ABSTRACT
It is well established that pig gut microbiota plays a critical role in maintaining metabolic homeostasis as well as in a myriad of physiological, neurological and immunological functions; including protection from pathogens and digestion of food materials – some of which would be otherwise indigestible by the pig. A rich and diverse gut microbial ecosystem (balanced microbiota) is the hallmark of good health; while qualitative and quantitative perturbations in the microbial composition can lead to development of various diseases. Alternatively, diseases caused by stressors or other factors have been shown to negatively impact the microbiota. This review focuses primarily on how commensal microorganisms in the gastrointestinal tract of pigs influence biochemical, physiological, immunological, and metabolic processes within the host animal.
KEYWORDS: Gut, host-microbe interactions, immunity, microbiome, pigs
Introduction
‘Microbiota’ is defined as all microbes, including their genomes and extra-chromosomal elements, present in and on the host animal; and their interactions within the gastrointestinal tract (GIT), skin, and genital environments.1,2 The terms “microbiota” (the microbial taxa associated with host) and “microbiome” (the catalog of these microbes and their genes) are often used interchangeably.3 The organisms comprising the gut microbiota reside outside the mucosal layer and play a role in triggering the host immune response and in communication between the gut and brain.4,5 The GIT is not only the largest interface between the external and internal environments of animals, but it also contains the largest amount, and the greatest diversity of microorganisms. The GIT microbiota is defined as an ecological community made up of commensal, symbiotic and pathogenic microorganisms, including bacteria, viruses, parasites, fungi, archaea and protists, that inhabit the mammalian gut.6,7 Such a mammalian GIT is estimated to host approximately 1014 bacterial organisms comprising 500–1000 unique species, which form a synergistic relationship with the host.8-10 Co-evolution of gut microorganisms with their hosts has led to the acquisition of microbial roles in digestion, nutrient utilization, toxin removal, protection against pathogens and regulation of the endocrine and immune systems.11-13 Hence, the abnormal intestinal functions observed in germ-free (GF) animals have been attributed to the absence of these essential GIT microbes.14
A healthy intestinal microbial community is diverse, stable, resistant (to minor changes) and resilient.15 Human and mouse studies have shown that dysbiosis, which consists of disequilibrium of the microbial community in the GIT microbiota is associated with the development of several acute and chronic inflammatory conditions, bowel diseases, metabolic syndromes and diabetes.16
As hosts mature, the organisms comprising the microbiota begin to sense their environment through toll-like receptors (TLR) and short chain fatty acid (SCFA) receptors on the gut epithelial surface enabling them to detect incoming nutrients and microbial components. Specifically, they sense structural molecules such as lipopolysaccharides (LPS), flagellins, and peptidoglycans; metabolites such as SCFAs and microbial secretions such as toxins or polyphosphate chains.17,18
In the animal husbandry sector, immense potential has been described for manipulating the GIT microbiota to improve nutritional and immunological activities within the host to boost livestock productivity. However, the GIT microbiota has not yet been fully explored in many of the prominent livestock species. The study of pigs has great potential to inform human research due to the many similarities identified in the physiological attributes of these two species. Moreover, pigs exhibit similar susceptibilities and clinical manifestations to pathogens that are the etiological agents in certain human intestinal disorders.19,20 Xiao et al.21 reported that although the homology between human and pig microbiomes was quite modest at the gene level, it was significant at the level of KEGG orthology functions. This same study identified more similarity between human and pig microbiomes than between human and mice microbiomes; and reported that approximately 96% of the functional pathways described in the human gut microbiome are common to the pig as well.21 Further, it has been shown that humans share more similarities with pigs in terms of anatomy, genetics, physiology, pharmaceutical bioavailability and nutrient digestibility than with rodents.7 Hence, the pig is a superior model to rodents for studying human physiology and pathology as it pertains to enteric health.
However, it has not yet been determined qualitatively or quantitatively what constitutes a healthy microbial community in the GIT of pigs or other mammals. It is, therefore, imperative to conduct statistically powerful studies to characterize the microbiome diversity in pigs and to determine how this diversity can be utilized to improve the performance and health of pigs. Toward this end, a study recently reported that the genome comprising the microbiome of pigs, at various stages of development, contained more than twice the number of genes as that of the actual pig genome.22
In this review, we examine how changes in the GIT microbiota may influence biochemical, physiological, immunological, and metabolic processes within the host and vice versa. Most of the microbiota studies in farm animals have focused on the effects that treatments with antibiotics, prebiotics, probiotics and feed additives have on the animals; while fundamental studies focusing on how the host-microbe relationships affect the physiology and immunology of farm animals, are scarce.
Gastrointestinal tract
The GIT is a functionally and anatomically diverse organ comprising the mouth, esophagus, stomach, small intestine, large intestine and anus.23 Culture-independent techniques have revealed that the GIT contains a dynamic microbial population with unique organisms located in the different organ sections,24,25 with the most diverse group of microbes inhabiting the colon in pigs.26 Specifically, within the colon of healthy adult pigs, the taxonomical composition of the bacterial community has been described as 35% Firmicutes, 21% Bacteroidetes, 3% Proteobacteria and 2% Spirochetes.27-29 In the jejunum and ileum, Proteobacteria account for about 70% of the microbes, followed by Firmicutes which are about 20%. In contrast, in the cecum and colon, the Firmicutes dominate with >75% and Proteobacteria are about 13%.24,30 Nevertheless, high microbial diversity identified within the small intestine is thought to minimize the negative effects of certain pathogenic bacterial strains that are often found within this organ. The microbiota colonizes the mucosal entry sites of pathogens, where it occupies biological niches and prevents invasion by foreign pathogens – known as colonization resistance.4
Brain-gut-microbiome axis
Several studies have shown that GIT microbiota contributes significantly to maintenance of normal physiological and metabolic functioning of mammalian hosts (Figure 1). Furthermore, the gut microbiota has been shown to contribute to neurophysiological regulation, which subsequently governs neurotransmission, cognition, and behavior. This is achieved through regulation of the immune and endocrine systems via release of bacterial metabolites.31 Although the brain-gut–microbiome axis has not yet been thoroughly examined in pigs, through analysis of this system in other mammalian species, we have hypothesized that this axis would also play a key role in pigs. Microbes in the gut communicate with the central nervous system through at least 3 parallel and interacting channels, which involve mechanisms encompassing the nervous system, endocrine system and immune signaling. The brain can affect the community structure and function of the gut microbiota via the autonomic nervous system, by modulation of local gut motility, intestinal transit and secretion, and gut permeability. It is thought to be accomplished through the luminal secretion of hormones that directly modulate microbial gene expression.32 Through release of metabolites, the gut microbiota communicates with a network of neuronal, glial, endocrine, and immune cells.33 Microbiota and their metabolites have, therefore, been described as associated with the modulation of behavior and brain processes, including emotional behavior, brain biochemistry, responses to stress and pain, and GIT functioning.34 The latter is due to changes in intestinal permeability, mucosal immune functioning, and activity of the enteric nervous system.
Figure 1.

A systems biology model for the brain-gut–microbiome interactions in mammals. The interconnected structural networks of the central nervous system influence via the autonomous nervous system to alter microbiota composition and function indirectly by regulating the microbial environment in the gut. The brain communicates with the gut microbiota indirectly through gut-derived molecules via afferent vagal and spinal nerve endings, or directly through microbe-generated signals. Alterations in these bidirectional interactions in response to perturbations like diet, medication, infections, and stress can alter the stability and behavior of this system resulting in brain-gut disorders.
A recent systems biology model postulated that circular communication loops exist between the brain and the gut microbiome, and that perturbations to any of these factors can lead to dysregulation of the circuit.32 Many studies, primarily in rats and mice, have implicated alterations in the brain-gut–microbiome axis in the pathogenesis and pathophysiology of irritable bowel syndrome (IBS), obesity, as well as several psychiatric and neurological disorders.32 Similar studies relating to the brain-gut–microbiome in pigs have not yet been reported.
Age, birth, and breed-related microbiota
Birth
The structure and composition of the gut microbiota in animals are determined by many factors (Figure 2), such as genetics, age, phylogeny, diet, and surrounding environmental conditions during birth.35,36 For example, in mammals, the initial exposure to microbes occurs at parturition in the birth canal. The mode of delivery, vaginal or cesarean section (CS), along with the nutrition provided during early stages of life, have a significant influence on the intestinal microbiota. Furthermore, a recent study on the microbial composition of the umbilical cord found that maternal transfer is possible and that it may occur during gestation. Additionally, pigs born vaginally have a higher bacterial density and higher concentration of SCFAs including acetate, propionate, and butyrate, compared to pigs born via CS.37 Moreover, a study examined the effects of inoculating CS-born piglets with either a placebo containing simple-composition microbiota, or complex fecal microbiota from adult sows, which would mimic the microbial environment acquired through vaginal deliveries. The results showed that the adult pigs inoculated with the placebo contained a less diverse fecal microbiota compared to piglets inoculated with complex microbiota.38 Further, the placebo pigs exhibited poor health and transient diarrhea. These results suggest that the adult pig microbial community is primarily influenced by the microbes that the piglet encounters during the earliest stages of its life. Pre-Labor CSs were seen to decrease bacterial diversity and density in piglets compared to normal vaginal delivery.39 Also, within the livers of piglets delivered via CS, lower expression of interferon (IFN), NKp80, and C-reactive protein (CRP) was observed.40 Microbial colonization, therefore, begins at birth and continues to diversify in the initial days of life based on exposure to environmental microbiota, which depends on the host habitat, diet, and physiology.41
Figure 2.

Factors affecting the gut microbiota in pigs. The diversity of gut microflora can be affected by a variety of factors. Sub-therapeutic doses of antibiotics used in commercial farming can negatively affect the microbiota. Similarly, different stressors such as high temperature, transportation, weaning, and overcrowding can also change the diversity of the microbiota for the worse. The inclusion of probiotics, prebiotics, and fiber appear to nullify these effects and improve diversity. Other factors, such as the age of the animal, its mode of the birth, breed and the environment it lives in, can influence the microbiota. Potentially pathogenic microbes are depicted in red and beneficial (and other commensals) microbes in green. The red arrows indicate a negative impact and a green arrow a positive impact. The gray arrows are an indication that different factors can affect the microbiota differently, either positively (e.g., if animals are bred in a healthy, growth-conducive environment) or negatively (e.g., following cesarean birth, devoid of any natural mother’s microbiota).
Breed
The prevalence of Firmicutes and Bacteroidetes within the fecal bacterial community varies between certain pig breeds. Specifically, in Chinese Jinhua pigs 70.4% of the fecal bacterial population is composed of Firmicutes whereas 14.4% are Bacteroidetes.42 Alternatively, western breeds such as the Duroc, Yorkshire, and Landrace contain 39.6%, 42.0%, and 45.6% Firmicutes and 57.0%, 51.4%, and 47.6% Bacteroidetes, respectively.43,44 The Firmicutes/Bacteroidetes ratio changes with increase in age45 and has an impact on the breakdown of polysaccharides, nutrient absorption, gut permeability, and inflammatory response.46 Bacteroidetes have been shown to participate in carbohydrate degradation, yet, in pigs, the proportion of bacterial species belonging to this phylum decreases with age, causing subsequent weight gain.47 Further, Prevotella spp. have been shown to account for 26% of the bacterial content in the feces of 10-week-old piglets, but only 4% in 22-week-old pigs. The fecal microbial composition continuously changes until the animal reaches 6 months of age, at which point it is seen to stabilize.45 However, as pigs move through their life, it is not only their age that changes but also the composition of their feed, weaning practices and the mixing patterns in their pens.48 Hence, the relationship between the growth of pigs and the diversity of the intestinal microbiota remains complex.
Age
The density and species diversity of the gut microbial population in the different compartments of the GIT are in constant flux as pigs develop.30,49 The composition of the microbiota also becomes increasingly diverse with progression through the pig GI tract.50,51 Lactobacilli, Bifidobacterium, Streptococcus, Bacteroides, Clostridium perfringes, and Escherichia coli are the major taxa identified in the pig GIT; however, the specific makeup changes with age.52 The earliest colonizers of the pig gut, between birth and 2 days, are primarily of the genera Escherichia, Clostridium, Fusobacterium, Streptococcus, and Enterococcus. It is further estimated that 34% of the total microbial population present at 6h of age is from the family Clostridiaceae, which is seen to reduce to 1% by 20 days, while Enterobacteriaceae are not detected during these early days.53 Rather, a steady increase in Enterobacteriaceae occurs from weaning (approximately 28 days) to 5 days post-weaning; however, they are seen to significantly decline after day 11 post-weaning.54 Thus, the microbial community may differ between siblings in the first two days following birth but begins to stabilize by day 28. Moreover, by day 36, substantial similarities are observed in the intestinal microbial community of cohabiting non-sibling piglets, however, not necessarily between siblings if separated from the sow within 3 days of birth.41 In the first 5 days after birth, the microbial community is dominated by strict aerobes and facultative anaerobes, which are gradually replaced almost entirely by strict anaerobes (starting from day 7 up to day 22).55 The first significant change in intestinal microbial diversity occurs in piglets on days 4–7 when the number of Clostridium perfringens organisms declines due to the activity of IgA inherited from the mother. The development of major immune system induction elements occurs approximately 2 weeks after birth, and by 4 weeks significant concentrations of sIgA are evident.55 Hence, microbial intestinal colonization affects susceptibility and tolerance to not only intestinal pathogens but also to systemic infectious and noninfectious diseases.
