Graphical abstract

Keywords: SARS-CoV-2, COVID-19, SARS-CoV-2 spike (S) protein, RNA dependent RNA polymerase, 3CL or Mpro Protease, Clinical trials
Highlights
-
•
COVID-19 created an unprecedented medical and economic crisis all over the world.
-
•
Spike protein, RNA dependent RNA polymerase and Mpro Protease are major targets for COVID-19.
-
•
Similarity and key amino acid interactions of these are compared with SARS-CoV.
-
•
It will help to understand key amino acids essential for interactions at the active site of target proteins in SARS-CoV-2.
Abstract
SARS-CoV-2 (COVID-19) epidemic has created an unprecedented medical and economic crisis all over the world. SARS-CoV-2 is found to have more contagious character as compared to MERS-CoV and is spreading in a very fast manner all around the globe. It has affected over 31 million people all over the world till date. This virus shares around 80% of genome similarity with SARS-CoV. In this perspective, we have explored three major targets namely; SARS-CoV-2 spike (S) protein, RNA dependent RNA polymerase, and 3CL or Mpro Protease for the inhibition of SARS-CoV-2. These targets have attracted attention of the medicinal chemists working on computer-aided drug design in developing new small molecules that might inhibit these targets for combating COVID-19 disease. Moreover, we have compared the similarity of these target proteins with earlier reported coronavirus (SARS-CoV). We have observed that both the coronaviruses share around 80% similarity in their amino acid sequence. The key amino acid interactions which can play a crucial role in designing new small molecule inhibitors against COVID-19 have been reported in this perspective. Authors believe that this study will help the medicinal chemists to understand the key amino acids essential for interactions at the active site of target proteins in SARS-CoV-2, based on their similarity with earlier reported viruses. In this review, we have also described the lead molecules under various clinical trials for their efficacy against COVID-19.
1. Introduction
Coronaviruses (CoVs) tend to have a high zoonotic potential and according to the World Health Organization (WHO), these viral diseases have emerged as a serious health issue to the world [1]. Two previously identified coronaviruses (CoVs), severe acute respiratory syndrome CoV (SARS-CoV) and Middle East respiratory syndrome CoV (MERS-CoV), have dominated medical, science, and media attention over the past two decades due to their dangerous epidemic potential [2], [3]. The first SARS case was found in Foshan, China, in November 2002 [4], and the first case of MERS died in June 2012 in a hospital in Jeddah, Saudi Arabia. Both zoonotic diseases remain on the list of priority diseases of the World Health Organization (WHO) as they pose a major threat to the global public health security [5], [6]. The novel coronavirus pneumonia of 2019, called COVID-19 by the World Health Organization (WHO), triggered by SARS-CoV-2, first emerged in Wuhan City in December 2019 and emerged as an epidemic overworld [7], [8], [9]. This new virus shares 80% of its genome with SARS-CoV but more contagious which leads to very fast spreading all over the globe and presented a great challenge to the health system of the whole world [10], [11]. Various studies on COVID-19 reported its epidemiological and clinical characteristics. Most patients effected from it were found to develop fever and cough while some patients experienced acute respiratory failure, ARDS, septic shock, and other serious complications. Critical patients tend to have poor outcome and high mortality as compared with other forms of CoV [12], [13], [14]. Currently, the diagnosed therapy of COVID-19 relies primarily on a consensus guideline involving epidemic communication history, laboratory testing, and CT imaging analysis [15], [16]. Since the first case in China, the epidemic has spread to a total of 213 countries and regions, with more than 31 million confirmed cases as reported till 23 September 2020.
Coronaviruses are Nidovirales single-strand RNA viruses which are spherical or pleomorphic in shape with bear’s club-shaped glycoprotein projections. These are classified into four genera: alpha, beta, gamma, and delta coronaviruses (Fig. 2 ) [17]. Gamma and delta CoVs are primarily associated with avian hosts, while alpha and beta CoVs contain many human and domestic pathogens that are likely to be associated with the transmission events of cross-species [18], [19]. Both SARS-CoV and MERS-CoV belong to the genus beta coronavirus and are associated with a severe infection of the respiratory tract with mortality rates of 10% and 35% respectively [20]. The SARS pandemic was managed rapidly through an unparalleled global containment campaign, and since May 2004, the virus has not been identified in humans [4]. Despite this rapid eradication, nearly 800 deaths in 27 countries were caused by SARS-CoV, with persistent outbreaks in 18 countries on three continents (WHO). Rhinolophid bats are gradually serving as natural reservoirs for SARS-related CoVs with a potential spill-over to the non-flying mammals. For example, as with the SARS coronavirus, some bat CoVs may use angiotensin-converting enzyme 2 (ACE2) as a cell receptor [21], [22], [23], [24]. In comparison, the role of bats in MERS-CoV epidemiology is less well explained, since this human viruse is mostly linked to the viruses found in dromedary camels [25]. In addition, while similar viruses have been identified in bats, they are divergent in their spike sequences and appear ineffective in the use of human dipeptidyl peptidase 4 (DPP4) as a receptor cell [23], [26], [27]. The MERS epidemic is ongoing in the Middle East, with travel-related cases recorded in 27 countries around the world [28]. Lastly, Alphacoronaviruses 229E and NL63, which induce a mild human influenza-like syndrome, share a common ancestor with Hipposideros and Triaenops bat-sampled viruses, respectively [29], [30], [31].
Fig. 2.
Classification of different coronaviruses.
Bats are considered to have high rates of CoV diversity with geographical range and prevalence in almost every species investigated. It supports the theory that bats played a major role in the evolution of CoV [18], [32]. Additionally, bat CoVs are interspersed taxonomically with other associated mammals including humans and domestic animals which is consistent with the theory that bats are a significant genetic reservoir of CoVs [31], [33]. The long-term evolutionary relations between bats and coronaviruses are also confirmed by phylogenetic evidence that CoVs display such tropisms unique to species and genus [34], [35], and those taxonomically-based viruses are identified independently of the sampling position in different bat species. Conversely, that CoVs are not always shared among bat species that co-roost suggests that there are some barriers to transmission across species [18], [26], [35], [36], [37], [38]. Due to the topological similarity between the phylogenetic trees of CoVs and their mammalian hosts, it has been proposed that the diversity of CoVs represents primarily the long-term co-divergence between bats and CoVs [35]. Indeed, recent studies of unique bat taxa from different locations indicate that the role of virus-host codivergence in the evolutionary history of CoVs may have been overestimated relative to other events like host-jumping [31], [32], [39].
In SARS-CoV-2 to date, three targets have been explored namely the SARS-CoV-2 spike (S) protein [40], RNA dependent RNA polymerase [41], and 3CL or Mpro Protease [42] with several mechanisms for the inhibition of the SARS-CoV-2. Out of these two targets, RNA dependent RNA polymerase and 3CL/Mpro Protease are reported with their inhibitors while the antibody-based inhibition was reported for the SARS-CoV-2 spike protein receptor-binding domain (RBD) [43]. Other than these three targets, some host-based targets, their role in SARS-CoV-2 transmission and problems in targeting them are also discussed in this manuscript. However it is also known that SARS-CoV-2, as well as SARS-CoV, are identical (80%) [44], [45], So the information about the structural similarity and identical amino acids of targets has unlocked the pathways of the medicinal chemists working in the area of computer-aided drug design in generating new small molecules and structural biologists to design the antibodies that might interact with these targets to combat the COVID-19 disease.
1.1. History of the coronavirus
In 1960, the coronavirus was first reported for causing the common cold [46], [47]. More than 500 patients have emerged with flu-like the symptoms in one study conducted in Canada in 2001. Virologic analysis has shown that 3.6 percent of these cases were polymerase chain reaction (PCR) positive for the HCoV- strain [48]. Coronavirus was considered as a fairly safe, non-virus until 2002. Before 2003, there were only 2 CoVs reported to cause human disease, human CoV 229E (HCoV-229E), and HCoV-OC43 [48] which present mild symptoms such as common cold in adults and more severe illness in children and elders along with the weakened immune system. In November 2002, rare cases of unknown cause of “atypical pneumonia” occurred in Foshan City, Guangdong Province, China, where many health care workers were infected [49]. The infection was brought to Hong Kong on 21 February 2003 by a physician who had looked after the similar cases of atypical pneumonia in mainland China, which resulted in serious pneumonia outbreaks in Hong Kong and on 15 March 2003, was named as “severe acute respiratory syndrome” by the WHO [50], [51], [52]. Several months were passed and several hundred cases of SARS were identified before discovering SARS-CoV. On 22 March 2003, a novel CoV (SARS CoV) of lineage B was identified as a cause of these cases of atypical pneumonia [51]. Therefore, a state of emergency was declared in 2004 by the Centers for Disease Control and Prevention (CDC) and WHO [50], [52], [53]. The evolution of this virus has shown that coronavirus is not a stable virus as well as it can adapt to humans to become more virulent, even lethal. Indeed, another outbreak in Saudi Arabia in 2012 resulted in numerous deaths. Afterwards it spreaded to other countries in the Middle East and then around the world, leading to renewed interest in studies of this new form of coronavirus (Fig. 1) [50], [51], [52], [53].
Fig. 1.
History of coronaviruses.
