Abstract
The development of broadly neutralizing antibodies (BNAbs) in HIV infection is a result of long-term coevolutionary interaction between viruses and antibodies. Understanding how this interaction promotes the increase of neutralization breadth during infection will improve the way in which AIDS vaccine strategies are designed. In this paper, we used SIV-infected rhesus macaques as a model to study the development of neutralization breadth by infecting rhesus macaques with longitudinal NAb escape variants and evaluating the kinetics of NAb response and viral evolution. We found that the infected macaques developed a stepwise NAb response against escape variants and increased neutralization breadth during the course of infection. Furthermore, the increase of neutralization breadth correlated with the duration of infection but was independent of properties of the inoculum, viral loads, or viral diversity during infection. These results imply that the duration of infection was the main factor driving the development of BNAbs. These data suggest the importance of novel immunization strategies to induce effective NAb response against HIV infection by mimicking long-term infection.
Keywords: AIDS/HIV, Virology
Keywords: Genetic variation, Immunoglobulins
Introduction
The induction of neutralizing antibodies (NAbs) is commonly used as the gold standard for evaluating clinical vaccine efficacy against infectious viral diseases, including influenza, smallpox, and polio (1). The failure of the Merck Ad5 gag/pol/nef vaccine in human clinical trials, which aimed at induction of cytotoxic T cell responses against HIV-1, has also driven the HIV vaccine field to reconsider the importance of generating NAb responses in generating an effective HIV vaccine. In the last decade, many broadly neutralizing antibodies (BNAbs), which are capable of neutralizing divergent HIV isolates, have been isolated from HIV-infected patients (2–8). Passive administration of BNAbs to macaques or humanized mice at appropriate doses conferred sterilizing protection against HIV or SHIV acquisition when given before challenge and reduced plasma viremia when given after infection (9–14). Critically, BNAbs were able to inhibit virus replication and reduce viral reservoirs when administered to HIV-infected patients in several clinical trials (15, 16). BNAbs are also being evaluated for prophylactic usage in several ongoing clinical trials (17). While it is generally accepted that a successful AIDS vaccine would need to induce potent BNAbs, no vaccine to date has been successful in inducing even low titers of BNAbs. During natural infection, most HIV-infected humans induce NAb response against their homologous viruses, but only few chronically infected patients, designated elite neutralizers, develop BNAbs (18). The mechanism that leads to the development of BNAbs in this underrepresented patient population remains unclear. Clearly understanding how BNAbs develop during HIV infection will improve the way in which we design AIDS vaccine strategies.
Longitudinal studies of NAb development in patients infected with HIV-1 indicated that the appearance of BNAbs was a result of long-term coevolution between antibodies and HIV viruses (19). The high rate of somatic mutation observed in BNAbs is consistent with the requirement for antibody maturation during the process of infection. Most HIV-infected individuals produced autologous NAb response against transmitted/founder viruses within weeks to months after primary infection. The primary NAb response was strain-specific but quickly selected virus escape mutants. The escape mutants in turn induced NAbs after their appearance in the infected individuals. This coevolutionary arms race between antibodies and viruses appears to stimulate the maturation of HIV-specific antibodies and to lead to the development of many BNAbs, including BNAbs targeting the CD4 binding site, variable regions 1 and 2 (V1/V2) glycans, variable region 3 (V3) glycans, and the membrane-proximal external region (MPER) (20–26). A proof-of-concept vaccine strategy, which aims to induce BNAbs through immunization with longitudinal envelope (Env) variants to recapitulate the coevolutionary interaction between viruses and antibodies, has been suggested based on these observations (19, 27). However, this vaccine strategy has not lived up to its promise when evaluated in animals. Immunization with a cocktail of variants still only induces autologous neutralization against the immunized Env or weak cross-neutralization against a small number of virus variants in animals (28–32).
In our studies, we used the SIVsmE660-infected rhesus macaque model to study the factors associated with NAb development during HIV infection. This model gives the advantage of uniformity in the virus, dose, route of inoculation, and control over the genetic background of the animals. Unlike commonly used SIVmac and SHIV viruses, SIVsmE660 clone viruses are neutralization sensitive but pathogenic to rhesus macaques (33). SIVsmE660-infected macaques developed high titers of autologous NAbs and selected for escape variants during infection. Similar to HIV-infected humans, the coevolution of NAbs and viruses was also observed in infected macaques with generation of sequential neutralization-escape variants and increasing neutralization breadth against virus variants (33). In the present study, we investigated the role of the neutralization sensitivity of viral escape variants in the development of NAbs during infection of macaques with SIV. Rather than studying antibody responses induced by vaccination, we constructed infectious viral clones carrying longitudinal Env escape variants and evaluated NAb responses in macaques infected with representative escape variants. We observed a stepwise development of NAb responses to the longitudinal escape viral variants and an increase of neutralization breadth in all infected animals. However, infection with escape variants did not promote an increase in the kinetics or breadth of neutralization. All the macaques developed NAb response with the same kinetics independent of the strains with which they were infected. The increase of neutralization breadth was only correlated with the duration of infection and not with the extent of viremia or the degree of viral diversity generated. These results revealed that viral characteristics, including viral sequences, neutralizing phenotype, viral loads, and extent of viral diversity are not the sole factors in determining the development of neutralization breadth. The results also suggested that the current effort to optimize Env antigens in HIV vaccine strategies may not be sufficient to induce BNAbs. A deeper understanding of virus-host interactions during virus infection is needed.
Results
Construction of longitudinal viral escape variants from macaques infected with SIVsmE660.
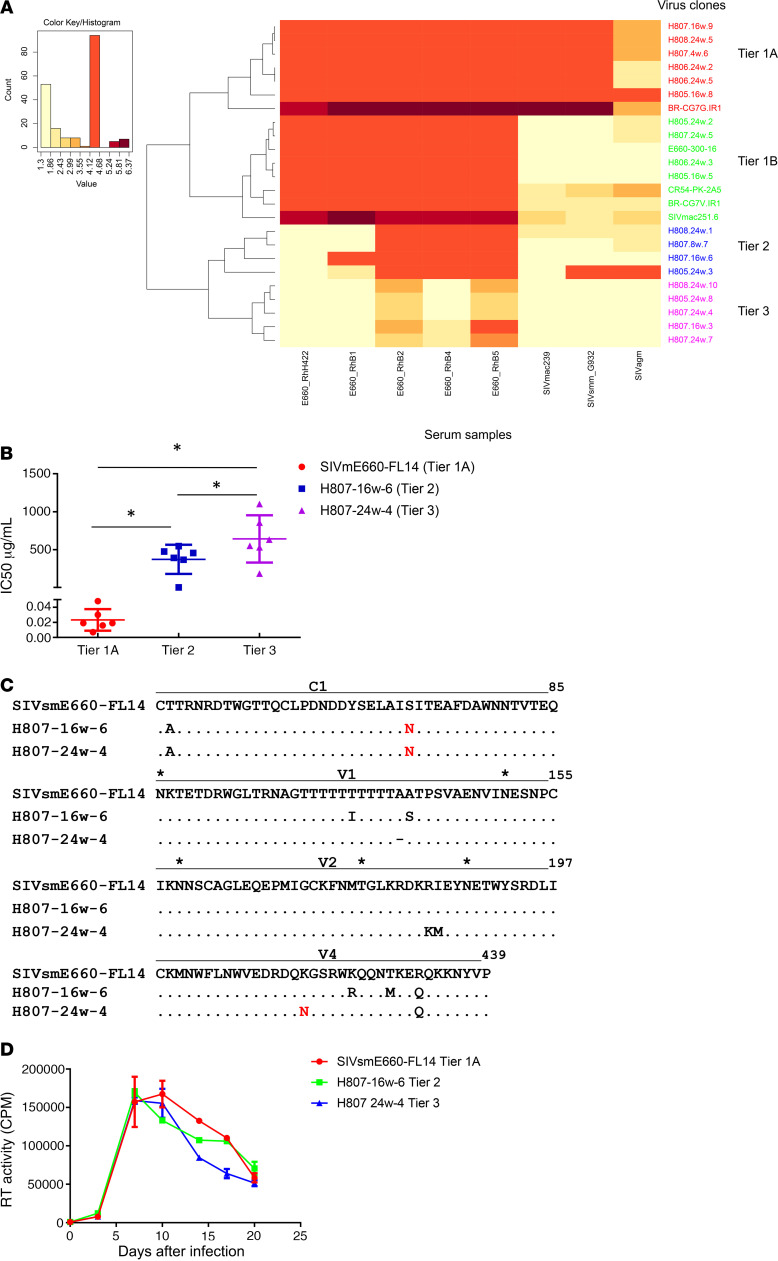
We previously reported that rhesus macaques infected with neutralization-sensitive SIVsmE660 clones, FL6 and FL14, developed high titers of autologous NAbs and continuously selected sequential NAb-escape variants (33), very similar to the pattern of neutralization escape seen in HIV-infected patients. Env NAb escape variants were amplified from plasma samples collected at 2, 4, 8, 16, and 24 weeks after infection, respectively, of these macaques. We constructed chimeric viruses by subcloning the env regions into the same SIVsmE660 backbone to generate a panel of infectious viruses carrying longitudinal env variants but isogenic in all other viral genes. Virus stocks were generated by transfection of 293T cells and amplification in rhesus macaque PBMCs. Neutralization sensitivity of these infectious clones was evaluated using the TZM-bl assay with a panel of antisera from chronically SIV-infected macaques as described in Methods. Neutralization results expressed as 50% inhibitory dose (IC50) titers of antisera are shown in Supplemental Table 1 (supplemental material available online with this article; https://doi.org/10.1172/JCI139123DS1). The infectious variants exhibited diverse sensitivity to a panel of sera in the neutralization assay. We used heatmap hierarchical clustering, which was previously used for tiered categorization of the neutralization sensitivity of HIV-1 viruses (34), to assess the neutralization sensitivity of these SIV variants. The heatmap was generated based on log10 values of IC50 titers using the web tool in the Los Alamos National Laboratory HIV sequence database with the Euclidean distance and complete clustering method. As shown in Figure 1A, SIV variants clustered into 4 subgroups based on their similarity in neutralization sensitivity. The tiered categorization for SIV variants followed a similar pattern to that seen with HIV-1. We designated the subgroups with the tiered categorization used to assess HIV neutralization sensitivity as tier 1A (highly sensitive), tier 1B (moderately sensitive), tier 2 (moderately resistant), and tier 3 (resistant) (34).
