Abstract
Psoriasis is a severe disease associated with the disturbance of metabolism and inflammation, but the molecular mechanisms underlying these aspects of psoriasis pathology are poorly understood. Here, we report that glutaminase 1–mediated (GLS1-mediated) glutaminolysis was aberrantly activated in patients with psoriasis and in psoriasis-like mouse models, which promoted Th17 and γδ T17 (IL-17A–producing γδ T) cell differentiation through enhancement of histone H3 acetylation of the Il17a promoter, thereby contributing to the immune imbalance and development of psoriasis. We further demonstrate that mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1) protease was constitutively active in psoriatic CD4+ and γδ T cells, thereby supporting GLS1 expression by stabilizing c-Jun, which directly binds to the GLS1 promoter region. Blocking the activity of either GLS1 or MALT1 protease resolved Th17 and γδ T17 cell differentiation and epidermal hyperplasia in the psoriasis-like mouse models. Finally, IL-17A enhanced GLS1 expression via the MALT1/cJun pathway in keratinocytes, resulting in hyperproliferation of and chemokine production by keratinocytes. Our findings identify the role of the MALT1/cJun/GLS1/glutaminolysis/H3 acetylation/T17 axis in psoriasis pathogenesis and reveal potential therapeutic targets for this disease.
Keywords: Autoimmunity, Dermatology
Keywords: Amino acid metabolism, T cells
Introduction
Psoriasis is a chronic inflammatory disease affecting up to 3% of the world’s population (1). Immune imbalance of T cells is believed to play a key role in psoriasis pathogenesis (2). A complex interplay between genetic and environmental factors, such as microorganism infection and physical trauma, initiates a cascade of events that lead to activation of plasmacytoid DCs (pDCs) (3). The activated DCs then produce IFN-α, IFN-β, IL-12, IL-23, IL-6, and TNF-α, which activate and polarize autoaggressive Th cells toward Th1, Th17, and Th22 cell subsets, as well as γδ T cells toward the γδ T17 (IL-17A–producing γδ T cells) subset, resulting in an immune imbalance of T cells (4–6). Release of proinflammatory cytokines from activated T cell subsets contributes to keratinocyte hyperproliferation and production of chemokines and defensins by keratinocytes, which leads to the recruitment of leukocytes and amplifies the immune response in psoriatic plaques (7, 8). Clinical studies demonstrated that blocking IL-23 p19 (9) or IL-17A (10–12) is highly effective for patients with psoriasis. This indicates that Th17 and γδ T17 cells, characterized by IL-17A as their major cytokine, are critical for the pathogenesis of psoriasis. Thus, a better understanding of the development and function of Th17 and γδ T17 cells will provide important insights and novel targets for therapeutic intervention in psoriasis.
Glutaminolysis is an important energy-yielding process and generates various substrates for anabolism in effector T cells. This process starts with the lysis of glutamine to glutamate, which later converts to α-ketoglutarate (α-KG) that then enters the citric acid cycle (TCA) and yields many metabolites such as acetyl-CoA (13). Glutaminase 1 (GLS1), the first enzyme in the glutaminolysis pathway and which converts glutamine to glutamate (14), has recently been shown to promote the generation of Th17 cells in mice (15, 16). But whether glutaminolysis participates in the pathogenesis of psoriasis is unknown. Recently, multivariate analyses of metabolomics profiles revealed that serum α-KG and glutamate are aberrantly elevated in patients with psoriasis (17–19). These observations suggest a potential regulatory role of glutaminolysis in this disease. Thus, the enzymatic activities, linkage to inflammation, regulatory mechanism, and in vivo physiological significance of glutaminolysis in psoriasis pathogenesis need to be investigated.
In this study, we report that increased expression of GLS1 aggravated Th17 and γδ T17 cell differentiation in patients with psoriasis and in psoriasis-like mouse models by augmenting intracellular acetyl-CoA, which contributed to histone H3 acetylation of the Il17a promoter and RORC transcriptional activity, thereby playing a much more important role in the pathogenesis of psoriasis. Pharmacological inhibition of GLS1 prevented the development of psoriasis-like inflammation in imiquimod-induced (IMQ-induced) psoriasis-like mouse models, indicating a potential therapeutic target for psoriasis. Furthermore, we reveal that consecutive activation of mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1) protease enhanced GLS1 expression through the stabilization of c-Jun in psoriatic CD4+ and γδ T cells. Increased expression of GLS1 induced by inflammatory cytokines such as TGF-β and IL-17A enhanced hyperproliferation of and chemokine production by keratinocytes. Our results indicate that GLS1-mediated glutaminolysis plays an essential role in psoriasis pathogenesis, including Th17 and γδ T17 cell differentiation and keratinocyte proliferation, highlighting a potential therapeutic target for psoriasis.
Results
GLS1-mediated glutaminolysis is associated with psoriasis pathogenesis.
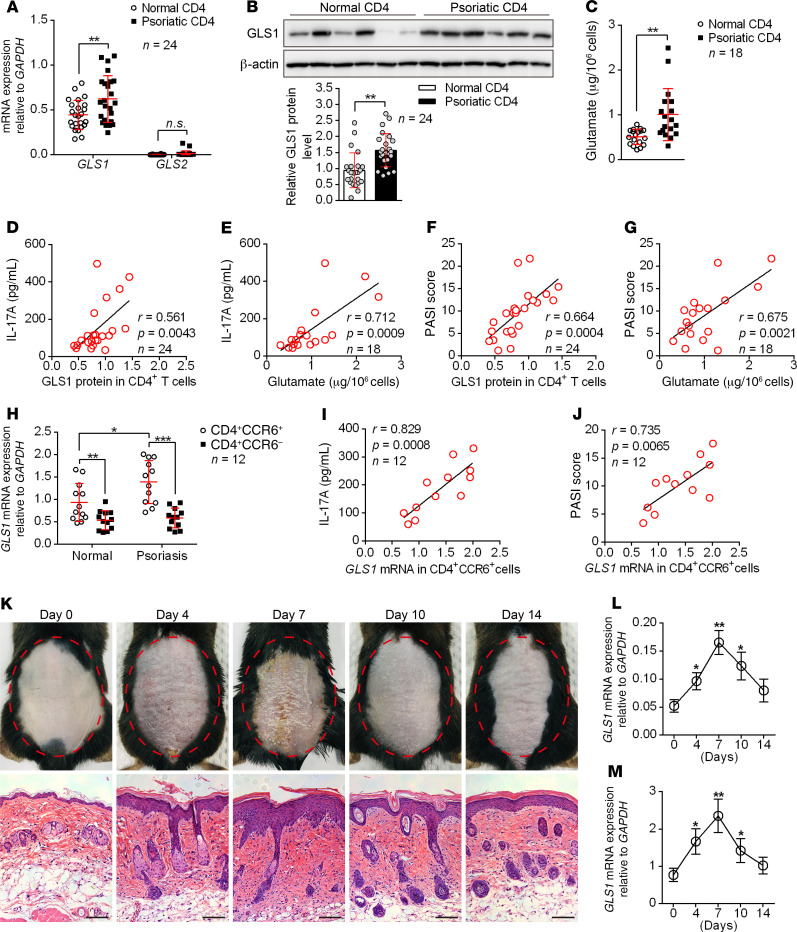
Glutaminolysis, initiated by glutaminase-mediated (GLS1- or GLS2-mediated) lysis of glutamine into glutamate, was reported to control the differentiation of Th17 cells in mice (20). Since Th17 cells play critical roles in the pathogenesis of human psoriasis, we speculated that glutaminolysis might also participate in the generation of human Th17 cells during disease development. Thus, we collected PBMCs, serum samples, and skin tissues from patients with psoriasis and from healthy donors and used these samples for glutaminolysis studies. Consistent with previous reports, patients with psoriasis showed elevated IL-17A production in serum (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI129269DS1), blood CD4+ T cells (Supplemental Figure 1B), and skin tissues (Supplemental Figure 1C), and IL-17A levels were positively correlated with disease severity (Supplemental Figure 1D). As speculated, glutaminolysis in CD4+ T cells was aberrantly activated in patients with psoriasis, as indicated by elevated mRNA and protein levels of GLS1 (Figure 1, A and B, and Supplemental Figure 1E) and increased production of glutamate (Figure 1C). The expression of GLS2 in CD4+ T cells was very low and unchanged (Figure 1A), suggesting that the robust glutaminolysis reactions were mainly mediated by GLS1. More important, both GLS1 protein levels and glutamate concentrations in CD4+ T cells were positively correlated with IL-17A production (Figure 1, D and E) and the psoriasis area and severity index (PASI) score for patients with psoriasis (Figure 1, F and G). For more specific cell populations, we found that the mRNA expression of GLS1 was higher in CD4+CCR6+ cells than in CD4+CCR6– cells from either healthy donors or donors with psoriasis (Figure 1H). In particular, mRNA levels of GLS1 in psoriatic CD4+CCR6+ cells were positively correlated with IL-17A production (Figure 1I) and the PASI score (Figure 1J) for patients with psoriasis. These results strongly suggested that GLS1-mediated glutaminolysis participated in Th17 cell differentiation and psoriasis pathogenesis. We also established IMQ-induced psoriasis-like mouse models (Supplemental Figure 2, A–H), which closely resemble human psoriasis. Consistent with the results seen in human samples, mice exposed to IMQ expressed significantly higher mRNA and protein levels of GLS1 (Supplemental Figure 2, I and J) and produced more glutamate (Supplemental Figure 2K) in splenic CD4+ T cells compared with matrix-exposed mice. Since dermal γδ T17 cells also play critical roles in the pathogenesis of IMQ-induced psoriasis-like disease, we further found that mRNA and protein levels of GLS1 (Supplemental Figure 2, L and M) were also significantly increased in dermal γδ T cells from IMQ-treated mice. In addition, the expression of GLS1 in both CD4+ and γδ T cells was highly correlated with the pathological changes over the progression of psoriasis (Figure 1, K–M). Collectively, these data demonstrated that GLS-mediated glutaminolysis was aberrantly activated and may be functionally involved in Th17 and γδ T17 cell–mediated immune dysfunction in psoriatic peripheral and skin lesions.
Figure 1. GLS1-mediated glutaminolysis is associated with psoriasis pathogenesis.
(A–C) Relative mRNA (A) (n = 24) and protein (B) (n = 24) levels of GLS1 and GLS2 and glutamate concentration (C) (n = 18) in CD4+ T cells derived from PBMCs of healthy controls and patients with psoriasis. Representative blots are shown (n = 6 of a total of 24 samples). (D–G) Correlation of GLS1 protein and glutamate concentration with IL-17A concentration (D and E) and GLS1 protein and glutamate concentration with PASI scores (F and G) in patients with psoriasis. n = 24 (D and F) and n = 18 (E and G). (H) Relative mRNA expression of GLS1 in CD4+CCR6+ and CD4+CCR6– cells derived from healthy controls and patients with psoriasis (n = 12). (I and J) Correlation of GLS1 mRNA in CD4+CCR6– cells with IL-17A concentration (I) and PASI scores (J) in patients with psoriasis (n = 12). (K–M) Phenotypic presentation and H&E staining of lesioned skin (K) (scale bars: 100 μm) and expression of Gls1 mRNA in splenic CD4+ T cells (L) and dermal γδ T cells (M) on days 4, 7, 10, and 14 during the application of IMQ in mice (n = 3). Data are presented as the mean ± SD and represent 1 of 3 independent experiments with consistent results. A 2-tailed, unpaired Student’s t test (A–C), Spearman’s r test (D–G, I, and J) or 1-way ANOVA with Tukey’s multiple comparisons test (H, L, and M) was used to determine statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001).
GLS1-mediated glutaminolysis favors Th17 and γδ T17 cell differentiation in psoriasis.