Genetics
It is well established that host genetics and the lean/obese nature of a particular breed of pig play an important role in the GIT microbiome/metabolome profiles. When maintained on the same diet, significant differences have been observed in Meihua piglets (fatty-type, slow-growing Chinese breed) and Landrace piglets (lean-type fast-growing European breed) in the production of SCFAs and secondary bile acids, including deoxycholic acid and lithocholic acid, all of which are naturally occurring within the GIT lumen, however, at high concentrations can cause oxidative/nitrosative stress, DNA damage, apoptosis, and mutation.56 Specifically, accumulation of SCFAs and secondary bile acids was found to be higher in the colon lumen of Landrace piglets.57 Moreover, a comparative study showed that Jinhua pigs exhibited better growth performance, lower diarrhea rates, and lower immune activation in response to challenge with an enterotoxigenic E. coli (ETEC) K88 species compared to Landrace pigs. The Landrace pigs also had a higher overall proportion of Lactobacilli spp., as well as a higher ratio of Lactobacilli to E. coli, and more tight junction proteins.58 The Lactobacillus, Bacteroides, Prevotella, and Ruminococcus species were found to increase in abundance throughout the colonization process; however, the exact proportions were dependent on the pig breed.59 Additionally, when different purebred pigs were cohabitated for several weeks, their gut microbial communities shared more similarities yet retained the distinguishable breed specific proportions.43 It has also been reported that the abundance of the methanogenic anaerobe, Methanobrevibacter smithii, significantly increased over the first 14 days of life in two pig breeds, namely, Meishan (obese) and Yorkshire (lean).60 However, the level in the lean breed was found to be significantly higher than that in the obese pigs. Methanogens such as M. smithii help to remove hydrogen and carbon dioxide, forming methane, thereby preventing the accumulation of hydrogen in the gut and the subsequent decrease in microbial fermentation efficiency and energy yield. Hence, methanogens are an important component of the gut microbial community.61 In a study by Guo et al.62 on Bama mini pigs, it was found that obese pigs contained approximately 61% fewer Bacteroidetes and approximately 56% fewer Bacteroides spp. than lean pigs. Hence, the authors concluded that elevated proportions of Bacteroidetes and Bacteroides species negatively impacted body weight.50,51
The housing conditions, rearing density, environmental temperature and time of sampling can also all significantly affect the intestinal gut microbiota diversity.63 For example, the intestinal microbiota diversity has been reported to change 3 times from birth to after weaning in young piglets.55
Effect of diet on the gut microbiome
The diet of the sow affects the piglet microbiota and the fermentation end-products profile.64 Diet significantly impacts gut microbial diversity and is extremely important in maintaining health by preventing the development of dysbiosis, an etiological factor in many chronic diseases.65 For dietary nutrients to be efficiently metabolized, a population of healthy GIT microbes is highly important, as this can lead to improved digestion and efficient absorption/utilization of nutrients via the pig gut mucosal membrane. When the GIT immune system, which accounts for approximately 70% of the total immune cell population, is activated in response to a stressor, a diverse set of specialized immune cells and signaling molecules are produced, sometimes at the expense of digestive efficiency.66 A recent study67 found that Ruminococcaceae spp., which produce SCFAs and Lactobacillus spp., which produce lactic acid, have an important role in suppressing swine feed intake, whereas Prevotella may have the opposite effect. These authors,67 therefore, postulated that Prevotella may be the keystone bacteria for porcine appetite control. These results suggest that the pig intestinal microbial community may contribute a vital role to the host’s feeding behavior; and thus, modulation of gut microbiota can be beneficial for the control of feed intake in the swine industry.67 Interestingly, a recent study showed that the largest change in bacterial composition occurs in pigs that are between 21 and 33 days of age, which is the period of time that the animal transitions from a primarily milk-based diet to one containing solid feed. These results were consistent across all examined GIT sites, namely, duodenum, ileum, cecum, and colon.68
Dietary fiber (DF)
Dietary fiber is a feed constituent that cannot be digested efficiently by monogastric digestive enzymes. However, it selectively stimulates the growth and activity of one or more bacteria within the GIT, resulting in microbiota-fermented DF in the distal aspect of the colon. The main products of such bacterial fermentation are short chain organic acids (SCOA) such as lactate, acetate, propionate, and butyrate. These SCOA assist in the development of the digestive tract by influencing gut epithelial cell proliferation.69 Further, the acidic properties associated with SCOA act to impede the growth of enteric bacterial pathogens such as Salmonella, E. coli, and Clostridia.70 Additionally, soluble non-starch polysaccharides stimulate the growth of commensal gut microorganisms, which increase SCOA production, thereby lowering the pH in the colon.71 Conversely, the inclusion of insoluble non-starch polysaccharides such as pectin, cellulose, gums, and hemicelluloses in the diet can serve to increase the villus length and delay GIT transit time, thereby allowing a longer period for degradation of fibrous material by microbiota in the colon.72 Lindberg, in 2014, having reviewed different references,71,73,74 suggested that the various types of plant carbohydrates behave differently in the GIT depending on their structural characteristics. Inclusion of soluble NSP (non-starch polysaccharides) in the diet can stimulate the growth of commensal gut microbes, leading to increased production of short chain organic acids (OA), and a lower pH in the large intestine. Insoluble NSP reduce the transit time and provide substrate that is slowly degradable by the microbiota in the distal large intestine and modulate gut morphology by increasing villus length.72
It is well established that lactobacilli supplementation in neonates aids in the early development of stable gut microflora, stimulates the immune system, and prevents diarrhea.52 Recently, it was also reported that by including xylanase in pig diet, the fecal and ileal counts of beneficial lactobacilli could be increased while simultaneously reducing the E. coli counts.75 The GIT microbial community adapts to variations in the host animal’s diet although the host diet also influences the distribution of microbiota within the GIT.76 The gut microbiota is an environmental regulator of fat storage and adiposity77. Bacteroidetes contain fewer genes for enzymes involved in carbohydrate and lipid metabolism, as compared to Firmicutes. Hence, the abundance of Firmicutes is higher is obese animals.77 Thus, reduction in their numbers following the feeding of DF consolidates our understanding of the positive effects that these fibers exhibit in controlling obesity in higher mammals such as the pig and humans. In another study, when pigs were fed ‘low fat and high fiber’ (LF) or ‘high fat and low fiber’ (HF) diets, the gene copy numbers for Lactobacilli spp., Bifidobacterium and Faecalibacterium prausnitzii were observed to be higher in LF-diet-fed pigs, while HF-diet-fed pigs contained more Enterobacteriaceae.78 The LF diet containing higher amounts of DF was able to stimulate the growth of beneficial bacteria in the microbiota and increase the production of SCFAs, especially butyrate. In contrast, the HF diet increased the number of potentially pathogenic organisms. Spurlock and Gabler (2008) presented a review of literature wherein swine were used as a model to study human obesity. Some breeds of swine such as the Ossabaw breed from the United States of America readily become obese in the absence of high-fiber diets.79
Dietary copper
In the swine industry, feed is commonly supplemented with copper (Cu) because of its antimicrobial properties and potential to promote growth. However, the nutritional requirement for Cu to swine varies from 5 mg/kg feed in piglets to 20 mg/kg in lactating sows.80 However, when weaned piglets were fed 175 mg/kg CuSO4, the populations of lactic acid bacteria, Lactobacilli and Streptococci in the GIT were reduced.81 Such high amounts of Cu in the feed can also function to increase the content of unsaturated fatty acids, which can result in softer pork fat82. High levels of dietary zinc (Zn) and Cu can also serve to decrease the commonly observed spike in plasma cortisol levels on day 9 and 19 when pigs are subjected to an LPS challenge. Further, high concentrations of dietary Zn, and particularly Cu, have been shown to significantly reduce the diversity of ileal microbiota. However, this effect was reversible, which suggests that microbiota diversity was restored following the removal of additional Zn and Cu from the diet.83 Enterococci has been shown to develop resistance to antibiotics such as macrolides and glycopeptides, including vancomycin, following exposure to high Cu concentrations. Such resistant enterococci, which are a part of the Lactobacillales order and are quite frequently found in the gut microbiota of mammals including pigs, may get transferred to humans that consume the meat of such animals.84
Prebiotics and probiotics
Probiotics (group of microorganisms which ‘when administered in adequate amounts, confer a health benefit on the host’85) can correct the imbalance of microbiota in the GIT and improve the overall health of humans and animals. Introduction of such beneficial microbes can serve to repair and reinforce the numbers of commensal microorganisms within the gut to restore or improve animal resistance to diseases. Simultaneously, probiotics also improve the efficacy of nutrient digestion, absorption, and utilization with subsequent improvement in production performance.86 However, the positive effects of probiotics in animals are strain-dependent.87 One study reported that in weaned pigs, the lactobacilli counts in the GIT increased while Clostridia, E. coli, and Enterobacterium spp. counts decreased, following administration of probiotic therapy.88
Lactobacillus, a component of the Firmicutes phylum, is a Gram-positive, facultative anaerobe or microaerophilic bacterium that improves feed conversion efficiency in animals. Furthermore, lactobacilli produce lactic acid, which elicits an inhibitory effect against E. coli and Enterobacteria.89 Administration of a cocktail of complex lactobacilli containing Lactobacillus johnsonii and L. mucosae, previously isolated from healthy pig feces was shown to promote a healthy gut by reducing the number of potential entero-pathogens such as Clostridia and E. coli.90 Similar effects were observed in weaned piglets administered lactic-acid bacteria (LAB) complexes containing Enterococcus faecium 6H2, Lactobacillus acidophilus C3, Pediococcus pentosaceus D7, L. plantarum 1K8 and L. plantarum 3K2.91 Moreover, the administration of L. salivarius UCC118 WT was found to significantly decrease the number of Spirochetes in the GIT of pigs. Moreover, administration of L. salivarius UCC118, which is well studied for its probiotic properties, positively influenced Firmicutes genus members, while production of bacteriocin Abp118 by L. salivarius affected gram-negative microorganisms, even though Abp118 is not normally active in vitro against this group of microorganisms. Hence, this strain has the potential to significantly affect pig microbiota through a partial bacteriocin-dependent mechanism.92 Lactobacillus reuteri is also a probiotic strain that has been shown to alter the abundance of several bacterial taxa, such as Enterobacteriaceae including E. coli. This lactobacilli strain, which produces reutericyclin, increases the abundance of two strict anaerobes of phylum Firmicutes, while production of reuteran affects colonization with ETEC without affecting other dominant members of the fecal microbiota.93
Supplementation of weaning pig diets with probiotics functions to compete with pathogenic bacteria for nutrition resulting in competitive exclusion of the harmful bacterial strains.94 The inclusion of specific probiotics, namely, L. casei ssp. casei, L. reuteri and L. acidophilus, during the suckling period and fortification of the piglet diet with probiotics and prebiotics during the post-weaning period serves to markedly improve growth rate and body weight gain.95 Higher counts of Lactobacillus spp. and lower E. coli counts in feces were also observed in these animals. Further, in pigs, the diversity of anaerobic bacteria was found to increase from day 13 to day 16 after birth, with detection of dominant anaerobes such as Eubacterium, Fusobacterium and Propionibacterium.55 This was attributed to the introduction of milk replacer from day 14 onwards. The same study also found changes in the intestinal microbiota after the introduction of the weaning diet beginning 35 days after birth.
Prebiotics (compounds found within foods which can induce the growth/activity of beneficial micro-organisms96), have been shown to promote the growth of specific groups of commensal gastrointestinal microbiota. Numerous metabolites are subsequently produced by these microorganisms, of which the SCFAs are transported across the epithelium by diffusion, low-affinity transport mechanism such as HCO3−/SCFA exchange, medium-affinity transport mechanism involving monocarboxylate transporter 1 [MCT1], or via high-affinity transport mediated by sodium-coupled monocarboxylate transporter 1 (SMCT1 or SLC5A8) into the colon97.There is also increased production of interleukin following supplementation of pig diets with prebiotics alone98,99 or in combination with probiotics, known as synbiotics.100,101 Further, lactulose (a prebiotic) supplementation of the feed of weaned piglets orally challenged with S. enterica subspecies enterica serovar Typhimurium served to improve the immunoglobulin IgG antibody responses as well as the total serum IgM and IgA levels.102 Lactose, which is a major sugar present in milk, acts as a prebiotic and can elicit the development of a highly diverse microbiota in the prenatal GIT of growing animals.103 Although the mechanisms by which lactulose and other prebiotics affect the immune system are not fully understood, it is postulated that they may act indirectly by altering the indigenous microbiota of the GIT and causing changes in microbial metabolite production.104
Numerous previous studies have identified Lactobacillus as one of the core genera in the GIT of pigs, accounting for approximately 15% of 16S rRNA gene sequences from swine intestinal samples, irrespective of age.105 Lactobacilli occur in both the proximal and distal regions of the swine digestive tract, and begin colonizing soon after birth.106 Improvement in overall health, growth performance and an increase in the productivity of swine husbandry are some of the key benefits of administration of probiotic lactobacilli to pigs.107 The LAB are also capable of suppressing microorganisms that are lethal to the host’s health. The lactic acid-related trophic chain in LAB, is one of the major metabolic pathways in the mammalian gut.108,109 Increased abundance of Lactobacillus spp. in the cecum of pigs directly correlates with high feed efficiency.110 Recent research has also shown that certain strains of lactobacilli, namely L. reuteri ZLR003 and L. salivarius ZLS006, can increase the average daily weight gain, feed conversion ratio and nitrogen digestibility in growing pigs. They also help to significantly reduce the total cholesterol, alanine transferase, aspartate transferase, blood urea nitrogen, and haptoglobin levels in serum.111
As has been suggested by several studies, administration of lactobacilli to pigs improves meat quality. Administration of L. plantarum ZJ316, which is a potential probiotic isolated from fecal samples of piglets, to newly weaned pigs had promising results. Most notably, it served to improve several meat texture indices, promoted increased villus height and also appeared to inhibit the growth of opportunistic pathogens.112 Moreover, the administration of probiotics containing L. amylovorus into post-weaning pigs has been shown to increase monosaturated and polyunsaturated fatty acids in muscles, suggesting potential usefulness of probiotic administration in improving the fatty acid profile of pig meat.113 In addition, analysis of the immune health-promoting properties elicited by L. jensenii TL2937 illustrated that the use of immunobiotic strains as supplemental additives to piglet feed significantly reduced tenderness while improving juiciness and palatability of pork meat, along with reducing backfat thickness.114 In pigs fed a diet high in calcium-phosphorus content, a 1.4-fold increase in lactobacilli was observed in the gastric pars nonglandularis of the stomach.115
Due to a high level of variation in growth and feed conversion between individual pigs in commercial production systems, it is difficult to accurately measure the impact of probiotics on gut health. To fully elucidate the effect of different variables, large-scale experiments are required; however, to date most of the studies have focused on assessing the effects that feed additives have on representatives of GIT health, including many immunological measures, in more controlled experiments (Table 1).116
Table 1.