2. Target proteins for COVID-19
In SARS-CoV-2 till date, three targets have been explored namely the SARS-CoV-2 spike (S) protein [40], RNA dependent RNA polymerase [41], and 3CL or Mpro Protease [42] with several mechanisms for the inhibition of SARS-CoV-2. Out of these two targets, RNA dependent RNA polymerase and 3CL or Mpro Protease are reported with their inhibitors while the antibody-based inhibition is reported for the SARS-CoV-2 spike protein receptor-binding domain (RBD) [43]. So the information about the structural similarity and identical amino acids of these targets has unlocked the pathways for the medicinal chemists working in the area of computer-aided drug design in generating new small molecules and structural biologists to design the antibodies that might interact with these targets for combating the COVID-19 disease. Each of these virus proteins has an important role in the viral life cycle. For targeting these virus proteins there is a need for the knowledge of the structural insights of these proteins. The structural insights of these proteins provide knowledge about the protein regions actively participating in the viral life cycle. The knowledge of the co-crystallized structures such as 3CL or Mpro Protease with inhibitor N3 (PDB ID: 6LU7), RNA dependent RNA polymerase with remdesivir (PDB ID: 7BV2), SARS-CoV-2 spike protein interaction with Human ACE2 (PDB ID: 6M0J) provides the knowledge of active sites and the important amino acids that are responsible for the inhibition of SARS-CoV-2. So the discussion about the amino acids interacting with the inhibitors provided the insights for targeting these different proteins viable for the viral replication.
2.1. Spike glycoprotein (S Protein)
The entry of the virus in the host cell is an important step towards infection of SARS-CoV and SARS-CoV-2. This entry is facilitated by the ACE 2 receptor. The Spike glycoprotein (S Protein) of these viruses is a trimer of about 1300 amino acids that splits into S1 (700 amino acids) and S2 (600 amino acids) subunits [54], [55], [56], [57]. The S1 contains the receptor-binding domain (RBD) that interacts with the peptidase domain (PD) of ACE 2 while S2 subunit is cleaved by the host proteases in post interaction and facilitates membrane fusion and hence important step in viral infection [55], [58], [59], [60]. The interaction with the ACE 2 is facilitated by polar interactions such as hydrogen bonding, pi-pi stacking, and salt bridge interactions. The PD arc-shaped helix of ACE 2 binds to the loop region of the RBD of S protein. The amino acid interactions that helped in binding were observed between the RBD of SARS-CoV-2 and PD of ACE 2 and are considered as important aspects for the interaction inhibitor design [61]. There are 17 amino acids of SARS-CoV-2 and 20 amino acids of ACE 2. Out of these 14 amino acid interactions of SARS-CoV-2 namely Tyr449, Tyr453, Leu455, Phe456, Phe486, Asn487, Tyr489, Gln493, Gly496, Gln498, Thr500, Asn501, Gly502, and Tyr505 are considered to be important. The 5 amino acid interactions are explored to be more critical as these amino acids interact with 18 amino acids of ACE 2. These critical amino acid interactions involved Leu455 interaction with Asp30, Lys31, and His34; Phe486 with Gln24, Leu79, Met82, Tyr83, and Leu472; Gln493 with ACE 2 Lys31, His34 and Glu35; GLN498 of SARS-CoV-2 with Asp38, Tyr41, Gln42, Leu45, and Lys353. Amino acid Asn501 has a similar type of interactions with ACE2 Lys353, Gly354, and Asp355 while H-bond interaction is observed with Tyr41. The binding affinity of the RBD domain of SARS-CoV-2 and PD of ACE2 is 4.7 nM which is strong as compared to the 31 nM in SARS-CoV. It was reported that in SARS-CoV-2 the amino acid Lys417 showed a salt bridge interaction with Asp30 of ACE-2. The positive patch on the surface of RBD of SARS-CoV-2 was added by Lys417 that contributed towards the electrostatic potential. The structure of the SARS-CoV-2 and ACE 2 interaction was reported (PDB ID: 6M0J) and the interactions of the residues are shown in Fig. 3 .
Fig. 3.
Interaction of the SARS-CoV-2 RBD domain (pink) with PD domain of Human ACE2 (blue) (PDB ID: 6M0J). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
This SARS-CoV-2 – ACE 2 interaction blocking agents is a novel approach as there is a no protein-protein interaction inhibitor available till date. However, Adedji et al. reported three mechanisms with their inhibitors for stopping the interaction in SARS-CoV with ACE 2 [62]. This approach could be applicable to the SARS-CoV-2 as well because of the identical (80%) structures of SARS-CoV and SARS-CoV-2 [44], [45]. The first sequence of RBD domain of the SARS-CoV (PDB ID: 2DD8) and SARS-CoV-2 (PDB ID: 6M0J) are aligned to show similar identical residues between them [63]. The residues that show interaction with ACE 2 are highlighted in the red boxes for SARS-CoV and SARS-CoV-2. The sequence alignment of the RBD domains of the SARS-CoV-2 and SARS-CoV showed that these are 70% identical and 80% similar to each other (Fig. 4 ).
Fig. 4.
Sequence alignment of fasta sequence of SARS-CoV-2 (PDB ID: 6M0J) and SARS-CoV (PDB ID: 2DD8) RBD domains with residues (highlighted in red) interacting with Human ACE 2. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.2. 3CL or Mpro proetease
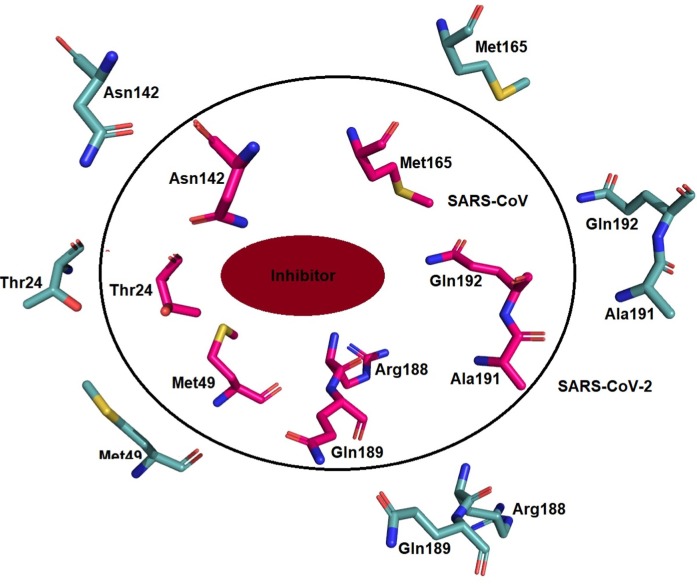
COVID-19 virus Mpro protease is a 33.79 kDa protein and its crystal structure was determined to elucidate the mechanism of inhibition by ligand N3 [42]. The co-crystallized structure of Mpro with N3 contains 303 amino acid residues that are divided into three domains. The first two domains contain the antiparallel ß sheets while the third domain consists of 5 α-helices connected to the second domain by a loop region. Domain I runs from the 8 to 101 residues which extend to domain II from 102 to184 residues. The loop region runs from 185 to 200 residues connecting domain III (201–303 residues) to domain II. The binding site for the substrate was located between domains I and II near to the Cys-His catalytic dyad. The substrate-binding pocket consists of backbone atoms with residues 164–168 (part of long strand 155–168) and 189–191 residues of loop region (connecting domain II to domain III) (Fig. 5 ) [64], [65], [66], [67].
Fig. 5.
3D crystal structure of SARS-CoV-2 Mpro with co-crystallized α-ketomide inhibitorN3 (PDB ID: 6LU7).
The co-crystallized ligand N3 is divided into 4 regions the first region contains the phenyl bulkier group that interacts with the Thr24 and Thr25 while O atom in the region interacts with Gly143. Region 2 contains lactam ring that interacts with the Phe140, Asn142, Glu166, His163, His172 via van der Waals, and H-bond interactions while the hydrophobic vinyl side chain binds to the Cys145 via covalent interactions. Region 3 of ligand consist of consists of the three amino acids leucine, valine, and alanine in which leucine interacts with the hydrophobic chain consisted of His41, Met49, Tyr54, and Met165 and its dimethyl side chain interacts with Asp187. Valine interacts with the Glu166, Leu167, and Gln189 via hydrogen bonding while alanine interacts with Thr190 via hydrogen bonding and fits into the cavity formed by Met165, Leu167, Phe185, Gln189, and Gln192. Region 4 contains an oxazole ring and showed van der Waals interaction with Thr190 and Ala191 (Fig. 6 ).
Fig. 6.
α-ketomide inhibitor four regions that interact with the different residues.
Moreover, the sequence alignment of SARS-CoV-2 and SARS-CoV Mpro has shown around 96% identical and 98% similar residues with no gaps. The similarity between the Mpro has suggested that there is no difference between the residues in the active site of SARS-CoV-2 and SARS-CoV [68] (Fig. 3). The interacting residues with the ketomide inhibitor N3 of SARS-CoV-2 and the residues interacting with an inhibitor in SARS-CoV are highlighted. The highlighted residues in different colors represent the interactions based on the region and the residues colored twice to show the interaction with both the regions (Fig. 7 ).
Fig. 7.
Sequence alignment of fasta sequence of SARS-CoV-2 (PDB ID: 6LU7) and SARS-CoV (PDB ID: 1WOF) Mpro protein with interacting residues (highlighted different regions of ligand).
