ABSTRACT
Colonization of the gastrointestinal tract with microorganisms during infancy represents a critical control point for shaping life-long immune-mediated disease susceptibility. Abnormal colonization or an imbalance of microbes, termed dysbiosis, is implicated in several diseases. Consequently, recent research has aimed at understanding ways to manipulate a dysbiotic microbiome during infancy to resemble a normal, healthy microbiome. However, one of the fundamental issues in microbiome research is characterizing what a “normal” infant microbiome is based on geography, ethnicity and cultural variations. This review provides a comprehensive account of what is currently known about the infant microbiome from a global context. In general, this review shows that the influence of cultural variations in feeding practices, delivery modes and hygiene are the biggest contributors to microbial variability. Despite geography or race, all humans have similar microbial succession during infancy.
KEYWORDS: Infant, gut microbiome, colonization, breastfed, full-term
Introduction
The symbiotic relationship between mammals and the trillions of microorganisms that reside in their gastrointestinal tract plays a critical role in host health and disease susceptibility. Collectively termed the gut microbiome, these organisms and their associated phenomes educate the immune system, protect against infectious diseases and contribute to host nutrient absorption. At parturition, a newborn’s immune system is not completely developed and requires signals from the gut microbiome trigger maturation.1 In the absence of microbial exposures, germ-free (sterile) mice show extensive immune and intestinal morphological defects.2 Reconstitution of germ-free mice with bacterial communities can restore many of these immunological abnormalities suggesting that if we can change our microbiome, we can alter host physiological deficiencies. However, appropriate induction of immune maturation in the gut extends beyond the mere presence of bacteria, and requires microbes, which have coevolved with the host.3 Therefore, the question becomes can we alter the gut microbiome and if so does time of induction matter? The microbial corrections may be age dependent and it has been hypothesized changes must occur within a certain time-frame during infancy to ensure appropriate intestinal immune development.4 In humans, current evidence indicates that microbial diversity increases in a stepwise manner during the first few years of life and stabilizes to an adultlike microbiome at approximately 3 y of age.5 Infants with an abnormal temporal progression of microbes during these first 3 y could be predisposed to negative health outcomes due to improper mucosal immune maturation as a result of dysbiosis. Consequently, there has been an increase in research aimed at defining the successive colonization stages of gut bacteria in infants responsible for maturation of a healthy host immune system.
While generalizing a stepwise progression in infant colonization may be a logical first step, this may not actually be meaningful across geographic locations given that microbes have coevolved with various human cultures and behaviors. As early human ancestors settled in various geographic locations, their gut microbial communities adapted to these new environments to optimize nutrient absorption from available food sources. Consistent with this notion, substantial divergences in adult gut microbial structures have been found in different populations of adults.6 Hunter-gatherer populations, such as the Hadza tribesmen of Tanzania, are characterized by high Prevotella spp. and Treponema spp., whereas urban populations are enriched in Bacteroides spp., Bifidobacteria spp. and Firmicutes. These divergences are often attributed to dietary habits whereby foraging populations consume diets consisting of fiber-rich foods, and urban populations consume more processed foods rich in simple sugars, emulsifiers and oxidizable fats. While Bifidobacteria spp. are often cited as health-promoting bacteria, lower abundances of these species in Hadza populations do not appear to negatively impact Hadza health.7 Instead, it appears that the Hadza people coevolved with their gut bacterial communities to allow for effective nutrient extraction from cellulose and fiber-rich foods, increasing their overall fitness and survival.8
Given the apparent geographic and genetic impact on the adult microbiota,9 one key question is whether lifestyle, geographic distance, sex and gender, cultural ethnicity and overall social economic development impacts the acquisition of gut microbes in infants. A number of reviews have been published summarizing the infant colonization process, many presuming that geography does a play a role in the infant bacteriome given variations seen in literature, but a comprehensive account of variations in the infant microbiome based on geography with consideration of cultural behaviors is long overdue. In this review, we describe the global range of profiles that constitute a healthy, full-term infant gut bacteriome, the bacterial community within the gut microbiome, and highlight what is currently known across 53 different countries (Table S1). We conclude that cultural variations in feeding practices, delivery modes and hygiene drive the succession of the infant bacteriome given there is more variation from person to person in any population than there is from breastfed vaginally birthed full-term infants from any ethnic group around the world. This means that human behavior is the biggest predictor of bacterial colonization in the infant gut (summarized in Figure 1). While we cannot change our own genotype easily, we can control our behavior and therefore understanding how to alter the gut bacteriome toward a healthy community may be one of the biggest contributions to promoting self-care this century.
Figure 1.

Human behavior and bacterial colonization in infants. Several features impact the establishment of bacterial communities in the infant gastrointestinal tract. Two of the most influential human behaviors that has affected infant gut colonization, globally are mode of delivery and diet. More subtle impacts may be driven by diet-driven changes to breast milk and hygiene practices.
Despite geography and race, interventions of birth mode reduce Bifidobacterium, Bacteroides and Lactobacillus spp. as primary colonizers in the infant gut
Until recently, the intrauterine environment was believed to be sterile. The consensus that the fetus is maintained in a sterile environment was established in the early 1980s when culture-based techniques failed to cultivate bacteria from amniotic fluid and meconium samples.10 However, recent studies using newer molecular techniques based on DNA methodology have questioned the validity of previous culture-based findings and report complex bacterial communities.11 Still, others maintain that any DNA detected using DNA-based methods may represent contaminants.12 Whether infants are colonized prior to birth or not remains unclear, most agree that mode of birth, vaginal or Cesarean section (C-section), impacts infant gut microbial communities. Numerous epidemiological studies illustrate that infants delivered by C-sections have increased risk of allergic rhinitis, type I diabetes, obesity, asthma and other immune deficiencies.13 This is concerning given the escalated frequency of non-medical C-section deliveries in recent years. China, similar to several developing nations, has seen C-sections rates increase from 5% in 1970s to more than 46.2% as their economy grows stronger.14 Given that increasing popularity of elective C-section deliveries parallels a rise in prevalence of non-communicable diseases, a contributing factor in this association may be attributable to differences in pioneering bacteria between vaginally birthed and C-section delivered infants.
When infants are vaginally birthed, they are inoculated with their mothers’ microbiota as they pass through the birth canal. This idea is supported through studies in both developed and developing countries. In Venezuela, vaginally delivered neonates contain established vaginal bacterial taxa like Lactobacillus, Prevotella, Atopobium or Sneathia spp.15 These infants have communities more similar to their own mother than to other vaginally delivered infants suggesting that vaginal communities unique to each mother are vertically transmitted to their infants. As expected, C-sectioned babies in this cohort lacked bacteria from mothers’ vaginal microbiome and were instead dominated by communities found on the skin surface.16 Interestingly, C-sectioned babies were no more similar to their own mothers’ skin bacterial communities than to others despite the fact that adult skin microbes are highly personalized. Similar cases of the maternal vaginal microbes colonizing the neonatal infant gut have been reported in other developing countries like Estonia17 and Puerto Rico18 and the same holds true in developed countries. More than 70% of the Swedish infants vaginally birthed are colonized by bacterial species found in their mothers’ stool, whereas only 41% of matching spp. were detected in C-sectioned infants.19 Given the vertical transmission between mothers and vaginally delivered infants, as well as the environmental impacts on C-sectioned infants, one would expect to see geographic divergences in infants born in developed and developing countries where adult and environmental microbial communities differ. However, current reports do not support this theory and instead, general trends can be found between C-sectioned and vaginally birthed infants, regardless of geographic location or even genetics.
In general, reports on C-section delivered infants show lower abundances of Lactobacillus spp. when compared to vaginally delivered infants. This has been shown in developing countries such as Venezuela15 and India,20 as well as developed countries such as Greece,21 Finland22 and the United Kingdom.23 Alongside Lactobacillus spp., Bifidobacteria spp. are often reported as decreased or having delayed colonization in C-sectioned infants. Bifidobacteria spp. are commonly cited as one of the major components of the infant intestinal microbiome. While the abundance of Bifidobacteria spp. is often attributed to diet, mode of delivery also appears to have an effect on early colonization. Mother-to-infant transmission of Bifidobacteria longum subsp. longum was first reported in vaginally delivered infants in Belgium.24 A follow-up study from Japan25 expanded on this finding and found that 92% of vaginally delivered infants had at least one monophyletic Bifidobacteria strain with their mother, whereas no monophyletic strains were identified in C-sectioned infants. Reduced transmission of Bifidobacteria spp. from mothers undergoing C-section procedures has been shown to decrease or delay Bifidobacteria spp. colonization in infants in several studies conducted in developed countries such as Finland,22,26 Sweden,27 Greece,21 Italy,28,29 France30 and the Netherlands31 as well as in developing countries such as Singapore32 and India.20,33 However, there are some exceptions reported in the literature. For example, Fallani et al. report no difference in the relative proportion of Bifidobacteria spp. by delivery mode (both were approximately 40% of the total), in infants born in Sweden, Scotland, Germany, Italy and Spain.34 Instead they found C-section delivery was associated with lower Bacteroides spp. and higher Streptococcus spp. and Clostridium coccoides. This reported decrease in Bacteroides spp. in infants delivered by C-section is another common finding. Like Bifidobacteria spp., Bacteroides spp. have become a major focus of human gut microbiology. Many studies have found that Bacteroides spp. are less abundant in C-sectioned infants in developed countries such as Sweden,19,27,35 France,30 the UK,23,36 Canada,37 Finland,22 the Netherland31 as well as in developing countries such as India20 and Brazil.38 Collectively, these studies report a global decreased or delayed presence of bacteria such as Bifidobacterium, Lactobacillus and Bacteroides spp. in C-sectioned infants, due to the lack of exposure to mothers’ vaginal microbes during the birthing process.
In leu of vaginal bacteria, neonates delivered by C-section are colonized by potentially pathogenic bacteria such as Clostridium and Klebsiella spp. A comparative study of five European countries including Sweden, Scotland, Germany, Italy and Spain found higher Clostridium spp. in C-sectioned infants compared to vaginally delivered infants.34 Using culture-based methods, a separate study comparing the fecal microbiota of infants from Sweden, the United Kingdom and Italy similarly found higher Clostridium and Klebsiella spp.36 Independent European studies in the Netherlands,31,39 Sweden27 and Finland22 likewise report increased Clostridium spp. colonization rates in C-sectioned infants. Therefore, in developed countries, it appears that Bifidobacterium spp. and Bacteroides spp. are largely replaced by Clostridium spp. and Klebsiella spp. There are, however, some exceptions reported in the literature. For example, a fluorescent in situ hybridization (FISH)-based study in Finland found that while Bifidobacteria spp. were 1,300-fold lower in C-section delivered infants at 1 month of age, there were no differences in Clostridium, Bacteroides and Lactobacillus spp. between vaginally and Cesarean-born infants.26 Instead, C-section deliveries resulted in a threefold lower total bacterial cell number in infants when compared to those born by vaginal deliveries. Nevertheless, the health impacts of higher Clostridium spp. and Klebsiella spp. reported in a majority of studies should be considered. In the United States, Goedert et al. found higher levels of certain Clostridium spp. in adults who had been born by C-section when compared with adults who had been born vaginally.40 This suggests that differences in initial communities may lead to life-long differences in the microbial communities. It is therefore an auspicious finding that exposure of newborns delivered by C-section to maternal vaginal fluids at birth can at least partially restore Lactobacillus spp. and Bacteroides spp.18 However, this “vaginal seeding” does not alleviate other factors accompanying C-section procedures that may drive microbial phenotypes such as labor and antibiotic administration.
While a global reduction in Bifidobacteria spp., Bacteroides spp. and to a lesser extent, Lactobacillus spp. in infants delivered by C-sections are replaced with potentially pathogenic bacteria such as Clostridium spp., and Klebsiella spp. in developed countries, the trend in developing countries remains to be fully elucidated due to the paucity of research and conflicting results. For example, a 16S rRNA cloning and sequencing study from India followed trends established in Europe, whereby C-sectioned infants were susceptible to colonization of potentially pathogenic bacteria from the environment, while vaginally delivered infants acquired their initial microbes from their mothers. In this study, 12 breastfed, C-section delivered infants had a predominance of Clostridium spp., Citrobacter spp. and E. coli at 1 week (65% of the clone library) and that the vaginally delivered infants had a predominance of Acinetobacter, Bifidobacteria and Staphylococcus spp.33 In stark contrast, a study in Brazil conducted on 13 C-sectioned and 46 vaginally delivered infants reported a decrease in the abundances of Clostridium spp. in the transitional stool of C-sectioned infants.38 In Venezuela, the lack of vaginal exposure resulted in early establishment of communities originating from the birth environment and skin surfaces.15 As a result, C-sectioned infants had a higher relative abundance of Staphylococcus, Corynebacterium and Propionibacterium spp. These discrepancies could be due to differences in methodologies, feeding practices, sampling age or place of delivery.39 Future studies are needed in to better understand the impact mode of birth has on pioneering bacteria in developing countries.
Diet is a strong predictor of infant microbial colonization
Alongside delivery mode, feeding practices can profoundly alter the trajectory of acquisition of the gut microbiota in infants. In fact, feeding practices can partially compensate for microbial differences between infants delivered vaginally or by C-section.23 There are two main nutritional sources available for infants: breast milk and formula. While breastfeeding is an unequaled way to ensure infant health and survival, there are several social and economic factors that influence the prevalence and duration of breastfeeding. For example, in Japan, breastfeeding has been a strong practice for centuries and Buddhist teachings promote extended breastfeeding in children to the age of 6.41 In stark contrast, Atlanta in the United States issued a law banning public breastfeeding for children over the age of two, citing public indecency. Similarly, in Ukraine, breastfeeding is viewed as socially unacceptable. Globally these social restrictions, religious viewpoints and cultural restraints have reduced breastfeeding practices. Data collected by UNICEF and the World Health Organization show that no country in the world fully meets the recommended standards for breastfeeding. In fact, only 23 countries were shown to have exclusive breastfeeding rates above 60% in the first 5 months postpartum.42 Instead of lactation, mothers elect to feed their baby using infant formulas. Disparities between the microbiome of breastfed and formula-fed infants are frequently cited in the literature. However, far fewer studies report differences in the infant microbiome based on breast milk bacterial and nutritional composition. As mentioned above, adult dietary habits vary by geographic region. It is possible that alterations in dietary intake during lactation may influence the composition of mothers’ breast milk, ultimately leading to altered colonization events in suckling infants. However, region-specific factors influencing breast milk properties are largely unexplored. The following discussion is organized into two main sections. First, the impact of geographic location on breast milk composition will be explored. Second, we will compare the impact of formula feeding and breast milk on the infant microbiome in various countries.
Global dietary habits alter breast milk lipid profile with limited impacts on the milk bacteria
Human breast milk is a complete source of nutrients and bioactive substrates required for optimal development and health in nursing infants. Human milk is composed of macronutrients such as protein, fats and carbohydrates, vitamins, minerals, digestive enzymes, immune cells and even bacteria. Theoretically, geography, ethnicity and cultural variations could alter any of these components. However, here we focus on the geographic impacts on breast milk macronutrients and bacteria, which are more frequently reported in the literature.
