ABSTRACT
Lactic acid bacteria (LAB) are the most frequently used probiotics in fermented foods and beverages and as food supplements for humans or animals, owing to their multiple beneficial features, which appear to be partially associated with their antioxidant properties. LAB can help improve food quality and flavor and prevent numerous disorders caused by oxidation in the host. In this review, we discuss the oxidative stress tolerance, the antioxidant capacity related herewith, and the underlying mechanisms and signaling pathways in probiotic LAB. In addition, we discuss appropriate methods used to evaluate the antioxidant capacity of probiotic LAB. The aim of the present review is to provide an overview of the current state of the research associated with the oxidative stress tolerance and antioxidant capacity of LAB.
KEYWORDS: Probiotic, lactic acid bacteria, oxidative stress, oxygen tolerance, antioxidant capacity, assessment method
Introduction
Lactic acid bacteria (LAB) are a diverse group of gram-positive bacteria that widely exist in nature, including plants and animals. Probiotic LAB strains, especially of the genera Lactobacillus and Bifidobacterium, have various health-promoting effects. Owing to their nutritional and functional benefits to humans and animals, public interest in the application of probiotic LAB in foods and feeds has increased. However, during industrial processing and in the gastrointestinal tract, probiotic LAB are exposed to unfavorable environments, including high or low temperature, low pH, bile salts, oxygen, or limited nutrition, inducing stress.1,2 These stressors affect LAB survival during processing and shelf-life during storage, as well as survival, proliferation, and functionality in the gastrointestinal tract. In order to guarantee a sufficient number of viable bacteria in the final product and effective health-promoting action in the host, it is critical to isolate strains that exhibit high viability and functionality as well as high stress resistance.
Among the above stressors, oxidative stress is of critical importance as it greatly influences viability and product quality.3 The oxygen sensitivity of probiotic LAB is a major factor limiting their viability, although LAB are regarded aerotolerant anaerobes. Anaerobic bacteria lack the capability to synthesize an active electron transport chain,4 which affects their survival in aerobic environments. High oxygen levels will lead the formation of reactive oxygen species (ROS), including the superoxide anion (O2–), hydrogen peroxide (H2O2), and the highly reactive hydroxyl radical (HO·). When accumulated, ROS cause oxidative stress, which results in damage to proteins, DNA, and lipids, and even cell death.5 Therefore, preventing oxidative stress in LAB cells by using O2-tolerant LAB strains and applying adequate production and storage techniques are important to ensure high bacterial viability during storage and in the gastrointestinal tract.2,6
Besides a rapid and sensitive oxidative stress response, probiotic LAB exhibit substantial antioxidant activity in the host intestine and promote the production of antioxidant enzymes to help remove ROS in the host intestine and thereby alleviate oxidative damage. When host defense is weakened, various stresses can readily induce ROS production, which may result in a redox imbalance and subsequent impairment of biomolecules, which can lead to various disorders. Evidence suggests that some probiotic LAB strains can increase the activity of antioxidative enzymes or modulate and relieve circulatory oxidative stress to protect cells from oxidative stress-induced damage.7 Although the antioxidant properties of probiotic LAB have been confirmed in vitro and in vivo, the mechanism by which they regulate oxidative stress tolerance is not fully understood.
Various methods have been developed and used to assess the antioxidant properties of probiotic LAB. These range from methods to detect free radicals and metal ions to end-product and enzymatic assays. However, the antioxidant mechanisms of probiotic LAB are complex, and different strains use different mechanisms. Currently, there are no uniform testing standards nor a comprehensive indicator, and thus, it is impossible to compare the antioxidant capacity of different probiotic strains. Various approaches have to be combined in order to identify and characterize novel probiotic LAB for food production and as effective food and feed additives.
LAB present in fermented foods and dietary supplements for humans and animals have been recognized to have beneficial effects on health and well-being, without having any obvious adverse effects. There are numerous mechanisms through which LAB can exert these beneficial effects. Recent studies have revealed the significant antioxidant abilities of LAB both in vivo and in vitro, which may contribute to their beneficial effects and have instigated a renewed interest. In this review, we discuss the redox system of LAB and their oxidative stress tolerance, with a focus on the LAB antioxidant properties and their mode of action. In addition, we present commonly used assays and methodologies to screen LAB antioxidant capacity. Our aim was to provide a comprehensive overview of LAB oxidative stress tolerance and antioxidant capacity, and their evaluation.
Oxidative stress tolerance
Oxygen radical formation and toxicity in probiotic LAB
Oxygen is considered one of the critical factors affecting the survival of anaerobic aerotolerant probiotic bacteria. An aerobic environment may stimulate the production of toxic oxygen byproducts, such as ROS, reactive nitrogen species (RNS), and reactive sulfur species, in probiotic LAB.5 H2O2 produced in such a condition can react with ferrous iron (Fe2+) salts and produce the extremely toxic HO· through the Fenton reaction (Figure 1).8 HO· can damage proteins, causing a reduction in ATP and resulting in a lower energy level within the bacterial cell. HO· can also break phosphodiester bonds in DNA molecules, which leads to DNA fragmentation, and damage lipid moieties within the plasma membrane. A high steady-state concentration of O2– can increase the release of Fe2+ from proteins containing iron-sulfur clusters, thus promoting the Fenton reaction (Figure 1). These radicals directly or indirectly damage proteins, DNA, and lipids, and thus eventually lead to low cell viability and cell death (Figure 1). Endogenous production of H2O2 and further reactive oxidants has been shown to be the main cause of oxidative stress in Lactobacillus johnsonii NCC 533 during aerobic growth.4 LAB lack dedicated enzymes that can eliminate HO·, but have developed other selective strategies to limit HO· formation through eliminating H2O2 and O2–9.
Figure 1.

Scheme summarizing the redox system in LAB. Oxygen within a LAB cell can be consumed by several oxidases (NADH oxidases (NOX), pyruvate oxidase (POX), and lactate oxidase (LOX)) to produce H2O2. H2O2 can react with Fe2+ to produce free radicals, which leads to protein, DNA, and lipid damage as well as cell death. H2O2-degrading enzymes in LAB cells, such as pseudocatalase (a manganese-containing enzyme, Mn-Kat) and heme-dependent catalase (Heme-Kat) can decrease the H2O2 level. In addition, LAB can chelate iron to reduce the level of Fe2+. Superoxide dismutases (SODs) in LAB cells, such as MnSODs, can reduce the level of O2–, thus preventing Fe2+ production. The thioredoxin-thioredoxin reductase system (Trxs) and glutathione-glutaredoxin system (Grxs) in LAB cells regulate the thiol-disulfide balance and thus contribute to maintaining redox homeostasis. Other protective systems in LAB cells may contribute to the repair of damaged protein and DNA.
Oxidative stress-related enzymes in probiotic LAB
O2-consuming enzymes
Some LAB have O2-consuming enzymes, such as NADH oxidases (NOX), pyruvate oxidase (POX), and lactate oxidase (LOX) (Figure 1).10 These enzymes are generally involved in the aerobic metabolism of bacteria, and especially, in microaerophilic bacteria such as LAB. NOX consumes O2 to form H2O or H2O2 in LAB.11,12 These O2-consuming enzymes are responsible for the rapid removal of O2 and play an important role in maintaining the intracellular redox balance. Research in NOX-defective Streptococcus mutants indicated that superoxide dismutase (SOD) and glutathione (GSH) reductase activities were increased to enhance oxidative stress tolerance, indicating that these enzymes have complementary actions.13 POX and LOX contribute to the formation of H2O2 in LAB.14,15 While both enzymes are expressed throughout growth, POX produces most of the H2O2 in the early and log phases, whereas LOX mainly contributes to H2O2 production in the stationary phase.16 Zotta and colleagues discovered that POX, but not NOX activities in Lactobacillus plantarum C17 were significantly affected by temperature and oxygen.17
Antioxidant enzymes
Superoxide dismutases (SODs) are among the most important antioxidant enzymes in LAB. They dismutate O2– and thus decrease the intracellular concentration of free metal cations and alleviate the damage caused by H2O2. Mn, Fe, and Cu are the major metal cofactors for the enzymatic function of SODs. MnSODs have been found in several LAB species (Figure 1), whereas FeSODs and Cu/ZnSODs are observed less in LAB. Further, Mn can serve as an O2– scavenger within SOD-deficient LAB cells (e.g., L. plantarum).18 Cu forms complexes with phosphates and other proteins that exhibit O2–- and H2O2-scavenging activities.19 Regular H2O2-degrading enzymes such as catalases and peroxidases scarcely exist in LAB, but they have other antioxidant enzymes. A Mn-containing pseudocatalase has been discovered in L. plantarum,20 and a heme-dependent catalase has been identified in Lactobacillus sakei.21 These enzymes provide protection against H2O2 toxicity (Figure 1). Knowledge about the regulation of antioxidant enzymes in LAB is very limited. It has been demonstrated that the activity of MnSOD is dependent on the intracellular concentration of Mn2+,22,23 whereas heme-Kat activity depends on the hematin concentration.24
Redox and repair systems in probiotic LAB
The thioredoxin-thioredoxin reductase and GSH-glutaredoxin systems, which maintain intracellular dithiol/disulfide homeostasis in both prokaryotic and eukaryotic cells, play an important role in the defense against oxidative stress.25
The thioredoxin system, comprising NADPH, thioredoxin reductase, and thioredoxin, shuttles electrons to thiol-dependent peroxidases to maintain redox homeostasis and protect probiotic bacteria from ROS and RNS damage. This system controls the thiol-disulfide balance and thus plays an essential role in DNA and protein repair by reducing ribonucleotide reductase and methionine sulfoxide reductases, and regulating the activity of numerous redox-sensitive transcription factors.26-28 Many LAB species have a thioredoxin-dependent reduction system (Figure 1). Overexpression of thioredoxin reductase in L. plantarum strain WCFS1 led to production of thioredoxin reductase, which improved the strain’s tolerance toward oxidative stress.29 A thioredoxin reductase mutant of Lactobacillus casei strain Shirota was not able to grow under aerobic conditions because it was deficient in this enzyme.27 Multiple thioredoxin genes have been reported in numerous bacterial species, and different levels of sensitivity to oxidative stress have been observed in strains lacking a thioredoxin gene.
Gram-negative bacteria such as Escherichia coli generally have a GSH-glutaredoxin-independent reduction system. Previously, it was thought that gram-positive bacteria cannot synthesize GSH and thus, do not have the GSH-glutaredoxin system, comprising NADPH, GSH, GSH reductase, and glutaredoxin, to serve as reducers.30 However, later studies revealed that some LAB, such as Streptococcus agalactiae and Lactobacillus fermentum E3 and E18, naturally synthesize GSH at a high level.31,32 Killisaar and colleagues also for the first time found that L. fermentum strain ME-3 has a fully functional GSH system comprising both GSH peroxidase and GSH reductase.33 GSH is oxidized by GSH peroxidase to a disulfide, which can be rapidly reduced back to GSH by GSH reductase in strain ME-3, suggesting that ME-3 harbors a complete GSH system (synthesis, transport, and redox recycling) that effectively protects the cells against oxidative stress. Studies on the precise physiological functions of GSH and the antioxidative role of the GSH system in gram-positive bacteria such as LAB are lacking.
Genes associated with redox in probiotic LAB
Complete genome sequencing has been used in recent years to identify genes related to antioxidant properties and to reveal the potential mechanisms of O2 tolerance and antioxidant activity of probiotic LAB. Genome analysis of Lactobacillus gasseri AL3 and AL5 revealed genes encoding NOX and NADH peroxidases, SOD, Dps-like peroxide resistance protein, and the complete thioredoxin reductase system.34 Five genes encoding proteins related to free-radical scavenging and O2 tolerance have been revealed in Bifidobacterium longum LTBL16 by genome analysis, including three peroxide oxidoreductases and one NOX.35 Genome analysis of Bifidobacterium animalis subsp. lactis 01 showed at least eight protein-coding genes are antioxidant-related genes, and qPCR results demonstrated that genes encoding thioredoxin system and non-enzyme factors of the divalent cation transporter were upregulated under H2O2 challenge.36 In B. longum LTBL16, a gene encoding SIR2, associated with antioxidant activity, has also been identified.37 Genes encoding the complete GSH system, including GSH peroxidase and GSH reductase, have been identified in L. plantarum ZLP001.38 L. plantarum ZLP001 also harbors genes for a complete thioredoxin system, including thioredoxin, thioredoxin reductase, and thiol peroxidase. However, L. plantarum ZLP001 does not harbor SOD genes, implying that different LAB species encode different redox-related genes and have different redox systems.
Strategies to increase oxidative stress tolerance in LAB
Coculture with starter strains
Coculture with starter strains has been demonstrated as a possible strategy to improve the survival of probiotic bacteria during fermentation. Some O2-depleting strains have been used as starter strains in coculture to exhaust O2 thus and improve the survival of O2-sensitive probiotic strains.39 However, if O2-sensitive probiotic species are cocultured with a strain that can produce a high level of H2O2, oxidative stress will occur and affect the viability of the cocultured microorganisms.40 Yeasts seem to have higher antioxidant activity than LAB. L. plantarum CCMA 0743 cocultured with the yeast Torulaspora delbrueckii CCMA 0235 exhibited increased antioxidant activity, as indicated by an α,α-diphenyl-β-picrylhydrazyl (DPPH) assay, as well as enhanced growth during fermentation, indicating the positive effect of this yeast strain on LAB proliferation and oxidative stress.41 Furthermore, catalase-expressing Streptococcus thermophilus improved the survival rate of Lactobacillus delbrueckii subsp. bulgaricus ATCC 11842 under H2O2 exposure and showed a protective effect against oxidative damage during milk fermentation.42
Addition of O2-consuming enzymes
The O2 level in a culture affects the rate of ROS production and the amounts of O2– versus H2O2 produced. Reducing and removing the O2 present in the medium is beneficial to probiotic fermentation. Sasaki et al.43 suggested that NOX is the major O2-consuming enzyme of S. thermophilus 1131, which plays an important role in yogurt fermentation mainly through removing dissolved O2. Supplementation of O2-consuming enzymes such as glucose oxidase has been demonstrated to alleviate oxidative stress in LAB in yogurt during refrigerated storage.44,45 Glucose oxidase supplementation combined with a suitable packaging system with low O2 permeability can increase the probiotic cell density.46 These results indicated that the addition of O2-consuming enzymes may be an effective way to avoid oxidative damage, but this requires further investigation.
