Abstract
During hemolysis, macrophages in the liver phagocytose damaged erythrocytes to prevent the toxic effects of cell-free hemoglobin and heme. It remains unclear how this homeostatic process modulates phagocyte functions in inflammatory diseases. Using a genetic mouse model of spherocytosis and single-cell RNA sequencing, we found that erythrophagocytosis skewed liver macrophages into an antiinflammatory phenotype that we defined as MarcohiHmoxhiMHC class IIlo erythrophagocytes. This phenotype transformation profoundly mitigated disease expression in a model of an anti-CD40–induced hyperinflammatory syndrome with necrotic hepatitis and in a nonalcoholic steatohepatitis model, representing 2 macrophage-driven sterile inflammatory diseases. We reproduced the antiinflammatory erythrophagocyte transformation in vitro by heme exposure of mouse and human macrophages, yielding a distinctive transcriptional signature that segregated heme-polarized from M1- and M2-polarized cells. Mapping transposase-accessible chromatin in single cells by sequencing defined the transcription factor NFE2L2/NRF2 as a critical driver of erythrophagocytes, and Nfe2l2/Nrf2 deficiency restored heme-suppressed inflammation. Our findings point to a pathway that regulates macrophage functions to link erythrocyte homeostasis with innate immunity.
Keywords: Hematology
Keywords: Innate immunity, Macrophages
Introduction
The archetypical functions of macrophages are their roles as sensors, effectors, and resolvers of inflammation and tissue damage (1). Historically defined as inflammatory M1-like and antiinflammatory M2-like, macrophage phenotypes and functions are regulated by ontogeny and by tissue- and disease-specific triggers that regulate innate immune responses and tissue repair following injury (2, 3). Macrophages also play a key role in physiological erythrocyte turnover by mediating phagocytic clearance of senescent and damaged red blood cells (RBCs). This homeostatic function of macrophages becomes imperative in conditions with enhanced erythrolytic stress, such as in patients with an acquired or genetic hemolytic anemia (4).
RBCs, or erythrocytes, typically compartmentalize hemoglobin and heme within their antioxidative intracellular space. However, in conditions with pathological erythrolysis (5–7), this protective compartmentalization is lost, and RBC toxins accumulate in tissue and plasma. Cell-free hemoglobin and heme cause adverse physiological effects by nitric oxide depletion, free radical reactions, and heme-triggered inflammation via Toll-like receptor (TLR) engagement (8–18). These disease-accelerating effects of uncontrolled erythrolysis are prevented by erythrophagocytosis, whereby macrophages recognize and phagocytose damaged RBCs before they disintegrate within the circulation (19–21). In conditions with systemic hemolysis such as in sickle cell disease, spherocytosis, or after the transfusion of stored blood or during sepsis, this function is provided by macrophages in the liver (19).
Cell culture experiments and immunophenotyping in human tissues have demonstrated that erythrophagocytosis supports the differentiation of specialized macrophages with high antioxidant and iron-metabolizing capacity, suggesting that adaptation of the myeloid cell compartment provides tolerance against the toxicity of hemolysis (22–32). In vitro studies with human monocyte-derived macrophages found that hemoglobin or heme exposure repressed inflammatory responses and immunophenotype markers of activation (22–25). So far, it remains unanswered whether these effects of altered RBC homeostasis and heme metabolism on phagocytes could also change regulatory macrophage functions in the context of inflammatory diseases. Here, we hypothesized that enhanced demand for RBC clearance in individuals with a genetic or acquired hemolysis changes the macrophage phenotype landscape in vivo and diverges the progression of macrophage-driven noninfectious inflammatory diseases.
Nuclear erythroid 2 p45-related factor 2 (NFE2L2, also known as NRF2) is a redox-sensitive basic leucine zipper transcription factor and the principal regulator of oxidative stress defense (33–36). Recently, NRF2 has gained attention as a negative regulator of inflammatory disease processes (37–40). NRF2 can suppress inflammation by at least 4 mutually interacting mechanisms including modulation of reactive oxygen species–dependent signaling (37, 41), transcriptional repression (42), interference with NF-κB (43), posttranscriptional modulation of inflammatory signaling genes (44, 45), or via enhancement of Hmox1 (46, 47).
In the present study, we provide a global in vivo definition of the phenotype of erythrophagocytic macrophages, which we termed erythrophagocytes. We define these cells as a unique macrophage type with an antiinflammatory gene signature driven by sustained activation of a heme/NRF2 signaling pathway. Using a mouse model of hereditary spherocytosis and single-cell RNA sequencing (scRNA-seq) of mouse liver macrophages, we discovered that erythrophagocytosis forces the transformation of liver macrophages into an antiinflammatory phenotype. As a result, we found in 2 models of macrophage-driven inflammatory liver disease that hemolytic stress profoundly attenuates disease expression.
Results
Identification of erythrophagocytes in the liver of hemolytic mice.
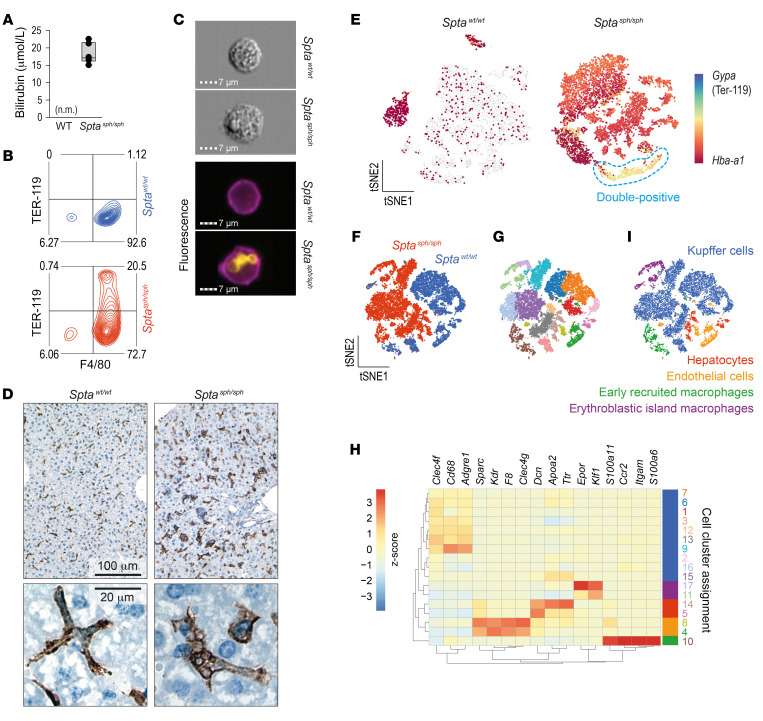
We used a mouse with a spontaneous truncation of the erythrocyte-specific cytoskeletal protein spectrin-α (Sptasph/sph) (48) as an in vivo model of erythrophagocytosis to characterize the phenotype and inflammatory function of erythrophagocytic macrophages, which we term here erythrophagocytes. Sptasph/sph mice exhibited chronic hemolytic anemia with hyperbilirubinemia (Figure 1A) resulting from a structural membrane instability leading to enhanced RBC clearance by phagocytes in the liver (49). Using flow cytometry with corresponding imaging, we identified a large population of F4/80+ macrophages in the livers of Sptasph/sph mice that were also positive for ingested TER-119+ erythrocytes (Figure 1, B and C). In a histological analysis, the large F4/80+ erythrophagocytes emerged as the predominant macrophage population in the liver of Sptasph/sph mice (Figure 1D). Automatic counting of F4/80+ cells in the liver sections revealed a higher number of macrophages in the hemolytic than in the WT mouse (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI137282DS1).
Figure 1. Identification of erythrophagocytes in the liver of hemolytic mice.
