Abstract
Immunotherapeutic strategies are increasingly important in neuro-oncology, and the elucidation of escape mechanisms that lead to treatment resistance is crucial. We investigated the impact of immune pressure on the clonal dynamics and immune escape signature by comparing glioma growth in immunocompetent versus immunodeficient mice. Glioma-bearing WT and Pd-1–/– mice survived significantly longer than immunodeficient Pfp–/– Rag2–/– mice. While tumors in Pfp–/– Rag2–/– mice were highly polyclonal, immunoedited tumors in WT and Pd-1–/– mice displayed reduced clonality with emergence of immune escape clones. Tumor cells in WT mice were distinguished by an IFN-γ–mediated response signature with upregulation of genes involved in immunosuppression. Tumor-infiltrating stromal cells, which include macrophages/microglia, contributed even more strongly to the immunosuppressive signature than the actual tumor cells. The identified murine immune escape signature was reflected in human patients and correlated with poor survival. In conclusion, immune pressure profoundly shapes the clonal composition and gene regulation in malignant gliomas.
Keywords: Immunology, Oncology
Keywords: Brain cancer
Introduction
Glioblastoma represents the most frequent aggressive intrinsic brain tumor, and patient outcome remains uniformly fatal despite improved treatment options (1). The current era of glioblastoma therapy is being redefined by immunotherapeutic approaches, including immune checkpoint inhibition, oncolytic virus therapy, vaccination strategies, and adoptive T cell transfer (2). While the results of several recent clinical trials are encouraging, the effects of immunotherapy have certainly not yet reached their full potential, and the profound tumor-induced immunosuppression and evasion of immune recognition by the tumor cells remain a major challenge for upcoming immunotherapeutic trials (3, 4).
The biology of cancer immune escape, which hinders the execution of efficient immune-mediated cancer elimination, is still incompletely understood. In healthy individuals, cancer cells are recognized and destroyed by the immune system. However, if eradication is unsuccessful, tumor cells can enter an equilibrium phase in which they progressively develop and refine mechanisms to evade immune recognition and elimination, a process conceptualized in the cancer immunoediting hypothesis (5). Initial studies in carcinogen-induced sarcoma models showed that the adaptive immune system and, in particular, interferon-producing lymphocytes are responsible for preventing tumor outgrowth and maintenance of the equilibrium state (6, 7). Tumors that manage to eventually escape immune control have been selected for phenotypic tumor cell variants invisible to the adaptive tumor immune response and fit to resist immune attack (6–8). While studies in other cancer types, such as sarcoma or lymphoma, pioneered our understanding of such processes, intracranial tumors are remarkably different. They arise in a partially immunoprivileged microenvironment, display a low mutational burden, and induce an exceptionally severe T cell dysfunction with sequestration of T cells in the bone marrow, which does not occur in peripheral tumors (9). It is therefore of particular importance to decipher the mechanisms by which cancer immunoediting shapes tumor biology in malignant gliomas.
In the present study, we leveraged orthotopic syngeneic immunocompetent and immunodeficient models to gain deeper insight into glioma immune escape mechanisms. We demonstrate that cancer immunoediting not only shapes the gene expression signature during glioma evolution, but the selective pressure of the immune system also has a fundamental impact on the clonal composition of tumors arising in the brain. Furthermore, we demonstrate that the identified immune escape signature is relevant in human glioma patients. A better understanding of the mechanisms of how immune escape clones are selected for can support the design of immunotherapeutic strategies that counteract these processes, in order to mount a more effective immune response against immunoresistant clones.
Results
The immune system impacts glioma survival and alters tumor morphology.
To investigate the impact of the immune system on the growth behavior of intracranial gliomas, we compared the survival and histomorphological characteristics of 2 syngeneic glioma cell lines in C57BL/6 WT mice and immunodeficient C57BL/6 Pfp–/– Rag2–/– mice (Figure 1A). The Pfp–/– Rag2–/– knockout results in complete T and B cell deficiency as well as in a severely compromised NK cell function due to the lack of perforin expression. Intracerebral injection of GL261 and CT2A glioma cells in Pfp–/– Rag2–/– mice resulted in significantly shorter survival in comparison with immunocompetent WT mice (median survival: GL261, 18 vs. 22 days, P = 0.012; CT2A, 14 vs. 20 days, P = 0.003), indicating that the functional immune system impairs tumor growth in the brain (Figure 1, B and C).
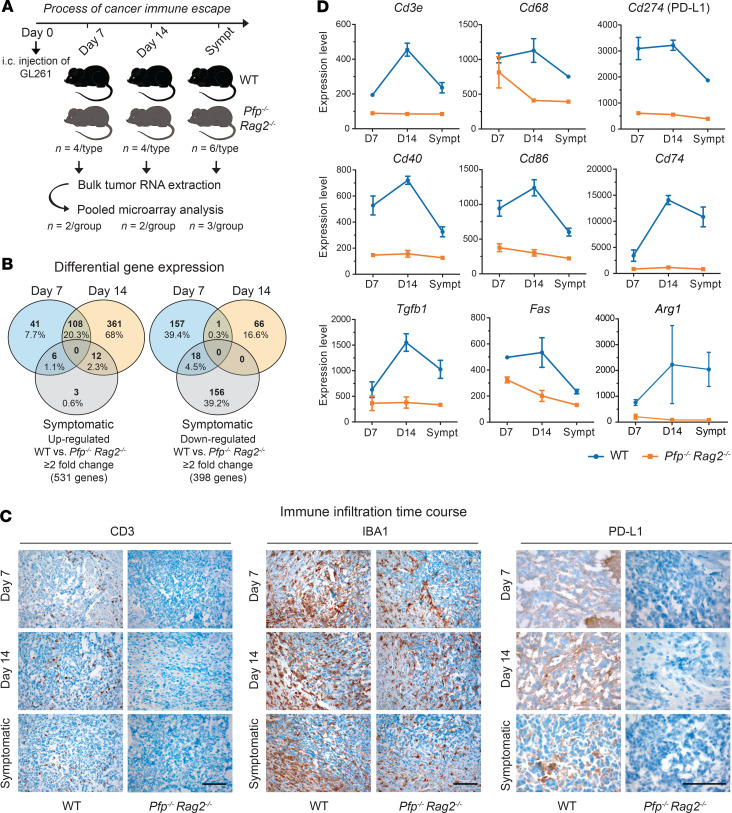
Figure 1. Glioma growth in immunocompetent versus immunodeficient mice.
(A) Schematic overview of the experimental setup. (B) Kaplan-Meier survival analysis of WT and Pfp–/– Rag2–/– mice injected intracerebrally with GL261 cells (n = 7 per group); log-rank test; *P = 0.0124. (C) Survival analysis of mice injected with CT2A cells; **P = 0.0029; n = 5 WT, 4 Pfp–/– Rag2–/–. (D) Macroscopic tumor appearance. (E) H&E staining of tumor sections. (F) Immunohistochemistry for CD3, CD8, IBA1, and CD34. (G) Quantification of macrophages/microglia (IBA1) and tumor microvessels (CD34). Bars represent means ± SEM; unpaired 2-tailed Student’s t test; for GL261, *P = 0.04, n = 4, and for CT2A, *P = 0.01, n = 3. Scale bars: 100 μm; hpf, high-power fields.
Gliomas in Pfp–/– Rag2–/– mice grew more invasive and typically also formed large extracerebral masses in addition to the main tumor mass located in the striatum, whereas extracerebral growth was uncommon in WT mice and instead tumors appeared to be more hemorrhagic on macroscopic inspection (Figure 1D). Histological analysis confirmed the increased invasiveness of gliomas in Pfp–/– Rag2–/– mice (Figure 1E). While GL261 cells tended to infiltrate the brain diffusely, CT2A cells preferentially exhibited a perivascular invasion pattern.
Immunohistochemistry confirmed the absence of infiltrating CD3+ T cells (Figure 1F) and B cells (not shown) in Pfp–/– Rag2–/– mice and showed that the majority of T cells in WT mice were CD8+. Infiltration with IBA1+ tumor-associated macrophages (TAMs), which can either be of peripheral origin or represent brain-intrinsic microglia, was strikingly increased in WT mice compared with Pfp–/– Rag2–/– mice (GL261: mean 11.88% vs. 5.98%, P = 0.039; CT2A: mean 35.42% vs. 23.43%, P = 0.010) (Figure 1, F and G). Intratumoral microvessel densities were not significantly different; however, blood vessels were more dilated in WT mice, possibly because of increased inflammatory signaling (Figure 1, F and G). Taken together, these findings suggest that the presence of a functional immune system shapes growth morphology during glioma evolution in murine brain.