Addition of antioxidants
The addition of O2 scavengers or antioxidant compounds has been suggested as a possible approach to temporarily reducing the O2 level and improving the survival of probiotic strains. Ascorbic acid, green tea extracts, and grape extract have been verified to improve the survival of Lactobacillus strains through their antioxidant action.47 Improved survival of Lactobacillus acidophilus in yogurt was achieved by cysteine supplementation.48 Catechin supplementation significantly improved the growth of Lactobacillus helveticus under aerobic conditions, likely through ROS and RNS scavenging or metal ion chelation.49,50 Mn2+ is an important metal in antioxidant enzymes and when intracellularly accumulated, it can help scavenge O2 in L. plantarum during aerobic growth.18,51 Mn2+ supplementation greatly promoted the viable count of L. plantarum under aerobic conditions.52 Most LAB species cannot synthesize GSH and can only accumulate it from the medium.31 GSH supplementation has been found to enhance growth as well as glucose consumption, and to increase soluble protein and amino acid concentrations in Lactobacillus reuteri strain ATCC 23272.53
Physicochemical methods
Novel packaging materials and encapsulation technologies for improving LAB viability in an O2-rich environment have been evaluated. High-impact polystyrene packaging combined with O2-scavenging material was found to not only prevent O2 diffusion, but also decrease dissolved O2 levels during storage, suggesting it provides a more favorable environment for the LAB in yogurt.54,55 Encapsulation, in which small particles that contain an active agent are produced by mechanical means, has been widely used to protect probiotic strains from adverse environmental conditions. The protective effect of encapsulation on probiotic viability in an aerobic environment has also been reported. L. acidophilus 2409 encapsulated with calcium alginate showed significantly enhanced viability when grown aerobically in reconstituted skim milk broth.56 However, not all strains show improved survival under anaerobic conditions after encapsulation.6 Further studies are required to reveal the mechanism underlying the O2 toxicity-protective effects of encapsulation on probiotics.
Adaption and modification
Microorganisms display the ability to adapt to unfavorable environments, which has been exploited to develop strains that can survive in adverse conditions. Exposing a probiotic strain to a sublethal level of oxidative stress will induce an adaptive response and improve the resistance of the strain toward potentially higher levels of oxidative stress. This may be explained by the fact that some silent gene clusters are activated to increase the antioxidant capacity. L. acidophilus and Bifidobacterium spp. adapted to oxidative stress when dissolved O2 was gradually increased from 0 to 210 ppm in yogurt and could survive well for more than 35 days of storage.57 Genetic modification is another strategy to improve probiotic survival and oxidative stress tolerance. Heterologous expression of genes such as KatA, Mn-Kat, and SodA has been demonstrated to markedly improve the oxidative stress resistance of Lactobacillus.23,58,59 Furthermore, starter strains that can produce catalases and MnSOD and thus improve the oxidative stress resistance of LAB have been studied. Starter strain S. thermophilus ST5 heterologously expressing KatE, encoding a heme-dependent catalase, effectively eliminated H2O2 and thus improved the survival of L. delbrueckii subsp. bulgaricus ATCC 11842 in yogurt.42
Antioxidant properties of probiotic LAB
In addition to their powerful redox systems, probiotics have strong antioxidant properties. When the body is in a state of oxidative stress, accumulated ROS will cause free-radical chain reactions through damaging biomolecules, resulting in harm to the organism. Oxidative stress is a major contributor to numerous disorders, such as cardiovascular, inflammatory, cerebrovascular, and degenerative diseases as well as aging and cancer.60,61 Young animals are readily exposed to oxidative damage because they lack a mature antioxidant system in their intestinal tract, leading to an imbalance in the oxidative and antioxidant systems as well as increased free radicals and malondialdehyde (MDA) and decreased antioxidant enzyme capacities.62,63 Multiple studies have demonstrated that probiotics, such as Lactobacillus and Bifidobacterium, possess excellent antioxidant capacity to provide a certain degree of protection against oxidative stress.64-67
Modes of action of probiotic LAB antioxidants
The mechanisms underlying the antioxidant activity of probiotic species are not completely understood; however, it has been suggested that LAB may play antioxidant roles through scavenging ROS, chelating metals, increasing antioxidant enzymes levels, and modulating the microbiota.68,69 The proposed modes of action of probiotic LAB antioxidants are shown in Figure 2.
Figure 2.
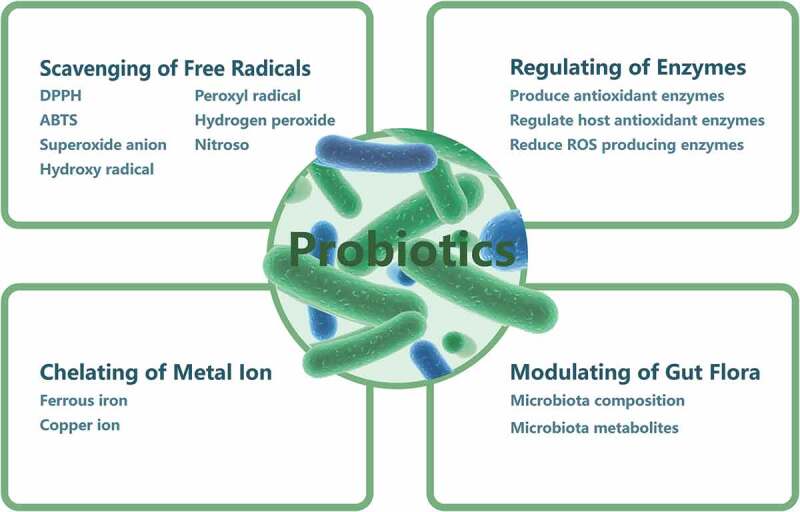
Proposed modes of action of probiotic LAB antioxidants. Probiotic LAB may exert antioxidative effects through the scavenging of free radicals, metal ion chelation, enzyme regulation, and modulation of the gut microbiota.
Radical scavenging
ROS and RNS are mainly produced under exposure to stress conditions that disturb bacterial or host cell metabolism. Most LAB have systems to scavenge O2 free radicals through which the risk of radical accumulation during food fermentation and the damage caused by such free radicals to the host organism are lowered. Many probiotic LAB strains and their metabolites exhibit high scavenging ability toward DPPH, O2–, and H2O2 in vitro.70-73 These scavenging activities generally increase with increasing bacterial cell concentration.71 In one study, culture supernatant of certain strains exhibited significantly higher free-radical-scavenging activity than intact cells did,74 whereas in another study, intact cells showed higher scavenging activity than supernatant.75 This difference is probably due to differences among probiotic LAB species.
Metal ion chelation
In addition to the production of antioxidative enzymes, microorganisms have a non-enzymatic oxidative stress defense mechanism relying on the chelation of metal ions.76 Fe2+ and Cu2+ are the most prevalent and active ions generated in free-radical formation. Fe2+ can produce HO· through the Fenton reaction, and Cu2+ released from chromatin can also catalyze HO· generation. Many probiotic LAB strains have been found to possess strong chelating ability for both Fe2+ and Cu2+,77-79 and probiotic LAB strains show a wide range of Fe2+- and Cu2+-chelating ability, indicating that chelation capacity is strain-specific. In a study by Lin and Yen, S. thermophilus 821 was found to show the best ion-chelating ability among 19 LAB strains tested.80 In another study, L. casei KCTC 3260 demonstrated high chelating activity for both Fe2+ and Cu2+, at 10.6 ppm and 21.8 ppm, respectively, but did not possess detectable SOD activity.77 The results suggested that metal chelation may have contributed more to the antioxidative capacity of L. casei KCTC 3260 than SOD activation.77
Enzymatic regulation
Antioxidant enzyme production
As mentioned above, LAB have their own antioxidant enzymatic system. Most LAB can scavenge free radicals by producing antioxidant enzymes that dismutate free radicals to O2 and H2O2.81 SOD activity has been reported in cell-free extracts of strains belonging to Lactococcus and S. thermophilus, with Lactococcus exhibiting higher activity than S. thermophilus.5 The antioxidant enzymes produced by these bacteria theoretically can help prevent free radical accumulation in the host. Furthermore, LAB strains expressing high levels of SOD or catalases could be developed as a strategy in traditional food applications and new therapeutic uses. de LeBlanc and colleagues proved that engineered Lactobacillus casei BL23 strains producing CAT were able to prevent or decrease the severity of intestinal pathologies caused by ROS.82 The same team later discovered that engineered L. casei BL23 strains producing either CAT or SOD promoted the recovery of initial weight loss in mice with trinitrobenzenesulfonic acid-induced Crohn’s disease, increased enzymatic activities in the gut, and reduced the extent of intestinal inflammation.83
Host antioxidant enzyme regulation
LAB can induce the activity of host antioxidative enzymes, thus regulating the antioxidant system and alleviating oxidative stress. In vitro experiments using human Caco-2 colorectal cells have shown that L. plantarum Y44 can elevate catalase expression in cells damaged by 2,2′-azobis (2-methylpropionamidine) dihydrochloride.84 Another study using an in vitro model of enterocytes investigated the modulation of L. casei Shirota on the expression of gastro-intestinal GSH peroxidase in enterocytes.85 Human patients with type 2 diabetes that consumed yogurt containing L. acidophilus LA5 and B. animalis subsp. lactis BB12 had increased erythrocyte SOD and GSH peroxidase activities as well as higher total antioxidant status.86 Furthermore, increased SOD, catalase, GSH S-transferase, GSH, and GSH peroxidase activities after Lactobacillus supplementation have been observed not only in serum, but also in diverse tissues, including the liver, in various animals,71,72,87 suggesting the great antioxidant properties of LAB.
ROS-producing enzyme regulation
LAB can exert antioxidant action to alleviate oxidative stress damage through regulating certain ROS-producing enzymes. NOX is considered to be a major source of ROS generation. A study using combined Lactobacillus strains demonstrated that LAB can decrease NOX activity and NOX-1 and NOX-4 mRNA expression in spontaneously hypertensive rats.88 Cyclo-oxygenase 2 is highly associated with ROS production, and they show a reciprocal relationship.89 Pretreatment with L. acidophilus significantly downregulated the expression of cyclo-oxygenase 2 in bovine thymic macrophages challenged by the pathogenic bacterium, Aeromonas hydrophila.90 Furthermore, cytochrome P450 (CYP), the terminal oxidase in the electron transfer chain, can induce continuous ROS production.91 L. casei reportedly decreased CYP1A1 expression in different parts of the jejunum, colon, ileum, and cecum in male rats.92 This downregulation of ROS-producing enzymes contributes to the antioxidant capacity of LAB.
Regulation of the gut microbiota
Under excessive proliferation of pathogens in the intestine, the intestinal epithelium produces and releases high levels of ROS, causing significant oxidative stress. It has been demonstrated that the gut microbiota can regulate redox signaling and affect redox homeostasis in the host.93 LAB supplementation can regulate the intestinal microbiota, and it has been speculated that LAB may exert their antioxidative effects partially through the reconstruction of the host intestinal microbiota composition.94 However, direct evidence of this is currently lacking. Dietary alteration of the gut microbiota is strongly linked to oxidative stress; in high-fat diet-fed mice treated with lipoic acid, decreased ROS and MDA and increased total antioxidant capacity showed a strong positive association with lactobacilli and a negative correlation with E. coli and enterococci.95 Supplementation of L. johnsonii BS15 alleviated high-fat diet-induced oxidative stress and changed the intestinal Firmicutes/Bacteroidetes ratio in mice,96 which suggested that modulation of the gut microbiota by LAB bacteria has the potential to improve the host redox state. However, further experiments are required to verify this speculation.
Potential signaling pathways underlying the antioxidant actions of LAB
A number of signaling pathways associated with the antioxidant mechanisms of LAB in the host, including nuclear factor erythroid-2-related factor 2 (Nrf2), silent information regulator 1 (SIRT1), mitogen-activated protein kinase (MAPK), and protein kinase C (PKC), have been identified to date (Figure 3). However, because they are generally strain-specific, likely not all signaling pathways related to LAB-induced antioxidant mechanisms have been identified, and further investigations are required.
Figure 3.