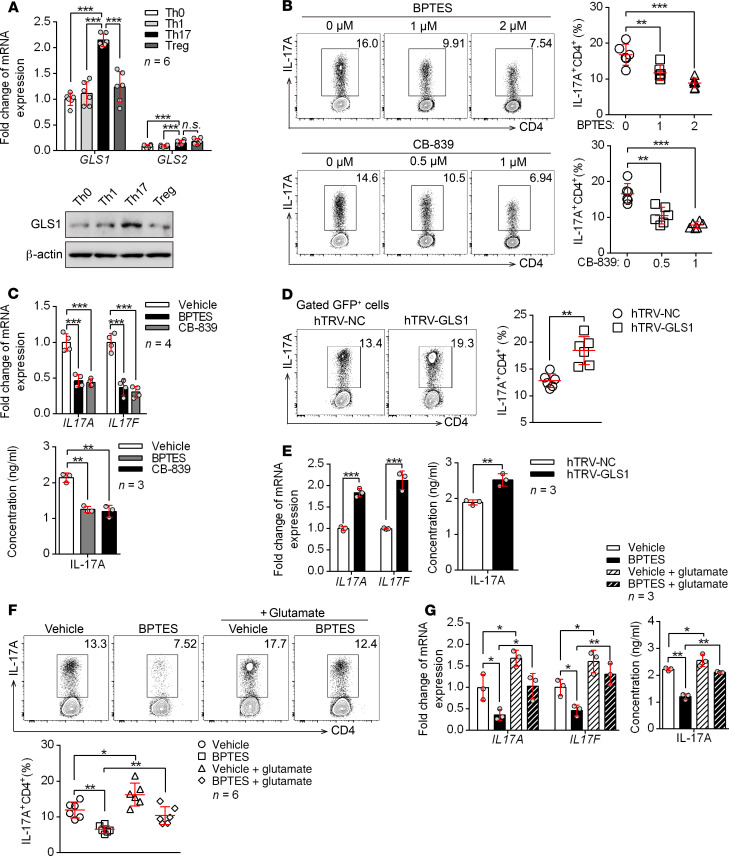
The imbalance of T cells was shown to play an important role in psoriasis pathogenesis (21). To determine whether the upregulation of GLS1 contributes to the immune imbalance of T cells in psoriasis, we first examined GLS1 expression in different T cell subsets. By activating human naive CD4+ T cells under different polarizing conditions in vitro (Supplemental Figure 3A), we found that both mRNA and protein expression levels of GLS1 were higher in Th17 cells than levels in other T cell subsets (Figure 2A). GLS2 was expressed at very low levels in all T cell subsets compared with GLS1 (Figure 2A), which was consistent with the results found in psoriatic CD4+ T cells (Figure 1A). These data suggested that GLS1-mediated glutaminolysis might favor Th17 cell differentiation. To directly assess the role of GLS1 in Th cell differentiation, we used 2 GLS1-specific inhibitors [bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl) ethyl sulfide (BPTES) and CB-839] in cultures of human naive CD4+ T cells undergoing Th0, Th1, Th17, and induced Treg (iTreg) differentiation in vitro (Supplemental Figure 3B). Both inhibitors effectively reduced the percentage of Th17 cells (Figure 2, B and C). In contrast, Th17 cell differentiation was significantly increased (Figure 2, D and E) by overexpression of GLS1 (Supplemental Figure 3C). In addition, glutamate supplementation corrected Th17 cell differentiation in BPTES-treated naive CD4+ T cells (Figure 2, F and G). However, GLS1 activity or expression level changes had no effect on the differentiation of other Th cell subsets, including Th1 cells (Supplemental Figure 3, D–G) and iTregs (Supplemental Figure 3, H and I). We also observed a similar effect of GLS1 on Th cell differentiation of mouse naive CD4+ T cells following treatment of the cells with BPTES or CB-839 (Supplemental Figure 4). To directly assess the role of GLS1 in γδ T17 cell differentiation, we also conducted experiments similar to those done in Th17 cells by activating mouse naive γδ T cells under γδ T17–polarizing conditions in vitro. As the results showed, both mRNA and protein levels of GLS1 were highly expressed in γδ T17 cells compared with levels in naive γδ T cells or γδ T1 cells (Supplemental Figure 5, A and B), but all these cells barely expressed GLS2. Furthermore, GLS1 inhibitors effectively weakened the differentiation of γδ T17 cells (Supplemental Figure 5, C and D), which was corrected by supplementation of glutamate (Supplemental Figure 5, E and F) in vitro. These findings suggested that GLS1-mediated glutaminolysis might favor γδ T17 differentiation. Together, our data indicate that GLS1-mediated glutaminolysis promoted the differentiation of Th17 and γδ T17 cells but had no effect on Th1 or iTreg subsets.
Figure 2. GLS1-mediated glutaminolysis favors human Th17 differentiation in psoriasis.
(A) Human naive CD4+ T cells were polarized under the indicated conditions in vitro for 5 days. Relative mRNA and protein expression levels of GLS1 and GLS2 (n = 6). (B and C) Vehicle-, BPTES-, or CB-839–treated human naive CD4+ T cells were polarized into Th17 cells for 5 days. (B) Flow cytometry and statistical analysis for the percentage of Th17 cells (n = 6). (C) Relative mRNA (n = 4) and protein (n = 3) levels of IL-17A and IL-17F. (D and E) Human naive CD4+ T cells were transduced with retrovirus carrying hTRV-GLS1 or control and then polarized into Th17 cells for 5 days. (D) Flow cytometry and statistical analysis for the percentage of Th17 cells (n = 6). (E) Relative mRNA and protein levels of IL-17A and IL-17F (n = 3). (F and G) Human naive CD4+ T cells were treated with vehicle or BPTES and polarized into Th17 cells for 3 days, followed by either no treatment or supplementation with glutamate for another 2 days. (F) Flow cytometry and statistical analysis for the percentage of Th17 cells (n = 6). (G) Relative mRNA expression and protein levels of IL-17A and IL-17F (n = 3). Data are presented as the mean ± SD and represent 1 of at least 2 independent experiments with consistent results. A 1-way ANOVA with Tukey’s multiple comparisons test (A–C, F, and G) or 2-tailed, unpaired Student’s t test (D and E) was used to determine statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001).
Aberrant activation of GLS1-mediated glutaminolysis in T cells assists the development of psoriasis.
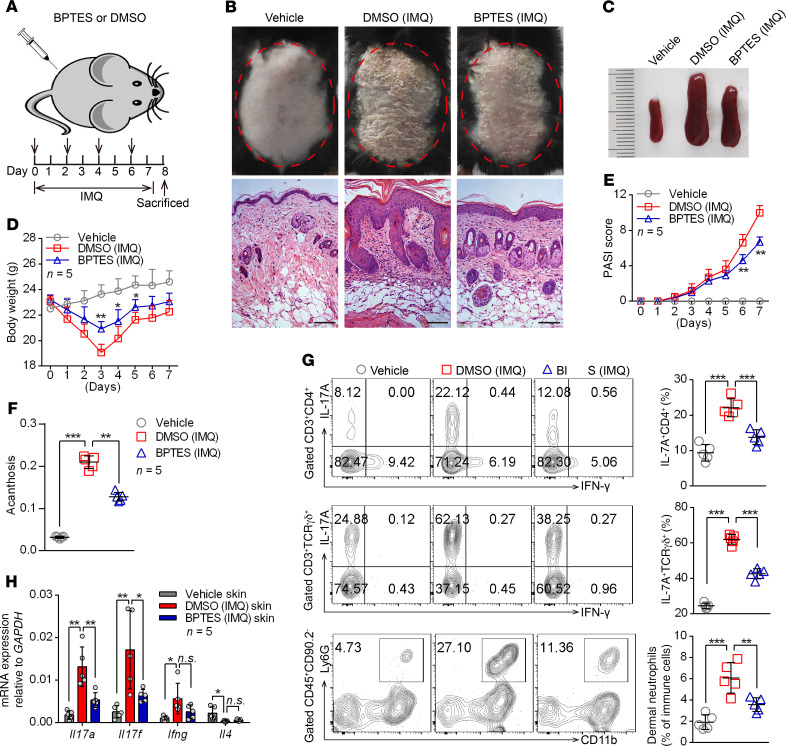
To clarify the functional relevance of GLS1-mediated glutaminolysis in psoriasis development, we intraperitoneally injected mice with BPTES during IMQ-induced psoriasis (Figure 3A). As expected, we observed significant improvement in both clinical and pathological characteristics following BPTES treatment (Figure 3B), including alleviation of splenomegaly (Figure 3C), body weight loss (Figure 3D), disease severity (Figure 3E and Supplemental Figure 6A), and acanthosis (Figure 3F). Notably, inhibition of GLS1 by BPTES markedly decreased the percentage of Th17 cells, γδ T17 cells, and neutrophils in dermal (Figure 3G) and splenic cells (Supplemental Figure 6B). Consistently, GLS1 inhibition significantly decreased mRNA levels of Il17a and Il17f in skin lesions (Figure 3H) as well as in splenic CD4+ and dermal γδ T cells (Supplemental Figure 6, C and D) from IMQ-treated mice, whereas Ifng expression was either unchanged or only marginally reduced.
Figure 3. Administration of GLS1 inhibitor ameliorates the pathological phenotype of IMQ-induced psoriasis-like mice.
(A) Schematic diagram of intraperitoneal administration of BPTES (100 μg/dose administration) or DMSO (5% DMSO in 200 μL PBS) on days 0, 2, 4, and 6 during the application of IMQ or not for 7 consecutive days. (B–F) Clinical manifestations and H&E staining of the back skin (B) (scale bars: 100 μm), splenomegaly (C), body weight (D), PASI scores (E), and acanthosis (F). n = 5. (G) Flow cytometry and statistical analysis of the percentage of IL-17A+ in dermal CD4+ T cells (gated on CD3+CD4+ cells) and γδ T cells (gated on CD3+TCR γδ+ cells) and of neutrophils in dermal CD45+CD90.2– lymphocytes (n = 5). (H) Relative mRNA expression of Il17a, Il17f, Ifng, and Il4 in skin lesions (n = 5). Data are presented as the mean ± SD and represent 1 of 3 independent experiments with consistent results. One-way ANOVA with Tukey’s multiple comparisons test (D–H) was used to determine statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001).
As we know, both activated immune cells and keratinocytes contribute to the psoriatic skin lesion. However, the ratio of GLS1 enzyme activity to GLS1 protein levels was dramatically reduced in splenic CD4+ or dermal γδ T cells but not in keratinocytes (Supplemental Figure 6E) following intraperitoneal treatment with BPTES, indicating that the inhibitory effect of the GLS1 inhibitor was mediated by T cells. Moreover, GLS1 inhibition also significantly reduced the expression of IL-17A in human psoriatic CD4+ T cells without affecting IFN-γ production (Supplemental Figure 7). In contrast, administration of glutamate during the development of IMQ-induced psoriasis-like disease promoted Th17 and γδ T17 cell generation and aggravated disease damage (Supplemental Figure 8). These results suggested that GLS1-mediated glutaminolysis promoted peripheral or dermal Th17 and γδ T17 cell differentiation and aggravated the pathogenesis of psoriasis.