While there is considerable temporal and inter-personal variability in breast milk,43 maternal nutritional status has little to no effect on the total amount of macronutrients (protein, carbohydrate, fat) in breast milk. Even if women consume less than the recommended dietary allowance, suckling infants will receive the needed macronutrients at the expense of maternal stores. In the Gambia, for example, the dietary mean intake is around 1,700 kcal/d.44 Despite this, the mean energy content of mother’s milk is maintained at 72 kcal/100 ml, which is comparable to well-nourished mother’s milk.45 It is not surprising then, that the macronutrient composition of human milk is remarkably conserved across populations.46 However, while the total macronutrient status is relatively stable, the vitamin/micronutrient content as well as biochemical makeup of macronutrients does vary with the mother’s nutrient intake. For example, veganism can result in vitamin B12 deficiency in the infant.47 Similarly, types of fatty acids like saturated, monounsaturated and polyunsaturated can vary due to mother’s diet.48 Given the geographic variations in diet, fatty acid proportions in breast milk should vary by country. Indeed, Kumar et al.49 show that breast milk fatty acid profiles differ between Europe (Spain and Finland), Africa (South Africa) and Asia (China). In this study, it was reported that the highest variability in milk samples was observed in the omega-6 polyunsaturated fatty acids, which were increased in both the triacylglycerides and phospholipids of Chinese milk samples, likely due to the high consumption of soy products in this country. A recent study in Denmark and the United States similarly shows national differences in milk fat content.43 These geographic variations in breast milk fat profiles are attributed to differences in diet and may affect colonization events in breastfed infants. In support of this notion, in nonhuman primates, it was shown that maternal high-fat diet (36% fat from animal fat and corn oil) consumed during gestation and lactation alters the composition of their offspring’s microbiome for up to 1 y, irrespective of the infants’ post-weaning diet.50 A follow-up study in the United States sought to understand if this finding could be recapitulated in a human cohort.51 To approach this, mother–infant dyads were separated into groups based on mother’s dietary fat intake during gestation. Culture-independent 16 S rRNA gene sequencing was used to characterize the neonates gut microbiome at the time of delivery. The authors found that the neonatal microbiome significantly clustered according to maternal diet, whereby Bacteroides spp. were depleted in infants exposed to a maternal high-fat diet in pregnancy. The authors concluded that maternal high-fat diet consumed during gestation can alter their offspring’s microbiome. However, this publication has several limitations with the major one being that maternal dietary intake of fat was estimated from a dietary screener questionnaire. The fat content in breast milk was never quantified and as a result, it is not possible to tell which fatty acids were altered. Given that the type of fatty acids in a high-fat diet uniquely alter the gut bacteriome,52 data from “high-fat” studies are hard to interpret without quantifying fatty acid profiles. Regardless, this evidence suggests that geographic- or diet-related factors could drive differences in fatty acid profiles in breast milk. However, more studies investigating the relationship between maternal dietary fat intake and associated changes to the milk lipid profiles are needed.
In addition to fats, it has been suggested that human milk oligosaccharides vary geographically.53 Human milk oligosaccharides represent the third largest portion of human milk following lactose and lipids.54 In a comparison of 109 human milk samples collected 1 month post-partum from Australia, Japan, the USA, Norway and South Africa55 it was shown that milk metabolites, such as human milk oligosaccharides, lactose and other milk sugars cluster by country of origin. Since many of these milk metabolites can be consumed by infant gut microbiota, ethnic differences in human milk metabolites may have a strong bearing on the bacterial community composition in infants or even, in the breast milk itself.
An interesting observation by West et al.56 in the 1970s suggested that breast milk may not be sterile. Today, numerous studies reported a highly diverse microbial community in human milk.57–60 Bacteria in human milk may constitute one of the main sources of gut bacteria in breastfed infants. Although it was originally thought that bacteria in breast milk are acquired from skin or fecal contamination, several recent studies support an entero-mammary pathway involving dendritic cells, macrophages and microfold cells. This controversial pathway is thought to allow for vertical transfer of bacteria from the maternal intestinal tract, to breast milk and finally to the infant’s gut. In support of this, Qutaishat et al. show transmission of Salmonella enterica subspecies enterica serovar Typhimurium DT104 from maternal gut to maternal mammary gland.61 Similarly, a study in Syria showed that identical random amplified polymorphic DNA (RAPD) genotypes of L. fermentum, L. plantarum. L brevis, Enterococcus faecium, E. faecalis and Pediococcus pentosaceus were isolated from mother’s stool, breast milk and corresponding infant stool.62 Bifidobacteria and Staphylococcus spp. have also been shown to transfer to infants.63,64 For example, one study investigating the potential mother-to-child transmission of Staphylococcus, Lactobacillus and Bifidobacteria spp. in 20 Spanish participants found the same bacterial strains in 11, 9 and 3 mother-infant pairs, respectively.63 Therefore, regional differences in mother’s gut microbial communities may impact the types of bacteria transferred from the maternal gut to their breast milk. Moreover, since maternal diet influences the biochemical makeup of macronutrients in human milk, geographic variations in dietary intake during lactation may indirectly select for bacterial species capable of utilizing various substrates in milk.
The idea that individual variations in breast milk bacteria may be influenced by geographic or dietary factors is supported by two independent studies. The first compared mature breast milk samples collected at 1 month of lactation from 20 participants in Europe (Spain and Finland), Asia (China) and Africa (South Africa) using the Illumina MiSeq sequencing platform.49 This study found that milk samples collected from South Africa had a predominance of Proteobacteria whereas samples from Spain, China and Finland were all enriched with Firmicutes. At the genus level, Pseudomonas spp. were prevalent in the milk samples collected from South Africa, followed by Staphylococcus and Streptococcus spp. whereas breast milk collected from Spain and Finland both had a predominance of Staphylococcus spp. followed by Streptococcus spp. In China, Streptococcus spp. predominated followed by Staphylococcus spp. In general, milk samples collected from South Africa had higher abundances of Proteobacteria when compared with the other countries, which could be due to high Enterobacteriaceae and Pseudomonadaceae found in South African and Spanish milk samples. Spanish samples had a high relative abundance of Propionibacterium and Pseudomonas spp. compared with the other countries. Finnish milk samples had the lowest relative abundance of Proteobacteria and Chinese samples had the highest Actinobacteria. Other noteworthy findings include the fact that Enterococcaceae spp. were found in breast milk collected from every geographic region except China. In contrast, Bifidobacteria spp. were only found in samples collected from South Africa and Lactobacillus spp. were only found in samples collected from Finland. When milk bacteria profiles were combined with fatty acid profiles, it was found that fatty acids profiles associated with differences in the milk bacteria.49 In all samples, monounsaturated fatty acids found in milk triacylglycerides were negatively associated with Proteobacteria. Monounsaturated fatty acids in triacylglycerides were also inversely correlated with abundances of Pseudomonas and Staphylococcus spp. Both monounsaturated fatty acids and polyunsaturated fatty acids in triacylglycerides positively associated with Streptococcus spp. whereas saturated fatty acids were negatively correlated with relative abundances of both Streptococcus and Acinetobacter spp. In contrast, saturated fatty acids found in both milk triacylglycerides and phospholipids were positively associated with Pseudomonas spp and n-3 polyunsaturated fatty acid concentrations in milk phospholipids were negatively correlated with Bifidobacteria spp. Taken together, this study supports the notion that geographic variations in breast milk lipid profiles align with differences in milk bacterial composition.
The second cross-cultural study supporting the notion that individual variations in breast milk bacteria may be influenced by geographic location compared milk samples collected from different regions in East Asia.65 In this study, milk samples were compared using 454 pyrosequencing of 16 S rRNA genes between Taiwan and six-regions of mainland China. The relative abundance of Pseudomonadaceae was found to be lower in samples from Taiwan (n = 31) compared to East (n = 34), Central (n = 24) and Northeast (n = 17) China. Similarly, North (n = 11) and Southwest (n = 9) China had lower relative abundance of Pseudomonadaceae than Central China. Finally, the relative abundance of Staphylococcaceae was higher in samples collected from Taiwan than East and Central China. Despite these notable differences, however, there was a high degree of overlap in the principal component analysis comparing the whole milk bacteriomes. Moreover, it seems that as with most natural assemblages in ecology,66 few species comprise the majority of the individuals in breast milk from both cross-cultural studies. This ‘core’ set of bacteria is consistently reported in independent studies worldwide.
Assessment of bacterial diversity in breast milk using culture-dependent and independent approaches shows that Streptococcus and Staphylococcus spp. are almost always among the most abundant genera in breast milk, irrespective of geographic or cultural variables. This includes findings reported from the United States,57 Canada,67 Switzerland,60,68 Spain,59,69,70 Slovenia,71 Mozambique,72 Taiwan65 and China.65,73 Other prevalent bacteria include lactic acid bacteria such as Lactobacillus, Weissella and Leuconostoc spp., certain Proteobacteria such as Pseudomonas spp. and certain Actinobacteria such as Propionibacterium and Corynebacterium, globally. While these frequently reported taxa appear to make up a common ‘core’ breast milk microbiome worldwide, data support a continuous gradient of these genera associated with interindividual variation.
High-interindividual variability in breast milk bacterial communities is commonly reported within a single population.57,59,70,73,74 For example, pyrosequencing of breast milk collected from 16 women in the United States found that in one subject, Staphylococcus spp. were the first or second most predominant genera accounting for 22-59% relative abundance over a four-week period whereas in another subject, Staphylococcus spp. only represented <5% of bacteria.57 Serial samples collected from a single individual tended to be more similar when compared to other individuals; however, while some participants had relatively stable milk bacterial communities over time, others had bacterial genera shifts. This high-interindividual variability within a single population makes it difficult to draw geographic conclusions from independent or underpowered studies. For instance, two independent studies in Spain assessing the bacterial communities in mature breast milk using qPCR found varying results. One found that the median count (log n° copies/ml) of Lactobacillus spp. (4.31) were highest in breast milk collected from 13 healthy women, followed by Enterococcus (3.95), Streptococcus (3.58) Staphylococcus (3.28) and Bifidobacteria spp (2.45).75 All bacteria were found at 100% prevalence except for Staphylococcus spp. which were only in 77% of samples collected. A second study found a predominance of Streptococcus (4.5 mean log10 copies/ml, 100% prevalence), followed by Lactobacillus (3.74, 100%), Bifidobacteria (3.56, 100%), Staphylococcus (3.55, 100%), Clostridium cluster XIVa-XIVb (3.32, 96%); Enterococcus (2.56, 76%), and Bacteroides spp. (2.02, 40%).69 Discrepancies between the abundance and prevalence of milk bacteria in these two studies within the same country could be attributed to methodical differences but the extraction kit and several of the primers used in this study were similar. Instead, differences between these studies may be due to high interindividual variability. While it is well recognized that low statistical power increases the probability of type II errors, paradoxically, low statistical power also increases the likelihood of finding a false positive.76 Therefore, the geographic differences reported in the two cross-cultural studies above may be falsely positive and instead, all breast milk samples contain a gradient of ‘core’ bacteria which varies between individuals.
The mechanisms driving high interindividual variation in breast milk remain poorly understood, but is presumably promoted by diet, genetics or possibly even mode of delivery. In Finland, higher Bifidobacteria spp. were reported in breast milk of women who delivered by vaginally at 6 months lactation when compared to mothers who delivered by C-section.58 In contrast, Streptococcus spp. were higher in the breast milk of mothers who delivered by C-section. In Spain,59 Staphylococcus spp. were marginally (P = .052) higher in the milk samples of women who delivered by C-section (n = 4). However, mode of delivery had no impact on milk samples collected in Canada67 or China.73
Collectively these studies show that the influence of geographic location and culture does not greatly influence the ‘core’ bacterial composition of human breast milk; however, variations in dietary habits or mode of delivery may shift the relative abundance of bacteria to favor one ‘core’ genera over another. The biological impact of this breast milk gradient on the establishment of the infant microbiome remains unclear. However, as the presence of these ‘core’ bacterial groups in human breast milk are among the first colonizers in the infant gut, it stands to reason that variations in breast milk bacteria would alter the colonization process in infants. Future large-scale studies examining geographic variations in milk bacteria and corresponding bacteria in infants may begin to address this notion.
Formula feeding alters normal microbial colonization patterns in the infant gut
While mothers are increasingly tasked to work and rear children simultaneously, as well as lack societal and economic support, one of the most influential human behavior that has negatively affected infant gut health is formula feeding. Infant formula is intended to be an effective substitute for breast milk and while mostly not true, is advertised to be formulated to mimic the composition of breast milk. Supplemental feeding with formulas is reported in over 80% of the infants in developing countries including Venezuela, Gabon, Chad, Somalia, Tunisia, Yemen, Romania and Belarus.42 The low prevalence of exclusive breastfeeding, especially in countries in Africa, coincides with the highest rates of infant malnutrition in the world.77 The complexities of the breast milk are not fully appreciated so while formulas are intended to mimic the properties of breast milk, epidemiologic studies consistently show that formula-fed infants are more susceptible to enteric infections.78 Differences in health outcomes between formula and breastfed children may be due to differences in early colonization events. Indeed, many reports have shown that breastfed and formula-fed infants have distinct gut colonization patterns, but these results are not without contradictions in literature.
One notable difference between breast milk and formula is the absence of bacteria. Supplementing infant formulas with bacteria is currently not recommended by The Codex Alimentarius Commission79 due to limited evidence of their clinical efficacy. Nevertheless, worldwide infant formulas supplemented with probiotics are commonly used. Even though Staphylococcus spp., Streptococcus spp. and Proteobacteria predominate breast milk globally, the typical probiotic strains included in formulas are Bifidobacterium and Lactobacillus spp. The supplementation is proposed to be necessary because formula-fed infants have lower abundances of Bifidobacteria spp. when compared to breastfed infants.80 This has been reported in developed countries such as the Netherlands,81,82 the United Kingdom,83 Sweden,19 Greece84 and Japan.80 Since Bifidobacteria spp. are associated with proxies for desirable health outcomes, it is thought that the lower abundances of these species in formula-fed infants may result in improper mucosal immune maturation. Indeed, administration of Bifidobacteria breve and oligosaccharides in C57BL/6 J male mouse pups enhanced gut immune and endocrine development in suckling mice.85 Therefore, investigating ways to increase proportions of Bifidobacteria spp. in formula-fed infants has become an active area of research. The increased proportion of Bifidobacterium spp. in breastfed infants could, in theory, be due to bifidobacteria in human breast milk. However, while breast milk has been shown to harbor several bifidobacterial species such as B. longum, B. breve and B. bifidum, most milk samples show low or absent Bifidobacteria spp. For example, in Japan, it was shown that no bifidobacteria were detectable in breast milk collected before delivery or colostrum, but could be detected 1-week post-partum or later.64 Similarly, in Spain, bifidobacteria were only isolated from 8 out of 23 milk samples collected between 3 and 7 d postpartum.86 Therefore, it is not likely that the abundance of Bifidobacteria spp. in breastfed infants is due to breast milk bacterial colonies. Nevertheless, some groups have tried to increase Bifidobacteria spp. in formula-fed infants by seeding formula with Bifidobacteria spp. In a double-blind, randomized, placebo-controlled study of newborn infants in Germany, formula containing 107 colony forming units/g of B. bifidum, B. breve, B. longum, and B. longum subsp. infantis did not result in long-term colonization of Bifidobacterium strains in the formula-fed infant gut.87 Another study sought to understand if probiotic supplements could ameliorate the effects of antibiotic use or C-section deliveries in Finland.88 In this study, both mothers and infants were given a multispecies probiotic, consisting of B. breve, Propionibacterium freundenreichii subsp. shermanii, L. rhamnosus, and L. rhamnosus GG. The results showed that probiotic supplementation increased Bifidobacteria spp. in breastfed, C-sectioned infants, but not in formula-fed infants. Based on the study design, however, it is not possible to determine if these effects were due to infant or maternal probiotic supplementation. An observational study corroborates these findings and shows maternal and infant supplementation with probiotics associated with higher abundances of Bifidobacteria spp. at 1 week of age.89 However, an inverse correlation between probiotic supplements during infancy and mucosal-associated illness in toddlers was reported. Given this, probiotic supplementation during infancy should be approached cautiously until rigorous controlled follow-up studies determining safety and efficacy have occurred.