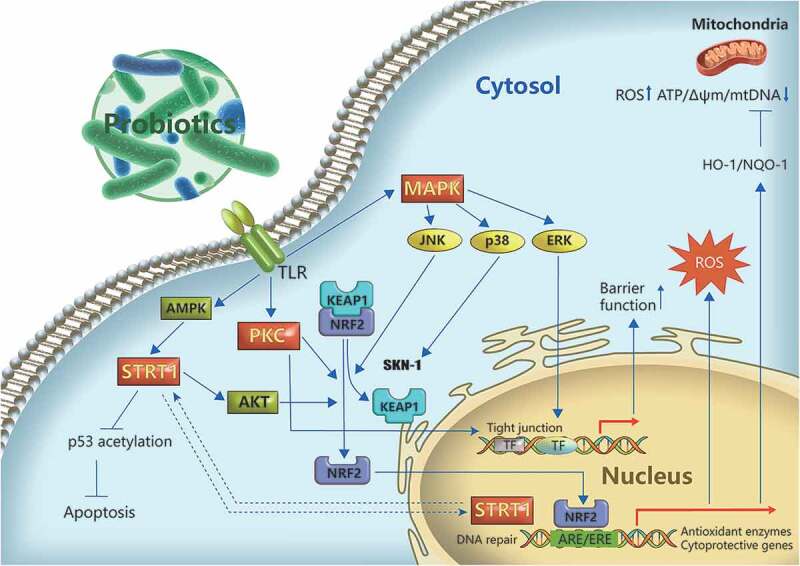
Potential regulatory pathways of LAB antioxidant action. The Nrf2/Keap1 signaling pathway plays a role in the antioxidant mechanisms of LAB in the host. In the host cells, Nrf2 is released from its cytosolic repressor Keap1 and translocates to the nucleus, where it binds to antioxidant response elements, thus enhancing the transcription of cytoprotective genes and alleviating ROS damage. LAB can activate AMPK to induce the phosphorylation and nuclear translocation of SIRT1, leading to AKT phosphorylation and Nrf2 activation. SIRT1 is required for DNA repair following H2O2-induced damage.97 In addition, it is involved in the protective action of LAB against p53-mediated apoptosis induced by oxidative damage.98 The MAPK pathway, including extracellular signal-regulated protein kinases (ERKs), c-jun N-terminal kinase (JNKs), and p38-MAPK, is involved in the regulation of antioxidant activity of LAB. JNKs and p38 are associated with the activation of Nrf2 or its ortholog, whereas ERK is related to the prevention of H2O2-induced disruption of epithelial barrier function. Protein kinase C (PKC) can also be regulated by LAB to alleviate oxidative damage. LAB can alleviate oxidative stress-induced mitochondrial dysfunction via Nrf2 signaling, strengthening the epithelial barrier function.99
Nrf2
Nrf2 is a member of the NF-E2 family of basic leucine zipper transcription factors.100 The Nrf2 system is well characterized as a ubiquitin-dependent signaling pathway that responds to oxidative stress.101 Modulation of Nrf2 signaling potentially is a novel therapeutic strategy against oxidative stress-induced complications.102 Under high levels of ROS, Nrf2 dissociates from its constitutive inhibitor Keap1, which contains redox-sensitive cysteine residues, and then translocates to the nucleus and binds to antioxidant response element (ARE) sequences to initiate the transcription of antioxidant and cytoprotective genes. Thus, Nrf2 activation leads to the upregulation of antioxidant and cytoprotective proteins, which play an important role in coping with oxidative stress and maintaining redox equilibrium. Nrf2 has been discovered to be one of the most important protective mechanisms against oxidant stress in probiotics.103,104 The involvement of Nrf2 in the activation of antioxidant cellular defenses by probiotics has been shown not only in human cells, but also in animal cell models in vitro.84,85,104 In vivo studies have focused mostly on different mouse models, including high cholesterol, high-fat diet, high oxidative stress, and aging models. Several studies have demonstrated that some probiotic LAB strains can activate Nrf2 signaling and upregulate downstream antioxidant enzymes including SOD, catalase, and heme oxygenase-1 in the mouse liver, thus enhancing antioxidative defense.66,105-107
SIRTs
SIRTs are an evolutionarily conserved family of NAD-dependent protein (histone/non-histone) deacetylases that play an important antioxidant role in mammals through regulating key genes and molecules integral to redox homeostasis.97 Recent studies have revealed that certain LAB strains harbor a Sir2 gene.37,108 However, functional studies of probiotic Sir2 are rare. It has been hypothesized that LAB Sir2 plays an antioxidant role in probiotics as well as in the host. Guo et al.37 discovered that Sir2 exists in B. longum and L. acidophilus and seems to play a role in aerotolerance by increasing antioxidant enzyme activity. Further study showed that Sir2 in B. longum upregulates the expression and activity of antioxidant enzymes by deacetylating the transcription protein σH. Moreover, in in vitro experiments using human 293 T cells, Sir2 of B. longum was verified to activate human MnSOD/SOD2 and catalase to reduce the cellular ROS level. Further, B. longum Sir2 deacetylates the forkhead transcription factor FOXO3a, which mediates antioxidant gene expression. Further studies are required to elucidate the functions of probiotic Sir2 proteins in different strains and to compare their mechanisms of antioxidant regulation. In another recent study, Bonfili and colleagues discovered that the administration of a probiotic formulation termed SLAB51 comprising bifidobacteria, lactobcilli, and S. thermophilus markedly alleviated oxidative stress in a mouse model of Alzheimer’s disease, and this was mediated mainly by activated SIRT1-dependent signaling as indicated by significantly increased SIRT1 activity and expression.98 Butyrate-producing probiotics can prevent the progression of nonalcoholic fatty liver disease involving oxidation damage through the activation of AMPK and AKT.109 The authors also confirmed that sodium butyrate-mediated AMPK activation induced the phosphorylation and nuclear translocation of SIRT1, leading to the phosphorylation of AKT and activation of Nrf2 in vitro.109 We speculate that probiotic strains may activate SIRT1 through this signaling pathway to induce Nrf2 expression and activation. A novel selenium-GSH-enriched probiotics strain reportedly can attenuate hepatic oxidative stress, ER stress, and inflammation caused by CCl4 via activating SIRT1 signaling.110
MAPKs
MAPKs, including ERKs, JNKs, and p38-MAPK, are involved in numerous signal transduction pathways. Diverse processes, including cell growth, proliferation, differentiation, inflammation, and immunization are associated with MAPK regulation. Some of these processes may be caused by oxidative stress. Several studies have demonstrated that probiotics can alleviate H2O2-induced disruption of barrier function and tight junctions in mammalian epithelial cells. The soluble proteins p40 and p75 produced by Lactobacillus rhamnosus GG prevent H2O2-induced disruption of human intestinal epithelial tight junctions and increase paracellular permeability, likely through the rapid activation of ERK1/2.111 In Caenorhabditis elegans fed L. gasseri SBT2055 (LG2055), SKN-1 (an Nrf ortholog) was activated, which induced the transcription of antioxidant genes via p38 MAPK signaling, thus enhancing the antioxidant defense response.112 In another study in mammalian cells, it was discovered that LG2055 treatment activated JNK, and inhibition of JNK activation suppressed Nrf-2 ARE signaling activation and the protective effect of LG2055 against oxidative stress.113 This indicated that LG2055 may activate Nrf2-ARE signaling through JNK activation, thus strengthening the antioxidant defense in mammalian cells.113
PKC
PKC is a family of protein kinases that control protein function through the phosphorylation of hydroxyl groups of serine and threonine residues. Evidence suggests that PKC is a target of redox modification because it contains unique structural features that are susceptible to oxidative modification,114,115 and this activity may be involved in various pathways that regulate cell growth and barrier function as well as stress responses.116-118 PKC-ζ improves microtubule and gut barrier integrity by preventing iNOS production caused by oxidants, and therefore, is considered as an endogenous stabilizer to prevent oxidative damage.119,120 Zhou et al.121 reported that administration of L. plantarum improved intestinal barrier function and oxidative stress in obstructive jaundice rats by enhancing the PKC pathway in terms of expression and activity. Furthermore, H2O2-induced epithelial barrier disruption can be ameliorated by the soluble proteins p40 and p75 produced by L. rhamnosus GG through a PKC- and MAPK-dependent mechanism.111
Antioxidant molecules produced by probiotic LAB
Exopolysaccharides (EPS)
EPS are group of carbohydrate polymers that play important roles in biofilm formation and cell adhesion, and are produced also by probiotics.122,123 EPS have various beneficial physiological functions in humans and animals, including the regulation of intestinal barrier function124 and the immune response.125,126 Moreover, EPS from Lactobacillus have been shown to exert excellent antioxidant activity in vitro and in vivo.127 In vitro, purified EPS improved the aerobic growth of Leuconostoc mesenteroides by ~10-fold.128 The aerobic growth of O2-sensitive probiotics was enhanced by EPS through the relief of O2 stress, which was achieved through the extrusion of dissolved O2 in biofilms and aggregations129 or in aqueous environments.128 EPS from different probiotics exhibit prominent and concentration-dependent free-radical-scavenging and metal-chelating activities.73,130,131 The protective effect of EPS from Lactobacillus against host oxidative stress have been evaluated in different cell lines, such as Caco-2 and PC12.132,133 EPS from L. plantarum LP6 and L. plantarum C88 exhibit antioxidant effects by improving cell viability and downregulating oxidative stress biomarkers.132,133 In vivo studies have shown that EPS from probiotics increase antioxidant enzyme activities and decrease end products of redox processes in the liver and in serum, indicating their excellent antioxidant effects.134,135 EPS may also have indirect effects via regulating the microbiota composition or shielding bacterial cell-wall surfaces.125,136 It is challenging to completely elucidate the molecular mechanism underlying the antioxidant action of EPS because EPS are highly diverse in terms of structure and physicochemical properties.137 Moreover, EPS production is affected by various conditions, including temperature, pH, and O2 strength.138,139
Carotenoids
Carotenoids, which are widespread in nature, have well-known antioxidant properties. Carotenoids are common in pigmented bacteria and their terpenoids possess 30, 40, or 50 carbons. C50 carotenoids are restricted to certain gram-positive bacteria, C40 are commonly present in photoautotrophic bacteria, and C30 are found in some unrelated genera and species.140 Some probiotic bacteria produce carotenoids, and this is likely associated with their antioxidant activity. Lactobacillus pentosus KCCP11226 harbors C30 carotenoid biosynthetic genes (crtM and crtN), shows high carotenoid production and survival under oxidative stress, and is considered a functional probiotic.141 Notably, aerobic growth conditions, while slowing down growth, significantly induced carotenoid production in Enterococcus gilvus,142 which indicated that aerobic culture conditions may contribute to conferring oxidative stress tolerance in carotenoid-producing LAB. Further research revealed that aerobic growth conditions not only affected crtN and crtM expression, but also the biosynthesis of the carotenoid precursor isoprenoid via mevalonate in E. gilvus.143 Transcriptome analysis of oxidative stress-response genes in E. gilvus corroborated that the regulation of isoprenoid biosynthetic genes is one of the potential mechanisms of the carotenoid-based oxidative stress response in LAB.144
Ferulic acid (FA)
FA is natural phenolic acid that is abundantly presented in many types of foods, such as cereals, fruits, and coffee. FA is a potent antioxidant that can eliminate free radicals through a neutralization reaction.145 Some probiotic bacteria produce feruloyl esterase (FE), which hydrolyzes and releases FA from its bound state146,147 and thus exerts health-beneficial antioxidant properties. Based on qualitative precipitation and quantitative HPLC assays, L. fermentum NCIMB 5221 was found to produce the most active FE among several bacteria tested,148 and antioxidant capacity tests verified its significant antioxidant activity. An in vivo study revealed that L. fermentum CRL1446 supplementation enhanced the production and bioavailability of FA in mice through increasing the activity of FE, exerting an antioxidant effect and improving the host oxidative status.149 FE activity was affected in a time- and dose-dependent manner, and optimal intestinal FE activity was observed on day 7 after supplementation of 107 CRL1446 cells per day. It can be inferred that probiotic strains might secrete FE enzymes into the intestine or regulate intestinal microbiota to directly stimulate FE activity. Further studies confirmed that FA-producing probiotics can induce metabolic changes and exert beneficial effects on the host metabolic state.150 This potent beneficial activity of FA can be explained by the modulation of certain metabolites and inflammatory markers linked to antioxidant activity.150,151
Histamine
Histamine produced by Lactobacillus species has been demonstrated to have immunoregulatory functions, comprising pro- as well as anti-inflammatory effects.152 Recent studies have revealed that histamine also plays a role in the antioxidant potential of lactobacilli. Histamine inhibits the generation of superoxide radicals by activated macrophages,153 and histamine dihydrochloride-treated human leucocytes showed increased catalase activity and decreased SOD activity.154 Histamine was found to be produced by L. reuteri strains, including L. reuteri E and L. reuteri KO5, under both aerobic and anaerobic conditions.154 The concentration of histamine increased along with an increase in lactobacilli cells, and the maximum concentration was reached not earlier than after 48 h.154 Cell culture supernatants of lactobacilli that produced histamine modulated the enzymatic activities of SOD and catalase.154 However, the presence of an adequate amount of precursor, i.e., biogenic amine, is required for LAB to produce histamine; thus, the effect of histamine produced by LAB on antioxidant capacity remains to be thoroughly studied.
Methods to assess the oxidative stress tolerance and antioxidative properties of probiotic LAB
Various methodologies have been used to assess the antioxidative properties of probiotics, and these can be classified based on the target of detection, mechanism of action, and measure adopted. Most of the methods used to assay common antioxidants can be applied to probiotics. Some procedures adopt biochemical approaches to test the radicals generated (external or cellular), some use specific techniques to detect the end products, and others involve eukaryotic-cell and animal testing. While the testing methods used in probiotics research are extremely versatile, it is important to choose the most appropriate assay to assess the antioxidant properties of probiotic strains. As the various antioxidative assays have their own characteristics, it is difficult to compare results among studies using different assays.155 Furthermore, various methods are often combined because no single method can provide unequivocal results.156
Assays of oxidative stress tolerance
O2 tolerance
The relative bacterial growth ratio (RBGR) method to evaluate the O2 tolerance of probiotics was originally developed by Kikuchi and Suziki157 and later optimized by Talwalkar and colleagues.3 The RBGR is calculated as the ratio of the absorbance (representing growth) of an aerobically shaken culture to that of an anaerobically shaken culture. The O2 tolerance of probiotic LAB strains can be measured quantitatively using the RBGR index, and an RBGR value close to 1 indicates good O2 tolerance. The RBGR method is a simple and easy approach that can be used for high-throughput screening of probiotic bacteria. Li and colleagues used this method to evaluate 10 strains of bifidobacteria from various sources in several O2 concentrations and to select strains exhibiting high O2 tolerance.158
Resistance to H2O2
Another method to evaluate the O2 resistance of probiotics is to evaluate their viability in H2O2. H2O2 is a weak oxidant that can permeate the cell membrane and subsequently cause damage.69 While probiotics are generally resistant to H2O2 as they produce H2O2 in the intestinal tract as an antimicrobial compound, this assay is commonly used to evaluate the viability of probiotics in an anaerobic environment.159,160 It is worth mentioning that H2O2 can be degraded by catalase; however, most Lactobacillus strains do not exhibit catalase activity. Some studies have suggested that the expression of trxB1 or uvrA may contribute to the survival of lactobacilli in the presence of H2O2.29,161 However, the precise mechanism requires further exploration.
Assays of antioxidant properties
Assays based on radical production or scavenging
Free radical detection is widely used to evaluate the antioxidant activity of various antioxidants.156 Most antioxidant evaluation methods based on reactive species can be applied to probiotics to evaluate their antioxidant capacity.69,162 Radical production and scavenging systems are the most straightforward methods, and most of them use biochemical methods without the requirement for eukaryotic cells. These biochemical assays are mostly based on fluorescence or chromophore reactions, and thus are straightforward and cheap. Radical production and scavenging systems are normally used to detect the antioxidant ability of probiotic strains in vitro, but they have also been used to evaluate radical production after probiotic treatment or supplementation in vivo.5,163 Further, these systems can be applied to intact cells as well as cell-free extracts and cell lysates or their metabolic products.72,75,164,165 Reactive species tests have also been applied to evaluate the antioxidant capacity of LAB-fermented foods, especially, milk.166 The methods used to assess the antioxidative capacity of probiotic LAB based on the detection of reactive species are summarized in Table 1. Most studies combine assays to evaluate the antioxidant capacity of probiotic strains.72,181
Table 1.