Figure 1. Characterization of infectious SIVsmE660 longitudinal escape variants.
(A) Tiered categorization of neutralization sensitivity of SIVsmE660 variants. A heatmap was generated based on log10 values of IC50 titers and was analyzed by hierarchical clustering. The SIVsmE660 variants clustered into 4 subgroups based on their similarity in neutralization sensitivity: tier 1A (highly sensitive), tier 1B (sensitive), tier 2 (moderately resistant), and tier 3 (resistant). (B) Neutralization sensitivity of the 3 representative clones. The 3 clones were evaluated in TZM-bl neutralization assay using 6 purified polyclonal immunoglobulins from rhesus macaques chronically infected with uncloned SIVsmE660. IC50 titers are shown as dot plots and median IC50 with 95% CI are shown for each group (n = 6). Significant differences were observed between the 3 clones (P < 0.05, 1-way ANOVA). (C) Alignment of Env amino acid sequences of the 3 representative SIVsmE660 variants: SIVsmE660-FL14 (tier 1A), H807-16w-6 (tier 2), and H807-24w-4 (tier 3). Identical amino acids are shown as a dot (.), deletions are shown as a dash (–). Amino acid substitutions are shown. Potential glycosylation sites are indicated with an asterisk (*). Only domains with sequence differences are shown. (D) Replication of the 3 clones in rhesus PBMCs. PHA-stimulated rhesus PBMCs were infected with the 3 clones at an MOI of 0.001. Virus production was quantified and shown as reverse transcriptase values in supernatants collected at 3-day intervals. The experiment was performed in duplicate, and median values with 95% CI are shown. Similar virus replication was observed in PBMCs from 2 separate macaques, and the results in a representative macaque are shown. No significant difference was observed between the replication of the 3 clones (P > 0.05, 2-way ANOVA).
We chose 3 virus clones for in vivo evaluation, the parental strain, SIVsmE660-FL14, and 2 clones derived from an SIVsmE660-FL14–inoculated macaque, H807, H807-16w-6 and H807 24w-4 isolated at 16 and 24 weeks after infection, respectively. These clones represented the majority of NAb escape variants at sequential times after infection. They showed significant differences in their sensitivity to polyclonal NAbs purified from the sera of rhesus macaques chronically infected with uncloned SIVsmE660 (Figure 1B). Although the viruses differed significantly in the sensitivity to neutralization, they were isogenic in all viral genes with the exception of a few amino acid substitutions and insertions/deletions located in their Env gp120 constant region 1 (C1), V1, V2, and variable region 4 (V4) domains (Figure 1C). The substitution in C1 of tier 2 and 3 viruses relative to tier 1 (T45A) was observed previously to be associated with neutralization resistance of SIVsmE660 transmitted/founder viruses (35). Both the tier 2 and tier 3 viruses had gained a potential N-linked glycosylation (PNG) site in C1 relative to the parental tier 1 virus. The tier 3 virus had also acquired an additional PNG in V4. Furthermore, none of these substitutions and deletions are located in known restricted SIV cytotoxic T lymphocytes (CTL) epitopes (36). Hence, the 3 infectious longitudinal escape variants provided a good model to evaluate NAb response without potential confounding effects from significant sequence variation in other genes or as a result of differences in CTL response.
Replication of SIV transmitted/founder and NAb escape variants in rhesus macaques.
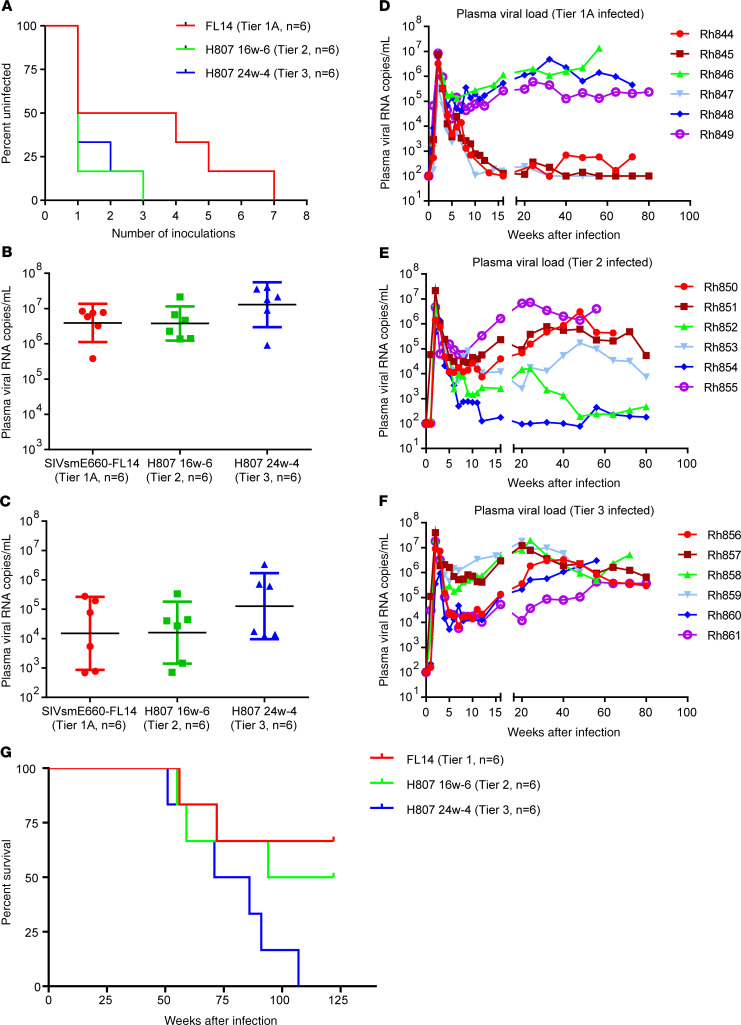
We evaluated the viral replication of the 3 clones both in vitro and in vivo. The 3 clones replicated efficiently and with similar kinetics in vitro in phytohemagglutinin-stimulated (PHA-stimulated) rhesus PBMCs of 2 separate naive macaques. As shown in Figure 1D, the extent and kinetics of replication (shown for a representative macaque) were similar (P > 0.05, 2-way ANOVA), consistent with similar replicative fitness. The in vivo infection and replication of these 3 clones were therefore evaluated in rhesus macaques that expressed the moderately susceptible TRIM-TFP/Q genotype to minimize the potential effect of host TRIM5α polymorphism. We divided 18 rhesus macaques into 3 groups with balanced distribution of restrictive MHC I genotypes (Supplemental Table 2). Each macaque was inoculated by repetitive intrarectal dosing with 1000 median tissue culture infectious dose (TCID50) of virus that had been expanded in rhesus PBMCs. Inoculations were continued until the animals were infected as described in Methods. As shown in the Kaplan-Meier curves in Figure 2A, although more inoculations were required to infect macaques with tier 1A viruses, the difference was not statistically significant (log-rank test, P = 0.0942). After the animals became infected, the kinetics of virus replication during primary infection were similar in the 3 groups. Thus, as shown in Figure 2, B and C, macaques infected with tier 1A and tier 2 viruses had similar peak (mean viral loads: 5.5 vs. 6.3 × 106 RNA copies/mL) and setpoint plasma viral loads (mean viral loads: 9.2 vs. 7.5 × 104 RNA copies/mL). The mean plasma viral loads in the tier 3 group at peak and setpoint were 3-fold and 6-fold higher, respectively, than those in the tier 1A and tier 2 groups (Figure 2, B and C), but the differences were not statistically significant (Kruskal-Wallis test, P > 0.05).
