Abstract
Pediatric and adult high-grade gliomas (HGGs) frequently harbor PDGFRA alterations. We hypothesized that cotreatment with everolimus may improve the efficacy of dasatinib in PDGFRα-driven glioma through combinatorial synergism and increased tumor accumulation of dasatinib. We performed dose-response, synergism, P-glycoprotein inhibition, and pharmacokinetic studies in in vitro and in vivo human and mouse models of HGG. Six patients with recurrent PDGFRα-driven glioma were treated with dasatinib and everolimus. We found that dasatinib effectively inhibited the proliferation of mouse and human primary HGG cells with a variety of PDGFRA alterations. Dasatinib exhibited synergy with everolimus in the treatment of HGG cells at low nanomolar concentrations of both agents, with a reduction in mTOR signaling that persisted after dasatinib treatment alone. Prolonged exposure to everolimus significantly improved the CNS retention of dasatinib and extended the survival of PPK tumor–bearing mice (mutant TP53, mutant PDGFRA, H3K27M). Six pediatric patients with glioma tolerated this combination without significant adverse events, and 4 patients with recurrent disease (n = 4) had a median overall survival of 8.5 months. Our results show that the efficacy of dasatinib treatment of PDGFRα-driven HGG was enhanced with everolimus and suggest a promising route for improving targeted therapy for this patient population.
Keywords: Oncology
Keywords: Brain cancer, Molecular biology, Pharmacology
Introduction
High-grade gliomas (HGGs) are common and aggressive pediatric and adult brain tumors. The median overall survival (OS) of adult patients diagnosed with glioblastoma (GBM), grade IV glioma, is 12.6 months (1), and the median OS for pediatric HGG is 14.1 months (2). Established therapies for adult and pediatric HGG are not currently targeted to the unique molecular attributes of each tumor, and patients frequently consider experimental therapies after up-front radiation and at recurrence.
One of the most frequently altered genes in HGG is platelet-derived growth factor receptor α (PDGFRA). PDGFRA is 1 of 2 PDGF receptor subunits, and it interacts with at least 4 PDGF ligands (3). It has been shown that PDGF activation induces multiple cellular activities including cell proliferation, transformation, migration, and survival (3). PDGFRA is mutated or amplified in 12% of adult GBMs and 21% of pediatric HGGs (4, 5). Within childhood HGGs, PDGFRA mutation is correlated with older age (10–30 years of age), whereas amplification is more commonly seen in younger patients (5–10 years of age) (5). PDGFRA alterations are drivers of aggressive glioma behavior and are associated with worse prognoses in pediatric non-brainstem HGG and adult anaplastic astrocytoma (WHO grade III glioma) (5, 6).
Preclinically, there are multiple effective agents targeting PDGFRA in glioma, including the tyrosine kinase inhibitor (TKI) dasatinib (5, 7). Unfortunately, monotherapy with dasatinib has failed to improve outcomes in adult HGG, even when gliomas were selected for by PDGFRα overexpression using IHC (8). The failure of dasatinib may be related to its use as a single agent. Recent data in other solid tumors have demonstrated synergism between TKIs and the mTOR inhibitor everolimus, including the clinical success of lenvatinib and everolimus for renal cell carcinoma (9). Osteosarcoma preclinical models treated with sorafenib overcame resultant upregulation of mTOR complex 1 (mTORC1) through cotreatment with everolimus (10).
Additionally, treatment with dasatinib for CNS tumors may be limited by CNS penetration. Given its intrinsic qualities (i.e., lipophilicity, size, and protein binding), dasatinib is a promising agent for CNS penetration (11). Although the ability of dasatinib to penetrate the blood-brain barrier (BBB) is promising, its retention is limited by the cerebrospinal fluid (CSF) efflux protein P-glycoprotein (P-gp) and breast cancer resistance protein (BCRP) (12). Recent work has demonstrated that coadministration of TKIs with agents that inhibit P-gp and BCRP1 can improve brain retention of the TKI (12–14). Everolimus has been shown to improve CNS retention of the TKI vandetanib (13).
To our knowledge, coadministration of dasatinib and everolimus has never been tested in CNS tumor models or administered to patients with PDGFRα-driven gliomas. In this study, we assessed whether cotreatment of dasatinib with everolimus could improve the efficacy and CNS retention of dasatinib in PDGFRα-driven gliomas in both preclinical models and patients. We found that everolimus acted synergistically with dasatinib in PDGFRα-driven glioma. We also found that sustained everolimus administration resulted in substantial increases in tumor concentrations of dasatinib in a genetically engineered HGG animal model. Our preliminary data on 6 patients who received dasatinib and everolimus combination therapy establish its promising feasibility. Furthermore, our findings represent a route for enhancing the efficacy of TKI precision therapy for pediatric and adult HGGs.
Results
Generation of TP53, PDGFRA, and H3K27M mutant glioma primary cell cultures and in vitro treatment with dasatinib.
In order to study the utility of dasatinib in PDGFRα-driven HGG, we adapted an intrauterine electroporation (IUE) mouse model (ref. 15 and Figure 1A). We induced glioma in mice by injecting plasmids encoding: (a) PBase; (b) PB-CAG-DNp53-Ires-luciferase (TP53); (c) PB-CAG-PdgfraD824V-Ires-EGFP (PDGFRA D842V); and (d) PB-CAG-H3.3 K27M-Ires-EGFP (H3K27M) into the lateral ventricles (forebrain) of E13.5 embryos in CD1 mice. Transfection efficiency and tumor growth were monitored by in vivo imaging of luminescence (Figure 1A). With the injection of PPK (mutant TP53, mutant PDGFRA, H3K27M) plasmids, mice developed high-grade, invasive glial tumors (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI133310DS1) with a high penetrance (>90%) and a median survival of 52 days. IHC analysis confirmed upregulation of total PDGFRα, tumor-specific expression of H3.3K27M, and reduced expression of H3.3K27me3, as expected for H3.3K27M-mutant tumors (ref. 16 and Supplemental Figure 1B). Multiple TP53, PDGFRA, and H3K27M (PPK) mutant glioma primary neurosphere cultures were generated from the dissociation of intracranial tumor tissue from these mice (Figure 1A), with confirmed mutations in cultured neurospheres (Supplemental Figure 1C).
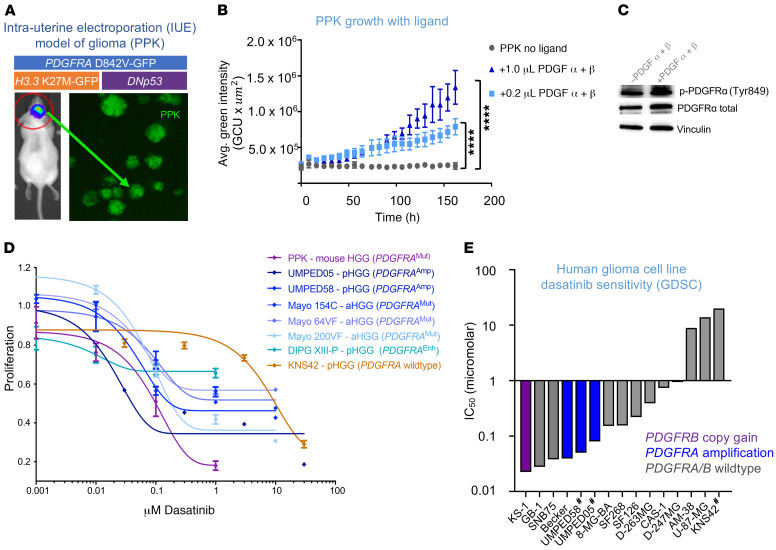
Figure 1. PDGFRα-driven glioma demonstrates sensitivity to dasatinib in vitro.
(A) HGG cells were generated from an IUE model with plasmids of TP53, PDGFRA, and H3K27M mutations. (B) Proliferation of IUE HGG cells in response to addition of PDGF α and β ligands in vitro (****P ≤ 0.0001, by Dunnett’s multiple comparisons test). n = 3 technical replicates. Avg., average. (C) Western blot analysis of p-PDGFRα and total PDGFRα in response to the addition of PDGF ligands. (D) Dose-response curve of PPK neurospheres, cultured cells from 6 human PDGFRα-driven glioma cell lines, and cells from 1 PDGFRA-WT glioma cell line following treatment with dasatinib. n = 3 technical replicates. (E) Plot of glioma cell culture sensitivity to dasatinib according to the GDSC database. Analysis of PDGFRA, PDGFA, PDGFRB, and PDGFB DNA alterations is referenced (#non-GDSC data generated from primary cell culture treatment for this study). Data represent the mean ± SEM for B and D.