Methods for the screening of the antioxidant capacity of LAB based on radical production or scavenging.
| Target radical | Method or solution | Principle | Probiotic LAB strains studied | Reference |
|---|---|---|---|---|
| ABTS | Trolox equivalent antioxidant capacity assay | Measures the ability of antioxidants to scavenge the stable radical cation 2,2′-azinobis (3-ethylbenzothiazoline-6-sulfonic acid), which is intensely colored. | 7 Bifidobacterium, 11 Lactobacillus, 6 Lactococcus, and 10 S. thermophilus | 5 |
| DPPH | DPPH radical solution | Antioxidants can reduce the free, stable, and purple-colored 2,2-diphenyl-1-picrylhydrazyl radical to the yellow-colored diphenylpicrylhydrazine. | L. plantarum FC225 | 167 |
| L. plantarum Y44 | 160 | |||
| L. helveticus KLDS1.8701 | 168 | |||
| Superoxide radical | Fluorescent dihydroethidium (DHE) | O2– production is measured based on reaction with the fluorescent dye dihydroethidium. | Probiotic formulation VSL#3 | 169 |
| Nitro blue tetrazolium (NBT) | The scavenging activity of O2– is analyzed based on the color reaction of NBT, NADH, and phenazine methosulfate. | Lactococcus lactis ssp. lactis CLFP 100, Leuconostoc mesenteroides CLFP 196, and L. sakei CLFP 202 | 170 | |
| L. reuteri SHA101 and Lactobacillus vaginalis SHA110 | 171 | |||
| Pyrogallol autoxidation | Pyrogallol can autoxidize in solutions to produce O2–. Antioxidants can affect the production of O2– by pyrogallol autoxidation. | L. plantarum FC225 | 167 | |
| Enterococcus faecium BDU7 | 172 | |||
| L. plantarum L.P2 | 79 | |||
| Hydroxyl radical | 1,10-phenanthroline/FeSO4 | HO· scavenging activity is analyzed based on the reaction of 1,10-phenanthroline, FeSO4, and H2O2, producing a colored product. | L. plantarum FC225 | 167 |
| L. plantarum LP6 | 132 | |||
| L. plantarum Y44 | 160 | |||
| E. faecium WEFA23 | 173 | |||
| Brilliant green | HO· levels in the Fenton system are indirectly detected based on the fact that HO· can make brilliant green fade. | 11 Lactobacillus strains | 174 | |
| Lactobacillus paraplantarum D-3 | 175 | |||
| Salicylic acid | HO· scavenging activity is analyzed based on the principle that salicylic acid can be used as trapping reagent of HO·. | L. acidophilus LA5 and B. animalis subsp. lactis BB12 | 176 | |
| L. mesenteroides S81 | 177 | |||
| Peroxyl radicals | Oxygen radical absorbance capacity (ORAC) assay | The fluorescence intensity of fluorescent molecules such as β-phycoerythrin decreases over time under reproducible and constant flux of peroxyl radicals. | Lactobacillus spp. | 164 |
| L. fermentum LF31 | 178 | |||
| Hydrogen peroxide | Horseradish peroxidase (HRP) | HRP mediates the oxidation of phenol red by H2O2, which results in the formation of a compound that absorbs at 610 nm. | Lactobacillus spp. | 179 |
| Nitroso | α-naphthylamine | The scavenging activity of nitroso is determined based on the color reaction of sulfanilic acid and α-naphthylamine. | Leuconostoc citreum B-2 | 180 |
Assays based on the dynamics or end products of redox processes
Lipid peroxidation is the best-studied biologically relevant free-radical chain reaction. Although lipid peroxidation generally occurs late in the oxidative damage process, after damage of proteins and DNA,182 lipid peroxidation detection is among the assays the most commonly used to assess the dynamics of isolated redox processes.162 The extent of lipid oxidation can be determined by measuring the loss of unsaturated fatty acids or the amount of peroxidation products.182 Several assays are available to measure lipid peroxidation; however, as with other free-radical assays, no single method can accurately account for the entire process. Thiobarbituric acid (TBA) and diene-conjugate assays are relatively simple, but nonspecific assays. In probiotic research, the TBA assay is one of the methods the most commonly used to evaluate antioxidant capacity. The suppression of lipid peroxidation by several L. acidophilus and B. longum strains was measured using the TBA method, and the result was confirmed by detecting the lipid peroxidation product-scavenging ability.183 Noureen and colleagues measured the levels of lipid peroxidation inhibition of 16 LAB strains from different sources using the TBA assay to compare their antioxidant potential.72 They found that intact cells (47–82.38%) and culture supernatants (41–74.34%) showed higher lipid peroxidation inhibition activity than did cell lysates (10–48.92%).72
End products of oxidative stress are regularly used to evaluate the oxidative damage caused by ROS. Oxidized products of proteins (nitrate tyrosine, protein carbonyls), nucleic acid bases (8-hydroxy-2-deoxyguanosine), carbohydrates (glycated products), and lipids (malondialdehyde, isoprostanes, lipoproteins) can all be used as biomarkers of oxidative damage.184 These molecules can be easily detected using specific techniques, such as ELISA, HPLC-UV, HPLC/UPLC-MS/MS, and GC-MS. Lipid peroxidation products are the most commonly detected. MDA is the breakdown product of major chain reactions leading to the oxidation of polyunsaturated fatty acids, and it causes severe oxidative stress as a mutagenic and carcinogenic reactive substance. It serves as a reliable marker of oxidative stress-mediated lipid peroxidation in biological systems and foods. MDA assays have been widely used to study the effects of probiotic LAB in mammalian cells as well as serum, the liver, and colonic mucosa.71,74,80,185 Isoprostanes, another specific ROS-induced lipid peroxidation product, have served as an oxidative status marker to evaluate the effect of dietary L. fermentum ME-3 supplementation in humans.186 Lipoproteins as another lipid oxidation product have been detected in the plasma and liver of cholesterol-fed rats to evaluate the antioxidant effect of Lactobacillus GG.187 Furthermore, oxidation products, protein carbonyls, and 8-hydroxy-2-deoxyguanosine have also been used as indicators in mice or piglet serum to evaluate the antioxidant response after LAB administration.188,189
Assays based on the reducing power
The reducing power of antioxidants can be measured through redox reactions with transition metal ions, such as Fe (ferric reducing antioxidant potential, FRAP) and Cu (cupric reducing antioxidant capacity, CUPRAC).162 FRAP evaluates the antioxidant ability based on the reduction of ferric 2,4,6-tripyridyl-s-triazine complex [Fe3+−(TPTZ)2]3+ to [Fe2+−(TPTZ)2]2+ depending on the available reducing species, along with a color change from yellow to blue in acidic condition.69 The FRAP test has been used to evaluate the antioxidant capacity of L. plantarum Y44 in Caco-2 cells damaged with 2,2′-azobis (2-methylpropionamidine) dihydrochloride in vitro, which revealed that L. plantarum Y44 exerted antioxidative effects in a dose-dependent manner.84 The total antioxidant capacity in the livers of hyperlipidemic rats significantly increased after L. casei supplementation as indicated by a FRAP assay.87 FRAP assays have been also used to compare the antioxidant capacities of fresh skimmed, pasteurized, and UHT milks before and after fermentation with several lactobacilli combined or not with the yeast Saccharomyces boulardii.190
Other methodologies
Assays based on DNA damage
Most types of environmental stress damage host biomolecules, including DNA. Cells with increased DNA damage display increased DNA migration from the nucleus toward the anode as indicated by photomicrography; thus, the extent of DNA migration in single-cell microgel electrophoresis under alkaline conditions can be used to estimate DNA damage.191 The prevention of oxidative stress-induced DNA damage by Lactobacillus has been evaluated in different mammalian cells (Caco-2 cells, HCT 116 cells, HT-29 cells, and lymphocytes) using a single-cell gel electrophoresis assay (Comet assay), which was developed based on DNA migration.192-195 The DNA-protective capacity of probiotic LAB can also be detected by molecular biology techniques to estimate oxidative stress protection.196 Nowak and colleagues used the DNA repair enzymes endonuclease III and formamidopyrimidine-DNA glycosylase to evaluate the antioxidant capacity of Lactobacillus toward H2O2 and several human carcinogens based on the recognition of oxidized DNA bases by DNA repair enzymes.192 Furthermore, the DNA-protective capacity of a LAB-fermented honey-based kefir beverage was assessed using a pPICZα C plasmid DNA, based on the supercoiled DNA cleavage level, indicating its DNA-protective effect against HO·-induced damage.196
Assays based on the biosensors
A biosensor is defined as an analytical device that combines a biological recognition component with a physicochemical detector and is used for the detection of a substance through generating a measurable signal.197,198 Microbial biosensors integrating microorganism(s) with a transducer have been also developed.199 In recent years, this approach has been adopted to evaluate the antioxidant properties of probiotics in vitro. Bacterial biosensors have been genetically modified to express the luxCDABE operon, encoding bioluminescence and luciferase, under the control of oxidation reaction-related gene promoters, such as SoxS or RecA promoters. The antioxidant activity of cell-free culture supernatant of lactobacilli has been evaluated using the biosensor strain E. coli MG1655, which harbors plasmids encoding the luminescent biosensors pSoxS-lux and pKatG-lux, which are inducible by O2– and H2O2, respectively.200 Eukaryotic cells have also been used to construct biosensing systems to assess the antioxidant capacity of LAB. Ge et al.201 immobilized RAW 264.7 macrophage cells using a one-step acidified MnO2-modified gold electrode and then encapsulated the cells in a 3D cell-culture system. This biosensor can be used to determine the flux of H2O2 released from RAW 264.7 macrophages after treatment with LAB to indirectly evaluate the antioxidant capacity of the latter. This biosensor platform demonstrates the potential for rapid, sensitive, and quantitative screening of the antioxidant properties of LAB.
Conclusion
In the last several decades, the oxidative stress tolerance and antioxidant capacity of probiotic LAB, as well as their health-promoting roles have been extensively investigated. The antioxidative property of probiotic strains has been confirmed in numerous studies, and the application of LAB in oxidative stress-related diseases has been investigated. Probiotic LAB strains have powerful redox systems associated with antioxidative enzymes and oxidative damage repair systems, which contribute to their O2 tolerance and functional roles. Probiotic LAB exhibit remarkable antioxidant capacity mainly by scavenging free radicals, chelating pro-oxidative metal ions, regulating relevant enzymes, and modulating the gut microbiota. As such, they can contribute to prolonging the shelf lives of food products and promoting health and redox equilibrium in the body. The antioxidant mechanisms of probiotic LAB involve a complex signaling network, mainly Nrf2 redox signaling. There are numerous assays for the antioxidative properties of probiotic LAB, based on different mechanisms and methodologies. However, many questions remain unanswered today. The principle and strategies of O2 resistance in probiotic LAB are not completely understood and require further studies. The mechanisms of antioxidant action of probiotic LAB have not been fully elucidated, and thorough pathway studies are needed to uncover the mode of action and achieve targeted use. Furthermore, the lack of a standardized and calibrated antioxidant capacity detection procedure and evaluation criteria makes it impossible to compare results among studies, and detection strategies and comparative methods need to be further investigated. However, we are hopeful that these questions will be answered in future studies.
Funding Statement
This study was financially supported by the Beijing Natural Science Foundation [grant nos. 6202004, 6202009]; the National Natural Science Foundation of China [grant nos. 31972576, 31972575]; and the Special Program on Science and Technology Innovation Capacity Building of BAAFS [grant no. KJCX20180414].
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
JW and TF contributed to the conception and design of the study. TF performed a systematic review and generated the figures. JW wrote the first draft of the manuscript and provided critical feedback while assisting with editing the subsequent drafts of the manuscript. All authors contributed to revisions and approved the final version.