(A) Plasma bilirubin in Sptasph/sph and Sptawt/wt (WT) mice (n = 4 for both groups, n.m. = not measurable). (B) Flow cytometry density plots of liver cell suspensions stained for F4/80 and intracellular TER-119 in Sptasph/sph mice (red) and Sptawt/wt littermates (blue). The displayed cells were gated from live CD45+ leukocytes. Data are representative of 3 independent experiments. Percentage proportions of cell counts are displayed in the plot. (C) Brightfield and fluorescence images showing intracellular TER-119+ erythrocytes (yellow) in F4/80+ Kupffer cells (KCs) (purple) from Sptawt/wt and Sptasph/sph mice obtained by imaging flow cytometry. (D) Top panel: Microphotographs of liver tissues stained for F4/80. Bottom panel: High-magnification microphotographs of F4/80+ macrophages from Sptawt/wt and Sptasph/sph mice. Data representative of 3 mice in each group. (E) Single-cell RNA sequencing (scRNA-seq) data of nonparenchymal liver cell suspensions enriched for macrophages with F4/80 antibody–coated magnetic Dynabeads from Sptasph/sph and Sptawt/wt mice. t-SNE plots showing color-coded Gypa/Hba-a1 gene expression in Sptawt/wt and Sptasph/sph mice. Double-positive cells are highlighted by the blue dashed line. Each dot represents a single cell. (F) Data sets from the 2 mice shown in E are merged and represented in a single t-SNE plot, showing 17,127 cells colored by cell origin. (G) Same t-SNE as in F. After an unsupervised clustering analysis, cells sharing similar transcriptome profiles were grouped by color. Seventeen clusters were identified. (H) Heatmap of the average expression of key marker genes for hepatocytes, endothelial cells, erythroblastic island macrophages (EBIs), early recruited macrophages, and KCs in each cluster. Data are standardized by row, Z scores are displayed by color code (red = high expression, blue = low expression). (I) Assigned cell populations defined in H. Cells are colored based on their identities.
Erythrophagocytes have an antioxidant and antiinflammatory transcriptional phenotype.
To assess the phenotype of erythrophagocytes in Sptasph/sph mice, we performed scRNA-seq analysis on macrophages that were enriched from nonparenchymal liver cell suspensions with anti-F4/80 antibody–coated Dynabeads. Figure 1E shows a t-distributed stochastic neighbor embedding (t-SNE) plot of scRNA-seq data from an Sptawt/wt mouse and an Sptasph/sph mouse. In these data, we could recapitulate the presence of a large population of erythrophagocytes in the Sptasph/sph hemolytic mouse by visualizing the mRNA expression of hemoglobin (Hba-a1), which stems from remnant mRNA in phagocytosed RBCs. To support this interpretation, we confirmed by RT-qPCR that Hba-a1 transcripts were highly abundant in leukocyte-depleted circulating RBCs of Sptasph/sph mice. Accordingly, while only a small population of Hba-a1–positive cells was observed in the Sptawt/wt mouse, almost all macrophages were Hba-a1 positive in the Sptasph/sph hemolytic mouse. In addition, we found a small population of Hba-a1/Gypa (TER-119) double-positive macrophages (Figure 1E), which likely represent erythroblastic island macrophages (EBIs) that phagocytosed the nuclei of RBC progenitors (50).
Further analysis and visualization of the liver macrophage scRNA-seq data was performed on the merged data sets from Sptawt/wt and Sptasph/sph mice (Figure 1F). Unsupervised graph-based clustering grouped the 17,127 cells into 17 populations (Figure 1G). The cell-type identity of each cell cluster was determined by matching its gene expression profile with ex ante–defined marker genes for Kupffer cells (KCs), EBIs (51, 52), and early-recruited macrophages as well as hepatocytes and endothelial cells (Figure 1, H and I, Supplemental Figure 1B, and refs. 53–59).
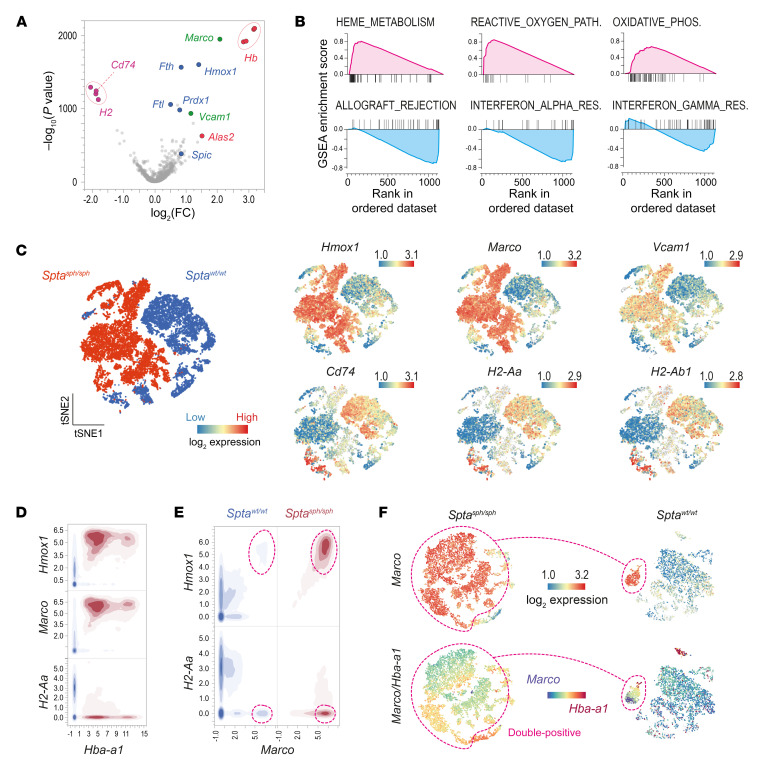
The volcano plot in Figure 2A illustrates the results of a differential gene expression analysis of KCs from the Sptasph/sph mouse (i.e., erythrophagocytes) and KCs from the Sptawt/wt littermate (Supplemental Table 1). Among the most overexpressed genes in liver macrophages from Sptasph/sph mice were marker genes of the erythrophagocytic process (Hb and Alas2), genes known to be induced by heme exposure (Hmox1 and Spic), oxidative stress–responsive genes (Fth1, Ftl1, and Prdx1), and cell surface receptor genes (Marco and Vcam1). In contrast, MHC class II–associated genes (Cd74 and several H2 genes) were most downregulated. Accordingly, gene set enrichment analysis (GSEA) using the hallmark gene sets of the Molecular Signature Database (MSigDB) (60) identified heme metabolism, the reactive oxygen species pathway, and oxidative phosphorylation as the most upregulated processes. In contrast, genes associated with allograft rejection and IFN-α/γ responses were the most downregulated, suggesting that erythrophagocytes in the livers of Sptasph/sph mice have an antiinflammatory phenotype (Figure 2B and Supplemental Figure 7A). Visualizations of the gene expression differences on a single-cell level between Sptasph/sph and Sptawt/wt mice for Hmox1, Marco, and Vcam1 as well as the MHC class II–associated genes Cd74, H2-Aa, and H2-Ab1 are shown in Figure 2C.
Figure 2. Antioxidant and antiinflammatory transcriptional phenotype of erythrophagocytes.
(A) Volcano plot demonstrating differential gene expression of KCs in Sptasph/sph versus Sptawt/wt mice [–log10(P value) versus log2(fold change)]. Among the positive differentially expressed genes (DEGs), marker genes of the erythrophagocytic process are highlighted in red, genes associated with heme exposure and oxidative stress are highlighted in blue, and macrophage receptors are highlighted in green. The downregulated MHC class II–associated genes are highlighted in purple. (B) DEGs were computed by gene set enrichment analysis (GSEA). Enrichment plots of the top 3 positively (magenta) and negatively (blue) enriched hallmark gene sets are shown. Plots display running enrichment score and position of gene set members on the rank-ordered list. (C) t-SNE plot showing all cells colored by cell origin. The expression of Hmox1, Marco, Vcam1, Cd74, H2-Aa, and H2-Ab1 is presented in global t-SNE plots. Legend for relative log2 expression of each gene from lowest expression (blue dots) to the highest expression (red dots) is displayed on the top right. (D) Contour plot showing the correlation of Hmox1, Marco, or H2-Aa and Hba-a1 count data from every cell in the Sptasph/sph (red) and Sptawt/wt (blue) mouse data sets. (E) Contour plot showing the correlation of Hmox1 or H2-Aa and Marco count data from every cell in the Sptasph/sph (red) and Sptawt/wt (blue) mouse data sets. (F) t-SNE plots with color-coded expression of Marco and Marco/Hba-a1 in Sptasph/sph (left) and Sptawt/wt (right) mice. The magenta dashed line highlights the Marco/Hba-a1 double-positive erythrophagocytes.