Previous studies have demonstrated that tumor cells driven by the PDGFRA D842V mutation may be relatively PDGF α and β ligand insensitive (as compared with PDGFRA amplification) (7). However, we found a dose-dependent increase in proliferation when the cells were supplemented with PDGF α and β ligands (Figure 1B). Cell cultures showed strong expression levels of phosphorylated [p-PDGFRα (Tyr849)] and total PDGFRα, which were not significantly different with the addition of ligand (Figure 1C and Supplemental Figure 1D). UMPED58, a brainstem HGG primary cell culture with PDGFRA amplification, also showed increased proliferation when grown with PDGF α and β ligands (P ≤ 0.0001) (Supplemental Figure 1E).
We tested in vitro dasatinib treatment on mouse PPK and several other human pediatric HGGs with PDGFRA alteration in the following cell cultures: 2 pediatric HGG (pHGG) cell cultures with PDGFRA amplification (UMPED58 and UMPED05; ref. 5); 3 PDGFRA-mutant adult HGG (aHGG) cultures derived from patient-derived xenografts (PDXs) (17); 1 pHGG cell culture with enhancer-mediated PDGFRA upregulation (DIPG-XIII-P; ref. 18); and 1 pediatric HGG cell culture without any growth factor receptor alteration (KNS42) as a control (5). PPK cells with a PDGFRA D842V mutation showed sensitivity to monotherapy with dasatinib, with an IC50 of 100 nM (Figure 1D). All the pHGG cell cultures also demonstrated different degrees of nanomolar sensitivity to dasatinib; UMPED05 (PDGFRA amplification) showed maximum sensitivity, and KNS42 with WT PDGFRA was the least sensitive (Figure 1D). We analyzed publicly available data on human glioma cell culture treated with dasatinib (n = 11) from the Genomics of Drug Sensitivity in Cancer (GDSC) database (19). Glioma cell cultures showed promising sensitivity to dasatinib in general (Supplemental Figure 2). Dasatinib demonstrated improved efficacy in glioma cell cultures with PDGFR alterations (Figure 1E). Collectively, these data showed that dasatinib effectively inhibits proliferation in mouse and human HGGs with various PDGFR alterations.
Addition of everolimus provides synergism with dasatinib treatment of pHGG cells.
Previous work has established that TKIs may have improved efficacy in combination with the mTOR inhibitor everolimus (9, 10). To determine whether the addition of everolimus can improve the efficacy of dasatinib, we treated PPK cells with varying concentrations of everolimus and dasatinib, which showed that at concentrations as low as 1 nM, everolimus resulted in a pronounced reduction in the dasatinib concentration required to achieve a reduction of mouse PPK cell proliferation in vitro, allowing for efficacy in PDGFRA-altered HGG cells at nanomolar concentrations of both drugs (Figure 2A). To assess whether this efficacy was improved via inhibition of the mTOR pathway, we performed Western blot analysis on mouse (PPK) and human (DIPG-XIII-P) cells treated with dasatinib and/or everolimus to measure the activity of RTK/MAPK/mTOR pathways (Figure 2B). Treatment with dasatinib alone resulted in reduced expression of p-PDGFRα/β (Tyr849/Tyr857) in a dose-dependent manner and of the RTK/MAPK targets p-Src (Tyr416) and p-ERK1/-2 (Thr202/Tyr204), even at the lowest dose (0.01 μM) (Figure 2B and Supplemental Figure 3). Everolimus effectively inhibited the mTOR pathway [p-S6 (Ser235–236)] at the lowest dose (0.01 μM). The combination of everolimus and dasatinib led to dual inhibition of mTOR (p-S6) and RTK/MAPK (p-Src) pathways at 0.01 nM dose (Figure 2B). Quantification of Western blot data showed a reduction in residual mTOR pathway (p-S6) pathway expression after dasatinib treatment alone (Figure 2C and Supplemental Figure 4). We performed a neurosphere limiting dilution assay on IUE PPK cells in the micromolar range for both dasatinib and everolimus, which indicated no enhancement of the effect beyond the cytotoxicity seen in proliferation assays at this dose, suggesting that these agents do not have an added impact on the stemness of cultured cells (Figure 2D and Supplemental Figure 5A). Using the Chou-Talalay median-effect method for drug combination analysis, everolimus and dasatinib demonstrated a synergistic effect at nearly all dose combinations, including single nanomolar doses of both agents (Figure 2E and ref. 20).
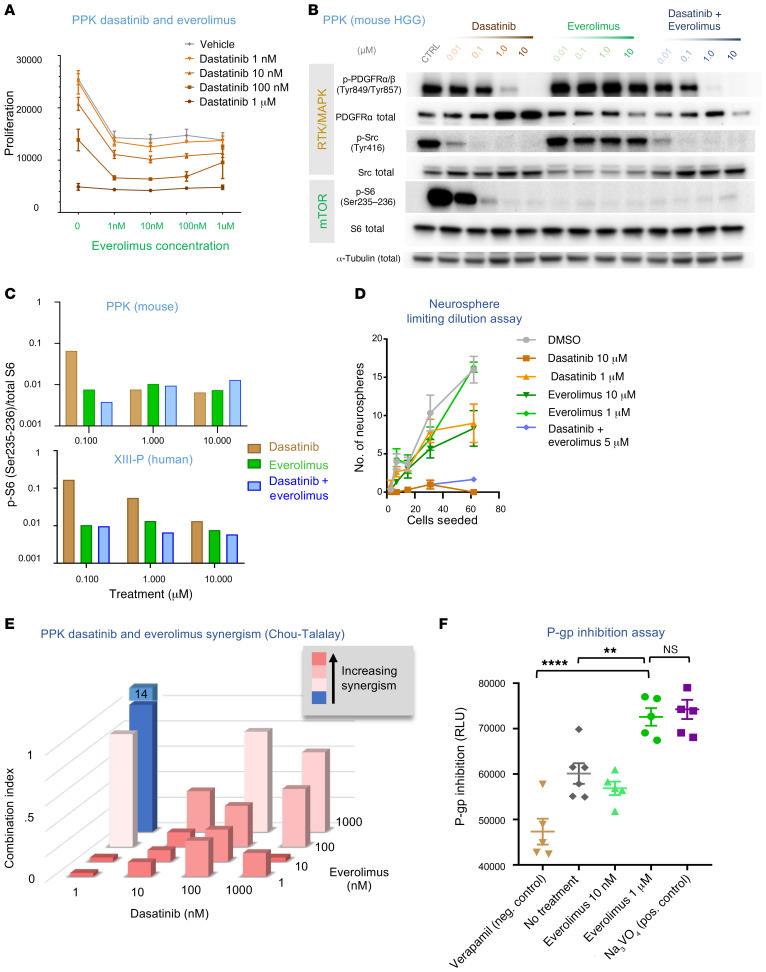
Figure 2. Addition of everolimus provides synergism and strong P-gp blockade.
(A) Viability of IUE HGG neurospheres in response to various combinations of dasatinib and everolimus. n = 3 technical replicates. (B) Western blot analysis of p-PDGFRα/β (Tyr849/Tyr857), p-Src (Tyr416), and p-S6 (Ser235–236) expression in PDGFRA-amplified mouse HGG cells treated with dasatinib or everolimus monotherapy or with coadministration of dasatinib and everolimus. In the coadministration condition, the drugs were administered at equal doses (doses are indicated at the top of the figure). CTRL, control. (C) Quantification of Western blot analysis for the mouse HGG cell line (PPK) and a human PDGFRA-enhanced cell line (XIII-P), demonstrating a greater reduction in p-S6 expression with coadministration of dasatinib and everolimus than with either drug alone at 0.1 μM in the mouse cell line and at all administered doses in the human cell line. (D) A limiting dilution assay was performed on IUE PPK neurospheres treated with either 1 μM or 10 μM doses of dasatinib or everolimus, or dasatinib and everolimus together (5 μM). n = 3 technical replicates. (E) Everolimus and dasatinib synergism CI assessment. (F) Plot of P-gp inhibition using an in vitro assay with controls (higher numbers = greater P-gp inhibition) shows higher P-gp inhibition with the 1 μM everolimus dose condition than with the negative control or no treatment conditions ( **P = 0.0045 and ****P ≤ 0.0001, by Tukey’s multiple comparisons test, respectively). Neg., negative; Pos., positive. Data represent the mean ± SEM (A, D, and F).
Everolimus provides P-gp blockade at a higher dose.
Previous work has established that dasatinib is a substrate for the efflux proteins P-gp and BCRP (21–24). As P-gp is probably the predominant substrate affecting CNS efflux (14), we focused our attention on P-gp. To assess the P-gp–inhibitory effects of everolimus, we performed an in vitro P-gp inhibition assay. Everolimus fully blocked P-gp activity at a dose of 1 μM at 4 hours (Figure 2F and Supplemental Figure 3B). Lower doses of everolimus (10 nM) did not produce an effect, which was significantly different from what was observed in nontreated cells (Figure 2F). The effect at 1 μM concentrations was similar to that seen with slightly lower-than-previous-levels of everolimus (3–5 μM) necessary for in vitro P-gp inhibition in Madin-Darby canine kidney (MDCK) cells overexpressing multidrug resistance genes (P-gp and BCRP) (25).