A better approach to increasing Bifidobacteria spp. in formula-fed infants is to provide appropriate conditions to promote the growth and productivity of Bifidobacteria spp. It is thought that the higher proportions of Bifidobacteria spp. in breastfed infants is due to selective pressures exerted by available dietary substrates. Bifidobacteria spp. have been shown to effectively metabolize human milk oligosaccharides through the bifidus pathway,90 giving them a competitive advantage over other bacterial species in the infant gut. There are three broad classes of human milk oligosaccharides, each containing various types of oligosaccharides, including neutral fucosylated, neutral nonfucosylated and acidic molecules.91 Certain types of these human milk oligosaccharides are utilized by various Bifidobacteria spp. more than others. For example, B. infantis strains are capable of metabolizing all types of milk oligosaccharides including sialylated and fucosylated molecules, whereas B. breve targets complex fucosylated molecules. In this sense, different human milk oligosaccharides give selective advantages to different bacterial species. The actual structures of human milk oligosaccharides are not easy to elucidate, and large-scale synthesis of these molecules is challenging. As a result, oligosaccharides in formulas do not effectively mimic human milk oligosaccharides, and other taxa which can effectively degrade these carbohydrates dominate. Therefore, differences in Bifidobacteria spp. between formula and breastfed infants can be attributable to differences in available oligosaccharides between the two nutrient sources.
Despite this, not all studies show decreased Bifidobacteria spp. in formula-fed infants. For instance, a study comparing Bifidobacteria, Escherichia and Clostridium spp. between 50 breastfed and 50 formula-fed infants in the Netherlands found that bifidobacterial counts were comparable using real-time PCR.92 A culture-dependent study in Sweden similarly found no differences between breastfed and formula-fed infants and reported that in both groups, Bifidobacteria spp. occurred in less than half of the samples collected.93 A pyrosequencing study in Europe likewise revealed a close relationship in the fecal microbial composition of breastfed and formula-fed infants born in Italy, Spain and Ireland.94 The authors concluded that bifidobacterial abundances may not be influenced by diet alone. In this vein, the age of infants should also be considered when comparing profiles between breastfed and formula-fed infants. A comprehensive study in Sweden reported no differences between exclusive breast-feeding or mixed feeding on the newborn microbiome.19 However, these infants diverged overtime and the gut microbiome in the infants exposed to formula “matured” quicker than the microbiome in those exclusively breastfed. Namely, at 4 months of age, formula-fed infants had elevated C. difficile, Granulicatella adiancens, Citrobacter spp. E. cloaceae and Bilophila wadsworthia spp.,19 whereas exclusively breastfed infants had increased abundances of L. johnsonii/L. gasseri, L. paracaseil/L. casei and B. longum. Developing countries show similar results. In Brazil, it was reported that Bifidobacteria spp. predominated the infant gut regardless of diet95 and in India, it was reported that supplemental feeding associated with a reduction in Enterobacteriaceae.20 Discrepancies between these findings could be attributed to differences in methodology, age, geographic location or mode of birth; however, a more likely explanation is differences in the formulas themselves. Formulas are available in various forms including powder, liquid or ready-to-feed, and are manufactured using various bases including cow-milk, goat-milk, soy, hypoallergenic, rice-milk and many more.96 While a detailed comparison between formulas extends beyond the scope of this review, it is important to acknowledge that formulas from a single manufacturer, in a single country will vary depending on the formula type purchased. This holds true in both developed countries like Italy,97 and developing countries like Brazil.98 Therefore, different formulas purchased within a single population may result in different microbial colonization patterns due to variations in product composition.
Interestingly, studies performed in both developed and developing countries show that the addition of a prebiotic compound to formula, such as galactooligosaccharides or fructooligosaccharides,99 can increase bifidobacteria proportions in formula-fed infants. In support of this, one study in Japan found that all major Bifidobacterial spp. were higher in mixed-fed infants compared to exclusively breastfed infants.100 A separate study in the United Kingdom also found that prebiotic-fed infants have more Bifidobacteria spp. than breastfed or non-prebiotic, formula-fed infants.101 These findings were recapitulated in developing countries such as Estonia where it was shown that the highest proportion of Bifidobacteria spp. were in infants receiving prebiotic formula.102 Similar results were seen in Indonesian infants fed probiotic/prebiotic formula.103 Collectively, this data shows that formula-fed infants often have lower bifidobacteria in their gut because many formulas do not adequately mimic the nutrient composition of breast milk. Formulas that are fortified with prebiotics can increase Bifidobacteria spp. counts reducing the variability between breastfed and formula-fed infants. This is applicable to infants in both developing and developed countries. Therefore, improving the oligosaccharides in formula to better mimic breast milk may be the better strategy to increase Bifidobacteria spp. in formula-fed infants. Collectively, these reports show that niche dynamics, or the competition for nutritional resources essential for growth, give rise to patterns in the abundance of species in the infant gut. Any geographic differences reported in formula-fed infants is likely because of the nutritional content of product consumed and is not likely attributable to genetic, cultural or environmental differences.
Hygiene practices impact the ecological succession of bacterial communities in infants
Early colonization studies characterized the infant microbiome using carefully performed, culture-based techniques. These early studies led to a widely accepted classical colonization dogma whereby Firmicutes, aerobic and facultative anaerobic bacteria such as enterobacteria are the first intestinal colonizers in infants. As the oxygen levels are depleted from the infant intestinal tract, strict anaerobes such as Bifidobacteria spp., Bacteroides spp., eubacteria and clostridia spp. gain a competitive advantage and predominate. However, recently a shift in this pattern has been observed whereby initial colonization with enterobacter and E. coli are diminished and colonization rates of Staphylococcus spp. are high.36,104 This shift may be a result of stricter hygiene in developed countries.104 Using this logic, developing countries which have not imposed these hygiene measures should, theoretically, have higher E. coli and other Enterobacteriaceae than developed countries, matching classical studies. This notion is supported by next-generation sequencing studies (Figure 2; Table S2) as well as culture-dependent studies. For instance, a culture-dependent study comparing fecal bacteria between Ethiopian and Swedish neonates found that the dominating spp. among the Swedish infants were S. epidermidis and Bacteroides spp.,105 whereas Bifidobacteria spp. and E. coli were only found in a minority of infants between 0 and 2 weeks of age. In contrast, the ‘classical’ aerobic dominance of E. coli and Enterococci spp. was common in Ethiopian neonates. These differences disappeared by 2 weeks of age. Similarly, a study on 10 neonates from Brazil106 reported a predominance by Escherichia and Clostridium spp., while Staphylococcus and Bacteroides spp. were only identified at low rates. The abundance of Escherichia spp. remained high throughout the first year of life,107 whereas Staphylococcus spp. was absent, and Bacteroides spp. was low from the third week onwards. In a separate comparison between Pakistani and Swedish infants, it was likewise found that infants from the developing country were more rapidly colonized with enterobacteria108 and had a predominance of E. coli. However, it should be noted that the Swedish infants were all breastfed and vaginally birthed whereas the Pakistani infants often started breastfeeding late and incompletely and were delivered both by vaginal and C-section. Still, other studies from Pakistan,109 Ethiopia,105 Brazil,95 Singapore,32 and Guatemala110 all similarly report ‘classical’ colonization patterns with high enterobacteria observed. While colonization of enterobacteria such as E. coli is thought to signify fecal contamination, Adlerberth et al.109 found that the high enterobacteria in Pakistani infants were likely derived from the environment as fewer than 50% of the Escherichia strains matched the mothers but could be found in the immediate home environment. Similarly, vertical transmission of E. coli was uncommon in infants born in hospitals in Japan.111 Therefore, the birth environment may play a role in the initial colonization of enterobacteria in infants.
Figure 2.

Neonate (<1 month) intestinal bacteria composition in diverse populations around the world. Pie chart color: bacterial taxa; color in map: human development index (http://hdr.undp.org/en/composite/HDI); *values were estimated from graph; a Bifidobacteria spp. were not detected using 16 S rRNA library technique; however, quantitative polymerase chain reaction detected Bifidobacteria spp. at 1 month which was the only tested timepoint; Country (age in d: days; w: weeks; 0: first stool).
Classical studies preformed in the 1970s and 1980s ubiquitously report E. coli as a pioneering bacteria in the gastrointestinal tract newborn infants in both developed and developing countries.109,110,112,113 However, as mentioned above, more recent studies in developed countries are trending away from this pattern and instead, report Staphylococcus spp. as the pioneering bacteria. Staphylococcus spp. were not previously been regarded as an intestinal commensal and were infrequent in infant stool from Australia,114 the United Kingdom115 and Scotland116 in the 1970s. However, in the 1980s and 1990s, S. epidermidis prevalence increased in industrialized countries such as Sweden93,105 and S. faecalis predominated neonates in the United Kingdom.112 A similar increase was described in France between 1975 and 1995.117 Since the millennium, studies in Sweden have asserted that enterobacteria are not predominant and found that fewer than 50% of the infants were colonized with Escherichia at 1 week of age104 and fewer than 61% at 2 months.118 In contrast, Staphylococci spp. colonized 99% of the infants from d 3 postpartum onwards.104 Another similar study showed staphylococci was positive in 73% of the infants by 2 months.119 Geographic comparisons of infant microbiome between Göteborg, London and Rome similarly showed staphylococci as the earliest facultative anaerobic colonizers, predominating in nearly 100% of the infants from d 3 postpartum onwards.36 In this study, E. coli and other Enterobacteriaceae which were traditionally recognized as early colonizers, appeared late. Similarly, a well-controlled study in Norway120 investigating exclusively breastfed, healthy, full-term infants who were not exposed to any medical interventions at d 4, 10, 30 and 120 found that while Enterobacteriaceae (E. coli and Shigella) were detected in over 70% of infants at 4 d post-partum, the prevalence of Staphylococcus was greater (90%). Collectively, these studies indicate that differences in hygiene between developed and developing countries impact initial colonization events in infants where enterobacteria are the pioneering bacteria in developing countries and Staphylococcus spp. predominate in developed countries.
While hygiene may impact initial colonization events, which would affect immune development, there are several exceptions to patterns described above. A comparison of Pakistani and Dutch infants matched by age, sex, mode of deliver, birth weight and breastfeeding practices found no differences in diversity, richness or evenness, but reported higher Proteobacteria in Dutch infants including bacteria such as Enterobacter spp., E.coli, Serratia spp. and Klebsiella spp.,121 which goes against the notion that developed countries have lower enterobacteria. Similarly in a study of 65–70 Nigerian neonates, rectal swabbed samples consisted mainly of coagulase-negative Staphylococcus (38.6%), followed by S. aureus (31.4%) and E. coli (30%) on the first day of life.122 Coagulase-negative Staphylococcus remained the most frequently isolated bacteria at d 3, 9 and 14 d postpartum. Interestingly, this study reported an absence of Actinobacteria such as Bifidobacteria spp., which is consistent with previous findings in Nigeria published in 1982.123 Therefore, not only does this publication contradict the notion that Staphylococcus spp. only predominate in developed countries but also challenges the dogma that Bifidobacteria spp. are the predominant anaerobe in healthy infants, globally.
While hygiene practices may impact early colonization events in infants, it is difficult to disentangle the impacts of hygiene in from other social and economic factors such as nutrition. As developing countries become urbanized, people’s diets change. They move away from starch-rich staples to high sugars, fats and animal-based foods. This “nutrition transition,”124 as opposed to improved hygiene, could account for the reported differences in breastfed infants. As discussed above, maternal dietary habits during lactation appear to shift the gradient of ‘core’ bacteria in breast milk. While there are currently too few studies to draw any definitive conclusions, we hypothesize that predominant microbial communities in breast milk would more effectively colonize the breastfed infants. In support of this, early colonization of Staphylococcus spp. in infants born in developed countries synergizes with increased Staphylococcus spp. in breast milk collected from Spain, China and Finland when compared with breast milk samples from South Africa, which were predominated by Proteobacteria.49
Collectively, ecological succession of microbial communities in infants start with pioneering bacteria adapted to an oxygen-rich environment, such as aerobes or facultative anaerobes. Hygiene and other social and economic factors associated with urbanization such as diet may contribute to taxonomic variations in these pioneer communities. Whether differences between these pioneering communities lead to functional health differences in infants remain to be determined.
Ecological succession of Bifidobacteria spp.
In general, the presence of early pioneering species such as Enterobacteriaceae, Streptococcus, and Staphylococcus spp. are succeeded by Enterococcus and Lactobacillus spp., which modify the infant gut habitat, generating an anaerobic environment that favors subsequent anaerobic colonizers. As mentioned above, niche dynamics in breastfed infants favor the predominance of Bifidobacteria spp. Bifidobacteria spp. have received a disproportionate amount of attention due to their reputed health benefits. It is thought that because Bifidobacteria spp. are a predominant component of the infant gut, they may be the main microbial candidate to influence the physiology/immunology of infants. As bifidobacteria are known producers of lactate and acetate in the gut, their abundance in breastfed infants may reduce risk of infections. In support of this, it has previously been shown that a low amount of acetate in infant stool is associated with infantile diarrhea and upper respiratory tract infections.125 However, the association between Bifidobacteria spp. and reduced infant morbidity was not observed in Gambian and Malawian infants, whereas there was a positive association with higher Prevotella spp.126 Nevertheless, identifying geographic differences in the succession of Bifidobacteria spp. in healthy infants may be useful in understanding global epidemiologic health differences.