References
- 1.Bron PA, Kleerebezem M.. Engineering lactic acid bacteria for increased industrial functionality. Bioeng Bugs. 2011;2:80–24. doi: 10.4161/bbug.2.2.13910. [DOI] [PubMed] [Google Scholar]
- 2.Mills S, Stanton C, Fitzgerald GF, Ross RP. Enhancing the stress responses of probiotics for a lifestyle from gut to product and back again. Microb Cell Fact. 2011;10:S19. doi: 10.1186/1475-2859-10-S1-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Talwalkar A, Kailasapathy K, Peiris P, Arumugaswamy R. Application of RBGR—a simple way for screening of oxygen tolerance in probiotic bacteria. Int J Food Microbiol. 2001;71:245–248. doi: 10.1016/s0168-1605(01)00563-3. [DOI] [PubMed] [Google Scholar]
- 4.Maresca D, Zotta T, Mauriello G. Adaptation to aerobic environment of Lactobacillus johnsonii/gasseri strains. Front Microbiol. 2018;9:157. doi: 10.3389/fmicb.2018.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amaretti A, Di Nunzio M, Pompei A, Raimondi S, Rossi M, Bordoni A. Antioxidant properties of potentially probiotic bacteria: in vitro and in vivo activities. Appl Microbiol Biotechnol. 2013;97:809–817. doi: 10.1007/s00253-012-4241-7. [DOI] [PubMed] [Google Scholar]
- 6.Talwalkar A, Kailasapathy K. A review of oxygen toxicity in probiotic yogurts: influence on the survival of probiotic bacteria and protective techniques. Curr Issues Intest Microbiol. 2004;3:117–124. doi: 10.1111/j.1541-4337.2004.tb00061.x. [DOI] [PubMed] [Google Scholar]
- 7.Dowarah R, Verma AK, Agarwal N, Singh P, Singh BR. Selection and characterization of probiotic lactic acid bacteria and its impact on growth, nutrient digestibility, health and antioxidant status in weaned piglets. PLoS ONE. 2018;13:e0192978. doi: 10.1371/journal.pone.0192978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang TS, Korber DR, Tanaka T. Influence of oxygen on NADH recycling and oxidative stress resistance systems in Lactobacillus panis PM1. AMB Express. 2013;3:10. doi: 10.1186/2191-0855-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fridovich I. Superoxide radical and superoxide dismutases. Annu Rev Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 10.Condon S. Responses of lactic acid bacteria to oxygen. FEMS Microbiol Rev. 1987;46:269–280. doi: 10.1016/0378-1097(87)90112-1. [DOI] [Google Scholar]
- 11.Tachon S, Chambellon E, Yvon M. Identification of a conserved sequence in flavoproteins essential for the correct conformation and activity of the NADH oxidase NoxE of Lactococcus lactis. J Bacteriol. 2011;193:3000–3008. doi: 10.1128/JB.01466-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jansch A, Freiding S, Behr J, Vogel RF. Contribution of the NADH-oxidase (Nox) to the aerobic life of Lactobacillus sanfranciscensis DSM20451T. Food Microbiol. 2011;28:29–37. doi: 10.1016/j.fm.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 13.Derr AM, Faustoferri RC, Betzenhauser MJ, Gonzalez K, Marquis RE, Quivey RG. Mutation of the NADH oxidase gene (nox) reveals an overlap of the oxygen- and acid-mediated stress responses in Streptococcus mutans. Appl Environ Microbiol. 2012;78:1215–1227. doi: 10.1128/AEM.06890-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pericone CD, Park S, Imlay JA, Weiser JN. Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the fenton reaction. J Bacteriol. 2003;185:6815–6825. doi: 10.1128/jb.185.23.6815-6825.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barre O, Mourlane F, Solioz M. Copper induction of lactate oxidase of Lactococcus lactis: a novel metal stress response. J Bacteriol. 2007;189:5947–5954. doi: 10.1128/JB.00576-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L, Tong H, Dong X. Function of the pyruvate oxidase-lactate oxidase cascade in interspecies competition between Streptococcus oligofermentans and Streptococcus mutans. Appl Environ Microbiol. 2012;78:2120–2127. doi: 10.1128/AEM.07539-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zotta T, Guidone A, Ianniello RG, Parente E, Ricciardi A. Temperature and respiration affect the growth and stress resistance of Lactobacillus plantarum C17. J Appl Microbiol. 2013;115:848–858. doi: 10.1111/jam.12285. [DOI] [PubMed] [Google Scholar]
- 18.Archibald F, Fridovich I. Manganese and defences against oxygen toxicity in Lactobacillus plantarum. J Bacteriol. 1981;145:442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Culotta VC, Daly MJ. Manganese complexes: diverse metabolic routes to oxidative stress resistance in prokaryotes and yeast. Antioxid Redox Signal. 2013;19:933–944. doi: 10.1089/ars.2012.5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kono Y, Fridovich I. Isolation and characterization of the pseudocatalase of Lactobacillus plantarum. J Biol Chem. 1883;258:6015–6019. doi: 10.1021/ja00040a064. [DOI] [PubMed] [Google Scholar]
- 21.Knauf H, Vogel R, Hammes W. Cloning, sequence, and phenotypic expression of kata, which encodes the catalase of Lactobacillus sake LTH677. Appl Environ Microbiol. 1992;58:832–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrus JM, Bowen SW, Klaenhammer TR, Hassan HM. Molecular characterization and functional analysis of the manganese-containing superoxide dismutase gene (sodA) from Streptococcus thermophilus AO54. Arch Biochem Biophys. 2003;420:103–113. doi: 10.1016/j.abb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Bruno-Barcena JM, Andrus JM, Libby SL, Klaenhammer TR, Hassan HM. Expression of a heterologous manganese superoxide dismutase gene in intestinal lactobacilli provides protection against hydrogen peroxide toxicity. Appl Environ Microbiol. 2004;70:4702–4710. doi: 10.1128/AEM.70.8.4702-4710.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolf G, Strahl A, Meisel J, Hammes WP. Heme-dependent catalase activity of lactobacilli. Int J Food Microbiol. 1991;12:133–140. doi: 10.1016/0168-1605(91)90062-t. [DOI] [PubMed] [Google Scholar]
- 25.Carmel-Harel O, Storz G. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu Rev Microbiol. 2000;54:439–461. doi: 10.1146/annurev.micro.54.1.439. [DOI] [PubMed] [Google Scholar]
- 26.Vido K, Diemer H, Van Dorsselaer A, Leize E, Juillard V, Gruss A, Gaudu P. Roles of thioredoxin reductase during the aerobic life of Lactococcus lactis. J Bacteriol. 2005;187:601–610. doi: 10.1128/JB.187.2.601-610.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Serata M, Iino T, Yasuda E, Sako T. Roles of thioredoxin and thioredoxin reductase in the resistance to oxidative stress in Lactobacillus casei. Microbiology. 2012;158:953–962. doi: 10.1099/mic.0.053942-0. [DOI] [PubMed] [Google Scholar]
- 28.Lu J, Holmgren A. The thioredoxin antioxidant system. Free Radic Biol Med. 2014;66:75–87. doi: 10.1016/j.freeradbiomed.2013.07.036. [DOI] [PubMed] [Google Scholar]
- 29.Serrano LM, Molenaar D, Wels M, Teusink B, Bron PA, de Vos WM, Smid EJ. Thioredoxin reductase is a key factor in the oxidative stress response of Lactobacillus plantarum WCFS1. Microb Cell Fact. 2007;6:29. doi: 10.1186/1475-2859-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fahey R, Brown W, Adams W, Worsham M. Occurrence of glutathione in bacteria. J Bacteriol. 1978;133:1126–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janowiak BE, Griffith OW. Glutathione synthesis in Streptococcus agalactiae. One protein accounts for gamma-glutamylcysteine synthetase and glutathione synthetase activities. J Biol Chem. 2005;280:11829–11839. doi: 10.1074/jbc.M414326200. [DOI] [PubMed] [Google Scholar]
- 32.Kullisaar T, Zilmer M, Mikelsaar M, Vihalemm T, Annuk H, Kairane C, Kilk A. Two antioxidative lactobacilli strains as promising probiotics. Int J Food Microbiol. 2002;72:215–224. doi: 10.1016/s0168-1605(01)00674-2. [DOI] [PubMed] [Google Scholar]
- 33.Kullisaar T, Songisepp E, Aunapuu M, Kilk K, Arend A, Mikelsaar M, Rehema A, Zilmer M. Complete glutathione system in probiotic Lactobacillus fermentum ME-3. Prikl Biokhim Mikrobiol. 2010;46:527–531. [PubMed] [Google Scholar]
- 34.Selle K, Klaenhammer TR. Genomic and phenotypic evidence for probiotic influences of Lactobacillus gasseri on human health. FEMS Microbiol Rev. 2013;37:915–935. doi: 10.1111/1574-6976.12021. [DOI] [PubMed] [Google Scholar]
- 35.Huang G, Pan H, Zhu Z, Li Q. The complete genome sequence of Bifidobacterium longum LTBL16, a potential probiotic strain from healthy centenarians with strong antioxidant activity. Genomics. 2020;112:769–773. doi: 10.1016/j.ygeno.2019.05.015. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Wang S, Zeng Z, Qin Y, Li P. The complete genome sequence of Bifidobacterium animalis subsp. lactis 01 and its integral components of antioxidant defense system. 3 Biotech. 2019;9:352. doi: 10.1007/s13205-019-1890-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo Q, Li S, Xie Y, Zhang Q, Liu M, Xu Z, Sun H, Yang Y. The NAD+-dependent deacetylase, Bifidobacterium longum Sir2 in response to oxidative stress by deacetylating SigH (σH) and FOXO3a in Bifidobacterium longum and HEK293T cell respectively. Free Radic Biol Med. 2017;108:929–939. doi: 10.1016/j.freeradbiomed.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 38.Zhang W, Ji H, Zhang D, Liu H, Wang S, Wang J, Wang Y. Complete genome sequencing of Lactobacillus plantarum ZLP001, a potential probiotic that enhances intestinal epithelial barrier function and defense against pathogens in pigs. Front Physiol. 2018;9:1689. doi: 10.3389/fphys.2018.01689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Odamaki T, Xiao JZ, Yonezawa S, Yaeshima T, Iwatsuki K. Improved viability of bifidobacteria in fermented milk by cocultivation with Lactococcus lactis subspecies lactis. J Dairy Sci. 2011;94:1112–1121. doi: 10.3168/jds.2010-3286. [DOI] [PubMed] [Google Scholar]
- 40.Ng EW, Yeung M, Tong P. Effects of yogurt starter cultures on the survival of Lactobacillus acidophilus. Int J Food Microbiol. 2011;145:169–175. doi: 10.1016/j.ijfoodmicro.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 41.Freire AL, Ramos CL, da Costa Souza PN, Cardoso MG, Schwan RF. Nondairy beverage produced by controlled fermentation with potential probiotic starter cultures of lactic acid bacteria and yeast. Int J Food Microbiol. 2017;248:39–46. doi: 10.1016/j.ijfoodmicro.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 42.Fu L, Kong J, Sun Z, Zhang L, Zhang X, Guo T. Enhancing the oxidative resistance of yoghurt starter bacteria with heterologous catalase expression in Streptococcus thermophilus. Int Dairy J. 2013;30:68–72. doi: 10.1016/j.idairyj.2012.11.012. [DOI] [Google Scholar]
- 43.Sasaki Y, Horiuchi H, Kawashima H, Mukai T, Yamamoto Y. NADH oxidase of Streptococcus thermophilus 1131 is required for the effective yogurt fermentation with Lactobacillus delbrueckii subsp. bulgaricus 2038. Biosci Microbiota Food Health. 2014;33:31–40. doi: 10.12938/bmfh.33.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cruz AG, Castro WF, Faria JAF, Bogusz S, Granato D, Celeguini RMS, Lima-Pallone J, Godoy HT. Glucose oxidase: A potential option to decrease the oxidative stress in stirred probiotic yogurt. LWT-Food Sci Technol. 2012;47:512–515. doi: 10.1016/j.lwt.2012.01.037. [DOI] [Google Scholar]
- 45.Cruz AG, Castro WF, Faria JA, Lollo PC, Amaya-Farfan J, Freitas MQ, Rodrigues D, Oliveira CA, Godoy HT. Probiotic yogurts manufactured with increased glucose oxidase levels: postacidification, proteolytic patterns, survival of probiotic microorganisms, production of organic acid and aroma compounds. J Dairy Sci. 2012;95:2261–2269. doi: 10.3168/jds.2011-4582. [DOI] [PubMed] [Google Scholar]
- 46.Cruz AG, Castro WF, Faria JAF, Bolini HMA, Celeghini RMS, Raices RSL, Oliveira CAF, Freitas MQ, Conte Júnior CA, Mársico ET. Stability of probiotic yogurt added with glucose oxidase in plastic materials with different permeability oxygen rates during the refrigerated storage. Food Res Int. 2013;51:723–728. doi: 10.1016/j.foodres.2013.01.028. [DOI] [Google Scholar]
- 47.Shah NP, Ding WK, Fallourd MJ, Leyer G. Improving the stability of probiotic bacteria in model fruit juices using vitamins and antioxidants. J Food Sci. 2010;75:M278–282. doi: 10.1111/j.1750-3841.2010.01628.x. [DOI] [PubMed] [Google Scholar]
- 48.Dave RI, Shah NP. Ingredient supplementation effects on viability of probiotic bacteria in yoghurt. J Dairy Sci. 1998;81:2804–2816. doi: 10.3168/jds.s0022-0302(98)75839-4. [DOI] [PubMed] [Google Scholar]
- 49.Frei B, Higdon JV. Antioxidant activity of tea polyphenols in vivo: evidence from animal studies. J Nutr. 2003;133:3275S–3284S. doi: 10.1093/jn/133.10.3275S. [DOI] [PubMed] [Google Scholar]
- 50.Gaudreau H, Champagne CP, Remondetto GE, Bazinet L, Subirade M. Effect of catechins on the growth of oxygen-sensitive probiotic bacteria. Food Res Int. 2013;53:751–757. doi: 10.1016/j.foodres.2012.10.014. [DOI] [Google Scholar]
- 51.Archibald FS, Fridovich I. Manganese, superoxide dismutase, and oxygen tolerance in some lactic acid bacteria. J Bacteriol. 1981;146:928–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watanabe M, van der Veen S, Nakajima H, Abee T. Effect of respiration and manganese on oxidative stress resistance of Lactobacillus plantarum WCFS1. Microbiology. 2012;158:293–300. doi: 10.1099/mic.0.051250-0. [DOI] [PubMed] [Google Scholar]
- 53.Lee K, Kim HJ, Rho BS, Kang SK, Choi YJ. Effect of glutathione on growth of the probiotic bacterium Lactobacillus reuteri. Biochemistry (Mosc). 2011;76:423–426. doi: 10.1134/s0006297911040043. [DOI] [PubMed] [Google Scholar]