2.3. RNA dependent RNA polymerase
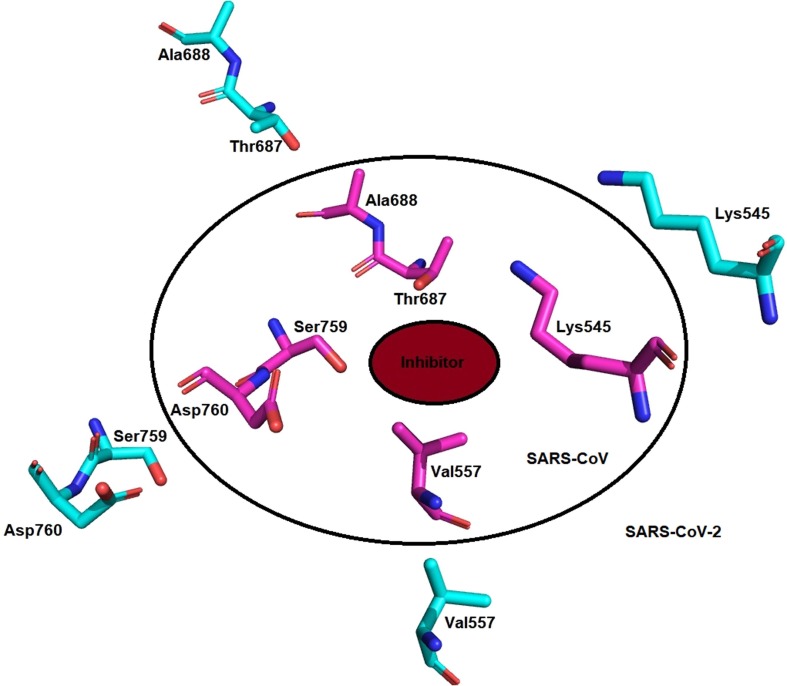
The transcription of the mRNA and replication is an important process in the viral life cycle that is carried out by the RNA dependent RNA polymerase (RdRp) [69]. The major part of the RdRp is viral non-structural proteins 12 (nsp12) which is a major catalytic subunit [70], [71]. Non-structural protein 12 (nsp12) itself is less active and require nsp7 and nsp8 for the binding of the template and processing of the mRNA [72], [73]. The structure of the complex of nsp12-nsp7-nsp8 has been determined (RdRp complex) [73], [74], [75], [76], [77]. Nsp 12 consists of β-hairpin with residues from 31 to 50, and is extended with seven helices and three β-strands [73], [77] to nidovirus RdRp-associated nucleotidyl-transferase domain (NiRAN) that contains residues from 115 to 250 [78]. The NiRAN domain connects to an interface domain that contains three helices and five β-strands from the residues 251–365 [79]. This interface domain is followed by the main RdRp domain (residues 366–920) (Fig. 7). The RdRp domain contains the finger subdomain that runs from 397 to 581 residues and 621–679 residues, followed by palm subdomain from 680 to 815 residues and connecting to the thumb subdomain from 819 to 920 residues. The complex of nsp12 was stabilized by the binding of nsp7 and nsp8 (Fig. 8 ).
Fig. 8.
X-ray structure of RNA dependent RNA polymerase (Rdrp) with the remdesvir tri-phosphate bound with nsp12, nsp7 and nsp8 regions (PDB ID: 7BV2).
nsp12 RdRp catalytic active site consists of 7 conserved motif A to G. The motifs ABCD are in the palm subdomain with an SDD (Ser759, Asp760, and Asp761) in C motif that is an active catalytic center. Asp760 and Asp761 are involved in the coordination with two magnesium ions. The motifs F and G in the finger subdomain interacts with the RNA with residues Lys545 and ARG 555. Remdesivir is a prodrug that gets converted to RTP (Remdesivir triphosphate) binds covalently to the mRNA strand and stops the mRNA transcription and replication of SARS-CoV-2. It also interacts with the magnesium ions present in the active site. The residues Asp760 and Asp761 also interact with the Remdesivir triphosphate via hydrogen bond interactions. The nsp12 of the SARS-CoV-2 and SARS-CoV are subjected to clustalW alignment and found to be 94% identical and 96% similar (Fig. 9 ).
Fig. 9.
ClustalW Sequence alignment of fasta sequence of SARS-CoV-2 (PDB ID: 7BV2) and SARS-CoV (PDB ID: 6NUR) RNA dependent RNA polymerase (RdRp) with interacting residues (highlighted in blue). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3. Specificity and key different amino acids residues in these target proteins
Table 1 shows the two types of columns, one column represents the binding residues and the other represents the residues that affect the specificity. The two columns are segregated based on differences/similarities in the binding site of SARS-CoV and SARS-CoV-2. It was observed that many of the residues in SARS-CoV and SARS-CoV-2 are similar in the case of S protein- ACE2 interaction, Mpro, and RNA dependent RNA polymerase (RdRp) i.e. more than 96%. So the specificity can be explained based on the difference between the active site residues, binding residues in case of (S1 subunit and ACE2 interaction), nature of residues, and the conformation of the residues in or near to the active site hence providing the insights to the specific inhibitor design for the SARS-CoV-2 as compared to SARS-CoV [80].
Table 1.
The important residues affecting the specificity and binding of the in SARS-CoV and SARS-CoV-2.
| Spike Protein (S1 Subunit interacting to ACE 2) | Spike protein residues affecting specificity | |
|---|---|---|
| SARS-CoV (PDB ID: 2DD8) | Tyr436, Tyr442, Leu443, Asn473, Tyr475, Asn479, Gly482, Tyr484, Thr486, Thr487, Gly488, Tyr491 | Lys439, Tyr442, Leu443, Trp476, Asn479, Asp480, Tyr484, Thr485, Thr487 |
| SARS-CoV-2 (PDB ID: 6M0J) | Tyr449, Leu455, Phe456, Phe486, Asn487, Tyr489, Gln493, Gly496, Gln498, Thr500, Asn501, Gly502, Tyr505 | Leu452, Leu455, Phe456, Phe486, Phe490, Gln493, Ser494, Gln498, Pro499, Asn501 |
| Mpro (Active site Residues) | Mpro (Residues affecting the specificity) | |
| SARS-CoV (PDB ID: 1WOF) | Thr24, Thr25, His41, Met49, Tyr54, Phe140, Asn142, Gly143, His163, Met165, Glu166, Leu167, His172, Phe185, Asp187, Gln189, Thr190, Ala191and Gln192 | Ala46, Ser65 |
| SARS-CoV-2 (PDB ID: 6LU7) | Thr24, Thr25, His41, Met49, Tyr54, Phe140, Asn142, Gly143, His163, Met165, Glu166, Leu167, Pro168, His172, Phe185, Asp187, Gln189, Thr190, Ala191and Gln192 | Ser46, Asn65 |
| RdRp (Binding to the ligand) | RdRp (Residues affecting specificity) | |
| SARS-CoV (PDB ID: 6NUR) | Lys545, Arg555, Ser759, Asp760, and Asp761 | Only conformational in the residues |
| SARS-CoV-2 (PDB ID: 7BV2) | Lys545, Arg555, Ser759, Asp760, and Asp761 | Only conformational in the residues |
Firstly the difference and similarities are highlighted in between the S1 subunits of the SARS-CoV and SARS-CoV-2 (Fig. 4). These helped in the shortlisting of the residues that are involved in the interaction of S1 with the ACE2. Although the binding is affected by the residues, some of the residues play an important role and contribute towards the binding affinities in case of SARS-CoV (31 nM) and SARS-CoV-2 (4.7 nM) [81], [82], [83]. The differences between the residues in the active site are important factors that affect the binding affinity of SARS-CoV and SARS-CoV-2. The important residue like Phe486 is present in the active site of SARS-CoV-2 while absent in the case of SARS-CoV. The other residues such as Tyr442, Leu443, Asn479, Tyr484, Thr487 are present in SARS-CoV while Leu455, Phe456, Gln493, Gln498, Asn501 are present in SARS-CoV-2 which contributes towards the binding affinities of the S1 subunits towards ACE2. These differences in the residues and nature also affect the specificity and binding towards ACE2 and useful for the medicinal chemists to design a strategy for inhibition of the interaction between S1 and ACE2. The representations of the difference between the active site residues, the nature of residues, and conformation (Fig. 10 ) depicts the specificity and may help in designing new inhibitors to stop S-protein-ACE2 interaction.
Fig. 10.
The difference between the residues affects the binding affinity of the SARS-CoV (Yellow, PDB ID: 2DD8, 31 nM) and SARS-CoV-2 (Red, PDB ID: 6M0J, 4.7 nM) towards the ACE2. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
On the other hand, 3CL or Mpro is another important target for the inhibitor design, when the active sites of the protease of SARS-CoV and SARS-CoV-2 are compared there is no difference between the residues in the active site and their nature but the conformation of the residues in the active site was different and the binding site entry residues such as Ala46 in SARS-CoV while Ser46 in SARS-CoV-2 that effects the entry as well as binding of ligands to the Mpro active site. The difference between active site residue and conformation of the active site residues are shown in Fig. 11, Fig. 12 .
Fig. 11.
The difference between the residues at the site of entry of inhibitor in Mpro of SARS-CoV (Magenta; PDB ID:6Y7M) and SARS-CoV-2 (Teal blue; PDB ID: 6LU7) affects the specificity. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 12.
There conformational changes of the residues affect the specificity and binding to the active site residues in Mpro of SARS-CoV (Magenta; PDB ID:6Y7M) and SARS-CoV-2 (Teal blue; PDB ID: 6LU7). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The RdRp (RNA dependent RNA polymerase) is another imperative target in SARS-CoV and SARS-CoV-2 and its active site also shares almost 100% similarity while there is only conformational difference of the residues in the active site. These conformational differences in the active site were highlighted in Fig. 13 .
Fig. 13.