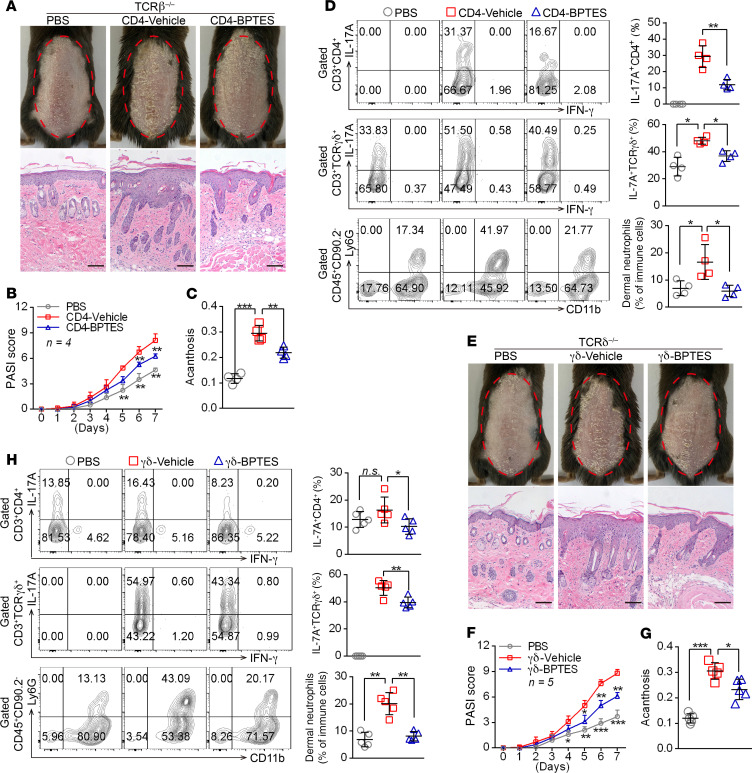
Given that previous studies have shown that αβ and γδ T cells cooperate to enhance IL-17A responses in inflammatory diseases such as experimental autoimmune uveitis (22), we next tested whether the interaction might be involved in the model of IMQ-induced psoriasis. Both Tcrb−/− and Tcrd−/− mice exhibited alleviative pathological characteristics of psoriasis (Supplemental Figure 9, A–E) and skin Il17a and Il17f expression (Supplemental Figure 9F) compared with WT mice treated with IMQ. Interestingly, Tcrb−/− mice treated with IMQ had significantly decreased percentages of dermal γδ T17 cells, whereas IMQ treatment of Tcrd −/− mice had no effect on dermal Th17 cells (Supplemental Figure 9G), but both showed comparatively decreased neutrophil infiltration (Supplemental Figure 9G). However, splenic Th17 or γδ T17 cells were largely unchanged in Tcrd−/− and Tcrb−/− mice treated with IMQ, respectively (Supplemental Figure 9H). These results suggested that IL-17A production from dermal γδ T cells was partially dependent on αβ T cells.
To investigate the role of GLS1-mediated glutaminolysis in CD4+ and γδ T cells in the development of psoriatic lesions, we transferred GLS1 inhibitor–pretreated (CD4-BPTES) or normal (CD4-vehicle) naive CD4+ T cells into Tcrb–/– mice, as well as GLS1 inhibitor–pretreated (γδ-BPTES) or normal (γδ-vehicle) naive γδ T cells into Tcrd–/– mice undergoing IMQ treatment, respectively. As expected, Tcrb–/– mice transferred with CD4-vehicle T cells developed typical psoriasis-like disease induced by IMQ, as indicated by aggravated pathological characteristics of psoriasis (Figure 4, A–C, and Supplemental Figure 10, A–C), higher Il17a and Il17f mRNA levels in skin (Supplemental Figure 10D), elevated percentages of dermal Th17 cells, γδ T17 cells, and neutrophil infiltration (Figure 4D). However, Tcrb–/– mice transferred with CD4-BPTES T cells had notably improved IMQ-induced skin lesions (Figure 4, A–C, and Supplemental Figure 10, A–C) and lower Il17a and Il17f mRNA levels in skin (Supplemental Figure 10D). More important, these mice showed decreased dermal Th17 cells, γδ T17 cells, and neutrophil infiltration (Figure 4D). Furthermore, splenic Th17 cell differentiation in mice transferred with CD4-BPTES T cells was impeded compared with those given CD4-vehicle T cells, but the differentiation of splenic γδ T17 cells was largely unaffected (Supplemental Figure 10, E and F). Similar to what was seen in Tcrb–/– mice, IMQ-induced skin lesions also significantly decreased in Tcrd–/– mice transferred with γδ-BPTES T cells (Figure 4, E–G, and Supplemental Figure 11, A–C), along with lower expression levels of Il17a and Il17f mRNA (Supplemental Figure 11D) and decreased dermal Th17 cells, γδ T17 cells, and neutrophil infiltration (Figure 4H). However, splenic Th17 and γδ T17 cell differentiation was not altered in these mice (Supplemental Figure 11, E and F). Taken together, these findings corroborated that the aberrant differentiation of Th17 and γδ T17 cells induced by GLS1-mediated glutaminolysis plays an important role in psoriasis pathogenesis, whereas inhibition of GLS1 activity in vivo can prevent the immunopathological changes in psoriasis and effectively ameliorate disease severity.
Figure 4. GLS1-mediated glutaminolysis in T cells is pivotal for IMQ-induced psoriasis-like skin inflammation.
(A–D) Tcrb–/– mice were transferred with PBS or naive CD4+ T cells pretreated with BPTES (CD4-BPTES) or not (CD4-Vehicle). (E–H) Tcrd–/– mice were transferred with PBS or naive γδ T cells pretreated with BPTES (γδ-BPTES) or not (γδ-Vehicle). The mice were then subjected to IMQ-induced psoriasis as part of the psoriasis-like mouse model. Representative phenotypic presentation and H&E staining of skin lesions (A and E) (scale bars: 100 μm), PASI scores (B and F), and acanthosis (C and G). n = 4 (B) and n = 5 (F). (D and H) Percentage of IL-17A+ cells in dermal CD4+ T cells (gated on CD3+CD4+ T cells) and γδ T cells (gated on CD3+ γδTCR+ T cells) and of neutrophils in dermal CD45+ lymphocytes. n = 4 (D) and n = 5 (H). Data are presented as the mean ± SD and represent 1 of at least 2 independent experiments with consistent results. A 1-way ANOVA with Tukey’s multiple-comparisons test (B–D and F–H) was used to determine statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001).
GLS1-mediated glutaminolysis enhances Th17 and γδ T17 cell differentiation via epigenetic regulation.
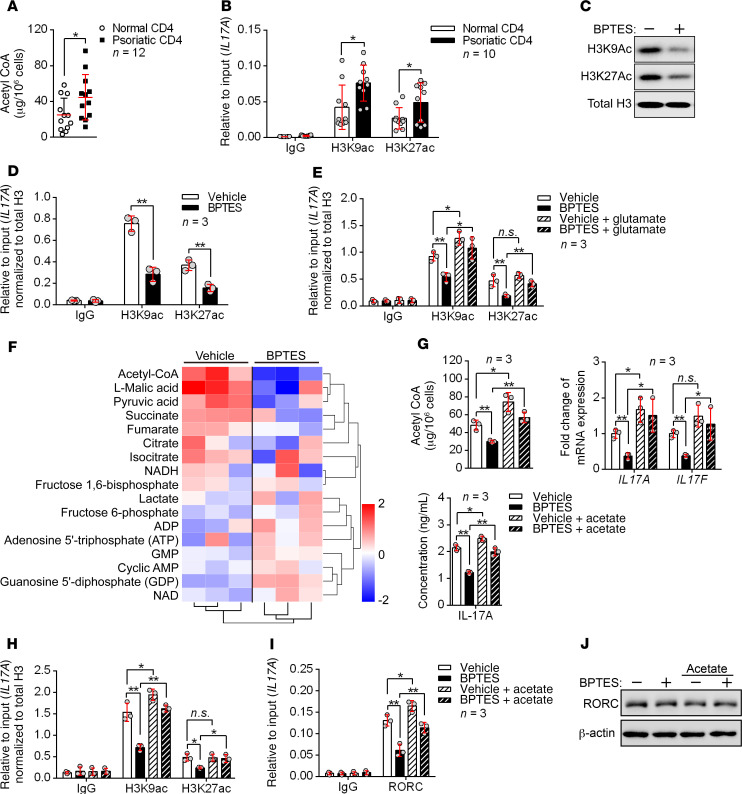
As mentioned above, GLS1 mediated the lysis of glutamine to glutamate, and the latter is further converted into α-KG and enters the citric acid cycle (TCA), which yields many intermediate metabolites and generates acetyl-CoA (13). Acetyl-CoA is the original substrate for lipid genesis and plasma membrane formation and is also the acetyl carrier for histone acetylation, which modulates gene expression (23). We hypothesized that glutaminolysis might promote the generation of psoriatic Th17 or γδ T17 cells via an increase in acetyl-CoA and histone acetylation. Thus, we detected the acetyl-CoA concentration in peripheral blood CD4+ T cells from patients with psoriasis and healthy donors and checked the acetylation status of histone 3 at the lysine 9 and 27 residues (H3K9Ac and H3K27Ac, 2 histone marks associated with active transcription) of the IL17A gene promoter region. Indeed, the psoriatic CD4+ T cells showed increased production of acetyl-CoA and elevated histone acetylation (Figure 5, A and B). We obtained similar results in psoriatic mouse CD4+ T cells (Supplemental Figure 12, A and B). To verify the connection between GLS1-mediated glutaminolysis and histone acetylation in human Th17 cells, we added a GLS1 inhibitor during Th17 cell differentiation and assessed the histone acetylation status. As expected, H3K9Ac and H3K27Ac acetylation was globally decreased in human Th17–polarizing cells with GLS1 inhibition (Figure 5C). More important, ChIP analysis showed that H3K9Ac and H3K27Ac levels in the IL17A gene promoter were also notably diminished (Figure 5D). In addition, glutamate supplementation reversed the suppression of H3K9Ac and H3K27Ac in response to GLS1 inhibition (Figure 5E). The inhibition of histone acetylation by BPTES (Supplemental Figure 12C) and the improvement effects of glutamate (Supplemental Figure 12D) were also confirmed in psoriatic mouse CD4+ T cells in vivo. These data demonstrated that GLS1-mediated glutaminolysis supported H3K9Ac and H3K27Ac in the IL17A gene promoter in Th17-polarizing cells. Facilitation of glutamine uptake by ASCT2 activates mTORC1 kinase (24), however, we found that GLS1-mediated glutaminolysis did not activate mTORC1 signaling in either human Th1 or Th17 cell differentiation in vitro (Supplemental Figure 12E).
Figure 5. GLS1-mediated glutaminolysis enhances Th17 cell differentiation via epigenetic regulation.
(A and B) Acetyl-CoA concentration (A) (n = 12) and histone acetylation of IL17A promoter regions (B) (n = 10) in CD4+ T cells from healthy controls and patients with psoriasis. (C–F) Human naive CD4+ T cells were polarized into Th17 cells for 5 days under the indicated conditions. (C) Representative blots for H3K9Ac and H3K27Ac histones. (D and E) Histone acetylation of IL17A promoter regions. Results were calculated relative to total chromatin input and normalized to total histone H3 levels to account for the nucleosomal occupancy at IL17A promoter regions (n = 3). (F) Heatmap analysis of LC-MS metabolomics. Rows display metabolites and columns represents the samples. Blue represents a decreasing trend; red represents an increasing trend (n = 3). (G–J) Vehicle- or BPTES-treated human naive CD4+ T cells were polarized into Th17 cells for 3 days and either left untreated or supplemented with 20 mM sodium acetate for another 2 days. (G) Acetyl-CoA concentration and mRNA and protein expression levels of IL-17A and IL-17F (n = 3). (H) Histone acetylation of IL17A promoter regions. Results were calculated relative to total chromatin input and normalized to total histone H3 levels to account for the nucleosomal occupancy at IL17A promoter regions (n = 3). (I) RORC occupancy at IL17A gene loci (n = 3). (J) Representative blots for RORC protein expression. Data are presented as the mean ± SD and represent 1 of at least 2 independent experiments with consistent results. A 2-tailed, unpaired Student’s t test (A, B, and D) or 1-way ANOVA with Tukey’s multiple comparisons test (E and G–I) was used to determine statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001).