While most researchers agree that Bifidobacteria spp. eventually predominates in breastfed infants, the rate of prevalence (Table 1), and overall abundance of Bifidobacteria spp. (Table 2) varies. For example, one study in Japan127 reported that newborns were predominated by anaerobic bacteria such as Streptococcus (100%) and Enterobacteriaceae (100%) with low initial colonization rates (0%) of Bifidobacteria spp. Overtime Bifidobacteria spp. proliferated and was present in 100% of infants 3–7 d postpartum. Other countries reporting low initial colonization rates of Bifidobacteria spp. include South Korea (2.1%),128 Greece (23%),21 Japan (21%),100 Nigeria (0%),122 the United Kingdom (27%),129 Italy (20%)130 and Australia (10%).131 However, while rates of Bifidobacteria spp. colonization are low in these neonates, counts are often high (Table 2). For example, in Japan, despite only 34% of the samples being positive for Bifidobacteria spp., mean counts were 5.7 ± 1.7 log10 CFU/g wet stool, which was higher than the more prevalent counts of Enterococcus (5.0 ± 1.8 log10 CFU/g; 57%) and Staphylococcus spp. (5.4 ± 1.5 log10 CFU/g; 60%). Similar results were reported in the United Kingdom where only 27% of the infants were colonized but had high numbers (10.24 log10 CFU/g wet stool).129 Moreover, the low reported prevalence of Bifidobacteria spp. colonization in neonates is not consistent. For instance, in Finland, 62% of the infants were colonized with a mean of 10.1 log10 CFU/g wet stool Bifidobacterium spp. 3 d post-partum.132 This exceeded the prevalence of other bacteria tested including lactobacillus-like bacteria, C. perfringens and B. fragilis, and was the highest reported count. Similarly, studies from Poland (73%),133 Sweden (76-91%),19,27 Brazil (80%),95 Norway (78–80)120 all report high colonization rates of Bifidobacteria spp. in neonates. By 1-week postpartum, culture and PCR-based counts of Bifidobacteria spp. were the most numerous taxa in infants born in Finland,132 Netherlands,31 Italy, London, Sweden,36 Indonesia,103 Brazil,95 Japan,127 and Armenia and Georgia134 despite not always being the highest % colonized (Tables 1 and 2). Studies using 16 S sequencing approaches (Table 2) yield similar results. By 1 month, the majority of studies analyzed globally by 16 S sequencing methods report a predominance of Bifidobacteria spp. (Figure 3), which persists through 4-month postpartum (Figure 4). However, while the large majority of published studies show Bifidobacteria spp. predominate the infant gut by 1-month postpartum, a select few do not. For example, in vaginally delivered Chinese infants, Proteobacteria such as Escherichia/Shigella and Klebsiella spp. still predominated in 2-month-old infants.135 A separate study using microarray assays and sequencing of cloned libraries of PCR-amplified small subunit ribosomal rDNA techniques in the United States found that Bifidobacteria spp. did not appear until several months after birth in 14 full-term infants and persisted only as a minority population.136 Moreover, despite high relative abundances, several papers still report low prevalence (<60% colonized) including papers from Belgium (57%),137 Greece (35%),21 Ireland (47%)101 the United Kingdom (21%),129 the United States (none detected [ND]),138 Ethiopia (35%)105 and Nigeria (ND).122 While these studies show that Bifidobacteria spp. are often only present in a small fraction of infants or are not numerically dominant, some argue that the paucity of Bifidobacteria spp. in these studies may be a result of technical bias relating to media selectivity, DNA extraction protocols and PCR primers used.94 Indeed, in Scotland, the prevalence of Bifidobacteria spp. largely depended on the method used.83
Table 1.
Frequency rates of bifidobacteria colonization by geographic region.
| Study | Year | Country | n | % BF | Method | <1 w | 1 w-1 m | 1 m | 2 m | 3 m | 4 m | 5 m | 6 m | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HDI >0.80 | ||||||||||||||
| 131a | 2013 | Australia | 6 V, 4 C | 50 | 16S rRNA | 10 | 100 | 100 | NA | NA | NA | NA | NA | |
| 114 | 1982 | Australia | 7 | 100 | Culture | NA | NA | 85.7 | NA | NA | NA | NA | NA | |
| 137 | 1995 | Belgium | 34 | 41 | Culture | NA | 21 | 57 | NA | NA | NA | NA | NA | |
| NA | NA | FF: 60 | NA | NA | NA | NA | NA | |||||||
| NA | NA | F: 20 | NA | NA | NA | NA | NA | |||||||
| 37 | 2013 | Canada | 18 V, 6 C | 42 | Illumina 16S rRNA; V5-V7 | NA | NA | NA | NA | NA | 75 | NA | NA | |
| 150 | 2009 | Czech Republic | 22 | 100 | Culture & FISH | NA | NA | NA | NA | 64 | NA | NA | NA | |
| 132 | 2000 | Finland | 34 V, 30 C | 64 | Culture | 62 | 78 | 90 | NA | NA | NA | NA | NA | |
| 21 | 2008 | Greece | 34 V | 23, 10 | Culture & PCR | 23 | NA | 35 | NA | NA | NA | NA | NA | |
| 101 | 2015 | Ireland | 11 V, 6 C | 100 | Culture & 16S rRNA-ITS + PFGE | 47 | NA | 47 | NA | NA | NA | NA | 58 | |
| 130 | 1992 | Italy | 12 | 100 | Culture | 20 | NA | 80b | NA | 50 | NA | NA | NA | |
| 36a | 2007 | Italy, Sweden, England | 216 V, 108 C | 23 | Culture & PCR, collectively | 63b | 80b | 90b | 95b | NA | NA | NA | NA | |
| 151 | 2012 | Japan | 132 | 32 | Reverse transcription qPCR | 34 | 73 | 88 | NA | 97 | NA | NA | 98 | |
| 100 | 2017 | Japan | 76 | 25 | Reverse transcription qPCR | 21 | 64 | 79 | NA | 97 | NA | NA | 99 | |
| 127 | 1985 | Japan | 6 V | 100 | Culture | NA | 100b | NA | NA | NA | NA | NA | NA | |
| 80 | 1983 | Japan | 5 V | 100 | Culture | 40 | 100 | 100 | NA | NA | NA | NA | NA | |
| 113 | 1984 | Japan | 35 V | 100 | Culture | NA | NA | 100b | NA | NA | NA | NA | NA | |
| 152 | 2015 | Korea, south | 143 | NS | Culture & T-RFLP | 2.1 | NA | NA | NA | NA | NA | NA | NA | |
| 31 | 2006 | Netherlands | 700 | 100 | qPCR | NA | NA | 99b | NA | NA | NA | NA | NA | |
| 92 | 2005 | Netherlands | 50 | 100 | qPCR | NA | NA | 100b | NA | NA | NA | NA | NA | |
| 120a | 2011 | Norway | 85 V | 100 | 16S rRNA clone libraries & PCR | 80 | NA | NA | NA | NA | NA | NA | NA | |
| 78 | NA | NA | NA | NA | NA | NA | NA | |||||||
| 133 | 2018 | Poland | 79 V | 100 | Culture | 73 | NA | NA | NA | 73 | NA | NA | NA | |
| 153 | 2017 | Slovenia | 49 V, 9 C | 67 | qPCR | 57 | NA | 94.7 | NA | 100 | NA | NA | NA | |
| 27 | 2014 | Sweden | 109 V, 19 C | 81 | 454 16S rRNA, V3-V4 | 91b,c | 91b | NA | 94b | NA | NA | NA | 98b | |
| 85b,d | 88b | NA | 92b | NA | NA | NA | 97b | |||||||
| 19 | 2015 | Sweden | 83 V | 74.4, 68.8 | Shotgun | 62 | NA | NA | NA | NA | 80 | NA | NA | |
| 129 | 1990 | United Kingdom | 15 V, 15 C | 100 | Culture & GC | 27 | NA | 21 | NA | NA | NA | NA | NA | |
| 138 | 2010 | United States | 1 | 100 | Sanger sequencing | NA | NA | ND | NA | NA | NA | NA | NA | |
| HDI <0.80 | ||||||||||||||
| 106 | 2012 | Brazil | 9 V | 100 | 16S rRNA & qPCR | NA | NA | 100 | NA | NA | NA | NA | NA | |
| 95 | 2017 | Brazil | 12 | 1 m: 100, 3 m: 75 | 16S rRNA qPCR | 80 | 90 | 100 | NA | 100 | NA | NA | NA | |
| 105a | 1991 | Ethiopiae | 43 V, 17 C | NS | Culture | NA | 25 | 35 | NA | NA | NA | NA | NA | |
| 122 | 2016 | Nigeria | 65 V, 5 C | 21.3 | Culture | ND | ND | ND | NA | NA | NA | NA | NA | |
Whenever possible, exclusively breastfed, vaginally birthed data were reported.
Abbreviations: % BF: percent exclusively breastfed; n: number of vaginally (V) and C-section (C) delivered infants; HDI: human developmental index score; FISH: Fluorescence in situ hybridization; qPCR: quantitative polymerase chain reaction; GC: gas chromatography; T-RFLP: Terminal restriction fragment length polymorphism; NA: not applicable; ND: none detected; FF: fortified formula; F: formula; m: month.
aIndicates values were estimated from available figures.
bIndicates Bifidobacteria spp. were predominant.
cAnthroposophic cohort.
dNon-anthroposophic cohort.
e21 infants receiving antibiotic treatment at the time of sampling.
Table 2.
Bifidobacteria abundance by geographic location.
| Counts (mean log10 CFU/g wet stool) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study | Year | Country | n | % BF | Method | <1 w | 1 w-1 m | 1 m | 2 m | 3 m | 4 m | 5 m | 6 m |
| HDI >0.80 | |||||||||||||
| 114 | 1982 | Australia | 7 | 100 | Culture | NA | NA | 10.6b | NA | NA | NA | NA | NA |
| 137 | 1995 | Belgium | 34 | 41 | Culture | NA | 8.51 | BF: 8.61 | NA | NA | NA | NA | NA |
| NA | NA | FF: 8.10 | NA | NA | NA | NA | NA | ||||||
| NA | NA | F: 9.75 | NA | NA | NA | NA | NA | ||||||
| 150 | 2009 | Czech Republic | 22 | 100 | Culture & FISH | NA | NA | NA | NA | 10.14b | NA | NA | NA |
| 132 | 2000 | Finland | 34 V, 30 C | 64 | Culture | 10.1b,c | 10.7b,c | 10.8b,c | NA | NA | NA | NA | NA |
| 154 | 2012 | Finland | 22 | 100i | FISH-FC & qPCR | NA | NA | NA | NA | NA | NA | NA | 10.7b |
| 154 | 2012 | Germany | 8 | 100i | FISH-FC & qPCR | NA | NA | NA | NA | NA | NA | NA | 9.9b |
| 21 | 2008 | Greece | 34 V, 48 C | 23, 20, 12 | Culture & PCR | 7.70 | NA | 7.47 | NA | NA | NA | NA | NA |
| 130a | 1992 | Italy | 12 | 100 | Culture | 8.9 | NA | 9b | NA | 9.8b | NA | NA | NA |
| 36a | 2007 | Italy, Sweden, England | 216 V, 108 C | 23j | Culture & PCR | NA | 9.5b | 9.6b | 9.8b | NA | NA | NA | NA |
| 151 | 2012 | Japan | 132 | 32 | RT qPCR | 5.7 | 8.2 | 9.0 | NA | 9.8b | NA | NA | 10.3b |
| 100 | 2017 | Japan | 76 | 25 | RT qPCR | 6.2 | 8.0 | 8.5 | NA | 9.3b | NA | NA | 9.6b |
| 127 | 1985 | Japan | 6 V | 100 | Culture | ND | 10.4b | NA | NA | NA | NA | NA | |
| 80 | 1983 | Japan | 5 V | 100 | Culture | 5.0 | 9.7b | 11.3b | NA | NA | NA | NA | NA |
| 113 | 1984 | Japan | 35 V | 100 | Culture | NA | NA | 10.74b | NA | NA | NA | NA | NA |
| 152 | 2015 | Korea, south | 143 | NS | Culture & T-RFLP | 2.24 | NA | NA | NA | NA | NA | NA | NA |
| 31 | 2006 | Netherlands | 1032 | 68 | qPCR | NA | NA | 10.68b,d | NA | NA | NA | NA | NA |
| 92 | 2005 | Netherlands | 50 | 100 | qPCR | NA | NA | 10.56b,d | NA | NA | NA | NA | NA |
| 133 | 2018 | Poland | 79 V | 100 | Culture | 9.14d | NA | NA | NA | 8.32d | NA | NA | NA |
| 153 | 2017 | Slovenia | 49 V, 9 C | 67 | qPCR | NA | NA | NA | NA | NA | 5.89d | NA | NA |
| 129 | 1990 | United Kingdom | 15 V, 15 C | 100 | culture of lacto and bifido +GC | 10.24b | NA | 9.48b | NA | NA | NA | NA | NA |
| HDI <0.80 | |||||||||||||
| 95 | 2017 | Brazil | 12 | 1 m: 100, 3 m: 75 | 16S rRNA NAqPCR | 10.9 | 12.98b | 11.68b | NA | 11.98b | NA | NA | NA |
| 102a | 2017 | Estonia | 4 V, 5 C | NSb | Illumina MiSeq 16S rRNA, V1-V2 & qPCR | ND | NA | NA | NA | 3.25e | NA | NA | 1.69f |
| 105a | 1991 | Ethiopia | 43 V, 17 C | NS | Culture | NA | 6.5f | 6.4b,f | NA | NA | NA | NA | NA |
| 155 | 2014 | Bangladesh | 8 V, 40 C | 6 w: 79.2, 11 w: 60.4, 15 w:45.8 | Illumina MiSeq 16S rRNA, V4 & T-RFLP, PCR | NA | NA | NA | NA | 7.48d | NA | NA | NA |
| Relative % Abundance | |||||||||||||
| 37 | 2013 | Canada | 18 V | 42 | Illumina 16S rRNA; V5-V7 | NA | NA | NA | NA | NA | 35.2b,c | NA | NA |
| 102 | 2017 | Estonia | 4 V, 5 C | NSb | Illumina MiSeq 16S rRNA, V1-V2 & qPCR | 0 | NA | NA | NA | NA | NA | NA | 5 |
| 154 | 2012 | Finland | 22 | 100i | NA | NA | NA | NA | NA | NA | NA | 68.1b | |
| 154 | 2012 | Germany | 8 | 100i | FISH-FC & qPCR | NA | NA | NA | NA | NA | NA | NA | 58.3b,g |
| 35 | 2014 | Sweden | 15 V | 73 | 454 16S rRNA, V3-V4 | NA | 11.6 | 34.4b | NA | 44.7b | NA | NA | 32.1b |
| 27 | 2014 | Sweden | 109 V, 19 C | 81 | 454 16S rRNA, V3-V4 | A: 35b | 39b | NA | 37b | NA | NA | NA | 45b |
| NA: 26b | 36b | NA | 42b | NA | NA | NA | 33b | ||||||
| 19 | 2015 | Sweden | 83 V, 15 C | 74.4, 68.8 | Shotgun | 20.2 | NA | NA | NA | NA | 42.2b | NA | NA |
| 23a | 2017 | United Kingdom | 83 V, 74 V, 74 V, 24 V | 100 | Illumina 16S rRNA, V4-V5 region | NA | 47.5b | 55.7b | 59b | NA | NA | NA | 49.1b |
| 138 | 2010 | United States | 1 | 100 | Sanger sequencing | NA | NA | ND | NA | NA | NA | NA | NA |
| 156a | 2015 | United States | 39 V | 70b | Illumina 16S rRNA, V4 | 23b | 30b | NA | 27b | NA | 45b | NA | NA |
| 139 | 2017 | United States | 71 (mixed VC) | 100 | 16S rRNA – V3-V5; 454 pyrosequencing | NA | NA | NA | NA | NA | NA | NA | 9.9 |
| HDI <0.80 | |||||||||||||
| 134 | 2017 | Armenia | 53 V | 100 | Illumina 16S rRNA, V4 | NA | 30b | 55b | 50b | NA | NA | NA | NA |
| 95 | 2017 | Brazil | 12 | 1 m: 100, 3 m: 75 | 16 S rRNA qPCR | 10.9 | 12.06b | 11.68b | NA | 11.98b | NA | NA | NA |
| 135 | 2016 | China | 7 V, 7 C | NS | 454 16S rRNA, V3-V5 | 0 | NA | NA | 2 | NA | NA | NA | NA |
| 139a | 2018 | Ecuador | 59 V, 11 C | 89 | Illumina MiSeq 16S rRNA | NA | NA | NA | NA | 50b | NA | NA | NA |
| 126 | 2017 | Gambia | 85 | NS | Illumina 16S rRNA, V4 | NA | NA | NA | NA | NA | NA | 68.3 | NA |
| 134 | 2017 | Georgia | 17 V, 11 C | 100 | Illumina 16S rRNA, V4 | NA | 40b | 40b | 55b | NA | NA | NA | NA |
| 20a | 2013 | India | 73 V | 1 w: 92, | qPCR | 3i | NA | 43h | NA | NA | NA | NA | NA |
| 103 | 2013 | Indonesia | 72 V, 8 C | 100 | qPCR | NA | BF: 45b,g | NA | NA | NA | NA | NA | NA |
| 67 V, 13 C | 0 | NA | FF: 45b,g | NA | NA | NA | NA | NA | NA | ||||
| 155 | 2014 | Bangladesh | 8 V, 40 C | 6 w: 79.2, 11 w: 60.4, 15 w:45.8 | Illumina MiSeq 16S rRNA, V4 & T-RFLP, PCR | NA | NA | NA | NA | 59.6b | NA | NA | NA |
Whenever possible, exclusively breastfed, vaginally birthed data were reported.