There conformational changes of the residues affect the specificity and binding though sharing the similarity between the active site residues in RdRp of SARS-CoV (pink; PDB ID:6NUR) and SARS-CoV-2 (cyan; PDB ID: 7BV2). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Host-based druggable target
4.1. Transmembrane serine protease 2 (TMPRSS2)
This type II transmembrane enzyme belongs to the serine protease family. Some studies reported that SARS-CoV-2 uses TMPRSS2 for S protein priming [84]. SARS-CoV-2 initiate the cleavage of S protein and these TMPRSS2 inhibitors such as camostat were found to prevent the coronavirus infection [85]. A study on the Vero E6 cell line, TMPRSS2 overexpressed cells, demonstrated higher chances of corona infection which further describes its role in the progression of SARS-CoV-2 in patients. Thus, overexpression of TMPRSS2 in lung tissues makes them more vulnerable to SARS-CoV-2 [86], [87]. These facts make TMPRSS2 another important target for the treatment of SARS-CoV-2. TMPRSS2 inhibitors can be repurposed for their role in the treatment of SARS-CoV-2. A study in TMPRSS2 knockout mice revealed that these knockout mice were immune to coronavirus [88]. Some studies suggest that TMPRSS2 expression is controlled by estrogens or androgen, thus to target by which pathway is still under consideration [89]. Moreover, there is no crystal structure of TMPRSS2 reported to date. It is another problem in target-based drug designing as it can be done only via homology modeling from other similar serine protease enzymes.
4.2. Cathepsin L
Cathepsin L is found to play a key role in the entry of SARS-CoV-2 to host cells via endocytosis [90]. Cathepsin L inhibitors prevent pulmonary fibrosis. This fact indicates the importance of Cathepsin L as a target for the treatment of SARS-CoV-2 [91]. A research study on HEK 293/hACE2 cells in which these cells were treated with specific Cathepsin L inhibitor demonstrated very low chances of SARS-CoV-2 entry to host cells. It decreased the entry rate by more than 76%. It showed the key role of Cathepsin L in S protein priming by SARS-CoV-2 in lysosomes [92]. Earlier, Cathepsin L inhibitors were also found effective in the treatment of SARS-CoV [62] which also describes it as a potential target for the treatment of novel coronavirus.
The main problem behind designing Cathepsin L inhibitors for the treatment of SARS-CoV-2 is their specificity. There are different types of cathepsins reported which can play a key role in entry of the virus to cells via endocytosis. Although, targeting Cathepsin L with other target proteins can provide some beneficial effects in SARS-CoV-2 patients.
4.3. Furin
Walls et al. reported an unexpected furin cleavage site as S1/S2 boundary of SARS-CoV-2. This site is cleaved during biosynthesis and is reported as a key feature to differentiate SARS-CoV-2 from SARS-CoV [40], [93]. Furin is found highly expressed in various tissues like lungs by which SARS-CoV-2 can successfully infect the respiratory system via this convertase and initiate activation of its surface glycoprotein [94]. Similarly, in some sample studies obtained from SARS-CoV-2 infected persons in China, various mutations in the furin cleavage site were observed which ultimately affect the binding ability of S-protein to surface [95]. Thus, this site is an important feature in the pathogenicity of SARS-CoV-2. As furin is also overexpressed in the liver, kidney, and glands, it may play a role in infecting these tissues too with the novel coronavirus.
The major issue in targeting furin for treatment of SARS-CoV-2 is their role in various cellular processes. Thus, inhibition of these furin-like enzymes in our body can lead to the various side or adverse effects. Different kinds of mechanisms of furin activation reported are also still confusing in designing novel furin inhibitors.
4.4. Other host-based targets
Other host-based targets that can play a key role in the treatment of SARS-CoV-2 in the future include Adaptor-Associated Kinase 1 (AAK1), Cyclin G-Associated Kinase (GAK), and Phosphatidylinositol 3-Phosphate 5-Kinase (PIKfyve). AAK1 and GAK are serine-threonine related proteins that play a key role in the endocytosis process of cells. These enzymes are reported to play important role in regulating the transport of virus entry, binding, and release of RNA in host cells in case of rabies, ebola, dengue etc. [96], [97]. Recently, an AAK1 and GAK inhibitor is reported to decrease the entry of SARS-CoV-2 to cells but still, no mechanism is reported for this study [98]. This inhibitor, baricitinib, is also beneficial in relief from inflammation which might be advantageous in the treatment of SARS-CoV-2 caused lung inflammation also [99]. For targeting AAK1 and GAK in COVID-19 patients still, a lot of clinical studies are required as these inhibitors are also found to increase the chances of lung infection in clinical trials which is too risky for COVID-19 patients [100].
Phosphatidylinositol 3-Phosphate 5-Kinase (PIKfyve) is another enzyme that sought to play a key role in the endocytosis process and its dynamical control [101], [102]. A study on treatment with PIKfyve, IL-12, and IL-23 inhibitors (apilimod and YM201636) of 293/hACE2 cells showed the reduced entry of SARS-CoV-2 virus to cells [103], [104]. Thus, PIKfyve can be an important target in the treatment of COVID-19 but major problem is that its 3D crystal structure is still unavailable. The target-based drug designing for this protein is still difficult.
5. Different repurposed drug molecules under clinical trials against COVID-19
The fast spread of infectious COVID-19 in the world outbroken the surge of clinical trials against it in search of treatment. The race to find a treatment of this deadly viral infectious disease has started all over the world [105], [106], [107]. More than one thousand trials of various synthetic small molecules, natural molecules, antibodies, or vaccines are under progress in search of effective treatment against this deadly disease (clinicaltrials.gov) [108], [109], [110], [111]. Most synthetic small molecules under clinical trials are repurposed for COVID-19 which are already reported for their efficiency against other disease states (Table 2 , Fig. 15, Fig. 16 ). Recently, remdesivir (RNA dependent RNA polymerase inhibitor) has been approved for the treatment of COVID-19. Many drugs are under trials that act at different phases of the viral lifecycle (Fig. 14).
Table 2.
Drugs under clinical trials against COVID-19.
| S. No. | Drugs | NCT No. | Status | Sponsor | Start date/Expected End date | Phase |
|---|---|---|---|---|---|---|
| 1. | Hydroxychloroquine | NCT04345692 | Recruiting patients | Queen's Medical Centre |
March 26, 2020 /December 31, 2021 |
Phase 3 |
| 2. | Hydroxychloroquine Sulfate |
NCT04342221 | Recruiting patients | University Hospital Tuebingen Robert Bosch Medical Center Universitätsklinikum Hamburg- Eppendorf Bernhard Nocht Institute for Tropical Medicine |
March 29, 2020 /February 2022 |
Phase 3 |
| 3. | Hydroxychloroquine Azithromycin |
NCT04329832 | Recruiting patients | Intermountain Health Care, Inc. University of Utah |
March 30, 2020 /December 31, 2021 |
Phase 2 |
| 4. | Lopinavir/ritonavir Hydroxychloroquine sulfate |
NCT04307693 | Recruiting patients | Asan Medical Center |
March 11, 2020 /May 2020 |
Phase 2 |
| 5. | Hydroxychloroquine, azithromycin |
NCT04322123 | Recruiting patients | Hospital do Coracao Hospital Israelita Albert Einstein Hospital Sirio- Libanes Brazilian Research In Intensive Care Network EMS |
April 1, 2020 /August 30, 2020 |
Phase 3 |
| 6. | Chloroquine Sulfate Hydroxychloroquine |
NCT04362332 | Recruiting patients | UMC Utrecht ZonMw: The Netherlands Organisation for Health Research and Development |
April 14, 2020 /May 14, 2021 |
Phase 4 |
| 7. | Chloroquine Diphosphate |
NCT04342650 | Recruiting patients | Fundação de Medicina Tropical Dr. Heitor Vieira Dourado |
April 8, 2020 /September 2020 |
Phase 2 |
| 8. | Chloroquine | NCT04349371 | Recruiting patients | Columbia University Study Start: |
April 2020 /April 2021 |
Phase 2 |
| 9. | Chloroquine phosphate | NCT04344951 | Recruiting patients | Uni-Pharma KleonTsetis Pharmaceutical Laboratories S.A. Athens General Hospital Hippokrateio Athens General Hospital of Thoracic Diseases SOTIRIA General Hospital of Athens Sismanoglio Divine Providence Hospital Pammakaristos |
April 6, 2020 /April 30, 2021 |
Phase 2 |