Microbiome outcomes in studies examining pigs exposed to dietary and environmental modulations.
| Experiment | Outcome | References |
|---|---|---|
| Piglets removed from the sow and reared on bovine-based milk formula | A higher concentration of desirable microbiomes evident in littermates reared with the sow Differences in mucosal immune system components (rapid recruitment of antigen presenting cells [APCs], fewer Treg cells, increased antibody responses) |
117,118 |
| Pigs reared indoors and outdoors | Differences in the microbiome between the groups Differential gene expressions of MCH-dependent antigen-presentation in intestinal mucosa |
119-121 |
Table 1 highlights pig studies in which the diet and environmental conditions were manipulated to determine their correlation with changes in the gut microbiota, and subsequent effects on the immune response.
Antibiotics
Pig feed and water on commercial farms are often supplemented with antibiotics to combat bacterial infections or promote growth. Although administration of antibiotics promote piglet growth, it has a negative effect on the commensal bacterial population as it often leads to increased proportions of pathogenic species that function to inhibit the normal intestinal function.122 Specifically, antibiotics such as penicillin, tylosin, sulfamethazine, and chlortetracycline have been shown to affect the composition of the gut microbiome in growing pigs.24,25,27,123 Moreover, simultaneous administration of multiple antibiotics, namely, chlortetracycline, sulfamethazine, and penicillin (ASP250), served to markedly increased the proportion of E. coli in the lumen and mucosa of the ileum compared to other gut compartments and feces in pigs.25 Many of the functional changes within the metagenome were also attributed to an increase in E. coli.25 Additionally, a decrease in the number of LAB Streptococcus organisms, and a simultaneous increase in Proteobacteria, specifically in the Escherichia population, was observed following administration of ASP250 antibiotics to weaned piglets.27,124 An additional study reported that short-term administration of low-dose antibiotics in feed caused an increase in the abundance and diversity of antibiotic-resistance genes specific for antibiotics that the animals had not previously been exposed to.29
Further, treatment with amoxicillin (600 mg/kg) was found to increase the abundance of fecal enterobacteria, while decreasing the proportion of LAB and the total bacterial viability as well as the total serum IgM concentrations within the jejunum.125 Additionally, Gao et al.126 showed that therapeutic antibiotic administration alters the composition and metabolism of the microbial communities within the ileum and feces. However, the ileal microbiota was found to be more susceptible to change than that of fecal microbiota. Specifically, Lactobacillus and Bifidobacterium spp. were found to decrease by an average of 3-fold and 508-fold respectively, in the ileum on days 2 and 13, and by an average of 45-fold and 72-fold, respectively, in the feces on days 7 and 13. Moreover, the proportion of Escherichia and Shigella spp. were found to increase by 265-fold in the ileum between days 2 and 13, and by 36-fold in feces between days 7 and 13.126 This study also suggested that changes in microbiota are closely associated with changes in production of specific microbial metabolites such as SCFAs, which can be used as biomarkers for determining the stability of the gut microbial community. The levels of total SCFA including acetate, propionate, butyrate, and valerate extracted from feces are regarded as effective indicators of intestinal health. In a recent study, pigs fed conventional diets (which included three types of antibiotics) contained 87 more antibiotic-resistant genes in the GIT compared to pigs fed organic diets, although the gut microbiota of both sets of pigs was not significantly different.127 Antibiotic-resistance virulence factors were identified in gene families unique to the swine fecal metagenome, exhibiting highest sequence homology to genes in Bacteroidetes, Clostridia, and Methanosarcina.28
Due to excessive antibiotic usage in pig production, an increase in the development of immune tolerance has been noted during the early stages of life against a range of pathogenic microbial species. Moreover, antibiotics have been shown to suppress the systemic immune response in mice.128 Development of immune tolerance early in life can lead to inefficient immune response later in life when similar pathogens are encountered.129
Gut microbiome and intestinal physiology
Animal gut microbiota influences many physiological functions necessary for the maintenance of a healthy GIT. Within the GIT lumen, the microbiota assists in converting bile acids into secondary forms via de-hydroxylation, dehydrogenation, and deconjugation.130 The gut microbiota is involved in digestion of otherwise indigestible carbohydrates to produce SCFAs, which protect against epithelial injury, as well as in the synthesis of essential amino acids, regulation of fat metabolism, induction of intestinal motility, improvement in intestinal angiogenesis, and regulation of immune system activation.131,132 The gut microbiota also serves to protect against colonization by pathogenic bacteria through the production of anti-microbial compounds, while also protecting the gut epithelial barrier from harmful effects of pathogens, thereby controlling the overgrowth of bacteria, and simultaneously reducing the susceptibility of pigs to enteric infections133.
Impact of the host on gut microbiome
The relationship between the microbiome and its mammalian host is one of the longest surviving symbioses, dating back to the beginning of multicellular life.35 The evolution of the microbiota within the host is driven by the need for each species to compete and survive within the host; natural selection alone will not make the microbiota useful to the host.134 At the same time, hosts, under natural selection, appear to select for organisms beneficial to them, and hence, the microbiota can be considered an ecosystem held on an ever-evolving leash by the host. The microbes are predictably controlled by the host, as there is a single host but many microbes. Thus, the host can influence the entire microbiome more readily while simultaneously benefiting from its components. Evolutionary theory has predicted that host-to-microbe effects are of larger importance for the microbiome form and function. The host exerts control over the microbiota through immigration, compartmentalization, monitoring and targeting. This has been previously covered in detail by Foster et al.135 However, a recent study has also suggested that bacterial biodiversity within the pig GIT may be influenced by the genetics of the host animal.22 The researchers, Lu et al., used paternal half-sib families, thus each family represented a breeding male pig that was mated with several female pigs to produce the offspring. The significant variation in alpha diversity observed among the families suggests bacterial biodiversity within the pig gut might be influenced by the host’s genetics.22
Stressors
The composition of the GIT microbiota changes when the host animal encounters stressful stimuli. Two-day-old piglets host a group of bacteria comprising primarily L. amylovorus, L. reuteri, E. coli, and L. acidophilus. Specifically, ileal samples of neonates and non-weaned pigs contain approximately 7 × 108 L. amylovorus and L. reuteri cells per gram of intestinal content.108
Weaning
At the weaning stage, the dietary changes constitute a major stressor and cause changes to the gut microbiota. Due to this weaning stress, the lactobacilli community of the pig ileum undergoes significant change.136 After weaning, the quantities of L. amylovorus and L. reuteri decrease significantly to less than 103 within the ileum; and thereafter Clostridia and E. coli appear along with changes in the composition and metabolic activities of the predominant microbiota.
In a study of porcine fecal microbiota, samples from 15 commercial pigs were collected during the pre-weaning and post-weaning periods.137 The pre-weaning microbial community consisted primarily of the phyla Firmicutes (54%) > Bacteroidetes (38.7%) > Proteobacteria (4.2%) > Spirochetes (0.7%) > Tenericutes (0.2%). Although the same major phyla prevailed post-weaning, the relative proportions varied, with Bacteroidetes (59.6%) > Firmicutes (35.8%) > Spirochetes (2.0%) > Proteobacteria (1%) and Tenericutes (1%). Thus, Firmicutes and Bacteroidetes accounted for more than 90% of the fecal bacterial community during both the pre-weaning and post-weaning stages. However, although Firmicutes accounted for the initial prominent phyla, a shift toward Bacteroidetes was observed after weaning. Among the genera, Bacteroides, Blautia, Dorea, Escherichia, and Fusobacterium were determined to be most abundant pre-weaning; however, Prevotella and Clostridia increased in the post-weaning pig with a corresponding decrease in Bacteroides.138
Mechanistically, during weaning, the piglet diet switches from easily digestible liquid milk to a less easily digestible, more complex solid feed. This change has significant consequences on the microbiota and the physiology of the GIT, which is still not fully mature. Other changes such as inflammatory response pathways are activated at this time in addition to hormonal changes, gastric motility reduction, small intestine atrophy, reduced height of villi, reduced absorption of nutrients, fluids, and electrolytes, and increased permeability to antigens and toxins.94 It is possible that at least some, or all of the above changes are linked to modifications in piglet intestinal microbiota since it has long been established that dietary change is responsible for the etiology of post-weaning diarrhea and enteric infections.139
Other stresses
In addition, growth of pathogenic E. coli occurs in pigs subjected to even mild handling stress140. The effect of stress on the microbiome is now recognized as a new field of study, called microbial endocrinology.141 These authors hypothesized a mechanism involving modulation of the transcription of virulence genes in a pathogen specifically via blocking with adrenergic antagonists.141 A key cascade of reactions occurs in the hypothalamic-pituitary-adrenal (HPA) axis in response to stress conditions resulting in release of glucocorticoids from the adrenal cortex.142 For example, when GF mice are subjected to restraint stress they exhibit elevated adrenocorticotropic hormone and corticosterone levels.143
Interestingly, this process can be largely reversed by just one commensal bacterium, namely, Bifidobacterium infantis.144 A crucial observation in this study was that the reversal of the HPA axis set-point was influenced by these commensal bacteria even in adults, however, this occurred only if the colonization had taken place before the host reached 6 weeks of age and not in animals where colonization had occurred after 14 weeks of age. It therefore appears that early life signals elicited by indigenous bacteria seem to exercise a long-lasting programming effect on the HPA axis to enable the host responses to better cope with stressful situations in later life.