- 54.Miller CW, Nguyen MH, Rooney M, Kailasapathy K. The influence of packaging materials on the dissolved oxygen content of probiotic yoghurt. Packag Technol Sci. 2002;15:133–138. doi: 10.1002/pts.578. [DOI] [Google Scholar]
- 55.Miller CW, Nguyen MH, Rooney M, Kailasapathy K. The control of dissolved oxygen content in probiotic yoghurts by alternative packaging materials. Packag Technol Sci. 2003;16:61–67. doi: 10.1002/pts.612. [DOI] [Google Scholar]
- 56.Rodrigues D, Sousa S, Rocha-Santos T, Silva JP, Sousa Lobo JM, Costa P, Amaral PH, Pintado MM, Gomes AM, Malcata FX, et al. Influence of l-cysteine, oxygen and relative humidity upon survival throughout storage of probiotic bacteria in whey protein-based microcapsules. Int Dairy J. 2011;21:869–876. doi: 10.1016/j.idairyj.2011.05.005. [DOI] [Google Scholar]
- 57.Talwalkar A, Kailasapathy K. Oxidative stress adaptation of probiotic bacteria. Milchwissenschaft. 2004;59:140–143. [Google Scholar]
- 58.Rochat T, Gratadoux JJ, Gruss A, Corthier G, Maguin E, Langella P. van de Guchte M. Production of a heterologous nonheme catalase by Lactobacillus casei: an efficient tool for removal of H2O2 and protection of Lactobacillus bulgaricus from oxidative stress in milk. Appl Environ Microbiol. 2006;72:5143–5149. doi: 10.1128/AEM.00482-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.An H, Zhou H, Huang Y, Wang G, Luan C, Mou J, Luo Y, Hao Y. High-level expression of heme-dependent catalase gene katA from Lactobacillus Sakei protects Lactobacillus rhamnosus from oxidative stress. Mol Biotechnol. 2010;45:155–160. doi: 10.1007/s12033-010-9254-9. [DOI] [PubMed] [Google Scholar]
- 60.Grajek W, Olejnik A, Sip A. Probiotics, prebiotics and antioxidants as functional foods. Acta Biochim Pol. 2005;52:665–671. doi: 10.18388/abp.2005_3428. [DOI] [PubMed] [Google Scholar]
- 61.Vasquez EC, Pereira TMC, Peotta VA, Baldo MP, Campos-Toimil M. Probiotics as beneficial dietary supplements to prevent and treat cardiovascular diseases: uncovering their impact on oxidative stress. Oxid Med Cell Longev. 2019;2019:3086270. doi: 10.1155/2019/3086270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song P, Zhang R, Wang X, He P, Tan L, Ma X. Dietary grape-seed procyanidins decreased postweaning diarrhea by modulating intestinal permeability and suppressing oxidative stress in rats. J Agric Food Chem. 2011;59:6227–6232. doi: 10.1021/jf200120y. [DOI] [PubMed] [Google Scholar]
- 63.Mueller K, Blum NM, Kluge H, Bauerfeind R, Froehlich J, Mader A. Effects of broccoli extract and various essential oils on intestinal and faecal microflora and on xenobiotic enzymes and the antioxidant system of piglets. OJAS. 2012;2:78–98. [Google Scholar]
- 64.Wang J, Ji HF, Wang SX, Zhang DY, Liu H, Shan DC, Wang YM. Lactobacillus plantarum ZLP001: in vitro assessment of antioxidant capacity and effect on growth performance and antioxidant status in weaning piglets. Asian-Australas J Anim Sci. 2012;25:1153–1158. doi: 10.5713/ajas.2012.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang BG, Xu HB, Xu F, Zeng ZL, Wei H. Efficacy of oral Bifidobacterium bifidum ATCC 29521 on microflora and antioxidant in mice. Can J Microbiol. 2016;62:249–262. doi: 10.1139/cjm-2015-0685. [DOI] [PubMed] [Google Scholar]
- 66.Qian Y, Zhang J, Zhou X, Yi R, Mu J, Long X, Pan Y, Zhao X, Liu W. Lactobacillus plantarum CQPC11 isolated from Sichuan pickled cabbages antagonizes d-galactose-induced oxidation and aging in mice. Molecules. 2018;23:3026. doi: 10.3390/molecules23113026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Y, Guo Y, Chen H, Wei H, Wan C. Potential of Lactobacillus plantarum ZDY2013 and Bifidobacterium bifidum WBIN03 in relieving colitis by gut microbiota, immune, and anti-oxidative stress. Can J Microbiol. 2018;64:327–337. doi: 10.1139/cjm-2017-0716. [DOI] [PubMed] [Google Scholar]
- 68.Wang Y, Wu Y, Wang Y, Xu H, Mei X, Yu D, Wang Y, Li W. Antioxidant properties of probiotic bacteria. Nutrients. 2017;9:521. doi: 10.3390/nu9050521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mishra V, Shah C, Mokashe N, Chavan R, Yadav H, Prajapati J. Probiotics as potential antioxidants: a systematic review. J Agric Food Chem. 2015;63:3615–3626. doi: 10.1021/jf506326t. [DOI] [PubMed] [Google Scholar]
- 70.Stranden I, Garrick DJ. Technical note: derivation of equivalent computing algorithms for genomic predictions and reliabilities of animal merit. J Dairy Sci. 2009;92:2971–2975. doi: 10.3168/jds.2008-1929. [DOI] [PubMed] [Google Scholar]
- 71.Wang AN, Yi XW, Yu HF, Dong B, Qiao SY. Free radical scavenging activity of Lactobacillus fermentum in vitro and its antioxidative effect on growing-finishing pigs. J Appl Microbiol. 2009;107:1140–1148. doi: 10.1111/j.1365-2672.2009.04294.x. [DOI] [PubMed] [Google Scholar]
- 72.Noureen S, Riaz A, Arshad M, Arshad N. In vitro selection and in vivo confirmation of the antioxidant ability of Lactobacillus brevis MG000874. J Appl Microbiol. 2019;126:1221–1232. doi: 10.1111/jam.14189. [DOI] [PubMed] [Google Scholar]
- 73.Li W, Ji J, Chen X, Jiang M, Rui X, Dong M. Structural elucidation and antioxidant activities of exopolysaccharides from Lactobacillus helveticus MB2-1. Carbohydr Polym. 2014;102:351–359. doi: 10.1016/j.carbpol.2013.11.053. [DOI] [PubMed] [Google Scholar]
- 74.Shen Q, Shang N, Li P. In vitro and in vivo antioxidant activity of Bifidobacterium animalis 01 isolated from centenarians. Curr Microbiol. 2011;62:1097–1103. doi: 10.1007/s00284-010-9827-7. [DOI] [PubMed] [Google Scholar]
- 75.Lin MY, Chang FJ. Antioxidative effect of intestinal bacteria Bifidobacterium longum ATCC 15708 and Lactobacillus acidophilus ATCC 4356. Dig Dis Sci. 2000;45:1617–1622. doi: 10.1023/a:1005577330695. [DOI] [PubMed] [Google Scholar]
- 76.Gavin IM, Melnik SM, Yurina NP, Khabarova MI, Bavykin SG. Zero-length protein-nucleic acid crosslinking by radical-generating coordination complexes as a probe for analysis of protein-DNA interactions in vitro and in vivo. Anal Biochem. 1998;263:26–30. doi: 10.1006/abio.1998.2827. [DOI] [PubMed] [Google Scholar]
- 77.Lee J, Hwang KT, Chung MY, Cho D, Park C. Resistance of Lactobacillus casei KCTC 3260 to reactive oxygen species (ROS): role for a metal ion chelating effect. J Food Sci. 2005;70:m388–m391. doi: 10.1111/j.1365-2621.2005.tb11524.x. [DOI] [Google Scholar]
- 78.Ahire JJ, Mokashe NU, Patil HJ, Chaudhari BL. Antioxidative potential of folate producing probiotic Lactobacillus helveticus CD6. J Food Sci Technol. 2013;50:26–34. doi: 10.1007/s13197-011-0244-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen Y, Li Q, Xia C, Yang F, Xu N, Wu Q, Hu Y, Xia L, Wang C, Zhou M. Effect of selenium supplements on the antioxidant activity and nitrite degradation of lactic acid bacteria. World J Microbiol Biotechnol. 2019;35:61. doi: 10.1007/s11274-019-2609-x. [DOI] [PubMed] [Google Scholar]
- 80.Lin MY, Yen CL. Antioxidative ability of lactic acid bacteria. J Agric Food Chem. 1999;47:1460–1466. doi: 10.1021/jf981149l. [DOI] [PubMed] [Google Scholar]
- 81.Stecchini ML, Del Torre M, Munari M. Derivation of equivalent computing algorithms for genomic predictions and reliabilities of animal merit. Int J Food Microbiol. 2001;64:183–188. doi: 10.1016/s0168-1605(00)00456-6. [DOI] [PubMed] [Google Scholar]
- 82.de LeBlanc AD, LeBlanc JG, Perdigon G, Miyoshi A, Langella P, Azevedo V, Sesma F. Oral administration of a catalase-producing Lactococcus lactis can prevent a chemically induced colon cancer in mice. J Med Microbiol. 2008;57:100–105. doi: 10.1099/jmm.0.47403-0. [DOI] [PubMed] [Google Scholar]
- 83.LeBlanc JG, Del Carmen S, Miyoshi A, Azevedo V, Sesma F, Langella P, Bermudez-Humaran LG, Watterlot L, Perdigon G, de LeBlanc AD. Use of superoxide dismutase and catalase producing lactic acid bacteria in TNBS induced Crohn’s disease in mice. J Biotechnol. 2011;151:287–293. doi: 10.1016/j.jbiotec.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 84.Mu G, Li H, Tuo Y, Gao Y, Zhang Y. Antioxidative effect of Lactobacillus plantarum Y44 on 2,2ʹ-azobis(2-methylpropionamidine) dihydrochloride (ABAP)-damaged Caco-2 cells. J Dairy Sci. 2019;102:6863–6875. doi: 10.3168/jds.2019-16447. [DOI] [PubMed] [Google Scholar]
- 85.Finamore A, Ambra R, Nobili F, Garaguso I, Raguzzini A, Serafini M. Redox role of Lactobacillus casei Shirota against the cellular damage induced by 2,2ʹ-Azobis (2-Amidinopropane) Dihydrochloride-induced oxidative and inflammatory stress in enterocytes-like epithelial cells. Front Immunol. 2018;9:1131. doi: 10.3389/fimmu.2018.01131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ejtahed HS, Mohtadi-Nia J, Homayouni-Rad A, Niafar M, Asghari-Jafarabadi M, Mofid V. Probiotic yogurt improves antioxidant status in type 2 diabetic patients. Nutrition. 2012;28:539–543. doi: 10.1016/j.nut.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 87.Zhang Y, Du R, Wang L, Zhang H. The antioxidative effects of probiotic Lactobacillus casei Zhang on the hyperlipidemic rats. Eur Food Res Technol. 2010;231:151–158. doi: 10.1007/s00217-010-1255-1. [DOI] [Google Scholar]
- 88.Gomez-Guzman M, Toral M, Romero M, Jimenez R, Galindo P, Sanchez M, Zarzuelo MJ, Olivares M, Galvez J, Duarte J. Antihypertensive effects of probiotics Lactobacillus strains in spontaneously hypertensive rats. Mol Nutr Food Res. 2015;59:2326–2336. doi: 10.1002/mnfr.201500290. [DOI] [PubMed] [Google Scholar]
- 89.Martinez-Revelles S, Avendano MS, Garcia-Redondo AB, Alvarez Y, Aguado A, Perez-Giron JV, Garcia-Redondo L, Esteban V, Redondo JM, Alonso MJ, et al. Reciprocal relationship between reactive oxygen species and cyclooxygenase-2 and vascular dysfunction in hypertension. Antioxid Redox Signal. 2013;18:51–65. doi: 10.1089/ars.2011.4335. [DOI] [PubMed] [Google Scholar]
- 90.Patel B, Kumar P, Banerjee R, Basu M, Pal A, Samanta M, Das S. Lactobacillus acidophilus attenuates Aeromonas hydrophila induced cytotoxicity in catla thymus macrophages by modulating oxidative stress and inflammation. Mol Immunol. 2016;75:69–83. doi: 10.1016/j.molimm.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 91.Zangar RC, Davydov DR, Verma S. Mechanisms that regulate production of reactive oxygen species by cytochrome P450. Toxicol Appl Pharmacol. 2004;199:316–331. doi: 10.1016/j.taap.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 92.Tan CW, Maziar S, Teo EHT, Tay BK. Microstructure and through-film electrical characteristics of vertically aligned amorphous carbon films. Diam Relat Mater. 2011;20:290–293. doi: 10.1016/j.diamond.2011.01.010. [DOI] [Google Scholar]
- 93.Jones RM, Neish AS. Redox signaling mediated by the gut microbiota. Free Radic Biol Med. 2017;105:41–47. doi: 10.1016/j.freeradbiomed.2016.10.495. [DOI] [PubMed] [Google Scholar]
- 94.Nardone G, Compare D, Liguori E, Di Mauro V, Rocco A, Barone M, Napoli A, Lapi D, Iovene MR, Colantuoni A. Protective effects of Lactobacillus paracasei F19 in a rat model of oxidative and metabolic hepatic injury. Am J Physiol Gastrointest Liver Physiol. 2010;299:G669–676. doi: 10.1152/ajpgi.00188.2010. [DOI] [PubMed] [Google Scholar]
- 95.Qiao Y, Sun J, Ding Y, Le G, Shi Y. Alterations of the gut microbiota in high-fat diet mice is strongly linked to oxidative stress. Appl Microbiol Biotechnol. 2012;97:1689–1697. doi: 10.1007/s00253-012-4323-6. [DOI] [PubMed] [Google Scholar]
- 96.Xin J, Zeng D, Wang H, Ni X, Yi D, Pan K, Jing B. Preventing non-alcoholic fatty liver disease through Lactobacillus johnsonii BS15 by attenuating inflammation and mitochondrial injury and improving gut environment in obese mice. Appl Microbiol Biotechnol. 2014;98:6817–6829. doi: 10.1007/s00253-014-5752-1. [DOI] [PubMed] [Google Scholar]
- 97.Singh CK, Chhabra G, Ndiaye MA, Garcia-Peterson LM, Mack NJ, Ahmad N. The role of sirtuins in antioxidant and redox signaling. Antioxid Redox Signal. 2018;28:643–661. doi: 10.1089/ars.2017.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bonfili L, Cecarini V, Cuccioloni M, Angeletti M, Berardi S, Scarpona S, Rossi G, Eleuteri AM. SLAB51 probiotic formulation activates sirt1 pathway promoting antioxidant and neuroprotective effects in an AD mouse model. Mol Neurobiol. 2018;55:7987–8000. doi: 10.1007/s12035-018-0973-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xu C, Qiao L, Ma L, Guo Y, Dou X, Yan S, Zhang B, Roman A. Biogenic selenium nanoparticles synthesized by Lactobacillus casei ATCC 393 alleviate intestinal epithelial barrier dysfunction caused by oxidative stress via Nrf2 signaling-mediated mitochondrial pathway. Int J Nanomedicine. 2019;14:4491–4502. doi: 10.2147/IJN.S199193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 101.Jones RM, Mercante JW, Neish AS. Reactive oxygen production induced by the gut microbiota: pharmacotherapeutic implications. Curr Med Chem. 2012;19:1519–1529. doi: 10.2174/092986712799828283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ashrafizadeh M, Ahmadi Z, Samarghandian S, Mohammadinejad R, Yaribeygi H, Sathyapalan T, Sahebkar A. MicroRNA-mediated regulation of Nrf2 signaling pathway: implications in disease therapy and protection against oxidative stress. Life Sci. 2020;244:117329. doi: 10.1016/j.lfs.2020.117329. [DOI] [PubMed] [Google Scholar]