Among the intrinsic macrophage genes, we identified macrophage receptor with collagenous structure (Marco) as the most upregulated gene in the Sptasph/sph mouse. Figure 2D shows a 2-dimensional density plot of mRNA count data summarizing the key phenotype characteristics that we identified for the erythrophagocytes in the Sptasph/sph mouse as MarcohiHmoxhiMHC class IIlo. Interestingly, we also detected a small population of HmoxhiMHC class IIlo macrophages in the Sptawt/wt mouse liver (encircled population in Figure 2, E and F). This population was also positive for Marco expression coinciding with the erythrophagocytosis marker Hba-a1 (double-positive in Figure 2F). To further characterize these physiological erythrophagocytes, we performed a differential gene expression analysis comparing Marco+ KCs with the major KC population (Marconeg) in the Sptawt/wt mouse liver (Supplemental Figure 2 and Supplemental Table 2). We found that these cells share their gene expression profile with the erythrophagocyte population of the Sptasph/sph hemolytic mouse.
In order to identify potential drivers of gene expression in erythrophagocytes, we performed an enrichment analysis of the upregulated genes identified in Sptasph/sph macrophages for transcription factor target motifs using EnrichR (61) and identified the transcription factor NRF2 as being the most significantly enriched (Supplemental Figure 1C).
To confirm the above scRNA-seq data, we performed bulk RNA-seq analysis of F4/80+ liver macrophages that were also isolated using anti-F4/80 antibody–coated Dynabeads from 3 Sptasph/sph mice and 3 Sptawt/wt mice, and identified the same transcriptional signature for erythrophagocytes with strongly increased expression of Hmox1, Marco, Spic, and Vcam1, while MHC class II–associated genes (Cd74, H2-Aa, H2-Ab1, and H2-Eb1) were profoundly suppressed (Supplemental Figure 3A). The top 2000 differentially expressed genes (DEGs) in Sptasph/sph versus Sptawt/wt macrophages (Supplemental Table 3) were subjected to hierarchical clustering to segregate them into 6 regulatory clusters defining hemolytic and nonhemolytic gene expression of liver macrophages (Supplemental Figure 3B, left). Enrichment analysis of transcription factor binding sites for each of the 6 gene clusters (Supplemental Figure 3B, right) revealed that among the genes upregulated in the Sptasph/sph macrophages, the largest cluster was most significantly enriched for genes known to be regulated by NRF2. A strong signal was also identified for GATA1, the master transcription factor of erythropoiesis. This is compatible with the detection of intrinsic erythrocyte mRNA after RBC phagocytosis by erythrophagocytes. The downregulated gene clusters were enriched for genes controlled by RFXAP, RFXANK, and RFX5, which are regulators of MHC class II–associated genes.
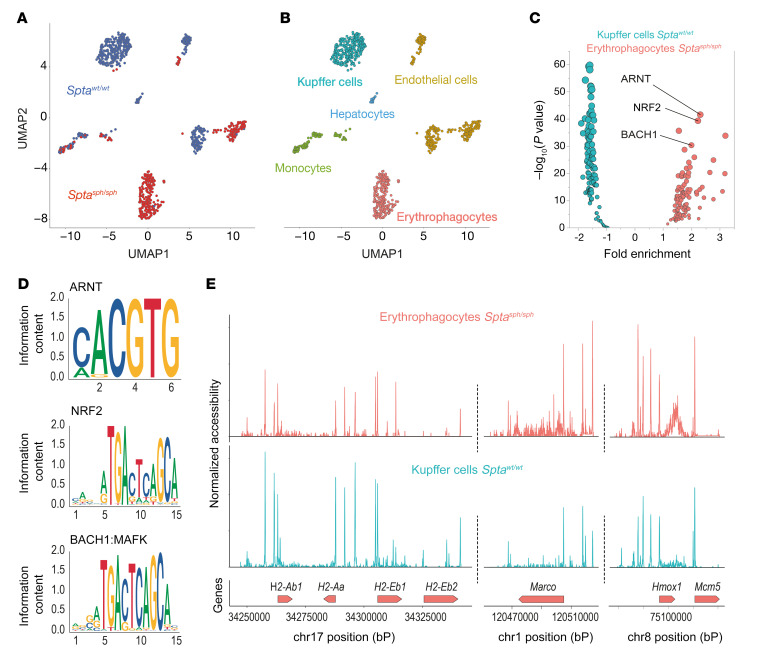
To further investigate mechanisms that may govern the differentiation of erythrophagocytes, we assessed genome-wide chromatin accessibility by sequencing transposase-accessible chromatin in single cells (scATAC-seq) of Sptasph/sph versus Sptawt/wt liver macrophages. Figure 3A shows an unsupervised clustering analysis of 862 nuclei that were isolated after enrichment of F4/80+ cells from mouse livers. The nuclei segregated into 10 clusters based on the different peak fragments they displayed (Supplemental Figure 4A). We assessed the pseudogene expression for specific genes in each cell by summing the reads containing the gene body with its associated upstream promoter region. After examining the accessibility of canonical marker genes (Supplemental Figure 4B), we could attribute cell-type identities to each cluster (Figure 3B). Although early-recruited macrophages of both genotypes appeared in the same cluster, KCs of Sptawt/wt and erythrophagocytes of Sptasph/sph were clearly separated from each other, indicating a significantly different overall accessibility of their chromatin. Differential analysis of accessible transcription factor motifs in these 2 cell types (Figure 3C) established the motif for NRF2 (MA0150.2) as the second most significantly overrepresented hit in erythrophagocytes (Figure 3D and Supplemental Tables 4 and 5). scATAC-seq also confirmed reduced accessibility for the MHC class II locus on chromosome 17 and enhanced accessibility of the Marco and Hmox1 genes, which is compatible with the scRNA-seq data (Figure 3E).
Figure 3. Chromatin accessibility and transcription factor motif enrichment in erythrophagocytes.
Single-cell ATAC sequencing data of nonparenchymal liver cell suspensions enriched for macrophages with F4/80 antibody–coated magnetic Dynabeads from Sptasph/sph and Sptawt/wt mice. (A) UMAP plots showing all cells colored by cell origin (Sptasph/sph red, Sptawt/wt blue). (B) UMAP plots showing all cells colored by cell type. Cell type was attributed based on the pseudogene expression of each cell for specific cell type markers displayed in Supplemental Figure 4B. (C) Volcano plot displaying the results of the motif enrichment analysis in Sptasph/sph erythrophagocytes compared with Sptawt/wt KCs (red, 119 motifs in total) and Sptawt/wt KCs compared with Sptasph/sph erythrophagocytes (blue, 119 motifs in total). (D) Position weight matrix plot of NRF2 motif (MA0150.2), BACH1 motif (MA0591.1), and ARNT motif (MA0004.1) from the JASPAR 2018 database with the corresponding P value from the motif enrichment analysis. (E) Coverage plots around the coding region for 4 MHC class II genes (H2-Ab1, H2-Aa, H2-Eb1, and H2-Eb2), Marco (with 20,000 kb upstream and downstream), and Hmox1 (with 20,000 kb upstream and downstream) in Sptasph/sph erythrophagocytes compared with Sptawt/wt KCs.
In summary, we identified a phenotype of MarcohiHmoxhiMHC class IIlo erythrophagocytes that is characterized by NRF2-driven gene expression. In the human liver, the expression of Marco defines a subset of antiinflammatory macrophages (62). Therefore, our data suggest the existence of a physiological population of antiinflammatory Marco+ erythrophagocytes that expands dramatically in response to hemolytic stress, becoming the most dominant macrophage phenotype in the liver and potentially modulating innate-immunity functions.
Erythrophagocytes have an antiinflammatory function.