To further confirm the role of glutaminolysis–TCA cycle–histone acetylation in the generation of human Th17 cells, we measured the absolute amounts of intermediate metabolites in human Th17–polarized cells cultured in the presence or absence of BPTES by liquid chromatography–mass spectrometry (LC-MS) analysis (Figure 5F). The inhibition of glutaminolysis by BPTES was confirmed by the reduction in glutamate concentration (Supplemental Figure 13A). In reference to the TCA cycle, we observed significant reductions in the levels of succinate, fumarate, malate, and acetyl-CoA, thus confirming the expected impairments in the TCA cycle (Supplemental Figure 13B). Consistent with the above observation, supplementation of glutamate in BPTES-treated human Th17 cells regenerated acetyl-CoA (Supplemental Figure 13C). We also confirmed the inhibition of acetyl-CoA by BPTES (Supplemental Figure 13D) and the improvement effects of glutamate (Supplemental Figure 13E) in psoriatic mouse CD4+ T cells in vivo. Besides, directly adding acetate, which could generate acetyl-CoA by acetyl-CoA synthetase independent of citrate (25), counteracts the suppressive effects of BPTES on histone acetylation and IL-17A production (Figure 5, G and H). More important, IL17A transcription is induced by RORC in Th17 cells (26), and this transcription can be enhanced by histone acetylation (27). RORC recruitment to the IL17A locus was reduced after GLS1 inhibition but could be reversed by supplementation of acetate (Figure 5I), however, the expression of RORC was not affected (Figure 5J). Furthermore, we observed similar effects of GLS1 on histone acetylation of the Il17a promoter in mouse γδ T17–polarized cells (Supplemental Figure 14, A–F). In mouse γδ T cells, AP-1 has been described as the transcription factor for Il17a and Il22 (28), but histone acetylation did not facilitate AP-1 recruitment to the promoter of Il17a (Supplemental Figure 14E). Collectively, the above results demonstrated that GLS1-mediated glutaminolysis promoted H3K9Ac and H3K27Ac epigenetic modification in the Il17a gene promoter of Th17 and γδ T17 cells, which enhanced chromatin accessibility of RORC and thereby aggravated IL-17A expression.
Interestingly, the amount of pyruvate was also significantly reduced in BPTES-treated Th17-polarizing cells (Figure 5F and Supplemental Figure 13F). Pyruvate is the metabolic product of glycolysis and mediates the translocation of acetyl-CoA (13), thus we also detected the intermediate metabolites upstream of glycolysis. Surprisingly, we found that the metabolites upstream of glycolysis were not significantly changed, neither was the generation of lactate (the main product in anaerobic glycolysis) (Figure 5F and Supplemental Figure 13G), which suggested that the reduction of pyruvate might be due to reduced transport demands of acetyl-CoA rather than to glycolysis.
MALT1 protease is consecutively activated and initiates aberrant GLS1-mediated glutaminolysis in psoriasis.
The generation of Th17 cells from naive CD4+ T cells requires a switching of oxidative phosphorylation metabolism to glycolysis and glutaminolysis. MALT1 protease is required for TCR-dependent metabolic changes in T cells (29) and promotes mouse Th17 cell polarization (30, 31). Recently, we found that MALT1 protease regulates GLS1 expression in B cell lymphoma (32). Thus, we wondered whether MALT1 regulates glutaminolysis-dependent Th17 cell generation in patients with psoriasis.
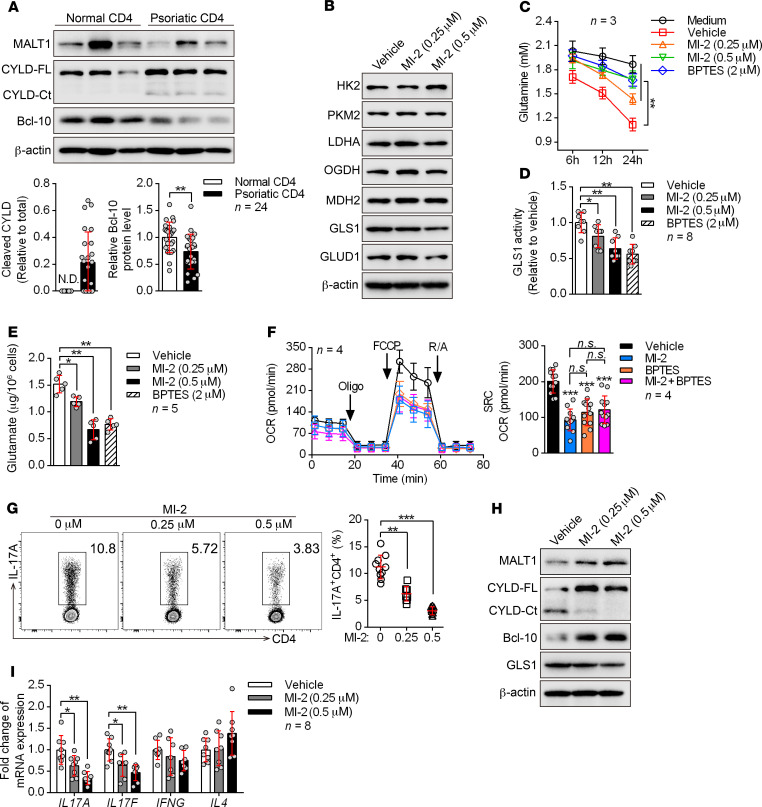
We first checked the proteolytic activity of MALT1 in peripheral blood CD4+ T cells from patients with psoriasis. Surprisingly, psoriatic CD4+ T cells showed remarkably elevated MALT1 activity, as indicated by increased cleavage of CYLD and BCL-10 (Figure 6A and Supplemental Figure 15). In addition, activation of MALT1 by brief stimulation of the TCR only induced GLS1 expression in human Th17 cells rather than in Th0, Th1, and Th2 cells and Tregs (Supplemental Figure 16A). We next examined the role of MALT1 in GLS1-mediated glutaminolysis through direct inhibition of MALT1 during human Th17 cell differentiation. Excitingly, we found that inhibition of MALT1 protease with MI-2 (a MALT1 protease–specific inhibitor) significantly reduced GLS1 activity, glutamine consumption, and glutamate production (Figure 6, B–E, and Supplemental Figure 16B). However, the levels of enzymes in glycolysis and the TCA pathway were largely unaffected upon MALT1 inhibition (Figure 6B). Consistently, inhibition of MALT1 protease decreased the oxygen consumption rate (OCR) (the OCR indicates aerobic respiration including oxidative phosphorylation, glutaminolysis, and fatty acid oxidation) (Supplemental Figure 16C) without affecting glycolysis (Supplemental Figure 16, D–F), and did not influence the number of mitochondria (Supplemental Figure 16G) in human Th17–polarizing cells. Interestingly, both MI-2 and BPTES significantly reduced the OCR, but the combination of both inhibitors did not further enhance this suppressive effect (Figure 6F), suggesting that MALT1 regulates metabolism mainly through GLS1-mediated glutaminolysis. Meanwhile, Th17 differentiation was significantly inhibited with MI-2 treatment (Figure 6G), and overexpression of GLS1 or supplementation of glutamate partially rescued the Th17 differentiation defects (Supplemental Figure 17). Similarly, we found that these metabolic changes also occurred in Th17- and γδ T17–polarizing cells from point mutation–knockin (KI) mice (Supplemental Figure 18, A–F) that have selectively reduced MALT1 protease activity (33) along with decreased Th17 or γδ T17 cell differentiation (Supplemental Figure 18G). These results substantiated the observation that MALT1 protease was upstream of GLS1-mediated glutaminolysis and promoted its effects on Th17 and γδ T17 differentiation.
Figure 6. MALT1 protease is consecutively activated and initiates aberrant GLS1-mediated glutaminolysis in psoriasis.
(A) Peripheral CD4+ T cells were collected from healthy donors and patients with psoriasis. Representative blots for the expression level and protease activity of MALT1 (cleavage of CYLD to CYLD-Ct, BCL-10) in CD4+ T cells are shown (n = 3 of a total of 24 samples). CYLD-FL, full-length CYLD; CYLD-Ct, cleaved CYLD. (B–G) Human naive CD4+ T cells subjected to the indicated treatments were polarized into Th17 cells for 5 days. (B) Representative blots show the protein levels of key metabolic enzymes. (C) Consumption of glutamine at the indicated time points. “Medium” refers to glutamine natural degradation (n = 3). (D and E) GLS1 activity (D) (n = 8) and glutamate concentration (E) (n = 5) were measured. (F) Cells were collected to detect the OCR using an extracellular flux analyzer. Cumulative data for the calculated spare respiratory capacity (SRC) are shown (n = 4). Oligo, oligomycin; FCCP, carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone; R/A, rotenone and antimycin. (G) Percentage of Th17 cells detected by flow cytometry and statistical analysis (n = 10). (H and I) Protein expression of GLS1 (H) and relative mRNA expression of IL17A, IL17F, IFNG, and IL4 (I) (n = 8) in human psoriatic CD4+ T cells treated with MI-2 for 48 hours. Data are presented as the mean ± SD and represent 1 of at least 2 independent experiments with consistent results. A 2-tailed, unpaired Student’s t test (A) or 1-way ANOVA with Tukey’s multiple comparisons test (C–G and I) was used to determine statistical significance. (*P < 0.05, **P < 0.01, ***P < 0.001).
To determine whether this regulatory mechanism also occurs in psoriasis, we isolated psoriatic CD4+ T cells from patients and treated the cells with a MALT1 protease inhibitor. Consistently, we found that inhibition of MALT1 protease pronouncedly interdicted GLS1 expression (Figure 6H) as well as expression of IL17A and IL17F (Figure 6I) in vitro. Collectively, the aforementioned findings converged to strongly support a critical role of MALT1 protease in initiating aberrant GLS1-mediated glutaminolysis in psoriasis, thereby promoting Th17 and γδ T17 cell generation and aggravating disease pathology.
MALT1 protease promotes GLS1-mediated glutaminolysis via c-Jun.
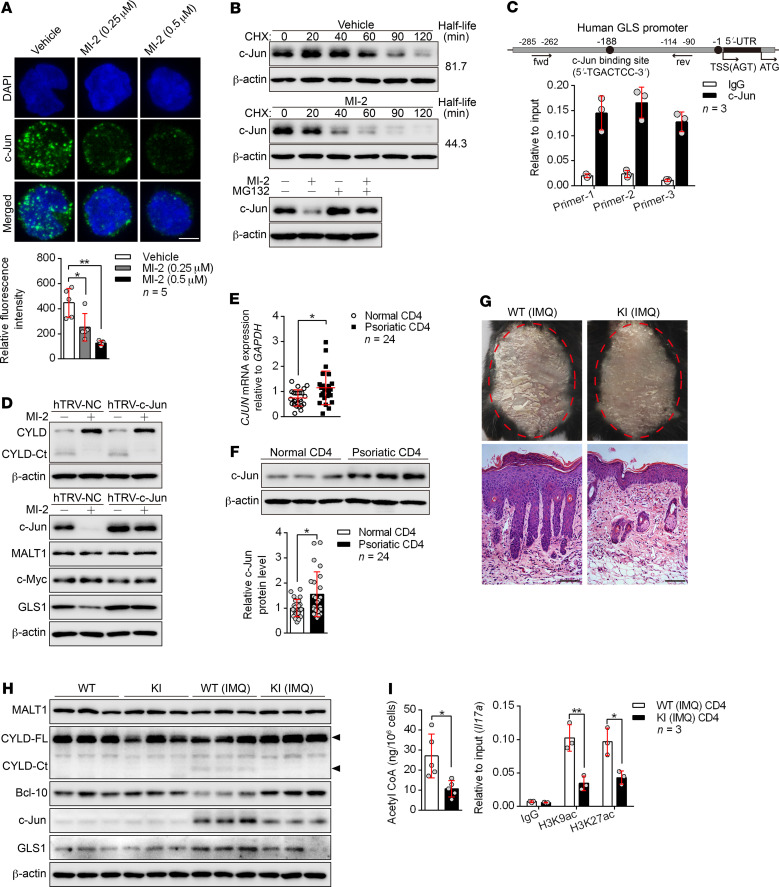
NF-κB signaling is the canonical target of MALT1 protease, however, we found that the expression of GLS1 was not affected in response to NF-κB inhibition (Supplemental Figure 19A), indicating that NF-κB was not involved in GLS1-mediated glutaminolysis. To demonstrate the regulatory mechanism of MALT1 protease on GLS1 expression, we investigated whether c-Jun or c-Myc, both of which have been found to initiate the transcription of GLS1 in tumor cells (34, 35), is a downstream target of MALT1 protease in Th17-polarizing cells. The protein level of endonuclear c-Jun rather than c-Myc was almost completely abolished by inhibition of MALT1 protease (Figure 7A and Supplemental Figure 19B), but the transcription of c-Jun was not affected (Supplemental Figure 19C). We further found that the degradation of c-Jun protein was significantly accelerated by inhibition of MALT1 protease (Figure 7B), whereas it was partially rescued by proteasome inhibition (Figure 7B). Importantly, we found the c-Jun binding motif in the promoter region of the GLS1 gene (Figure 7C), suggesting that GLS1 expression was regulated by c-Jun. Indeed, overexpression of c-Jun remarkably potentiated GLS1 expression (Supplemental Figure 19D) and intensified the conversion of glutamine into glutamate (Supplemental Figure 19E). Finally, c-Jun was indispensable for MALT1 protease–enhanced GLS1 expression (Figure 7D). Consistently, c-Jun was significantly elevated at both mRNA (Figure 7E) and protein levels (Figure 7F) in psoriatic CD4+ T cells derived from patients with psoriasis.