Figure 2. Acquisition and replication of SIVsmE660 escape variants in rhesus macaques.
(A) The acquisition of infection in macaques challenged with the 3 clones by repeated intrarectal inoculations. The acquisition of infection in each group, shown as an uninfected percentage after each inoculation, was not significantly different between the groups (P > 0.05, log-rank test, n = 6 for each group). (B) Plasma viremia at peak infection is shown as dot plots with a geometric mean with 95% CI for each group (n = 6). Plasma viremia at peak infection was not significantly different between the 3 groups (P > 0.05, Kruskal-Wallis test). (C) Plasma viremia at peak infection is shown as dot plots with a geometric mean with 95% CI for each group (n = 6). Plasma viremia at setpoint was not significantly different between groups (P > 0.05, Kruskal-Wallis test). (D) Sequential plasma viremia of the 6 macaques infected with SIVsmE660-FL14, tier 1A virus. (E) Plasma viremia of the 6 macaques infected with H807-16w-6, tier 2 virus. (F) Plasma viremia of the 6 macaques infected with the H807-24w-4, tier 3 virus. (G) The cumulative survival of macaques infected with SIVsmE660-FL14, tier 1A (red); H807-16w-6, tier 2 (green); and H807-24w-4, tier 3 (blue) are shown as Kaplan-Meier curves and compared by log-rank test for trend (P = 0.0379, n = 6 for each group).
However, the kinetics of viremia during the chronic phase of infection differed between the 3 cohorts (Figure 2, D–F). Although all macaques in the tier 3 group maintained plasma viremia above 104 copies/mL throughout the course of infection, viral loads were more inconsistent in the cohorts inoculated with tier 1 and tier 2 viruses. Three macaques in the tier 1A group suppressed plasma viremia to below 103 copies/mL, and 2 of them controlled viremia to below the detection limit of the assay (100 copies/mL). Two macaques in the tier 2 group also suppressed plasma viremia to below 103 copies/mL. Consistent with higher viremia, all macaques inoculated with the tier 3 variant progressed to AIDS within 2 years of infection and had a median survival time of 78.5 weeks after infection. In contrast, the median survival time of macaques infected with the tier 2 and tier 1A viruses was significantly longer, 108 and greater than 120 weeks after infection, respectively. A significant difference was observed when the survival rates in the 3 groups were compared in the order of increased or decreased neutralization resistance (Figure 2G, P = 0.0379, log-rank test for trend). Similar acquisition and plasma viral loads in the acute phase of infection indicated that the 3 clones appeared to have similar ability to establish an infection in the absence of antibody responses. This, in combination with equivalent in vitro replication phenotypes, suggests that the viruses did not differ significantly in their ability to establish an infection or in their replicative fitness. We postulate that the different viral loads during the chronic phase of infection and substantially different disease progression could be due to differences in the humoral response in the 3 groups of macaques.
Kinetics of NAb response in infected rhesus macaques.
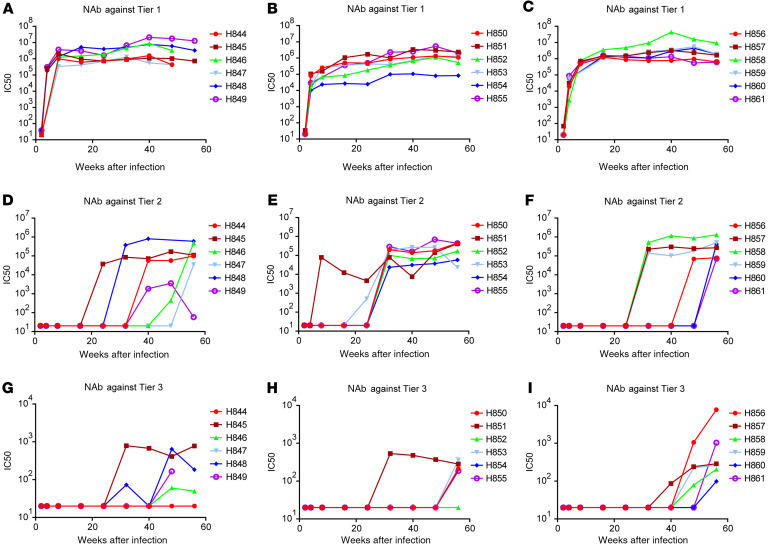
Next, we evaluated the kinetics of NAb response in the 3 groups of macaques. We collected serum samples at sequential time points after infection and evaluated them in TZM-bl assays against the 3 tier 1A, tier 2, and tier 3 inocula. Neutralization results are expressed as IC50 titers of antisera and the development of NAbs after infection is shown as IC50 kinetics in Figure 3. NAbs against tier 1A, tier 2, and tier 3 viruses developed sequentially with similar kinetics and strength in all the macaque cohorts. Thus, NAbs against the tier 1A clone SIVsmE660-FL14 were the first to be detected by 4 weeks after infection in all the macaques (Figure 3, A–C). The IC50 titers ranged from 2 × 103 to 3 × 105 at 4 weeks after infection, increased to 105 and 106 by 16 weeks after infection, and were maintained throughout chronic infection. NAbs against tier 2 viruses were not detectable until 24 weeks after infection in all the macaques, with the exception of macaque Rh851 from the tier 2 group (Figure 3, D–F). Some macaques, including Rh846 and Rh847 from the tier 1A group and Rh856, Rh860, and Rh861 from the tier 3 group, did not induce NAbs against tier 2 virus until 48 weeks after infection. In addition to the delay in development, the IC50 titers against the tier 2 clone were 10-fold lower than NAbs against the tier 1A clone. Most macaques did not develop detectable NAbs against the tier 3 clone until 48 weeks after infection (Figure 3, G–I), and the IC50 titers were significantly lower than titers against tier 1A and tier 2 clones, ranging from 100 to 1000.
Figure 3. Sequential development of neutralizing antibody responses against tier 1A, tier 2, and tier 3 clones during infection.
Sera samples were collected at sequential time points after infection and evaluated against the 3 clones, SIVsmE660-FL14 (tier 1A), H807-16w-6 (tier 2), and H807-24w-4 (tier 3), in TZM-bl assays. IC50 titers were calculated and shown as kinetics for individual macaques. (A–C) IC50 titers against the tier 1A clone in the sera of macaques infected with the tier 1A virus (A), tier 2 virus (B), and tier 3 virus (C). (D–F) IC50 titers against the tier 2 clone in the sera of macaques infected with the tier 1A virus (D), tier 2 virus (E), and tier 3 virus (F). (G–I) IC50 titers against the tier 3 clone in the sera of macaques infected with the tier 1A virus (G), tier 2 virus (H), and tier 3 virus (I).