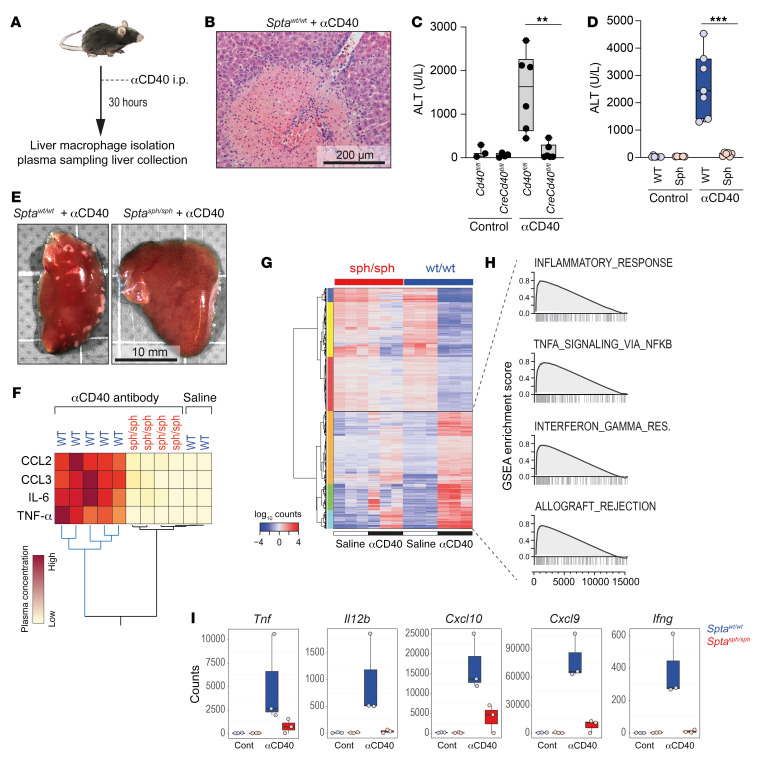
We examined erythrophagocyte functions in an in vivo mouse model of inflammation. To establish a suitable model where inflammation is driven by macrophages, we decided to target CD40 (Figure 4A), an NF-κB–activating receptor on mononuclear phagocytes. Thirty hours after injection of an agonistic anti-CD40 antibody, WT mice developed a systemic inflammatory syndrome reminiscent of a macrophage activation syndrome (63, 64), which was characterized by a cytokine storm, activated coagulation with thrombosis in the liver vasculature, and multifocal liver necrosis (Figure 4B). As a result of this liver necrosis, the mice had increased plasma levels of alanine aminotransferase (ALT), which we used as a biomarker for the disease process. Depletion of macrophages by clodronate liposomes completely prevented the disease phenotype, as illustrated by the lack of ALT increase after antibody treatment (Supplemental Figure 3D), suggesting that the inflammatory condition seen in the model represents a macrophage-driven process. To conclusively validate the specificity of macrophage-driven inflammatory disease in this model, we generated a macrophage-specific conditional Cd40–knockout mouse (Cd40fl/fl LysM-Cre). In comparison with Cd40fl/fl littermates that exhibited increased ALT levels upon treatment with anti-CD40 antibody, plasma ALT levels in Cd40fl/fl LysM-Cre mice remained unchanged with anti-CD40 antibody treatment, indicating lack of inflammatory disease in the absence of CD40-expressing macrophages (Figure 4C).
Figure 4. Antiinflammatory function of erythrophagocytes.
(A) Schematic representation of experimental design: mice were treated with an agonistic anti-CD40 antibody for 30 hours (for macrophage isolation and blood removal) or 48 hours (for liver collection). (B) Representative histology image for H&E staining of a thrombus with an ischemic liver infarct after treatment of Sptawt/wt mouse with anti-CD40 antibody. Scale bar: 200 μm. (C) Plasma alanine transaminase (ALT) concentrations in LysM-Cre Cd40fl/fl (Cre+/–, knockout) or Cd40fl/fl (Cre–/–, control) mice treated with saline or anti-CD40 antibody (n = 3–6). (D) Plasma ALT concentrations in Sptawt/wt (WT, blue) and Sptasph/sph (Sph, red) mice treated with saline or anti-CD40 antibody (n = 6–8). (E) Photographs of representative liver lobes from Sptasph/sph and Sptawt/wt mice treated with anti-CD40 antibody. (F) Hierarchical clustering analysis of plasma cytokines from Sptasph/sph and WT littermates treated with anti-CD40 antibody (yellow = low concentration, red = high concentration) or saline. (G) Bulk RNA-seq data of F4/80-enriched nonparenchymal liver cell suspensions from 3 Sptasph/sph and Sptawt/wt mice treated with anti-CD40 antibody or saline. Hierarchical clustering analysis of the 2000 top significantly DEGs (rows) between the anti-CD40 and saline-treated Sptawt/wt (column). In total, 5803 DEGs were identified based on an absolute log2(ratio) > 0.5 and P value < 0.01 threshold. (H) GSEA of DEGs of KCs in Sptawt/wt mice treated with anti-CD40 antibody versus saline. Enrichment plots of the top 4 positively enriched hallmark gene sets are shown. Plots display running enrichment score and position of gene set members on the rank-ordered list. (I) Normalized count data for key inflammatory genes expressed in KCs from anti-CD40– or saline-treated (cont) Sptawt/wt (blue) and Sptasph/sph (red) mice. Each data point represents a single mouse. ***P < 0.001; **P < 0.01 by ANOVA with Tukey’s post hoc test for C and D; counts were normalized using DEseq2 for I.
Based on the above results, we concluded that the anti-CD40 model would be suitable to evaluate the specific effects of hemolytic stress and erythrophagocytosis on inflammatory macrophage functions in vivo, and accordingly administered anti-CD40 antibody to Sptasph/sph and Sptawt/wt mice. Compared with Sptawt/wt littermates, the anti-CD40 antibody–induced disease expression was significantly attenuated in Sptasph/sph mice. No ALT elevation (Figure 4D) or ischemic infarcts (Figure 4E) were observed in the livers of anti-CD40 antibody–treated Sptasph/sph animals. Although anti-CD40 antibody–treated Sptawt/wt mice exhibited increased plasma concentrations of CCL2, CCL3, IL-6, and TNF-α in comparison with saline-treated mice, plasma concentrations of these cytokines remained almost unchanged in anti-CD40 antibody–treated Sptasph/sph mice (Figure 4F).
We also characterized the CD40 ligation–induced transcriptome changes in F4/80+ liver macrophages isolated from anti-CD40 antibody–treated Sptasph/sph and Sptawt/wt mice by bulk RNA-seq. Macrophages from Sptawt/wt mice exhibited a broad inflammatory response upon anti-CD40 treatment, which was characterized by GSEA enrichment of hallmark gene sets associated with inflammation, TNF signaling, IFN signaling, and allograft rejection (Figure 4, G and H, and Supplemental Figure 7B), as well as by expression of key inflammatory genes (Figure 4I and Supplemental Table 6). This CD40-stimulated macrophage transcriptional response was markedly suppressed in erythrophagocytes isolated from Sptasph/sph mice (Figure 4I and Supplemental Table 7). At baseline, Sptasph/sph macrophages displayed a slight, statistically not significant reduction in Cd40 expression compared with WT mice (Supplemental Figure 3C). Cd40 expression was induced in Sptawt/wt and in Sptasph/sph macrophages after antibody treatment, suggesting that the broad antiinflammatory effect of erythrophagocytosis occurs downstream of the CD40 signaling pathway. This is also consistent with the observation that anti-CD40 antibody treatment further stimulated erythrophagocytosis by liver macrophages in Sptasph/sph mice (Supplemental Figure 3E).
Taken together, these results suggest that erythrophagocytosis skews liver macrophages into an antiinflammatory functional state.
Heme exposure of BM-derived macrophages in vitro replicates antiinflammatory erythrophagocyte polarization.
Erythrophagocytosis leads to a steep increase in the intracellular concentration of the porphyrin metabolite heme, which is a known modulator of gene expression in macrophages and other cell types. Therefore, we performed a series of studies to test whether heme exposure of macrophages in vitro could mimic the effects of hemolytic stress on macrophage phenotype and inflammatory function.
