1042/BCJ20160510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aminov RI. A brief history of the antibiotic era: lessons learned and challenges for the future. Front Microbiol. 2010;1:134. doi: 10.3389/fmicb.2010.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wright GD, Poinar H. Antibiotic resistance is ancient: implications for drug discovery. Trends Microbiol. 2012;20(4):157–159. doi: 10.1016/j.tim.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Karst SM. The influence of commensal bacteria on infection with enteric viruses. Nat Rev Microbiol. 2016;14(4):197–204. doi: 10.1038/nrmicro.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotwal G, Cannon JL. Environmental persistence and transfer of enteric viruses. Curr Opin Virol. 2014;4:37–43. doi: 10.1016/j.coviro.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 10.Pramanik PK, Alam MN, Roy Chowdhury D, Chakraborti T. Drug resistance in protozoan parasites: an incessant wrestle for survival. J Glob Antimicrob Resist. 2019;18:1–11. doi: 10.1016/j.jgar.2019.01.023. [DOI] [PubMed] [Google Scholar]
- 11.Revie NM, Iyer KR, Robbins N, Cowen LE. Antifungal drug resistance: evolution, mechanisms and impact. Curr Opin Microbiol. 2018;45:70–76. doi: 10.1016/j.mib.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sukhum KV, Diorio-Toth L, Dantas G. Genomic and metagenomic approaches for predictive surveillance of emerging pathogens and antibiotic resistance. Clin Pharmacol Ther. 2019;106(3):512–524. doi: 10.1002/cpt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Theuretzbacher U, Outterson K, Engel A, Karlén A. The global preclinical antibacterial pipeline. Nat Rev Microbiol. 20. 20;18:275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang Y, Fang L, Xu C, Zhang Q. Antibiotic resistance trends and mechanisms in the foodborne pathogen, Campylobacter. Anim Health Res Rev. 2017;18(2):87–98. doi: 10.1017/S1466252317000135. [DOI] [PubMed] [Google Scholar]
- 15.Yao H, Shen Z, Wang Y, Deng F, Liu D, Naren G, Dai L, Su CC, Wang B, Wang S, et al. Emergence of a potent multidrug efflux pump variant that enhances campylobacter resistance to multiple antibiotics. MBio. 2016;7(5). doi: 10.1128/mBio.01543-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sproston EL, Wimalarathna HML, Sheppard SK. Trends in fluoroquinolone resistance in campylobacter. Microb Genom. 2018;4(8):e000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehtopolku M, Kotilainen P, Haanperä-Heikkinen M, Nakari UM, Hänninen ML, Huovinen P, Siitonen A, Eerola E, Jalava J, Hakanen AJ. Ribosomal mutations as the main cause of macrolide resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother. 2011;55(12):5939–5941. doi: 10.1128/AAC.00314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolinger H, Kathariou S. The current state of macrolide resistance in Campylobacter spp.: trends and impacts of resistance mechanisms. Appl Environ Microbiol. 2017;83(12). doi: 10.1128/AEM.00416-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen JC, Tagg KA, Joung YJ, Bennett C, Francois Watkins L, Eikmeier D, Folster JP. Report of erm(B)+ Campylobacter jejuni in the United States. Antimicrob Agents Chemother. 2018;62(6). doi: 10.1128/AAC.02615-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Zhang M, Deng F, Shen Z, Wu C, Zhang J, Zhang Q, Shen J. Emergence of multidrug-resistant Campylobacter species isolates with a horizontally acquired rRNA methylase. Antimicrob Agents Chemother. 2014;58(9):5405–5412. doi: 10.1128/AAC.03039-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ranjbar R, Farahani A. Shigella: antibiotic-resistance mechanisms and new horizons for treatment. Infect Drug Resist. 2019;12:3137–3167. doi: 10.2147/IDR.S219755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Z, Cao M, Zhou X, Li B, Zhang J. Epidemic characterization and molecular genotyping of Shigella flexneri isolated from calves with diarrhea in Northwest China. Antimicrob Resist Infect Control. 2017;6:92. doi: 10.1186/s13756-017-0252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu BT, Yang QE, Li L, Sun J, Liao XP, Fang LX, Yang SS, Deng H, Liu YH. Dissemination and characterization of plasmids carrying oqxAB-bla CTX-M genes in Escherichia coli isolates from food-producing animals. PLoS One. 2013;8(9):e73947. doi: 10.1371/journal.pone.0073947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao YS, Liu YY, Lo YC, Chiou CS. Azithromycin-nonsusceptible Shigella flexneri 3a in men who have sex with men, Taiwan, 2015-2016. Emerg Infect Dis. 2016;23(2):345–346. doi: 10.3201/eid2302.161260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crump JA, Sjölund-Karlsson M, Gordon MA, Parry CM, Epidemiology CP, Diagnosis L. Antimicrobial resistance, and antimicrobial management of invasive salmonella infections. Clin Microbiol Rev. 2015;28:901–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carraro N, Durand R, Rivard N, Anquetil C, Barrette C, Humbert M, Burrus V. Salmonella genomic island 1 (SGI1) reshapes the mating apparatus of IncC conjugative plasmids to promote self-propagation. PLoS Genet. 