To detect whether the macaques developed cross-clade NAbs, we evaluated the neutralization of serum samples against 2 genetically divergent tier 3 SIV clones, SIVsmE543-3 and SIVmac239. The Env diversity between SIVsmE543-3 and the SIVsmE660 clones was 7.2% versus 19.4% between SIVmac239 and the SIVsmE660 clones. Weak cross-neutralizing antibodies against heterologous tier 3 SIV clone SIVsmE543-3 were detectable by 56 weeks after infection in 1 macaque of each group (Rh845 from tier 1A, Rh853 from tier 2, and Rh861 from tier 3) (Supplemental Figure 1, A–C). Development of detectable NAbs against the even more heterologous tier 3 virus, SIVmac239 (IC50: 1500) was also observed in 1 macaque at 80 weeks after infection (Rh853 from the tier 2 group) (Supplemental Figure 1, D–F). Both SIVsmE543-3 and SIVmac239 were very resistant to neutralization in general. Even macaques infected with these 2 viruses rarely developed a detectable autologous NAb response.
Factors associated with the increase of neutralization breadth.
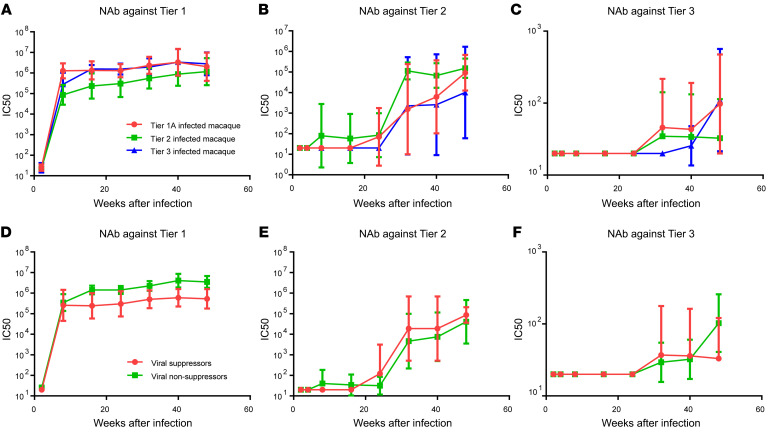
With the above information on NAb response, we statistically evaluated the effect of different factors on the increase of neutralization breadth by categorizing the macaques into different subgroups and comparing the kinetics of NAb response between the groups. First, we evaluated the effect of inoculum on NAb response. The kinetics of NAb response in macaques infected with the 3 variants were compared with each other by repeated-measures 2-way ANOVA. As shown in Figure 4, A–C, the SIVsmE660-FL14 clone and NAb escape variants induced similar NAb responses against tier 1A, tier 2, and tier 3 viruses despite differences in sensitivity to neutralization. Neither the time to appearance of detectable NAbs nor the NAb titers significantly differed between the 3 groups. NAb titers against tier 1 (P = 0.3353, Figure 4A), tier 2 (P = 0.9261, Figure 4B), or tier 3 (P = 0.7609, Figure 4C) viruses did not differ significantly in the 3 groups. However, statistical analysis revealed that the increase of neutralization breadth in all 3 groups was significantly correlated with the duration of infection (P < 0.001).
Figure 4. Statistical analysis of factors associated with the increase of neutralization breadth during infection.
(A–C) Macaques divided into 3 groups based on the inoculated viruses and the kinetics of NAb titers against SIVsmE660-FL14 tier 1 (A), H807-16w-6 tier 2 (B), and H807-24w-4 tier 3 (C) are shown as the geometric mean with 95% CI (n = 6 for each group). The different inoculated viruses did not make a significant difference on NAb titers against tier 1 (P = 0.3353, A), tier 2 (P = 0.9261, B), or tier 3 (P = 0.7609, C) viruses, whereas infection time made a significant difference on NAb titers against all 3 viruses (P < 0.001, repeated-measures 2-way ANOVA). To analyze the effect of viral loads on NAb titers, macaques were divided into suppressors or nonsuppressors based on the plasma viremia at the chronic phase of infection. Macaques Rh844, Rh845, Rh847, Rh852, Rh853, and Rh854, whose plasma viremia was below 104 copies, were designated as suppressors (n = 6), and the other macaques, whose plasma viremia was above 104 copies, were designated as nonsuppressors (n = 12). (D–F) The kinetics of NAb titers against SIVsmE660-FL14 tier 1 (D), H807-16w-6 tier 2 (E), and H807-24w-4 tier 3 (F) in suppressors and nonsuppressors are shown as geometric mean with 95% CI. The different plasma viremia did not make a significant difference on NAb titers against tier 1 (P = 0.0762, D), tier 2 (P = 0.2614, E), or tier 3 (P = 0.9592, F) viruses, whereas infection time made a significant difference on NAb titers against all 3 viruses (P < 0.01, repeated-measures 2-way ANOVA).
Second, we evaluated the effect of plasma viral loads on NAb response. Several studies on HIV-infected human cohorts indicated that high plasma viral loads were positively associated with an increase of neutralization breadth (37–39). To explore the effect of viral loads on the development of NAb, we divided the 18 macaques into 2 groups based on the level of plasma viremia during the chronic phase of infection. Six macaques that suppressed plasma viremia to below 104 copies/mL, including macaques Rh844, Rh845, Rh847, Rh852, Rh853, and Rh854, were designated as viral suppressors, and the other macaques were designated as viral nonsuppressors. The kinetics of NAb response against tier 1A, tier 2, and tier 3 viruses were compared between the 2 groups by repeated-measures 2-way ANOVA. As shown in Figure 4, D–F, the nonsuppressor macaques induced higher titers of NAbs against tier 1A viruses, but the difference was not significant (P = 0.0762, Figure 4D). The extent of plasma viremia did not correlate with NAb titers against tier 2 (P = 0.2614, Figure 4E) or tier 3 (P = 0.9592, Figure 4F) viruses. These results suggested that the plasma viral load was not a major determinant of NAb development in those macaques.
Since only 3 macaques developed cross-neutralization against SIVsmE543-3, and only 1 of them (macaque Rh853) developed NAbs against SIVmac239, we were unable to statistically analyze the viral and host factors associated with development of cross-neutralizing antibodies. The 3 macaques did not show any common features in terms of plasma viral loads, disease progression, inocula, or restrictive MHC genotypes. However, these macaques did not develop detectable NAbs against SIVsmE543-3 or SIVmac239 until 2 years after infection, further confirming that duration of infection was the important factor associated with the increase of NAb breadth.
The relation between NAb and viral Env diversity.