Immunoediting of the gene expression signature during tumor development.
To determine how gene expression is regulated over time by the persistent challenge of an immunocompetent microenvironment, we performed microarray analyses of GL261 tumors at 3 different time points: days 7 and 14 after injection and the survival endpoint (Figure 2A). Tumors were excised from brains of WT and Pfp–/– Rag2–/– mice, and tumor RNA from 2 different mice was pooled for expression profiling. Unsupervised cluster analysis showed that tumor samples separated mainly according to mouse type rather than time point (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI138760DS1). The similarity between the expression profiles on day 14 and the endpoint indicates that by day 14 immune escape mechanisms were stably established in WT mice.
Figure 2. Gene regulation during tumor evolution.
(A) Workflow of GL261 tumor analysis. RNA from 2× 2 mice per mouse type (days 7 and 14) or 3× 2 mice per mouse type (symptomatic endpoint [Sympt]) was pooled for expression profiling by Illumina WG-6 v2.0 arrays. i.c., intracerebral. (B) Venn diagrams of genes over- and underexpressed in WT versus Pfp–/– Rag2–/– mice. (C) Immunohistochemistry for CD3, IBA1, and PD-L1. Scale bars: 100 μm. (D) Expression values of selected immune-related genes over time. Data are presented as mean ± SEM; day 7, n = 2; day 14, n = 2; symptomatic, n = 3.
In total, 531 genes were overexpressed and 398 genes were underexpressed at least 2-fold in WT versus Pfp–/– Rag2–/– mice (Figure 2B). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed that genes upregulated on day 7 in tumors in WT mice were most significantly enriched in the term “immune response” and were further strongly enriched in terms related to antigen processing and presentation as well as inflammation (Supplemental Table 1). On day 14, “immune response” continued to be the predominant pathway, but upregulated genes were now increasingly enriched in adaptive immune response pathways, including T and B cell activation. Overexpression of immune-related genes was decreased when mice became symptomatic, indicating that once tumors have successfully escaped immune control, high-level expression of immune regulators is no longer essential.
The infiltration of tumors with immune cells followed a time course similar to that of the regulation of immune-related genes. Infiltration with CD3+ T cells, including CD8+ T cells (Supplemental Figure 2) and macrophages/microglia, was maximal on day 14 and decreased when mice became symptomatic (Figure 2C). Concomitantly, expression of PD-L1 in tumors in WT mice was strongest on day 14, while PD-L1 was undetectable in Pfp–/– Rag2–/– mice at any time point. These dynamics were reflected at the transcriptional level, with Cd3e, Cd68, and Cd274 expression peaking on day 14 in WT mice (Figure 2D). T cell infiltration was further accompanied by increased expression of costimulatory molecules, such as Cd40 and Cd86, as well as Cd74, which is involved in antigen presentation. Moreover, prominent genes associated with immunosuppression beyond Cd274, such as Tgfb1, Fas, and Arg1, displayed similar expression kinetics (Figure 2D). To obtain a more comprehensive picture of immune gene regulation, we further assessed the expression of a set of key immunomodulators that were identified by Thorsson et al. through a meta-analysis of more than 10,000 tumors across 33 diverse cancer types, including gliomas (10), as well as the expression time course of cyto- and chemokines (Supplemental Figure 3). Heatmap representation of the immunomodulators contained in our data set highlighted overexpression of nearly all stimulatory and inhibitory modulators as well as antigen presentation molecules in WT mice, mostly with a peak of expression activity on day 14 (Figure 3). Collectively, these findings indicate that the immune pressure has a maximum effect on adaptive gene regulation around this time point, which may reflect a critical period in cancer immunoediting, after which the tumor cells have successfully overwhelmed the immune system.
Figure 3. Regulation of immune modulators.
Gene expression of stimulatory and inhibitory immune modulators identified by Thorsson et al. (10) in our data set. NA, not applicable.
RNA sequencing of isolated tumor cells identifies an immune escape gene expression signature.
While the microarray analysis had revealed that the immune system has a strong dynamic impact on gene regulation in developing tumors over time, this approach had not allowed us to determine what part of the expression signature was derived from the actual glioma cells versus from intermixed stromal cells, such as macrophages and microglia, which can account for 30%–50% of the tumor mass in malignant gliomas (11). To address this question, we next marked GL261 cells with green fluorescent protein (GFP) and injected them into WT and Pfp–/– Rag2–/– mice (Figure 4A). Mice were sacrificed when they became symptomatic, and GL261 GFP+ tumor cells as well as nontumor stromal cells were isolated by flow cytometric cell sorting. Separated cells as well as unsorted bulk tumor samples and in vitro cultured GL261 cells were analyzed by RNA sequencing (Figure 4A).
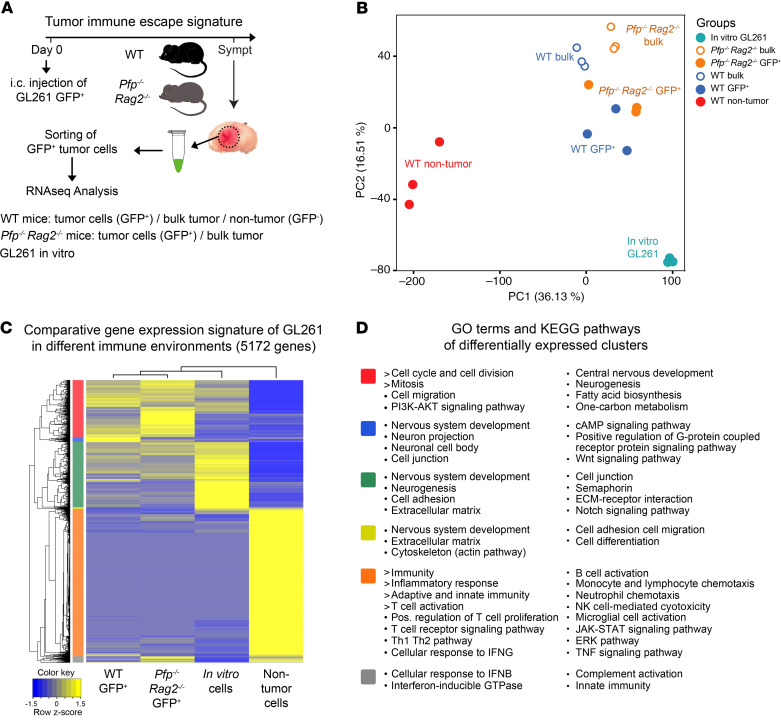
Figure 4. Expression signatures in tumor and nontumor cell populations.
(A) Schematic workflow of GFP-marked GL261 cell injection, flow cytometric sorting, and RNA sequencing analysis. Stromal cells from Pfp–/– Rag2–/– mice could not be analyzed because of very low RNA yield from the sparse nontumor population (see Figure 1E). (B) Principal component analysis of gene expression profiles of GFP+ tumor cells from WT and Pfp–/– Rag2–/– mice, nontumor stromal cells from WT mice, bulk tumors from both mouse types, and in vitro cultured cells. All samples were analyzed in triplicate. (C) Unsupervised cluster analysis of differentially expressed genes (2-fold cutoff, P < 0.05). (D) GO and KEGG pathway analysis of differentially expressed gene clusters (colors correspond to clusters in C).
Principal component analysis showed that tumor cells grown in vivo clustered apart from the in vitro cell line and that sorted GFP+ tumor cells separated from bulk tumors, confirming the successful isolation process (Figure 4B). Differences between sorted tumor cells from WT and Pfp–/– Rag2–/– mice were less pronounced, while the tumor cells clustered widely apart from nontumor stromal cells in WT mice, underlining the necessity for dissecting the microenvironmental background. Stromal cells from Pfp–/– Rag2–/– mice could not be analyzed since the RNA yield was too low, presumably because of the significantly reduced TAM infiltration in these mice (Figure 1, F and G).