| 10. | Favipiravir | NCT04336904 | Recruiting patients | Giuliano Rizzardini ASST Fatebenefratelli Sacco |
March 25, 2020 /July 2020 |
Phase 3 |
| 11. | Favipiravir | NCT04358549 | Recruiting patients | Fujifilm Pharmaceuticals U.S.A., Inc. |
April 17, 2020 /December 2020 |
Phase 2 |
| 12. | Lopinavir/ritonavir | NCT04330690 | Recruiting patients | Sunnybrook Health Sciences Centre |
March 18, 2020 /May 18, 2022 |
Phase 2 |
| 13. | Lopinavir/ritonavir Hydroxychloroquine sulfate |
NCT04307693 | Recruiting patients | Asan Medical Center |
March 11, 2020 /May 2020 |
Phase 2 |
| 14. | Remdesivir Lopinavir/ritonavir Interferon Beta-1A Hydroxychloroquine |
NCT04315948 | Recruiting patients | Institut National de la Santé Et de la Recherche Médicale, France |
March 22, 2020 /March 2023 |
Phase 3 |
| 15. | Remdesivir | NCT04292899 | Recruiting patients | Gilead Sciences | March 6, 2020 /May 2020 |
Phase 3 |
| 16. | Deferoxamine | NCT04333550 | Recruiting patients | Kermanshah University of Medical Sciences | April 2020/March 2021 | Phase 1 Phase 2 |
| 17. | Sarilumab | NCT04315298 | Recruiting patients | Regeneron Pharmaceuticals Sanofi |
March 18, 2020/April 1, 2021 | Phase 2 Phase 3 |
| 18. | Sarilumab | NCT04324073 | Active/not recruting | Assistance Publique - Hôpitaux de Paris | March 27, 2020/December 31, 2021 | Phase 2 Phase 3 |
| 19. | Amiodarone Verapamil |
NCT04351763 | Recruiting patients | Nicolaus Copernicus University | April 27, 2020/April 10, 2021 | Phase 2 Phase 3 |
| 20. | Ruxolitinib | NCT04334044 | Recruiting patients | Grupo Cooperativo de Hemopatías Malignas | April 15, 2020/June 1, 2020 | Phase 1 Phase 2 |
| 21. | Ruxolitinib | NCT04348071 | Active | University of Colorado, Denver | April 15, 2020/October 2020 | Phase 2 Phase 3 |
| 22. | Ruxolitinib plus simvastatin | NCT04348695 | Recruiting patients | Fundación de investigación HM Apices Soluciones S.L. |
April 12, 2020/May 13, 2020 | Phase 2 |
| 23. | Losartan | NCT04335123 | Recruiting patients | University of Kansas Medical Center | March 25, 2020/October 2020 | Phase 1 |
| 24. | Dapagliflozin | NCT04350593 | Recruiting patients | Saint Luke's Health System AstraZeneca George Clinical Pty Ltd |
April 15, 2020/December 2020 | Phase 3 |
| 25. | Tocilizumab | NCT04317092 | Recruiting patients | National Cancer Institute, Naples | March 19, 2020/December 19, 2022 | Phase 2 |
| 26. | Anakinra Siltuximab Tocilizumab |
NCT04330638 | Recruiting patients | University Hospital, Ghent Belgium Health Care Knowledge Centre |
April 2020/December 2020 | Phase 3 |
| 27. | Tocilizumab | NCT04331795 | Recruiting patients | University of Chicago | April 4, 2020/December 2020 | Phase 2 |
| 28. | Tocilizumab | NCT04346355 | Recruiting patients | Azienda Unità Sanitaria Locale Reggio Emilia | March 31, 2020/May 30, 2020 | Phase 2 |
| 29. | Tocilizumab | NCT04320615 | Recruiting patients | Hoffmann-La Roche | April 3, 2020/September 30, 2020 | Phase 3 |
| 30. | Sirolimus | NCT04341675 | Recruiting patients | University of Cincinnati | April 24, 2020/September 2020 | Phase 2 |
| 31. | Clazakizumab | NCT04343989 | Recruiting patients | NYU Langone Health | March 31, 2020/July 1, 2020 | Phase 2 |
| 32. | Pyridostigmine Bromide | NCT04343963 | Recruiting patients | Instituto Nacional de Ciencias Medicas y Nutricion Salvador Zubiran | April 4, 2020/April 30, 2021 | Phase 2 |
| 33. | Danoprevir/Ritonavir | NCT04345276 | Recruiting patients | Huoshenshan Hospital Ascletis Pharmaceuticals Co., Ltd. |
March 18, 2020/May 31, 2020 | Phase 4 |
| 34. | Atovaquone/Azithromycin | NCT04339426 | Recruiting patients | HonorHealth Research Institute | April 20, 2020/April 2021 | Phase 2 |
| 35. | Fluvoxamine | NCT04342663 | Recruiting patients | Washington University School of Medicine | April 10, 2020/July 1, 2020 | Phase 2 |
| 36. | Eculizumab | NCT04346797 | Recruiting patients | Assistance Publique - Hôpitaux de Paris | April 16, 2020/December 31, 2020 | Phase 2 |
| 37. | Canakinumab | NCT04365153 | Recruiting patients | The Cleveland Clinic | April 24, 2020/December 31, 2020 | Phase 2 |
| 38. | Valsartan | NCT04335786 | Recruiting patients | Radboud University | April 2020/August 2021 | Phase 4 |
| 39. | Anakinra Drug: trimethoprim/sulfamethoxazole |
NCT04357366 | Recruiting patients | Hellenic Institute for the Study of Sepsis | April 15, 2020/April 15, 2022 | Phase 2 |
| 40. | Camostat Mesilate | NCT04321096 | Recruiting patients | University of Aarhus | March 31, 2020/May 1, 2021 | Phase 1 |
| 41. | Sevoflurane Propofol |
NCT04359862 | Recruiting patients | Fundación para la Investigación del Hospital Clínico de Valencia | April 16, 2020/September 16, 2020 | Phase 4 |
| 42. | Bevacizumab | NCT04275414 | Recruiting patients | Qilu Hospital of Shandong University Renmin Hospital of Wuhan University Moriggia-Pelascini Gravedona Hospital |
February 15, 2020/May 2020 | Phase 2 Phase 3 |
| 43. | Leronlimab | Recruiting patients | CytoDyn, Inc. | April 1, 2020/April 4, 2021 | Phase 2 | |
| 44. | Selinexor | NCT04349098 | Recruiting patients | Karyopharm Therapeutics Inc | April 17, 2020/August 31, 2020 | Phase 2 |
| 45. | Abidol hydrochloride Oseltamivir Lopinavir/ritonavir |
NCT04255017 | Recruiting patients | Tongji Hospital | February 1, 2020/July 1, 2020 | Phase 4 |
| 46. | Methylprednisolone Sodium Succinate | NCT04343729 | Recruiting patients | Fundação de Medicina Tropical Dr. Heitor Vieira Dourado | April 18, 2020/September 2020 | Phase 2 |
| 47. | Colchicine | NCT04322682 | Recruiting patients | Montreal Heart Institute DACIMA Software |
March 23, 2020/September 2020 | Phase 3 |
| 48. | Cholecalciferol | NCT04344041 | Recruiting patients | University Hospital, Angers Mylan Laboratories |
April 15, 2020/July 2020 | Phase 3 |
| 49. | Honey Nigella Sativa/Black Cumin |
NCT04347382 | Recruiting patients | Sohaib Ashraf Sheikh Zayed Federal Postgraduate Medical Institute |
April 20, 2020/May 30, 2020 | Phase 3 |
| 50. | Vitamin C | NCT04264533 | Recruiting patients | ZhiYong Peng Zhongnan Hospital |
February 14, 2020/September 30, 2020 | Phase 2 |
Fig. 15.
Small molecules or drug molecules under various clinical trials against COVID-19.
Fig. 16.
Some synthetic and natural molecules under various clinical trials against COVID-19.
Fig. 14.
Various inhibitors in trials act at different stages of the life cycle of SARS-CoV-2.
Hydroxychloroquine, an antimalarial drug, is sought to play a key role in treated patients of COVID-19 [112]. This drug molecule is under multiple phase-III clinical trials (NCT04345692, NCT04342221, NCT04322123) against this disease by different health improvement agencies and companies (Table 2). In combination with antibiotic azithromycin, hydroxychloroquine is under phase-II (NCT04329832) and phase-III (NCT04322123) clinical trials. Hydroxychloroquine is found to show a significant reduction in the viral carriage at D6-post inclusion with lower carrying duration as compared to control and untreated patients. In combination with azithromycin, the efficiency was increased and the viral elimination rate was significantly higher [113]. Lopinavir and ritonavir, anti-HIV drugs, are two other molecules that are sought as future treatment of this COVID-19 disease. These drug molecules are also in alone or in combination with other molecules are under different clinical trials for determining their effectiveness in COVID-19 patients (NCT04330690, NCT04307693, NCT04315948). These studies will also determine if these drugs reduce the viral load from the respiratory system in COVID-19 patients or not. Similarly, in various studies effectiveness of Lopinavir/ritonavir is reported against COVID-19 [114], [115]. Remdesivir, drug for ebola virus, is the first drug which is observed in clinical trials to show effectiveness in COVID-19 patients and treated patients with 30% more rapidly as compared to placebo and treated the first patient in US successfully [116]. It has shown possibility in various in vitro studies against COVID-19 and MERS viral infections [117], [118], [119]. The efficiency of this drug against SARS-CoV and MERS-CoV also increased expectations from it against COVID-19 [118], [120]. During some in vitro studies, remdesivir is efficiently found to inhibit this novel coronavirus COVID-19 [121]. Further, it is under phase-III clinical trials against COVID-19 to determine its efficiency concerning clinical status assessed by a 7-point ordinal scale on Day 14 (NCT04315948, NCT04292899).