Figure 7. MALT1 protease promotes GLS1-mediated glutaminolysis via c-Jun.
(A) Human naive CD4+ T cells were polarized into Th17 cells with the indicated treatment for 5 days. Confocal microscopy of cells stained with c-Jun antibody (green) and the DNA-binding dye DAPI (blue). Scale bars: 5 μm. Graph shows relative intensity of c-Jun expression in (n = 5). (B) Representative blots for c-Jun expression in cells from A that were restimulated with anti-CD3 and anti-CD28 antibodies followed by cycloheximide (CHX) treatment at the indicated time points (upper panel), or that were treated with MG132 or not for 12 hours. (C) ChIP for the c-Jun binding site on the proximal region of the GLS promoter (n = 3). fwd, forward; rev, reverse; TSS, transcription start site. (D) Cells in A were transduced with a concentrated retrovirus carrying hTRV–c-Jun or control (hTRV-NC). Representative blots for MALT1 protease activity following restimulation with anti-CD3 and anti-CD28 antibodies and expression of c-Jun, MALT1, c-Myc, and GLS1. (E and F) Relative mRNA (E) (n = 24) and protein (F) (n = 3 of a total of 24 samples) levels of c-Jun in psoriatic CD4+ T cells. (G–I) WT or MALT1 protease-deficient (KI) mice were painted with IMQ cream for 7 consecutive days. Clinical manifestations and H&E staining of the back skin (G) (scale bars: 100 μm). Representative blots for MALT1 protease activity and GLS1 and c-Jun expression in splenic CD4+ T cells (H) (n = 3). Acetyl-CoA concentration (n = 5) and histone acetylation of Il17a promoter regions (n = 3) in splenic CD4+ T cells (I). Data are presented as the mean ± SD and represent 1 of at least 2 independent experiments with consistent results. One-way ANOVA with Tukey’s multiple comparisons test (A) or 2-tailed, unpaired Student’s t test (E, F, and I) was used to determine statistical significance. (*P < 0.05, **P < 0.01).
In order to further verify the effect of the MALT1/c-Jun/GLS1 axis in the pathogenesis of psoriasis, we adopted MALT1 protease–KI mice were adopted for use in an IMQ-induced psoriasis-like mouse model. We found that MALT1 protease–KI mice were unable to develop typical psoriasis-like dermatitis with regard to both clinical manifestations and pathological changes (Figure 7G and Supplemental Figure 20, A–D). Meanwhile, MALT1 protease–KI mice exhibited lower expression levels of c-Jun and GLS1 (Figure 7H and Supplemental Figure 20, E and F) as well as decreased GLS1 activity and glutamate production (Supplemental Figure 20G) and lower levels of acetyl-CoA and histone H3 acetylation in the Il17a promoter (Figure 7I) in splenic CD4+ or dermal γδ T cells. Furthermore, these mice showed limited dermal or splenic Th17 and γδ T17 cell differentiation (Supplemental Figure 21, A and B) and elimination of Il17a and Il17f expression in splenic CD4+ T cells, skin lesions, and dermal γδ T cells (Supplemental Figure 21, C–E) in response to IMQ treatment in vivo. Taken together, we found that the MALT1/c-Jun/GLS1 axis was activated in the pathogenesis of psoriasis and promoted Th17 and γδ T17 differentiation.
GLS1-mediated glutaminolysis contributes to excessive proliferation of and chemokine production by keratinocytes.
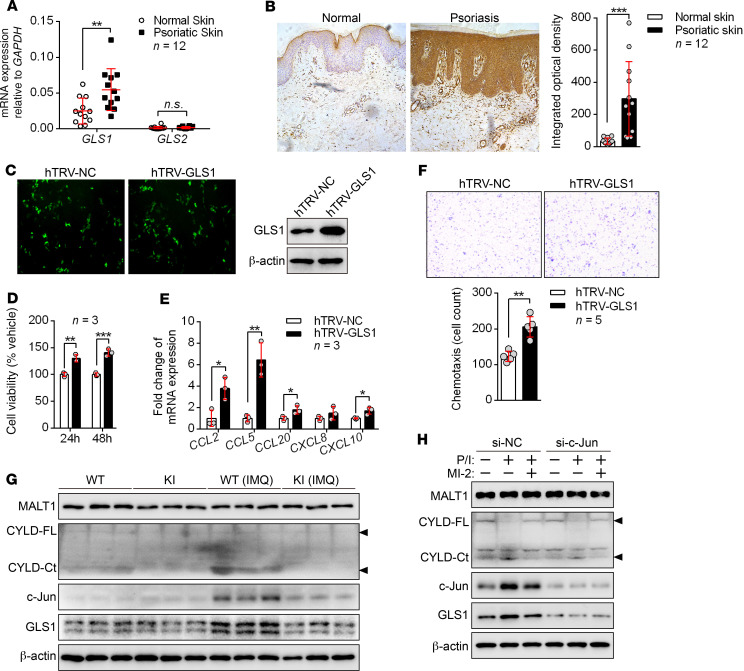
In addition to immune cells, keratinocytes also play an indispensable role in the pathogenesis of psoriasis. The mRNA and protein levels of GLS1 were elevated in the psoriatic epidermis of both psoriasis patients (Figure 8, A and B) and IMQ-induced psoriasis-like mice (Supplemental Figure 22, A and B). Next, we investigated the effects of GLS1 on keratinocytes using the HaCaT cell line, which is an immortal keratinocyte cell line from adult human skin. Overexpression of GLS1 (Figure 8C) promoted the proliferation of keratinocytes (Figure 8D) as well as the secretion of chemokines such as CCL2, CCL5, CCL20, and CXCL10 (Figure 8E), resulting in enhanced chemotaxis of CD4+ T lymphocytes to keratinocytes in vitro (Figure 8F). In contrast, inhibition of GLS1-mediated glutaminolysis restrained the proliferation of keratinocytes (Supplemental Figure 23A), the secretion of chemokines (Supplemental Figure 23B) by keratinocytes, and the chemotaxis of CD4+ T lymphocytes to keratinocytes (Supplemental Figure 23C). However, other immune modulators such as IL36G, DEFB4, and IL17C were not affected by inhibition of GLS1-mediated glutaminolysis in keratinocytes (Supplemental Figure 23D). We further found that inhibition of GLS1-mediated glutaminolysis significantly suppressed the expression of chemokines in epidermis from the IMQ-induced psoriasis-like mouse model (Supplemental Figure 23E) and decreased the number of dermis-infiltrating CD4+ and γδ T cells in vivo (Supplemental Figure 23F). Unfortunately, GLS1-mediated glutaminolysis did not affect histone acetylation at the promoter regions of CCL2, CCL5, CCL20, or CXCL10 (Supplemental Figure 23G). These data indicated that GLS1-mediated glutaminolysis promoted hyperproliferation of and chemokine production by keratinocytes in psoriasis. Interestingly, keratinocytes stimulated with IL-23, TGF-β, or IL-17A had significantly increased GLS1 expression (Supplemental Figure 24, A and B), suggesting a potential positive feedback between immune cells and keratinocytes.
Figure 8. GLS1-mediated glutaminolysis by the MALT1/c-Jun axis contributes to excessive proliferation of and chemokine production by keratinocytes.
(A and B) qPCR (A) and IHC (B) for GLS1 and GLS2 expression in skin from healthy controls and patients with psoriasis (n = 12). (C–F) HaCaT cells were transfected with the control (hTRV-NC) or hTRV-GLS1. (C) Detection of transfection efficiency of hTRV-GLS1. (D) HaCaT cell proliferation was analyzed by CCK8 assay (n = 3). (E) qPCR was performed to determine the mRNA levels of chemokines secreted by HaCaT cells (n = 3). (F) Culture supernatants were collected 48 hours later and added to the lower chambers, and normal human CD4+ T cells were added to the upper chambers of Transwell plates for 2 hours. The upper chamber was removed and stained with crystal violet, and cells were counted (n = 5). (G) Representative blots for MALT1 protease activity and GLS1 and c-Jun expression in keratinocytes from mice. (H) HaCaT cells were transfected with an siRNA against c-Jun (si–c-Jun) or a control siRNA (si-NC) and then primed with MI-2 or stimulated with PMA/ionomycin (P/I) for 2 hours. Representative blots for MALT1 protease activity and GLS1 and c-Jun expression. Data are presented as the mean ± SD and represent 1 of at least 2 independent experiments with consistent results. A 2-tailed, unpaired Student’s t test (A, B, and D–F) was used to determine statistical significance. (*P < 0.05, **P < 0.01, ***P < 0.001).
To determine whether MALT1 also plays a similar role in GLS1 induction in keratinocytes, we detected GLS1 expression in keratinocytes from IMQ-treated MALT1-KI mice. As expected, keratinocytes from IMQ-treated WT mice showed active MALT1 protease and high protein levels of c-Jun and GLS1, whereas these activations had vanished in IMQ-treated MALT1-KI mice (Figure 8G). In addition, we found that c-Jun was indispensable for MALT1 protease–enhanced GLS1 expression in keratinocytes (Figure 8H). The CARD14-BCL10-MALT1 (CBM) complex promotes IL-17A signaling and skin inflammation in the IMQ model (36, 37). We further discovered that MALT1 protease activity was needed for IL-17A–induced GLS1 expression in both HaCaT and primary murine keratinocytes (Supplemental Figure 24, C and D). Together, these data suggested that IL-17A/MALT1/c-Jun axis–induced GLS1-mediated glutaminolysis in keratinocytes also contributes to the pathogenesis of psoriasis.
Discussion
It is well established that the disturbance of T cell–mediated inflammation plays a key role in psoriasis pathogenesis (38). Recently, multivariate analyses of metabolomic profiles revealed that metabolic restructuring is also involved in psoriasis pathology (17–19). However, the involved enzymes, the linkage to inflammation, the regulatory mechanism, as well as the in vivo physiological significance of metabolic restructuring in psoriasis pathogenesis are poorly understood. Here, we showed that GLS1, but not GLS2, the first enzymes in the glutaminolysis pathway, was upregulated in CD4+ and γδ T cells derived from patients with psoriasis and in IMQ-induced psoriasis-like mice. Inhibition of GLS1-mediated glutaminolysis substantially prevented the formation of skin lesions induced by IMQ. Thus, our finding broadens the knowledge regarding the precise metabolic enzymes that link metabolism with psoriasis pathogenesis.
Cell metabolism fuels the bioenergetic and biosynthetic demands of T cells and paves the way to their delicate differentiation. For Th17 cells, the energy-yielding metabolism switches from oxidative phosphorylation to glycolysis and glutaminolysis immediately after TCR and cytokine engagement (39, 40), which supports the biomass required for robust cytokine production (41, 42). Deprivation of glutamine in culture media, deficiency in the glutamine transporter ASCT2, or inhibition of GLS1 activity impedes the differentiation of mouse Th17 cells (15, 16, 24). Glutaminolysis also favors mouse Th17cells over iTregs partially via a reduction of N-glycan branching (20). In our study, we discovered that GLS1-mediated glutaminolysis contributed to the immune imbalance of psoriasis not only by promoting the differentiation of Th17 but also of γδ T17 cells. Although the role of GLS1 specifically in Th17 cell differentiation has already been shown before (15, 16), we present new data about the regulation of γδ T17 cell differentiation and psoriasis by GLS1. In addition, we investigated the in vitro and in vivo effects of GLS1-mediated glutaminolysis on the Th1 cell and iTreg response, which has also been reported to be involved in the pathogenesis of psoriasis (43). However, we observed no significant change in these T cell subsets with fluctuation of GLS1 expression, which was consistent with a recent report (15). The results suggested that GLS1-mediated glutaminolysis contributes to psoriasis through induction of Th17 and γδ T17 cell differentiation, but not Th1 cell or iTreg differentiation. Unexpectedly, there are some discrepancies in the data from another recent study, which reported that GLS deficiency increased T-bet to promote the differentiation and effector function of Th1 cells (16). This discordance might be due to distinct effects of GLS1 protein and its enzymic activity or to the distinction between murine and human cells. Nonetheless, our work sheds light on how GLS1 regulates psoriasis and demonstrates that GLS1-mediated glutaminolysis supports psoriatic Th17 and γδ T17 cell differentiation, especially in humans, and that this process integrates the disturbed metabolism and disordered inflammation in psoriasis pathogenesis.