Pharmacokinetic analysis of dasatinib and everolimus CNS retention.
We then proceeded to test the effect of everolimus on CNS concentrations of dasatinib in nontumor-bearing mice. Mice were treated with dasatinib (10 mg/kg via tail vein injection) with or without a single dose of everolimus (5 mg/kg via oral gavage) 2 hours prior. Mice were sacrificed at 1-, 2-, 4-, and 7-hour time points (n = 3 each) for collection of serum, brainstem, and cortex (Supplemental Figure 6A). We performed a separate analysis of brain and brainstem pharmacokinetics (PK), because previous literature and clinical experience suggest that the BBB is more restrictive in the brainstem (26). We found no statistically significant differences for dasatinib concentrations between the 2 conditions (dasatinib alone and dasatinib plus everolimus) within the plasma, cortex, and brainstem (Supplemental Figure 6, B–F). Average plasma concentrations of dasatinib were higher in the cotreated mice, although this finding was not statistically significant. PK analysis of everolimus monotherapy in nontumor-bearing mice revealed an increase in everolimus concentrations (ng/g) within mouse brain tissue over time (Supplemental Figure 7, A and B).
Sustained exposure of everolimus improves dasatinib tumor delivery and increases the survival of mice bearing PPK tumors.
We then proceeded to study our IUE murine model of pHGG to assess whether the intratumoral concentration of dasatinib would be affected by cotreatment with everolimus. We performed treatment studies in mice with large cortical PPK tumors that met the enrollment luminescence criteria (average of 32 dpi). As seen in our nontumor-bearing mice, single-dose treatment of everolimus did not influence the tumor concentration of dasatinib (Figure 3A). However, with the addition of a second dose of everolimus 24 hours before dasatinib administration, tumor concentrations of dasatinib were increased in proportion to the dasatinib dose (Figure 3A). In additional tumor-bearing mice treated with 2 doses of everolimus before dasatinib administration, immediately before sacrifice, we detected reduced expression of P-gp by IHC (Supplemental Figure 8, A and B).
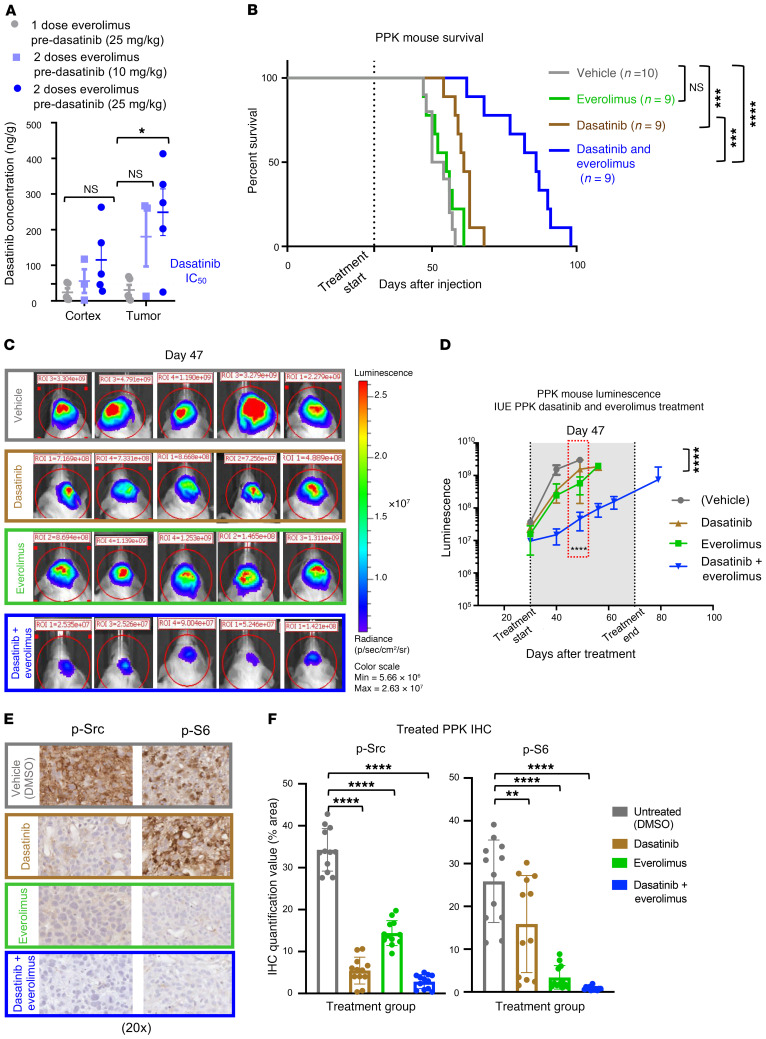
Figure 3. Sustained exposure of everolimus improves dasatinib tumor concentration and efficacy in a mouse model.
(A) PK data documenting an increase in dasatinib levels detected in tumors with an additional 24-hour everolimus pre-dasatinib dose of either 5 mg/kg or 10 mg/kg (*P ≤ 0.05, by Dunnett’s multiple comparisons test). n = 5 replicates. (B) Survival curve data for mice with PDGFRA-amplified HGG via IUE shows that the median survival for the control condition was 52 days after IUE injection (n = 10), 55 days for the everolimus condition (5 mg/kg, n = 9), 61 days for the dasatinib condition (10 mg/kg, n = 9), and 86 days for the dasatinib plus everolimus condition (10 mg/kg and 5 mg/kg, respectively, n = 9) (NS P = 0.3378, ***P ≤ 0.0005, and ****P ≤ 0.0001, by log-rank test). (C) Representative bioluminescence images of PPK mouse tumors (vehicle, n = 10; dasatinib, n = 9; everolimus, n = 9; dasatinib + everolimus, n = 10) 47 days after IUE injection, displaying lower average luminescence in the cotreatment group than in the 2 groups receiving monotherapy of dasatinib or everolimus. (D) IUE PPK bioluminescence tumor monitor growth data with statistical significance between dasatinib plus everolimus and DMSO treatment groups 47 days after IUE injection (****P ≤ 0.0001, by Tukey’s multiple comparisons test.). (E) In vivo IHC images of p-Src and p-S6 expression levels, documenting a reduction with everolimus and dasatinib treatment in comparison with nontreated tumors. Images are representative of 3 biological replicates. Original magnification, ×20. (F) IHC quantification data for treated PPK mice show statistical significance of p-Src and p-S6 expression levels between tumors treated with dasatinib and everolimus and nontreated tumors (**P = 0.0065 and ****P ≤ 0.0001, by Dunnett’s multiple comparisons test). n = 3 animals per treatment group and 4 images per animal. Data represent the mean ± SEM (A, D, and F).
To assess the effect of sustained cotreatment on survival, we treated IUE PPK mice under 4 separate conditions: vehicle, daily treatment with dasatinib alone (10 mg/kg), daily treatment with everolimus alone (5 mg/kg), or daily cotreatment with dasatinib and everolimus (10 mg/kg and 5 mg/kg, respectively). Cotreatment with dasatinib and everolimus significantly improved median survival (86 days) compared with dasatinib alone (61 days), everolimus alone (55 days), and vehicle alone (52 days) (Figure 3B). Forty-seven days after IUE injection, the dasatinib- and everolimus-treated PPK mice demonstrated reduced luminescent tumor signals when compared with control mice (Figure 3, C and D). We further analyzed tumors from each treatment group for p-Src and p-S6 expression. IHC analysis and quantification showed reductions in p-Src and p-S6 expression after dasatinib and everolimus monotherapy, with retained p-S6 expression in tumors treated with dasatinib alone (Figure 3, E and F, and Supplemental Figure 9, A and B).
Treatment of children and young adults with relapsed PDGFRα-driven glioma with dasatinib and everolimus.