2017;13(3):e1006705. doi: 10.1371/journal.pgen.1006705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carretto E, Brovarone F, Nardini P, Russello G, Barbarini D, Pongolini S, Gagliotti C, Carattoli A, Sarti M. Detection of mcr-4 positive Salmonella enterica serovar typhimurium in clinical isolates of human origin, Italy, October to November 2016. Euro Surveill. 2018;23(2). doi: 10.2807/1560-7917.ES.2018.23.2.17-00821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morita Y, Li X-Z. Antimicrobial resistance and drug efflux pumps in vibrio and legionella. Efflux-mediated antimicrobial resistance in bacteria. In: Li XZ, Elkins C, Zgurskaya H, editors, Efflux-Mediated Antimicrobial Resistance in Bacteria. Adis, Cham. 2016;307–328. doi:10.1007/978-3-319-39658-3_12. [Google Scholar]
- 29.Spagnoletti M, Ceccarelli D, Rieux A, Fondi M, Taviani E, Fani R, Colombo MM, Colwell RR, Balloux F. Acquisition and evolution of SXT-R391 integrative conjugative elements in the seventh-pandemic Vibrio cholerae lineage. MBio. 2014;5(4). doi: 10.1128/mBio.01356-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitaoka M, Miyata ST, Unterweger D, Pukatzki S. Antibiotic resistance mechanisms of Vibrio cholerae. J Med Microbiol. 2011;60(Pt 4):397–407. doi: 10.1099/jmm.0.023051-0. [DOI] [PubMed] [Google Scholar]
- 31.Peng Z, Jin D, Kim HB, Stratton CW, Wu B, Tang YW, Sun X. Update on antimicrobial resistance in Clostridium difficile: resistance mechanisms and antimicrobial susceptibility testing. J Clin Microbiol. 2017;55(7):1998–2008. doi: 10.1128/JCM.02250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moura I, Monot M, Tani C, Spigaglia P, Barbanti F, Norais N, Dupuy B, Bouza E, Mastrantonio P. Multidisciplinary analysis of a nontoxigenic Clostridium difficile strain with stable resistance to metronidazole. Antimicrob Agents Chemother. 2014;58(8):4957–4960. doi: 10.1128/AAC.02350-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuehne SA, Dempster AW, Collery MM, Joshi N, Jowett J, Kelly ML, Cave R, Longshaw CM, Minton NP. Characterization of the impact of rpoB mutations on the in vitro and in vivo competitive fitness of Clostridium difficile and susceptibility to fidaxomicin. J Antimicrob Chemother. 2018;73(4):973–980. doi: 10.1093/jac/dkx486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwanbeck J, Riedel T, Laukien F, Schober I, Oehmig I, Zimmermann O, Overmann J, Groß U, Zautner AE, Bohne W. Characterization of a clinical Clostridioides difficile isolate with markedly reduced fidaxomicin susceptibility and a V1143D mutation in rpoB. J Antimicrob Chemother. 2019;74(1):6–10. doi: 10.1093/jac/dky375. [DOI] [PubMed] [Google Scholar]
- 35.Snydman DR, Jacobus NV, McDermott LA, Goldstein EJ, Harrell L, Jenkins SG, Newton D, Patel R, Hecht DW. Trends in antimicrobial resistance among bacteroides species and parabacteroides species in the United States from 2010-2012 with comparison to 2008-2009. Anaerobe. 2017;43:21–26. doi: 10.1016/j.anaerobe.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 36.Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev. 2007;20(4):593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pumbwe L, Chang A, Smith RL, Wexler HM. BmeRABC5 is a multidrug efflux system that can confer metronidazole resistance in Bacteroides fragilis. Microb Drug Resist. 2007;13(2):96–101. doi: 10.1089/mdr.2007.719. [DOI] [PubMed] [Google Scholar]
- 38.Zirakzadeh A, Patel R. Epidemiology and mechanisms of glycopeptide resistance in enterococci. Curr Opin Infect Dis. 2005;18(6):507–512. doi: 10.1097/01.qco.0000186849.54040.2a. [DOI] [PubMed] [Google Scholar]