In total, 5172 genes were differentially expressed between purified GFP+ tumor cells from WT mice, GFP+ tumor cells from Pfp–/– Rag2–/– mice, stromal cells from WT mice, or in vitro cultured cells, using a FDR of less than 0.05 and a log2 fold change greater than 2 (Figure 4C). Unsupervised clustering confirmed the relative similarity between tumor cells in WT and Pfp–/– Rag2–/– mice and their separation from nontumor stromal cells (Figure 4C). The majority of genes overexpressed in stromal cells were significantly enriched in pathways associated with immune and inflammatory responses, including adaptive and innate immunity, and, in particular, T cell activation (Figure 4D). In contrast, expression signatures of tumor cells in vivo were mainly related to cell cycling and division. To obtain a more detailed view on immune-activating as opposed to immunosuppressive responses, we compiled a list of immunosuppressive genes from the literature (4, 12–15) to determine their origin of expression. Interestingly, the vast majority of immunosuppressive cytokines, enzymes, checkpoint ligands, and cell surface molecules as well as signaling pathways were overexpressed in the stromal cells rather than in the tumor cells themselves (Figure 5), indicating that the immunosuppressive signature in gliomas mainly originates from tumor-associated stromal cells, the majority of which consist of macrophages and microglia.
Figure 5. Expression intensity of immunosuppressive genes in tumor cells versus nontumor cells.
Expression levels were determined by RNA-Seq as shown in Figure 4A.
In order to determine how the actual tumor cells adapt to the enduring immune challenge in WT mice and establish mechanisms of immune escape, we excluded the dominant signature of the stromal cells and restricted the analysis to the comparison of tumor cells only. Clustering based on 4148 differentially expressed genes identified 6 main clusters, of which 4 represented transcripts differentially up- or downregulated in the different types of mice (Figure 6A and Supplemental Table 2). Genes upregulated in WT mice were mainly related to an immune response signature, including IFN-γ, interleukin, and cytokine signaling pathways (Figure 6B, cluster 1). Querying the Interferome database (www.interferome.org), we found that 57% of the genes overexpressed in cluster 1 are induced by interferons. Network assembly of the regulatory interactions between differentially expressed genes in clusters 1–4 identified interferon regulatory factor 1 (Irf1) as a key transcriptional regulator of the interferon response signature (Figure 7). Upregulated genes linked with this factor include prominent candidates involved in immune escape, such as Cd274 (PD-L1), Ctla4, Socs1, Arg1, Tnfrsf14, and Il18bp, as well as in immune stimulation, including Cxcl9, Cxcl10, Ciita, Ccl6, Ccl8, Ccl24, and Tnfsf13b (Figure 7 and Supplemental Figure 4). A number of the genes upregulated in tumor cells have been previously associated primarily with TAMs, including Arg1, Cd274, Ccl24, Ciita, Cd74, and other MHC class II pathway–related genes, or with T cells (Ctla4, Il12rb1), suggesting that tumor cells can in part recapitulate the expression patterns of immune cells when exposed to immunoselective pressure. Collectively, these findings demonstrate that the selective pressure of the immune system shapes the gene expression signature of glioma cells mainly by activating interferon-regulated pathways, which results in upregulation of potent immunosuppressive genes.
Figure 6. Identification of an immunoedited tumor cell expression signature.
(A) Unsupervised hierarchical clustering of differentially expressed genes identifies 6 main clusters, of which 4 represent genes differentially up- or downregulated in tumors in WT mice versus Pfp–/– Rag2–/– mice. (B) GO and KEGG pathway analysis of genes upregulated in the 4 clusters.
Figure 7. Network analysis of the genes contained in clusters 1 to 4 (see Figure 6) that are annotated in the STRING database.
Comparisons with human gene expression data highlight the relevance of the immune escape signature.
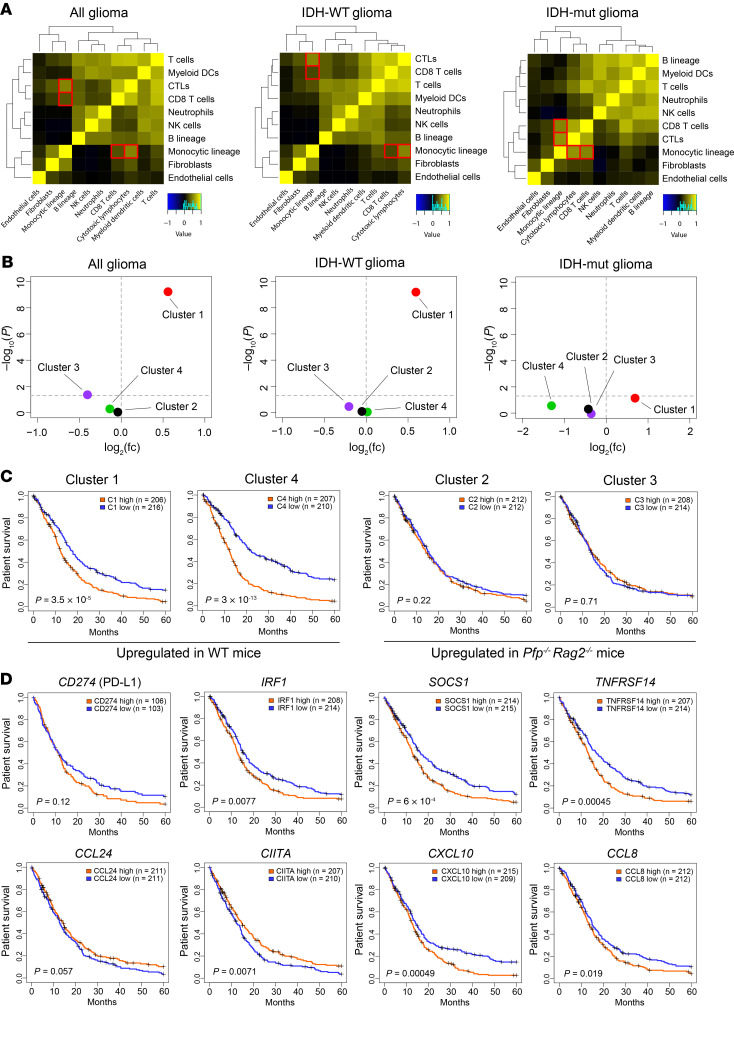
After identifying an immunoedited gene expression signature in mice, we next assessed its translational relevance in human gliomas. To this end, we analyzed a data set of 1135 high-grade gliomas that was compiled by Bockmayr et al. (16) through a meta-analysis of previously published glioma microarray data sets (17–25). First, we evaluated our finding of a correlative infiltration of tumor tissue with T cells and microglia/macrophages (Figure 2C). In human malignant gliomas the “cytotoxic lymphocyte” and “CD8+ T cell” signatures correlated with the infiltration of “monocytic lineage” cells (both P < 0.001), and these correlations were particularly pronounced in IDH-mutated tumors (Figure 8A), supporting our observation that TAM infiltration increases in tumors under immune attack.
Figure 8. Comparison with human gene expression data.
(A) Correlations between different tumor-infiltrating immune and stromal cell subsets in human malignant gliomas. Cytotoxic lymphocytes and CD8+ T cells correlate with monocytic lineage in all gliomas (R = 0.52 and 0.31, respectively, P < 0.001), and in particular in IDH-mutated (IDH-mut) gliomas (R = 0.52 and 0.57, respectively, P < 0.001). (B) Overlap between genes overexpressed in the 20% of human gliomas with the highest T cell gene expression signature (fold change and P value, 2-sided Fisher test) and the 4 murine gene clusters identified in Figure 4A. (C) Patient survival according to genes over- or underexpressed in the 4 murine gene clusters (top vs. bottom 20% tumors). (D) Kaplan-Meier analysis for individual genes, comparing the 20% of tumors with the highest versus the lowest expression; log-rank test. See Methods for detailed statistics.