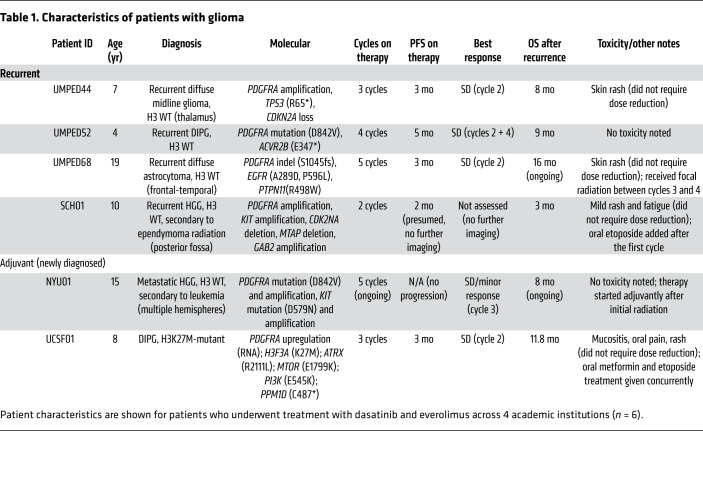
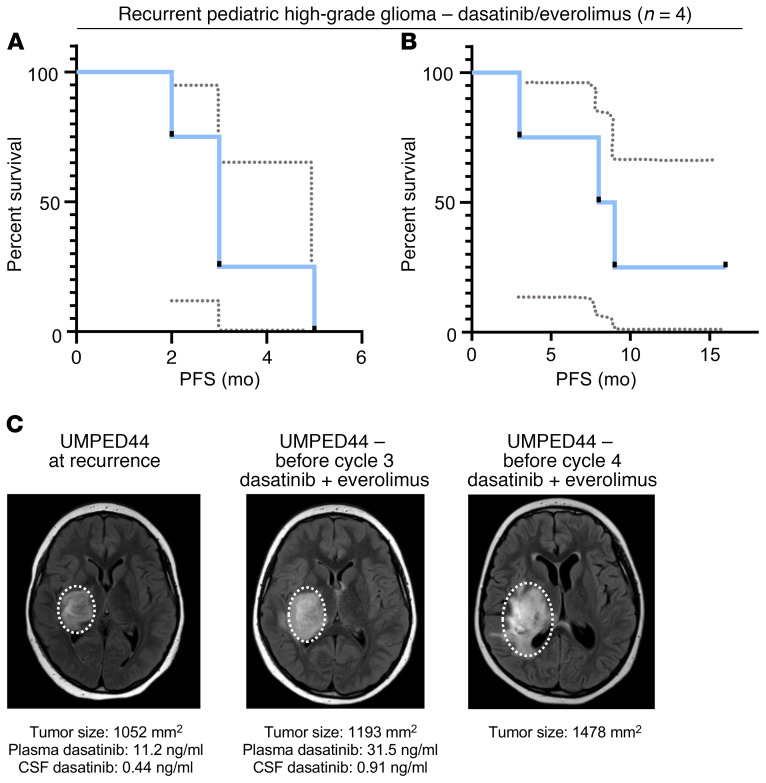
Patients with recurrent HGG have no proven therapies and an expected OS of 4 to 6 months (27, 28). Given the promising preclinical data on dasatinib and everolimus, we moved forward in our pursuit of clinical treatment with daily dasatinib and everolimus for PDGFRα-driven glioma in patients. We treated a series of children and young adults with confirmed PDGFRα-driven glioma with continuous dasatinib (60 mg/m2 orally, twice daily) and everolimus (3 mg/m2 orally, once daily) at 4 academic institutions. We treated 4 patients with recurrent grade II–IV glioma and 2 patients with HGG after initial radiation (Table 1). All patients tolerated the combination with the exception of rash (n = 4 of 6), mucositis (n = 1), and fatigue (n = 1), none of which required a dose reduction. In patients with recurrent glioma (n = 4), the median progression-free survival (PFS) was 3 months (range, 2–5 months), and the median OS was 8.5 months (range, 3–16 months and ongoing) (Figure 4, A and B), although some patients received additional concurrent therapies (Table 1).
Table 1. Characteristics of patients with glioma.
Figure 4. Patient survival curves and treatment of UMPED44 with relapsed PDGFRA-amplified HGG.
(A) PFS curve for patients with recurrent PDGFRα-driven HGG treated with dasatinib and everolimus (n = 4). Dashed lines represent a 95% confidence interval. (B) OS curve for patients with recurrent PDGFRα-driven HGG treated with dasatinib and everolimus (n = 4). Dashed lines represent a 95% confidence interval. (C) UMPED44 axial FLAIR MRI at baseline, before cycle 3, and before cycle 4 (MR spectroscopy in cycle 4 confirmed tumor progression).
Additionally, to measure CSF and plasma levels of dasatinib with and without concurrent everolimus treatment, we collected paired CSF/plasma samples from 2 patients after 1 week of dasatinib monotherapy and then after 2 cycles of dual therapy with dasatinib and everolimus, following an IRB-approved protocol (Supplemental Figure 10A). Patient UMPED44 had an increase in both plasma (11.2 ng/mL to 31.5 ng/mL) and CSF (0.44 ng/mL to 0.91 mg/mL) dasatinib levels (Figure 4C and Supplemental Figure 10, A–C). Patient UMPED52 had an increase in plasma (26.4 ng/mL to 165 ng/mL) dasatinib levels, but we were unable to collect CSF samples (Supplemental Figure 11, A–D).
P-gp expression in human HGG RNA-Seq data sets.
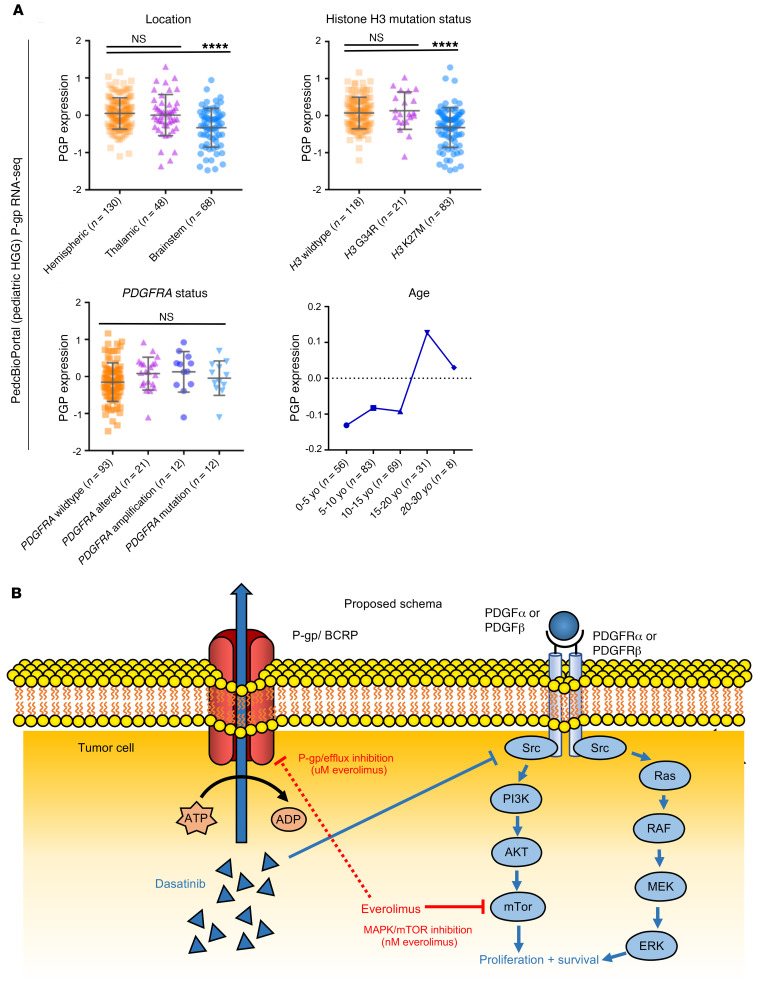
As P-gp is expressed in both brain and tumor cells and overall expression at the protein level was reduced in our mouse HGG models after sustained treatment with everolimus, we were interested in exploring whether P-gp expression correlated with clinical or genomic features in human data sets. Previous data have shown a correlation between P-gp mRNA and protein expression in human liver samples, with both predicting organ penetration of a Pgp-effluxed compound (29). We analyzed RNA-Seq data from pediatric HGGs within the PedcBioPortal (Pediatric HGG ICR London data set, n = 247 with RNA-Seq data; ref. 2) and found that P-gp expression was highest in (a) pediatric hemispheric HGG, (b) tumors with WT H3F3A (gain of function mutations are found in half of pediatric HGGs), and (c) young adult–aged patients (10–15 years old) (Figure 5A). Importantly, P-gp expression was higher in pediatric HGG with PDGFRA alterations (Figure 5A, n = 147). In an analysis of adult HGGs (cBioPortal, The Cancer Genome Atlas [TCGA] data set, n = 585) (30), none of the clinical parameters showed a correlation with P-gp expression, including PDGFRA status, methylguanine-DNA methyltransferase (MGMT) status, and age at diagnosis (Supplemental Figure 12A). Future studies involving P-gp IHC of human tissue are needed to further explore possible correlations between P-gp expression and clinical or genomic features.
Figure 5. P-gp expression in human pediatric HGG and proposed schema.