Infection and inflammation
Salmonella enterica is a pathogen that can induce substantial changes in the composition of the intestinal microbiota. For instance, disturbances in the porcine colon and cecal microbiota occur when challenged with S. enterica.145 The microbiota profiles in the S. enterica-challenged pigs were similar to each other yet varied markedly from the non-challenged controls. Statistically significant increases were observed in proportions of Anaerobacter, Prevotella, Barnesiella, Pediococcus, Sporacetigenium, Turicibacter, Catenibacterium, Xylanibacter and Pseudobutyrivibrio in the challenged pigs. Furthermore, in mice studies, inflammation has been shown to be induced in response to bacterial infection by species such as Citrobacter rodentium or S. enterica subspecies enterica serovar Typhimurium, or by chemical inducers such as dextran sulfate sodium (DSS, or in response to genetic deficiencies such as in the interleukin-10-deficient (IL-10−/-) mouse model.146-148 These factors function to change the composition of the intestinal microbiota by reducing both the quantity and diversity of resident intestinal bacteria. Similarly, the Enterobacteriaceae count has been shown to increase in mice following the induction of colitis by treatment with DSS.149 Enteric infections caused by pathogens such as the porcine epidemic diarrhea viruses, Brachyspira hampsonii and Lawsonia intracellularis also influence the gut microbial composition and cause dysbiosis.145,150,151 These viruses cause substantial reduction in the pig microbiota diversity to one dominated by the bacterial phylum Fusobacteria. In contrast, control pigs that were not exposed to the virus exhibited a rich microbial diversity with Firmicutes in the majority.150
Impact of the gut microbiome on the host immune system
The host animal is subjected to many internal and external stresses during its lifetime. In pigs, defects in genes encoding various innate and adaptive immune cells result in dysbiosis and can induce development of pathogenic disorders of the GIT such as IBD.152 Further, external pressures including infections or exposure to antibiotics can introduce major disturbances to the microbiota, as is observed in IBD development,153 which is now believed to occur as the result of disruption in communication between the host and intestinal microbiota. However, the molecular mechanism responsible for this communication breakdown is not fully understood. Nevertheless, it is has been hypothesized to involve genetic susceptibility of the epithelial barrier and innate immunity, both of which are vital components in the host–microbiota relationship.154
Many approaches have been examined, including the use of GF animals, to demonstrate the critical link between gut microbiota and the host innate and acquired immune system. Since GF animals are reared in sterile conditions from birth and are not exposed to microbes during their life, changes that occur in the body on exposure to microorganisms from the external environment can be accurately monitored. The microbiota is known to influence not only the local intestinal immune system, but also systemic immunity.155,156
Gut bacteria monitor and regulate the immune system in a way that allows the immune system to distinguish between commensal microbes and pathogenic bacteria. A healthy GIT results from positive interactions between the microbiome and host. In this context, the epithelial barrier function and the mucosal immune system are vital components.157,158 The innate immune system and the gut are interdependent and therefore can be influenced by a system of interactions.159 The innate immune system relays signals to the host animal for functional adaption at the tissue level including influencing the composition and functional capabilities of the microbiota.160 It also acts to promote the growth of beneficial species to help maintain a stable community of microbes. Further, during an intestinal infection, fucosylated proteins are shed into the lumen of the intestine which serve as an energy source for the GIT microbiota.161 Thus, the innate immune system diverts its resources to aid the microbiota in times of perturbations of the intestinal ecosystem. As an example, in Yersinia enterocolitica infection, signaling from TLR1 is necessary to preserve the composition of the commensal microbial community.162
Antigen presenting cells (APCs) help protect the host against infections, while simultaneously maintaining immune tolerance to the commensal gut microbiota. The dendritic cells (DCs) of Peyer’s patches in the gut wall generate high levels of IL-10 compared to DCs in the spleen when subjected to similar conditions.163 Moreover, in GF animals, a reduction in the number of intestinal DCs is observed, however, not in splenic DCs. Escherichia coli (107 CFU of O83:K24: H31 E. coli and O86:K24: H31 E. coli) alone are sufficient to elicit a DC response in the GIT. In addition, fewer intestinal and systemic macrophages are observed in GF pigs resulting in reduced chemotaxis, phagocytosis, and microbiocidal activities.164,165
Commensal bacteria function to regulate the immune response in host cells, primarily through the inflammatory cascade via the nuclear factor-kappa B (NF-κB) pathway.166 NF-κB regulates transcription by translocating to the nucleus and stimulating the production of inflammatory cytokines and recruitment of immune cells. This occurs only when NF-κB is unbound from IκB (inhibitor of κB). However, resident bacteria in the gut inhibit the NF-κB–IκB dissociation thereby halting the cascade since NF-κB is no longer able to enter the nucleus.166 Thus, commensal microbes and their products may be useful in therapeutics for inflammatory-based diseases such as IBD.
It is well-established that gut commensal microbes, upon colonizing the neonatal mammals, activate the systemic immune system. This is achieved primarily by increasing the number of circulating antimicrobial specific antibodies.167 Since the food ingested by animals contains intact molecules that can retain their antimicrobial activity even after irradiation or autoclaving, it is difficult to generate antigen-naïve pigs. Moreover, the formation and development of the mucosal immune system in GF animals is very limited as compared to that in conventional animals that possess hypoplastic Peyer’s patches. GF animals also lack Treg cells, and express minimal levels of heat shock proteins (HSPs).168 The intraepithelial T lymphocytes in the GIT play a vital role in the defense system of the host. As compared to conventional pigs, GF pigs only have a fraction (approximately 35%) of the normal level of T-lymphocytes in the jejunum and ileum.55 When the balance between host immunity and microbiota is disrupted, dysbiosis is created, which is a vital step in the progression of diseases such as diarrhea and swine dysentery.169 However, when GF pigs are colonized with even a limited, defined microbiota, most of the functional immune system components including APCs, T-cells, and B-cells develop similar to that in conventional pigs.170,171
Additionally, different strains of lactobacilli possess varied capacities to modulate the expression of host immune pathways. The presence of many lactobacilli species can lead to greater cross-talk between these microorganisms and the host immune cells. For proper development and function of the immune system to occur, communication between the microbiota and intestinal cells is pivotal. In piglets challenged with E. coli K88ac, one probiotic strain, L. fermentum, enhanced T-cell differentiation, increased pro-inflammatory cytokines as well as the proportion of CD4+ lymphocytes in the ileum.172,173 Several studies have utilized different Lactobacillus spp. and strains in pigs to demonstrate their effects on the intestinal microbial communities and their beneficial activities following ETEC, Salmonella or rotavirus challenges. The specific Lactobacillus spp. studied include L. plantarum, L. amylovorus DSM 16698, and L. reuteri or Lactobacillus rhamnosus GG. The strain L. amylovorus DSM 16698 was employed in an experiment conducted on pig intestinal explants, where ETEC induced a higher level of TLR4, P-IKKα, P-IκBα, and P-p65; while L. amylovorus functioned to eliminate all these variations and simultaneously upregulated the expression of TLR4 regulators Tollip and IRAK-M.116
Pigs are the only animals that are susceptible to HRV (human rotavirus) initiated diarrhea. A study was performed with neonatal gnotobiotic pigs (born from near-term sows via CS, lacking a well-established microbiota) inoculated orally with probiotics, namely, Lactobacillus rhamnosus strain GG and Bifidobacterium animalis lactis Bb12, which are the primary bacterial species found in the gut of breastfed infants. This study sought to determine the impact that an attenuated (Att) HRV Wa strain vaccine had on B-cell responses. The AttHRV-vaccinated piglets colonized with probiotics were found to exhibit considerably lower fecal scores and reduced HRV shedding titers as compared to the uncolonized, AttHRV-vaccinated piglets. Further, a reduction in HRV-associated diarrhea was noted, which was correlated with the presence of a high number of intestinal IgA HRV antibodies and intestinal HRV-specific IgA antibody-secreting cells in probiotic-treated piglets compared to uncolonized, vaccinated pigs.174 Moreover, in an additional study, elevated levels of IL-6 and IL-10 were observed in ileal mononuclear cells.175 in gnotobiotic pigs inoculated with healthy human infant gut microbiota (Firmicutes and Proteobacteria accounting for approximately 98%). Colonization with this microbiota also promoted development of the neonatal immune system by substantially enhancing IFN-γ producing T-cell responses and by reducing Treg cell differentiation and their associated cytokine production in the AttHRV-vaccinated pigs. Wu and Wu have further summarized many additional studies in GF rats, mice and humans, that highlight the importance of a healthy microbiota for proper development and functioning of the acquired immune system in mammals.155
Many studies have been reported on colonization of gnotobiotic pigs with intestinal microbiota. In a study performed by Laycock et al.,176 24 gnotobiotic pigs were inoculated with Bristol microbiota (a novel simple porcine microbiota). These pigs exhibited no significant health problems and the Bristol microbiota successfully induced up-regulation in the expression of serum immunoglobulins IgA and IgM. However, the level of IgG2 was much lower than in conventional pigs that have access to colostrum, which suggests a maternal influence on IgG2 phroduction. Thus, the Bristol microbiota may be used to improve the formation, and subsequent development, of the intestinal mucosa and general immune system in neonatal pigs.
Mechanisms of interactions between microbiota and host components
It has been suggested that mammals possess a developmental window (2–3 weeks in pigs and a similar duration in other mammals) in which the developing host–gut microbiota interactions are easiest to manipulate and during which time it is most susceptible to major disturbances.41 Although the molecular mechanisms of these interactions are not clearly defined, limited data in pigs suggest that intestinal alkaline phosphatase (IAP) and inducible HSPs (iHSPs) have important roles in this process, especially in controlling inflammation and modulating gut function. These two components are involved in regulating antioxidant and anti-inflammatory reactions, by which they confer protection to the GIT epithelium.177
Inducible heat shock proteins
Pigs have been shown to express high levels of iHSP in both the small and large intestines178-180. However, the distal ileum was noted as having a higher relative concentration of iHSP proteins than the proximal colon in growing pigs, suggesting that higher microbial stimulation occurs in the distal region of the ileum.168 Oral administration of the broad-spectrum antibiotic amoxicillin to sows during parturition affected the sow’s fecal, and the piglet’s gut microbiota as well as the level of gut epithelial iHSPs.178 The association of gut commensal microbiota with iHSPs in growing pigs demonstrates that colonic iHSP70 correlates negatively with Bacteroidetes and Prevotella brevis colonization, and positively with that of Faecalibacterium prausnitzii, the latter of which exhibits anti-inflammatory properties and has been found to be depleted in pig IBD.181 These individual correlations are often difficult to interpret as direct cause-and-effect relationships; however, they all suggest intimate associations between iHSPs and the GIT microbiota in pigs. A systematic study carried out by Lallès and David182 showed that subjecting growing pigs to feeding or fasting for 1.5 days, or to fasting for 1.5 days followed by re-feeding for 2.5 days, that fasting induced an increase in iHSP27, but not iHSP70, throughout the small and large intestines. However, as soon as feeding was restored, so too were the intestinal and colonic concentrations of iHSP27, however, still with no influence on iHSP70.
Figure 3 is a diagrammatic representation of how dietary nutrients and components from the commensal microbiota can function to induce production of HSPs, while reinforcing protection to the host against various stressors.
Figure 3.

Inducible heat shock proteins. Dietary nutrients, gut microbiota components, and certain diseases can induce the formation of iHSPs (HSP25 and HSP70) in the GIT epithelium either directly from the microbiota or indirectly through the microbiota secretions and/or metabolites. This increases protection for the host against stressors like oxidation or inflammation in the epithelium.
Many different HSPs have been described as associated with the GIT mucosa and its function. A transient reduction is expression of HSP70, and in crypt depth was noted in a study performed by Liu et al.,168 who reported that modifications to bacterial colonization during early life functions to control the intestinal architecture and function, at least for a short period. Furthermore, longterm site- and diet-specific effects are observed in the major immune components that serve to control intestinal homeostasis. The same study reported an association between the cytoprotective HSP72 and the relative abundance of Lactobacillus spp. in the small intestine, together with specific members of clostridial clusters IV and XIVa in the large intestine of pigs.168
Heat shock proteins can be induced by a broad spectrum of stimuli, including commensal microbes.183 The physiological expression of molecular chaperones and HSPs is dependent on dietary components, commensal microbes, and resulting metabolites to which the mucosal surface is exposed.168 For example, the expression of ileal HSP27 has been correlated with inclusion of fiber in the diet. HSPs, in addition to being protein chaperones within cells, also function in immune responses, cell proliferation, apoptosis, and control of oxidation and inflammation. However, one of the most relevant functions of HSPs is their ability to regulate barrier function and minimize the adverse effects associated with inflammation and oxidative stresses on host cells. These proteins regulate the GIT barrier by controlling the expression of tight junction proteins such as occludins184. In pigs specifically, a high concentration of HSPs is present in the small and large intestines; while an increase in duodenal and jejunal HSP70 is highly associated with fetal stress.178
Intestinal alkaline phosphatase (IAP)
The IAP is produced by enterocytes in the small intestine and secreted into the lumen and subsequently into the circulatory system, where it is involved in detoxification of microbial components by dephosphorylation (Figure 4). IAP confers many physiological properties including absorption of minerals and nutrients such as calcium and fatty acids, and control of GIT and systemic inflammation through detoxification of pro-inflammatory components produced by the microbiota, such as LPS and flagellin.185 It is also directly involved in the control of the gut barrier.186 Moreover, when IAP is bound to enterocytes it can function to delay the growth of potential pathogens such as E. coli and, hence, influence the microbiota composition while restricting the translocation of E. coli into the body.187,188 Stressful conditions, such as weaning, significantly inhibit IAP production in pigs, which is responsible for development of many post-weaning disorders as well as an increased sensitivity of pigs to enteric infections.179
Figure 4.
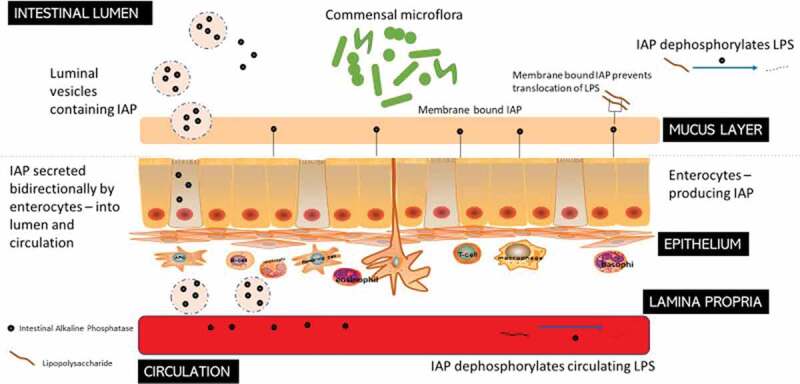
Intestinal Alkaline Phosphatase (IAP) – roles in the GIT. IAPs secreted by the enterocytes travel bi-directionally into the blood circulation and to the intestinal lumen where they act to dephosphorylate LPS from Gram-negative bacteria. Membrane-bound IAPs also prevent the translocation of the LPS through the mucus layer.
Both the gene expression and enzymatic activity of IAP are influenced by the GIT microbiota, while IAP simultaneously impacts the composition of the gut microbiota by removing proinflammatory free luminal adenosine triphosphate (ATP), by reducing the level of inflammation, and by regulating intestinal surface pH.189 Thus, the expression of iHSPs in gut epithelial cells is proportionate to the number of microbes present along the GIT.