Favipiravir, an RNA polymerase inhibitor [122], is under phase-III clinical trial (NCT04336904) to determine its safety and performance in the treatment of COVID-19 patients by Giuliano Rizzardini, ASST Fatebenefratelli Sacco. Fujifilm Pharmaceuticals U.S.A., Inc. is conducting another phase-II trial of Favipiravir in hospitalized patients of COVID-19 for 14 days treatment (NCT04358549). In one study, against lopinavir, ritonavir, and control groups, Favipiravir receiving patients showed shorter viral clearance time along with improvement in chest imaging [123]. Ruxolitinib, a Janus kinase inhibitor approved for the treatment of myelofibrosis, polycythemia vera, and graft-versus-host disease [124], is under different phase-I, II, and III trials (NCT04348071, NCT04334044) by various agencies to learn its effect on the progression of COVID-19, inflammation caused by this virus and its severity. Losartan, an orally available angiotensin-receptor antagonist [125], is under phase-I clinical trials (NCT04335123) for evaluation of its safety in the failure of respiratory system of COVID-19 patients. Dapagliflozin, an antidiabetic drug [126], is under phase-III studies (NCT04350593) by Saint Luke's Health System in US and high prevalence countries to determine its role in reducing the progression of COVID-19, its complication and mortality due to this disease. Tocilizumab, a recombinant human monoclonal antibody [127], is gone to phase-III study on 310 patients (NCT04317092) for determining its clinical benefits in COVID-19 patients with risk of the cytokine storm. Although in initial studies, tocilizumab treatment was found associated with an increase in CRP (C-reactive proteins) in few patients, it was also found to clinically stabilize the patients with risk of complications of cytokine storm [128]. Zhang et al. reported the successful treatment of COVID-19 patients with multiple myeloma with tocilizumab treatment [129]. Further, tocilizumab is under various phase-II and III studies by various health agencies to determine its efficacy in the early treatment of COVID-19 patients with evaluation of its safety parameters and role in the prevention of inflammation (NCT04331795, NCT04346355, NCT04320615). Sirolimus, an immunosuppressive agent [130], under phase-II, double-blind, placebo controlled trial to determine its benefits in hospitalized patients with COVID-19 pneumonia (NCT04341675). Clazakizumab is another human monoclonal antibody under phase-II clinical trial to study its efficiency against COVID-19 (NCT04343989). Eculizumab (NCT04346797) and canakinumab (NCT04365153) are other monoclonal antibodies under phase-II studies separately for determining their efficiency against COVID-19. Camostat mesylate, a serine protease inhibitor, is under phase-I and II (NCT04321096) studies to determine efficacy against COVID-19 infection in patients.
Some food supplements like Vitamin C and D (Cholecalciferol) are under different clinical investigations as interventions against COVID-19. Various studies evidenced that a high dose of vitamin C and supplementation of vitamin D reduces the chance of viral infection and COVID-19 [131], [132]. Thus, these supplements are also sought to play a key role in the intervention of COVID-19 and need to be evaluated. To determine the short-term effect of colchicine in COVID-19 patients, it is under phase-III trial by Montreal Heart Institute (NCT04322682).
6. Perspective and conclusion
SARS-CoV-2 (COVID-19) has emerged as a more contagious virus compared to earlier found CoVs and was declared as pandemic by World Health Organization. It is challenging the health system of the world. The information about its structural similarity with SARS-CoV has unlocked pathways for the rapid development of new molecules for targeting this virus. The exploration of the X-ray crystal structure of SARS-CoV-2 spike (S) protein, RNA dependent RNA polymerase, and 3CL or Mpro Protease has provided critical information for developing small molecule inhibitors for the SARS-CoV-2. The sequencing of SARS-CoV-2 and SARS-CoV was found around 80% similar and key amino acids responsible for the interactions of these proteins have been reported. In case of spike proteins, the important residue like Phe486 is present in the active site of SARS-CoV-2 while absent in the case of SARS-CoV. The other residues such as Tyr442, Leu443, Asn479, Tyr484, Thr487 are present in SARS-CoV while Leu455, Phe456, Gln493, Gln498, Asn501 are present in SARS-CoV-2 which contributes towards the binding affinities of the S1 subunits towards ACE2. Similarly, in 3CL or Mpro target, the conformation of the residues in the active site was found different and the binding site entry residues such as Ala46 in SARS-CoV while Ser46 in SARS-CoV-2 were found to affect the entry as well as binding of ligands to the Mpro active site. Further, in case of RdRp (RNA dependent RNA polymerase), the only conformational difference of the residues in the active site. These findings will help medicinal chemists to understand the binding mechanism and key interactions of these proteins responsible for the virus cycle. Further, it will help the medicinal chemists to design new small molecules to target these enzymes of COVID-19 and help in its treatment. Authors also believe that this perspective will help the chemists and biologists to understand the key proteins to target for finding the cure of this epidemic.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors are thankful to Chairman and Director of ISF College of Pharmacy, Moga, Punjab for supporting this work. Sourav is thankful to ICMR for RA fellowship. Author’s are also thankful to Dr. Vinod Kumar, Associate Professor, Central University of Punjab for his valuable inputs and guidance and Ms. Karamjeet Kaur, Assistant Professor, ISF College of Pharmacy for help in the manuscript.
References
- 1.Cascella M., Rajnik M., Cuomo A., Dulebohn S.C., Di Napoli R. StatPearls Publishing; 2020. Features, Evaluation and Treatment Coronavirus (COVID-19) [PubMed] [Google Scholar]
- 2.Chan J.F., Lau S.K., To K.K., Cheng V.C., Woo P.C., Yuen K.-Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microb. Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee P.-I., Hsueh P.-R. Emerging threats from zoonotic coronaviruses-from SARS and MERS to 2019-nCoV. J. Microbiol. Immunol. Infect. 2020;53:365–367. doi: 10.1016/j.jmii.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microb. 2013;11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hui D.S., Zumla A. Severe acute respiratory syndrome: historical, epidemiologic, and clinical features. Infect. Dis. Clin. 2019;33:869–889. doi: 10.1016/j.idc.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheahan T., Rockx B., Donaldson E., Sims A., Pickles R., Corti D., Baric R. Mechanisms of zoonotic severe acute respiratory syndrome coronavirus host range expansion in human airway epithelium. J. Virol. 2008;82:2274–2285. doi: 10.1128/JVI.02041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. The Lancet. 2019;395(2020):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. The Lancet. 2020;395:689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mokhtari R.B., Homayouni T.S., Baluch N., Morgatskaya E., Kumar S., Das B., Yeger H. Combination therapy in combating cancer. Oncotarget. 2017;8:38022. doi: 10.18632/oncotarget.16723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du Toit A. Outbreak of a novel coronavirus. Nat. Rev. Microb. 2020;18 doi: 10.1038/s41579-020-0332-0. 123-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bassetti M., Vena A., Giacobbe D.R. The novel Chinese coronavirus (2019-nCoV) infections: challenges for fighting the storm. Eur. J. Clin. Invest. 2020;50:e13209. doi: 10.1111/eci.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. New Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson R. Pandemic potential of 2019-nCoV. Lancet. Infect. Dis. 2020;20:280. doi: 10.1016/S1473-3099(20)30068-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai Y., Yao L., Wei T., Tian F., Jin D.-Y., Chen L., Wang M. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020;323:1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan W.-J., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X., Liu L., Shan H., Lei C.-L., Hui D.S. Clinical characteristics of coronavirus disease 2019 in China. New Eng. J. Med. 2019;382(2020):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., Fan Y., Zheng C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet. Infect. Dis. 2020;20:425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu X., Fang B., Liu Y., Cai M., Jun J., Ma J., Bu D., Wang L., Zhou P., Wang H. Newly emerged porcine enteric alphacoronavirus in southern China: identification, origin and evolutionary history analysis. Infect. Genet. Evol. 2018;62:179–187. doi: 10.1016/j.meegid.2018.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drexler J.F., Corman V.M., Drosten C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Res. 2014;101:45–56. doi: 10.1016/j.antiviral.