(A) PedcBioPortal (pediatric HGG) P-gp RNA-Seq data by location (upper left), histone H3 mutation status (upper right), PDGFRA status (lower left), and age (lower right) (****P ≤ 0.0001, by Tukey’s multiple comparisons test). Data presented in the upper 2 graphs and lower left graph reflect the mean ± SEM. (B) Proposed schema of dasatinib and everolimus targeting of PDGF-driven HGG. Red line depicting inhibition of P-gp/BCRP by everolimus is dashed to reflect the higher concentration (μM) required to achieve P-gp inhibition than to achieve MAPK/mTOR inhibition (nM). yo, years old.
Discussion
Our study illustrates that everolimus improves the efficacy of dasatinib in the treatment of PDGFRα-driven glioma. Although response attribution is difficult for our heterogeneous consecutive cases, our early clinical experience with this treatment combination demonstrated excellent tolerance of the regimen and OS following disease progression that compared favorably with that of historical controls (27, 28). To our knowledge, cotreatment with everolimus and dasatinib had not previously been tested in glioma tumor models or patients. More broadly, our results demonstrate that TKI treatment targeted to alterations in HGG may have renewed clinical relevance when combined with mTOR inhibitors.
Importantly, our murine IUE tumor model experiments demonstrate that everolimus and dasatinib had improved combinatorial activity in the treatment of PDGFR-driven glioma cells. This may be due to the presence of dual inhibition within the MAPK/mTOR cell proliferation pathway, with everolimus reducing residual mTOR activation from dasatinib treatment alone (refs. 5, 31 and Figure 5B). Attribution to tumor cell toxicity from an improved intracellular dasatinib concentration versus direct mTOR inhibition was difficult to fully distinguish. However, we found no in vitro P-gp inhibition with everolimus treatment at a low dose (10 nM), whereas our treatment studies showed a marked contribution of everolimus to reduced tumor cell viability at doses as low as 1 nM. Murine PPK cells displayed dasatinib/everolimus synergism via the Chou-Talalay median-effect method for drug combination analysis (ref. 20 and Figure 2E). Future mechanistic studies may further clarify whether the benefit is additive (as opposed to synergistic).
In our in vivo model, administration of everolimus increased tumor concentrations of dasatinib above therapeutic (IC50) levels, and this increase required sustained (>24 hours) everolimus exposure. This requirement may be related to the time required for protein-level changes (i.e., reduced P-gp expression and/or activity) to occur. Interestingly, dasatinib concentrations increased more prominently in the CNS tumor samples than in the brain itself. This may be due to the disruption of the BBB in high-grade tumors, allowing for greater dasatinib influx (32). Indeed, our murine tumor had high-grade features, including necrosis and vascular proliferation, that may have influenced BBB integrity (Supplemental Figure 1A).
A concern with P-gp inhibition strategies that remains in our study is whether the inhibitor itself can penetrate the CNS at high enough concentrations to achieve P-gp inhibition (33). Our data showed a concentration of everolimus required for P-gp inhibition that was achievable in the plasma (~10 μM) and brain (~1 μM) of our mouse models, but may be more difficult to achieve in human brains with the current clinical trough goals (5–15 ng/mL, or ~5–15 nM). Interestingly, early adult phase I studies of everolimus involved weekly higher dosing of everolimus (up to 70 mg), which resulted in mean peak levels of everolimus of 175 ng/mL (190 nM) and a tolerability that was similar to or better than that seen with daily 10 mg dosage (34). Future studies may clarify whether everolimus inhibits P-gp at this dosage, or whether higher dosing of everolimus should be reconsidered. P-gp also promotes efflux on the luminal side of blood vessels (Figure 5B), and so future clinical efforts to achieve P-gp inhibition may be based on plasma (rather than brain parenchyma) concentrations of everolimus. At a minimum, based on our in vitro models, everolimus should contribute to synergistic cytotoxicity of PDGFR-driven HGG at clinically relevant (~low nM) concentrations of everolimus and dasatinib.
Although everolimus may help prevent efflux of dasatinib from brain parenchyma and tumor cells, the efficacy of dasatinib will ultimately rely on the cells’ sensitivity to it, which may be variable based on the type of PDGFRA alteration expressed by the cells. The PDGFRA alteration in each glioma can strongly contribute to the sensitivity to dasatinib (7). Our PPK cells carried the recurrent PDGFRA D842V mutation (as did patient UMPED52) and displayed nanomolar reductions in proliferation in response to dasatinib. Previous studies have shown mixed sensitivity of the D842V mutant to dasatinib, including reduced sensitivity to dasatinib in PDGFRA D842V-mutant normal human astrocytes (NHAs) (7). However, nanomolar reductions in D842V-mutant kinase activity have been demonstrated in gastrointestinal stromal tumors (GISTs) (35). Of note, our IUE-based PPK cells retained endogenous, WT PDGFRA, which may have contributed to tumor proliferation and dasatinib sensitivity. Future studies will continue to clarify the responsiveness of various PDGFRA alterations to dasatinib sensitivity, but we believe our data demonstrate the promise of this dual therapy in both PDGFRA-mutant and -amplified gliomas.
Our clinical case series (n = 6) confirms the feasibility of this dual therapy in this very high-risk patient population. The patients tolerated this combination without significant adverse events. Efficacy is difficult to fully assess, given the heterogeneity and concurrent therapies in this group, but the median OS (8.5 months) for the 4 patients with recurrent pediatric glioma was slightly higher than that for historical controls for relapsed pediatric glioma (4–6 months) (27, 28). While we did find an increase in CSF dasatinib levels after everolimus coadministration in UMPED44, the overall CSF dasatinib concentration (~2 nM) still remained near or below the IC50 of most human PDGFRα-driven glioma cell cultures. We used CSF as the best proxy for dasatinib concentrations within the tumor, and it may not be an accurate reflection of the tumor dasatinib concentration. Indeed, in our murine model, cortex concentrations of dasatinib were consistently lower than tumor concentrations. Future patient data will help clarify the CSF retention of dasatinib, and future clinical studies may consider a further dose escalation of this drug.
Thus far, targeted treatments for PDGFRα-driven HGG have failed to improve outcomes in both adult and pediatric patients. This shortcoming has exposed the need for new precision medicine approaches for these patient populations. Our study confirms that dasatinib effectively inhibited PDGFRA, and the sustained coadministration of dasatinib and everolimus allowed for additional combinatorial efficacy and the potential for improved CNS penetration. The combination of dasatinib and everolimus represents a new avenue for adult and pediatric HGG treatment, and the coadministration of these drugs reopens the possibility of dasatinib being an effective therapy for PDGFRα-driven glioma.
Methods
Murine model of HGG using IUE
IUE was performed using sterile technique on isoflurane/oxygen-anesthetized pregnant CD1 female mice at E13.5 (cortex), according to established methodology (36). All tumors for this study were generated by injecting plasmids into either the lateral ventricle (forebrain) or the fourth ventricle (hindbrain). In this study, we injected the following 4 plasmids together: (a) PBase, (b) PB-CAG-DNp53-Ires-luciferase (dominant-negative TP53, referred to herein as just TP53), (c) PB-CAG-PdgfraD824V-Ires-EGFP (PDGFRA D842V), and (d) PB-CAG-H3.3 K27M-Ires-EGFP (H3K27M), referred to herein as the PPK model.
Following anesthesia induction, carprofen was administered subcutaneously for additional analgesia. Uterine horns were exposed through a 1 cm incision, and individual embryos were digitally manipulated into the correct orientation for intraventricular injection. A pulled capillary needle was loaded with endotoxin-free DNA and Fast Green Dye (0.05%, MilliporeSigma) for visualization, and a microinjector was used to inject either the lateral or fourth ventricles with the DNA-dye mixture. Three to 4 plasmids were injected simultaneously, each at a final concentration of 1 μg/μL, and 1–2 μL total solution was injected per embryo. DNA was then electroporated into cortical neural progenitors using 5 mm tweezertrodes (BTX), applying 5 square pulses at 35 V, 50 ms each at 950 ms intervals. The embryos were then returned into the abdominal cavity, the muscle and skin were sutured, and the animal was monitored until fully recovered from the procedure.
After delivery, the efficacy of plasmids uptake was monitored by bioluminescence using the IVIS Spectrum (Caliper Life Sciences) (tumors express luciferase). Pups that did not display bioluminescence (approximately 5%–10% of the pups) were euthanized. After 3 weeks, the juveniles were weaned and separated by sex. The mice with a positive signal were monitored for tumors every day by observation and biweekly by bioluminescence imaging on an IVIS Spectrum imaging system until the signs of intracranial tumor burden ensued.