- 103.Jones RM, Desai C, Darby TM, Luo L, Wolfarth AA, Scharer CD, Ardita CS, Reedy AR, Keebaugh ES, Neish AS. Lactobacilli modulate epithelial cytoprotection through the Nrf2 pathway. Cell Rep. 2015;12:1217–1225. doi: 10.1016/j.celrep.2015.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang Y, Wu Y, Wang Y, Fu A, Gong L, Li W, Li Y. Bacillus amyloliquefaciens SC06 alleviates the oxidative stress of IPEC-1 via modulating Nrf2/Keap1 signaling pathway and decreasing ROS production. Appl Microbiol Biotechnol. 2017;101:3015–3026. doi: 10.1007/s00253-016-8032-4. [DOI] [PubMed] [Google Scholar]
- 105.Wang LX, Liu K, Gao DW, Hao JK. Protective effects of two Lactobacillus plantarum strains in hyperlipidemic mice. World J Gastroenterol. 2013;19:3150–3156. doi: 10.3748/wjg.v19.i20.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lin X, Xia Y, Wang G, Xiong Z, Zhang H, Lai F, Ai L. Lactobacillus plantarum AR501 alleviates the oxidative stress of D-galactose-induced aging mice liver by upregulation of Nrf2-mediated antioxidant enzyme expression. J Food Sci. 2018;83:1990–1998. doi: 10.1111/1750-3841.14200. [DOI] [PubMed] [Google Scholar]
- 107.Zhao Z, Wang C, Zhang L, Zhao Y, Duan C, Zhang X, Gao L, Li S. Lactobacillus plantarum NA136 improves the non-alcoholic fatty liver disease by modulating the AMPK/Nrf2 pathway. Appl Microbiol Biotechnol. 2019;103:5843–5850. doi: 10.1007/s00253-019-09703-4. [DOI] [PubMed] [Google Scholar]
- 108.Olesen SV, Rajabi N, Svensson B, Olsen CA, Madsen AS. An NAD+-dependent sirtuin depropionylase and deacetylase (Sir2La) from the probiotic bacterium Lactobacillus acidophilus NCFM. Biochemistry. 2018;57:3903–3915. doi: 10.1021/acs.biochem.8b00306. [DOI] [PubMed] [Google Scholar]
- 109.Endo H, Niioka M, Kobayashi N, Tanaka M, Watanabe T. Butyrate-producing probiotics reduce nonalcoholic fatty liver disease progression in rats: new insight into the probiotics for the gut-liver axis. PLoS ONE. 2013;8:e63388. doi: 10.1371/journal.pone.0063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu Y, Liu Q, Hesketh J, Huang D, Gan F, Hao S, Tang S, Guo Y, Huang K. Protective effects of selenium-glutathione-enriched probiotics on CCl4-induced liver fibrosis. J Nutr Biochem. 2018;58:138–149. doi: 10.1016/j.jnutbio.2018.04.011. [DOI] [PubMed] [Google Scholar]
- 111.Seth A, Yan F, Polk DB, Rao RK. Probiotics ameliorate the hydrogen peroxide-induced epithelial barrier disruption by a PKC- and MAP kinase-dependent mechanism. Am J Physiol Gastrointest Liver Physiol. 2008;294:G1060–1069. doi: 10.1152/ajpgi.00202.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nakagawa H, Shiozaki T, Kobatake E, Hosoya T, Moriya T, Sakai F, Taru H, Miyazaki T. Effects and mechanisms of prolongevity induced by Lactobacillus gasseri SBT2055 in Caenorhabditis elegans. Aging Cell. 2016;15:227–236. doi: 10.1111/acel.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pizzo SV, Kobatake E, Nakagawa H, Seki T, Miyazaki T. Protective effects and functional mechanisms of Lactobacillus gasseri SBT2055 against oxidative stress. Plos ONE. 2017;12:e0177106. doi: 10.1371/journal.pone.0177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gopalakrishna R, Chen ZH, Gundimeda U. Modification of cysteine-rich regions in protein kinase C induced by oxidant tumor promoters and the enzyme specific inhibitors. Methods Enzymol. 1995;252:132–146. doi: 10.1016/0076-6879(95)52016-3. [DOI] [PubMed] [Google Scholar]
- 115.Gopalakrishna R, Jaken S. Protein kinase C signaling and oxidative stress. Free Radic Biol Med. 2000;28:1349–1361. doi: 10.1016/s0891-5849(00)00221-5. [DOI] [PubMed] [Google Scholar]
- 116.Blumberg PM. Complexities of the protein kinase c pathway. Mol Carcinog. 1991;4:339–344. doi: 10.1002/mc.2940040502. [DOI] [PubMed] [Google Scholar]
- 117.Stabel S, Parker PJ. Protein kinase C. Pharmacol Therapeut. 1991;51:71–95. doi: 10.1016/0163-7258(91)90042-k. [DOI] [PubMed] [Google Scholar]
- 118.Farhadi A, Keshavarzian A, Ranjbaran Z, Fields JZ, Banan A. The role of protein kinase c isoforms in modulating injury and repair of the intestinal barrier. J Pharmacol Exp Ther. 2006;316:1–7. doi: 10.1124/jpet.105.085449. [DOI] [PubMed] [Google Scholar]
- 119.Banan A, Fields JZ, Talmage DA, Zhang L, Keshavarzian A. PKC-zeta is required in EGF protection of microtubules and intestinal barrier integrity against oxidant injury. Am J Physiol Gastrointest Liver Physiol. 2002;282:G794–808. doi: 10.1152/ajpgi.00284.2001. [DOI] [PubMed] [Google Scholar]
- 120.Banan A, Zhang L, Fields JZ, Farhadi A, Talmage DA, Keshavarzian A. PKC-zeta prevents oxidant-induced iNOS upregulation and protects the microtubules and gut barrier integrity. Am J Physiol Gastrointest Liver Physiol. 2002;283:G909–922. doi: 10.1152/ajpgi.00143.2002. [DOI] [PubMed] [Google Scholar]
- 121.Zhou YK, Qin HL, Zhang M, Shen TY, Chen HQ, Ma YL, Chu ZX, Zhang P, Liu ZH. Effects of Lactobacillus plantarum on gut barrier function in experimental obstructive jaundice. World J Gastroenterol. 2012;18:3977–3991. doi: 10.3748/wjg.v18.i30.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bowen WH, Koo H. Biology of Streptococcus Mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 2011;45:69–86. doi: 10.1159/000324598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Villena J, Kitazawa H. Probiotic microorganisms: A closer look. Microorganisms. 2017;5:7. doi: 10.3390/microorganisms5020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhou X, Qi W, Hong T, Xiong T, Gong D, Xie M, Nie S. Exopolysaccharides from Lactobacillus plantarum NCU116 regulate intestinal barrier function via STAT3 signaling pathway. J Agric Food Chem. 2018;66:9719–9727. doi: 10.1021/acs.jafc.8b03340. [DOI] [PubMed] [Google Scholar]
- 125.Lebeer S, Claes IJJ, Verhoeven TLA, Vanderleyden J, De Keersmaecker SCJ. Exopolysaccharides of Lactobacillus rhamnosus GG form a protective shield against innate immune factors in the intestine. Microb Biotechnol. 2011;4:368–374. doi: 10.1111/j.1751-7915.2010.00199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kanmani P, Albarracin L, Kobayashi H, Iida H, Komatsu R, AKM HK, Ikeda-Ohtsubo W, Suda Y, Aso H, Makino S, et al. Exopolysaccharides from Lactobacillus delbrueckii OLL1073R-1 modulate innate antiviral immune response in porcine intestinal epithelial cells. Mol Immunol. 2018;93:253–265. doi: 10.1016/j.molimm.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 127.Polak-Berecka M, Wasko A, Szwajgier D, Chomaz A. Bifidogenic and antioxidant activity of exopolysaccharides produced by Lactobacillus rhamnosus E/N cultivated on different carbon sources. Pol J Microbiol. 2013;62:181–188. [PubMed] [Google Scholar]
- 128.Yan M, Wang BH, Xu X, der Meister T, Tabgammaac HT, Hwang FF, Liu Z. Extrusion of dissolved oxygen by exopolysaccharide from Leuconostoc mesenteroides and its implications in relief of the oxygen stress. Front Microbiol. 2018;9:2467. doi: 10.3389/fmicb.2018.02467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yan M, Han J, Xu X, Liu L, Gao C, Zheng H, Chen Y, Tao Y, Zhou H, Li Y, et al. Gsy, a novel glucansucrase from Leuconostoc mesenteroides, mediates the formation of cell aggregates in response to oxidative stress. Sci Rep. 2016;6:38122. doi: 10.1038/srep38122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Trabelsi I, Ktari N, Ben Slima S, Triki M, Bardaa S, Mnif H, Ben Salah R. Evaluation of dermal wound healing activity and in vitro antibacterial and antioxidant activities of a new exopolysaccharide produced by Lactobacillus sp.Ca6. Int J Biol Macromol. 2017;103:194–201. doi: 10.1016/j.ijbiomac.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 131.Rani RP, Anandharaj M, Ravindran AD. Characterization of a novel exopolysaccharide produced by Lactobacillus gasseri FR4 and demonstration of its in vitro biological properties. Int J Biol Macromol. 2018;109:772–783. doi: 10.1016/j.ijbiomac.2017.11.062. [DOI] [PubMed] [Google Scholar]
- 132.Li JY, Jin MM, Meng J, Gao SM, Lu RR. Exopolysaccharide from Lactobacillus planterum LP6: antioxidation and the effect on oxidative stress. Carbohydr Polym. 2013;98:1147–1152. doi: 10.1016/j.carbpol.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 133.Zhang L, Liu C, Li D, Zhao Y, Zhang X, Zeng X, Yang Z, Li S. Antioxidant activity of an exopolysaccharide isolated from Lactobacillus plantarum C88. Int J Biol Macromol. 2013;54:270–275. doi: 10.1016/j.ijbiomac.2012.12.037. [DOI] [PubMed] [Google Scholar]
- 134.Pan D, Mei X. Antioxidant activity of an exopolysaccharide purified from Lactococcus lactis subsp. lactis 12. Carbohyd Polym. 2010;80:908–914. doi: 10.1016/j.carbpol.2010.01.005. [DOI] [Google Scholar]
- 135.Guo Y, Pan D, Li H, Sun Y, Zeng X, Yan B. Antioxidant and immunomodulatory activity of selenium exopolysaccharide produced by Lactococcus lactis subsp. lactis. Food Chem. 2013;138:84–89. doi: 10.1016/j.foodchem.2012.10.029. [DOI] [PubMed] [Google Scholar]
- 136.Ruas-Madiedo P, Medrano M, Salazar N, De Los Reyes-Gavilan CG, Perez PF, Abraham AG. Exopolysaccharides produced by Lactobacillus and Bifidobacterium strains abrogate in vitro the cytotoxic effect of bacterial toxins on eukaryotic cells. J Appl Microbiol. 2010;109:2079–2086. doi: 10.1111/j.1365-2672.2010.04839.x. [DOI] [PubMed] [Google Scholar]
- 137.Salazar N, Ruas-Madiedo P, Kolida S, Collins M, Rastall R, Gibson G, de Los Reyes-gavilan CG. Exopolysaccharides produced by Bifidobacterium longum IPLA E44 and Bifidobacterium animalis subsp. lactis IPLA R1 modify the composition and metabolic activity of human faecal microbiota in pH-controlled batch cultures. Int J Food Microbiol. 2009;135:260–267. doi: 10.1016/j.ijfoodmicro.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 138.Ma L, Wang J, Wang S, Anderson EM, Lam JS, Parsek MR, Wozniak DJ. Synthesis of multiple Pseudomonas aeruginosa biofilm matrix exopolysaccharides is post-transcriptionally regulated. Environ Microbiol. 2012;14:1995–2005. doi: 10.1111/j.1462-2920.2012.02753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bafana A. Characterization and optimization of production of exopolysaccharide from Chlamydomonas reinhardtii. Carbohydr Polym. 2013;95:746–752. doi: 10.1016/j.carbpol.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 140.Steiger S, Perez-Fons L, Fraser PD, Sandmann G. Biosynthesis of a novel C30 carotenoid in Bacillus firmus isolates. J Appl Microbiol. 2012;113:888–895. doi: 10.1111/j.1365-2672.2012.05377.x. [DOI] [PubMed] [Google Scholar]
- 141.Kim M, Seo DH, Park YS, Cha IT, Seo MJ. Isolation of Lactobacillus plantarum subsp. plantarum producing C30 carotenoid 4,4ʹ-diaponeurosporene and the assessment of its antioxidant activity. J Microbiol Biotechnol. 2019;29:1925–1930. doi: 10.4014/jmb.1909.09007. [DOI] [PubMed] [Google Scholar]
- 142.Hagi T, Kobayashi M, Nomura M. Aerobic condition increases carotenoid production associated with oxidative stress tolerance in Enterococcus gilvus. FEMS Microbiol Lett. 2014;350:223–230. doi: 10.1111/1574-6968.12341. [DOI] [PubMed] [Google Scholar]
- 143.Hagi T, Kobayashi M, Nomura M. Aerobic conditions increase isoprenoid biosynthesis pathway gene expression levels for carotenoid production in Enterococcus gilvus. FEMS Microbiol Lett. 2015;362:fnv075. doi: 10.1093/femsle/fnv075. [DOI] [PubMed] [Google Scholar]
- 144.Ohki S, Hagi T, Nakano K, Shiroma A, Tamotsu H, Shimoji M, Shinzato M, Ashimine N, Minami M, Nakanishi T, et al. Complete genome sequence of carotenoid-producing Enterococcus gilvus CR1, isolated from raw cow’s milk. Microbiol Resour Announc. 2018;7:e00988–00918. doi: 10.1128/MRA.00988-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rice-Evans CA, Miller NJ, Paganga G. Structure-antioxidant activity relationships of flavonoids and phenolic acids. Free Radical Bio Med. 1996;20:933–956. doi: 10.1016/0891-5849(95)02227-9. [DOI] [PubMed] [Google Scholar]
- 146.Bhathena J, Kulamarva A, Urbanska AM, Martoni C, Prakash S. Microencapsulated bacterial cells can be used to produce the enzyme feruloyl esterase: preparation and in-vitro analysis. Appl Microbiol Biotechnol. 2007;75:1023–1029. doi: 10.1007/s00253-007-0908-x. [DOI] [PubMed] [Google Scholar]
- 147.Bhathena J, Kulamarva A, Martoni C, Urbanska AM, Prakash S. Preparation and in vitro analysis of microencapsulated live Lactobacillus fermentum 11976 for augmentation of feruloyl esterase in the gastrointestinal tract. Biotechnol Appl Biochem. 2008;50:1–9. doi: 10.1042/BA20070007. [DOI] [PubMed] [Google Scholar]
- 148.Tomaro-Duchesneau C, Saha S, Malhotra M, Coussa-Charley M, Al-Salami H, Jones ML, Labbé A, Prakash S. Lactobacillus fermentum NCIMB 5221 has a greater ferulic acid production compared to other ferulic acid esterase producing Lactobacilli. Int J Probiotics Prebiotics. 2012;7:23–32. [Google Scholar]
- 149.Abeijón Mukdsi MC, Gauffin Cano MP, González SN, Medina RB. Administration of Lactobacillus fermentum CRL1446 increases intestinal feruloyl esterase activity in mice. Lett Appl Microbiol. 2012;54:18–25. doi: 10.1111/j.1472-765X.2011.03166.x. [DOI] [PubMed] [Google Scholar]
- 150.Tomaro-Duchesneau C, Saha S, Malhotra M, Jones ML, Labbe A, Rodes L, Kahouli I, Prakash S. Effect of orally administered L. fermentum NCIMB 5221 on markers of metabolic syndrome: an in vivo analysis using ZDF rats. Appl Microbiol Biotechnol. 2014;98:115–126. doi: 10.1007/s00253-013-5252-8. [DOI] [PubMed] [Google Scholar]
- 151.Adisakwattana S, Moonsan P, Yibchok-Anun S. Insulin-releasing properties of a series of cinnamic acid derivatives in vitro and in vivo. J Agric Food Chem. 2008;56:7838–7844. doi: 10.1021/jf801208t. [DOI] [PubMed] [Google Scholar]