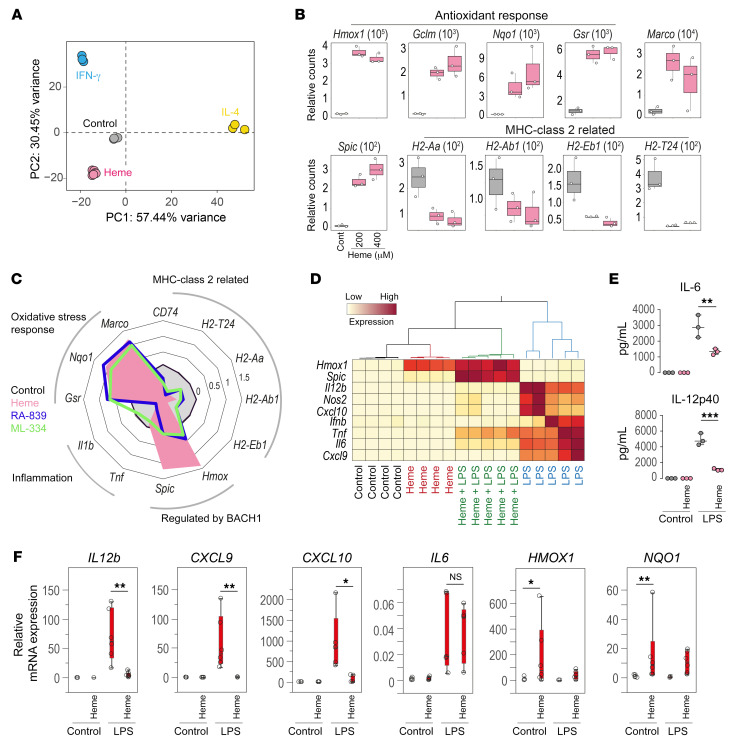
We exposed BM-derived macrophages (BMDMs) to heme-albumin complexes, IFN-γ, or IL-4 for 72 hours, and studied their transcriptome profile by bulk RNA-seq (Supplemental Tables 8–10). Principal component analysis demonstrated that heme-polarized macrophages clearly segregated from IFN-γ– and IL-4–polarized cells (Figure 5A), suggesting that heme promotes an idiosyncratic macrophage phenotype that is different from classical M1 (IFN-γ–polarized) and M2 (IL-4–polarized) macrophages. GSEA for heme-treated BMDMs identified the strongest signals for upregulated reactive oxygen species pathway and glycolysis, while IFN responses were strongly suppressed (Supplemental Figure 5B and Supplemental Figure 7C), as seen previously in the erythrophagocytes isolated from livers of Sptasph/sph mice. Macrophages stimulated with IL-4 or IFN-γ exhibited distinct transcriptional profiles, indicating that they differentiated into divergent macrophage subsets, as illustrated by comparative enrichment analysis of key gene sets and transcription factors (Supplemental Figure 5A). mRNA expression count data for selected heme-response genes confirmed the induction of genes involved in the antioxidant response and suppression of MHC class II–associated genes (Figure 5B). In addition, a strong signal for the previously identified marker gene for erythrophagocytes, Marco, and for the gene expressing the heme-inducible transcription factor, Spic, was observed. Consistent with an authentic cell differentiation process, we found a progressive change in gene induction and suppression over 72 hours (Supplemental Figure 5C).
Figure 5. Replication of the antiinflammatory erythrophagocyte polarization by heme exposure of mouse BM-derived macrophages in vitro.
(A) Mouse BM-derived macrophages (BMDMs) were exposed to albumin (control), heme-albumin (200 and 400 μM), IFN-γ (10 ng/mL), or IL-4 (10 ng/mL) for 72 hours and then processed for bulk RNA-seq. Samples were plotted based on the first 2 principal components (n = 3). (B) Normalized count data for the expression of antioxidant genes and the MHC class II–associated genes Marco, Hmox1, and Spic in heme-albumin–treated (200 and 400 μM) (magenta) and control BMDMs (gray). (C) Radar plot of relative mRNA gene expression of antioxidant genes, the MHC class II–associated genes Marco, Hmox1, and Spic, and inflammatory genes measured by RT-qPCR in BMDMs treated with albumin (control; gray), heme-albumin (200 μM; magenta), RA-839 (15 μM; blue), or ML-334 (50 μM; green) for 72 hours (n = 4–8). (D) Hierarchical clustering analysis of the relative mRNA expression of key inflammatory genes, Hmox1 and Spic, measured by RT-qPCR in BMDMs treated or not with heme-albumin for 72 hours with and without LPS for an additional 4 hours (yellow = low expression, red = high expression) (n = 4–5). (E) Concentrations of IL-12p40 and IL-6 in cell culture medium measured by Bio-Plex in BMDMs treated or not with heme-albumin for 72 hours with and without LPS for an additional 4 hours (n = 3). (F) Relative mRNA expression of key inflammatory genes, HMOX1 and NQO1, measured by RT-qPCR in PBMC-derived human macrophages treated or not with heme-albumin for 72 hours with and without LPS for an additional 4 hours (n = 6). ***P < 0.001; **P < 0.01; *P < 0.05 by ANOVA with Tukey’s post hoc test for E and Kruskal-Wallis with Wilcoxon’s test for F.
As in erythrophagocytes of hemolytic mice, an EnrichR analysis of upregulated genes in heme-albumin–treated BMDMs for transcription factor target genes provided the most significant hit for genes regulated by NRF2 (Supplemental Figure 5A). To test whether NRF2 activation alone could reproduce the phenotype transition induced by heme-albumin, BMDMs were differentiated in parallel with heme-albumin or with either of the 2 pharmacological NRF2 activators, RA-839 and ML-334, for 72 hours. The radar chart in Figure 5C illustrates that key phenotype features of heme-polarized macrophages are mimicked by the NRF2 activators, including the upregulation of oxidative stress–response genes and Marco, as well as profound downregulation of MHC class II–associated genes. Hmox1 and Spic are less induced by the NRF2 activators compared with heme. This is congruent with the established function of the heme-responsive transcriptional repressor BACH1 in the regulation of these genes (Figure 5C and refs. 26, 65). Collectively, these data point to NRF2 as a strong candidate signaling pathway transmitting key phenotypic features of erythrophagocytes.
To establish that heme also drives antiinflammatory erythrophagocyte function, we measured the mRNA expression of select inflammatory response genes by RT-qPCR using in vitro heme-polarized BMDMs stimulated with LPS. The clustering analysis of mRNA gene expression data revealed a broad inflammatory response of BMDMs that were stimulated with LPS. In contrast, this LPS response was markedly suppressed in heme-polarized macrophages (Figure 5D). However, under the same conditions the expression of heme-inducible genes, Hmox1 and Spic, was significantly enhanced (Figure 5D), indicating the target specificity of the heme-transmitted signal. The antiinflammatory effect of heme-albumin was also reflected by the reduced secretion of IL-12p40 and IL-6 into the cell culture medium (Figure 5E). To exclude cytotoxicity of heme-albumin, such as that resulting from oxidative stress or ferroptosis, as a cause of the antiinflammatory effect, we measured mitochondrial function and the glycolytic rate of BMDMs using a Seahorse metabolic flux analyzer. Heme exposure for 72 hours did not change mitochondrial function, and enhanced, rather than suppressed, glycolytic activity (Supplemental Figure 5D), a finding that is consistent with the increased expression of glycolysis-related genes identified in bulk RNA-seq data (Supplemental Figure 7C).
Collectively, these results demonstrate that heme drives differentiation of BMDMs into erythrophagocyte-like cells, supporting a strong antiinflammatory function.
To translate our findings to human physiology, we performed heme-exposure experiments with human macrophages that were differentiated from PBMCs of healthy blood donors and stimulated with LPS. As in murine BMDMs, heme exposure induced HMOX1 and the NRF2 target gene NQO1, and the LPS-induced inflammatory response was significantly attenuated (Figure 5F). However, the antiinflammatory effect appeared to be more selective. While CXCL9, CXCL10, and IL12b were strongly suppressed by heme-albumin, we found only a weak effect on IL6 expression, with high interindividual variability. This difference may be related to the more differentiated in vitro phenotype of PBMCs compared with mouse BMDMs.
Heme/NRF2 signaling drives BMDM polarization into erythrophagocyte-like antiinflammatory cells.