Next, we analyzed whether the 4 differentially regulated gene expression clusters (Figure 6A) are enriched in human T cell–rich gliomas versus T cell–depleted gliomas, i.e., the 20% of tumors with the highest expression versus the 20% of tumors with the lowest T cell signature (16). Cluster 1, which represents genes overexpressed in tumors under immune attack, was found to be highly enriched in T cell–rich gliomas, especially in IDH-WT tumors (P < 0.001; Figure 8B). Conversely, cluster 3, which includes genes downregulated in tumors grown in WT mice, was downregulated in T cell–rich gliomas (P < 0.041, Figure 8B). Our analysis further revealed a strong impact of clusters 1 and 4 on patient survival. Genes from these “immune escape” and “cell cycle” clusters, respectively, that were overexpressed in WT mice (Figure 6B) were associated with significantly poorer survival (Figure 8C). We then focused on selected genes present in cluster 1 that may contribute to the immune escape of gliomas and analyzed their impact on survival, by comparing the 20% of samples with the highest versus lowest expression. As expected, high expression of Cd274 tended to be associated with shorter survival (P = 0.120). In addition, high expression of Irf1 (P = 0.008), Socs1 (P < 0.001), and Tnfrsf14 (P < 0.001) was associated with significantly shorter survival (Figure 8D). Consistent with their function in immune activation, overexpression of Ccl24 and Ciita tended to be associated with prolonged survival. However, some other immune activation genes were associated with shorter survival — for example, Cxcl10 and Ccl8 — suggesting that they may serve additional functions, such as autocrine stimulation of tumor cell growth. Notably, Apol6 was the second most highly overexpressed gene in cluster 1 (330-fold), and this gene, which has a largely unknown function, also tended to be associated with a worse prognosis (Supplemental Figures 4 and 5), suggesting a potential role in immunosuppression.
The immune environment shapes glioma heterogeneity in vivo.
The upregulation of immunosuppressive genes in tumors grown in WT mice is consistent with the concept of cancer immunoediting and the paradigm that during tumor evolution only cells that are capable of resisting immune attack survive. To investigate how the immunoactive environment affects the clonal composition during tumor evolution, we used a multicolor cell tracking system (26, 27). GL261 cells were simultaneously transduced with lentiviral vectors that mediate highly variable expression of random combinations of the 3 basic colors red, green, and blue (RGB), thereby displaying all perceivable colors, comparable to the rainbow spectrum (Figure 9A). RGB-labeled GL261 cells were injected into the brains of WT and Pfp–/– Rag2–/– mice, and tumors were analyzed when mice became symptomatic. Microscopically, tumors in WT mice were composed of fewer different clones than tumors in Pfp–/– Rag2–/– mice (Figure 9B). To parameterize and quantify this difference, tumors were dissociated, and the clonal composition was analyzed by flow cytometry (Figure 9C). Chromaticity data were represented as 3D plots, and the quantitative analysis of color distributions revealed a significantly higher color contraction in tumors in WT mice compared with Pfp–/– Rag2–/– mice and compared with the preinjection cell mix (Figure 9, C and D). These findings demonstrate that tumor clonality is reduced in immunocompetent mice, indicating that immune selection pressure restricts the survival and expansion of a number of tumor subclones, while others escape immune control and gain dominance.
Figure 9. Clonal heterogeneity in RGB-marked tumors.
(A) Schematic representation of lentiviral RGB marking of GL261 cells. Fluorescent proteins of the 3 basic colors are mixed at different but highly stable expression intensities so that all perceivable colors are generated. (B) Confocal fluorescence microscopy of RGB-marked GL261 tumors in WT and Pfp–/– Rag2–/– mice. Scale bar: 50 μm. (C) Spherical scatter plot of cells analyzed by flow cytometry. Each data point designates the chromaticity value of a cell. Note the reduced occupancy, i.e., the plot area occupied with data points in WT mice compared with Pfp–/– Rag2–/– mice and compared with the preinjection mix. (D) Quantification of occupancy after multistep dimensionality reduction validates significantly higher clonal contraction in tumors in WT mice than in Pfp–/– Rag2–/– mice, compared with the multiclonal preinjection mix. Box plots with IQR (box), mean (line), and maximum/minimum (whiskers). One-way ANOVA (P = 0.001) with Tukey’s post hoc test; *P < 0.05, ***P = 0.001; n = 4 per mouse group, n = 2 for the preinjection mix.
Optical barcoding reveals immunoediting of the clonal architecture.
While RGB tracking allows assessment of the overall degree of tumor heterogeneity, it is not designed for precise quantification of individual clones because of spectral overlap. To overcome this limitation and assess more precisely how the immune system quantitatively shapes the clonal composition in glioma, we leveraged optical barcoding (OBC) as another technique (28). OBC employs a combinatorial binary approach in which cells are separately transduced with definite combinations of well-distinguishable fluorescent proteins to mark individual clones. By using 6 different input colors and allowing a maximum of 2 output colors to be expressed per cell, we generated 21 distinctly color-coded GL261 clones that could stably be reidentified and quantified in mixed populations (Figure 10A).
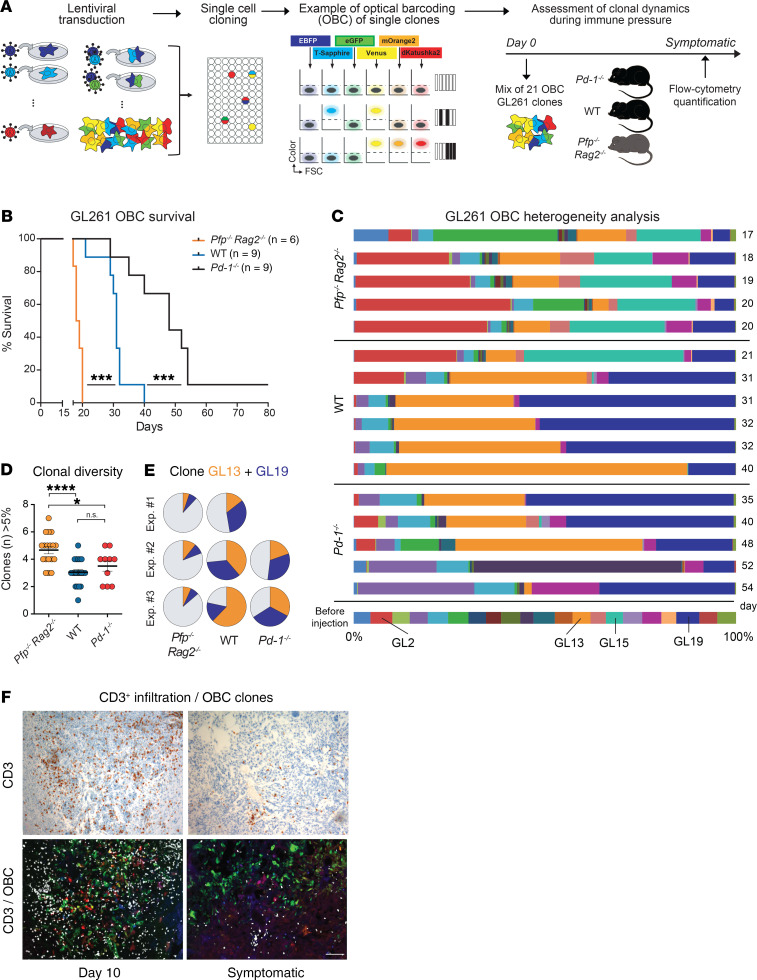
Figure 10. Clonal composition in OBC-marked tumors.
(A) Workflow of GL261 optical barcoding (OBC) and analysis of tumors in mice. (B) Survival of mice implanted with OBC-labeled GL261 cells; Kaplan-Meier analysis, log-rank test; ***P < 0.001. (C) Flow cytometry analysis of the clonal tumor composition in mice that became symptomatic at the indicated time points (days). (D) Number of clones that contribute more than 5% to the total number of tumor cells. One-way ANOVA (P = 0.0001) with Tukey’s post hoc test; *P < 0.05, ****P < 0.0001; Pfp–/– Rag2–/–, n = 18; WT, n = 20; Pd-1–/–, n = 10. (E) Mean dominance of clones GL13 and GL19 in repeat experiments. (F) Tumor infiltration with CD3+ T cells is maximal on day 10 and parallels the progressive loss of clonal heterogeneity in WT mice. Scale bar: 100 μm.