- 152.Thomas CM, Hong T, van Pijkeren JP, Hemarajata P, Trinh DV, Hu W, Britton RA, Kalkum M, Versalovic J. Histamine derived from probiotic Lactobacillus reuteri suppresses TNF via modulation of PKA and ERK signaling. PLoS ONE. 2012;7:e31951. doi: 10.1371/journal.pone.0031951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Azuma Y, Shinohara M, Wang P-L, Hidaka A, Ohura K. Histamine inhibits chemotaxis, phagocytosis, superoxide anion production, and the production of TNFα and IL-12 by macrophages via H2-receptors. Int Immunopharmacol. 2001;1:1867–1875. doi: 10.1016/s1567-5769(01)00112-6. [DOI] [PubMed] [Google Scholar]
- 154.Greifová G, Body P, Greif G, Greifová M, Dubničková M. Human phagocytic cell response to histamine derived from potential probiotic strains of Lactobacillus reuteri. Immunobiology. 2018;223:618–626. doi: 10.1016/j.imbio.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 155.Fardet A, Rock E. In vitro and in vivo antioxidant potential of milks, yoghurts, fermented milks and cheeses: a narrative review of evidence. Nutr Res Rev. 2018;31:52–70. doi: 10.1017/S0954422417000191. [DOI] [PubMed] [Google Scholar]
- 156.Carocho M, Ferreira ICFR. A review on antioxidants, prooxidants and related controversy: natural and synthetic compounds, screening and analysis methodologies and future perspectives. Food Cheml Toxicol. 2013;51:15–25. doi: 10.1016/j.fct.2012.09.021. [DOI] [PubMed] [Google Scholar]
- 157.Kikuchi HE, Suzuki T. Quantitative method for measurement of aerotolerance of bacteria and its application to oral indigenous anaerobes. Appl Environ Microbiol. 1986;52:971–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Li Q, Chen Q, Ruan H, Zhu D, He G. Isolation and characterisation of an oxygen, acid and bile resistant Bifidobacterium animalis subsp. lactis Qq08. J Sci Food Agric. 2010;90:1340–1346. doi: 10.1002/jsfa.3942. [DOI] [PubMed] [Google Scholar]
- 159.Yang X, Li L, Duan Y, Yang X. Antioxidant activity of JM113 in vitro and its protective effect on broiler chickens challenged with deoxynivalenol. J Anim Sci. 2017;95:837–846. doi: 10.2527/jas2016.0789. [DOI] [PubMed] [Google Scholar]
- 160.Mu G, Gao Y, Tuo Y, Li H, Zhang Y, Qian F, Jiang S. Assessing and comparing antioxidant activities of lactobacilli strains by using different chemical and cellular antioxidant methods. J Dairy Sci. 2018;101:10792–10806. doi: 10.3168/jds.2018-14989. [DOI] [PubMed] [Google Scholar]
- 161.Cappa F, Cattivelli D, Cocconcelli PS. The uvrA gene is involved in oxidative and acid stress responses in Lactobacillus helveticus CNBL1156. Res Microbiol. 2005;156:1039–1047. doi: 10.1016/j.resmic.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 162.Zolotukhin PV, Prazdnova EV, Chistyakov VA. Methods to assess the antioxidative properties of probiotics. Probiotics Antimicrob Proteins. 2018;10:589–599. doi: 10.1007/s12602-017-9375-6. [DOI] [PubMed] [Google Scholar]
- 163.Harikrishnan R, Balasundaram C, Heo MS. Effect of probiotics enriched diet on Paralichthys olivaceus infected with lymphocystis disease virus (LCDV). Fish Shellfish Immunol. 2010;29:868–874. doi: 10.1016/j.fsi.2010.07.031. [DOI] [PubMed] [Google Scholar]
- 164.Saide JAO, Gilliland SE. Antioxidative activity of Lactobacilli measured by oxygen radical absorbance capacity. J Dairy Sci. 2005;88:1352–1357. doi: 10.3168/jds.S0022-0302(05)72801-0. [DOI] [PubMed] [Google Scholar]
- 165.Ou CC, Lu TM, Tsai JJ, Yen JH, Chen HW, Lin MY. Antioxidative effect of lactic acid bacteria: intact cells vs. Intracellular Extracts. J Food Drug Anal. 2009;17:209–216. [Google Scholar]
- 166.Virtanen T, Pihlanto A, Akkanen S, Korhonen H. Development of antioxidant activity in milk whey during fermentation with lactic acid bacteria. J Appl Microbiol. 2007;102:106–115. doi: 10.1111/j.1365-2672.2006.03072.x. [DOI] [PubMed] [Google Scholar]
- 167.Gao D, Gao Z, Zhu G. Antioxidant effects of Lactobacillus plantarum via activation of transcription factor Nrf2. Food Funct. 2013;4:982–989. doi: 10.1039/c3fo30316k. [DOI] [PubMed] [Google Scholar]
- 168.Li B, Du P, Smith EE, Wang S, Jiao Y, Guo L, Huo G, Liu F. In vitro and in vivo evaluation of an exopolysaccharide produced by Lactobacillus helveticus KLDS1.8701 for the alleviative effect on oxidative stress. Food Funct. 2019;10:1707–1717. doi: 10.1039/c8fo01920g. [DOI] [PubMed] [Google Scholar]
- 169.Rashid SK, Idris-Khodja N, Auger C, Alhosin M, Boehm N, Oswald-Mammosser M, Schini-Kerth VB. Probiotics (VSL#3) prevent endothelial dysfunction in rats with portal hypertension: role of the angiotensin system. PLoS ONE. 2014;9:e97458. doi: 10.1371/journal.pone.0097458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Balcazar JL, de Blas I, Ruiz-Zarzuela I, Vendrell D, Girones O, Muzquiz JL. Enhancement of the immune response and protection induced by probiotic lactic acid bacteria against furunculosis in rainbow trout (Oncorhynchus mykiss). FEMS Immunol Med Microbiol. 2007;51:185–193. doi: 10.1111/j.1574-695X.2007.00294.x. [DOI] [PubMed] [Google Scholar]
- 171.Rajoka MSR, Mehwish HM, Hayat HF, Hussain N, Sarwar S, Aslam H, Nadeem A, Shi J. Characterization, the antioxidant and antimicrobial activity of exopolysaccharide isolated from poultry origin Lactobacilli. Probiotics Antimicrob Proteins. 2019;11:1132–1142. doi: 10.1007/s12602-018-9494-8. [DOI] [PubMed] [Google Scholar]
- 172.Abdhul K, Ganesh M, Shanmughapriya S, Kanagavel M, Anbarasu K, Natarajaseenivasan K. Antioxidant activity of exopolysaccharide from probiotic strain Enterococcus faecium (BDU7) from Ngari. Int J Biol Macromol. 2014;70:450–454. doi: 10.1016/j.ijbiomac.2014.07.026. [DOI] [PubMed] [Google Scholar]
- 173.Jia K, Tao X, Liu Z, Zhan H, He W, Zhang Z, Zeng Z, Wei H. Characterization of novel exopolysaccharide of Enterococcus faecium WEFA23 from infant and demonstration of its in vitro biological properties. Int J Biol Macromol. 2019;128:710–717. doi: 10.1016/j.ijbiomac.2018.12.245. [DOI] [PubMed] [Google Scholar]
- 174.Li S, Zhao Y, Zhang L, Zhang X, Huang L, Li D, Niu C, Yang Z, Wang Q. Antioxidant activity of Lactobacillus plantarum strains isolated from traditional Chinese fermented foods. Food Chem. 2012;135:1914–1919. doi: 10.1016/j.foodchem.2012.06.048. [DOI] [PubMed] [Google Scholar]
- 175.Arasu MV, Al-Dhabi NA. In vitro antifungal, probiotic, and antioxidant functional properties of a novel Lactobacillus paraplantarum isolated from fermented dates in Saudi Arabia. J Sci Food Agric. 2017;97:5287–5295. doi: 10.1002/jsfa.8413. [DOI] [PubMed] [Google Scholar]
- 176.Amiri S, Rezaei Mokarram R, Sowti Khiabani M, Rezazadeh Bari M, Alizadeh Khaledabad M. Exopolysaccharides production by Lactobacillus acidophilus LA5 and Bifidobacterium animalis subsp. lactis BB12: optimization of fermentation variables and characterization of structure and bioactivities. Int J Biol Macromol. 2019;123:752–765. doi: 10.1016/j.ijbiomac.2018.11.084. [DOI] [PubMed] [Google Scholar]
- 177.Taylan O, Yilmaz MT, Dertli E. Partial characterization of a levan type exopolysaccharide (EPS) produced by Leuconostoc mesenteroides showing immunostimulatory and antioxidant activities. Int J Biol Macromol. 2019;136:436–444. doi: 10.1016/j.ijbiomac.2019.06.078. [DOI] [PubMed] [Google Scholar]
- 178.Persichetti E, De Michele A, Codini M, Traina G. Antioxidative capacity of Lactobacillus fermentum LF31 evaluated in vitro by oxygen radical absorbance capacity assay. Nutrition. 2014;30:936–938. doi: 10.1016/j.nut.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 179.Rodríguez C, Cofré JV, Sánchez M, Fernández P, Boggiano G, Castro E. Lactobacilli isolated from vaginal vault of dairy and meat cows during progesteronic stage of estrous cycle. Anaerobe. 2011;17:15–18. doi: 10.1016/j.anaerobe.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 180.Wang Y, Du R, Qiao X, Zhao B, Zhou Z, Han Y. Optimization and characterization of exopolysaccharides with a highly branched structure extracted from Leuconostoc citreum B-2. Int J Biol Macromol. 2020;142:73–84. doi: 10.1016/j.ijbiomac.2019.09.071. [DOI] [PubMed] [Google Scholar]
- 181.Cuevas-González PF, Aguilar-Toalá JE, García HS, González-Córdova AF, Vallejo-Cordoba B, Hernández-Mendoza A. Protective effect of the intracellular content from potential probiotic bacteria against oxidative damage induced by acrylamide in human erythrocytes. Probiotics Antimicrob Proteins. 2020. January 22. doi: 10.1007/s12602-020-09636-9 [DOI] [PubMed] [Google Scholar]
- 182.Halliwell B, Chirico S. Lipid peroxidation: its mechanism, measurement, and significance. Am J Clin Nutr. 1993;57:715S–724S. doi: 10.1093/ajcn/57.5.715S. [DOI] [PubMed] [Google Scholar]
- 183.Lin MY, Yen CL. Inhibition of lipid peroxidation by Lactobacillus acidophilus and Bifidobacterium longum. J Agric Food Chem. 1999;47:3661–3664. doi: 10.1021/jf981235l. [DOI] [PubMed] [Google Scholar]
- 184.Czerska M, Mikołajewska K, Zieliński M, Gromadzińska J, Wąsowicz W. Today’s oxidative stress markers. Med Pr. 2015;66:393–405. doi: 10.13075/mp.5893.00137. [DOI] [PubMed] [Google Scholar]
- 185.Sun J, Hu XL, Le GW, Shi YH. Inhibition of Fe-induced colon oxidative stress by lactobacilli in mice. World J Microbiol Biotechnol. 2013;29:209–216. doi: 10.1007/s11274-012-1172-5. [DOI] [PubMed] [Google Scholar]
- 186.Kullisaar T, Songisepp E, Mikelsaar M, Zilmer K, Vihalemm T, Zilmer M. Antioxidative probiotic fermented goats’ milk decreases oxidative stress-mediated atherogenicity in human subjects. Br J Nutr. 2007;90:449–456. doi: 10.1079/bjn2003896. [DOI] [PubMed] [Google Scholar]
- 187.Broccali G, Berti M, Pistolesi E, Cestaro B. Study of the effect of lactobacillus GG supplementation in combination with and without arginine aspartate on lipoproteins and liver peroxidation in cholesterol-fed rats. Int J Food Sci Nutr. 2000;51:475–482. doi: 10.1080/09637480050208080. [DOI] [PubMed] [Google Scholar]
- 188.Li Y, Hou S, Peng W, Lin Q, Chen F, Yang L, Li F, Huang X. Oral administration of Lactobacillus delbrueckii during the suckling phase improves antioxidant activities and immune responses after the weaning event in a piglet model. Oxid Med Cell Longev. 2019;2019:6919803. doi: 10.1155/2019/6919803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kaushal D, Kansal VK. Probiotic Dahi containing Lactobacillus acidophilus and Bifidobacterium bifidum alleviates age-inflicted oxidative stress and improves expression of biomarkers of ageing in mice. Mol Biol Rep. 2012;39:1791–1799. doi: 10.1007/s11033-011-0920-1. [DOI] [PubMed] [Google Scholar]
- 190.Parrella A, Caterino E, Cangiano M, Criscuolo E, Russo C, Lavorgna M, Isidori M. Antioxidant properties of different milk fermented with lactic acid bacteria and yeast. Int J Food Sci Tech. 2012;47:2493–2502. doi: 10.1111/j.1365-2621.2012.03127.x. [DOI] [Google Scholar]
- 191.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 192.Nowak A, Czyżowska A, Stańczyk M. Protective activity of probiotic bacteria against 2-amino-3-methyl-3H-imidazo[4,5-f]quinoline (IQ) and 2-amino-1-methyl-6-phenyl-1H-imidazo[4,5-b]pyridine (PhIP) – an in vitro study. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2015;32:1927–1938. doi: 10.1080/19440049.2015.1084651. [DOI] [PubMed] [Google Scholar]
- 193.de Oliveira Coelho B, Fiorda-Mello F, de Melo Pereira GV, Thomaz-Soccol V, Rakshit SK, de Carvalho JC, Soccol CR. In Vitro probiotic properties and DNA protection activity of yeast and lactic acid bacteria isolated from a honey-based kefir beverage. Foods. 2019;8:485. doi: 10.3390/foods8100485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Yamamoto N, Shoji M, Hoshigami H, Watanabe K, Watanabe K, Takatsuzu T, Yasuda S, Igoshi K, Kinoshita H. Antioxidant capacity of soymilk yogurt and exopolysaccharides produced by lactic acid bacteria. Biosci Microbiota Food Health. 2019;38:97–104. doi: 10.12938/bmfh.18-017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Chang JH, Shim YY, Cha SK, Chee KM. Probiotic characteristics of lactic acid bacteria isolated from kimchi. J Appl Microbiol. 2010;109:220–230. doi: 10.1111/j.1365-2672.2009.04648.x. [DOI] [PubMed] [Google Scholar]
- 196.Fiorda FA, de Melo Pereira GV, Thomaz-Soccol V, Medeiros AP, Rakshit SK, Soccol CR. Development of kefir-based probiotic beverages with DNA protection and antioxidant activities using soybean hydrolyzed extract, colostrum and honey. LWT-Food Sci Technol. 2016;68:690–697. doi: 10.1016/j.lwt.2016.01.003. [DOI] [Google Scholar]
- 197.Turner A, Wilson G, Kaube I. Biosensors: fundamentals and applications. Oxford (UK): Oxford University Press; 1987. [Google Scholar]
- 198.Bănică F-G. Chemical sensors and biosensors: fundamentals and applications. Hoboken (NJ): John Wiley & Sons; 2012. [Google Scholar]
- 199.Su L, Jia W, Hou C, Lei Y. Microbial biosensors: A review. Biosens Bioelectron. 2011;26:1788–1799. doi: 10.1016/j.bios.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 200.Marsova M, Abilev S, Poluektova E, Danilenko V. A bioluminescent test system reveals valuable antioxidant properties of lactobacillus strains from human microbiota. World J Microbiol Biotechnol. 2018;34:27. doi: 10.1007/s11274-018-2410-2. [DOI] [PubMed] [Google Scholar]
- 201.Ge Q, Ge P, Jiang D, Du N, Chen J, Yuan L, Yu H, Xu X, Wu M, Zhang W, et al. A novel and simple cell-based electrochemical biosensor for evaluating the antioxidant capacity of Lactobacillus plantarum strains isolated from Chinese dry-cured ham. Biosens Bioelectron. 2018;99:555–563. doi: 10.1016/j.bios.2017.08.037. [DOI] [PubMed] [Google Scholar]