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Osail A.M., Al-Wazzah M.J. The history and epidemiology of Middle East respiratory syndrome corona virus. Multidiscip. Respir. Med. 2017;12:20. doi: 10.1186/s40248-017-0101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu B., Ge X., Wang L.-F., Shi Z. Bat origin of human coronaviruses. Virol. J. 2015;12:221. doi: 10.1186/s12985-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ge X.-Y., Li J.-L., Yang X.-L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menachery V.D., Yount B.L., Sims A.C., Debbink K., Agnihothram S.S., Gralinski L.E., Graham R.L., Scobey T., Plante J.A., Royal S.R. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. 2016;113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X.-L., Hu B., Wang B., Wang M.-N., Zhang Q., Zhang W., Wu L.-J., Ge X.-Y., Zhang Y.-Z., Daszak P. Isolation and characterization of a novel bat coronavirus closely related to the direct progenitor of severe acute respiratory syndrome coronavirus. J. Virol. 2016;90:3253–3256. doi: 10.1128/JVI.02582-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng L.-P., Gao Y.-T., Ge X.-Y., Zhang Q., Peng C., Yang X.-L., Tan B., Chen J., Chmura A.A., Daszak P. Bat severe acute respiratory syndrome-like coronavirus WIV1 encodes an extra accessory protein, ORFX, involved in modulation of the host immune response. J. Virol. 2016;90:6573–6582. doi: 10.1128/JVI.03079-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sabir J.S., Lam T.T.-Y., Ahmed M.M., Li L., Shen Y., Abo-Aba S.E., Qureshi M.I., Abu-Zeid M., Zhang Y., Khiyami M.A. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science. 2016;351:81–84. doi: 10.1126/science.aac8608. [DOI] [PubMed] [Google Scholar]
- 26.Anthony S.J., Gilardi K., Menachery V., Goldstein T., Ssebide B., Mbabazi R., Navarrete-Macias I., Liang E., Wells H., Hicks A. Further evidence for bats as the evolutionary source of Middle East respiratory syndrome coronavirus. MBio. 2017;8:e00373–00317. doi: 10.1128/mBio.00373-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reusken C.B., Raj V.S., Koopmans M.P., Haagmans B.L. Cross host transmission in the emergence of MERS coronavirus. Curr. Opin. Virol. 2016;16:55–62. doi: 10.1016/j.coviro.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Organization W.H. Middle East respiratory syndrome coronavirus (MERS-CoV)—update. Disease Outbreak News. 2014;14 [Google Scholar]
- 29.Corman V.M., Baldwin H.J., Tateno A.F., Zerbinati R.M., Annan A., Owusu M., Nkrumah E.E., Maganga G.D., Oppong S., Adu-Sarkodie Y. Evidence for an ancestral association of human coronavirus 229E with bats. J. Virol. 2015;89:11858–11870. doi: 10.1128/JVI.01755-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corman V.M., Eckerle I., Memish Z.A., Liljander A.M., Dijkman R., Jonsdottir H., Ngeiywa K.J.J., Kamau E., Younan M., Al Masri M. Link of a ubiquitous human coronavirus to dromedary camels. Proc. Natl. Acad. Sci. 2016;113:9864–9869. doi: 10.1073/pnas.1604472113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tao Y., Shi M., Chommanard C., Queen K., Zhang J., Markotter W., Kuzmin I.V., Holmes E.C., Tong S. Surveillance of bat coronaviruses in Kenya identifies relatives of human coronaviruses NL63 and 229E and their recombination history. J. Virol. 2017;91:e01953–01916. doi: 10.1128/JVI.01953-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anthony S.J., Johnson C.K., Greig D.J., Kramer S., Che X., Wells H., Hicks A.L., Joly D.O., Wolfe N.D., Daszak P. Global patterns in coronavirus diversity. Virus Evol. 2017;3:vex012. doi: 10.1093/ve/vex012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen H., Guan Y., Vijaykrishna D., Smith G., Zhang J., Peiris J. Evolutionary Insights into the Ecology of. J. Virol. 2007;81:4012. doi: 10.1128/JVI.02605-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui J., Han N., Streicker D., Li G., Tang X., Shi Z., Hu Z., Zhao G., Fontanet A., Guan Y. Evolutionary relationships between bat coronaviruses and their hosts. Emerg. Infect. Dis. 2007;13:1526. doi: 10.3201/eid1310.070448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corman V.M., Rasche A., Diallo T.D., Cottontail V.M., Stöcker A., Souza B.F.D.C.D., Corrêa J.I., Carneiro A.J.B., Franke C.R., Nagy M. Highly diversified coronaviruses in neotropical bats. J. Gen. Virol. 2013;94:1984–1994. doi: 10.1099/vir.0.054841-0. [DOI] [PubMed] [Google Scholar]
- 37.Smith C., De Jong C., Meers J., Henning J., Wang L.-F., Field H. Coronavirus infection and diversity in bats in the Australasian region. EcoHealth. 2016;13:72–82. doi: 10.1007/s10393-016-1116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang X., Zhang J., Zhang S., Wang P., Fan X., Li L., Li G., Dong B., Liu W., Cheung C. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 2006;80:7481–7490. doi: 10.1128/JVI.00697-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin X.-D., Wang W., Hao Z.-Y., Wang Z.-X., Guo W.-P., Guan X.-Q., Wang M.-R., Wang H.-W., Zhou R.-H., Li M.-H. Extensive diversity of coronaviruses in bats from China. Virology. 2017;507:1–10. doi: 10.1016/j.virol.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yin W., Mao C., Luan X., Shen D.-D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science Accepted. 2020 doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature Accepted. 2020 doi: 10.1038/s41586-41020-42223-y. [DOI] [PubMed] [Google Scholar]
- 43.Yuan M., Wu N.C., Zhu X., Lee C.-C.D., So R.T., Lv H., Mok C.K., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H., Liu S.M., Yu X.H., Tang S.L., Tang C.K. Coronavirus disease 2019 (COVID-19): current status and future perspectives. Int. J. Antimicrob. Agents. 2020;55:105951. doi: 10.1016/j.ijantimicag.2020.105951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loeffelholz M.J., Tang Y.-W. Laboratory diagnosis of emerging human coronavirus infections—the state of the art. Emerg. Microbes. Infect. 2020;9:747–756. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Su S., Wong G., Shi W., Liu J., Lai A.C., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.G. Eysenbach, Infodemiology: tracking flu-related searches on the web for syndromic surveillance, in: AMIA Annual Symposium Proceedings, American Medical Informatics Association, 2006, pp. 244. [PMC free article] [PubMed]
- 49.Zhao Z., Zhang F., Xu M., Huang K., Zhong W., Cai W., Yin Z., Huang S., Deng Z., Wei M. Description and clinical treatment of an early outbreak of severe acute respiratory syndrome (SARS) in Guangzhou, PR China. J. Med. Microbiol. 2003;52:715–720. doi: 10.1099/jmm.0.05320-0. [DOI] [PubMed] [Google Scholar]
- 50.W.H. Organization, Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003, 2003. http://www.who.int/csr/sars/country/table2004_04_21/en/index. html.
- 51.Peiris J., Lai S., Poon L., Guan Y., Yam L., Lim W., Nicholls J., Yee W., Yan W., Cheung M. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsang T., Lai-Yin T., Pak-Yin L., Lee M. Update: outbreak of severe acute respiratory syndrome-worldwide. MMWR Morb. Mortal. Wkly Rep. 2003;52(2003) 241-241. [PubMed] [Google Scholar]
- 53.W. Update, 31-Coronavirus never before seen in humans is the cause of SARS, Unprecedented Collaboration Identifies New Pathogen in Record Time 16, 2003. https://www.who.int/csr/sars/archive/2003_2004_2016/en/.
- 54.Simmons G., Zmora P., Gierer S., Heurich A., Pöhlmann S. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 2013;100:605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simmons G., Reeves J.D., Rennekamp A.J., Amberg S.M., Piefer A.J., Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. 2004;101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathogen. 2018;14:e1007236. doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 59.Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 62.Adedeji A.O., Severson W., Jonsson C., Singh K., Weiss S.R., Sarafianos S.G. Novel inhibitors of severe acute respiratory syndrome coronavirus entry that act by three distinct mechanisms. J. Virol. 2013;87:8017–8028. doi: 10.1128/JVI.00998-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prabakaran P., Gan J., Feng Y., Zhu Z., Choudhry V., Xiao X., Ji X., Dimitrov D.S. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J. Biol. Chem. 2006;281:15829–15836. doi: 10.1074/jbc.M600697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anand K., Palm G.J., Mesters J.R., Siddell S.G., Ziebuhr J., Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra α-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xue X., Yu H., Yang H., Xue F., Wu Z., Shen W., Li J., Zhou Z., Ding Y., Zhao Q. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J. Virol. 2008;82:2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang F., Chen C., Tan W., Yang K., Yang H. Structure of main protease from human coronavirus NL63: insights for wide spectrum anti-coronavirus drug design. Sci. Rep. 2016;6:22677. doi: 10.1038/srep22677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z., Pei D., Ziebuhr J. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3:e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.J. Ziebuhr, The coronavirus replicase, in: Coronavirus Replication and Reverse Genetics, Springer, 2005, pp. 57–94.