Metabolism is implicated in the control of gene expression through epigenetic mechanisms that are frequently mediated by certain metabolites (44). The glutaminolysis process generates an abundance of metabolites such as glutamate, α-ketoglutaric acid, 2-hydroxyglutarate, succinyl-CoA, fumaric acid, acetyl-CoA, citric acid, 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA), and pyruvic acid. These metabolites are precursors for nucleotide, amino acid, and lipid biosynthetic pathways, and several of these metabolites also play critical roles in mediating epigenetic modification such as methylation and acetylation (45). But it is unclear whether glutaminolysis controls Th17 and γδ T17 cell differentiation via an epigenetic mechanism. Recent studies demonstrated that the mechanism of GLS1 in regulating Th17 cell differentiation is associated with ROS or the mTORC1 signaling pathway (15, 16). In this study, we identified acetyl-CoA as a functional metabolite of GLS1-mediated glutaminolysis, which enhanced acetylation of histone 3 in the IL17A promoter and RORC recruitment to the IL17A locus. A previous study suggested that aberrant epigenetic modifications were involved in the pathogenesis of psoriasis (46). Consistently, we found that both acetyl-CoA and the acetylation of histone 3 in the IL17A promoter were significantly elevated in CD4+ T cells of patients with psoriasis. Thus, our finding reveals a regulatory mechanism by which GLS1-mediated glutaminolysis regulates Th17 and γδ T17 cell differentiation in psoriasis.
Our previous study suggested that MALT1 protease regulates GLS1 expression in B cell lymphoma (32). Here, we also found that MALT1 protease was involved in the initialization of GLS1-mediated glutaminolysis during Th17 and γδ T17 cell differentiation in psoriasis. Consistent with the upregulation of GLS1, we also detected increased MALT1 protease activity in psoriatic CD4+ T cells, as indicated by enhanced cleavage of CYLD and BCL-10. This indicated that MALT1 protease activity was not specific for Th17 cells in psoriasis, because only a small proportion of psoriatic CD4+ T cells in PBMCs is expected to be a Th17 subset. In other inflammatory diseases, MALT1 protease activity was found to be dramatically evoked in T cells in an experimental autoimmune encephalomyelitis (EAE) model (47) and in PBMCs from a spinal cord injury (33). Although MALT1-mediated canonical activation of NF-κB plays a critical role in Th17 cell differentiation (30), it was independent of NF-κB for the promotion of GLS1-mediated glutaminolysis by MALT1 protease. Previous reports have proven that the transcription of GLS1 is initiated by c-Myc and c-Jun in tumor cells (34, 35). In our report, we found markedly reduced c-Jun expression in response to MALT1 inhibition, whereas c-Myc was largely unchanged in Th17-polarized cells. Moreover, the reduced c-Jun levels were due to aggravated protein degradation and not to transcription, and c-Jun indeed bound to the promoter region of the GLS1 gene. Notably, expression of c-Jun was markedly elevated in psoriatic CD4+ and dermal γδ T cells. We found decreased expression levels of c-Jun and GLS1 in splenic CD4+ and dermal γδ T cells from MALT1-KI mice in response to IMQ treatment in vivo. Given that MALT1 protease is required for TCR-initiated signal transduction (48), we believe that active MALT1 protease is induced by pathogenic signals to evoke and stabilize c-Jun, thereby promoting GLS1-mediated glutaminolysis in IL-17–producing T cells in psoriasis.
In addition to immune cells, keratinocytes also play an indispensable role in the pathogenesis of psoriasis. Activated by proinflammatory factors, keratinocytes exhibit excessive proliferation and secretion of chemokines, which play a crucial role in amplifying the immune response in psoriatic plaques (49, 50). Our studies found that GLS1 expression was also elevated in the epidermis of psoriatic skin from both patients with psoriasis and IMQ-induced psoriasis-like mouse models. In addition, GLS1-mediated glutaminolysis promoted hyperproliferation and chemotaxis of psoriatic keratinocytes. Interestingly, HaCaT cells stimulated with IL-23, TGF-β, or IL-17A had substantially increased GLS1 expression levels, suggesting a potential positive feedback between immune cells and keratinocytes. Since the CBM complex promotes IL-17A signaling and skin inflammation in the IMQ model, we further demonstrated that MALT1 protease activity was needed for IL-17A/TNF-α–induced expression of GLS1 in keratinocytes. However, IL-17A/TNF-α failed to induce MALT1 protease activity, as indicated by the unchanged levels of cleaved CYLD. According to a previous report (51), keratinocyte-specific MALT1 substrates that might contribute to regulation of the IL-17A/TNF-a pathway by MALT1 remain to be identified. Together, these data suggest that aberrant GLS1-mediated glutaminolysis in keratinocytes might also contribute to the pathogenesis of psoriasis. A recent study indicated that glucose metabolism is selectively required for injury- and inflammation-associated keratinocyte proliferation (52), which further supports the idea that cell metabolism plays an essential role in pathological hyperproliferation of psoriasis. However, identification of the enzymes involved, the molecular mechanism, as well as the physiological significance of keratinocyte metabolism contributing to the pathogenesis of psoriasis requires further investigation.
Although several drugs, such as ustekinumab (53) and secukinumab (54), have been developed for the treatment of psoriasis, remaining issues include the single effect, the exorbitant price of these therapeutics, and the uncertainty of long-term antagonism of just 1 cytokine. GLS1 inhibitors are being considered for the treatment of various cancers in mice (55, 56), and some of them have entered clinical trials (57, 58). A recent study also revealed that the treatment of mice with a GLS1 inhibitor ameliorated EAE (15). Here, we succeeded in suppressing psoriasis-like disease in mice with only 10% of the dose of BPTES used in the treatment of cancer. MALT2 protease inhibitors selectively suppress the growth of MALT2-dependent ABC-DLBCL but not GCB-DLBCL both in vitro and in vivo (59, 60). In addition, they effectively reduced EAE symptoms without influencing Treg numbers (61). Therefore, our findings indicate that either GLS1 or MALT1 protease inhibitors are attractive potential drugs for the treatment of psoriasis.
In conclusion, the present study provides evidence that GLS1-mediated glutaminolysis, as a downstream target of MALT1 protease, promotes Th17 and γδ T17 cell differentiation through the promotion of acetyl-CoA–induced acetylation of histone 3 in the Il17a promoter, thereby causing the immune imbalance in peripheral blood and skin lesions of psoriasis. On the other hand, IL-17A/MALT1/c-Jun axis–induced GLS1-mediated glutaminolysis also enhances the proliferation of and chemokine secretion by keratinocytes, which further promotes the formation of psoriatic skin lesions (Figure 9). Taking together, our studies uncover the function and mechanism of GLS1-mediated glutaminolysis in the formation of immune imbalance and skin lesions in psoriasis, provide new information for a better understanding of the linkage of metabolism and inflammation in the pathogenesis of psoriasis, and identify a potential target for psoriasis treatment.
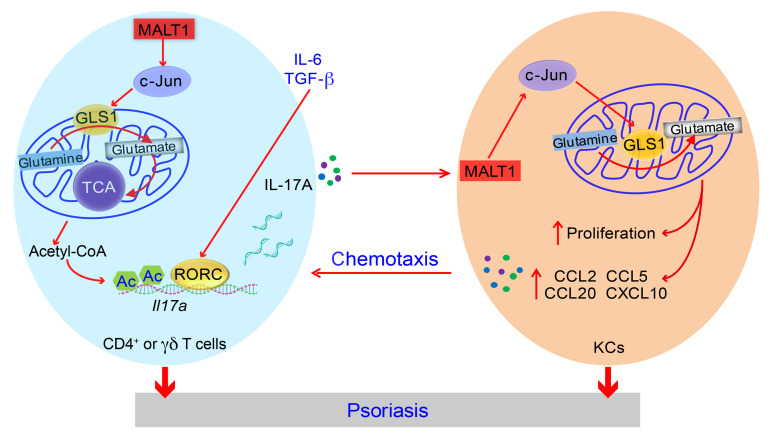
Figure 9. Schematic illustration of GLS1-mediated glutaminolysis contributing to the pathogenesis of psoriasis.
In patients with psoriasis, consecutive activation of MALT1 protease stabilizes c-Jun, which binds to the GLS1 promoter region and increases expression of GLS1, thus leading to aberrant glutaminolysis. GLS1-mediated glutaminolysis augments intracellular acetyl-CoA via the TCA cycle, which contributes to histone H3 acetylation of the IL17A promoter and RORC transcriptional activity, thereby aggravating T17 cell differentiation in psoriasis. On the other hand, IL-17A/MALT1/c-Jun axis–induced GLS1-mediated glutaminolysis also enhances the proliferation of and chemokine secretion by keratinocytes, which increases the recruitment of activated T cells in skin lesions. Together, the effects of the aberrant MALT1/c-Jun/GLS1 pathway on CD4+ and γδ T cells, together with keratinocytes contribute to the formation of psoriatic skin lesions. KCs, keratinocytes.
Methods
Further information can be found in the Supplemental Methods.
Human subjects.
A total of 36 patients with psoriasis who were diagnosed with psoriasis vulgaris by pathologic examination were recruited from outpatient clinics of the First Affiliated Hospital of Jinan University. Psoriasis disease activity was assessed using the PASI score. Patient information is provided in Supplemental Table 1. A total of 36 sex- and age-matched healthy controls were recruited from the medical staff at the First Affiliated Hospital of Jinan University. Psoriatic skin samples were obtained by punch biopsy under local lidocaine anesthesia. Normal skin specimens were taken from healthy adults undergoing plastic surgery. The fresh tissue samples were snap-frozen in liquid nitrogen and stored at –80°C or fixed in formalin and embedded in paraffin.
Mice.
WT, Tcrd−/−, and Tcrb−/− mice on a C57BL/6J background were purchased from The Jackson Laboratory and inbred at our facility. MALT1 protease–deficient (KI) mice on a C57BL/6J background were generated by Cyagen Biosciences and have been previously described (33). All mice were housed and maintained under specific pathogen–free conditions at Jinan University (Guangzhou, China).
IMQ-induced psoriasis-like mouse model.
Female WT, Tcrd−/−, Tcrb−/−, and MALT1-KI mice (8–10 weeks of age) were treated with a daily topical dose of 62.5 mg IMQ cream (5%) (Sichuan Med-shine Pharmaceuticals) on the shaved back for 7 consecutive days. Control mice were treated with the same dose of vehicle cream. For in vivo administration of the GLS1 inhibitor, mice were injected intraperitoneally with a total of 100 μg BPTES (Selleckchem) or DMSO in PBS every other day, 4 times, during the application of IMQ. For adoptive transfer of naive CD4+ or γδ T cells, Tcrb−/− or Tcrd−/− recipient mice were administered naive CD4+ or γδ T cells (2 × 106) pretreated or not with BPTES (suspended in 200 μL sterile PBS) via tail vein injection. Control Tcrb−/− or Tcrd−/− mice were injected with 200 μL PBS. Five days later, the reconstituted mice were subjected to induction of psoriasis-like disease for the mouse model. All mice were sacrificed, and samples were collected for analysis on day 8. For histopathology, tissues from the euthanized animals were fixed in formalin and embedded in paraffin. Sections (5 μm thickness) were stained with H&E. For skin inflammation, an objective scoring system was developed to evaluate the severity of back skin inflammation in the IMQ-induced psoriasis-like mouse model, and this system was based on the clinical PASI score for psoriasis patients. Erythema, scaling, and thickening were scored independently on a scale of 0 to 4: 0, none; 1, slight; 2, moderate; 3, marked; 4, very marked. The total score was obtained by adding the 3 index scores (score of 0–12). For the measurement of acanthosis, the epidermal area was outlined, and its pixel size was measured using the lasso tool in Adobe Photoshop CS4. The relative area of the epidermis was calculated using the following formula: area = pixels/(horizontal resolution × vertical resolution). For counting of dermis-infiltrating CD4+ or γδ T cells, we used the following formula: absolute cell numbers = percentage of CD3+ cells × percentage of CD4+ or γδ + T cells × total cell numbers.