Conclusion
The resident GIT microbiota is unique for each species and has continually evolved over generations to become more functional and relevant to their current local environment and the host. Its prominent role in stimulating the maturation of the GIT and regulating the gut–brain axis, especially in young pigs, represents opportunities to design effective strategies to increase animal robustness.207 There is, hitherto, no consensus on the definition of balanced or favorable microbiota even though knowledge on host–microbiota cross-talk is constantly being updated, revealing a highly complex scenario. Moreover, studies attempting to define the factors affecting the GIT microbiota and their subsequent roles in pig physiology and immunity are still in progress. Future studies will serve to inform the development of hypothesizes for effective strategies to manage and restore intestinal homeostasis after an external perturbation, such as stress, early administration of an antibiotic, or a bacterial infection, all of which ultimately could be utilized to improve productivity, minimize stress and prevent diseases.
Acknowledgments
Not applicable.
Funding Statement
This work was supported by the National Natural Science Foundation of China [grant nos. 31101862, 31472243] and Shenzhen Projects for Basic Research [JCYJ20170306162414058]
Abbreviations
- APCs
antigen presenting cells
- DC
dendritic cells
- DF
dietary fiber
- GF
germ free
- GIT
gastrointestinal tract
- HSPs
heat shock proteins
- HRV
human rotavirus
- IAP
intestinal alkaline phosphatase
- IBD
inflammatory bowel disease
- IFN
interferon
- iHSP
inducible heat shock proteins
- LPS
lipopolysaccharides
- NF-κB
nuclear factor-kappa B
- SCFA
short chain fatty acids
- SCOA
short chain organic acids.
Authors Contributions
XHJ and RG conceived the study. YP performed a systematic review and wrote the first draft of the manuscript and generated the figures. XHJ and RG assisted in reviewing literature, guided the analyses, and provided critical feedback while assisting with editing the subsequent drafts of the manuscript. All authors reviewed the final draft of the manuscript, offered critical feedback, and approved the final version.
Conflict of Interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Data Availability Statement
No datasets were generated for this study.
References
- 1.Dominguez-Bello MG, Godoy-Vitorino F, Knight R, Blaser MJ.. Role of the microbiome in human development. Gut. 2019;68(6):1108. doi: 10.1136/gutjnl-2018-317503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim HB, Isaacson RE. The pig gut microbial diversity: understanding the pig gut microbial ecology through the next generation high throughput sequencing. Vet Microbiol. 2015;177(3):242–251. doi: 10.1016/j.vetmic.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 3.Ursell LK, Metcalf JL, Parfrey LW, Knight R. Defining the human microbiome. Nutr Rev. 2012;70(1):S38–S44. doi: 10.1111/nure.2012.70.issue-s1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. 2016;535(7610):65. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 5.Min YW, Rhee P-L. The role of microbiota on the gut immunology. Clin Ther. 2015;37(5):968–975. doi: 10.1016/j.clinthera.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 6.Peterson J, Garges S, Giovanni M, McInnes P, Lu W, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, et al. The NIH human microbiome project. (Report). Genome Res. 2009;19(12):2317–2333. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang M, Monaco MH, Donovan SM. Impact of early gut microbiota on immune and metabolic development and function. Seminars in Fetal and Neonatal Medicine. 2016;21(6):380–387. doi: 10.1016/j.siny.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102(31):11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31(1):107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 10.Xu J, Gordon JI. Honor thy symbionts. Proc Natl Acad Sci U.S.A. 2003;100(18):10452–10459. doi: 10.1073/pnas.1734063100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 12.Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu Rev Immunol. 2009;28:623–667. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ley RE, Hamady M, Lozupone C, Turnbaugh P, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, et al. Evolution of mammals and their gut microbes. Science (New York, NY). 2008;320(5883):1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu H-J, Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes. 2012;3(1):4–14. doi: 10.4161/gmic.19320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levy M, Blacher E, Elinav E. Microbiome, metabolites and host immunity. Curr Opin Microbiol. 2017;35::8–15. doi: 10.1016/j.mib.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Brown K, Decoffe D, Molcan E, Gibson DL. Diet-induced dysbiosis of the intestinal microbiota and the effects on immunity and disease. Nutrients. 2012;4:1095–1119. doi: 10.3390/nu4081095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madsen KL. Interactions between microbes and the gut epithelium. J Clin Gastroenterol. 2011;45:S111–S114. doi: 10.1097/MCG.0b013e3182274249. [DOI] [PubMed] [Google Scholar]
- 18.Yu S, Gao N. Compartmentalizing intestinal epithelial cell toll-like receptors for immune surveillance. Cell Mol Life Sci. 2015;72(17):3343–3353. doi: 10.1007/s00018-015-1931-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meurens F, Summerfield A, Nauwynck H, Saif L, Gerdts V. The pig: a model for human infectious diseases. Trends Microbiol. 2012;20(1):50–57. doi: 10.1016/j.tim.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q, Widmer G, Tzipori S. A pig model of the human gastrointestinal tract. Gut Microbes. 2013;4(3):193–200. doi: 10.4161/gmic.23867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao L, Estellé J, Kiilerich P, Ramayo-Caldas Y, Xia Z, Feng Q, Liang S, Pedersen AØ, Kjeldsen NJ, Liu C, et al. A reference gene catalogue of the pig gut microbiome. Nat Microbiol. 2016;16161(1):1–6. [DOI] [PubMed] [Google Scholar]
- 22.Lu D, Tiezzi F, Schillebeeckx C, McNulty NP, Schwab C, Shull C, Maltecca C. Host contributes to longitudinal diversity of fecal microbiota in swine selected for lean growth. Microbiome. 2018;6(1):4. doi: 10.1186/s40168-017-0384-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saffrey M. Aging of the mammalian gastrointestinal tract: a complex organ system. The Official J Am Aging Assoc. 2014;36:1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim B, Borewicz K, White BA, Singer RS, Sreevatsan S, Tu ZJ, Isaacson RE. Microbial shifts in the swine distal gut in response to the treatment with antimicrobial growth promoter, tylosin. Proc Natl Acad Sci U.S.A. 2012;109(38):15485–15490. doi: 10.1073/pnas.1205147109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Looft T, Allen HK, Cantarel BL, Levine UY, Bayles DO, Alt DP, Henrissat B, Stanton TB. Bacteria, phages and pigs: the effects of in-feed antibiotics on the microbiome at different gut locations. Isme J. 2014;8:1566. doi: 10.1038/ismej.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maradiaga N, Aldridge B, Zeineldin M, Lowe J. Gastrointestinal microbiota and mucosal immune gene expression in neonatal pigs reared in a cross-fostering model. Microb Pathog. 2018;121:27–39. doi: 10.1016/j.micpath.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Allen HK, Looft T, Bayles DO, Humphrey S, Levine UY, Alt D, Stanton TB. Antibiotics in feed induce prophages in swine fecal microbiomes. Antibiot in Feed Induce Prophages in Swine Fecal Microbiomes. 2011;2(6):1–9:e00260-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamendella R, Santo Domingo JW, Ghosh S, Martinson J, Oerther DB. Comparative fecal metagenomics unveils unique functional capacity of the swine gut. BMC Microbiol. 2011;11(1):103. doi: 10.1186/1471-2180-11-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Looft T, Johnson TA, Allen HK, Bayles DO, Alt DP, Stedtfeld RD, Sul WJ, Stedtfeld TM, Chai B, Cole JR, et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc Natl. Acad Sci U.S.A. 2012;109(5):1691. doi: 10.1073/pnas.1120238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao W, Wang Y, Liu S, Huang J, Zhai Z, He C, Ding J, Wang J, Wang H, Fan W, et al. The dynamic distribution of porcine microbiota across different ages and gastrointestinal tract segments. PLoS One. 2015;10(2):e0117441. doi: 10.1371/journal.pone.0117441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandhu KV, Sherwin E, Schellekens H, Stanton C, Dinan TG, Cryan JF. Feeding the microbiota-gut-brain axis: diet, microbiome, and neuropsychiatry. Transl Res. 2017;179:223–244. doi: 10.1016/j.trsl.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Martin CR, Osadchiy V, Kalani A, Mayer EA. The brain-gut-microbiome axis. Cell Mol Gastroentero Hepatol. 2018. doi: 10.1016/j.jcmgh.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bohórquez DV, Liddle RA. The gut connectome: making sense of what you eat. J Clin Invest. 2015;125(3):888–890. doi: 10.1172/JCI81121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayer EA, Tillisch K, Gupta A. Gut/brain axis and the microbiota. J Clin Invest. 2015;125(3):926–938. doi: 10.1172/JCI76304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. (ANALYSIS)(Report). Nat Rev Microbiol. 2008;6(10):776. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leser TD, Amenuvor JZ, Jensen TK, Lindecrona RH, Boye M, Møller K. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl Environ Microbiol. 2002;68(2):673–690. doi: 10.1128/AEM.68.2.673-690.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang M, Radlowski EC, Monaco MH, Fahey GC, Gaskins HR, Donovan SM. Mode of delivery and early nutrition modulate microbial colonization and fermentation products in neonatal piglets. J Nutr. 2013;143(6):795–803. doi: 10.3945/jn.112.173096. [DOI] [PubMed] [Google Scholar]
- 38.Jansman AJM, Zhang J, Koopmans SJ, Dekker RA, Smidt H. Effects of a simple or a complex starter microbiota on intestinal microbiota composition in caesarean derived piglets1. J Anim Sci. 2012;90(4):433–435. doi: 10.2527/jas.53850. [DOI] [PubMed] [Google Scholar]
- 39.Siggers RH, Thymann T, Jensen BB, Mølbak L, Heegaard PMH, Schmidt M, Buddington RK, Sangild PT. Elective cesarean delivery affects gut maturation and delays microbial colonization but does not increase necrotizing enterocolitis in preterm pigs. Am J Physiol-Regul, Integr and Comp Physiol. 2008;294(3):R929–R938. doi: 10.1152/ajpregu.00705.2007. [DOI] [PubMed] [Google Scholar]
- 40.Hyde Matthew J, Griffin Julian L, Herrera E, Byrne Christopher D, Clarke L, Kemp Paul R. Delivery by Caesarean section, rather than vaginal delivery, promotes hepatic steatosis in piglets. Clin Sci. 2010;118(1):47. doi: 10.1042/CS20090169. [DOI] [PubMed] [Google Scholar]
- 41.Thompson CL, Wang B, Holmes AJ. The immediate environment during postnatal development has long-term impact on gut community structure in pigs. Isme J. 2008;2:739. doi: 10.1038/ismej.2008.29. [DOI] [PubMed] [Google Scholar]
- 42.Yang H, Xiao Y, Wang J, Xiang Y, Gong Y, Wen X, Li D. Core gut microbiota in Jinhua pigs and its correlation with strain, farm and weaning age. Journal of Microbiology. 2018;56(5):346–355. doi: 10.1007/s12275-018-7486-8. [DOI] [PubMed] [Google Scholar]
- 43.Pajarillo EAB, Chae JP, Balolong MP, Kim HB, Seo K-S, Kang D-K. Pyrosequencing-based analysis of fecal microbial communities in three purebred pig lines. Journal of Microbiology. 2014a;52(8):646–651. doi: 10.1007/s12275-014-4270-2. [DOI] [PubMed] [Google Scholar]
- 44.Pajarillo EAB, Chae JP, Balolong MP, Kim HB, Seo K-S, Kang D-K. Characterization of the fecal microbial communities of Duroc pigs using 16S rRNA gene pyrosequencing. Asian-Australas J Anim Sci. 2015;28:584+. doi: 10.5713/ajas.14.0651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim HB, Borewicz K, White BA, Singer RS, Sreevatsan S, Tu ZJ, Isaacson RE. Longitudinal investigation of the age-related bacterial diversity in the feces of commercial pigs. Vet Microbiol. 2011;153(1):124–133. doi: 10.1016/j.vetmic.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 46.Mathur R, Barlow GM. Obesity and the microbiome. Expert Review of Gastroenterology & Hepatology. 2015;9:1087–1099. Informa Healthcare. doi: 10.1586/17474124.2015.1051029. [DOI] [PubMed] [Google Scholar]