OBC-labeled GL261 cells were injected into the brains of WT and Pfp–/– Rag2–/– mice, and we additionally included programmed cell death 1 (Pdcd1) knockout (Pd-1–/–) mice in the experiment as a model of increased immunoselective pressure. Confirming our previous finding (Figure 1B), WT mice survived significantly longer (median 31 days) than Pfp–/– Rag2–/– mice (median 18.5 days), and Pd-1–/– mice survived even longer (median 48 days), suggesting that deficiency of the PD-1 checkpoint further enhances tumor control by the immune system (Figure 10B). To assess the clonal tumor composition, we sacrificed mice when they developed tumor-related symptoms and quantified the OBC-marked tumor cells by flow cytometry. Consistent with the RGB-labeling results, tumors in Pfp–/– Rag2–/– mice exhibited a highly polyclonal composition with multiple different clones contributing to the tumor mass (Figure 10, C–E). In contrast, tumors in WT mice displayed a striking reduction in clonal diversity (mean number of clones greater than 5%: 4.67 in Pfp–/– Rag2–/– vs. 3.05 in WT [P < 0.001] and vs. 3.50 in Pd-1–/– [P = 0.016]), with predominance of 2 major clones, GL13 and GL19. Interestingly, in Pd-1–/– mice, clones GL13 and GL19 presented the same growth advantage as in the WT mice at earlier time points but were later partly eliminated, and instead several other clones successfully escaped immune control and expanded. Repeat experiments confirmed that the clonal diversity was greater in Pfp–/– Rag2–/– mice than in immunocompetent mice (Figure 10D and Supplemental Figure 6). Moreover, the same 2 clones were again dominant in tumors in WT and Pd-1–/– mice (Figure 10E), suggesting that they share an extraordinary capacity to escape immune control compared with others. T cell infiltration in WT tumors was high at day 10 when tumors were still polyclonal, whereas few T cells remained when mice became symptomatic and when tumor cell heterogeneity was reduced to 2 major clones (Figure 10F).
Since interferon secretion by tumor-infiltrating lymphocytes has a major impact on gene regulation in GL261 tumors, and is known to have cytotoxic effects, we analyzed whether IFN-γ itself alters the clonal composition in vitro. Incubation of OBC-labeled GL261 cells with IFN-γ increased the expression of PD-L1; however, long-term incubation with either 1 or 10 ng/mL IFN-γ had only a minor effect on the clonal composition (Supplemental Figure 7, A and B). In contrast to our in vivo findings, clone GL13, which was dominant in immunocompetent mice, showed a lower prevalence in IFN-γ–treated cultures compared with controls. Conversely, clone GL15, which had a relatively high prevalence in tumors in Pfp–/– Rag2–/– mice, expanded relatively more in IFN-γ–treated than in untreated cultures. These findings suggest that the presence or absence of IFN-γ alone cannot explain the clonal development in mice in vivo and that interferon-induced effects are highly dependent on the tumor microenvironment.
Discussion
While earlier studies on cancer immunoediting primarily focused on carcinogen-induced or spontaneously formed tumors in mice (5), our study investigated the impact of the immune system on tumor development by implanting syngeneic glioma cells into the brain. The major findings of our study are as follows: (a) immunocompetent glioma-bearing mice survive significantly longer than immunodeficient mice; (b) tumors grow more invasive in immunodeficient mice; (c) immune pressure shapes the clonal architecture and profoundly reduces clonal diversity in immunocompetent mice; (d) glioma cells respond to immune selection pressure with activation of interferon response pathways; (e) nontumor stromal cells are the major origin of the immunosuppressive gene signature in gliomas; and (f) the immune escape signature identified in mice is reflected in human patients and correlates with poor survival.
Our finding that WT mice survived significantly longer than Pfp–/– Rag2–/– mice indicates that the functional immune system can limit glioma growth in the brain to some extent. Loss of the PD-1 checkpoint conferred an additional survival advantage, consistent with studies in mice and in human glioblastoma patients showing that PD-1 blockade can prolong survival (29, 30). Morphologically, we discovered striking alterations in tumor growth characteristics. Both GL261 and CT2A glioma cell lines grew far more invasive in Pfp–/– Rag2–/– mice, with tumors typically extending also extracerebrally, in contrast to the well-contained intracerebral growth in WT mice. This suggests that local adaptive and innate immune cells may restrict brain invasion in WT mice by tackling glioma cells at the leading edge. In addition, systemic immune control may prevent tumor outgrowth from the relatively protected brain microenvironment into meningeal spaces, supporting the concept of the brain as immunoprivileged site for tumor formation (31).
Our gene expression analysis in bulk tumors over time showed that the majority of genes overexpressed in WT mice were related to an immune response and that differential gene expression in comparison with Pfp–/– Rag2–/– mice was maximal on day 14. By the time when mice became symptomatic, downregulation of key immune activators as well as inhibitors had occurred, indicating that the immune response is exhausted and lower levels of endogenous immunosuppressors are required to facilitate tumor progression. T cell infiltration was also maximal around day 14, consistent with a similar time course described for the T cell response after infection or vaccination, which peaks about 7–15 days after antigen stimulation with a decrease thereafter (32). Tumor infiltration with T cells was further associated with an increased infiltration by macrophages/microglia that was strikingly more pronounced in WT mice than in Pfp–/– Rag2–/– mice. Accordingly, in human glioma patients, “cytotoxic lymphocyte” and “CD8+ T cell” signatures correlated with the infiltration by “monocytic lineage” cells, underlining the importance of the interplay between tumor-targeting and immunosuppressive cell populations, as well as of the continuous stimulatory presence of cytotoxic T cells for the attraction and activation of TAMs.
Macrophages/microglia constitute up to 50% of the cells in human gliomas and thus represent the majority of non-neoplastic cells in the tumors (11). As our bulk tumor analyses had not allowed us to determine to what extent the immune response signature was derived from the actual glioma cells as opposed to the nontumor cells, we injected GFP-marked glioma cells to compare the expression profiles of sorted cell populations. Interestingly, the immunosuppressive expression signature mainly originated from the stromal cells. The common perception of immunosuppressive mechanisms in gliomas is that tumor cells secrete cytokines, enzymes, checkpoint ligands, and other surface molecules, which suppress the adaptive immune response and reprogram macrophages/microglia toward an immunosuppressive M2-like phenotype (4). The reprogrammed TAMs then also contribute to the attenuation of an antitumor immune response by expressing additional immunosuppressive factors. Our results demonstrate that the impact of this contribution has likely been underestimated and that TAMs are in fact the major origin of the immunosuppressive expression signature in gliomas. This conclusion is supported by a transgenic model in which microglia depletion resulted in strongly reduced growth of orthotopic GL261 gliomas (33). It remains to be determined whether stromal cell–derived factors are also primarily responsible for the T cell sequestration in bone marrow that is observed when glioma cells or other cancer cells are implanted into the brain, but not into the subcutaneous microenvironment (9). Our results strongly support the concept that the tumor stroma must be a major target for counteracting immunosuppression in malignant gliomas (11, 34–36).
To determine how immune pressure alters gene regulation in the actual tumor cells and identify mechanisms of the tumor cell–specific immune escape, we specifically compared GFP+ tumor cells in WT mice versus Pfp–/– Rag2–/– mice. This comparison revealed that the gene expression signature of immunoedited glioma cells is dominated by interferon-regulated genes. IFN-γ, which is secreted by tumor-infiltrating T cells and NK cells, is known to play a dual role in cancer immunology (37, 38). On the one hand, it is an antitumor cytokine that is necessary for tumor elimination, and IFN-γ–deficient mice are more susceptible to tumor formation than WT mice (37, 39). IFN-γ causes upregulation of MHC class I on tumor cells, increasing their antigenicity, and it can have antiproliferative and proapoptotic effects on tumor cells (40). Hence, the recent success of neoadjuvant pembrolizumab in recurrent glioblastoma patients was largely attributed to downregulation of cell cycle activity by T cell–derived interferon (29). On the other hand, it was shown for other types of cancer that tumor cells exposed to IFN-γ over prolonged times exhibit greater immunoevasive properties with suppression of cytotoxic T cell– and NK cell–mediated immune responses (40). Consistent with this dual role, our study demonstrates that interferon-regulated genes that were overexpressed in gliomas in WT mice include prominent candidates involved in immunosuppression as well as in immune activation. Beyond upregulation of the well-known checkpoint-related genes Cd274 (9-fold) and Ctla4 (5-fold), we identified Socs1 (7-fold), Tnfrsf14 (6-fold), Il18bp (5-fold), and Arg1 (78-fold) as immunosuppressors overexpressed in glioma cells. SOCS1 (suppressor of cytokine signaling 1) is a potent inhibitor of IFN-γ–mediated JAK/STAT signaling that takes part in a negative-feedback loop to attenuate cytokine signaling (40). TNFRSF14 (tumor necrosis factor receptor superfamily member 14), also known as herpes virus entry mediator (HVEM), is a surface molecule with checkpoint function that was recently discovered to be overexpressed and associated with a poor prognosis in glioblastoma patients (41). IL18bp (IL-18–binding protein) is an inhibitor of the proinflammatory cytokine IL-18 that inhibits IL-18–induced IFN-γ production (42). Arg1 encodes the enzyme arginase, which is known as a marker of protumorigenic (M2-polarized) macrophages/microglia in gliomas (43) and has not yet been reported to be expressed by tumor cells at significant levels. Arginase depletes l-arginine in the local tumor environment, resulting in the inhibition of T and NK cell activation (44). By upregulating Arg1, glioma cells thus recapitulate an immunosuppressive mechanism used by TAMs, similarly to what has been described for PD-L1 (CD274) (45). Notably, the second most highly overexpressed gene in WT tumors was Apol6 (330-fold). Little is known about the function of the cytoplasmic protein apolipoprotein L6, which is encoded by this gene. It is known to be induced by interferon, and its expression in HIV-infected T cells can limit the production of infectious virus, suggesting a potential involvement in immunoregulatory processes (46). High expression of Apol6 in human glioma patients is associated with shorter survival, supporting the idea that it may contribute to immunosuppression and mandating future studies to address this question.