Generation of IUE primary HGG cell cultures
Mouse HGG primary cell cultures were generated by harvesting IUE tumors at the time of euthanasia. Tumors were located by GFP expression under an epifluorescence microscope (Olympus CKX41) at the time of resection. The tumor mass was gently homogenized and dissociated with nonenzymatic cell dissociation buffer (Gibco, Thermo Fisher Scientific). The cell suspension was filtered through a 70 μm cell strainer, centrifuged at 300 g for 4 minutes, and resuspended into 7 mL Neurobasal-A Medium (1X) (base medium) (Invitrogen, Thermo Fisher Scientific). Each base stock of medium was supplemented with 10 mL B27 with vitamin A, 5 mL N2, 5 mL sodium pyruvate (1 mM), 5 mL nonessential amino acids (100 μM), 7.5 mL l-glutamine (2 mM), 1 mL normocin, 5 mL antibiotic-antimycotic, and 1 μL EGF (20 ng/mL) per 1 mL medium every 3 days.
In vitro treatment and synergism calculations
IUE-generated PDGFRα-driven HGG primary (PPK) cells were cultured in the conditions described above. Cell viability in vitro was monitored either with an XTT Cell Proliferation Assay Kit (Cayman Chemical) using the included protocol, or by bioluminescence readings collected using the Synergy HTX Multi-Mode Microplate Reader (BioTek Instruments). Additionally, cell proliferation was monitored via IncuCyte ZOOM (Sartorius) live-cell analysis. Through use of the IncuCyte ZOOM live imager, the growth of 3000 PPK primary cells, plated in 96-well plates, was monitored according to units of average green fluorescence intensity (green calibrated unit [GCU] × μm2). PPK primary cell culture was grown with 200 μL media and 1 μL PDGF-AA and PDGF-BB at a working concentration of 10 ng/mL in each 96-well plate (Shenandoah Biotechnology). All in vitro treatments were completed with research-grade dasatinib and everolimus (Selleck Chemicals). UMPED58 is a primary human cell line generated from a 7-year-old male with diffuse intrinsic pontine glioma (DIPG) at autopsy. At diagnosis, the tumor was sequenced and found to carry H3F3A K27M, ATRX pQ119*, PDGFRA, KIT, and KDR amplifications. UMPED05 is a primary cell line generated from a 2-year-old female with thalamic HGG with PDGFRA amplification at autopsy, as previously described by Koschmann et al. (5). UMPED58, UMPED05, and KNS42 were grown in 10 mL l-glutamine (200 mM), 5 mL 100× penicillin-streptomycin (pen-strep), and 10% FBS. KNS42 was provided by Alan Meeker (Johns Hopkins University, Baltimore, Maryland, USA). Mayo 154C, Mayo 64VF, and Mayo 200VF were derived from PDX models provided by Jann Sarkaria (Mayo Clinic College of Medicine, Rochester, Minnesota, USA) (37), and DIPG-XIII-P was provided by Michele Monje (Stanford University, Stanford, California, USA) (38). All synergism calculations were performed by the Chou-Talalay method for drug combination based on the median-effect equation (20). Briefly, we entered normalized proliferation assay data for combinations of no treatment and 4 serial dilutions of each compound (D1 and D2) to generate linear regression curves for each compound, resulting in the generation of theoretic doses (Dx1 and Dx2). Comparison of the actual dose (D1) and the theoretic dose (D1/Dx1) + (D2/Dx2) provides a combination index (CI). The Chou-Talalay theorem offers a quantitative definition for additive effect (CI = 1), synergism (CI <1), and antagonism (CI >1) in drug combinations (20).
Murine IUE HGG treatment studies
Mice harboring IUE-generated PPK HGG tumors were treated with everolimus and/or dasatinib. Mice were treated when tumors reached a logarithmic growth phase (minimum 2 × 105 photons/second via bioluminescence imaging), and confirmation was made that the treatment groups had equivalent average luminescence at the time of treatment.
Mice litters from each experimental group were randomized to (a) treatment with 10 mg/kg dasatinib or (b) 5 mg/kg everolimus alone (10% dasatinib or everolimus suspended in DMSO, 80% ultrapure water, 10% Tween-80), (c) combination treatment with 10 mg/kg dasatinib and 5 mg/kg everolimus, or (d) control treatment (10% DMSO, 80% ultrapure water, 10% Tween-80). Results from initial studies with the high-dose combination (25 mg/kg dasatinib and 10 mg/kg everolimus) were associated with early treatment-related morbidity/mortality; therefore, this arm was excluded from further studies. Mice were treated every other week on this schedule until morbidity.
Animals displaying symptoms of morbidity after treatment were euthanized for immunohistochemical or pharmacodynamic/PK (PD/PK) analysis of tumors. Some mice (n = 3) were given an initial treatment dose for their assigned arm 4 hours before euthanization. For intratumoral concentration studies, all doses of everolimus were administered to mice 24 hours before dasatinib administration, consistent with previous methods used for in vitro analysis of mTOR/MAPK regulation after treatment with everolimus (10). For immunohistochemical analysis, mice were perfused with Tyrode’s Solution followed by 4% paraformaldehyde fixative solution to preserve the structures of the brain.
PK analysis
Mouse PK sample procurement.
Drug administration to nontumor-bearing C57/Bl6 mice for PK studies was performed by oral gavage of everolimus 2 hours before a tail vein injection of dasatinib. One, 2, 4, and 7 hours after the dasatinib injection, the mice were anesthetized with isoflurane/oxygen, and 500 μL to 1 mL blood was drawn from the apex of the heart within the mouse’s enclosed cavity. Immediately, the withdrawn blood was centrifuged within a microvette EDTA-coated conical tube for 10 minutes at 2000g, and the plasma was separated and stored at –80°C until PK analysis was performed. Following the blood draw, the mouse was sacrificed, and the brain, brainstem, and/or tumor were extracted separately and stored at –80°C until PK analysis was performed.
Chemicals and reagents.
For PK studies, dasatinib powder was procured from MilliporeSigma. Liquid chromatography–mass spectrometry–grade (LC-MS–grade) acetonitrile was purchased from MilliporeSigma. Formic acid (98%, LC-MS grade) was obtained from Fluka. A Milli-Q water system from MilliporeSigma was used to obtain ultrapure deionized water.
Stock solutions, working solutions, and quality control samples
Dasatinib and the internal standard were individually weighed and dissolved in acetonitrile to stock solutions and then stored at −20°C. The dasatinib stock solution was then diluted with acetonitrile to a series of working solutions from 2.5 to 5000 ng/mL The quality control working solutions at low, medium, and high concentrations were prepared using a separately prepared stock solution. For sample preparation, the dasatinib stock solution was diluted to 1000 ng/mL with acetonitrile. Quality control samples were evenly distributed among samples from each batch.
Sample preparation.
Plasma (40 μL) was dispensed into a Fisher Scientific 96-well plate, to which 40 μL ice-cold acetonitrile (100%) and 120 μL internal standard solution (1000 ng/mL) were added. Next, the plate was vortexed for 10 minutes. The plate was then centrifuged at 3500 rpm for 10 minutes at 4°C to precipitate the protein. LC–tandem mass spectrometry (LC-MS/MS) was used to analyze 5 μL of the supernatant. The plasma samples were sonicated before being transferred into the 96-well plates. Tissue samples were weighed and suspended in 20% acetonitrile (80% water; 1:5 wt/vol). The samples were then homogenized 4 times for 20 seconds each time at 6500 rpm in a Precellys Evolution system (Bertin Instruments). For LC-MS/MS analysis, the dasatinib in brain tissue homogenates was extracted from the samples in the same manner as for the dasatinib in plasma. Prior to extraction, samples that were above the upper limit of qualification were diluted with the same matrix. Calibrator-standard samples and quality control samples were prepared by mixing 40 μL blank bio matrix, 40 μL working solution, and 120 μL internal standard solution.
Calibration curve.
Analytical curves were made with 12 nonzero standards by plotting the peak area ratio of dasatinib to the internal standard versus the concentration. The curve was created with linear regression and weighted (1/x2). The correlation coefficient demonstrated the linearity of the relationship between peak area ratio and concentration.
LC-MS/MS.
The concentrations of dasatinib were determined with a SCIEX AB-5500 QTRAP mass spectrometer with an electrospray ionization source, interfaced with a Shimadzu high-performance LC system. The LC-MS/MS system was controlled with Analyst Software, version 1.6 (Applied Biosystems), which was also used for data acquisition and processing. Separation was performed on a Waters Xbridge C18 column (50 × 2.1 mm ID, 3.5 μm); the flow rate was 0.4 mL/min. A (100% H2O with 0.1% formic acid) and B (100% acetonitrile with 0.1% formic acid) comprised the mobile phase. The gradient began with 5% B for 30 seconds and then linearly increased to 99% B at 2 minutes, and was then reduced to 5% B at 4.1 minutes to 5.5 minutes with a runtime of 6 minutes in total. The mass spectrometer was operated in positive mode, and multiple reaction monitoring was used for analysis. The Q1 m/z and Q3 m/z were 487.9 and 401.1, respectively.