- 47.Zhou J, He Z, Yang Y, Deng Y, Tringe SG, Alvarez-Cohen L. High-throughput metagenomic technologies for complex microbial community analysis: open and closed formats. mBio. 2015;6(1). doi: 10.1128/mBio.02288-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han GG, Lee J-Y, Jin G-D, Park J, Choi YH, Kang S-K, Chae BJ, Kim EB, Choi Y-J. Tracing of the fecal microbiota of commercial pigs at five growth stages from birth to shipment. Sci Rep. 2018;8(1):6012. doi: 10.1038/s41598-018-24508-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holman DB, Brunelle BW, Trachsel J, Allen HK, Bik H. Meta-analysis to define a core microbiota in the swine gut. mSystems. 2017;2(3):1-14:e00004-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pryde SE, Richardson AJ, Stewart CS, Flint HJ. Molecular analysis of the microbial diversity present in the colonic wall, colonic lumen, and cecal lumen of a pig. Appl Environ Microbiol. 1999;65:5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simpson JM, McCracken VJ, White BA, Gaskins HR, Mackie RI. Application of denaturant gradient gel electrophoresis for the analysis of the porcine gastrointestinal microbiota. J Microbiol Methods. 1999;36(3):167. doi: 10.1016/S0167-7012(99)00029-9. [DOI] [PubMed] [Google Scholar]
- 52.Dowarah R, Verma AK, Agarwal N, Patel BHM, Singh P. Effect of swine based probiotic on performance, diarrhoea scores, intestinal microbiota and gut health of grower-finisher crossbred pigs. Livestock Sci. 2017;195:74–79. doi: 10.1016/j.livsci.2016.11.006. [DOI] [Google Scholar]
- 53.Petri D, Hill JE, Van Kessel AG. Microbial succession in the gastrointestinal tract (GIT) of the preweaned pig. Livestock Sci. 2010;133(1):107–109. doi: 10.1016/j.livsci.2010.06.037. [DOI] [Google Scholar]
- 54.Dou S, Gadonna-Widehem P, Rome V, Hamoudi D, Rhazi L, Lakhal L, Larcher T, Bahi-Jaber N, Pinon-Quintana A, Guyonvarch A, et al. Characterisation of early-life fecal microbiota in susceptible and healthy pigs to post-weaning diarrhoea. PLoS One. 2017;12(1):e0169851. doi: 10.1371/journal.pone.0169851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Inoue R, Tsukahara T, Nakanishi N, Ushida K. Development of the intestinal microbiota in the piglet. J Gen Appl Microbiol. 2005;51(4):257–265. doi: 10.2323/jgam.51.257. [DOI] [PubMed] [Google Scholar]
- 56.Ajouz H, Mukherji D, Shamseddine A. Secondary bile acids: an underrecognized cause of colon cancer. World J Surg Oncol. 2014;12:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yan S, Zhu C, Yu T, Huang W, Huang J, Kong Q, Shi J, Chen Z, Liu Q, Wang S, et al. Studying the differences of bacterial metabolome and microbiome in the colon between landrace and meihua piglets. Front Microbiol. 2017;8:1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao Y, Han F, Huang X, Rong Y, Yi H, Wang Y. Changes in gut microbial populations, intestinal morphology, expression of tight junction proteins, and cytokine production between two pig breeds after challenge with Escherichia coli K88: A comparative study1. J Anim Sci Champaign. 2013;91:13. [DOI] [PubMed] [Google Scholar]
- 59.Bian G, Ma S, Zhu Z, Su Y, Zoetendal EG, Mackie R, Liu J, Mu C, Huang R, Smidt H, et al. Age, introduction of solid feed and weaning are more important determinants of gut bacterial succession in piglets than breed and nursing mother as revealed by a reciprocal cross-fostering model. Environ Microbiol. 2016;18(5):1566–1577. doi: 10.1111/1462-2920.13272. [DOI] [PubMed] [Google Scholar]
- 60.Su Y, Bian G, Zhu Z, Smidt H, Zhu W. Early methanogenic colonisation in the faeces of meishan and yorkshire piglets as determined by pyrosequencing analysis. Archaea. 2014;2014:10. doi: 10.1155/2014/547908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Armougom F, Henry M, Vialettes B, Raccah D, Raoult D. Monitoring Bacterial community of human gut microbiota reveals an increase in lactobacillus in obese patients and methanogens in anorexic patients. PLoS One. 2009;4(9):e7125. doi: 10.1371/journal.pone.0007125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo X, Xia X, Tang R, Zhou J, Zhao H, Wang K. Development of a real-time PCR method for Firmicutes and Bacteroidetes in faeces and its application to quantify intestinal population of obese and lean pigs. Lett Appl Microbiol. 2008;47(5):367–373. doi: 10.1111/j.1472-765X.2008.02408.x. [DOI] [PubMed] [Google Scholar]
- 63.Liu H, Guo X, Gooneratne R, Lai R, Zeng C, Zhan F, Wang W. The gut microbiome and degradation enzyme activity of wild freshwater fishes influenced by their trophic levels. Sci Rep. 2016;6:24340. doi: 10.1038/srep24340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leblois J, Massart S, Li B, Wavreille J, Bindelle J, Everaert N. Modulation of piglets’ microbiota: differential effects by a high wheat bran maternal diet during gestation and lactation. Sci Rep. 2017;7(1):7426. doi: 10.1038/s41598-017-07228-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Doré J, Blottière H. The influence of diet on the gut microbiota and its consequences for health. Curr Opin Biotechnol. 2015;32:195–199. doi: 10.1016/j.copbio.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 66.Liao SF, Nyachoti M. Using probiotics to improve swine gut health and nutrient utilization. Anim Nutr. 2017;3(4):331–343. doi: 10.1016/j.aninu.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang H, Yang M, Fang S, Huang X, He M, Ke S, Gao J, Wu J, Zhou Y, Fu H, et al. Evaluating the profound effect of gut microbiome on host appetite in pigs. BMC Microbiol. 2018;18(1):215. doi: 10.1186/s12866-018-1364-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.De Rodas B, Youmans BP, Danzeisen JL, Tran H, Johnson TJ. Microbiome profiling of commercial pigs from farrow to finish. J Anim Sci. 2018;96(5):1778–1794. doi: 10.1093/jas/sky109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Montagne L, Pluske JR, Hampson DJ. A review of interactions between dietary fibre and the intestinal mucosa, and their consequences on digestive health in young non-ruminant animals. Anim Feed Sci Technol. 2003;108(1):95–117. doi: 10.1016/S0377-8401(03)00163-9. [DOI] [Google Scholar]
- 70.Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev. 2017;279:70–89. doi: 10.1111/imr.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bach K, Erik K, Hedemann MS, Lærke HN. The role of carbohydrates in intestinal health of pigs. Anim Feed Sci Technol. 2012;173(1):41–53. doi: 10.1016/j.anifeedsci.2011.12.020. [DOI] [Google Scholar]
- 72.Lindberg JE. Fiber effects in nutrition and gut health in pigs. J Anim Sci Biotechnol. 2014;5(1):15. doi: 10.1186/2049-1891-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Freire JPB, Guerreiro AJG, Cunha LF, Aumaitre A. Effect of dietary fibre source on total tract digestibility, caecum volatile fatty acids and digestive transit time in the weaned piglet. Anim Feed Sci Technol. 2000;87(1–2):71–83. doi: 10.1016/S0377-8401(00)00183-8. [DOI] [Google Scholar]
- 74.Hedemann MS, Eskildsen M, Laerke HN, Pedersen C, Lindberg JE, Laurinen P, Knudsen KEB. Intestinal morphology and enzymatic activity in newly weaned pigs fed contrasting fiber concentrations and fiber properties. J Anim Sci. 2006;84(6):1375. doi: 10.2527/2006.8461375x. [DOI] [PubMed] [Google Scholar]
- 75.Balasubramanian B, Lee SI, Kim I-H. Inclusion of dietary multi-species probiotic on growth performance, nutrient digestibility, meat quality traits, faecal microbiota and diarrhoea score in growing–finishing pigs. Ital J Anim Sci. 2018;17(1):100–106. doi: 10.1080/1828051X.2017.1340097. [DOI] [Google Scholar]
- 76.Leser TD, Lindecrona RH, Jensen TK, Jensen BB, Møller K. Changes in bacterial community structure in the colon of pigs fed different experimental diets and after infection with brachyspira hyodysenteriae. Appl Environ Microbiol. 2000;66(8):3290–3296. doi: 10.1128/AEM.66.8.3290-3296.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stephens RW, Arhire L, Covasa M. Gut microbiota: from microorganisms to metabolic organ influencing obesity. Obesity. 2018;26(5):801–809. doi: 10.1002/oby.v26.5. [DOI] [PubMed] [Google Scholar]
- 78.Heinritz SN, Weiss E, Eklund M, Aumiller T, Louis S, Rings A, Messner S, Camarinha-Silva A, Seifert J, Bischoff SC, et al. Intestinal microbiota and microbial metabolites are changed in a pig model fed a high-fat/low-fiber or a low-fat/high-fiber diet. PLoS One. 2016;11(4):e0154329. doi: 10.1371/journal.pone.0154329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Spurlock ME, Gabler NK. The development of porcine models of obesity and the metabolic syndrome. J Nutr. 2008;138(2):397–402. doi: 10.1093/jn/138.2.397. [DOI] [PubMed] [Google Scholar]
- 80.National Research Council. Committee on Nutrient Requirements of S, National Research Council. Committee on Nutrient Requirements of Swine ib, National Research Council. Board on A, Natural Resources ib . Nutrient requirements of swine 11th. Washington (D.C.): National Academies Press; 2012. [Google Scholar]
- 81.Hojberg O, Canibe N, Poulsen HD, Hedemann MS, Jensen BB. Influence of Dietary Zinc Oxide and Copper Sulfate on the gastrointestinal ecosystem in newly weaned piglets. Appl Environ Microbiol. 2005;71(5):2267. doi: 10.1128/AEM.71.5.2267-2277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Debski B. Supplementation of pigs diet with zinc and copper as alternative to conventional antimicrobials. Pol J Vet Sci. 2016;19(4):917–924. doi: 10.1515/pjvs-2016-0113. [DOI] [PubMed] [Google Scholar]
- 83.Namkung H, Gong J, Yu H, de Lange CFM. Effect of pharmacological intakes of zinc and copper on growth performance, circulating cytokines and gut microbiota of newly weaned piglets challenged with coliform lipopolysaccharides. Can J Anim Sci. 2006;86(4):511–522. doi: 10.4141/A05-075. [DOI] [Google Scholar]
- 84.Yazdankhah S, Rudi K, Bernhoft A. Zinc and copper in animal feed - development of resistance and co-resistance to antimicrobial agents in bacteria of animal origin. Microb Ecol Health Dis. 2014;25:11. https://doi.org/10.3389/fmicb.2017.00846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Daniel ML, Carolina G, Erica R, José MF, María ET, Ross RP, Catherine S. Lactic Acid bacteria and bifidobacteria with potential to design natural biofunctional health-promoting dairy foods. Front Microbiol. 2017. May;8:1–11. https://doi.org/10.3389/fmicb.2017.00846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kenny M, Smidt H, Mengheri E, Miller B. Probiotics – do they have a role in the pig industry? Animal. 2010;5(3):462–470. doi: 10.1017/S175173111000193X. [DOI] [PubMed] [Google Scholar]
- 87.Galdeano CM, de Leblanc ADM, Dogi C, Perdigón G. Lactic acid bacteria as immunomodulators of the gut-associated immune system. Biotechnol Lactic Acid Bacteria. 2010. doi: 10.1002/9780813820866.ch7 [DOI] [Google Scholar]
- 88.Wang M, Donovan SM. Human microbiota-associated swine: current progress and future opportunities. ILAR Journal. 2015;56(1):63–73. doi: 10.1093/ilar/ilv006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li D, Ni K, Pang H, Wang Y, Cai Y, Jin Q. Identification and antimicrobial activity detection of lactic acid bacteria isolated from corn stover silage. Asian-Australas J Anim Sci. 2015;28:620+. doi: 10.5713/ajas.14.0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chiang M-L, Chen H-C, Chen K-N, Lin Y-C, Lin Y-T, Chen M-J. Optimizing production of two potential probiotic lactobacilli strains isolated from piglet feces as feed additives for weaned piglets. Asian-Australas J Anim Sci. 2015;28:1163+. doi: 10.5713/ajas.14.0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Giang HH, Viet TQ, Ogle B, Lindberg JE. Growth performance, digestibility, gut environment and health status in weaned piglets fed a diet supplemented with potentially probiotic complexes of lactic acid bacteria. Livestock Sci. 2010;129(1):95–103. doi: 10.1016/j.livsci.2010.01.010. [DOI] [Google Scholar]
- 92.Riboulet-Bisson E, Sturme MHJ, Jeffery IB, O’Donnell MM, Neville BA, Forde BM, Claesson MJ, Harris H, Gardiner GE, Casey PG, et al. Effect of lactobacillus salivarius bacteriocin abp118 on the mouse and pig intestinal microbiota. PLoS One. 2012;7(2):e31113. doi: 10.1371/journal.pone.0031113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang Y, Zhao X, Le MHA, Zijlstra R, Gänzle M. Reutericyclin producing Lactobacillus reuteri modulates development of fecal microbiota in weanling pigs. Front Microbiol. 2015;6:762. doi: 10.3389/fmicb.2015.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lallès J-P, Bosi P, Smidt H, Stokes CR. Nutritional management of gut health in pigs around weaning. Proc Nutr Soc. 2007;66(2):260–268. doi: 10.1017/S0029665107005484. [DOI] [PubMed] [Google Scholar]