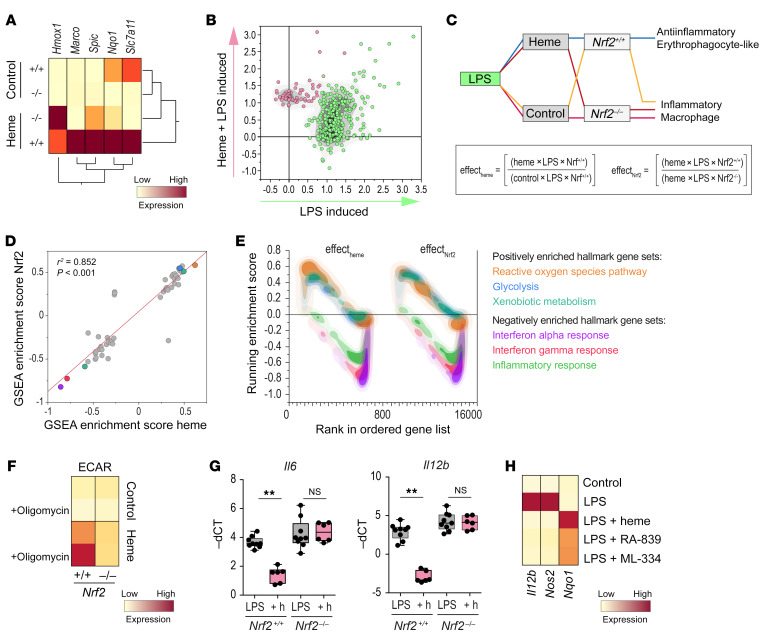
To further explore the potential role of NRF2-mediated inflammatory suppression in response to heme we performed studies with BMDMs from Nrf2-knockout (Nrf2–/–) mice. We first established that the absence of the transcription factor in BMDMs markedly reduced heme-induced mRNA expression of the NRF2 target genes Marco, Spic, Nqo1, and Slc7a11 (Figure 6A). In contrast, Hmox1 expression was not attenuated, reinforcing the critical function of BACH1 rather than NRF2 in its regulation by heme. Heme-induced NRF2 target gene expression and inflammatory suppression could be observed as early as after 4 hours of heme treatment. This short exposure period was chosen for experiments with Nrf2–/– macrophages to minimize confounding effects that may occur due to unopposed oxidative stress during longer heme exposures. Figure 6B shows a scatter plot of mRNA expression data from gene array experiments with WT (Nrf2+/+) BMDMs treated with heme-albumin for 4 hours and LPS for an additional 4 hours. Based on unobserved clustering analysis, the genes upregulated in response to LPS treatment (green, total 271 genes) comprised a population that was distinct from the genes upregulated only in response to combined LPS and heme-albumin treatment (red, 57 genes) (Figure 6B and Supplemental Table 11). An EnrichR analysis of these 57 heme-induced genes found again the most significant enrichment for genes that were regulated by NRF2 (Supplemental Figure 6A). Experimentally, this gene cluster was less upregulated in BMDMs from Nrf2–/– mice. Supplemental Figure 6B shows the cumulative effect of heme exposure on the LPS-induced inflammatory response in Nrf2+/+ and Nrf2–/– BMDMs. The data suggest that the suppression of the LPS-induced genes by heme was less pronounced in the absence of NRF2.
Figure 6. Heme/NRF2 signaling drives erythrophagocyte-like polarization of BMDMs.
(A) RT-qPCR expression data for heme-response genes in Nrf2+/+ and Nrf2–/– BMDMs after heme-albumin (200 μM) or albumin (control) treatment for 6 hours (mean of n = 3 replicates). (B) Scatter plot of differentially expressed genes (logratio > 0.5) of heme-albumin– versus albumin-treated BMDMs followed by LPS (4 hours). Data represent the log10 of the expression ratio as mean from n = 4 gene array experiments. A transcription factor enrichment analysis for the red-labeled gene cluster is displayed in Supplemental Figure 6A. (C) Factorial layout and predicted phenotype outcomes of gene array experiments with BMDMs from Nrf2+/+ and Nrf2–/– mice that were treated with or without heme-albumin and LPS. Effects on differential gene expression for heme in WT cells (effectheme) and for the Nrf2 gene in heme-exposed cells (effectNrf2) were calculated by the displayed formulae. (D) Correlation of the GSEA hallmark gene set enrichment scores for effectheme and effectNrf2. The top 3 hallmark gene sets are highlighted (P < 0.001). (E) Overlaid GSEA enrichment score plots for the hallmark gene sets highlighted in D. (F) Heatmap of the ECAR of Nrf2+/+ and Nrf2–/– BMDMs. BMDMs were treated for 4 hours with albumin (control) or with 200 μM heme-albumin before the Seahorse experiment. Cells were analyzed at baseline and after treatment with oligomycin. Mean values from 3 measurements per condition (yellow = low ECAR, red = high ECAR). (G) LPS-stimulated Il6 and Il12b mRNA expression in Nrf2+/+ and Nrf2–/– BMDMs after pretreatment with albumin or with heme-albumin (+h) for 4 hours (n = 6–9), measured by RT-qPCR. (H) Il12b, Nos2, and Nqo1 mRNA in WT BMDMs treated with LPS, LPS+heme, or with the Nrf2 activators RA-839 and ML-334 measured by RT-qPCR. Mean of 3 independent experiments. **P < 0.01 by ANOVA with Tukey’s post hoc test for G. NS, not significant.
To further test the role of NRF2 in heme-induced inflammatory suppression, we performed a factorial analysis of these experiments with LPS-stimulated macrophages from Nrf2+/+ or Nrf2–/– mice in the presence or absence of heme (Figure 6C). The individual effects of heme and NRF2 in this model were calculated from the differential gene expression data (Supplemental Tables 12 and 13) and the output data were analyzed by GSEA (Supplemental Figure 8, A and B). A correlation plot of the GSEA enrichment scores (ESs) for all 50 hallmark gene sets suggested that heme (in WT macrophages) and NRF2 (in heme-exposed macrophages) have almost identical qualitative effects on macrophage polarization, which agrees with our hypothesis of a linked effector pathway (Figure 6D; a correlation plot of the same effects on the individual gene level is shown in Supplemental Figure 6C). Overlay density plots of the running ESs for the most significant gene sets highlighted in Figure 6D illustrate equal induction of antioxidant pathways and glycolysis alongside a strong suppression of LPS-induced inflammatory responses by heme and NRF2 (Figure 6E). In accordance with the predicted positive effect of heme/NRF2 signaling on glycolysis-related gene expression, metabolic flux analysis revealed that in comparison with heme-albumin–treated Nrf2+/+ BMDMs, the extracellular acidification rate (ECAR) was strongly attenuated in Nrf2–/– BMDMs even after maximal stimulation with oligomycin (Figure 6F).
We confirmed the role of NRF2 in the antiinflammatory function of heme in an independent set of experiments with BMDMs from Nrf2+/+ and Nrf2–/– mice. Heme-albumin–mediated suppression of Il12b and Il6 mRNA after LPS stimulation was not observed in Nrf2–/– BMDMs (Figure 6G). The key role of NRF2 is further supported by the observation that also the antiinflammatory effect of heme in BMDMs is mimicked by pharmacological activators of NRF2 (Figure 6H).
Collectively, these results indicate that NRF2 is an important driver of heme-induced gene expression and antiinflammatory polarization in macrophages, and may have a role in metabolic adaptation.
Hemolytic mice exhibit tolerance against diet-induced nonalcoholic fatty liver disease.