- 39.Arias CA, Panesso D, McGrath DM, Qin X, Mojica MF, Miller C, Diaz L, Tran TT, Rincon S, Barbu EM, et al. Genetic basis for in vivo daptomycin resistance in enterococci. N Engl J Med. 2011;365(10):892–900. doi: 10.1056/NEJMoa1011138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diaz L, Kiratisin P, Mendes RE, Panesso D, Singh KV, Arias CA. Transferable plasmid-mediated resistance to linezolid due to cfr in a human clinical isolate of Enterococcus faecalis. Antimicrob Agents Chemother. 2012;56(7):3917–3922. doi: 10.1128/AAC.00419-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oberoi L, Singh N, Sharma P, Aggarwal A, ESBL MBL. Ampc β lactamases producing superbugs - havoc in the intensive care units of Punjab India. J Clin Diagn Res. 2013;7(1):70–73. doi: 10.7860/JCDR/2012/5016.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing enterobacteriaceae. Emerg Infect Dis. 2011;17(10):1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swick MC, Morgan-Linnell SK, Carlson KM, Zechiedrich L. Expression of multidrug efflux pump genes acrAB-tolC, mdfA, and norE in Escherichia coli clinical isolates as a function of fluoroquinolone and multidrug resistance. Antimicrob Agents Chemother. 2011;55(2):921–924. doi: 10.1128/AAC.00996-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skov RL, Monnet DL. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Surveill. 2016;21(9):30155. doi: 10.2807/1560-7917.ES.2016.21.9.30155. [DOI] [PubMed] [Google Scholar]
- 45.Carroll LM, Gaballa A, Guldimann C, Sullivan G, Henderson LO, Wiedmann M. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype typhimurium isolate. mBio. 2019;10(3). doi: 10.1128/mBio.00853-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Furuya-Kanamori L, Marquess J, Yakob L, Riley TV, Paterson DL, Foster NF, Huber CA, Clements AC. Asymptomatic Clostridium difficile colonization: epidemiology and clinical implications. BMC Infect Dis. 2015;15:516. doi: 10.1186/s12879-015-1258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun D. Pull in and push out: mechanisms of horizontal gene transfer in bacteria. Front Microbiol. 2018;9:2154. doi: 10.3389/fmicb.2018.02154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balloux F, Brønstad Brynildsrud O, van Dorp L, Shaw LP, Chen H, Harris KA, Wang H, Eldholm V. From theory to practice: translating whole-genome sequencing (WGS) into the clinic. Trends Microbiol. 2018;26(12):1035–1048. doi: 10.1016/j.tim.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cooley L, Teng J. Anaerobic resistance: should we be worried? Curr Opin Infect Dis. 2019;32(6):523–530. doi: 10.1097/QCO.0000000000000595. [DOI] [PubMed] [Google Scholar]
- 50.Shane AL, Mody RK, Crump JA, Tarr PI, Steiner TS, Kotloff K, Langley JM, Wanke C, Warren CA, Cheng AC, et al. 2017 Infectious diseases society of america clinical practice guidelines for the diagnosis and management of infectious diarrhea. Clin Infect Dis. 2017;65(12):e45–e80. doi: 10.1093/cid/cix669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collaborators GDD. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: a systematic analysis for the global burden of disease study 2016. Lancet Infect Dis. 2018;18(11):1211–1228. doi: 10.1016/S1473-3099(18)30362-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaakoush NO, Castaño-Rodríguez N, Mitchell HM, Man SM. Global epidemiology of campylobacter infection. Clin Microbiol Rev. 2015;28:687–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitehouse CA, Young S, Li C, Hsu CH, Martin G, Zhao S. Use of whole-genome sequencing for campylobacter surveillance from NARMS retail poultry in the United States in 2015. Food Microbiol. 2018;73:122–128. doi: 10.1016/j.fm.2018.01.018. [DOI] [PubMed] [Google Scholar]
- 54.CDC National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Human Isolates Surveillance Report for 2015 (Final Report); Atlanta (GA), 2018. [Google Scholar]
- 55.Grinnage-Pulley T, Mu Y, Dai L, Zhang Q. Dual repression of the multidrug Efflux pump CmeABC by CosR and CmeR in campylobacter jejuni. Front Microbiol. 2016;7:1097. doi: 10.3389/fmicb.2016.01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bae J, Oh E, Jeon B. Enhanced transmission of antibiotic resistance in Campylobacter jejuni biofilms by natural transformation. Antimicrob Agents Chemother. 2014;58(12):7573–7575. doi: 10.1128/AAC.04066-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baker KS, Dallman TJ, Ashton PM, Day M, Hughes G, Crook PD, Gilbart VL, Zittermann S, Allen VG, Howden BP, et al. Intercontinental dissemination of azithromycin-resistant shigellosis through sexual transmission: a cross-sectional study. Lancet Infect Dis. 2015;15(8):913–921. doi: 10.1016/S1473-3099(15)00002-X. [DOI] [PubMed] [Google Scholar]
- 58.Shiferaw B, Solghan S, Palmer A, Joyce K, Barzilay EJ, Krueger A, Cieslak P. Antimicrobial susceptibility patterns of Shigella isolates in foodborne diseases active surveillance network (FoodNet) sites, 2000-2010. Clin Infect Dis. 2012;54(Suppl 5):S458–63. doi: 10.1093/cid/cis230. [DOI] [PubMed] [Google Scholar]