- 95.Wells JE, Yen JT, Miller DN. Impact of dried skim milk in production diets on Lactobacillus and pathogenic bacterial shedding in growing-finishing swine1. J Appl Microbiol. 2005;99(2):400–407. doi: 10.1111/jam.2005.99.issue-2. [DOI] [PubMed] [Google Scholar]
- 96.Hutkins RW, Krumbeck JA, Bindels LB, Cani PD, Fahey G, Goh YJ, Hamaker B, Martens EC, Mills DA, Rastal RA, et al. Prebiotics: why definitions matter. Curr Opin Biotechnol. 2016;37:1–7. doi: 10.1016/j.copbio.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sivaprakasam S, Prasad PD, Singh N. Benefits of short-chain fatty acids and their receptors in inflammation and carcinogenesis. Pharmacol Ther. 2016;164:144–151. doi: 10.1016/j.pharmthera.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Çeti̇n N, Güçlü BK, Çeti̇n E. The effects of probiotic and mannanoligosaccharide on some haematological and immunological parameters in Turkeys. J Vet Med Series A. 2005;52(6):263–267. doi: 10.1111/j.1439-0442.2005.00736.x. [DOI] [PubMed] [Google Scholar]
- 99.Yin YL, Tang ZR, Sun ZH, Liu ZQ, Li TJ, Huang RL, Ruan Z, Deng ZY, Gao B, Chen LX, et al. Effect of galacto-mannan-oligosaccharides or chitosan supplementation on cytoimmunity and humoral immunity in early-weaned piglets. (Report). Asian-Australas J Anim Sci. 2008;21(5):723. doi: 10.5713/ajas.2008.70408. [DOI] [Google Scholar]
- 100.Smith HW, Jones JET. Observations on the alimentary tract and its bacterial flora in healthy and diseased pigs. J Pathol Bacteriol. 1963;86(2):387–412. doi: 10.1002/(ISSN)1555-2039. [DOI] [PubMed] [Google Scholar]
- 101.Krause DO, Bhandari SK, House JD, Nyachoti CM. Response of nursery pigs to a synbiotic preparation of starch and an anti-escherichia coli K88 probiotic. Appl Environ Microbiol. 2010;76(24):8192. doi: 10.1128/AEM.01427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pié S, Awati A, Vida S, Falluel I. Effects of added fermentable carbohydrates in the diet on intestinal proinflammatory cytokine-specific mRNA content in weaning piglets1. J Anim Sci. 2007;85(3):673–683. doi: 10.2527/jas.2006-535. [DOI] [PubMed] [Google Scholar]
- 103.Call L, Stoll B, Oosterloo B, Ajami N, Sheikh F, Wittke A, Waworuntu R, Berg B, Petrosino J, Olutoye O, et al. Metabolomic signatures distinguish the impact of formula carbohydrates on disease outcome in a preterm piglet model of NEC. Microbiome. 2018;6(1):111. doi: 10.1186/s40168-018-0498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Naqid IA, Owen JP, Maddison BC, Gardner DS, Foster N, Tchórzewska MA, La Ragione RM, Gough KC. Prebiotic and probiotic agents enhance antibody-based immune responses to Salmonella Typhimurium infection in pigs. Anim Feed Sci Technol. 2015;201:57–65. doi: 10.1016/j.anifeedsci.2014.12.005. [DOI] [Google Scholar]
- 105.Qing N, Pinghua L, Shuaishuai H, Yeqiu Z, Sung Woo K, Huizhi L, Xiang M, Shuo G, Lichun H, Wangjun W, et al. Dynamic distribution of the gut microbiota and the relationship with apparent crude fiber digestibility and growth stages in pigs. Sci Rep. 2015;5(1):1–7. DOI: 10.1038/srep09938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Etleva D, Männerin K. Efficiency of probiotics in farm animals. In:Rigobelo EC, editors. Probiotic in Animals.Rigobelo EC: IntechOpen; 2012. [Google Scholar]
- 107.Kenny M, Smidt H, Mengheri E, Miller B. Probiotics – do they have a role in the pig industry? Animal. 2011;5(3):462–470. doi: 10.1017/S175173111000193X. [DOI] [PubMed] [Google Scholar]
- 108.Konstantinov SR, Awati AA, Williams BA, Miller BG, Jones P, Stokes CR, Akkermans ADL, Smidt H, De Vos WM. Post-natal development of the porcine microbiota composition and activities. Environ Microbiol. 2006;8(7):1191–1199. doi: 10.1111/emi.2006.8.issue-7. [DOI] [PubMed] [Google Scholar]
- 109.Katouli M, Wallgren P. Chapter 2 Metabolism and population dynamics of the intestinal microflora in the growing pig. In editors, Holzapfel WH, Naughton PJ, Pierzynowski SG, Zabielski R, Salek E. Biology of Growing Animals. Vol. 2. The Netherlands: Elsevier; 2005. p. 21–53. [Google Scholar]
- 110.Stafford V, John VOD, Alan KK, Cormac JOS, Torres S. The effect of divergence in feed efficiency on the intestinal microbiota and the intestinal immune response in both unchallenged and lipopolysaccharide challenged ileal and colonic explants. PLoS One. 2016;11(2):e0148145. doi: 10.1371/journal.pone.0148145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang DY, Ji HF, Wang SX, Liu H, Wang J, Wang YM. In vitro characterisation of two Lactobacillus strains and evaluation of their suitability as probiotics for growing-finishing pigs. Anim Prod Sci. 2018;59(8):1537–1545. [Google Scholar]
- 112.Suo C, Yin Y, Wang X, Lou X, Song D, Wang X, Gu Q. Effects of lactobacillus plantarum ZJ316 on pig growth and pork quality. BMC Vet Res. 2012;8(1):89. doi: 10.1186/1746-6148-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ross GR, Van Nieuwenhove CP, González SN. Fatty acid profile of pig meat after probiotic administration. J Agric Food Chem. 2012;60(23):5974. doi: 10.1021/jf205360h. [DOI] [PubMed] [Google Scholar]
- 114.Suda Y, Villena J, Takahashi Y, Hosoya S, Tomosada Y, Tsukida K, Shimazu T, Aso H, Tohno M, Ishida M, et al. Immunobiotic Lactobacillus jensenii as immune-health promoting factor to improve growth performance and productivity in post-weaning pigs. BMC Immunol. 2014;15(1):24. doi: 10.1186/1471-2172-15-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mann E, Schmitz-Esser S, Zebeli Q, Wagner M, Ritzmann M, Metzler-Zebeli BU. Mucosa-associated bacterial microbiome of the gastrointestinal tract of weaned pigs and dynamics linked to dietary calcium-phosphorus. PLoS One. 2014;9(1):e86950. doi: 10.1371/journal.pone.0086950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Roselli M, Pieper R, Rogel-Gaillard C, de Vries H, Bailey M, Smidt H, Lauridsen C. Immunomodulating effects of probiotics for microbiota modulation, gut health and disease resistance in pigs. Anim Feed Sci Technol. 2017;233(C):104–119. doi: 10.1016/j.anifeedsci.2017.07.011. [DOI] [Google Scholar]
- 117.Inman CF, Haverson K, Konstantinov SR, Jones PH, Harris C, Smidt H, Miller B, Bailey M, Stokes C. Rearing environment affects development of the immune system in neonates. Clin Exp Immunol. 2010;160(3):431–439. doi: 10.1111/cei.2010.160.issue-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lewis MC, Inman CF, Patel D, Schmidt B, Mulder I, Miller B, Gill BP, Pluske J, Kelly D, Stokes CR, et al. Direct experimental evidence that early-life farm environment influences regulation of immune responses. Pediatr Allergy Immunol. 2012;23(3):265–269. doi: 10.1111/pai.2012.23.issue-3. [DOI] [PubMed] [Google Scholar]
- 119.Mulder IE, Schmidt B, Lewis M, Delday M, Stokes CR, Bailey M, Aminov RI, Gill BP, Pluske JR, Mayer C-D, et al. Restricting microbial exposure in early life negates the immune benefits associated with gut colonization in environments of high microbial diversity. PLoS One. 2011;6(12):e28279. doi: 10.1371/journal.pone.0028279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mulder IE, Schmidt B, Stokes CR, Lewis M, Bailey M, Aminov RI, Prosser JI, Gill BP, Pluske JR, Mayer C-D, et al. Environmentally-acquired bacteria influence microbial diversity and natural innate immune responses at gut surfaces. BMC Biol. 2009;7(1):79. doi: 10.1186/1741-7007-7-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schmidt B, Mulder IE, Musk CC, Aminov RI, Lewis M, Stokes CR, Bailey M, Prosser JI, Gill BP, Pluske JR, et al. Establishment of normal gut microbiota is compromised under excessive hygiene conditions. PLoS One. 2011;6(12):e28284. doi: 10.1371/journal.pone.0028284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dibner JJ, Richards JD. Antibiotic growth promoters in agriculture: history and mode of action. Poult Sci. 2005;84(4):634. doi: 10.1093/ps/84.4.634. [DOI] [PubMed] [Google Scholar]
- 123.Kim J, Guevarra RB, Nguyen SG, Lee J-H, Jeong DK, Unno T. Effects of the antibiotics growth promoter tylosin on swine gut microbiota. J Microbiol Biotechnol. 2016;26(5):876. doi: 10.4014/jmb.1512.12004. [DOI] [PubMed] [Google Scholar]
- 124.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. U.S.A. 2011;108(13):5354. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bosi P, Merialdi G, Scandurra S, Messori S, Bardasi L, Nisi I, Russo D, Casini L, Trevisi P. Feed supplemented with 3 different antibiotics improved food intake and decreased the activation of the humoral immune response in healthy weaned pigs but had differing effects on intestinal microbiota. J Anim Sci. 2011;89(12):4043. doi: 10.2527/jas.2010-3311. [DOI] [PubMed] [Google Scholar]
- 126.Gao K, Pi Y, Peng Y, Mu C-L, Zhu W-Y. Time-course responses of ileal and fecal microbiota and metabolite profiles to antibiotics in cannulated pigs. Appl Microbiol Biotechnol. 2018;102(5):2289–2299. doi: 10.1007/s00253-018-8774-2. [DOI] [PubMed] [Google Scholar]
- 127.DebRoy C, Hegde NV, Schilling KV, Katani R. Gut microbiomes of pigs grown in organic and conventional dietary regimens. 2017;5. [Google Scholar]
- 128.Hill DA, Hoffmann C, Abt MC, Du Y, Kobuley D, Kirn TJ, Bushman FD, Artis D. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2009;3(2):148. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Schokker D, Zhang J, L-l Z, Vastenhouw SA, Heilig HGHJ, Smidt H, Rebel JMJ, Smits MA. Early-life environmental variation affects intestinal microbiota and immune development in new-born piglets. PLoS One. 2014;9(6):e100040. doi: 10.1371/journal.pone.0100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shapiro H, Thaiss CA, Levy M, Elinav E. The cross talk between microbiota and the immune system: metabolites take center stage. Curr Opin Immunol. 2014;30:54–62. doi: 10.1016/j.coi.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 131.Hooper LV. Bacterial contributions to mammalian gut development. Trends Microbiol. 2004;12(3):129–134. doi: 10.1016/j.tim.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 132.Hooper LV, Gordon JI. Commensal host- bacterial relationships in the gut. (statistical data included). Science. 2001;292(5519):1115. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 133.Frick J-S, Autenrieth IB. The gut microflora and its variety of roles in health and disease. In: Dobrindt U, Hacker JH, Svanborg C, editors. Between Pathogenicity and Commensalism. Berlin (Heidelberg): Springer Berlin Heidelberg; 2013. p. 273–289. [DOI] [PubMed] [Google Scholar]
- 134.Carey DN, Knut D, Kevin RF. Spatial structure, cooperation and competition in biofilms. Nat Rev Microbiol. 2016;14(9). [DOI] [PubMed] [Google Scholar]
- 135.Foster KR, Schluter J, Coyte KZ, Rakoff-Nahoum S. The evolution of the host microbiome as an ecosystem on a leash. Nature. 2017;548:43. doi: 10.1038/nature23292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Janczyk P, Pieper R, Smidt H, Souffrant WB. Changes in the diversity of pig ileal lactobacilli around weaning determined by means of 16S rRNA gene amplification and denaturing gradient gel electrophoresis. FEMS Microbiol Ecol. 2007;61(1):132–140. doi: 10.1111/fem.2007.61.issue-1. [DOI] [PubMed] [Google Scholar]
- 137.Pajarillo E, Chae J-P, Balolong MP, Bum Kim H, Kang D-K. Assessment of fecal bacterial diversity among healthy piglets during the weaning transition. J Gen Appl Microbiol. 2014;60(4):140. doi: 10.2323/jgam.60.140. [DOI] [PubMed] [Google Scholar]
- 138.Pajarillo E, Chae J, Balolong M, Kim H, Seo K-S, Kang D-K. Pyrosequencing-based analysis of fecal microbial communities in three purebred pig lines. Journal of Microbiology. 2014;52(8):646–651. doi: 10.1007/s12275-014-4270-2. [DOI] [PubMed] [Google Scholar]
- 139.Bomba L, Minuti A, Moisá SJ, Trevisi E, Eufemi E, Lizier M, Chegdani F, Lucchini F, Rzepus M, Prandini A, et al. Gut response induced by weaning in piglet features marked changes in immune and inflammatory response. Funct Integr Genomics. 2014;14(4):657–671. doi: 10.1007/s10142-014-0396-x. [DOI] [PubMed] [Google Scholar]
- 140.Dowd S,E, Callaway T,R, Morrow-Tesch J. Handling May Cause Increased Shedding of Escherichia coli And Total Coliforms in Pigs. Foodborne Pathog Dis. 2007;4(1):99–102. [DOI] [PubMed] [Google Scholar]