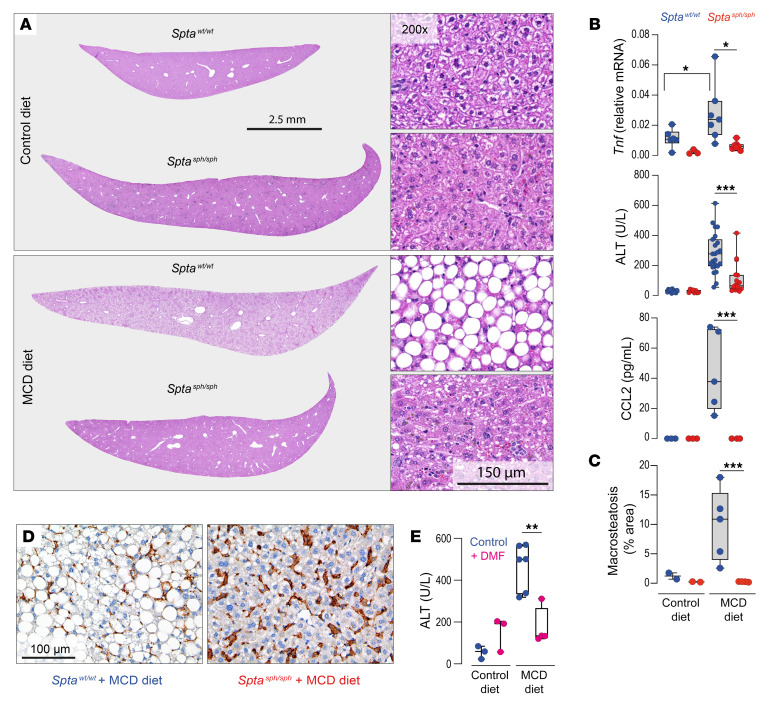
We aimed to establish whether the antiinflammatory function of erythrophagocytes has broader implications beyond specific exogenous inflammatory triggers such as anti-CD40 antibodies and LPS, to impact more complex and multifactorial inflammatory disease processes. To this end, we examined the effect of erythrophagocyte expansion in the liver on the development of diet-induced nonalcoholic fatty liver disease (NAFLD), a disease caused by metabolic dysregulation that induces KC activation with Tnf expression, inflammation, steatosis, and hepatocellular damage (66–68). NAFLD was induced in Sptawt/wt and Sptasph/sph mice by feeding them a methionine- and choline-deficient (MCD) diet (69–71). On day 12 of the MCD diet, Sptawt/wt mice exhibited enhanced mRNA expression of Tnf in the isolated F4/80+ liver macrophages, as well as increased plasma concentrations of ALT and the macrophage chemokine CCL2 (Figure 7B). Compared with Sptawt/wt mice, these changes were mostly repressed in hemolytic Sptasph/sph mice. Liver histology revealed extensive steatosis in Sptawt/wt, but not in Sptasph/sph mice fed the MCD diet (Figure 7, A and C). In comparison with Sptawt/wt mice fed the MCD diet, a markedly expanded pool of F4/80+ erythrophagocytes was found in the livers of Sptasph/sph mice fed the MCD diet, which indicated that disease tolerance was a function of not a reduced number of macrophages but of an altered functional state (Figure 7D). The treatment of WT mice with the NRF2 activator dimethyl fumarate (DMF), an approved drug for relapsing-remitting multiple sclerosis (35, 38), mimicked the heme response and attenuated the increase in liver enzymes in MCD diet–fed mice (Figure 7E).
Figure 7. Tolerance against diet-induced nonalcoholic fatty liver disease in hemolytic mice.
(A) Scans of H&E-stained liver lobe sections at ×15 magnification (left) or ×200 magnification (right) from Sptawt/wt (blue) and Sptasph/sph (red) mice fed a methionine- and choline-deficient (MCD) or control diet for 12 days. The images displayed are representative of 5 animals in each group. (B) Sptawt/wt (blue) and Sptasph/sph (red) mice that were fed an MCD or control diet for 12 days. Top to bottom: mRNA expression of Tnf in F4/80+ liver macrophages measured by RT-qPCR (n = 3–7), plasma ALT (n = 8–23), and CCL2 (n = 3–5) concentrations. (C) Quantitative morphometric analysis of the area with macrosteatosis in liver tissues (percentage of the total tissue area) based on liver histology images of the 5 mice mentioned in A (n = 2–5). (D) Microphotographs of liver tissues from MCD diet–fed Sptawt/wt and Sptasph/sph mice stained for F4/80+ macrophages. Scale bar: 100 μm. (E) Plasma ALT concentrations from Sptawt/wt mice treated with dimethyl fumarate (DMF) at 100 mg/kg (or vehicle control) for 7 days and then fed control or MCD diet for 12 days (n = 3–6). Individual symbols represent 1 mouse. ***P < 0.001; **P < 0.01; *P < 0.05 by ANOVA with Tukey’s post hoc test for all panels.
Discussion
Using scRNA-seq studies of liver macrophages, we discovered a population of erythrophagocytes with an antiinflammatory phenotype and function. Under hemolytic stress, these antiinflammatory erythrophagocytes become the predominant macrophage population leading to a pronounced attenuation in 2 models of inflammatory liver disease. In vitro heme exposure of BMDMs reproduced the MarcohiHmoxhiMHC class IIlo erythrophagocyte profile, yielding a transcriptional signature that segregated the heme-polarized macrophages from IFN-γ–polarized (M1-like) and IL-4–polarized (M2-like) cells. This suggests that heme-polarized macrophages represent an idiosyncratic phenotype, providing on-demand adaptation to hemolytic stress.
A defining feature of erythrophagocytes in our studies was the strong expression of the scavenger receptor Marco. In the human liver, the expression of Marco segregates inflammatory (Marcolo) macrophages from antiinflammatory (Marcohi) macrophages, which are characterized by an attenuated Tnf expression response upon in vitro stimulation with LPS and IFN-γ (62). In alveolar macrophages, Marco has a nonredundant antiinflammatory function protecting against oxidative lung injury (72). Enhanced Marco expression and the diminished expression of MHC class II genes are also phenotypic characteristics of tumor-associated macrophages linked to reduced immunogenicity and immunosuppression in cancer (73). MarcohiHmoxhiMHC class IIlo erythrophagocytes in the liver appear to acquire similar antiinflammatory functions. To support this assumption, we used an in vivo model of anti-CD40 antibody–induced systemic inflammation that had the advantages of a macrophage-driven disease process with a primarily liver-based pathology. By transforming inflammatory KCs into antiinflammatory erythrophagocytes, mice with spherocytosis displayed a markedly attenuated disease response. Macrophages were numerous in the livers of Sptasph/sph mice before and after anti-CD40 antibody treatment. They exhibited enhanced erythrophagocytic activity alongside a gene expression profile that did not indicate nonspecific cell stress or activation of controlled-death pathways, suggesting that the profound antiinflammatory function does not reflect a toxic response, but instead results from a divergent macrophage phenotype.
Supporting our hypothesis of a broader role for the antiinflammatory impact of heme-mediated macrophage polarization, we found in a model of NAFLD that erythrophagocytes in the MCD diet–fed Sptasph/sph mice expressed less Tnf, which coincided with low plasma levels of the disease-associated macrophage-derived chemokine CCL2 (68), markedly reduced hepatocellular damage, and almost absent liver steatosis compared with WT mice on the same diet. As key integrators of endogenous and exogenous proinflammatory signals, KCs and recruited liver macrophages play a central role in the initiation and progression of NAFLD and nonalcoholic steatohepatitis (NASH) (74). Several macrophage-specific interventions involving depletion of KCs (69, 71, 75), macrophage-selective knockout of pattern recognition receptor pathways (76, 77), or macrophage-targeted delivery of antiinflammatory drugs (78) have successfully been used to interrupt NAFLD progression, with many of these approaches being particularly effective during the early disease phase (69).
The antiinflammatory phenotype and function of erythrophagocytes are supported by the finding of NRF2-driven gene expression across our studies, which was consistent with the open chromatin landscape defined by scATAC-seq. Heme-induced antioxidant gene expression was significantly attenuated in Nrf2-knockout macrophages. This finding supports the idea that heme acts as an activator of Nrf2 in our model, as has been shown in human macrophages (79) and in other cell types (80–82). Furthermore, the suppressive effect of heme on LPS-triggered inflammatory gene expression was rescued in Nrf2-knockout macrophages.
A limitation of our study is that we could not directly investigate the contribution of NRF2 to the antiinflammatory heme polarization of macrophages in vivo. This is due to the impossibility of experimentally dissociating the adaptive function of NRF2 activation from its antiinflammatory activity. In other words, because NRF2 is indispensable to resist oxidative heme stress in vivo, systemic toxicity with multiorgan failure overwhelms the antiinflammatory effects of heme after experimental heme exposure or during hemolysis in an Nrf2-knockout mouse. Nevertheless, we found that the NRF2-activating drug DMF mimicked the effect of chronic hemolysis on NAFLD progression in our MCD model.