- 59.Gal-Mor O, Boyle EC, Grassl GA. Same species, different diseases: how and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front Microbiol. 2014;5:391. doi: 10.3389/fmicb.2014.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Browne AJ, Kashef Hamadani BH, Kumaran EAP, Rao P, Longbottom J, Harriss E, Moore CE, Dunachie S, Basnyat B, Baker S, et al. Drug-resistant enteric fever worldwide, 1990 to 2018: a systematic review and meta-analysis. BMC Med. 2020;18(1):1. doi: 10.1186/s12916-019-1443-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cohen E, Davidovich M, Rokney A, Valinsky L, Rahav G, Gal-Mor O. Emergence of new variants of antibiotic resistance genomic islands among multidrug-resistant Salmonella enterica in poultry. Environ Microbiol. 2020;22(1):413–432. doi: 10.1111/1462-2920.14858. [DOI] [PubMed] [Google Scholar]
- 62.Lekshmi N, Joseph I, Ramamurthy T, Thomas S. Changing facades of Vibrio cholerae: an enigma in the epidemiology of cholera. Indian J Med Res. 2018;147(2):133–141. doi: 10.4103/ijmr.IJMR_280_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haycocks JRJ, Warren GZL, Walker LM, Chlebek JL, Dalia TN, Dalia AB, Grainger DC. The quorum sensing transcription factor AphA directly regulates natural competence in Vibrio cholerae. PLoS Genet. 2019;15(10):e1008362. doi: 10.1371/journal.pgen.1008362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Metzger LC, Blokesch M. Regulation of competence-mediated horizontal gene transfer in the natural habitat of Vibrio cholerae. Curr Opin Microbiol. 2016;30:1–7. doi: 10.1016/j.mib.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 65.Federspiel F, Ali M. The cholera outbreak in Yemen: lessons learned and way forward. BMC Public Health. 2018;18(1):1338. doi: 10.1186/s12889-018-6227-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hebbard AI, Slavin MA, Reed C, Teh BW, Thursky KA, Trubiano JA, Worth LJ. The epidemiology of Clostridium difficile infection in patients with cancer. Expert Rev Anti Infect Ther. 2016;14(11):1077–1085. doi: 10.1080/14787210.2016.1234376. [DOI] [PubMed] [Google Scholar]
- 67.Theriot CM, Bowman AA, Young VB. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere. 2016;1(1). doi: 10.1128/mSphere.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spigaglia P. Recent advances in the understanding of antibiotic resistance in Clostridium difficile infection. Ther Adv Infect Dis. 2016;3(1):23–42. doi: 10.1177/2049936115622891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeño-Tárraga AM, Wang H, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet. 2006;38(7):779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 70.Chow J, Tang H, Mazmanian SK. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol. 2011;23(4):473–480. doi: 10.1016/j.coi.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sóki J, Eitel Z, Urbán E, Nagy E, Infections ESGOA. Molecular analysis of the carbapenem and metronidazole resistance mechanisms of bacteroides strains reported in a Europe-wide antibiotic resistance survey. Int J Antimicrob Agents. 2013;41(2):122–125. doi: 10.1016/j.ijantimicag.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 72.Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012;10(4):266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Depardieu F, Bonora MG, Reynolds PE, Courvalin P. The vanG glycopeptide resistance operon from Enterococcus faecalis revisited. Mol Microbiol. 2003;50(3):931–948. doi: 10.1046/j.1365-2958.2003.03737.x. [DOI] [PubMed] [Google Scholar]
- 74.Palmer KL, Kos VN, Gilmore MS. Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr Opin Microbiol. 2010;13(5):632–639. doi: 10.1016/j.mib.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS, Conway T. Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157: H7 in the mouse intestine. PLoS One. 2013;8(1):e53957. doi: 10.1371/journal.pone.0053957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bailey JK, Pinyon JL, Anantham S, Hall RM. Commensal Escherichia coli of healthy humans: a reservoir for antibiotic-resistance determinants. J Med Microbiol. 2010;59(Pt 11):1331–1339. doi: 10.1099/jmm.0.022475-0. [DOI] [PubMed] [Google Scholar]
- 77.Rozwandowicz M, Brouwer MSM, Fischer J, Wagenaar JA, Gonzalez-Zorn B, Guerra B, Mevius DJ, Hordijk J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother. 2018;73(5):1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 78.Brolund A, Rajer F, Giske CG, Melefors O, Titelman E, Sandegren L. Dynamics of resistance plasmids in extended-spectrum-beta-lactamase-producing enterobacteriaceae during postinfection colonization. Antimicrob Agents Chemother. 2019;63(4). doi: 10.1128/AAC.02201-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mulvey MR, Mataseje LF, Robertson J, Nash JH, Boerlin P, Toye B, Irwin R, Melano RG. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16(3):289–290. doi: 10.1016/S1473-3099(16)00067-0. [DOI] [PubMed] [Google Scholar]