- 141.Freestone PPE, Sandrini SM, Haigh RD, Lyte M. Microbial endocrinology: how stress influences susceptibility to infection. Trends Microbiol. 2008;16(2):55–64. doi: 10.1016/j.tim.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 142.Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu X-N, Kubo C, Koga Y. Postnatal microbial colonization programs the hypothalamic–pituitary–adrenal system for stress response in mice. J Physiol. 2004;558(1):263–275. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lupien SJ, McEwen BS, Gunnar MR, Heim C. Effects of stress throughout the lifespan on the brain, behaviour and cognition. Nat Rev Neurosci. 2009;10:434+. doi: 10.1038/nrn2639. [DOI] [PubMed] [Google Scholar]
- 144.O’Mahony L, McCarthy J, Kelly P, Hurley G, Luo F, Chen K, O’Sullivan GC, Kiely B, Collins JK, Shanahan F, et al. Lactobacillus and bifidobacterium in irritable bowel syndrome: symptom responses and relationship to cytokine profiles. Gastroenterology. 2005;128(3):11. doi: 10.1053/j.gastro.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 145.Borewicz KA, Kim HB, Singer RS, Gebhart CJ, Sreevatsan S, Johnson T, Isaacson RE. Changes in the Porcine intestinal microbiome in response to infection with salmonella enterica and lawsonia intracellularis. PLoS One. 2015;10(10):e0139106. doi: 10.1371/journal.pone.0139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Drumo R, Pesciaroli M, Ruggeri J, Tarantino M, Chirullo B, Pistoia C, Petrucci P, Martinelli N, Moscati L, Manuali E, et al. Salmonella enterica Serovar Typhimurium Exploits Inflammation to Modify Swine Intestinal Microbiota. Front Cell Infect Microbiol. 2016;5:106. doi: 10.3389/fcimb.2015.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hoffmann C, Hill DA, Minkah N, Kirn T, Troy A, Artis D, Bushman F. Community-wide response of the gut microbiota to enteropathogenic citrobacter rodentium infection revealed by deep sequencing. Infect Immun. 2009;77(10):4668. doi: 10.1128/IAI.00493-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota (inflammation impairs colonization resistance). PLoS Biol. 2007;5(10):e244. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay B. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of enterobacteriaceae. Cell Host Microbe. 2007;2(2):119–129. [DOI] [PubMed] [Google Scholar]
- 150.Koh H-W, Kim MS, Lee J-S, Kim H, Park S-J. Changes in the swine gut microbiota in response to porcine epidemic diarrhea infection. Microbes Env. 2015;30(3):284–287. doi: 10.1264/jsme2.ME15046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Niederwerder MC. Role of the microbiome in swine respiratory disease. Vet Microbiol. 2017;209:97–106. doi: 10.1016/j.vetmic.2017.02.017. [DOI] [PubMed] [Google Scholar]
- 152.Knights D, Lassen KG, Xavier RJ. Advances in inflammatory bowel disease pathogenesis: linking host genetics and the microbiome. Gut. 2013;62(10):1505. doi: 10.1136/gutjnl-2012-303954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Keeney KM, Yurist-Doutsch S, Arrieta M-C, Finlay BB. Effects of antibiotics on human microbiota and subsequent disease. Annu Rev Microbiol. 2014;68(1):217–235. doi: 10.1146/annurev-micro-091313-103456. [DOI] [PubMed] [Google Scholar]
- 154.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;1476–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wu H-J, Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Micr. 2012;3(1):4–14. DOI: 10.4161/gmic.19320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19(2):59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 157.Burkey TE, Skjolaas KA, Minton JE. BOARD-INVITED REVIEW: porcine mucosal immunity of the gastrointestinal tract1. J Anim Sci. 2009;87:1493–1501. doi: 10.2527/jas.2008-1330. [DOI] [PubMed] [Google Scholar]
- 158.Pluske JR, Turpin DL, Kim J-C. Gastrointestinal tract (gut) health in the young pig. Anim Nutr. 2018. doi: 10.1016/j.aninu.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Thaiss CA, Levy M, Suez J, Elinav E. The interplay between the innate immune system and the microbiota. Curr Opin Immunol. 2014;26:1879–0372. (Electronic):8. doi: 10.1016/j.coi.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 160.Levy M, Thaiss CA, Elinav E. Metagenomic cross-talk: the regulatory interplay between immunogenomics and the microbiome. Genome Med. 2015;7(1):120. doi: 10.1186/s13073-015-0249-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV, Bogatyrev SR, Ismagilov RF, Pamer EG, Turnbaugh PJ, et al. Rapid fucosylation of intestinal epithelium sustains host–commensal symbiosis in sickness. Nature. 2014;514:638. doi: 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kamdar K, Khakpour S, Chen J, Leone V, Brulc J, Mangatu T, Antonopoulos Dionysios A, Chang Eugene B, Kahn Stacy A, Kirschner Barbara S, et al. Genetic and metabolic signals during acute enteric bacterial infection alter the microbiota and drive progression to chronic inflammatory disease. Cell Host Microbe. 2016;19(1):21–31. doi: 10.1016/j.chom.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Iwasaki A, Kelsall BL. Freshly isolated peyer’s patch, but not spleen, dendritic cells produce interleukin 10 and induce the differentiation of T Helper Type 2 Cells. J Exp Med. 1999;190(2):229–240. doi: 10.1084/jem.190.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Haverson K, Rehakova Z, Sinkora J, Sver L, Bailey M. Immune development in jejunal mucosa after colonization with selected commensal gut bacteria: A study in germ-free pigs. Vet Immunol Immunopathol. 2007;119(3):243–253. doi: 10.1016/j.vetimm.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 165.Zhang W, Wen K, Azevedo MSP, Gonzalez A, Saif LJ, Li G, Yousef AE, Yuan L. Lactic acid bacterial colonization and human rotavirus infection influence distribution and frequencies of monocytes/macrophages and dendritic cells in neonatal gnotobiotic pigs. Vet Immunol Immunopathol. 2008;121(3):222–231. doi: 10.1016/j.vetimm.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Neish AS, Gewirtz A, Fau - Zeng H, Zeng H, Fau - Young AN, An Y, Fau - Hobert ME, Hobert M, Fau - Karmali V, Karmali V, et al. Prokaryotic regulation of epithelial responses by inhibition of IkappaB-alpha ubiquitination. Science. 2000;289(5484). (0036-8075 (Print)):4. doi: 10.1126/science.289.5484.1560. [DOI] [PubMed] [Google Scholar]
- 167.Cebra JJ. Influences of microbiota on intestinal immune system development. Am J Clin Nutr. 1999;69(5):1046s–1051s. doi: 10.1093/ajcn/69.5.1046s. [DOI] [PubMed] [Google Scholar]
- 168.Liu H, Dicksved J, Lundh T, Lindberg J. Expression of heat shock proLEteins 27 and 72 correlates with specific commensal microbes in different regions of porcine gastrointestinal tract. Am J Physiol-Gastroint Liver Physiol. 2014;306(12):C1033–C1041. doi: 10.1152/ajpgi.00299.2013. [DOI] [PubMed] [Google Scholar]
- 169.Tamboli C, Fau - Neut C, Neut C, Fau - Desreumaux P, Desreumaux P, Fau - Colombel JF, Colombel JF. Dysbiosis as a prerequisite for IBD. Gut. 2004;53(7):1057. [PMC free article] [PubMed] [Google Scholar]
- 170.Inman CF, Laycock GM, Mitchard L, Harley R, Warwick J, Burt R, van Diemen PM, Stevens M, Bailey M. Neonatal Colonisation Expands a Specific Intestinal Antigen-Presenting Cell Subset Prior to CD4 T-Cell Expansion, without Altering T-Cell Repertoire. PLoS One. 2012;7(3):e33707. doi: 10.1371/journal.pone.0033707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sun J, Hayward C, Shinde R, Christenson R, Ford SP, Butler JE. Antibody repertoire development in fetal and neonatal piglets. I. Four VH Genes account for 80 percent of VH Usage during 84 days of fetal life. J Immunol. 1998;161:5070. [PubMed] [Google Scholar]
- 172.Wang A, Yu H, Gao X, Li X, Qiao S. Influence of Lactobacillus fermentum I5007 on the intestinal and systemic immune responses of healthy and E. coli challenged piglets. Antonie Van Leeuwenhoek. 2009;96(1):89–98. doi: 10.1007/s10482-009-9339-2. [DOI] [PubMed] [Google Scholar]
- 173.Wang AN, Yi XW, Yu HF, Dong B, Qiao SY. Free radical scavenging activity of Lactobacillus fermentum in vitro and its antioxidative effect on growing–finishing pigs. J Appl Microbiol. 2009;107(4):1140–1148. doi: 10.1111/j.1365-2672.2009.04294.x. [DOI] [PubMed] [Google Scholar]
- 174.Kandasamy S, Chattha KS, Vlasova AN, Rajashekara G, Saif LJ. Lactobacilli and Bifidobacteria enhance mucosal B cell responses and differentially modulate systemic antibody responses to an oral human rotavirus vaccine in a neonatal gnotobiotic pig disease model. Gut Microbes. 2014;5(5):639–651. doi: 10.4161/19490976.2014.969972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhang H, Wang H, Shepherd M, Wen K, Li G, Yang X, Kocher J, Giri-Rachman E, Dickerman A, Settlage R, et al. Probiotics and virulent human rotavirus modulate the transplanted human gut microbiota in gnotobiotic pigs. Gut Pathog. 2014;6(1):39. doi: 10.1186/s13099-014-0039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Laycock G, Sait L, Inman C, Lewis M, Smidt H, van Diemen P, Jorgensen F, Stevens M, Bailey M. A defined intestinal colonization microbiota for gnotobiotic pigs. Vet Immunol Immunopathol. 2012;149(3):216–224. doi: 10.1016/j.vetimm.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 177.Lallès J. Microbiota-host interplay at the gut epithelial level, health and nutrition. J Anim Sci Biotechnol. 2016;7(1):66. doi: 10.1186/s40104-016-0123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Arnal M-E, Zhang J, Erridge C, Smidt H, Lallès J-P. Maternal antibiotic-induced early changes in microbial colonization selectively modulate colonic permeability and inducible heat shock proteins, and digesta concentrations of alkaline phosphatase and TLR-stimulants in swine offspring. PLoS One. 2015;10(2):e0118092. doi: 10.1371/journal.pone.0118092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Arnal M-E, Zhang J, Messori S, Bosi P, Smidt H, Lallès J-P. Early changes in microbial colonization selectively modulate intestinal enzymes, but not inducible heat shock proteins in young adult swine. PLoS One. 2014;9(2):e87967. doi: 10.1371/journal.pone.0087967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lallès J-P, Lessard M, Oswald IP, David J-C. Consumption of fumonisin B 1 for 9 days induces stress proteins along the gastrointestinal tract of pigs. Toxicon. 2010;55(2):244–249. doi: 10.1016/j.toxicon.2009.07.027. [DOI] [PubMed] [Google Scholar]
- 181.Malago JJ, Van Dijk JE. The heat shock response and cytoprotection of the intestinal epithelium. Cell Stress Chaperones. 2002;7(2):191–199. doi:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lallès JP, David JC. Fasting and refeeding modulate the expression of stress proteins along the gastrointestinal tract of weaned pigs. Fasting and Refeeding Modulate the Expression of Stress Proteins along the Gastrointestinal Tract of Weaned Pigs. 2011;95:478–488. [DOI] [PubMed] [Google Scholar]
- 183.Arvans DL, Vavricka S, Ren H, Musch M, Kang L, Rocha F, Lucioni A, Turner J, Alverdy J, Chang E. Luminal bacterial flora determines physiological expression of intestinal epithelial cytoprotective heat shock proteins 25 and 72. Am J Physiol-Gastroint Liver Physiol. 2005;288(4):G696–G704. doi: 10.1152/ajpgi.00206.2004. [DOI] [PubMed] [Google Scholar]
- 184.Willem van E. Diet and the Anti-inflammatory effect of heat shock proteins. Endocrine, Metabolic & Immune Disorders - Drug Targets. 2015;15(1):31–36. doi: 10.2174/1871530314666140922145333. [DOI] [PubMed] [Google Scholar]
- 185.Lallès J-P:. Intestinal alkaline phosphatase: novel functions and protective effects. Nutr Rev. 2014;72(2):82–94. doi: 10.1111/nure.12082. [DOI] [PubMed] [Google Scholar]
- 186.Geddes K, Philpott DJ. A new role for intestinal alkaline phosphatase in gut barrier maintenance. Gastroenterology. 2008;135(1):8–12. doi: 10.1053/j.gastro.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 187.Bates JM, Akerlund J, Mittge E, Guillemin K. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe. 2007;2(6):371–382. doi: 10.1016/j.chom.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Malo MS, Alam SN, Mostafa G, Zeller SJ, Johnson PV, Mohammad N, Chen KT, Moss AK, Ramasamy S, Faruqui A, et al. Intestinal alkaline phosphatase preserves the normal homeostasis of gut microbiota. Gut. 2010;59(11):1476. doi: 10.1136/gut.2010.211706. [DOI] [PubMed] [Google Scholar]
- 189.Mizumori M, Ham M, Guth PH, Engel E, Kaunitz JD, Akiba Y. Intestinal alkaline phosphatase regulates protective surface microclimate pH in rat duodenum. J Physiol. 2009;587(14):3651–3663. doi: 10.1113/jphysiol.2009.172270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Trevisi P, Pérez JF. Diets and pig gut health: preface. Anim Feed Sci Technol. 2017;233:87–88. doi: 10.1016/j.anifeedsci.2017.11.005. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated for this study.