Human treatment studies
All patients received dasatinib 60 mg/m2 orally twice daily and everolimus 3 mg/m2 orally once daily continuously, with titration of dosing to maintain an everolimus trough level of 5–15 ng/mL. Four patients were treated off-therapy (patients UMPED68, SCH01, NYU01, and UCSF01) and 2 patients (UMPED44 and UMPED52) were treated in an IRB-approved clinical trial. Enrollment required Clinical Laboratory Improvement Amendments–certified (CLIA-certified) confirmation of a DNA (mutation, amplification) or RNA (fusion) alteration in a PDGF-related gene (PDGFRA, PDGFA, PDGFRB, PDGFB). Paired CSF and plasma samples (before and after addition of everolimus) were collected from enrolled patients when clinically feasible. Treatment cycles were repeated every 28 days for up to 24 cycles. CSF and serum dasatinib PK analyses were performed by the University of Michigan Pharmacokinetic Core Facility (see Supplemental Methods for details) to determine whether blood-CSF permeability of dasatinib was affected by dual therapy with everolimus. Prior to the first cycle, dasatinib was taken as monotherapy for 1 week followed by CSF collection in order to establish a monotherapy dasatinib CSF level. After the second cycle, CSF was collected to establish a dual-therapy dasatinib level (while on everolimus). Additionally, all CSF samples were assessed for cell-free tumor DNA (cf-tDNA) using previous methods (39), when sample remained.
Human HGG clinical sequencing and PDGFRA alteration confirmation
For confirmation of PDGFRA status in HGG patients, tumor (formalin-fixed, paraffin-embedded [FFPE] or frozen) and normal (cheek swab) samples were submitted for whole-exome (paired tumor and germline DNA) and transcriptome (tumor RNA) sequencing. Clinically integrated sequencing was performed after enrollment in the University of Michigan’s IRB-approved Pediatric MiOncoSeq study using standard protocols, in adherence with CLIA guidelines. All somatic mutation information generated from this study has been uploaded to the Database of Genotypes and Phenotypes (dbGaP) (https://www.ncbi.nlm.nih.gov/gap) under accession number phs000673.v1.p1.
Human P-gp expression
Pediatric and adult human P-gp expression data set analysis was performed using RNA-Seq data accessed from the PedcBioPortal (https://d3b.center/our-research/pedcbioportal/). For pediatric HGG, data were accessed from the Pediatric HGG ICR London (2). We performed comparisons of P-gp expression by location of tumor, H3 (H3F3A or HIST1H3B) mutation status, PDGFRA mutation status, and age. For adult HGG, data were accessed from TCGA data set (30). For TCGA (adult HGG) P-gp RNA, we compared P-gp expression levels by PDGFRA mutation status, age, and MGMT status.
Statistics
For comparison of dose-response curves using DNA-damaging agents, a nonlinear regression curve [log (agent) versus normalized response with variable slope] was generated. Statistical significance in all experiments was defined as a 2-sided P value of 0.05 or less. All analyses were conducted with GraphPad Prism, version 7.0 (GraphPad Software). Expression levels of p-Src and p-S6 were quantified using IHC performed on mouse brain sections treated with everolimus and/or dasatinib. Four randomly selected JPEG images were captured from each IHC slide at ×12.3 magnification on an Aperio ImageScope (Leica Biosystems) and quantified using ImageJ software (NIH). ImageJ software was also used to quantify IHC and Western blot expression levels as well as the PPK neurosphere area in squared pixel units. Comparisons between treatment groups in experiments with only 2 groups were completed using a 2-tailed Welch’s t test. Comparisons between treatment groups in experiments with more than 2 groups were done using Tukey’s or Dunnett’s multiple comparisons test. Survival curve comparisons between treatment groups were performed using the log-rank test.
Study approval
Animal studies.
All animal studies were conducted according to guidelines approved by the University Committee on the Use and Care of Animals (UCUCA) of the University of Michigan.
Human studies.
Four patients (UMPED68, SCH01, NYU01, and UCSF01) were treated off-therapy according to IRB-approved protocols for molecular tumor assessment. Two patients (UMPED44 and UMPED52) were treated in an IRB-approved clinical trial with dasatinib and everolimus for children and adults with newly diagnosed (after radiation) high-grade (grade III–IV) glioma or recurrent grade II–IV glioma with PDGF alterations (NCT03352427).
Author contributions
ZM, VNY, BM, RTC, CCT, and C. Koschmann designed the in vitro experiments and performed experiments and data analysis. ZM, VNY, BM, TG, TNP, TY, RTC, CCT, JRC, DRW, and JNS designed the in vivo experiments and conducted data analysis. ZM, VNY, RS, BLM, KW, AKB, SS, HJLG, AP, IW, ML, PLR, HP, SESL, SV, TN, CG, C. Kline, SM, CKS, AMC, RM, and C. Koschmann performed human clinical studies. ZM, VNY, MPP, BLM, and C. Koschmann performed PK analyses. ZM, VNY, BLM, JRC, C. Koschmann, RS, RTC, and SV prepared the manuscript. All authors discussed and reviewed the manuscript and approved the manuscript for publication.
Supplementary Material
Acknowledgments
The authors thank the patients and their families for participation in this study. C. Koschmann is supported by the National Institute of Neurological Disorders and Stroke (NINDS), NIH (K08-NS099427-01), the University of Michigan Chad Carr Pediatric Brain Tumor Center, the Chad Tough Foundation, Hyundai Hope on Wheels, Catching up With Jack, the Prayers from Maria Foundation, the U CAN-CER VIVE Foundation, the Morgan Behen Golf Classic, and the DIPG Collaborative. The Peds-MiOncoSeq study was supported by a Clinical Sequencing Exploratory Research Award from the NIH (1UM1HG006508; principal investigator, Arul Chinnaiyan).
Version 1. 06/30/2020
In-Press Preview
Version 2. 08/31/2020
Electronic publication
Version 3. 10/01/2020
Print issue publication
Footnotes
Conflict of interest: AMC is on the Scientific Advisory Board of Tempus.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(10):5313–5325.https://doi.org/10.1172/JCI133310.
Contributor Information
Zachary Miklja, Email: miklja@umich.edu.
Viveka Nand Yadav, Email: vyadav@umich.edu.
Rodrigo T. Cartaxo, Email: rteodoro@umich.edu.
Ruby Siada, Email: rubsiada@umich.edu.
Chase C. Thomas, Email: ccthomas@med.umich.edu.
Jessica R. Cummings, Email: jessrcu@umich.edu.
Brendan Mullan, Email: brendanfmullan@gmail.com.
Stefanie Stallard, Email: stefst@umich.edu.
Alyssa Paul, Email: arpaul@med.umich.edu.
Amy K. Bruzek, Email: abruzek@med.umich.edu.
Kyle Wierzbicki, Email: kylewier@umich.edu.
Tao Yang, Email: taoyang@umich.edu.
Taylor Garcia, Email: taygarc@umich.edu.
Ian Wolfe, Email: iawolfe@med.umich.edu.
Patricia L. Robertson, Email: prob@umich.edu.
Hugh J.L. Garton, Email: hgarton@med.umich.edu.
Daniel R. Wahl, Email: dwahl@med.umich.edu.
Jann N. Sarkaria, Email: Sarkaria.Jann@mayo.edu.
Cassie Kline, Email: Cassie.Kline@ucsf.edu.
Sabine Mueller, Email: Sabine.Mueller@ucsf.edu.
Theodore Nicolaides, Email: Theodore.Nicolaides@nyulangone.org.
Chana Glasser, Email: Chana.Glasser@nyulangone.org.
Sriram Venneti, Email: svenneti@med.umich.edu.
Chandan Kumar-Sinha, Email: chakumar@umich.edu.
Arul M. Chinnaiyan, Email: arul@med.umich.edu.
Rajen Mody, Email: Rmody@med.umich.edu.
Manjunath P. Pai, Email: amitpai@med.umich.edu.
Timothy N. Phoenix, Email: timothy.phoenix@cchmc.org.
Bernard L. Marini, Email: bernmari@med.umich.edu.
Carl Koschmann, Email: ckoschma@umich.edu.