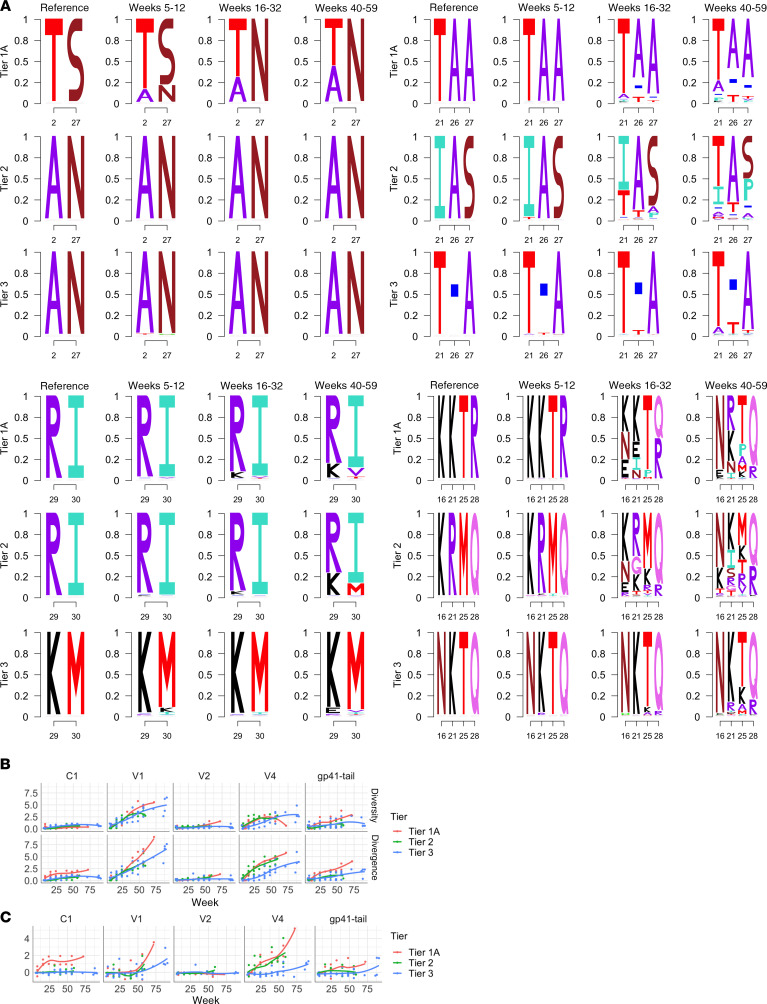
An increase of viral diversity was also observed to be associated with development of BNAbs during HIV infection (23, 40). Because of the error-prone replication of HIV-1, the appearance of NAbs, including the most broadly NAbs, would inevitably drive and select out viral escape variants, which increases the viral diversity during infection. However, whether the increase of viral diversity would stimulate the increase of neutralization breadth is still open to debate (39). To study the relationship between viral diversity and NAb development in the SIV model, we used Illumina next generation sequencing (NGS) to trace the evolution of the viral env gene during infection. We collected longitudinal samples of plasma from each macaque from 2 to 80 weeks after infection and amplified a 2.7 kb amplicon containing all of env by reverse transcription PCR (RT-PCR). After tagmentation and indexing, all samples were pooled and run on a MiSeq, generating 150-bp paired-end reads at roughly 12,500 times coverage. As described in Methods, we split the whole Env into 20 nonoverlapping regions composed of 40 codons, and we calculated the diversity and divergence levels at each region of each longitudinal sample. For a particular longitudinal sample, diversity measured the genetic variation of the viral population, and divergence measured the evolution of the viral population away from the inoculum. Four distinct regions, codons 34–73 (C1), codons 114–153 (V1), codons 401–440 (V4), and codons 801–840 (Gp41 tail), showed significantly higher divergence of nonsynonymous mutations than others (Supplemental Figure 2), indicating that these regions were under positive selection. The V1 and V4 loops, which are known to be under NAb selection (15, 26, 27), showed the highest diversity and divergence rates overall of Env in all the macaques (Supplemental Figures 3 and 4). A comparison of the codons that distinguished the 3 inocula in the C1, V1, V2, and V4 regions showed that divergence in these regions was associated with evolution in the tier 1A and tier 2 viruses toward the tier 3 viral sequence, and the timing with which tier 1A and 2 viral populations evolved toward the tier 3 viral sequence correlated with the appearance of NAbs in the macaques (Figure 5A). During the initial phase of infection (5–12 weeks after infection), when NAbs against tier 1 viruses appeared, the viral sequences of the C1 region in the tier 1A group evolved toward the tier 2 and 3 groups. During the early chronic part of infection (16–32 weeks after infection), when NAbs against tier 2 viruses appeared in all groups, elevated divergence and diversity were observed at the V1 and V4 regions in both the tier 1A and tier 2 groups, but not in the tier 3 group, and the sequences of the V1 and V4 regions in the tier 1A and 2 groups evolved toward the tier 3 group. In later parts of chronic infection (40–59 weeks after infection), the sequences of the V2 region in tier 1A and 2 began to evolve toward tier 3 groups. Elevated divergence and diversity were observed in tier 3 viruses only at the later phase (24 to 48 weeks after infection), coincident with the appearance of NAbs against tier 3 virus. However, the sequence analysis did not support the role for viral diversity in driving the increase of neutralization breadth. First, there was no trend, across all loci and tiers, of diversity rising before the rise of divergence. Instead, in C1 and the Gp41 tail, diversity was low through all longitudinal samples, whereas divergence rose as infection duration progressed in tier 1A. In V1 and V4, diversity and divergence grew at similar rates until divergence dynamics increased faster than diversity (Figure 5, B and C). Second, the divergence and diversity rates in the tier 1A–infected group were higher than those in the tier 2 group, which were in turn higher than in the tier 3 group (Figure 5, B and C), but all the macaques developed a NAb response with similar kinetics. Furthermore, the sequence divergence and diversity in each group were elevated after the appearance of NAbs against the inoculated viruses, suggesting that in these infected macaques, the increase of viral diversity was a result of neutralization breadth, but not the reverse.
Figure 5. Sequence divergence and diversity of viral Env in macaques infected with the 3 clones.
(A) Codons that distinguish the 3 inoculated SIVsmE660 variants in C1 (34–73; top left), V1 (113–154; top right), V2 (154–194; bottom left), and V4 (405–440; bottom right) regions evolved toward the tier 3 viral sequence correlated with the appearance of NAbs in the macaques. Sequence logos that were generated based on the diversity of viral sequences at different time points in the group of macaques are shown. (B) The dynamics of Env diversity (upper) and divergence (lower) at C1, V1, V4, and the Gp41 tail across the 3 tiers are shown. Diversity was calculated as the average number of amino acid differences among the pooled sequences in each sample. Divergence was calculated as the average number of amino acid differences between the sampled sequence and the inoculated reference. (C) The difference between Env divergence and diversity was calculated by subtracting the diversity number from the divergence number.
Discussion
Deciphering the key determinants of BNAb development would greatly improve the design of effective vaccines against HIV infection. Although several factors, including viral loads, viral diversity, infection time, virus subtypes, and some immune parameters have been reported to be associated with BNAb development in HIV-infected cohorts (37, 38, 40–44), the key factors determining whether an individual infected with HIV will develop BNAbs have remained unknown. None of the vaccine strategies attempted so far has successfully induced the breadth of BNAb responses seen in some infected individuals. Thus, natural infection is still the only system in which we can study how BNAbs develop. We used SIV-infected rhesus macaques as an animal model to study the development of NAb responses during AIDS virus infection. Although the sequences and antigenic property of SIV viruses are different from HIV, the structure of their envelope proteins share common features, and SIV-infected macaques exhibit a disease progression that closely mirrors HIV infection. Longitudinal studies on B cell lineages in HIV-infected patients suggest that the development of BNAbs is the result of an arms race between viruses and NAbs, with increased affinity and breadth developing as a result of somatic mutations of the immunoglobulin genes responding to changes in the circulating HIV antigens (20–23). This coevolutionary interaction was also observed in SIV-infected rhesus macaques (33, 45, 46).
In this study, we found that the coevolutionary interaction between viruses and antibodies during SIV infection was far more complex than the classical antigen-B cell interaction induced by vaccination. Whereas the presence of antigen administered after vaccination stimulated antigen-specific B cells to produce type-specific antibodies, infection resulted in progressive broadening of NAb responses. We previously found that SIV-infected macaques also developed NAb response against escape variants a few weeks after their appearance in macaques. However, when inoculated into naive macaques, the viral escape variants did not accelerate the induction of NAbs against the variants. The increase of neutralization breadth did not correlate with the viral antigen levels either. High levels of viremia induced only minor elevation in NAb response to the tier 1 virus, but had no effect on development of the NAb response against tier 2 or tier 3 viruses when compared with responses in macaques with low viremia. The kinetics of the broadening of NAb responses were similar in all macaques, and the increase of neutralization breadth only correlated with the duration of infection; i.e., NAbs to tier 1 virus appearing at 4 weeks after infection, NAbs to tier 2 virus at 6 months after infection, and NAbs to tier 3 virus at 1 year after infection. It appears likely that the type of broad neutralization achieved after long-term infection would be protective if evaluated by passive transfer to naive macaques before virus exposure, similar to studies using BNAb monoclonal Abs in the SHIV-infected rhesus macaque model. A correlation between neutralization breadth and infection time has also been commonly observed in HIV-infected cohorts (37, 40, 42). In HIV-infected humans, autologous strain-specific NAbs usually develop within a few weeks after infection, but cross-neutralization does not appear until 1–2 years after infection. Here we also found that this correlation could not be explained by the increase of viral diversity during the course of infection. Actually, we found that the increase of viral diversity was more likely to be a result of immune selection by NAbs in the macaques. Viral sequences in macaques infected with neutralization-sensitive virus changed more rapidly than those in macaques infected with neutralization-resistant virus, and the sequence divergence rates were elevated after the appearance of NAbs against the inoculated viruses, but the kinetics of NAb development were the same in these macaques.