The fact that the immune escape signature that we identified in murine gliomas is specifically enriched in human T cell–rich gliomas and is associated with significantly shorter patient survival highlights the translational relevance of our findings. The comparison with human expression data is particularly important, since our models do not recapitulate spontaneous glioma formation in mice, which might be associated with different immune responses. A number of genes that are part of the identified murine escape signature have a known immunosuppressive function and are associated with worse overall patient survival (Socs1, Irf1, Tnfrsf14, and Cd274). Conversely, several immune-activating genes tend to be associated with better survival (Ccl24, Ciita). However, there are exceptions to this scheme; for example, high expression of the activating chemokine genes Cxcl10 and Ccl8 is associated with a worse prognosis, while Il18bp expression is linked with a better prognosis (not shown). These seemingly paradoxical results indicate that these immune modulators most likely have additional functions in glioma biology and may, for example, also exert autocrine effects on tumor cell growth or vascular cells by which they promote glioma growth.
In addition to identifying an immune escape signature, our study demonstrates that immune selection pressure profoundly affects tumor heterogeneity and sculpts the clonal tumor architecture. Tumors in Pfp–/– Rag2–/– mice exhibited a highly polyclonal composition, whereas immunoedited tumors in WT and Pd-1–/– mice displayed greatly reduced clonal diversity, indicating that immunoselective pressure restricts the survival and expansion of certain tumor subclones in WT mice while selecting for escape clones that are capable of resisting immune attack. These findings demonstrate that the local microenvironment, even in a partially immunoprivileged organ such as the brain, is capable of controlling clonal evolution. In multiple repeat experiments we found that always the same 2 clones (GL13, GL19) became dominant in WT mice. In vitro, these clones did not have a growth advantage over others, and in Pfp–/– Rag2–/– mice they were outgrown by yet different clones, highlighting the importance of the individual microenvironment in shaping the clonal composition. Since gene expression profiles of tumor cells in WT mice were mainly distinguished by an interferon response signature and IFN-γ can have cytotoxic effects on cells, we hypothesized that IFN-γ by itself may exert clonal selection pressure. However, long-term incubation with IFN-γ in vitro had little effect on the clonal composition of GL261 cultures, indicating that the complex in vivo microenvironment with its multilateral interactions between IFN-γ–secreting T cells, TAMs, tumor cells, and other cells is necessary to achieve the particular selection of immune escape clones that we observed.
Studies in other types of cancer, such as non–small cell lung cancer (NSCLC) or melanoma, showed that neoantigen load is a major factor determining an efficient immune response against tumors (47). In NSCLC patients, high clonal mutational burden and low subclonal mutational heterogeneity correlate with a superior prognosis, and these patients also respond better to immune checkpoint blockade (48). Local mutational burden is associated with local T cell clonal expansion, suggesting that tumor–immune cell interactions impose negative selection by pruning out tumor cell populations carrying neoepitopes (49). GL261 cells are hypermutated, and the majority of nonsynonymous mutations is subclonal (50, 51). It is therefore possible that the 2 dominant clones in WT mice might display a lower mutational burden and reduced neoantigenicity in comparison with other clones. In addition, the nature of presented tumor antigens determines the extent of T cell recognition and activation, and a higher “antigen fitness” may have resulted in depletion of more immunogenic clones during tumor evolution (52). Heterogeneous expression of costimulatory or inhibitory ligands, the ability to recruit immunoregulatory cells, or other differences in tumor–immune cell interaction may have added to differences in immunogenicity between individual clones. Further studies are necessary to elucidate the precise mechanisms by which certain clones escape immune control while others become eliminated. To this end, tumor cell clones need to be captured and analyzed at early time points from nascent tumors when immunosensitive clones have not yet been eliminated and can be directly compared with those that later gain dominance.
In conclusion, our study demonstrates that glioma immunoediting is defined not only by alterations in gene expression profiles and growth patterns, but also by distinct clonal selection that reduces tumor heterogeneity. The mechanisms of immune escape identified here can inform translational studies aiming to target tumor-induced immunosuppression, in order to advance the efficacy of tumor-specific immune activation in glioma patients.
Methods
Cell culture.
GL261 and CT2A murine malignant glioma cell lines (provided by Darrell Bigner and Matthias Gromeier, Duke University, Durham, North Carolina, USA) as well as 293T cells (ATCC CRL-3216) used for production of lentiviral particles were propagated in DMEM containing 4.5 g/L glucose, 4 mM l-glutamine, and 1 mM pyruvate (31966-021, Gibco) supplemented with 10% FBS, 25 mM HEPES, 100 U/mL penicillin, and 100 mg/mL streptomycin.
Optical barcoding and RGB labeling.
Fluorescent optical barcoding (OBC) was performed as described previously (28). Briefly, cells were plated in a 24-well plate at a density of 50,000 cells per well in 500 μL medium. Cells were transduced with lentiviral gene ontology (LeGO) vectors expressing 6 different fluorescent proteins, allowing a maximum of 2 colors per well. Transduced cultures were single-cell-sorted into separate wells in a 96-well plate using a BD FACSAria IIIu cell sorter. Clones grown from single cells were reanalyzed and expanded separately. Red, green, and blue (RGB) marking was performed as described before (26, 27). Cells were transduced simultaneously with equal amounts of 3 LeGO vectors (LeGO-C2-Puro+, LeGO-V2-Puro+, and LeGO-Cer2-Puro+; www.LentiGO-Vectors.de), each expressing 1 fluorescent protein. After the optimal MOI was defined (to obtain about 50%–70% transduction rate per color), cells were selected with 1 μg/mL puromycin and expanded for experiments. The clonal composition of tumors generated in vivo from OBC- or RGB-marked cells was analyzed by flow cytometry (BD LSRFortessa). RGB marking FACS data were visualized and quantified for clonal composition using an in-house MATLAB-based program adapted from Wu et al. (53). Data sets were filtered based on viability and fluorescence intensities and normalized to in vitro controls. 3D spherical scatter plots were created by normalizing all data point vectors to a length of 1. Chromaticity values were calculated by assigning a radius to all data points and selecting all points on a spherical theta-phi grid within this distance to any data point. Areas between all grid points were calculated and compared with the total surface area of the plot.
Orthotopic in vivo models.
GL261 or CT2A cells (1 × 105 cells/μL in 2 μL DMEM) were injected into 8- to 14-week-old female C57BL/6 WT mice (Charles River), C57BL/6 Pfp–/– Rag2–/– mice (B6.129S6-Rag2tm1FwaPrf1tm1Clrk N12, Taconic), or C57BL/6 Pd-1–/– mice (B6.Cg-Pdcd1tm1.1Shr/J, provided by Tasuku Honjo, Kyoto University, Kyoto, Japan; ref. 54) as described previously (28). In experiments with OBC-labeled cells, clones were mixed in equal proportions to a final cell concentration of 1 × 105 cells/μL and were injected into the right striatum. Animals were euthanized when they developed tumor-related symptoms, and either brains were formalin-fixed and paraffin-embedded (FFPE) or tumors were removed, dissociated, and processed for flow cytometry.
Microarray analysis.
GL261 cells were injected into WT C57BL/6 or Pfp–/– Rag2–/– mice, and tumors were collected on day 7 (n = 4 per mouse type) or day 14 (n = 4 per type) or when mice became symptomatic (n = 6 per type). RNA was extracted from tumor tissue using Nucleospin columns (Macherey Nagel), pooled from 2 mice (day 7 or 14) or from 3 mice (symptomatic endpoint), and analyzed on Illumina microarray mouse WG-6 v2.0 chips. Raw data were preprocessed using GenePattern (Broad Institute, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA) and with cubic spline normalization (55, 56).