In vitro human T cell differentiation.
Human naive CD4+ T cells were isolated with an EasySep Human Naive CD4+ T Cell Isolation Kit (STEMCELL Technologies). The purity of the enriched subset was validated by flow cytometry and was generally higher than 95%. Cells were stimulated with plate-bound anti–human CD3 (5 μg/mL, Sungene Biotech, H10034-14B) and anti–human CD28 (1 μg/mL, Sungene Biotech, H10281-14B) for 5 days under different polarizing conditions. For Th0 polarization, anti–IFN-γ (10 μg/mL, BioLegend, 506513) and anti–IL-4 (10 μg/mL, BioLegend, 500707) were added. For Th1 polarization, IL-12 (10 ng/mL, PeproTech) and anti–IL-4 (10 μg/mL) were added. For Th17 polarization, IL-1β (10 ng/mL, PeproTech), IL-6 (20 ng/mL, PeproTech), IL-23 (20 ng/mL, PeproTech), TGF-β 1.2 (5 ng/mL, R&D Systems), anti–IFN-γ (10 μg/mL, BioLegend, 506513), and anti–IL-4 (10 μg/mL, BioLegend, 500707) were added. For iTreg polarization, IL-2 (10 ng/mL, PeproTech) and TGF-β1.2 (5 ng/mL) were added. Cell culturing was performed in 48-well plates with a total volume of 0.5 mL/well culture medium with 2.5 × 105 naive CD4+ T cells. The medium was refreshed on day 3.
In vitro mouse T cell differentiation.
Mouse naive splenic CD4+ T cells were prepared using the Naive CD4+ T Cell Isolation Kit (STEMCELL Technologies). Mouse naive γδ T cells were isolated as previously described (62). The purity of the enriched subset was validated by flow cytometry and was generally higher than 95%. Naive CD4+ T cells were activated by plate-bound anti–mouse CD3 (10 μg/mL, Sungene Biotech, M10034-14B) and anti–mouse CD28 (1 μg/mL, Sungene Biotech, M10282-14B) for 5 days under conditions with the following cytokines and antibodies: anti–IFN-γ (10 μg/mL, BioLegend, 517904) and anti–IL-4 (10 μg/mL, BioLegend, 504122) for Th0; IL-12 (10 ng/mL, PeproTech) and anti–IL-4 (10 μg/mL, BioLegend, 504122) for Th1; TGF-β1.2 (2 ng/mL, R&D Systems), IL-6 (10 ng/mL, PeproTech), IL-1β (10 ng/mL, PeproTech), IL-23 (20 ng/mL, PeproTech), anti–IFN-γ (10 μg/mL, BioLegend, 517904) and anti–IL-4 (10 μg/mL, BioLegend, 504122) for Th17; TGF-β1.2 (5 ng/mL, R&D Systems), IL-2 (10 ng/mL, PeproTech), and anti–IFN-γ (5 μg/mL, BioLegend, 517904) and anti–IL-4 (5 μg/mL, BioLegend, 504122) for iTregs. Naive γδ T cells were cultured in anti–mouse γδ TCR–coated plates (10 μg/mL, BioLegend, 107502) with soluble anti–mouse CD28 (1 μg/mL, Sungene Biotech, M10282-14B) and IL-2 (2 ng/mL, PeproTech). IL-12 (5 ng/mL, Peprotech) and anti–IL-4 (10 μg/mL, BioLegend, 504122) were added for γδ T1-skewing conditions, and IL-1β (5 ng/mL, PeproTech, 504122), IL-23 (5 ng/mL, PeproTech), and anti-IFN-γ (5 μg/mL, BioLegend, 517904) were added for γδ T17-skewing conditions. We performed cell culturing in 48-well plates with a total volume of 0.5 ml/well of culture medium with 2.5 × 105 cells. The medium was refreshed on day 3.
LC-MS.
Metabolites were prepared as described previously (63). Briefly, cells were frozen in liquid N2 and extracted with 40% acetonitrile/40% methanol/20% H2O. Extracts were vortexed at 4 °C for 1 hour and centrifuged at 4 °C for 10 minutes at approximately 18,000 g in an Eppendorf microfuge. Supernatants were evaporated in a Speedvac (Acid-Resistant CentriVap Vacuum Concentrators, Labconco), and samples were resolved by Agilent 1290 Infinity LC. Analysis was performed in negative modes at the optimum polarity in MRM mode on an electrospray ionization (ESI) triple-quadrupole mass spectrometer (AB Sciex 5600 QTRAP). Principal component analysis and heatmaps were generated using Mass Profiler Professional (Agilent Technologies).
RNA extraction, cDNA synthesis, and quantitative real-time PCR.
Cells or skin tissue samples were collected in 500 μL TRIzol (Invitrogen, Thermo Fisher Scientific), and RNA was extracted with a RNA Simple Total RNA Kit (TIANGEN Biotech) according to the manufacturer’s protocols. RNA from each sample (300–500 ng) was reverse transcribed with the PrimeScript RT Reagent Kit (Takara). The cDNA was diluted 1:10 in RNase/DNAse-free water for quantitative real-time PCR (qRT-PCR). qRT–PCR reactions were run on a CFX Connect System (Bio-Rad) in duplicate. 2× SYBR Green qPCR Master Mix (Bimake) was used according to the manufacturer’s instructions, and each PCR reaction had a final volume of 20 μL and 5 μL diluted cDNA. The running conditions were 5 minutes at 95°C, followed by 45 cycles of 5 seconds at 95°C and 30 seconds at 60°C, and then 15 seconds at 95°C, 1 minute at 60°C, and 15 seconds at 95°C. A melting curve was obtained for each PCR product after each run to control for primer dimmers and gene-specific peaks. GAPDH was used as the housekeeping gene. The list of primer sequences is provided in Supplemental Table 2.
ChIP-qPCR.
ChIPs were performed according to the protocol for the SimpleCHIP Enzymatic Chromatin IP Kit (Cell Signaling Technology [CST]). Briefly, cells (4 × 106 to 1 × 107) were fixed for 10 minutes at 25°C with 1% formaldehyde, followed by addition of glycine to achieve a final concentration of 0.125 M to quench formaldehyde. Subsequently, cells were lysed and chromatin was harvested and fragmented (150–900 bp) using enzymatic digestion. The chromatin was then subjected to immunoprecipitation with anti-H3K9Ac (CST, 9649), anti-H3K27Ac (CST, 8173), anti-H3 (CST, 4620), anti–c-Jun (CST, 9165), anti–c-Fos (CST, 2250), and anti-RORC (Abcam, ab80690) antibodies at 4°C overnight, followed by incubation with protein G magnetic beads at 4°C for 2 hours. Normal rabbit IgG (CST, 2729) was used as a negative control. The immune complexes were washed and eluted. Eluted DNA and 2% input DNA were incubated at 65°C to reverse the cross-linking and then purified with spin columns. The abundance of precipitated DNA fragments was quantified by qPCR using primers for the indicated loci. The results are presented as being relative to the total chromatin input or normalized to total histone H3 levels to account for the nucleosomal occupancy at Il17a promoter regions in some experiments. The primer sequences are provided in Supplemental Table 3.
Statistics.
Statistical analysis was performed using GraphPad Prism 6.0 (GraphPad Software). Data are presented as the mean ± SD. Data were assessed for normal distribution and similar variances between groups. Statistical significance was assessed using a 2-tailed, unpaired Student’s t test for comparisons between 2 groups and 1-way ANOVA with relevant post hoc tests for multiple comparisons. When the data were not normally distributed or displayed unequal variances between 2 groups, the 2-tailed Mann-Whitney U test was used for statistical analysis. Correlation analysis was performed using Spearman’s r test (for abnormally distributed data). No statistical method was used to predetermine the sample size. Animals were randomly allocated to the treatment groups.
Study approval.
All human studies were performed in accordance with Declaration of Helsinki principles and approved by the ethics committee of the First Affiliated Hospital of Jinan University. Written informed consent was obtained from all study subjects. All animal care protocols and experiments were approved by and performed in accordance with the guidelines of the IACUC of Jinan University.
Author contributions
YG and ZY conceived the project and designed the study. XX, GC, GS, LZ, YT, YS, and CG performed the experiments and acquired data. XX, GC, LZ, WZ, PL, and HZ performed computational analyses. LD, GS, JZ, ZL, and XW provided the peripheral blood and skin tissue samples from healthy subjects and patients with psoriasis. YG, JH, and ZY contributed to preparation of the manuscript, and XX and GC wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported by the Postdoctoral Fund of the First Affiliated Hospital of Jinan University (grant 809007, to XX); the National Natural Science Foundation of China (grant 31770964, to YG, 31830021, to ZY, 31700753, to GC, 31970862, to GS, and 31970830, to JH); the Science and Technology Program of Guangzhou (grant 201604020162, to YG); and the 111 Project (grant B16021, to ZY and YG).
Version 1. 08/24/2020
Electronic publication
Version 2. 10/01/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interests exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(10):5180–5196.https://doi.org/10.1172/JCI129269.
Contributor Information
Xichun Xia, Email: season_xia@126.com.
Guangchao Cao, Email: ershao@mail.nankai.edu.cn.
Guodong Sun, Email: sgd96@jnu.edu.cn.
Leqing Zhu, Email: zhuleqing1@163.com.
Yixia Tian, Email: 786750241@qq.com.
Yueqi Song, Email: 13631342368@163.com.
Chengbin Guo, Email: 609250340@qq.com.
Xiao Wang, Email: 673249535@qq.com.
Jingxiang Zhong, Email: zjx85221206@163.com.
Wei Zhou, Email: 291446761@qq.com.
Peng Li, Email: 474029194@qq.com.
Hua Zhang, Email: 945786272@qq.com.
Jianlei Hao, Email: haojianlei@jnu.edu.cn.
Zhizhong Li, Email: tzzli@jnu.edu.cn.
Liehua Deng, Email: Liehuadeng@126.com.
Zhinan Yin, Email: zhinan.yin@yale.edu.
Yunfei Gao, Email: tyunfeigao@jnu.edu.cn.