The link between neutralization breadth and duration of infection also suggests that conventional vaccine strategies such as the use of recombinant antigen or nonreplicating viral vectors may not be sufficient to induce BNAbs against HIV. The effort of current HIV vaccine strategies is heavily focused on redesigning the viral antigen in an attempt to generate BNAbs; such strategies include the use of Env protein trimer with native structure, mosaic antigens with consensus sequences, and antigen carrying conserved epitopes. Except for the viral antigen optimization, long infection time, which means not only long-term antigen exposure but also the change in immunological parameters such as cytokines and immune cell subsets, may also play an important role in the increase of neutralization breadth. Novel vaccination strategies may be required to mimic the nature of long-term virus infection, either by the use of persistent viral vectors such as adeno-associated virus to express HIV antigen or new immunization strategies to extend antigen persistence. For example, a recent study has shown that slow-delivery immunizations with native-like HIV Env trimer BG505 enhanced humoral response and antibody diversity in rhesus macaques (47). This current study focuses on only virological factors in determining the neutralization breadth. However, the development of neutralization breadth is a highly regulated process that involves complex interactions between B cells and CD4 helper T cells (48). Further studies are required to explore molecular and immunological mechanisms in the increase of neutralization breadth during infection, for example, the dynamics of follicular CD4 T cells in germinal centers and the pathway of B cell maturation, all of which may reveal the key determinants of BNAb development and could be readily performed with this SIV-macaque model.
Methods
Animals.
Colony-bred rhesus macaques of Indian origin (Macaca mulatta) were from an NIH animal facility and housed in a BSL2 facility using BSL3 practices. The TRIM5α genotype of rhesus macaques was determined as previously described (49), and MHC I genotypes were determined by the Rhesus Macaque MHC Typing Core facility at the University of Miami Miller School of Medicine.
Viruses.
Infectious SIVsmE660 variants were isolated and constructed as previously described (33). Briefly, plasma samples were collected from rhesus macaques infected with SIVsmE660-FL6 and SIVsmE660-FL14 at different time points after infection. Viral RNA was isolated from plasma using the QiaAmp Viral RNA kit (QIAGEN). Reverse transcription was performed using SuperScript III Reverse Transcriptase (Thermo Fisher Scientific) with primer Env-R, 5′-catcatccacatcatccatg-3′. The Env region was amplified with primers Env-S, 5′-ggtgttgctatcattgtcagc-3′ and Env-R, and the PCR products were cloned into PCR4-TOPO vector with TOPO-TA cloning kit (Thermo Fisher Scientific) and sequenced. Env variants were cut with BsmI and BglII, and subcloned into the backbone of parental SIVsmE660 clones SIVsmE660-FL6 and SIVsmE660-FL14 to make infectious chimeric virus variants.
Virus stocks were made by transfection of 293T cells for neutralization assays. The 293T cells were maintained in Gibco GlutaMAX DMEM plus 10% FCS, 100 U/mL penicillin, and 100 μg/mL streptomycin and transfected with 10 μg plasmid using FuGENE 6 transfection reagent (Roche Diagnostics). Virus stocks were collected from the supernatant of transfected cells after 48 hours and filtered with a 0.22 μm filter. The TCID50 of virus stocks was tested on TZM-bl cells (NIH AIDS Reagent Program) and calculated by the Reed–Muench method.
Viruses SIVsmE660-FL14, H807-16w-6, and H807-24w-4 were expanded on rhesus PBMCs before inoculation into rhesus macaques. PBMCs were isolated by Ficoll gradient density centrifugation and cultured in complete RPMI 1640 media containing 10% IL-2. After stimulation with 2 μg/mL PHA for 72 hours, activated PBMCs were infected with the 3 SIVsmE660 variants at an MOI of 0.001. Infected PBMCs were washed and cultured in complete RPMI 1640 media containing 10% IL-2. Virus stocks were collected at day 6 and filtered, followed by titration on TZM-bl cells and quantitation by real-time PCR and p27 test. The env regions were amplified from virus PBMC stocks by RT-PCR, and PCR products were sequenced as described above.
Neutralization assay.
SIV-specific NAbs were evaluated using the TZM-bl neutralization assay as previously described (33, 50). Briefly, serial diluted heat-inactivated serum or purified anti-SIV antibody samples were mixed with 100 TCID50 viruses and incubated at 37oC for 90 minutes. After incubation, 104 TZM-bl cells were added to each well with DEAE-dextran at a final concentration of 12.5 μg /mL to enhance virus infectivity. After 40 hours of culture at 5% CO2 and 37oC, luciferase activity was measured with the Luciferase Assay Kit (Promega) and read on either a Mithras LB940 (Berthold Technologies) or Victor 3 Multilabel Plate Counter (PerkinElmer Life Sciences). Average relative luminescence units (RLU) of cell controls were subtracted as background. The IC50 was calculated with nonlinear regression by GraphPad Prism 6 (GraphPad Software) or Microsoft Excel macro and expressed as the highest dilution of sera, which resulted in a 50% reduction of RLU compared with the virus control.
Tiered categorization of neutralization sensitivity of SIV viruses.
SIV viruses were generated by transfection of 293T cells, and neutralization sensitivity was evaluated using the TZM-bl assay with a panel of antisera from SIV-infected macaques consisting of 5 individual serum samples from rhesus macaques infected with the uncloned SIVsmE660 virus, 1 pooled serum sample from 6 SIVmac251-infected rhesus macaques, 1 pooled serum sample from SIVsmmG932-infected rhesus macaques, and 1 pooled serum sample from SIVagm-infected African green monkeys. IC50 titers were calculated and a heatmap was generated based on log10 values of IC50 titers by using the web tool in the Los Alamos National Laboratory HIV sequence database with the Euclidean distance and complete clustering method (http://www.hiv.lanl.gov/content/sequence/HEATMAP/heatmap.html). Viruses were clustered according to their IC50 neutralization profiles, and those with similar sensitivity were grouped together. The representative clones with different neutralization sensitivity were tested with polyclonal NAbs purified from rhesus macaques chronically infected with uncloned SIVsmE660, which was generated as previously described (51).
In vitro PBMC replication assay.
The in vitro virus fitness was evaluated in PHA-stimulated rhesus PBMCs as previous described (33). Briefly, PBMCs were isolated by Ficoll gradient density centrifugation and cultured in complete RPMI 1640 media containing 10% IL-2. After stimulation with 2 μg/mL PHA for 72 hours, activated PBMC were infected with the 3 SIVsmE660 variants at an MOI of 0.001. Infected PBMCs were washed and cultured in complete RPMI 1640 media containing 10% IL-2. Virus production was monitored by quantifying the reverse transcriptase activity of supernatant collected at 3-day intervals with radioactive [3H] labeled dTTP as previously described (52). Reverse transcriptase values were quantified with a phosphorimaging plate (FujiFilm).
Animal inoculations.
Eighteen STLV-, SRV-, and SIV-seronegative rhesus macaques with TRIM5-TFP/Q genotypes were divided into 3 groups. The distribution of MHC class I alleles Mamu-A*01, Mamu-B*08, and Mamu-B*17 was balanced between the groups. Each macaque was inoculated intrarectally with a 1:50 dilution of SIVsmE660-FL14, H807-16w-6, or H807-24w-4 PBMC virus stocks (1000 TCID50, 107 RNA copies, 0.5 ng P27). After inoculation, viral RNA levels in plasma were determined by quantitative PCR. Four weeks later, any of the macaques that remained uninfected were inoculated intrarectally on a weekly schedule with the same amount of virus until viral RNAs became detectable in plasma. After infection, blood and plasma were collected, and plasma viral RNA levels were determined. Early endpoint criteria, as specified by the IACUC-approved parameters, were used to determine when animals should be humanely euthanized. Macaques were euthanized if they lost more than 20% of their body weight or developed intractable diarrhea that was unresponsive to supportive or antibiotic treatment, respiratory signs with radiographic evidence of pneumonia, persistent anorexia and lethargy, or neurological signs.