RNA sequencing.
Tumors were excised from symptomatic mice (C57BL/6 WT or Pfp–/– Rag2–/–, 6 mice each) previously injected with GL261 GFP+ cells. Single-cell suspensions were obtained by collagenase digestion (1.5 mg/mL; Biochrom). After filtration and washing, the cells were sorted using a BD FACSAria IIIu cell sorter into GFP+ tumor cells and GFP– nontumor cells. These samples were compared with bulk unsorted cells that contained both populations and to GL261 in vitro cultured cells used for mice injection (all in triplicate). RNA was extracted using Nucelospin columns and quantified by the Agilent Bioanalyzer RNA 6000 kit. All samples were sequenced in the genomic core facility of Münster University on a NextSeq 500 v2 sequencer, using the NEBNext mRNA isolation module and NEBNext RNA Ultra directional library prep kit. Sequencing reads were aligned to the mouse reference assembly (GRCm38.87) using STAR version 2.5.2b (https://github.com/alexdobin/STAR/releases). Differential expression analysis was carried out using DESeq2 (https://bioconductor.org/packages/release/bioc/html/DESeq2.html) (57). Genes were considered differentially expressed if the FDR was below 0.05 and the log2 fold change was above 2.
Immunohistochemistry.
Immunohistochemistry was performed on FFPE tissue using a Ventana BenchMark XT stainer (Roche) and the UtlraView DAB detection kit (Roche). The following primary antibodies were used: IBA1 (019-19741, Wako), CD3 (ab16669, Abcam), CD8a (361003, Synaptic Systems), PD-L1 (Lola 6H6, provided by Thomas Jacobs, Bernhard-Nocht-Institut für Tropenmedizin, Hamburg, Germany), and CD34 (ab8158, Abcam). Blood vessel density was determined by counting of the number of CD34 immunoreactive blood vessels in 5 high-power fields (0.031 mm2 each) in the most densely vascularized tumor hotspot area.
Statistics.
Comparison of human and murine data was performed with the statistical programming language R (www.r-project.org), using the packages survival, gplots, and biomaRt. Processed gene expression and clinical data of 1135 high-grade gliomas (WHO grade III–IV) as well as signatures of 10 immune and stromal cell populations were obtained from Bockmayr et al. (16). Pearson’s correlation coefficient was used for correlation of human immune-stromal signatures. Significance of correlation was assessed using the R-function cor.test. Matching of human and mouse genes was performed with biomaRt using the Ensembl gene annotation. T cell–rich and –depleted tumors were defined as the 20% of tumors with the highest or lowest expression, respectively, of the signature “T cells” (16).
Other statistical tests were applied as indicated in the figure legends. Statistical analyses were performed using GraphPad Prism and IBM SPSS v22. All t tests were unpaired and 2-tailed. Tests for multiple comparisons were performed using 1-way ANOVA with Tukey’s post hoc test. P values less than 0.05 were considered significant. Plots were graphed using GraphPad Prism, R Foundation’s R v2.12, and Adobe Illustrator CC 2017.
Databases.
Murine gene expression profiles were investigated using publicly available databases. Functional annotation and identification of overrepresented GO terms and KEGG pathways were performed using the DAVID 6.7 database (https://david-d.ncifcrf.gov/summary.jsp) (58). Network analysis was performed using Cytoscape version 3.7.1. (https://github.com/cytoscape/cytoscape/releases/3.7.1) (59). Protein Ensembl identifiers were used to build the network using the STRING database version 11.0 (https://string-db.org) (60). Genes involved in IFN-γ regulation and pathways were extracted from the Interferome database (www.interferome.org) (61). Gene expression profiles from murine experiments were submitted to the NCBI’s Gene Expression Omnibus (GEO) database with accession number GSE151285 (https://www.ncbi.nlm.nih.gov/geo). Human microarray gene expression data sets are publicly available at cBioPortal (http://www.cbioportal.org) (62) under the following accession numbers: GSE83130, GSE108474, GSE50161, GSE13041, GSE16011, GSE36245, GSE73038, GSE19578, and GSE26576. Additional information on the analysis of human gene expression profiles can be found in the publication by Bockmayr et al. (16).
Study approval.
All animal experiments were performed in accordance with the animal welfare guidelines and with the approval of the authority for health and consumer protection in Hamburg, Germany.
Author contributions
CLM and MM performed the experiments, collected and analyzed the data, and prepared the manuscript. MB performed the in silico analysis of published gene expression profiles and prepared the manuscript. KDF analyzed the RNA gene expression data and applied and adapted the bioinformatic tool to quantify RGB heterogeneity experiments. KR and BF generated RGB and OBC labeling constructs, labeled the cell lines, and helped with the manuscript. DB and MA performed the bioinformatic analysis. AF helped perform the microscopic analysis of OBC-labeled cells. KK, SZ, and MH provided technical support, prepared cell lines, and assisted in every aspect of the performed experiments. KN and TL provided additional mouse strains. LD assisted in the mouse experiments and interpreted the human data sets. MW supervised the studies, provided scientific input, and critically revised the manuscript. KL supervised the study, analyzed the data, and prepared the manuscript.
Supplementary Material
Acknowledgments
We thank the FACS core facility and the Mouse Pathology Core Facility at the University Medical Center Hamburg-Eppendorf for their excellent support. We are grateful to Darell D. Bigner and Matthias Gromeier (Duke University) for providing the GL261 and CT2A cell lines. We thank Anika Witten, Institute for Human Genetics, University Medical Center Münster, for the excellent support with the RNA sequencing. This study was supported by grants from the Anni Hofmann Stiftung (to KL), the Else Kröner-Fresenius-Stiftung (to MM), the Johannes Bauer Stiftung (to MW), and the Deutsche Forschungsgemeinschaft (SFB841 to KR).
Version 1. 06/30/2020
In-Press Preview
Version 2. 08/31/2020
Electronic publication
Version 3. 10/01/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(10):5257–5271.https://doi.org/10.1172/JCI138760.
Contributor Information
Cecile L. Maire, Email: cmaire@uke.de.
Malte Mohme, Email: m.mohme@uke.de.
Michael Bockmayr, Email: m.bockmayr@uke.de.
Krystian D. Fita, Email: k.fita@uke.de.
Kristoffer Riecken, Email: k.riecken@uke.de.
Daniela Börnigen, Email: d.boernigen@uke.de.
Malik Alawi, Email: m.alawi@uke.de.
Katharina Kolbe, Email: k.kolbe@uke.de.
Svenja Zapf, Email: s.zapf@uke.de.
Mareike Holz, Email: ma.holz@uke.de.
Katrin Neumann, Email: kat.neumann@uke.de.
Lasse Dührsen, Email: l.duehrsen@uke.de.
Tobias Lange, Email: to.lange@uke.de.
Boris Fehse, Email: fehse@uke.de.
Manfred Westphal, Email: westphal@uke.de.
Katrin Lamszus, Email: lamszus@uke.uni-hamburg.de.