Abbreviations: %BF: percent exclusively breastfed; n: number of vaginally (V) and C-section (C) delivered infants; HDI: human development index score; FISH: Fluorescence in situ hybridization; qPCR: quantitative polymerase chain reaction; RT reverse transcription; GC: gas chromatography; T-RFLP: Terminal restriction fragment length polymorphism; NA: not applicable; ND: none detected; FF: fortified formula; F: formula; w: week; m: month.
aIndicates values were estimated from available figures.
bIndicates Bifidobacteria spp. were the most abundant taxa at this point.
cExpressed as median value.
dLog10 value calculated from mean copies/g feces.
eValues expressed as mean log10 mg of stool.
f21 infants receiving antibiotic treatment at the time of sampling.
gValues expressed as mean percentage of bacteria compared to total bacteria analyzed.
hValues expressed as relative difference to ‘universal’ bacteria domain sequence.
iValue presented at d 1 postpartum, value increased 10-fold on d 4.
j% Breastfed at 6 months .
Figure 3.
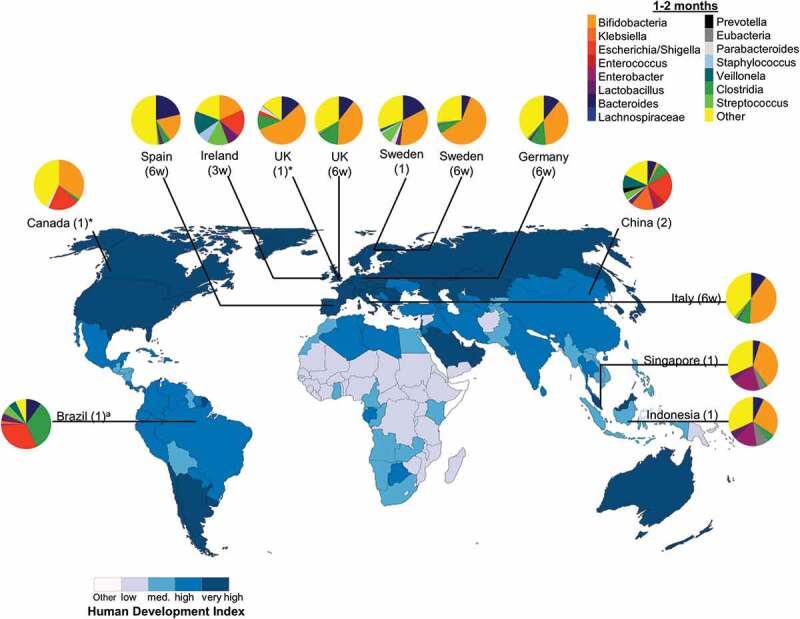
Intestinal bacteria composition in infants aged 1–2 months from diverse populations around the world. Pie chart color: bacterial taxa: human development index (http://hdr.undp.org/en/composite/HDI); *values were estimated from graph; a Bifidobacteria spp. were not detected using 16 S rRNA library technique; however, quantitative polymerase chain reaction detected Bifidobacteria spp. at 1 month which was the only tested timepoint; Country (age in months unless otherwise stated; w: weeks).
Figure 4.

Intestinal bacteria composition in infants aged 3–4 months from diverse populations around the world. Pie chart color: bacterial taxa; color in map: human development index (http://hdr.undp.org/en/composite/HDI); *values were estimated from graph; #values were calculated from provided data; a Bifidobacteria spp. were not detected using 16 S rRNA library technique; however, quantitative polymerase chain reaction detected Bifidobacteria spp. at 1 month which was the only tested timepoint; Country (age in months).
Collectively, these independent studies support the ecological succession from aerobic and facultative anaerobic pioneering bacteria to a community rich in species like Bifidobacteria spp. which can replace or reduce pioneering species abundance through competition. The rate of this succession varies but does not appear coincide with any patterns based on geography.
The influence of geography on infant fecal microbiota does not supersede classical patterns of bacterial colonization
While many studies have reported on bacterial species in infants within one population, far fewer studies have attempted to compare the temporal progression of bacterial communities across cultural and geographic settings. A recent study compared the bacterial composition of healthy 3-month-old Ecuadorian infants with data previously collected in Canada140 and found infants from both countries were dominated by Bifidobacteria spp., followed by either Bacteroides or Veillonella spp. However, Canadian infants were found to have comparably higher abundances of Bacteroides spp., and lower abundances of Veillonella spp. Unfortunately in this study, methodical differences between the Canadian (CHILD) and Ecuadorian (ECUAVIDA) study make comparisons difficult and interpretations should be taken cautiously. A similar approach was used by Kuang et al.,135 who compared the fecal microbial composition of 29 healthy, 2-month-old Chinese infants to studies previously published in Brazil, the United States, Sweden, Canada and Bangladesh. They reported marked differences among countries at the phylum level, which broadly separated into three clusters. All the infants from China and 70% of the infants from Brazil fell into a high Proteobacteria cluster. Additionally, 82% of the infants from the United States, 54% of the infants from Sweden and 79% of the infants from Canada contained a high prevalence of Actinobacteria, whereas 70% of the infants from Bangladesh and 33% of the infants from Sweden were highly abundant in Firmicutes. Using a PERMANOVA, they found geographic location accounted for 19.6% of variation in infant fecal microbiota, which was larger than age, delivery mode and feeding patterns and concluded that geographical factors have a strong impact on infant colonization. However, as they utilized data extracted from other studies there are limitations, such as differences in primers, DNA extraction methodology, hypervariable region amplified as well as the sequencing platforms. To eliminate methodological biases between independent studies, large cross-cultural studies are needed.
In an effort to approach this, one cross-cultural study comparing the 6-week fecal bacterial composition of infants born in Sweden, Scotland, Germany, Italy and Spain reported that geography had a high impact on levels of Bifidobacteria spp., Bacteroides spp., and enterobacteria.34 In this study, infants born in Spain had higher proportions of Bacteroides spp. and enterobacteria and lower Bifidobacteria spp. compared with infants born in Sweden, Scotland, Germany and Italy. In contrast, infants in Sweden had higher proportions of Bifidobacteria spp. While the authors reported that Swedish and Italian bacterial communities did not cluster as a function of feeding method, there were notable differences in breastfeeding rates between countries. For example, in Spain breastfeeding rates were 43.1% at the time of sample collection whereas in Sweden breastfeeding rates were 75.9% and Germany had rates of 45%. In a separate analysis, the authors reported that breastfed infants had higher proportions of Bifidobacteria spp. and lower proportions of Bacteroides spp., which corresponded with varying rates of breastfeeding between the countries. Regardless of geographic location and breastfeeding practices, all infants had a predominance of Bifidobacteria spp., followed by Bacteroides spp. then enterobacteria, supporting previous findings.92,93,113,114 A separate study by Turroni et al.94 similarly compared fecal samples of infants born in Italy, Ireland and Spain. In contrast to the geographic impacts reported by Fallani et al., this study found that infants from within a given population displayed widely dissimilar microbial patterns and concluded that infant gut colonization was not influenced by the geographic origin to which the infant was born. Another European study compared the infant microbial colonization process between Armenia and Georgia.134 While these are neighboring countries, they possess distinct language and cultural differences. This study found that country of origin accounted for 8.4% of the total variation between samples, delivery mode accounted for 3.1% and mother’s secretor status accounted for 1.6%. However, despite these factors, the overall patterns of colonization were similar between the two countries. In both countries, B. breve predominated. The major non-Bifidobacteria taxa included enterobacteria, streptococci, lactobacilli, enterococci, Bacteroides spp., Clostridia spp. and staphylococci, in agreement with other studies.
While differences between developed and developing countries have been reported, some groups propose that the influence of lifestyle is subtle compared with the variability between infants within a single population, at least until weaning. For example, Yatsunenko et al. show that major geographic differences between the fecal microbial communities of breastfed infants in rural Malawi, Venezuela (Amerindians) and metropolitan America do not appear until 3 y of age.141 This study reported three notable findings. The first was that bacterial diversity increased with age in all infants and that all infants diverged toward an adultlike bacterial community within a 3-y period. The second was that interpersonal variations are significantly greater between infants than between adults. The authors confirmed that this finding is robust to geography. Finally, they reported differences in the phylogenetic composition of fecal microbiota between infants living in different countries. Using shotgun and 16 S rRNA-V4 sequences it was found that infants from all three populations had a predominance (75 ± 20%) of Bifidobacteria spp. throughout their first year of life, which declined as the infants aged. There were several operational taxonomic units which could be used to distinguish between Malawian, Venezuelan and American infant microbiomes. In general, the American babies had an overrepresentation of Prevotella spp. compared to the Malawian and Venezuelan infants. While a total of 28 operational taxonomic units distinguished American infants from those born in Malawi and Venezuela, there were 21 operational taxonomic units discriminating between Malawian and Venezuelan infants. The Venezuelan infants were found to have higher Enterococcaceae compared to the Malawian. The authors concluded that a Western lifestyle appears to systematically affect the bacterial component of the intestinal microbiome, but the influence is subtle compared to the high degree of variability observed in infants within each population. Insights from independent studies corroborate these findings. Using 16 S rRNA gene-based microarrays, it was shown that there is considerable intra- and interpersonal variability in the fecal bacteria of 12 unrelated American children in the first year of life.136 This significant level of inter-individual variation among unweaned infants has also been reported within infants born in Italy, Ireland and Spain94 as well as Japan.142 Similar to the results obtained by Yatsunenko et al., this variability decreased and converged toward an adultlike microbiome as the infants age. When looking at population-specific differences in the microbiome functions, it was observed that Malawian and Venezuelan infants had higher genes involved in riboflavin (vitamin B2) biosynthesis suggesting an adaptive response to vitamin availability. Similarly, genes involved in utilizing host glycans differed suggesting differences in the glycan content of breast milk. Both these findings are not surprising given that human milk vitamin content and oligosaccharides cluster by country of origin. What was somewhat surprising was that Bifidobacteria spp. may not be pivotal to biosynthetic pathways in infants and that these effects remained when all the Bifidobacteria spp. reads were excluded.
Cross-cultural comparisons between developing countries show similar trends to those reported in Europe. For example, a comparison of infants from Bangladesh, the Gambia, Kenya and Mali found that healthy infants in these countries followed the stereotypical136,143 colonization process described in developed countries. During the first year of life, the “healthy” microbial community was characterized by low overall diversity and high proportions of facultative anaerobes and potentially pathogenic organisms such as Escherichia/Shigella spp.144 As these infants were weaned, microbial diversity increased as did the abundance of Prevotella spp. While these observations broadly held true when stratifying by geographic location, some country-specific effects were observed. For example, infants in Bangladesh were characterized by lower proportions of Prevotella spp. and higher Escherichia/Shigella and Streptococcus spp. over the study period (up to 5 y of age).144 A separate study comparing the microbiota colonization in two partially breastfed Asian populations, Singapore and Indonesia, likewise found a similar trend in bacterial colonization over the first year of life, despite differences in geography, cultural ethnicity and overall social economic development.145 They reported that both populations had Enterobacteriaceae and Bifidobacteria spp. predominating the stool microbiota of infants under 3 months of age. However, the relative abundance of Bifidobacteria spp. was lower in the Indonesian group, who had higher relative abundances of C. leptum and Atopobium compared to the Singapore population.
Clinical implications and limitations
The aim of this review was to synthesize data across studies on the healthy infant gut colonization process, globally. The trajectory of the infant colonization process has lifelong clinical implications as discussed previously.146,147 The results from this review emphasize that human behaviors and not genetics are the largest predictor of microbial variability in infants and can be used as a guideline for behavioral changes. This includes increasing global breastfeeding rates by providing education about breastfeeding and reducing societal stigma. For mothers who are unable to breastfeed, it includes having access to formulas that truly mimic breast milk composition.148 With respect to cesarian section deliveries, every effort should be made to reduce non-medical cesarian deliveries as there is strong evidence of overuse in all parts of the world.149
There are several limitations in comparing data extracted from independent studies. This includes differences in DNA extraction methodology, hypervariable region amplified, reference databases, primers and sequencing platforms. These differences in experimental protocols may produce variation that undermines subtle biological differences. Furthermore, there are inherent shortcomings of molecular approaches used in studies. For instance, differences in bacterial communities cannot be distinguished at the species level using operational taxonomic units.150 Finally, we did not merge and statistically analyze the summarized data. Merging and analyzing from different studies is limited in practice. Current and future developments, such as amplicon sequence variants methods and standardization of procedures, will better allow for data to be compared across studies.150 With these caveats stated, studies of the microbiota based on 16 S rRNA gene targets can, at least in principal, be compared to gain insight into broad evolutionary patterns.