Illumina MiSeq deep sequencing.
Total plasma RNA was extracted with the QiaAmp Viral RNA kit (QIAGEN), and DNase treated per the manufacturer’s instructions (Invitrogen DNase I, Amplification Grade, Thermo Fisher Scientific). For animals with plasma viral loads of greater than 1 × 105 copies/mL, 8 μL of RNA were used; for animals with plasma loads of 1 × 104 to 1 × 105 copies/mL, 16 μL of RNA were used. After DNase treatment, 8 μL/16 μL RNA (high viral load/low viral load) were used as template for reverse-transcribing PCR, and the remaining 2.5 μL RNA were used in a DNA-contamination negative-control PCR. Env-specific 2.7 kb amplicons were generated using the QIAGEN One-Step RT-PCR kit with the following primers: 5′-/5AmMC6/GTA TGG GAT GTC TTG GGA ATC AGC TGC TTA TCG-3′ and 5′-/5AmMC6/GGC TCA CAA GAG AGT GAG CTC AAG CC-3′, and the following touchdown PCR conditions: 45°C for 2 hours, 95°C for 15 minutes, 6 cycles of 94°C for 30 seconds, 68°C–1°C for 20 seconds, 68°C for 4 minutes; 35 cycles of 94°C for 15 seconds, 62°C for 20 seconds, 68°C for 4 minutes; 68°C for 15 minutes, 4°C hold. DNA-contamination control PCRs were done using either TaKaRa Ex Taq DNA Polymerase (Takara Bio Company) or Invitrogen Platinum PCR SuperMix (Thermo Fisher Scientific), and run as follows: 95°C for 1 minute, 40 cycles of 94°C for 15 seconds, 62°C for 30 seconds, 68°C for 4 minutes; 68°C for 10 minutes, 4°C hold.
RT-PCR products were purified with the Agencourt AMPure XP PCR Purification system (Beckman Coulter, Inc.) or with Axygen AxyPrep (Thermo Fisher Scientific), eluted in nuclease-free water, and concentrations were determined with the Qubit dsDNA HS Assay Kit on the Qubit 3.0 Fluorometer (Life Technologies; Thermo Fisher Scientific).
The Illumina Nextera XT DNA Library Preparation Kit was used to tagment the amplicons per the manufacturer’s instructions, and each library was uniquely indexed with the Illumina Nextera XT Index Kit V2 Set A.
After pooling all individual libraries, the pooled sample was analyzed on a 2200 TapeStation (Agilent Technologies) and Qubit to check the size distribution and total amount of DNA. Illumina PhiX Control v3 was added at 15% of the total amount of DNA to create the final library, which was run on an Illumina MiSeq with an Illumina MiSeq Reagent Kit v2 (300-cycle). The NGS sequencing data was deposited into GenBank with BioProject number PRJNA637366.
NGS data analysis.
The data were initially analyzed with the following tools: Vicuna (http://www.broadinstitute.org/scientific-community/science/projects/viral-genomics/vicuna; 06/15/2016) was used to assemble and reassemble reads, Mosaik was used to align reads back to the assembly, Picard script (http://broadinstitute.github.io/picard/; 06/15/2016) was used to remove duplicate reads, and V-FAT (http://www.broadinstitute.org/scientific-community/science/projects/viral-genomics/v-fat; 06/15/2016) was used to clean up the assemblies. Additional analyses and visualization were performed using Geneious version R8 (Biomatters Ltd.).
Analysis of viral evolution.
To analyze viral evolution, we used the tier 1A, tier 2, and tier 3 inoculum consensus sequences as references. Each reference was split into 20 nonoverlapping but contiguous loci composed of 40 codons: the first locus consisted of nucleotide position 100–219, the second locus consisted of positions 220–339, and so on. The first 100 and last 140 nucleotides on the references were ignored because of low coverage. To calculate divergence and diversity at a specific locus, we considered only those reads that spanned the whole locus. We calculated divergence as the average hamming distance between the reads spanning the locus and the reference, and we calculated diversity as the pairwise average hamming distance between reads spanning the locus.
To produce the sequence logo figures, we calculated codon frequencies of the viral sequences at positions that aligned to the variable codon positions across the 3 inoculum references. Codon frequencies were then used to determine amino acid frequencies. The logo figures were generated using the Logolas R package.
Statistics.
All statistical analyses and graphical analyses were performed using GraphPad Prism 6. Kaplan-Meier curves were plotted based on the inoculation number before infection and a log-rank test was used to compare the acquisition of infection. A nonparametric Kruskal-Wallis test was used for the comparison of viral loads between the 3 groups, and Dunn’s multiple-comparisons test was used to correct for multiple comparisons. The cumulative survival rates of infected macaques were plotted as Kaplan-Meier curves and were compared by log-rank test. Repeated-measures 2-way ANOVA was used to compare the kinetics of viral replication in PBMCs or NAb development. A P value less than 0.05 was considered statistically significant.
Study approval.
All the macaques were maintained in accordance with the guidelines of the Committee on the Care and Use of Laboratory Animals under an NIAID-approved animal study protocol.
Author contributions
FW and VMH conceived and designed the experiments; FW, IO, SW, and KM performed in vivo experiments; FW and IO performed in vitro experiments; CL and DM performed neutralization sensitivity screen. AK and SL performed NGS sequencing analyses; FW, AK, SL, CL, JH, and VMH analyzed data and provided feedback. FW and VMH wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Alicia Buckler-White and Ronald Plishka in the Laboratory of Molecular Microbiology, NIAID, NIH for determining plasma viral RNA loads. We also thank Heather Cronise-Santis, Joanne Swerczek, and Richard Herbert in the NIH Animal Center for care of the study animals. This work was supported by the intramural program of NIAID, NIH. Fan Wu and Jinghe Huang were supported by National Science and Technology Major Project 2018ZX10301403, 2017ZX10202102. Celia LaBranche and David Montefiori were supported by NIH/NIAID contract HHSN27201100016C. Jinghe Huang was supported by National Natural Science Foundation of China 31771008. Andrea Kirmaier was sponsored by the Boston College Ignite Award.
Version 1. 07/14/2020
In-Press Preview
Version 2. 09/14/2020
Electronic publication
Version 3. 10/01/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(10):5413–5424.https://doi.org/10.1172/JCI139123.
Contributor Information
Fan Wu, Email: wufan@fudan.edu.cn.
Ilnour Ourmanov, Email: IOURMANOV@niaid.nih.gov.
Andrea Kirmaier, Email: andrea.kirmaier@bc.edu.
Sivan Leviyang, Email: sivan.leviyang@georgetown.edu.
Celia LaBranche, Email: celia.labranche@duke.edu.
Jinghe Huang, Email: Jinghehuang@fudan.edu.cn.
Sonya Whitted, Email: swhitted@niaid.nih.gov.
Kenta Matsuda, Email: kenta.matsuda@nih.gov.
David Montefiori, Email: monte@duke.edu.
Vanessa M. Hirsch, Email: vhirsch@niaid.nih.gov.