- 80.Cox G, Wright GD. Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int J Med Microbiol. 2013;303(6–7):287–292. doi: 10.1016/j.ijmm.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 81.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lopatkin AJ, Meredith HR, Srimani JK, Pfeiffer C, Durrett R, You L. Persistence and reversal of plasmid-mediated antibiotic resistance. Nat Commun. 2017;8(1):1689. doi: 10.1038/s41467-017-01532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Corona F, Martinez JL. Phenotypic resistance to antibiotics. Antibiotics (Basel). 2013;2(2):237–255. doi: 10.3390/antibiotics2020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lewis K, Shan Y. Why tolerance invites resistance. Science. 2017;355(6327):796. doi: 10.1126/science.aam7926. [DOI] [PubMed] [Google Scholar]
- 85.Wozniak RA, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. 2010;8(8):552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- 86.Norman A, Hansen LH, Sørensen SJ. Conjugative plasmids: vessels of the communal gene pool. Philos Trans R Soc Lond B Biol Sci. 2009;364(1527):2275–2289. doi: 10.1098/rstb.2009.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ghaly TM, Chow L, Asher AJ, Waldron LS, Gillings MR. Evolution of class 1 integrons: mobilization and dispersal via food-borne bacteria. PLoS One. 2017;12(6):e0179169. doi: 10.1371/journal.pone.0179169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rubio-Cosials A, Schulz EC, Lambertsen L, Smyshlyaev G, Rojas-Cordova C, Forslund K, Karaca E, Bebel A, Bork P, Barabas O. Transposase-DNA complex structures reveal mechanisms for conjugative transposition of antibiotic resistance. Cell. 2018;173(1):208–220.e20. doi: 10.1016/j.cell.2018.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Seitz P, Blokesch M. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol Rev. 2013;37(3):336–363. doi: 10.1111/j.1574-6976.2012.00353.x. [DOI] [PubMed] [Google Scholar]
- 90.Sitaraman R. Prokaryotic horizontal gene transfer within the human holobiont: ecological-evolutionary inferences, implications and possibilities. Microbiome. 2018;6(1):163. doi: 10.1186/s40168-018-0551-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Balcazar JL. Bacteriophages as vehicles for antibiotic resistance genes in the environment. PLoS Pathog. 2014;10(7):e1004219. doi: 10.1371/journal.ppat.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Enault F, Briet A, Bouteille L, Roux S, Sullivan MB, Petit MA. Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. Isme J. 2017;11(1):237–247. doi: 10.1038/ismej.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gomez-Gomez C, Blanco-Picazo P, Brown-Jaque M, Quiros P, Rodriguez-Rubio L, Cerda-Cuellar M, Muniesa M. Infectious phage particles packaging antibiotic resistance genes found in meat products and chicken feces. Sci Rep. 2019;9(1):13281. doi: 10.1038/s41598-019-49898-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Frazao N, Sousa A, Lassig M, Gordo I. Horizontal gene transfer overrides mutation in Escherichia coli colonizing the mammalian gut. Proc Natl Acad Sci U S A. 2019;116(36):17906–17915. doi: 10.1073/pnas.1906958116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mourkas E, Florez-Cuadrado D, Pascoe B, Calland JK, Bayliss SC, Mageiros L, Meric G, Hitchings MD, Quesada A, Porrero C, et al. Gene pool transmission of multidrug resistance among campylobacter from livestock, sewage and human disease. Environ Microbiol. 2019;21:4597–4613. doi: 10.1111/1462-2920.14760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen Y, Mukherjee S, Hoffmann M, Kotewicz ML, Young S, Abbott J, Luo Y, Davidson MK, Allard M, McDermott P, et al. Whole-genome sequencing of gentamicin-resistant Campylobacter coli isolated from U.S. retail meats reveals novel plasmid-mediated aminoglycoside resistance genes. Antimicrob Agents Chemother. 2013;57(11):5398–5405. doi: 10.1128/AAC.00669-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Seiffert SN, Carattoli A, Schwendener S, Collaud A, Endimiani A, Perreten V. Plasmids carrying blaCMY −2/4 in Escherichia coli from poultry, poultry meat, and humans belong to a novel IncK subgroup designated IncK2. Front Microbiol. 2017;8:407. doi: 10.3389/fmicb.2017.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ahmed HA, El-Hofy FI, Shafik SM, Abdelrahman MA, Elsaid GA. Characterization of virulence-associated genes, antimicrobial resistance genes, and Class 1 integrons in Salmonella enterica serovar typhimurium isolates from chicken meat and humans in Egypt. Foodborne Pathog Dis. 2016;13(6):281–288. doi: 10.1089/fpd.2015.2097. [DOI] [PubMed] [Google Scholar]