Conclusion
Microbial communities in the infant gut exhibit orderly and predictable patterns of succession beginning with relatively few pioneering bacteria which develop through increasing complexity until they reach a relatively stable “climax” (adult) community. The trajectory of successional change appears to be influenced by human behaviors that might influence the availability of colonists. In newborn infants, the gastrointestinal site is oxygenated, and only aerobic and facultative anaerobic bacteria are capable of thriving in this habitat. The availability of initial colonists is dependent on delivery mode whereby vaginally delivered infants are exposed to maternal vaginal and fecal microbial communities and C-section delivered infants are exposed to skin and environmental microbes. These pioneering bacteria modify the habitat, generating an anaerobic environment which favors subsequent anaerobic colonizers. While site conditions allow anaerobes to colonize, nutrient availability from feeding practices predicts dominating species. Breast milk, despite maternal diet, increases Bifidobacteria in the infant gut likely due to their ability to metabolize human milk oligosaccharides, whereas the predominant anaerobes in formula-fed infants vary depending on the available dietary substrates in the formula used. In addition to delivery mode and feeding practices, environmental factors such as hygiene also impacts infant microbial colonization with hygiene resulting in higher Staphylococcus spp. and lower enterobacteria. Overall, however, independent studies investigating the gut microbiota at various points in postnatal development are limited either by low sample sizes, analytic methods used, or the scope of the population surveyed. While these studies provide important insights, large cross-cultural studies are needed given the variability between infants within a single population are higher than any other factor including geography, genetics and environment. Despite this, the general succession of microbial colonization is similar among vaginally birthed and exclusively breastfed infants globally. We conclude that human behavior including cultural variations in feeding practices, delivery modes and hygiene, over geography or race, are the largest predictors of microbial variability in the infant gut.
Methods
Literature search strategy
For this literature review, we searched Web of Science (1864–2019), PubMed (1966 to 2019), Google Scholar and hand searches for primary studies profiling the microbiome in infants aged 0 to 6 months. The search strategy was based on a clear and careful selection of key words including combinations of: (microbiome* OR bacteriome* OR commensal* OR flora* OR microorganism* OR coloniz* OR colonis*) AND (neonatal* OR infant* OR baby*) AND [country] where we systematically searched for publications stating one of the 195 counties in the world today.
Study eligibility
The included primary studies had to report a healthy infant gut bacterial community. Healthy was defined as having no preexisting or diagnosed conditions such as allergic diseases, autoimmune diseases or known infections at the time of sample collection. The methods for assessing bacterial composition could include sequencing or culture-based methods. All levels of reported phylogeny were included. Studies were excluded if they did not report on the gastrointestinal microbiome, the full text was not available, the study population was not less than 6 months of age, or there was no means to quantify the findings.
Supplementary Material
Acknowledgments
We thank Helena C. Nunes for data entry.
Funding Statement
CQ is funded through a CAG and CIHR Doctoral Research Award. DLG is funded through grants from the Natural Sciences and Engineering Research Council and Crohn’s and Colitis Canada.
Authors’ contribution
Critically evaluated the data: CQ. Wrote the paper: CQ, DLG
Competing Interests
The authors declare no conflicts of interest.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Levy O. Innate immunity of the newborn: basic mechanisms and clinical correlates. Nat Rev Immunol. 2007;7:379–390. doi: 10.1038/nri2075. [DOI] [PubMed] [Google Scholar]
- 2.Ivanov II, Littman DR. Modulation of immune homeostasis by commensal bacteria. Curr Opin Microbiol. 2011;14:106–114. doi: 10.1016/j.mib.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung H, Pamp S, Hill J, Surana N, Edelman S, Troy E, Reading N, Villablanca E, Wang S, Mora J, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El Aidy S, Hooiveld G, Tremaroli V, Backhed F, Kleerebezem M. The gut microbiota and mucosal homeostasis: colonized at birth or at adulthood, does it matter? Gut Microbes. 2013;4:118–124. doi: 10.4161/gmic.23362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, Basaglia G, Turroni S, Biagi E, Peano C, Severgnini M, et al. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. 2014;5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett FJ, Barnicot NA, Woodburn JC, Pereira MS, Henderson BE. Studies on viral, bacterial, rickettsial and treponemal diseases in the Hadza of Tanzania and a note on injuries. Hum Biol. 1973;45:243–272. [PubMed] [Google Scholar]
- 8.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta VK, Paul S, Dutta C. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front Microbiol. 2017;8:1162. doi: 10.3389/fmicb.2017.01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez-Munoz ME, Arrieta MC, Ramer-Tait AE, Walter J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome. 2017;5:48. doi: 10.1186/s40168-017-0268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep. 2016;6:23129. doi: 10.1038/srep23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jervis-Bardy J, Leong LEX, Marri S, Smith RJ, Choo JM, Smith-Vaughan HC, Nosworthy E, Morris PS, O’Leary S, Rogers GB, et al. Deriving accurate microbiota profiles from human samples with low bacterial content through post-sequencing processing of Illumina MiSeq data. Microbiome. 2015;3:19. doi: 10.1186/s40168-015-0083-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thavagnanam S, Fleming J, Bromley A, Shields MD, Cardwell CR. A meta-analysis of the association between Caesarean section and childhood asthma. Clin Exp Allergy. 2008;38:629–633. doi: 10.1111/j.1365-2222.2007.02780.x. [DOI] [PubMed] [Google Scholar]
- 14.Lumbiganon P, Laopaiboon M, Gülmezoglu AM, Souza JP, Taneepanichskul S, Ruyan P, Attygalle DE, Shrestha N, Mori R, Hinh ND, et al. Method of delivery and pregnancy outcomes in Asia: the WHO global survey on maternal and perinatal health 2007-08. Lancet. 2010;375:490–499. doi: 10.1016/S0140-6736(09)61870-5. [DOI] [PubMed] [Google Scholar]
- 15.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin H, Pei Z, Martinez KA, Rivera-Vinas JI, Mendez K, Cavallin H, Dominguez-Bello MG. The first microbial environment of infants born by C-section: the operating room microbes. Microbiome. 2015;3:59. doi: 10.1186/s40168-015-0126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandar R, Mikelsaar M. Transmission of mother’s microflora to the newborn at birth. Biol Neonate. 1996;69:30–35. doi: 10.1159/000244275. [DOI] [PubMed] [Google Scholar]
- 18.Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, Cox LM, Amir A, Gonzalez A, Bokulich NA, Song SJ, Hoashi M, Rivera-Vinas JI, et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat Med. 2016;22:250–253. doi: 10.1038/nm.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Kabeerdoss J, Ferdous S, Balamurugan R, Mechenro J, Vidya R, Santhanam S, AK Jana, BS Ramakrishna, et al. Development of the gut microbiota in southern Indian infants from birth to 6 months: a molecular analysis. J Nutr Sci. 2013;2:e18. doi: 10.1017/jns.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitsou EK, Kirtzalidou E, Oikonomou I, Liosis G, Kyriacou A. Fecal microflora of Greek healthy neonates. Anaerobe. 2008;14:94–101. doi: 10.1016/j.anaerobe.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 22.Gronlund MM, Lehtonen OP, Eerola E, Kero P. Fecal microflora in healthy infants born by different methods of delivery: permanent changes in intestinal flora after cesarean delivery. J Pediatr Gastroenterol Nutr. 1999;28:19–25. doi: 10.1097/00005176-199901000-00007. [DOI] [PubMed] [Google Scholar]
- 23.Hill CJ, Lynch DB, Murphy K, Ulaszewska M, Jeffery IB, O’Shea CA, Watkins C, Dempsey E, Mattivi F, Tuohy K, et al. Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET Cohort. Microbiome. 2017;5:4. doi: 10.1186/s40168-016-0213-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makino H, Kushiro A, Ishikawa E, Muylaert D, Kubota H, Sakai T, Oishi K, Martin R, Ben Amor K, Oozeer R, et al. Transmission of intestinal Bifidobacterium longum subsp. longum strains from mother to infant, determined by multilocus sequencing typing and amplified fragment length polymorphism. Appl Environ Microbiol. 2011;77:6788–6793. doi: 10.1128/AEM.05346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makino H, Kushiro A, Ishikawa E, Kubota H, Gawad A, Sakai T, Oishi K, Martin R, Ben-Amor K, Knol J, et al. Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant’s microbiota. PLoS One. 2013;8:e78331. doi: 10.1371/journal.pone.0078331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huurre A, Kalliomäki M, Rautava S, Rinne M, Salminen S, Isolauri E. Mode of delivery - effects on gut microbiota and humoral immunity. Neonatology. 2008;93:236–240. doi: 10.1159/000111102. [DOI] [PubMed] [Google Scholar]
- 27.Hesla HM, Stenius F, Jäderlund L, Nelson R, Engstrand L, Alm J, Dicksved J. Impact of lifestyle on the gut microbiota of healthy infants and their mothers-the ALADDIN birth cohort. FEMS Microbiol Ecol. 2014;90:791–801. doi: 10.1111/1574-6941.12434. [DOI] [PubMed] [Google Scholar]
- 28.Biasucci G, Benenati B, Morelli L, Bessi E, Boehm G. Cesarean delivery may affect the early biodiversity of intestinal bacteria. J Nutr. 2008;138:1796S–1800S. doi: 10.1093/jn/138.9.1796S. [DOI] [PubMed] [Google Scholar]
- 29.Biasucci G, Rubini M, Riboni S, Morelli L, Bessi E, Retetangos C. Mode of delivery affects the bacterial community in the newborn gut. Early Hum Dev. 2010;86(Suppl 1):13–15. doi: 10.1016/j.earlhumdev.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 30.Neut C, Bezirtzoglou E, Romond C, Beerens H, Delcroix M, Noel AM. Bacterial colonization of the large intestine in newborns delivered by cesarean section. Zentralbl Bakteriol Mikrobiol Hyg A. 1987;266:330–337. doi: 10.1016/s0176-6724(87)80213-4. [DOI] [PubMed] [Google Scholar]
- 31.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, Stobberingh EE. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 32.Dogra S, Sakwinska O, Soh S-E, Ngom-Bru C, Brück WM, Berger B, Brüssow H, Lee YS, Yap F, Chong Y-S, et al. Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. MBio. 2015;6. doi: 10.1128/mBio.02419-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pandey PK, Verma P, Kumar H, Bavdekar A, Patole MS, Shouche YS. Comparative analysis of fecal microflora of healthy full-term Indian infants born with different methods of delivery (vaginal vs cesarean): acinetobacter sp. prevalence in vaginally born infants. J Biosci. 2012;37:989–998. doi: 10.1007/s12038-012-9268-5. [DOI] [PubMed] [Google Scholar]
- 34.Fallani M, Young D, Scott J, Norin E, Amarri S, Adam R, Aguilera M, Khanna S, Gil A, Edwards CA, et al. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastroenterol Nutr. 2010;51:77–84. doi: 10.1097/MPG.0b013e3181d1b11e. [DOI] [PubMed] [Google Scholar]
- 35.Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, Björkstén B, Engstrand L, Andersson AF. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. 2014;63:559–566. doi: 10.1136/gutjnl-2012-303249. [DOI] [PubMed] [Google Scholar]
- 36.Adlerberth I, Strachan DP, Matricardi PM, Ahrné S, Orfei L, Åberg N, Perkin MR, Tripodi S, Hesselmar B, Saalman R, et al. Gut microbiota and development of atopic eczema in 3 European birth cohorts. J Allergy Clin Immunol. 2007;120:343–350. doi: 10.1016/j.jaci.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 37.Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Chari RS, Sears MR, Becker AB, Scott JA, Kozyrskyj AL, et al. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ. 2013;185:385–394. doi: 10.1503/cmaj.121189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mueller NT, Shin H, Pizoni A, Werlang I, Matte U, Goldani M, Goldani H, Dominguez-Bello M. Delivery mode and the transition of pioneering gut-microbiota structure, composition and predicted metabolic function. Genes (Basel). 2017;8:364. doi: 10.3390/genes8120364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Nimwegen FA, Penders J, Stobberingh EE, Postma DS, Koppelman GH, Kerkhof M, Reijmerink NE, Dompeling E, van den Brandt PA, Ferreira I, et al. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J Allergy Clin Immunol. 2011;128:948. doi: 10.1016/j.jaci.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 40.Goedert JJ, Hua X, Yu G, Shi J. Diversity and composition of the adult fecal microbiome associated with history of cesarean birth or appendectomy: analysis of the American gut project. EBioMedicine. 2014;1:167–172. doi: 10.1016/j.ebiom.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segawa M. Buddhism and breastfeeding. Breastfeed Med. 2008;3:124–128. doi: 10.1089/bfm.2008.0104. [DOI] [PubMed] [Google Scholar]
- 42.WHO . Global breastfeeding collective: rates of exclusive breastfeeding at 0-5 months. 2017.