References
- 1.Lowes MA, Bowcock AM, Krueger JG. Pathogenesis and therapy of psoriasis. Nature. 2007;445(7130):866–873. doi: 10.1038/nature05663. [DOI] [PubMed] [Google Scholar]
- 2.Mak RK, Hundhausen C, Nestle FO. Progress in understanding the immunopathogenesis of psoriasis. Actas Dermosifiliogr. 2009;100(Suppl 2):2–13. doi: 10.1016/s0001-7310(09)73372-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becher B, Pantelyushin S. Hiding under the skin: Interleukin-17-producing γδ T cells go under the skin? Nat Med. 2012;18(12):1748–1750. doi: 10.1038/nm.3016. [DOI] [PubMed] [Google Scholar]
- 4.Gutcher I, Becher B. APC-derived cytokines and T cell polarization in autoimmune inflammation. J Clin Invest. 2007;117(5):1119–1127. doi: 10.1172/JCI31720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krueger JG. Hiding under the skin: A welcome surprise in psoriasis. Nat Med. 2012;18(12):1750–1751. doi: 10.1038/nm.3025. [DOI] [PubMed] [Google Scholar]
- 6.Duhen T, Geiger R, Jarrossay D, Lanzavecchia A, Sallusto F. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat Immunol. 2009;10(8):857–863. doi: 10.1038/ni.1767. [DOI] [PubMed] [Google Scholar]
- 7.Kagami S, Rizzo HL, Lee JJ, Koguchi Y, Blauvelt A. Circulating Th17, Th22, and Th1 cells are increased in psoriasis. J Invest Dermatol. 2010;130(5):1373–1383. doi: 10.1038/jid.2009.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greb JE, et al. Psoriasis. Nat Rev Dis Primers. 2016;2:16082. doi: 10.1038/nrdp.2016.82. [DOI] [PubMed] [Google Scholar]
- 9.Papp K, et al. Tildrakizumab (MK-3222), an anti-interleukin-23p19 monoclonal antibody, improves psoriasis in a phase IIb randomized placebo-controlled trial. Br J Dermatol. 2015;173(4):930–939. doi: 10.1111/bjd.13932. [DOI] [PubMed] [Google Scholar]
- 10.Gordon KB, et al. Adalimumab retreatment successfully restores clinical response and health-related quality of life in patients with moderate to severe psoriasis who undergo therapy interruption. J Eur Acad Dermatol Venereol. 2015;29(4):767–776. doi: 10.1111/jdv.12677. [DOI] [PubMed] [Google Scholar]
- 11.Griffiths CE, et al. Comparison of ixekizumab with etanercept or placebo in moderate-to-severe psoriasis (UNCOVER-2 and UNCOVER-3): results from two phase 3 randomised trials. Lancet. 2015;386(9993):541–551. doi: 10.1016/S0140-6736(15)60125-8. [DOI] [PubMed] [Google Scholar]
- 12.Lebwohl M, et al. Phase 3 studies comparing brodalumab with ustekinumab in psoriasis. N Engl J Med. 2015;373(14):1318–1328. doi: 10.1056/NEJMoa1503824. [DOI] [PubMed] [Google Scholar]
- 13.Yang C, et al. Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol Cell. 2014;56(3):414–424. doi: 10.1016/j.molcel.2014.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheong H, Lu C, Lindsten T, Thompson CB. Therapeutic targets in cancer cell metabolism and autophagy. Nat Biotechnol. 2012;30(7):671–678. doi: 10.1038/nbt.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kono M, Yoshida N, Maeda K, Tsokos GC. Transcriptional factor ICER promotes glutaminolysis and the generation of Th17 cells. Proc Natl Acad Sci USA. 2018;115(10):2478–2483. doi: 10.1073/pnas.1714717115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson MO, et al. Distinct regulation of Th17 and Th1 cell differentiation by glutaminase-dependent metabolism. Cell. 2018;175(7):1780–1795.e19. doi: 10.1016/j.cell.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Armstrong AW, et al. Metabolomics in psoriatic disease: pilot study reveals metabolite differences in psoriasis and psoriatic arthritis. F1000Res. 2014;3:248. doi: 10.12688/f1000research.4709.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamleh MA, et al. LC-MS metabolomics of psoriasis patients reveals disease severity-dependent increases in circulating amino acids that are ameliorated by anti-TNFα treatment. J Proteome Res. 2015;14(1):557–566. doi: 10.1021/pr500782g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang H, et al. Exploration of candidate biomarkers for human psoriasis based on gas chromatography-mass spectrometry serum metabolomics. Br J Dermatol. 2017;176(3):713–722. doi: 10.1111/bjd.15008. [DOI] [PubMed] [Google Scholar]
- 20.Araujo L, Khim P, Mkhikian H, Mortales CL, Demetriou M. Glycolysis and glutaminolysis cooperatively control T cell function by limiting metabolite supply to N-glycosylation. Elife. 2017;6:e21330. doi: 10.7554/eLife.21330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diani M, Altomare G, Reali E. T helper cell subsets in clinical manifestations of psoriasis. J Immunol Res. 2016;2016:7692024. doi: 10.1155/2016/7692024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui Y, et al. Major role of gamma delta T cells in the generation of IL-17+ uveitogenic T cells. J Immunol. 2009;183(1):560–567. doi: 10.4049/jimmunol.0900241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389(6649):349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 24.Nakaya M, et al. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity. 2014;40(5):692–705. doi: 10.1016/j.immuni.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng M, Yin N, Chhangawala S, Xu K, Leslie CS, Li MO. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science. 2016;354(6311):481–484. doi: 10.1126/science.aaf6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruan Q, et al. The Th17 immune response is controlled by the Rel-RORγ-RORγ T transcriptional axis. J Exp Med. 2011;208(11):2321–2333. doi: 10.1084/jem.20110462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu HP, et al. TGF-β converts Th1 cells into Th17 cells through stimulation of Runx1 expression. Eur J Immunol. 2015;45(4):1010–1018. doi: 10.1002/eji.201444726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valeri M, Raffatellu M. Cytokines IL-17 and IL-22 in the host response to infection. Pathog Dis. 2016;74(9):ftw111. doi: 10.1093/femspd/ftw111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamilton KS, et al. T cell receptor-dependent activation of mTOR signaling in T cells is mediated by Carma1 and MALT1, but not Bcl10. Sci Signal. 2014;7(329):ra55. doi: 10.1126/scisignal.2005169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brüstle A, et al. The NF-κB regulator MALT1 determines the encephalitogenic potential of Th17 cells. J Clin Invest. 2012;122(12):4698–4709. doi: 10.1172/JCI63528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeltsch KM, et al. Cleavage of roquin and regnase-1 by the paracaspase MALT1 releases their cooperatively repressed targets to promote T(H)17 differentiation. Nat Immunol. 2014;15(11):1079–1089. doi: 10.1038/ni.3008. [DOI] [PubMed] [Google Scholar]
- 32.Xia X, et al. Glutaminolysis mediated by MALT1 protease activity facilitates PD-L1 expression on ABC-DLBCL cells and contributes to their immune evasion. Front Oncol. 2018;8:632. doi: 10.3389/fonc.2018.00632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H, et al. Inhibition of MALT1 paracaspase activity improves lesion recovery following spinal cord injury. Sci Bull (Beijing) 2019;64(16) doi: 10.1016/j.scib.2019.04.026. [DOI] [PubMed] [Google Scholar]
- 34.Lukey MJ, Greene KS, Erickson JW, Wilson KF, Cerione RA. The oncogenic transcription factor c-Jun regulates glutaminase expression and sensitizes cells to glutaminase-targeted therapy. Nat Commun. 2016;7:11321. doi: 10.1038/ncomms11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458(7239):762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang M, et al. Gain-of-function mutation of Card14 leads to spontaneous psoriasis-like skin inflammation through enhanced keratinocyte response to IL-17A. Immunity. 2018;49(1):66–79.e5. doi: 10.1016/j.immuni.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 37.Mellett M, et al. CARD14 gain-of-function mutation alone is sufficient to drive IL-23/IL-17-mediated psoriasiform skin inflammation in vivo. J Invest Dermatol. 2018;138(9):2010–2023. doi: 10.1016/j.jid.2018.03.1525. [DOI] [PubMed] [Google Scholar]
- 38.Nestle FO, Kaplan DH, Barker J. Psoriasis. N Engl J Med. 2009;361(5):496–509. doi: 10.1056/NEJMra0804595. [DOI] [PubMed] [Google Scholar]
- 39.Barbi J, Pardoll D, Pan F. Metabolic control of the Treg/Th17 axis. Immunol Rev. 2013;252(1):52–77. doi: 10.1111/imr.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12(5):325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 42.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu R, et al. MicroRNA-210 overexpression promotes psoriasis-like inflammation by inducing Th1 and Th17 cell differentiation. J Clin Invest. 2018;128(6):2551–2568. doi: 10.1172/JCI97426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab. 2012;16(1):9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chisolm DA, Weinmann AS. Connections between metabolism and epigenetics in programming cellular differentiation. Annu Rev Immunol. 2018;36:221–246. doi: 10.1146/annurev-immunol-042617-053127. [DOI] [PubMed] [Google Scholar]
- 46.Zhang P, Su Y, Zhao M, Huang W, Lu Q. Abnormal histone modifications in PBMCs from patients with psoriasis vulgaris. Eur J Dermatol. 2011;21(4):552–557. doi: 10.1684/ejd.2011.1383. [DOI] [PubMed] [Google Scholar]
- 47.Mc Guire C, et al. Paracaspase MALT1 deficiency protects mice from autoimmune-mediated demyelination. J Immunol. 2013;190(6):2896–2903. doi: 10.4049/jimmunol.1201351. [DOI] [PubMed] [Google Scholar]
- 48.Thome M. Multifunctional roles for MALT1 in T-cell activation. Nat Rev Immunol. 2008;8(7):495–500. doi: 10.1038/nri2338. [DOI] [PubMed] [Google Scholar]
- 49.Nograles KE, et al. Th17 cytokines interleukin (IL)-17 and IL-22 modulate distinct inflammatory and keratinocyte-response pathways. Br J Dermatol. 2008;159(5):1092–1102. doi: 10.1111/j.1365-2133.2008.08769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Büchau AS, Gallo RL. Innate immunity and antimicrobial defense systems in psoriasis. Clin Dermatol. 2007;25(6):616–624. doi: 10.1016/j.clindermatol.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Israël L, et al. A CARD10-dependent tonic signalosome activates MALT1 paracaspase and regulates IL-17/TNF-α-driven keratinocyte inflammation. J Invest Dermatol. 2018;138(9):2075–2079. doi: 10.1016/j.jid.2018.03.1503. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Z, et al. Differential glucose requirement in skin homeostasis and injury identifies a therapeutic target for psoriasis. Nat Med. 2018;24(5):617–627. doi: 10.1038/s41591-018-0003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bartlett BL, Tyring SK. Ustekinumab for chronic plaque psoriasis. Lancet. 2008;371(9625):1639–1640. doi: 10.1016/S0140-6736(08)60702-3. [DOI] [PubMed] [Google Scholar]
- 54.Speeckaert R, van Geel N, Lambert J, Claeys L, Delanghe JR, Speeckaert MM. Secukinumab: IL-17A inhibition to treat psoriatic arthritis. Drugs Today. 2016;52(11):607–616. doi: 10.1358/dot.2016.52.11.2526754. [DOI] [PubMed] [Google Scholar]
- 55.Okazaki A, et al. Glutaminase and poly(ADP-ribose) polymerase inhibitors suppress pyrimidine synthesis and VHL-deficient renal cancers. J Clin Invest. 2017;127(5):1631–1645. doi: 10.1172/JCI87800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elgogary A, et al. Combination therapy with BPTES nanoparticles and metformin targets the metabolic heterogeneity of pancreatic cancer. Proc Natl Acad Sci USA. 2016;113(36):E5328–E5336. doi: 10.1073/pnas.1611406113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shukla K, et al. Design, synthesis, and pharmacological evaluation of bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide 3 (BPTES) analogs as glutaminase inhibitors. J Med Chem. 2012;55(23):10551–10563. doi: 10.1021/jm301191p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeLaBarre B, Hurov J, Cianchetta G, Murray S, Dang L. Action at a distance: allostery and the development of drugs to target cancer cell metabolism. Chem Biol. 2014;21(9):1143–1161. doi: 10.1016/j.chembiol.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 59.Fontan L, et al. MALT1 small molecule inhibitors specifically suppress ABC-DLBCL in vitro and in vivo. Cancer Cell. 2012;22(6):812–824. doi: 10.1016/j.ccr.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagel D, et al. Pharmacologic inhibition of MALT1 protease by phenothiazines as a therapeutic approach for the treatment of aggressive ABC-DLBCL. Cancer Cell. 2012;22(6):825–837. doi: 10.1016/j.ccr.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 61.Mc Guire C, et al. Pharmacological inhibition of MALT1 protease activity protects mice in a mouse model of multiple sclerosis. J Neuroinflammation. 2014;11:124. doi: 10.1186/1742-2094-11-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang Q, et al. Roles of mTORC1 and mTORC2 in controlling γδ T1 and γδ T17 differentiation and function. Cell Death Differ. 2020;27(7):2248–2262. doi: 10.1038/s41418-020-0500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banh RS, et al. PTP1B controls non-mitochondrial oxygen consumption by regulating RNF213 to promote tumour survival during hypoxia. Nat Cell Biol. 2016;18(7):803–813. doi: 10.1038/ncb3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.