- 99.Belaynehe KM, Shin SW, Yoo HS. Interrelationship between tetracycline resistance determinants, phylogenetic group affiliation and carriage of class 1 integrons in commensal Escherichia coli isolates from cattle farms. BMC Vet Res. 2018;14(1):340. doi: 10.1186/s12917-018-1661-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stecher B, Denzler R, Maier L, Bernet F, Sanders MJ, Pickard DJ, Barthel M, Westendorf AM, Krogfelt KA, Walker AW, et al. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal enterobacteriaceae. Proc Natl Acad Sci U S A. 2012;109(4):1269–1274. doi: 10.1073/pnas.1113246109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Florez-Cuadrado D, Ugarte-Ruiz M, Meric G, Quesada A, Porrero MC, Pascoe B, Saez-Llorente JL, Orozco GL, Dominguez L, Sheppard SK. Genome comparison of erythromycin resistant campylobacter from Turkeys identifies hosts and pathways for horizontal spread of erm(B) genes. Front Microbiol. 2017;8:2240. doi: 10.3389/fmicb.2017.02240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lebreton F, Valentino MD, Schaufler K, Earl AM, Cattoir V, Gilmore MS. Transferable vancomycin resistance in clade B commensal-type Enterococcus faecium. J Antimicrob Chemother. 2018;73(6):1479–1486. doi: 10.1093/jac/dky039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Werner G, Freitas AR, Coque TM, Sollid JE, Lester C, Hammerum AM, Garcia-Migura L, Jensen LB, Francia MV, Witte W, et al. Host range of enterococcal vanA plasmids among gram-positive intestinal bacteria. J Antimicrob Chemother. 2011;66(2):273–282. doi: 10.1093/jac/dkq455. [DOI] [PubMed] [Google Scholar]
- 104.Jasni AS, Mullany P, Hussain H, Roberts AP. Demonstration of conjugative transposon (Tn5397)-mediated horizontal gene transfer between Clostridium difficile and Enterococcus faecalis. Antimicrob Agents Chemother. 2010;54(11):4924–4926. doi: 10.1128/AAC.00496-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cyoia PS, Koga VL, Nishio EK, Houle S, Dozois CM, de Brito KCT, de Brito BG, Nakazato G, Kobayashi RKT. Distribution of ExPEC virulence factors, bla CTX-M, fosA3, and mcr-1 in Escherichia coli isolated from commercialized chicken carcasses. Front Microbiol. 2018;9:3254. doi: 10.3389/fmicb.2018.03254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang CZ, Ding XM, Lin XL, Sun RY, Lu YW, Cai RM, Webber MA, Ding HZ, Jiang HX. The emergence of chromosomally located. Front Microbiol. 2019;10:1268. doi: 10.3389/fmicb.2019.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Amy J, Bulach D, Knight D, Riley T, Johanesen P, Lyras D. Identification of large cryptic plasmids in Clostridioides (Clostridium) difficile. Plasmid. 2018;96-97:25–38. doi: 10.1016/j.plasmid.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 108.Sanchez-Romero MA, Casadesus J. Contribution of phenotypic heterogeneity to adaptive antibiotic resistance. Proc Natl Acad Sci U S A. 2014;111(1):355–360. doi: 10.1073/pnas.1316084111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rego EH, Audette RE, Rubin EJ. Deletion of a mycobacterial divisome factor collapses single-cell phenotypic heterogeneity. Nature. 2017;546(7656):153–157. doi: 10.1038/nature22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Conlon BP, Rowe SE, Gandt AB, Nuxoll AS, Donegan NP, Zalis EA, Clair G, Adkins JN, Cheung AL, Lewis K. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat Microbiol. 2016;1:16051. doi: 10.1038/nmicrobiol.2016.51. [DOI] [PubMed] [Google Scholar]
- 111.Jiang X, Hall AB, Arthur TD, Plichta DR, Covington CT, Poyet M, Crothers J, Moses PL, Tolonen AC, Vlamakis H, et al. Invertible promoters mediate bacterial phase variation, antibiotic resistance, and host adaptation in the gut. Science. 2019;363(6423):181–187. doi: 10.1126/science.aau5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kang KN, Klein DR, Kazi MI, Guerin F, Cattoir V, Brodbelt JS, Boll JM. Colistin heteroresistance in Enterobacter cloacae is regulated by PhoPQ-dependent 4-amino-4-deoxy-l-arabinose addition to lipid A. Mol Microbiol. 2019;111(6):1604–1616. doi: 10.1111/mmi.14240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Weston N, Sharma P, Ricci V, Piddock LJV. Regulation of the AcrAB-TolC efflux pump in enterobacteriaceae. Res Microbiol. 2018;169(7–8):425–431. doi: 10.1016/j.resmic.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 114.Chancey ST, Zähner D, Stephens DS. Acquired inducible antimicrobial resistance in Gram-positive bacteria. Future Microbiol. 