- 43.Robinson DT, Caplan MS. Linking fat intake, the intestinal microbiome, and necrotizing enterocolitis in premature infants. Pediatr Res. 2015;77:121–126. doi: 10.1038/pr.2014.155. [DOI] [PubMed] [Google Scholar]
- 44.Prentice AM, Roberts S, Watkinson M, Whitehead RG, Paul A, Prentice A, Watkinson A. Dietary supplementation of Gambian nursing mothers and lactational performance. Lancet. 1980;2:886–888. pii S0140-6736(80)92048-6. [DOI] [PubMed] [Google Scholar]
- 45.Jenness R. The composition of human milk. Semin Perinatol. 1979;3:225–239. [PubMed] [Google Scholar]
- 46.Prentice A, Handbook of milk composition. San Diego (CA): Jensen RG; 1995. ed Inc Academic Press . [Google Scholar]
- 47.Roed C, Skovby F, Lund AM. Severe vitamin B12 deficiency in infants breastfed by vegans. Ugeskr Laeger. 2009;171:3099–3101. pii. VP05070024. [PubMed] [Google Scholar]
- 48.Tian H-M, Wu Y-X, Lin Y-Q, Chen X-Y, Yu M, Lu T, Xie L. Dietary patterns affect maternal macronutrient intake levels and the fatty acid profile of breast milk in lactating Chinese mothers. Nutrition. 2019;58:83–88. doi: 10.1016/j.nut.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 49.Kumar H, Du Toit E, Kulkarni A, Aakko J, Linderborg KM, Zhang Y, Nicol MP, Isolauri E, Yang B, Collado MC, et al. Distinct patterns in human milk microbiota and fatty acid profiles across specific geographic locations. Front Microbiol. 2016;7:1619. doi: 10.3389/fmicb.2016.01619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma J, Prince AL, Bader D, Hu M, Ganu R, Baquero K, Blundell P, Alan Harris R, Frias AE, Grove KL, et al. High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nat Commun. 2014;5:3889. doi: 10.1038/ncomms4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chu DM, Antony KM, Ma J, Prince AL, Showalter L, Moller M, Aagaard KM. The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med. 2016;8:77. doi: 10.1186/s13073-016-0330-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abulizi N, Quin C, Brown K, Chan Y, Gill S, Gibson D. Gut mucosal proteins and bacteriome are shaped by the saturation index of dietary lipids. Nutrients. 2019;11:418. doi: 10.3390/nu11020418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gomez-Gallego C, Morales J, Monleón D, Du Toit E, Kumar H, Linderborg K, Zhang Y, Yang B, Isolauri E, Salminen S, et al. Human breast milk NMR metabolomic profile across specific geographical locations and its association with the milk microbiota. Nutrients. 2018;10:1355. doi: 10.3390/nu10101355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zivkovic AM, German JB, Lebrilla CB, Mills DA. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4653–4658. doi: 10.1073/pnas.1000083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gay MCL, Koleva P, Slupsky C, Toit E, Eggesbo M, Johnson C, Wegienka G, Shimojo N, Campbell D, Prescott S, et al. Worldwide variation in human milk metabolome: indicators of breast physiology and maternal lifestyle? Nutrients. 2018;10:1151. doi: 10.3390/nu10091151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.West PA, Hewitt JH, Murphy OM. Influence of methods of collection and storage on the bacteriology of human milk. J Appl Bacteriol. 1979;46:269–277. doi: 10.1111/j.1365-2672.1979.tb00820.x. [DOI] [PubMed] [Google Scholar]
- 57.Hunt KM, Foster JA, Forney LJ, Schütte UME, Beck DL, Abdo Z, Fox LK, Williams JE, McGuire MK, McGuire MA, et al. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS One. 2011;6:e21313. doi: 10.1371/journal.pone.0021313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr. 2012;96:544–551. doi: 10.3945/ajcn.112.037382. [DOI] [PubMed] [Google Scholar]
- 59.Cabrera-Rubio R, Mira-Pascual L, Mira A, Collado MC. Impact of mode of delivery on the milk microbiota composition of healthy women. J Dev Orig Health Dis. 2016;7:54–60. doi: 10.1017/S2040174415001397. [DOI] [PubMed] [Google Scholar]
- 60.Jost T, Lacroix C, Braegger CP, Rochat F, Chassard C. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ Microbiol. 2014;16:2891–2904. doi: 10.1111/1462-2920.12238. [DOI] [PubMed] [Google Scholar]
- 61.Qutaishat SS, Stemper ME, Spencer SK, Borchardt MA, Opitz JC, Monson TA, Anderson JL, Ellingson JLE. Transmission of Salmonella enterica serotype typhimurium DT104 to infants through mother’s breast milk. Pediatrics. 2003;111:1442–1446. doi: 10.1542/peds.111.6.1442. [DOI] [PubMed] [Google Scholar]
- 62.Albesharat R, Ehrmann MA, Korakli M, Yazaji S, Vogel RF. Phenotypic and genotypic analyses of lactic acid bacteria in local fermented food, breast milk and faeces of mothers and their babies. Syst Appl Microbiol. 2011;34:148–155. doi: 10.1016/j.syapm.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 63.Martin V, Maldonado-Barragán A, Moles L, Rodriguez-Baños M, Campo RD, Fernández L, Rodríguez JM, Jiménez E. Sharing of bacterial strains between breast milk and infant feces. J Hum Lact. 2012;28:36–44. doi: 10.1177/0890334411424729. [DOI] [PubMed] [Google Scholar]
- 64.Makino H, Martin R, Ishikawa E, Gawad A, Kubota H, Sakai T, Oishi K, Tanaka R, Ben-Amor K, Knol J, et al. Multilocus sequence typing of bifidobacterial strains from infant’s faeces and human milk: are bifidobacteria being sustainably shared during breastfeeding? Benef Microbes. 2015;6:563–572. doi: 10.3920/BM2014.0082. [DOI] [PubMed] [Google Scholar]
- 65.Li S-W, Watanabe K, Hsu -C-C, Chao S-H, Yang Z-H, Lin Y-J, Chen -C-C, Cao Y-M, Huang H-C, Chang C-H, et al. Bacterial composition and diversity in breast milk samples from mothers living in Taiwan and Mainland China. Front Microbiol. 2017;8:965. doi: 10.3389/fmicb.2017.00965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Preston FW. The commonness, and rarity, of species. Ecology. 1948;29:254–283. doi: 10.2307/1930989. [DOI] [Google Scholar]
- 67.Urbaniak C, Angelini M, Gloor GB, Reid G. Human milk microbiota profiles in relation to birthing method, gestation and infant gender. Microbiome. 2016;4:1. doi: 10.1186/s40168-015-0145-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jost T, Lacroix C, Braegger C, Chassard C. Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br J Nutr. 2013;110:1253–1262. doi: 10.1017/S0007114513000597. [DOI] [PubMed] [Google Scholar]
- 69.Collado MC, Delgado S, Maldonado A, Rodriguez JM. Assessment of the bacterial diversity of breast milk of healthy women by quantitative real-time PCR. Lett Appl Microbiol. 2009;48:523–528. doi: 10.1111/j.1472-765X.2009.02567.x. [DOI] [PubMed] [Google Scholar]
- 70.Jimenez E, de Andrés J, Manrique M, Pareja-Tobes P, Tobes R, Martínez-Blanch JF, Codoñer FM, Ramón D, Fernández L, Rodríguez JM, et al. Metagenomic analysis of milk of healthy and mastitis-suffering women. J Hum Lact. 2015;31:406–415. doi: 10.1177/0890334415585078. [DOI] [PubMed] [Google Scholar]
- 71.Obermajer T, Lipoglavšek L, Tompa G, Treven P, Lorbeg PM, Matijašić BB, Rogelj I. Colostrum of healthy Slovenian mothers: microbiota composition and bacteriocin gene prevalence. PLoS One. 2014;10:e0123324. doi: 10.1371/journal.pone.0123324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gonzalez R, Mandomando I, Fumadó V, Sacoor C, Macete E, Alonso PL, Menendez C. Breast milk and gut microbiota in African mothers and infants from an area of high HIV prevalence. PLoS One. 2013;8:e80299. doi: 10.1371/journal.pone.0080299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sakwinska O, Moine D, Delley M, Combremont S, Rezzonico E, Descombes P, Vinyes-Pares G, Zhang Y, Wang P, Thakkar SK, et al. Microbiota in breast milk of chinese lactating mothers. PLoS One. 2016;11:e0160856. doi: 10.1371/journal.pone.0160856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moossavi S, Sepehri S, Robertson B, Bode L, Goruk S, Field CJ, Lix LM, de Souza RJ, Becker AB, Mandhane PJ, et al. Composition and variation of the human milk microbiota are influenced by maternal and early-life factors. Cell Host Microbe. 2019;25:324–335 e324. doi: 10.1016/j.chom.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 75.Khodayar-Pardo P, Mira-Pascual L, Collado MC, Martinez-Costa C. Impact of lactation stage, gestational age and mode of delivery on breast milk microbiota. J Perinatol. 2014;34:599–605. doi: 10.1038/jp.2014.47. [DOI] [PubMed] [Google Scholar]
- 76.Christley R. Power and error: increased risk of false positive results in underpowered studies. Open Epidemiol J. 2010;3:16–19. doi: 10.2174/1874297101003010016. [DOI] [Google Scholar]
- 77.UNICEF . Overview of breastfeeding patterns. 2008. Accessed 6 Feb 2019. http://www.childinfo.org/breastfeeding_ghana.html
- 78.Wright AL, Bauer M, Naylor A, Sutcliffe E, Clark L. Increasing breastfeeding rates to reduce infant illness at the community level. Pediatrics. 1998;101(5):837–844. [DOI] [PubMed] [Google Scholar]
- 79.CODEX . Codex Alimentarius. International food standards. 2016.
- 80.Yoshioka H, Iseki K, Fujita K. Development and differences of intestinal flora in the neonatal period in breast-fed and bottle-fed infants. Pediatrics. 1983;72:317–321. [PubMed] [Google Scholar]
- 81.Favier CF, Vaughan EE, De Vos WM, Akkermans AD. Molecular monitoring of succession of bacterial communities in human neonates. Appl Environ Microbiol. 2002;68:219–226. doi: 10.1128/AEM.68.1.219-226.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Harmsen HJ, Wildeboer–Veloo ACM, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, Welling GW. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr. 2000;30:61–67. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 83.Hopkins MJ, Macfarlane GT, Furrie E, Fite A, Macfarlane S. Characterisation of intestinal bacteria in infant stools using real-time PCR and northern hybridisation analyses. FEMS Microbiol Ecol. 2005;54:77–85. doi: 10.1016/j.femsec.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 84.Bezirtzoglou E, Tsiotsias A, Welling GW. Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (FISH). Anaerobe. 2011;17:478–482. doi: 10.1016/j.anaerobe.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 85.Izumi H, Ehara T, Sugahara H, Matsubara T, Mitsuyama E, Nakazato Y, Tsuda M, Shimizu T, Odamaki T, Xiao J-Z, et al. The combination of Bifidobacterium breve and three prebiotic oligosaccharides modifies gut immune and endocrine functions in neonatal MiceWe. J Nutr. 2019;149:344–353. doi: 10.1093/jn/nxy248. [DOI] [PubMed] [Google Scholar]
- 86.Martin R, Jimenez E, Heilig H, Fernandez L, Marin ML, Zoetendal EG, Rodriguez JM. Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR. Appl Environ Microbiol. 2009;75:965–969. doi: 10.1128/AEM.02063-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bazanella M, Maier TV, Clavel T, Lagkouvardos I, Lucio M, Maldonado-Gòmez MX, Autran C, Walter J, Bode L, Schmitt-Kopplin P, et al. Randomized controlled trial on the impact of early-life intervention with bifidobacteria on the healthy infant fecal microbiota and metabolome. Am J Clin Nutr. 2017;106:1274–1286. doi: 10.3945/ajcn.117.157529. [DOI] [PubMed] [Google Scholar]
- 88.Korpela K, Salonen A, Vepsäläinen O, Suomalainen M, Kolmeder C, Varjosalo M, Miettinen S, Kukkonen K, Savilahti E, Kuitunen M, et al. Probiotic supplementation restores normal microbiota composition and function in antibiotic-treated and in caesarean-born infants. Microbiome. 2018;6:182. doi: 10.1186/s40168-018-0567-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Quin C, Estaki M, Vollman DM, Barnett JA, Gill SK, Gibson DL. Probiotic supplementation and associated infant gut microbiome and health: a cautionary retrospective clinical comparison. Sci Rep. 2018;8:8283. doi: 10.1038/s41598-018-26423-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Palframan RJ, Gibson GR, Rastall RA. Carbohydrate preferences of Bifidobacterium species isolated from the human gut. Curr Issues Intest Microbiol. 2003;4:71–75. [PubMed] [Google Scholar]
- 91.Smilowitz JT, Lebrilla CB, Mills DA, German JB, Freeman SL. Breast milk oligosaccharides: structure-function relationships in the neonate. Annu Rev Nutr. 2014;34:143–169. doi: 10.1146/annurev-nutr-071813-105721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Penders J, Vink C, Driessen C, London N, Thijs C, Stobberingh EE. Quantification of Bifidobacterium spp., Escherichia coli and Clostridium difficile in faecal samples of breast-fed and formula-fed infants by real-time PCR. FEMS Microbiol Lett. 2005;243:141–147. doi: 10.1016/j.femsle.2004.11.052. [DOI] [PubMed] [Google Scholar]
- 93.Lundequist B, Nord CE, Winberg J. The composition of the faecal microflora in breastfed and bottle fed infants from birth to eight weeks. Acta Paediatr Scand. 1985;74:45–51. doi: 10.1111/apa.1985.74.issue-1. [DOI] [PubMed] [Google Scholar]
- 94.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, et al. Diversity of bifidobacteria within the infant gut microbiota. PLoS One. 2012;7:e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Talarico ST, Santos FE, Brandt KG, Martinez MB, Taddei CR. Anaerobic bacteria in the intestinal microbiota of Brazilian children. Clinics (Sao Paulo). 2017;72:154–160. doi: 10.6061/clinics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Carver JD. Advances in nutritional modifications of infant formulas. Am J Clin Nutr. 2003;77:1550S–1554S. doi: 10.1093/ajcn/77.6.1550S. [DOI] [PubMed] [Google Scholar]
- 97.Riva E, Verduci E, Agostoni C, Giovannini M. Comparison of the nutritional values of follow-on formulae available in Italy. J Int Med Res. 2007;35:20–37. doi: 10.1177/147323000703500102. [DOI] [PubMed] [Google Scholar]
- 98.Kus MMM, Silva SAD, Aued-pimentel, S, Mancino-filho, J. Nutrition facts of infant formulas sold in São Paulo state: assessment of fat and fatty acid contents. Rev Nutr. 2011;24:209–218. doi: 10.1590/S1415-52732011000200002. [DOI] [Google Scholar]
- 99.Vandenplas Y, Zakharova I, Dmitrieva Y. Oligosaccharides in infant formula: more evidence to validate the role of prebiotics. Br J Nutr. 2015;113:1339–1344. doi: 10.1017/S0007114515000823. [DOI] [PubMed] [Google Scholar]
- 100.Nagpal R, Kurakawa T, Tsuji H, Takahashi T, Kawashima K, Nagata S, Nomoto K, Yamashiro Y, et al. Evolution of gut Bifidobacterium population in healthy Japanese infants over the first three years of life: a quantitative assessment. Sci Rep. 2017;7:10097. doi: 10.1038/s41598-017-10711-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Barrett E, Deshpandey AK, Ryan CA, Dempsey EM, Murphy B, O’Sullivan L, Watkins C, Ross RP, O’Toole PW, Fitzgerald GF, et al. The neonatal gut harbours distinct bifidobacterial strains. Arch Dis Child Fetal Neonatal Ed. 2015;100:F405–410. doi: 10.1136/archdischild-2014-306110. [DOI] [PubMed] [Google Scholar]