References
- 1.Liu J, et al. An integrated TCGA Pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400–416.e11. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mackay A, et al. Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell. 2017;32(4):520–537.e5. doi: 10.1016/j.ccell.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farahani RM, Xaymardan M. Platelet-derived growth factor receptor alpha as a marker of mesenchymal stem cells in development and stem cell biology. Stem Cells Int. 2015;2015:362753. doi: 10.1155/2015/362753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mackay A, et al. Molecular, pathological, radiological, and immune profiling of non-brainstem pediatric high-grade glioma from the HERBY phase II randomized trial. Cancer Cell. 2018;33(5):829–842.e5. doi: 10.1016/j.ccell.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koschmann C, et al. Characterizing and targeting PDGFRA alterations in pediatric high-grade glioma. Oncotarget. 2016;7(40):65696–65706. doi: 10.18632/oncotarget.11602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alentorn A, et al. Prevalence, clinico-pathological value, and co-occurrence of PDGFRA abnormalities in diffuse gliomas. Neuro-oncology. 2012;14(11):1393–1403. doi: 10.1093/neuonc/nos217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paugh BS, et al. Novel oncogenic PDGFRA mutations in pediatric high-grade gliomas. Cancer Res. 2013;73(20):6219–6229. doi: 10.1158/0008-5472.CAN-13-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lassman AB, et al. Phase 2 trial of dasatinib in target-selected patients with recurrent glioblastoma (RTOG 0627) Neuro-oncology. 2015;17(7):992–998. doi: 10.1093/neuonc/nov011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Motzer RJ, et al. Lenvatinib, everolimus, and the combination in patients with metastatic renal cell carcinoma: a randomised, phase 2, open-label, multicentre trial. Lancet Oncol. 2015;16(15):1473–1482. doi: 10.1016/S1470-2045(15)00290-9. [DOI] [PubMed] [Google Scholar]
- 10.Pignochino Y, et al. The combination of sorafenib and everolimus abrogates mTORC1 and mTORC2 upregulation in osteosarcoma preclinical models. Clin Cancer Res. 2013;19(8):2117–2131. doi: 10.1158/1078-0432.CCR-12-2293. [DOI] [PubMed] [Google Scholar]
- 11.Porkka K, et al. Dasatinib crosses the blood-brain barrier and is an efficient therapy for central nervous system Philadelphia chromosome-positive leukemia. Blood. 2008;112(4):1005–1012. doi: 10.1182/blood-2008-02-140665. [DOI] [PubMed] [Google Scholar]
- 12.Agarwal S, et al. Active efflux of dasatinib from the brain limits efficacy against murine glioblastoma: broad implications for the clinical use of molecularly targeted agents. Mol Cancer Ther. 2012;11(10):2183–2192. doi: 10.1158/1535-7163.MCT-12-0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minocha M, Khurana V, Qin B, Pal D, Mitra AK. Enhanced brain accumulation of pazopanib by modulating P-gp and Bcrp1 mediated efflux with canertinib or erlotinib. Int J Pharm. 2012;436(1-2):127–134. doi: 10.1016/j.ijpharm.2012.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oberoi RK, Mittapalli RK, Elmquist WF. Pharmacokinetic assessment of efflux transport in sunitinib distribution to the brain. J Pharmacol Exp Ther. 2013;347(3):755–764. doi: 10.1124/jpet.113.208959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel SK, et al. Generation of diffuse intrinsic pontine glioma mouse models by brainstem-targeted in utero electroporation. Neuro-oncology. 2020;22(3):381–392. doi: 10.1093/neuonc/noz197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis PW, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340(6134):857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlson BL, Pokorny JL, Schroeder MA, Sarkaria JN. Establishment, maintenance and in vitro and in vivo applications of primary human glioblastoma multiforme (GBM) xenograft models for translational biology studies and drug discovery. Curr Protoc Pharmacol. 2011;14(14):1–14.16. doi: 10.1002/0471141755.ph1416s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagaraja S, et al. Transcriptional dependencies in diffuse intrinsic pontine glioma. Cancer Cell. 2017;31(5):635–652.e6. doi: 10.1016/j.ccell.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang W, et al. Genomics of drug sensitivity in cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41(Database issue):D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70(2):440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 21.Mittapalli RK, et al. ABCG2 and ABCB1 limit the efficacy of dasatinib in a PDGF-B-driven brainstem glioma model. Mol Cancer Ther. 2016;15(5):819–829. doi: 10.1158/1535-7163.MCT-15-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Vries NA, Zhao J, Kroon E, Buckle T, Beijnen JH, van Tellingen O. P-glycoprotein and breast cancer resistance protein: two dominant transporters working together in limiting the brain penetration of topotecan. Clin Cancer Res. 2007;13(21):6440–6449. doi: 10.1158/1078-0432.CCR-07-1335. [DOI] [PubMed] [Google Scholar]
- 23.Dash RP, Jayachandra Babu R, Srinivas NR. Therapeutic potential and utility of elacridar with respect to P-glycoprotein inhibition: an insight from the published in vitro, preclinical and clinical studies. Eur J Drug Metab Pharmacokinet. 2017;42(6):915–933. doi: 10.1007/s13318-017-0411-4. [DOI] [PubMed] [Google Scholar]
- 24.Lagas JS, et al. Brain accumulation of dasatinib is restricted by P-glycoprotein (ABCB1) and breast cancer resistance protein (ABCG2) and can be enhanced by elacridar treatment. Clin Cancer Res. 2009;15(7):2344–2351. doi: 10.1158/1078-0432.CCR-08-2253. [DOI] [PubMed] [Google Scholar]
- 25.Minocha M, Khurana V, Qin B, Pal D, Mitra AK. Co-administration strategy to enhance brain accumulation of vandetanib by modulating P-glycoprotein (P-gp/Abcb1) and breast cancer resistance protein (Bcrp1/Abcg2) mediated efflux with m-TOR inhibitors. Int J Pharm. 2012;434(1-2):306–314. doi: 10.1016/j.ijpharm.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warren KE. Beyond the blood:brain barrier: the Importance of central nervous system (CNS) pharmacokinetics for the treatment of CNS tumors, including diffuse intrinsic pontine glioma. Front Oncol. 2018;8:239. doi: 10.3389/fonc.2018.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruggiero A, et al. Phase II trial of temozolomide in children with recurrent high-grade glioma. J Neurooncol. 2006;77(1):89–94. doi: 10.1007/s11060-005-9011-2. [DOI] [PubMed] [Google Scholar]
- 28.Narayana A, et al. Bevacizumab in recurrent high-grade pediatric gliomas. Neuro-oncology. 2010;12(9):985–990. doi: 10.1093/neuonc/noq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, Chen XP, Qiu FZ. Correlation of expression of multidrug resistance protein and messenger RNA with 99mTc-methoxyisobutyl isonitrile (MIBI) imaging in patients with hepatocellular carcinoma. World J Gastroenterol. 2004;10(9):1281–1285. doi: 10.3748/wjg.v10.i9.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minocha M, Khurana V, Qin B, Pal D, Mitra AK. Co-administration strategy to enhance brain accumulation of vandetanib by modulating P-glycoprotein (P-gp/Abcb1) and breast cancer resistance protein (Bcrp1/Abcg2) mediated efflux with m-TOR inhibitors. Int J Pharm. 2012;434(1-2):306–314. doi: 10.1016/j.ijpharm.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolburg H, Noell S, Fallier-Becker P, Mack AF, Wolburg-Buchholz K. The disturbed blood-brain barrier in human glioblastoma. Mol Aspects Med. 2012;33(5-6):579–589. doi: 10.1016/j.mam.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Kalvass JC, et al. Why clinical modulation of efflux transport at the human blood-brain barrier is unlikely: the ITC evidence-based position. Clin Pharmacol Ther. 2013;94(1):80–94. doi: 10.1038/clpt.2013.34. [DOI] [PubMed] [Google Scholar]
- 34.O’Donnell A, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral mammalian target of rapamycin inhibitor everolimus in patients with advanced solid tumors. J Clin Oncol. 2008;26(10):1588–1595. doi: 10.1200/JCO.2007.14.0988. [DOI] [PubMed] [Google Scholar]
- 35.Dewaele B, et al. Activity of dasatinib, a dual SRC/ABL kinase inhibitor, and IPI-504, a heat shock protein 90 inhibitor, against gastrointestinal stromal tumor-associated PDGFRAD842V mutation. Clin Cancer Res. 2008;14(18):5749–5758. doi: 10.1158/1078-0432.CCR-08-0533. [DOI] [PubMed] [Google Scholar]
- 36.Patel SK, et al. Generation of diffuse intrinsic pontine glioma mouse models by brainstem-targeted in utero electroporation. Neuro-oncology. 2020;22(3):381–392. doi: 10.1093/neuonc/noz197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vaubel RA, et al. Genomic and phenotypic characterization of a broad panel of patient-derived xenografts reflects the diversity of glioblastoma. Clin Cancer Res. 2020;26(5):1094–1104. doi: 10.1158/1078-0432.CCR-19-0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagaraja S, et al. Transcriptional dependencies in diffuse intrinsic pontine glioma. Cancer Cell. 2017;31(5):635–652.e6. doi: 10.1016/j.ccell.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stallard S, et al. CSF H3F3A K27M circulating tumor DNA copy number quantifies tumor growth and in vitro treatment response. Acta Neuropathol Commun. 2018;6(1):80. doi: 10.1186/s40478-018-0580-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.