2012;7(8):959–978. doi: 10.2217/fmb.12.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Crabbe A, Jensen PO, Bjarnsholt T, Coenye T, Tolerance A. Metabolic adaptations in microbial biofilms. Trends Microbiol. 2019;27(10):850–863. doi: 10.1016/j.tim.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 116.Hirt H, Greenwood-Quaintance KE, Karau MJ, Till LM, Kashyap PC, Patel R, Dunny GM. Enterococcus faecalis sex pheromone cCF10 enhances conjugative plasmid transfer in Vivo. MBio. 2018;9(1). doi: 10.1128/mBio.00037-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Band VI, Weiss DS. Heteroresistance: A cause of unexplained antibiotic treatment failure? PLoS Pathog. 2019;15(6):e1007726. doi: 10.1371/journal.ppat.1007726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Band VI, Hufnagel DA, Jaggavarapu S, Sherman EX, Wozniak JE, Satola SW, Farley MM, Jacob JT, Burd EM, Weiss DS. Antibiotic combinations that exploit heteroresistance to multiple drugs effectively control infection. Nat Microbiol. 2019;4(10):1627–1635. doi: 10.1038/s41564-019-0480-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Band VI, Crispell EK, Napier BA, Herrera CM, Tharp GK, Vavikolanu K, Pohl J, Read TD, Bosinger SE, Trent MS, et al. Antibiotic failure mediated by a resistant subpopulation in Enterobacter cloacae. Nat Microbiol. 2016;1(6):16053. doi: 10.1038/nmicrobiol.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Peláez T, Cercenado E, Alcalá L, Marín M, Martín-López A, Martínez-Alarcón J, Catalán P, Sánchez-Somolinos M, Bouza E. Metronidazole resistance in Clostridium difficile is heterogeneous. J Clin Microbiol. 2008;46(9):3028–3032. doi: 10.1128/JCM.00524-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Boolchandani M, D’Souza AW, Dantas G. Sequencing-based methods and resources to study antimicrobial resistance. Nat Rev Genet. 2019;20(6):356–370. doi: 10.1038/s41576-019-0108-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, Folster JP, Ayers SL, Lam C, Tate HP, Zhao S. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal salmonella. Antimicrob Agents Chemother. 2016;60(9):5515–5520. doi: 10.1128/AAC.01030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wardenburg KE, Potter RF, D’Souza AW, Hussain T, Wallace MA, Andleeb S, Burnham CD, Dantas G. Phenotypic and genotypic characterization of linezolid-resistant Enterococcus faecium from the USA and Pakistan. J Antimicrob Chemother. 2019;74(12):3445–3452. doi: 10.1093/jac/dkz367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gasparrini AJ, Wang B, Sun X, Kennedy EA, Hernandez-Leyva A, Ndao IM, Tarr PI, Warner BB, Dantas G. Persistent metagenomic signatures of early-life hospitalization and antibiotic treatment in the infant gut microbiota and resistome. Nat Microbiol. 2019;4:2285–2297. doi: 10.1038/s41564-019-0550-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Douglas GM, Langille MGI. Current and promising approaches to identify horizontal gene transfer events in metagenomes. Genome Biol Evol. 2019;11(10):2750–2766. doi: 10.1093/gbe/evz184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Durrant MG, Li MM, Siranosian BA, Montgomery SB, Bhatt AS. A bioinformatic analysis of integrative mobile genetic elements highlights their role in bacterial adaptation. Cell Host Microbe. 2020;27(1):140–153 e9. doi: 10.1016/j.chom.2019.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Podell S, Gaasterland T. DarkHorse: a method for genome-wide prediction of horizontal gene transfer. Genome Biol. 2007;8(2):R16. doi: 10.1186/gb-2007-8-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Stalder T, Press MO, Sullivan S, Liachko I, Top EM. Linking the resistome and plasmidome to the microbiome. Isme J. 2019;13(10):2437–2446. doi: 10.1038/s41396-019-0446-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sydenham TV, Overballe-Petersen S, Hasman H, Wexler H, Kemp M, Justesen US. Complete hybrid genome assembly of clinical multidrug-resistant Bacteroides fragilis isolates enables comprehensive identification of antimicrobial-resistance genes and plasmids. Microb Genom. 2019;5(11). doi: 10.1099/mgen.0.000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Doyle RM, O’Sullivan DM, Aller SD, Bruchmann S, Clark T, Coello Pelegrin A, Cormican M, Diez Benavente E, Ellington MJ, McGrath E, et al. Discordant bioinformatic predictions of antimicrobial resistance from whole-genome sequencing data of bacterial isolates: an inter-laboratory study. Microb Genom. 2020;6(2):e000335. doi:10.1099/mgen.0.000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gibson MK, Wang B, Ahmadi S, Burnham CA, Tarr PI, Warner BB, Dantas G. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat Microbiol. 2016;1:16024. doi: 10.1038/nmicrobiol.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen AV, Cheng AA, Liu S, et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48(D1):D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Leggett RM, Alcon-Giner C, Heavens D, Caim S, Brook TC, Kujawska M, Martin S, Peel N, Acford-Palmer H, Hoyles L, et al. Rapid MinION profiling of preterm microbiota and antimicrobial-resistant pathogens. Nat Microbiol. 2020;5(3):430–442. doi: 10.1038/s41564-019-0626-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Serazin AC, Shackelton LA, Wilson C, Bhan MK. Improving the performance of enteric vaccines in the developing world. Nat Immunol. 2010;11(9):769–773. doi: 10.1038/ni0910-769. [DOI] [PubMed] [Google Scholar]
- 136.Riddle MS, Chen WH, Kirkwood CD, MacLennan CA. Update on vaccines for enteric pathogens. Clin Microbiol Infect. 2018;24(10):1039–1045. doi: 10.1016/j.cmi.2018.06.023. [DOI] [PubMed] [Google Scholar]
- 137.Sorbara MT, Dubin K, Littmann ER, Moody TU, Fontana E, Seok R, Leiner IM, Taur Y, Peled JU, van den Brink MRM, et al. Inhibiting antibiotic-resistant enterobacteriaceae by microbiota-mediated intracellular acidification. J Exp Med. 2019;216(1):84–98. doi: 10.1084/jem.20181639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Baumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535(7610):85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chan AP, Choi Y, Brinkac LM, Krishnakumar R, DePew J, Kim M, Hinkle MK, Lesho EP, Fouts DE. Multidrug resistant pathogens respond differently to the presence of co-pathogen, commensal, probiotic and host cells. Sci Rep. 2018;8(1):8656. doi: 10.1038/s41598-018-26738-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Taft DH, Liu J, Maldonado-Gomez MX, Akre S, Huda MN, Ahmad SM, Stephensen CB, Mills DA. Bifidobacterial dominance of the gut in early life and acquisition of antimicrobial resistance. mSphere. 2018;3(5). doi: 10.1128/mSphere.00441-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Casaburi G, Duar RM, Vance DP, Mitchell R, Contreras L, Frese SA, Smilowitz JT, Underwood MA. Early-life gut microbiome modulation reduces the abundance of antibiotic-resistant bacteria. Antimicrob Resist Infect Control. 2019;8:131. doi: 10.1186/s13756-019-0583-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kim P, Gadani A, Abdul-Baki H, Mitre R, Mitre M. Fecal microbiota transplantation in recurrent clostridium difficile infection: A retrospective single-center chart review. JGH Open. 2019;3(1):4–9. doi: 10.1002/jgh3.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yelin I, Flett KB, Merakou C, Mehrotra P, Stam J, Snesrud E, Hinkle M, Lesho E, McGann P, McAdam AJ, et al. Genomic and epidemiological evidence of bacterial transmission from probiotic capsule to blood in ICU patients. Nat Med. 2019;25:1728–1732. doi: 10.1038/s41591-019-0626-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ghimire S, Roy C, Wongkuna S, Antony L, Maji A, Keena MC, Foley A, Scaria J. Identification of Clostridioides difficile-inhibiting gut commensals using culturomics, phenotyping, and combinatorial community assembly. mSystems. 2020;5(1). doi: 10.1128/mSystems.00620-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517(7533):205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rohde C, Wittmann J, Kutter E. Bacteriophages: A therapy concept against multi-drug-resistant bacteria. Surg Infect (Larchmt). 2018;19(8):737–744. doi: 10.1089/sur.2018.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Brown Gandt A, Griffith EC, Lister IM, Billings LL, Han A, Tangallapally R, Zhao Y, Singh AP, Lee RE, LaFleur MD. In vivo and in vitro effects of a ClpP-activating antibiotic against vancomycin-resistant enterococci. Antimicrob Agents Chemother. 2018;62(8). doi: 10.1128/AAC.00424-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sassone-Corsi M, Chairatana P, Zheng T, Perez-Lopez A, Edwards RA, George MD, Nolan EM, Raffatellu M. Siderophore-based immunization strategy to inhibit growth of enteric pathogens. Proc Natl Acad Sci U S A. 2016;113(47):13462–13467. doi: 10.1073/pnas.1606290113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cherian PT, Wu X, Yang L, Scarborough JS, Singh AP, Alam ZA, Lee RE, Hurdle JG. Gastrointestinal localization of metronidazole by a lactobacilli-inspired tetramic acid motif improves treatment outcomes in the hamster model of Clostridium difficile infection. J Antimicrob Chemother. 2015;70(11):3061–3069. doi: 10.1093/jac/dkv231. [DOI] [PMC free article] [PubMed] [Google Scholar]