- 102.Drell T, Štšepetova J, Simm J, Rull K, Aleksejeva A, Antson A, Tillmann V, Metsis M, Sepp E, Salumets A, et al. The influence of different maternal microbial communities on the development of infant gut and oral microbiota. Sci Rep. 2017;7:9940. doi: 10.1038/s41598-017-09278-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Oswari H, Prayitno L, Dwipoerwantoro PG, Firmansyah A, Makrides M, Lawley B, Kuhn-Sherlock B, Cleghorn G, Tannock GW. Comparison of stool microbiota compositions, stool alpha1-antitrypsin and calprotectin concentrations, and diarrhoeal morbidity of Indonesian infants fed breast milk or probiotic/prebiotic-supplemented formula. J Paediatr Child Health. 2013;49:1032–1039. doi: 10.1111/jpc.12307. [DOI] [PubMed] [Google Scholar]
- 104.Adlerberth I, Lindberg E, Åberg N, Hesselmar B, Saalman R, Strannegård I-L, Wold AE. Reduced enterobacterial and increased staphylococcal colonization of the infantile bowel: an effect of hygienic lifestyle? Pediatr Res. 2006;59:96–101. doi: 10.1203/01.pdr.0000191137.12774.b2. [DOI] [PubMed] [Google Scholar]
- 105.Bennet R, Eriksson M, Tafari N, Nord CE. Intestinal bacteria of newborn Ethiopian infants in relation to antibiotic treatment and colonisation by potentially pathogenic gram-negative bacteria. Scand J Infect Dis. 1991;23:63–69. doi: 10.3109/00365549109023376. [DOI] [PubMed] [Google Scholar]
- 106.Brandt K, Taddei CR, Takagi EH, Oliveira FF, Duarte R, Irino I, Martinez MB, Carneiro-Sampaio M. Establishment of the bacterial fecal community during the first month of life in Brazilian newborns. Clinics (Sao Paulo). 2012;67:113–123. pii. doi:S1807-59322012000200005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Taddei CR, Oliveira FF, Duarte RTD, Talarico ST, Takagi EH, Ramos Carvalho II, Gomes FMS, Brandt K, Martinez MB. High abundance of escherichia during the establishment of fecal microbiota in Brazilian children. Microb Ecol. 2014;67:624–634. doi: 10.1007/s00248-014-0381-x. [DOI] [PubMed] [Google Scholar]
- 108.Adlerberth I, CARLSSON B, Man PD, JALIL F, Khan SR, LARSSON P, MELLANDER L, Svanborg C, WOLD AE, HANSON LÅ, et al. Intestinal colonization with enterobacteriaceae in Pakistani and Swedish hospital-delivered infants. Acta Paediatr Scand. 1991;80:602–610. doi: 10.1111/apa.1991.80.issue-6-7. [DOI] [PubMed] [Google Scholar]
- 109.Adlerberth I, Jalil F, Carlsson B, Mellander L, Hanson LÅ, Larsson P, Khalil K, WOLD AE. High turnover rate of escherichia coli strains in the intestinal flora of infants in Pakistan. Epidemiol Infect. 1998;121:587–598. doi: 10.1017/S0950268898001484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mata LJ, Mejicanos ML, Jimenez F. Studies on the indigenous gastrointestinal flora of guatemalan children. Am J Clin Nutr. 1972;25:1380–1390. doi: 10.1093/ajcn/25.12.1380. [DOI] [PubMed] [Google Scholar]
- 111.Murono K, Fuiita K, Yoshikawa M, Saijo M, Inyaku F, Kakehashi H, Tsukamoto T. Acquisition of nonmaternal Enterobacteriaceae by infants delivered in hospitals. J Pediatr. 1993;122:120–125. pii. .S0022-3476(05)83504-8. [DOI] [PubMed] [Google Scholar]
- 112.Rotimi VO, Duerden BI. The development of the bacterial flora in normal neonates. J Med Microbiol. 1981;14:51–62. doi: 10.1099/00222615-14-1-51. [DOI] [PubMed] [Google Scholar]
- 113.Benno Y, Sawada K, Mitsuoka T. The intestinal microflora of infants: composition of fecal flora in breast-fed and bottle-fed infants. Microbiol Immunol. 1984;28:975–986. doi: 10.1111/mim.1984.28.issue-9. [DOI] [PubMed] [Google Scholar]
- 114.Stark PL, Lee A. The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year of life. J Med Microbiol. 1982;15:189–203. doi: 10.1099/00222615-15-2-189. [DOI] [PubMed] [Google Scholar]
- 115.Bullen CL, Tearle PV, Willis AT. Bifidobacteria in the intestinal tract of infants: an in-vivo study. J Med Microbiol. 1976;9:325–333. doi: 10.1099/00222615-9-3-325. [DOI] [PubMed] [Google Scholar]
- 116.McAllister TA, Givan J, Black A, Turner MJ, Kerr MM, Hutchison JH. The natural history of bacterial colonization of the newborn in a maternity hospital. I Scott Med J. 1974;19:119–124. doi: 10.1177/003693307401900304. [DOI] [PubMed] [Google Scholar]
- 117.Borderon JC, Lionnet C, Rondeau C, Suc AL, Laugier J, Gold F. Current aspects of the fecal flora of the newborn without antibiotherapy during the first 7 days of life: enterobacteriaceae, enterococci, staphylococci. Pathol Biol (Paris). 1996;44:416–422. [PubMed] [Google Scholar]
- 118.Nowrouzian F, Hesselmar B, Saalman R, Strannegård I-L, Åberg N, Wold AE, Adlerberth I. Escherichia coli in infants’ intestinal microflora: colonization rate, strain turnover, and virulence gene carriage. Pediatr Res. 2003;54:8–14. doi: 10.1203/01.PDR.0000069843.20655.EE. [DOI] [PubMed] [Google Scholar]
- 119.Lindberg E, Nowrouzian F, Adlerberth I, Wold AE. Long-time persistence of superantigen-producing staphylococcus aureus strains in the intestinal microflora of healthy infants. Pediatr Res. 2000;48:741–747. doi: 10.1203/00006450-200012000-00007. [DOI] [PubMed] [Google Scholar]
- 120.Eggesbo M, Moen B, Peddada S, Baird D, Rugtveit J, Midtvedt T, Bushel PR, Sekelja M, Rudi K. Development of gut microbiota in infants not exposed to medical interventions. APMIS. 2011;119:17–35. doi: 10.1111/j.1600-0463.2010.02688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Harris V, Ali A, Fuentes S, Korpela K, Kazi M, Tate J, Parashar U, Wiersinga WJ, Giaquinto C, de Weerth C, et al. Rotavirus vaccine response correlates with the infant gut microbiota composition in Pakistan. Gut Microbes. 2018;9:93–101. doi: 10.1080/19490976.2017.1376162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kigbu A, Orimadegun AE, Tongo OO, Odaibo GN, Olaleye DO, Akinyinka OO. Intestinal bacterial colonization in the first 2 weeks of life of nigerian neonates using standard culture methods. Front Pediatr. 2016;4:139. doi: 10.3389/fped.2016.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ekwempu C, Lawande R, Egler L. Bacterial colonisation of various sites at birth of babies born in Zaria. J Infect. 1982;5:177–181. doi: 10.1016/S0163-4453(82)91859-X. [DOI] [Google Scholar]
- 124.Popkin B. Nutritional patterns and transitions. Popul Dev Rev. 1993;19:138–157. doi: 10.2307/2938388. [DOI] [Google Scholar]
- 125.Siigur U, Ormisson A, Tamm A. Faecal short-chain fatty acids in breast-fed and bottle-fed infants. Acta Paediatr. 1993;82:536–538. doi: 10.1111/apa.1993.82.issue-6-7. [DOI] [PubMed] [Google Scholar]
- 126.Davis JC, Lewis ZT, Krishnan S, Bernstein RM, Moore SE, Prentice AM, Mills DA, Lebrilla CB, Zivkovic AM. Growth and morbidity of Gambian infants are influenced by maternal milk oligosaccharides and infant gut microbiota. Sci Rep. 2017;7:40466. doi: 10.1038/srep40466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sakata H, Yoshioka H, Fujita K. Development of the intestinal flora in very low birth weight infants compared to normal full-term newborns. Eur J Pediatr. 1985;144:186–190. doi: 10.1007/BF00451911. [DOI] [PubMed] [Google Scholar]
- 128.Lee EK, Ahn YT, Huh CS, Soo Kim H, Kim E, Chun YH, Yoon J-S, Kim HH, Tack Kim J. The early intestinal microbiota of healthy korean newborns. Iran J Pediatr. 2015;25:e2079. doi: 10.5812/ijp.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hall MA, Cole CB, Smith SL, Fuller R, Rolles CJ. Factors influencing the presence of faecal lactobacilli in early infancy. Arch Dis Child. 1990;65:185–188. doi: 10.1136/adc.65.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Roberts AK, Chierici R, Sawatzki G, Hill MJ, Volpato S, Vigi V. Supplementation of an adapted formula with bovine lactoferrin: 1. Effect on the infant faecal flora. Acta Paediatr. 1992;81:119–124. doi: 10.1111/apa.1992.81.issue-2. [DOI] [PubMed] [Google Scholar]
- 131.Hallab JC, Leach ST, Zhang L, Mitchell HM, Oei J, Lui K, Day AS. Molecular characterization of bacterial colonization in the preterm and term infant’s intestine. Indian J Pediatr. 2013;80:1–5. doi: 10.1007/s12098-012-0753-5. [DOI] [PubMed] [Google Scholar]
- 132.Gronlund MM, Arvilommi H, Kero P, Lehtonen OP, Isolauri E. Importance of intestinal colonisation in the maturation of humoral immunity in early infancy: a prospective follow up study of healthy infants aged 0-6 months. Arch Dis Child Fetal Neonatal Ed. 2000;83:F186–192. doi: 10.1136/fn.83.3.F186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gabriel I, Olejek A, Stencel-Gabriel K, Wielgos M. The influence of maternal vaginal flora on the intestinal colonization in newborns and 3-month-old infants. J Matern Fetal Neonatal Med. 2018;31:1448–1453. doi: 10.1080/14767058.2017.1319352. [DOI] [PubMed] [Google Scholar]
- 134.Lewis ZT, Sidamonidze K, Tsaturyan V, Tsereteli D, Khachidze N, Pepoyan A, Zhgenti E, Tevzadze L, Manvelyan A, Balayan M, et al. The fecal microbial community of breast-fed infants from Armenia and Georgia. Sci Rep. 2017;7:40932. doi: 10.1038/srep40932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kuang Y-S, Li S-H, Guo Y, Lu J-H, He J-R, Luo B-J, Jiang F-J, Shen H, Papasian CJ, Pang H, et al. Composition of gut microbiota in infants in China and global comparison. Sci Rep. 2016;6:36666. doi: 10.1038/srep36666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Langhendries JP, Detry J, Hees JV, Lamboray JM, Darimont J, Mozin MJ, Secretin MC, Senterre J. Effect of a fermented infant formula containing viable bifidobacteria on the fecal flora composition and pH of healthy full-term infants. J Pediatr Gastroenterol Nutr. 1995;21:177–181. doi: 10.1097/00005176-199508000-00009. [DOI] [PubMed] [Google Scholar]
- 138.Vaishampayan PA, Kuehl JV, Froula JL, Morgan JL, Ochman H, Francino MP. Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. Genome Biol Evol. 2010;2:53–66. doi: 10.1093/gbe/evp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sordillo JE, Zhou Y, McGeachie MJ, Ziniti J, Lange N, Laranjo N, Savage JR, Carey V, O’Connor G, Sandel M, et al. Factors influencing the infant gut microbiome at age 3-6 months: findings from the ethnically diverse vitamin D Antenatal Asthma Reduction Trial (VDAART). J Allergy Clin Immunol. 2017;139:482–491 e414. doi: 10.1016/j.jaci.2016.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Arrieta M-C, Arévalo A, Stiemsma L, Dimitriu P, Chico ME, Loor S, Vaca M, Boutin RCT, Morien E, Jin M, et al. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J Allergy Clin Immunol. 2018;142:424–434 e410. doi: 10.1016/j.jaci.2017.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivastava TP, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. 1999;69:1035S–1045S. doi: 10.1093/ajcn/69.5.1035s. [DOI] [PubMed] [Google Scholar]
- 144.Pop M, Walker AW, Paulson J, Lindsay B, Antonio M, Hossain M, Oundo J, Tamboura B, Mai V, Astrovskaya I, et al. Diarrhea in young children from low-income countries leads to large-scale alterations in intestinal microbiota composition. Genome Biol. 2014;15:R76. doi: 10.1186/gb-2014-15-6-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yap GC, Chee KK, Hong P-Y, Lay C, Satria CD, Soenarto Y, Haksari EL, Aw M, Shek LPC, et al. Evaluation of stool microbiota signatures in two cohorts of Asian (Singapore and Indonesia) newborns at risk of atopy. BMC Microbiol. 2011;11:193. doi: 10.1186/1471-2180-11-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Walker WA. The importance of appropriate initial bacterial colonization of the intestine in newborn, child, and adult health. Pediatr Res. 2017;82:387–395. doi: 10.1038/pr.2017.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis. 2015;26:26191. doi: 10.3402/mehd.v26.26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Quin C, Gibson DL. Dietary fatty acids and host-microbial crosstalk in neonatal enteric infection. Nutrients. 2019;11:2064. doi: 10.3390/nu11092064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Delport S. Global epidemiology of use of and disparities in caesarean sections. Lancet. 2019;394:23–24. doi: 10.1016/S0140-6736(19)30717-2. [DOI] [PubMed] [Google Scholar]
- 150.Callahan BJ, McMurdie PJ, Holmes SP. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. Isme J. 2017;11:2639–2643. doi: 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nevoral J, Rada V, Vlková E, Bláhová K, Bronský J, Bubáková D, Killer J. Intestinal microbiota in exclusively breast-fed infants with blood-streaked stools. Folia Microbiol (Praha). 2009;54:167–171. doi: 10.1007/s12223-009-0026-5. [DOI] [PubMed] [Google Scholar]
- 152.Tsuji H, Oozeer R, Matsuda K, Matsuki T, Ohta T, Nomoto K, Tanaka R, Kawashima M, Kawashima K, Nagata S, et al. Molecular monitoring of the development of intestinal microbiota in Japanese infants. Benef Microbes. 2012;3:113–125. doi: 10.3920/BM2011.0038. [DOI] [PubMed] [Google Scholar]
- 153.Obermajer T, Grabnar I, Benedik E, Tušar T, Robič Pikel T, Fidler Mis N, Bogovič Matijašić B, Rogelj I. Microbes in infant gut development: placing abundance within environmental, clinical and growth parameters. Sci Rep. 2017;7:11230. doi: 10.1038/s41598-017-10244-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Grzeskowiak Ł, Grönlund -M-M, Beckmann C, Salminen S, von Berg A, Isolauri E. The impact of perinatal probiotic intervention on gut microbiota: double-blind placebo-controlled trials in Finland and Germany. Anaerobe. 2012;18:7–13. doi: 10.1016/j.anaerobe.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 155.Huda MN, Lewis Z, Kalanetra KM, Rashid M, Ahmad SM, Raqib R, Qadri F, Underwood MA, Mills DA, Stephensen CB, et al. Stool microbiota and vaccine responses of infants. Pediatrics. 2014;134:e362–372. doi: 10.1542/peds.2013-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lewis ZT, Totten SM, Smilowitz JT, Popovic M, Parker E, Lemay DG, Van Tassell ML, Miller MJ, Jin Y-S, German JB, et al. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome. 2015;3:13. doi: 10.1186/s40168-015-0071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- UNICEF . Overview of breastfeeding patterns. 2008. Accessed 6 Feb 2019. http://www.childinfo.org/breastfeeding_ghana.html