References
- 1.Zinkernagel RM. On natural and artificial vaccinations. Annu Rev Immunol. 2003;21:515–546. doi: 10.1146/annurev.immunol.21.120601.141045. [DOI] [PubMed] [Google Scholar]
- 2.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329(5993):856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pejchal R, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334(6059):1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang J, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491(7424):406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheid JF, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458(7238):636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 6.Huang J, et al. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature. 2014;515(7525):138–142. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang J, et al. Identification of a CD4-binding-site antibody to HIV that evolved near-pan neutralization breadth. Immunity. 2016;45(5):1108–1121. doi: 10.1016/j.immuni.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schoofs T, et al. Broad and potent neutralizing antibodies recognize the silent face of the HIV envelope. Immunity. 2019;50(6):1513–1529. doi: 10.1016/j.immuni.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shingai M, et al. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J Exp Med. 2014;211(10):2061–2074. doi: 10.1084/jem.20132494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saunders KO, et al. Broadly neutralizing human immunodeficiency virus type 1 antibody gene transfer protects nonhuman primates from mucosal simian-human immunodeficiency virus infection. J Virol. 2015;89(16):8334–8345. doi: 10.1128/JVI.00908-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, et al. Antibody-mediated protection against SHIV challenge includes systemic clearance of distal virus. Science. 2016;353(6303):1045–1049. doi: 10.1126/science.aag0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishimura Y, et al. Early antibody therapy can induce long-lasting immunity to SHIV. Nature. 2017;543(7646):559–563. doi: 10.1038/nature21435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Julg B, et al. Protection against a mixed SHIV challenge by a broadly neutralizing antibody cocktail. Sci Transl Med. 2017;9(408):eaao4235. doi: 10.1126/scitranslmed.aao4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halper-Stromberg A, et al. Broadly neutralizing antibodies and viral inducers decrease rebound from HIV-1 latent reservoirs in humanized mice. Cell. 2014;158(5):989–999. doi: 10.1016/j.cell.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caskey M, et al. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature. 2015;522(7557):487–491. doi: 10.1038/nature14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bar KJ, et al. Effect of HIV Antibody VRC01 on viral rebound after treatment interruption. N Engl J Med. 2016;375(21):2037–2050. doi: 10.1056/NEJMoa1608243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caskey M, Klein F, Nussenzweig MC. Broadly neutralizing anti-HIV-1 monoclonal antibodies in the clinic. Nat Med. 2019;25(4):547–553. doi: 10.1038/s41591-019-0412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landais E, Moore PL. Development of broadly neutralizing antibodies in HIV-1 infected elite neutralizers. Retrovirology. 2018;15(1):61. doi: 10.1186/s12977-018-0443-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonsignori M, et al. Antibody-virus co-evolution in HIV infection: paths for HIV vaccine development. Immunol Rev. 2017;275(1):145–160. doi: 10.1111/imr.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao HX, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496(7446):469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao F, et al. Cooperation of B cell lineages in induction of HIV-1-broadly neutralizing antibodies. Cell. 2014;158(3):481–491. doi: 10.1016/j.cell.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonsignori M, et al. Maturation pathway from germline to broad HIV-1 neutralizer of a CD4-mimic antibody. Cell. 2016;165(2):449–463. doi: 10.1016/j.cell.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doria-Rose NA, et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509(7498):55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonsignori M, et al. Staged induction of HIV-1 glycan-dependent broadly neutralizing antibodies. Sci Transl Med. 2017;9(381):eaai7514. doi: 10.1126/scitranslmed.aai7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacLeod DT, et al. Early antibody lineage diversification and independent limb maturation lead to broad HIV-1 neutralization targeting the Env high-mannose patch. Immunity. 2016;44(5):1215–1226. doi: 10.1016/j.immuni.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krebs SJ, et al. Longitudinal analysis reveals early development of three MPER-directed neutralizing antibody lineages from an HIV-1-infected individual. Immunity. 2019;50(3):677–691. doi: 10.1016/j.immuni.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore PL, Gorman J, Doria-Rose NA, Morris L. Ontogeny-based immunogens for the induction of V2-directed HIV broadly neutralizing antibodies. Immunol Rev. 2017;275(1):217–229. doi: 10.1111/imr.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams WB, et al. Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations. Nat Commun. 2017;8(1):1732. doi: 10.1038/s41467-017-01336-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pauthner M, et al. Elicitation of robust tier 2 neutralizing antibody responses in nonhuman primates by HIV envelope trimer immunization using optimized approaches. Immunity. 2017;46(6):1073–1088.e6. doi: 10.1016/j.immuni.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saunders KO, et al. Vaccine induction of heterologous tier 2 HIV-1 neutralizing antibodies in animal models. Cell Rep. 2017;21(13):3681–3690. doi: 10.1016/j.celrep.2017.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voss JE, et al. Elicitation of neutralizing antibodies targeting the V2 apex of the HIV envelope trimer in a wild-type animal model. Cell Rep. 2017;21(1):222–235. doi: 10.1016/j.celrep.2017.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malherbe DC, et al. Sequential immunization with a subtype B HIV-1 envelope quasispecies partially mimics the in vivo development of neutralizing antibodies. J Virol. 2011;85(11):5262–5274. doi: 10.1128/JVI.02419-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu F, et al. Sequential evolution and escape from neutralization of simian immunodeficiency virus SIVsmE660 clones in rhesus macaques. J Virol. 2012;86(16):8835–8847. doi: 10.1128/JVI.00923-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seaman MS, et al. tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol. 2010;84(3):1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roederer M, et al. Immunological and virological mechanisms of vaccine-mediated protection against SIV and HIV. Nature. 2014;505(7484):502–508. doi: 10.1038/nature12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Loffredo JT, Valentine LE, Watkins DI. Beyond Mamu-A*01+ Indian rhesus macaques: continued discovery of new MHC class I molecules that bind epitopes from the simian AIDS viruses. In: Korber B, et al, eds. HIV Molecular Immunology. Los Alamos National Laboratory, Theoretical Biology and Biophysics;2006/2007:29–51. [Google Scholar]
- 37.Sather DN, et al. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol. 2009;83(2):757–769. doi: 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gray ES, et al. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J Virol. 2011;85(10):4828–4840. doi: 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore PL, Williamson C, Morris L. Virological features associated with the development of broadly neutralizing antibodies to HIV-1. Trends Microbiol. 2015;23(4):204–211. doi: 10.1016/j.tim.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rusert P, et al. Determinants of HIV-1 broadly neutralizing antibody induction. Nat Med. 2016;22(11):1260–1267. doi: 10.1038/nm.4187. [DOI] [PubMed] [Google Scholar]
- 41.Doria-Rose NA, et al. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J Virol. 2010;84(3):1631–1636. doi: 10.1128/JVI.01482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Landais E, et al. Broadly neutralizing antibody responses in a large longitudinal sub-Saharan HIV primary infection cohort. PLoS Pathog. 2016;12(1):e1005369. doi: 10.1371/journal.ppat.1005369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mikell I, Sather DN, Kalams SA, Altfeld M, Alter G, Stamatatos L. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog. 2011;7(1):e1001251. doi: 10.1371/journal.ppat.1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piantadosi A, et al. Breadth of neutralizing antibody response to human immunodeficiency virus type 1 is affected by factors early in infection but does not influence disease progression. J Virol. 2009;83(19):10269–10274. doi: 10.1128/JVI.01149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sato S, Johnson W. Antibody-mediated neutralization and simian immunodeficiency virus models of HIV/AIDS. Curr HIV Res. 2007;5(6):594–607. doi: 10.2174/157016207782418515. [DOI] [PubMed] [Google Scholar]
- 46.Yeh WW, et al. Autologous neutralizing antibodies to the transmitted/founder viruses emerge late after simian immunodeficiency virus SIVmac251 infection of rhesus monkeys. J Virol. 2010;84(12):6018–6032. doi: 10.1128/JVI.02741-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cirelli KM, et al. Slow delivery immunization enhances HIV neutralizing antibody and germinal center responses via modulation of immunodominance. Cell. 2019;177(5):1153–1171.e28. doi: 10.1016/j.cell.2019.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 49.Kirmaier A, et al. TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol. 2010;8(8):e1000462. doi: 10.1371/journal.pbio.1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Montefiori DC. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol Biol. 2009;485:395–405. doi: 10.1007/978-1-59745-170-3_26. [DOI] [PubMed] [Google Scholar]
- 51.Haigwood NL, et al. Passive immune globulin therapy in the SIV/macaque model: early intervention can alter disease profile. Immunol Lett. 1996;51(1-2):107–114. doi: 10.1016/0165-2478(96)02563-1. [DOI] [PubMed] [Google Scholar]
- 52.Lee MH, Sano K, Morales FE, Imagawa DT. Sensitive reverse transcriptase assay to detect and quantitate human immunodeficiency virus. J Clin Microbiol. 1987;25(9):1717–1721. doi: 10.1128/JCM.25.9.1717-1721.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.