- 70.Ahn D.-G., Choi J.-K., Taylor D.R., Oh J.-W. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch. Virol. 2012;157:2095–2104. doi: 10.1007/s00705-012-1404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Te Velthuis A.J., Arnold J.J., Cameron C.E., van den Worm S.H., Snijder E.J. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acid Res. 2010;38:203–214. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Comm. 2019;10:1–9. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhai Y., Sun F., Li X., Pang H., Xu X., Bartlam M., Rao Z. Insights into SARS-CoV transcription and replication from the structure of the nsp7–nsp8 hexadecamer. Nat. Struct. Mol. Biol. 2005;12:980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peti W., Johnson M.A., Herrmann T., Neuman B.W., Buchmeier M.J., Nelson M., Joseph J., Page R., Stevens R.C., Kuhn P. Structural genomics of the severe acute respiratory syndrome coronavirus: nuclear magnetic resonance structure of the protein nsP7. J. Virol. 2005;79:12905–12913. doi: 10.1128/JVI.79.20.12905-12913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Johnson M.A., Jaudzems K., Wüthrich K. NMR structure of the SARS-CoV nonstructural protein 7 in solution at pH 6.5. J. Mol. Biol. 2010;402:619–628. doi: 10.1016/j.jmb.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lehmann K.C., Gulyaeva A., Zevenhoven-Dobbe J.C., Janssen G.M., Ruben M., Overkleeft H.S., van Veelen P.A., Samborskiy D.V., Kravchenko A.A., Leontovich A.M. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acid Res. 2015;43:8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McDonald S.M. RNA synthetic mechanisms employed by diverse families of RNA viruses. Wiley Interdiscip Rev. RNA. 2013;4:351–367. doi: 10.1002/wrna.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kalra S., Joshi G., Munshi A., Kumar R. Structural insights of cyclin dependent kinases: implications in design of selective inhibitors. Eur. J. Med. Chem. 2017;142:424–458. doi: 10.1016/j.ejmech.2017.08.071. [DOI] [PubMed] [Google Scholar]
- 81.Chauhan G., Madou M.J., Kalra S., Chopra V., Ghosh D., Martinez-Chapa S.O. Nanotechnology for COVID-19: therapeutics and vaccine research. ACS Nano. 2020;14:7760–7782. doi: 10.1021/acsnano.0c04006. [DOI] [PubMed] [Google Scholar]
- 82.He J., Tao H., Yan Y., Huang S.-Y., Xiao Y. Molecular mechanism of evolution and human infection with sars-cov-2. Virus. 2020;12:428. doi: 10.3390/v12040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mishra S.S., Ranjan S., Sharma C.S., Singh H.P., Kalra S., Kumar N. Computational investigation of potential inhibitors of novel coronavirus 2019 through structure-based virtual screening, molecular dynamics and density functional theory studies. J. Biomol. Struct. Dyn. 2019;2020:1–13. doi: 10.1080/07391102.2020.1791957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rahman N., Basharat Z., Yousuf M., Castaldo G., Rastrelli L., Khan H. Virtual screening of natural products against Type II transmembrane serine protease (TMPRSS2), the priming agent of coronavirus 2 (SARS-CoV-2) Molecule. 2020;25:2271. doi: 10.3390/molecules25102271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., Winter H., Meister M., Veith C., Boots A.W. SARS-CoV-2 receptor ACE 2 and TMPRSS 2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39:e105114. doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J. Virol. 2019;93:e01815–18. doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang X., Dhindsa R., Povysil G., Zoghbi A., Motelow J., Hostyk J., Goldstein D. Transcriptional inhibition of host viral entry proteins as a therapeutic strategy for SARS-CoV-2. Preprints. 2020:2020030360. [Google Scholar]
- 90.Zhou Y., Vedantham P., Lu K., Agudelo J., Carrion R., Jr, Nunneley J.W., Barnard D., Pöhlmann S., McKerrow J.H., Renslo A.R. Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res. 2015;116:76–84. doi: 10.1016/j.antiviral.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yuan L., Zou C., Ge W., Liu Y., Hu B., Wang J., Lin B., Li Y., Ma E. A novel cathepsin L inhibitor prevents the progression of idiopathic pulmonary fibrosis. Bioorg. Chem. 2020;94:103417. doi: 10.1016/j.bioorg.2019.103417. [DOI] [PubMed] [Google Scholar]
- 92.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1–12. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xi J., Xu K., Jiang P., Lian J., Hao S., Jia H., Yao H., Zhang Y., Zheng R., Chen D. Virus strain of a mild COVID-19 patient in Hangzhou representing a new trend in SARS-CoV-2 evolution related to Furin cleavage site. medRxiv. 2020 doi: 10.1101/2020.03.10.20033944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bekerman E., Neveu G., Shulla A., Brannan J., Pu S.-Y., Wang S., Xiao F., Barouch-Bentov R., Bakken R.R., Mateo R. Anticancer kinase inhibitors impair intracellular viral trafficking and exert broad-spectrum antiviral effects. J. Clin. Investig. 2017;127:1338–1352. doi: 10.1172/JCI89857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang C., Wang J., Shuai L., Ma X., Zhang H., Liu R., Chen W., Wang X., Ge J., Wen Z. The serine/threonine kinase AP2-associated kinase 1 plays an important role in rabies virus entry. Viruses. 2020;12:45. doi: 10.3390/v12010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., Stebbing J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;395:e30. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stebbing J., Phelan A., Griffin I., Tucker C., Oechsle O., Smith D., Richardson P. COVID-19: combining antiviral and anti-inflammatory treatments. Lancet. Infect. Dis. 2020;20:400–402. doi: 10.1016/S1473-3099(20)30132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Praveen D., Puvvada R.C., Aanandhi V. Janus kinase inhibitor baricitinib is not an ideal option for management of COVID-19. Int. J. Antimicrob. 2020;55:105967. doi: 10.1016/j.ijantimicag.2020.105967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.He K., Marsland R., III, Upadhyayula S., Song E., Dang S., Capraro B.R., Wang W., Skillern W., Gaudin R., Ma M. Dynamics of phosphoinositide conversion in clathrin-mediated endocytic traffic. Nature. 2017;552:410–414. doi: 10.1038/nature25146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.de Lartigue J., Polson H., Feldman M., Shokat K., Tooze S.A., Urbé S., Clague M.J. PIKfyve regulation of endosome-linked pathways. Traffic. 2009;10:883–893. doi: 10.1111/j.1600-0854.2009.00915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ikonomov O.C., Sbrissa D., Shisheva A. YM201636, an inhibitor of retroviral budding and PIKfyve-catalyzed PtdIns (3, 5) P2 synthesis, halts glucose entry by insulin in adipocytes. Biochem. Biophys. Res. Commun. 2009;382:566–570. doi: 10.1016/j.bbrc.2009.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wada Y., Cardinale I., Khatcherian A., Chu J., Kantor A.B., Gottlieb A.B., Tatsuta N., Jacobson E., Barsoum J., Krueger J.G. Apilimod inhibits the production of IL-12 and IL-23 and reduces dendritic cell infiltration in psoriasis. PLoS ONE. 2012;7:e35069. doi: 10.1371/journal.pone.0035069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Guo Y.-R., Cao Q.-D., Hong Z.-S., Tan Y.-Y., Chen S.-D., Jin H.-J., Tan K.-S., Wang D.-Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Mil. Med. Res. 2019;7(2020):1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rabby M.I.I. Current drugs with potential for treatment of COVID-19: a literature review. J. Pharm. Pharm. Sci. 2020;23:58–64. doi: 10.18433/jpps31002. [DOI] [PubMed] [Google Scholar]
- 107.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2019;19(2020):149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 108.Dong L., Hu S., Gao J. Discovering drugs to treat coronavirus disease 2019 COVID-19. Drug Discov. Ther. 2019;14(2020):58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- 109.Lythgoe M.P., Middleton P. Ongoing clinical trials for the management of the COVID-19 pandemic. Trends Pharmacol. Sci. Accepted. 2020 doi: 10.1016/j.tips.2020.1003.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Qiu R., Wei X., Zhao M., Zhong C., Zhao C., Hu J., Li M., Huang Y., Han S., He T. Outcome reporting from protocols of clinical trials of Coronavirus Disease 2019 (COVID-19): a review. MedRxiv Accepted. 2020 doi: 10.1101/2020.1103.1104.20031401. [DOI] [Google Scholar]
- 111.Zhu R.-f., Gao R.-l., Robert S.-H., Gao J.-p., Yang S.-g., Zhu C. Systematic review of the registered clinical trials of coronavirus diseases 2019 (COVID-19) MedRxiv Accepted. 2020 doi: 10.1101/2020.1103.1101.20029611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:1–4. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gautret P., Lagier J.-C., Parola P., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Dupont H.T. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agent Accepted. 2020 doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 114.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. New Eng. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lim J., Jeon S., Shin H.-Y., Kim M.J., Seong Y.M., Lee W.J., Choe K.-W., Kang Y.M., Lee B., Park S.-J. Case of the index patient who caused tertiary transmission of COVID-19 infection in Korea: the application of lopinavir/ritonavir for the treatment of COVID-19 infected pneumonia monitored by quantitative RT-PCR. J. Korean Med. Sci. 2020;35:e79. doi: 10.3346/jkms.2020.35.e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A. First case of 2019 novel coronavirus in the United States. New Eng. J. Med. 2019;382(2020):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Feldt T., Green G., Green M.L., Lescure F.-X. Compassionate use of remdesivir for patients with severe Covid-19. New Eng. J. Med. Accepted. 2020 doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T., Scott D., Cihlar T., Feldmann H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. 2020;117:6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Al-Tawfiq J.A., Al-Homoud A.H., Memish Z.A. Remdesivir as a possible therapeutic option for the COVID-19. Travel Med. Infect. Dis. 2020;34:101615. doi: 10.1016/j.tmaid.2020.101615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Furuta Y., Gowen B.B., Takahashi K., Shiraki K., Smee D.F., Barnard D.L. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antiviral Res. 2013;100:446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cai Q., Yang M., Liu D., Chen J., Shu D., Xia J., Liao X., Gu Y., Cai Q., Yang Y. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering Accepted. 2020 doi: 10.1016/j.eng.2020.1003.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mesa R.A., Yasothan U., Kirkpatrick P. Ruxolitinib. Nat. Rev. Drug Discov. 2012;11:103–104. doi: 10.1038/nrd3652. [DOI] [PubMed] [Google Scholar]
- 125.Sica D.A., Gehr T.W., Ghosh S. Clinical pharmacokinetics of losartan. Clin. Pharmacokinet. 2005;44:797–814. doi: 10.2165/00003088-200544080-00003. [DOI] [PubMed] [Google Scholar]
- 126.Plosker G.L. Dapagliflozin. Drugs. 2012;72:2289–2312. doi: 10.2165/11209910-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 127.Oldfield V., Dhillon S., Plosker G.L. Tocilizumab. Drugs. 2009;69:609–632. doi: 10.2165/00003495-200969050-00007. [DOI] [PubMed] [Google Scholar]
- 128.Luo P., Liu Y., Qiu L., Liu X., Liu D., Li J. Tocilizumab treatment in COVID-19: a single center experience. J. Med. Virol. Accepted. 2020 doi: 10.1002/jmv.25801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang X., Song K., Tong F., Fei M., Guo H., Lu Z., Wang J., Zheng C. First case of COVID-19 in a patient with multiple myeloma successfully treated with tocilizumab. Blood Adv. 2020;4:1307. doi: 10.1182/bloodadvances.2020001907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sehgal S. Transplantation Proceedings. Elsevier; 2003. Sirolimus: its discovery, biological properties, and mechanism of action; pp. S7–S14. [DOI] [PubMed] [Google Scholar]
- 131.Cheng R.Z. Can early and high intravenous dose of vitamin C prevent and treat coronavirus disease 2019 (COVID-19)? Med. Drug Discov. 2020;5:100028. doi: 10.1016/j.medidd.2020.100028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Grant W.B., Lahore H., McDonnell S.L., Baggerly C.A., French C.B., Aliano J.L., Bhattoa H.P. Evidence that vitamin D supplementation could reduce risk of influenza and COVID-19 infections and deaths. Nutrients. 2020;12:988. doi: 10.3390/nu12040988. [DOI] [PMC free article] [PubMed] [Google Scholar]