References
- 1.Westphal M, Lamszus K. The neurobiology of gliomas: from cell biology to the development of therapeutic approaches. Nat Rev Neurosci. 2011;12(9):495–508. doi: 10.1038/nrn3060. [DOI] [PubMed] [Google Scholar]
- 2.Lim M, Xia Y, Bettegowda C, Weller M. Current state of immunotherapy for glioblastoma. Nat Rev Clin Oncol. 2018;15(7):422–442. doi: 10.1038/s41571-018-0003-5. [DOI] [PubMed] [Google Scholar]
- 3.Mohme M, Neidert MC, Regli L, Weller M, Martin R. Immunological challenges for peptide-based immunotherapy in glioblastoma. Cancer Treat Rev. 2014;40(2):248–258. doi: 10.1016/j.ctrv.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 4.Mangani D, Weller M, Roth P. The network of immunosuppressive pathways in glioblastoma. Biochem Pharmacol. 2017;130:1–9. doi: 10.1016/j.bcp.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol. 2004;22:329–360. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- 6.Koebel CM, et al. Adaptive immunity maintains occult cancer in an equilibrium state. Nature. 2007;450(7171):903–907. doi: 10.1038/nature06309. [DOI] [PubMed] [Google Scholar]
- 7.Shankaran V, et al. IFNγ and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410(6832):1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 8.Matsushita H, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482(7385):400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chongsathidkiet P, et al. Sequestration of T cells in bone marrow in the setting of glioblastoma and other intracranial tumors. Nat Med. 2018;24(9):1459–1468. doi: 10.1038/s41591-018-0135-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorsson V, et al. The immune landscape of cancer. Immunity. 2018;48(4):812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hambardzumyan D, Gutmann DH, Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci. 2016;19(1):20–27. doi: 10.1038/nn.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doucette T, et al. Immune heterogeneity of glioblastoma subtypes: extrapolation from the cancer genome atlas. Cancer Immunol Res. 2013;1(2):112–122. doi: 10.1158/2326-6066.CIR-13-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Platten M, Ochs K, Lemke D, Opitz C, Wick W. Microenvironmental clues for glioma immunotherapy. Curr Neurol Neurosci Rep. 2014;14(4):440. doi: 10.1007/s11910-014-0440-1. [DOI] [PubMed] [Google Scholar]
- 14.Nduom EK, Weller M, Heimberger AB. Immunosuppressive mechanisms in glioblastoma. Neuro Oncol. 2015;17(suppl 7):vii9–vii14. doi: 10.1093/neuonc/nov151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grauer OM, Wesseling P, Adema GJ. Immunotherapy of diffuse gliomas: biological background, current status and future developments. Brain Pathol. 2009;19(4):674–693. doi: 10.1111/j.1750-3639.2009.00315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bockmayr M, et al. Immunologic profiling of mutational and transcriptional subgroups in pediatric and adult high-grade gliomas. Cancer Immunol Res. 2019;7(9):1401–1411. doi: 10.1158/2326-6066.CIR-18-0939. [DOI] [PubMed] [Google Scholar]
- 17.Ceccarelli M, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, Buetow K. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7(2):157–167. doi: 10.1158/1541-7786.MCR-08-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griesinger AM, et al. Characterization of distinct immunophenotypes across pediatric brain tumor types. J Immunol. 2013;191(9):4880–4888. doi: 10.4049/jimmunol.1301966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y, et al. Gene expression analysis of glioblastomas identifies the major molecular basis for the prognostic benefit of younger age. BMC Med Genomics. 2008;1:52. doi: 10.1186/1755-8794-1-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gravendeel LA, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69(23):9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 22.Sturm D, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 23.Sturm D, et al. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell. 2016;164(5):1060–1072. doi: 10.1016/j.cell.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paugh BS, et al. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol. 2010;28(18):3061–3068. doi: 10.1200/JCO.2009.26.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paugh BS, et al. Genome-wide analyses identify recurrent amplifications of receptor tyrosine kinases and cell-cycle regulatory genes in diffuse intrinsic pontine glioma. J Clin Oncol. 2011;29(30):3999–4006. doi: 10.1200/JCO.2011.35.5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weber K, et al. RGB marking facilitates multicolor clonal cell tracking. Nat Med. 2011;17(4):504–509. doi: 10.1038/nm.2338. [DOI] [PubMed] [Google Scholar]
- 27.Weber K, Thomaschewski M, Benten D, Fehse B. RGB marking with lentiviral vectors for multicolor clonal cell tracking. Nat Protoc. 2012;7(5):839–849. doi: 10.1038/nprot.2012.026. [DOI] [PubMed] [Google Scholar]
- 28.Mohme M, et al. Optical barcoding for single-clone tracking to study tumor heterogeneity. Mol Ther. 2017;25(3):621–633. doi: 10.1016/j.ymthe.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cloughesy TF, et al. Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat Med. 2019;25(3):477–486. doi: 10.1038/s41591-018-0337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reardon DA, et al. Glioblastoma eradication following immune checkpoint blockade in an orthotopic, immunocompetent model. Cancer Immunol Res. 2016;4(2):124–135. doi: 10.1158/2326-6066.CIR-15-0151. [DOI] [PubMed] [Google Scholar]
- 31.Mrass P, Weninger W. Immune cell migration as a means to control immune privilege: lessons from the CNS and tumors. Immunol Rev. 2006;213:195–212. doi: 10.1111/j.1600-065X.2006.00433.x. [DOI] [PubMed] [Google Scholar]
- 32.Pennock ND, White JT, Cross EW, Cheney EE, Tamburini BA, Kedl RM. T cell responses: naive to memory and everything in between. Adv Physiol Educ. 2013;37(4):273–283. doi: 10.1152/advan.00066.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markovic DS, et al. Gliomas induce and exploit microglial MT1-MMP expression for tumor expansion. Proc Natl Acad Sci U S A. 2009;106(30):12530–12535. doi: 10.1073/pnas.0804273106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szulzewsky F, et al. Human glioblastoma-associated microglia/monocytes express a distinct RNA profile compared to human control and murine samples. Glia. 2016;64(8):1416–1436. doi: 10.1002/glia.23014. [DOI] [PubMed] [Google Scholar]
- 35.Roesch S, Rapp C, Dettling S, Herold-Mende C. When immune cells turn bad—tumor-associated microglia/macrophages in glioma. Int J Mol Sci. 2018;19(2):E436. doi: 10.3390/ijms19020436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glass R, Synowitz M. CNS macrophages and peripheral myeloid cells in brain tumours. Acta Neuropathol. 2014;128(3):347–362. doi: 10.1007/s00401-014-1274-2. [DOI] [PubMed] [Google Scholar]
- 37.Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat Rev Immunol. 2006;6(11):836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 38.Zaidi MR, Merlino G. The two faces of interferon-γ in cancer. Clin Cancer Res. 2011;17(19):6118–6124. doi: 10.1158/1078-0432.CCR-11-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benci JL, et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell. 2016;167(6):1540–1554.e12. doi: 10.1016/j.cell.2016.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaidi MR. The interferon-gamma paradox in cancer. J Interferon Cytokine Res. 2019;39(1):30–38. doi: 10.1089/jir.2018.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han MZ, et al. Immune checkpoint molecule herpes virus entry mediator is overexpressed and associated with poor prognosis in human glioblastoma. EBioMedicine. 2019;43:159–170. doi: 10.1016/j.ebiom.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dinarello CA. Novel targets for interleukin 18 binding protein. Ann Rheum Dis. 2001;60(suppl 3):iii18–iii24. doi: 10.1136/ard.60.90003.iii18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang I, et al. Characterization of arginase expression in glioma-associated microglia and macrophages. PLoS One. 2016;11(12):e0165118. doi: 10.1371/journal.pone.0165118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez PC, et al. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 2004;64(16):5839–5849. doi: 10.1158/0008-5472.CAN-04-0465. [DOI] [PubMed] [Google Scholar]
- 45.Berghoff AS, et al. Programmed death ligand 1 expression and tumor-infiltrating lymphocytes in glioblastoma. Neuro Oncol. 2015;17(8):1064–1075. doi: 10.1093/neuonc/nou307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McLaren PJ, et al. Identification of potential HIV restriction factors by combining evolutionary genomic signatures with functional analyses. Retrovirology. 2015;12:41. doi: 10.1186/s12977-015-0165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 48.McGranahan N, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jia Q, et al. Local mutational diversity drives intratumoral immune heterogeneity in non-small cell lung cancer. Nat Commun. 2018;9(1):5361. doi: 10.1038/s41467-018-07767-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aslan K, et al. Heterogeneity of response to immune checkpoint blockade in hypermutated experimental gliomas. Nat Commun. 2020;11(1):931. doi: 10.1038/s41467-020-14642-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johanns TM, et al. Endogenous neoantigen-specific CD8 T cells identified in two glioblastoma models using a cancer immunogenomics approach. Cancer Immunol Res. 2016;4(12):1007–1015. doi: 10.1158/2326-6066.CIR-16-0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luksza M, et al. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature. 2017;551(7681):517–520. doi: 10.1038/nature24473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu JW, Turcotte R, Alt C, Runnels JM, Tsao H, Lin CP. Defining clonal color in fluorescent multi-clonal tracking. Sci Rep. 2016;6:24303. doi: 10.1038/srep24303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nishimura H, Nose M, Hiai H, Minato N, Honjo T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity. 1999;11(2):141–151. doi: 10.1016/S1074-7613(00)80089-8. [DOI] [PubMed] [Google Scholar]
- 55.Workman C, et al. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 2002;3(9):research0048. doi: 10.1186/gb-2002-3-9-research0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38(5):500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 57.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 59.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Szklarczyk D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rusinova I, et al. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41(Database issue):D1040–D1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.