In human macrophage cell cultures, heme activates liver X receptor–coordinated (LXR-coordinated) gene expression, driving the differentiation of macrophages with a CD163hiHmox1hiMHC class IIlo immunophenotype that resembles the Mhem macrophages identified in microhemorrhage areas of atherosclerotic plaques (23–25, 83). As a central integrator of metabolic and antiinflammatory signaling, LXR activation may help consolidate the antiinflammatory phenotype of erythrophagocytes (84). In addition, heme and other porphyrins inhibit the proteasome and could, therefore, directly interfere with a key process of proinflammatory NF-κB activation (85, 86). Thus, it is likely that erythrophagocyte polarization by heme-induced NRF2 signaling is aided by a synergistic network that enables immune adaptation to hemolytic stress in a disease state– and tissue type–specific way.
The heme-driven differentiation of macrophages into the stable antiinflammatory state that we describe here needs to be distinguished from an intrinsic inflammatory activity of free heme that can be mediated by ligation of TLR4 on leukocytes and endothelial cells (9, 10, 87). Our previous observations suggested that the way heme is presented to a cell may determine its ultimate effect on the innate immune system. In the case of erythrophagocytosis or with a protein-bound heme (e.g., as hemoglobin or albumin-bound heme) the metalloporphyrin is preferentially delivered for intracellular antiinflammatory signaling. In contrast, free heme in the extracellular space may more avidly interact with pattern recognition receptors on the cell surface (88).
In conclusion, our findings suggest the existence of a pathway that links hemolysis and erythrophagocytosis with the regulation of inflammatory phagocyte functions. This concept may provide a basis for further research to understand immune dysfunction accompanying hemolysis such as after massive blood transfusion or in patients with sickle cell disease.
Methods
See the Supplemental Methods section for further details.
Animal models.
WT C57BL/6J mice were obtained from Charles River. Nrf2–/– mice were obtained from Yuet Wai Kan (UCSF). Interbreeding of heterozygous Sptasph mice (B6.C3-Spta1sph/BrkJ × WB.C3-Spta1sph/BrkJ mice, The Jackson Laboratory) yielded spherocytic and WT offspring. The Cd40-targeted mouse strain used for this research project was created from ES cell clone EPD0901_3_A02, obtained from the KOMP Repository (www.komp.org) and generated by the Wellcome Trust Sanger Institute (WTSI). Targeting vectors used were generated by the WTSI and the Children’s Hospital Oakland Research Institute as part of the Knockout Mouse Project (3U01HG004080). We crossed the gene-targeted Cd40fl/fl mice with FLP recombinase mice that recognize the FRT site to remove the LacZ reporter and neomycin selection cassette. The spliced exons, which are flanked by loxP sites before Cd40 exon 2 and after Cd40 exon 3, were removed by crossing to the LysM-Cre mouse, resulting in macrophage-specific Cd40-knockout mice. Littermates without the LysM-Cre driver were used as controls (89). Strain-appropriate controls were used across all the other experiments. All mice were housed and bred under specific pathogen–free conditions in the Laboratory Animal Services Center (LASC) of the University of Zürich. Eight- to 10-week-old males and females were used.
Experimental details for the different animal models and treatments can be found in the Supplemental Methods (agonistic anti-CD40 antibody–induced systemic inflammation and hepatitis model, liposomal clodronate injections, MCD diet model, DMF treatment).
Heme preparation for cell culture.
Hemin (heme-chloride) was obtained from MilliporeSigma (catalog 51280). The porphyrin was dissolved in 10 mL of 100 mM NaOH at 37°C, followed by addition of 10 mL of 20% human serum albumin (CSL Behring AG). After 1 hour of incubation at 37°C, the solution was adjusted to pH 7.4 using ortho-phosphoric acid, and the final volume was brought to 25 mL with saline solution. The heme-albumin solutions were sterile filtered (0.22 μm) and used immediately.
Mouse BMDM and human monocyte-derived macrophage isolation and culture.
Mouse and human macrophages were isolated and cultured according to standard protocols. Details can be found in the Supplemental Methods.
In situ liver digestion of mouse livers and macrophage enrichment.
Liver digestion was performed with a protocol modified from Cabral et al. (90). The abdominal cavity of a living, deeply anesthetized mouse was opened, and the portal vein was catheterized for in situ liver perfusion and digestion with collagenase B buffered solution (Roche, 11088815001). Once digested, livers were dissected out and the mouse was sacrificed. The digested livers were then mechanically disaggregated in a Petri dish on ice and filtered through a 100-μm-pore cell strainer. The cell suspensions were centrifuged twice at 60 g for 2 minutes at 4°C, and the pellets were discarded. The supernatants were then centrifuged at 300 g for 5 minutes at 4°C to obtain a pellet of nonparenchymal liver cells containing liver macrophages. To select for F4/80+ macrophages from liver cell suspensions, Dynabeads Sheep anti-Rat IgG (Invitrogen, 11035), in combination with purified rat anti–mouse F4/80 antibody (BD Pharmingen, 0.5 mg/mL, 565409), and a DynaMag-2 Magnet (Thermo Fisher Scientific, 12321D) were used according to the manufacturer’s positive-selection protocol (Invitrogen).
Flow cytometry sample preparation and analysis.
Flow cytometry was performed according to standard protocols. Cells were analyzed on an LSRFortessa (BD) and on an ImageStream X Mk II Imaging Flow Cytometer (Amnis). Details (including antibodies) can be found in the Supplemental Methods.
scRNA-seq.
F4/80+ nonparenchymal liver cells were processed for scRNA-seq according to the 10× Genomics Chromium Single Cell 3′ v3 workflow. Libraries were sequenced using the Illumina NovaSeq 6000 system according to 10× Genomics recommendations. Details on cell processing, library production, sequencing, data analysis, and visualization can be found in the Supplemental Methods.
Bulk RNA-seq and data analysis.
Sequencing was performed using a standard workflow with an Illumina NovaSeq 6000 sequencer. Details can be found in the Supplemental Methods.
Assay for transposase-accessible chromatin.
ATAC-seq was performed using the Chromium Single Cell ATAC workflow (10× Genomics). Details on the isolation of nuclei, library preparation, sequencing, and data analysis are described in the Supplemental Methods.
Microarray experiments and data analysis.
We used whole-mouse or whole-human genome oligo microarrays (G4846A, 4 × 44 K, Agilent Technologies) as described previously (29). Statistical analysis and visualization were performed with JMP Genomics 7.0 (SAS Institute). Details are provided in the Supplemental Methods.
Real-time PCR.
RT-qPCR was performed according to a standard workflow on a 7500 Fast Real-Time PCR System (Applied Biosystems). Protocol details and a complete list of human and mouse primers can be found in the Supplemental Methods.
Bio-Plex cytokine assays.
Concentrations of cytokines were determined with Bio-Plex Cytokine Assays (Bio-Rad). The assay was analyzed with a Bio-Plex 200 system (Bio-Rad). The results were analyzed using Bio-Plex Data Pro software (Bio-Rad).
Mouse tissue histology.
Details on tissue preparation microscopy, imaging, and data analysis can be found in the Supplemental Methods.
Data sharing.
For original data, please contact florence.vallelian@usz.ch. All next-generation sequencing and microarray data can be accessed via the following link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145244 All Supplemental Tables described in the text and the GSEA full results can be accessed via the following link: https://www.dropbox.com/sh/1qvwj3c5ylpvoow/AAAVTJ_JS8OvRKKxPCC9TmWza?dl=0
Statistics.
Data plotting and statistical analysis were performed with Prism 8 (GraphPad), JMP 13 (SAS), Bioturing SingleCell browser (v2.4.21), or R (v3.6.1) (https://www.R-project.org/). RNA count data were normalized and compared using either DESseq2 R package (v1.26.0) (91) or EdgeR (v 3.30.0) (92, 93). For group comparisons of other data types, we used ANOVA with Tukey’s post hoc test as indicated in the figure legends. For the analysis of human macrophage gene expression data, we used Kruskal-Wallis with Wilcoxon’s post hoc test to account for the high variability with occasional outliers. All data points are displayed in the graphs. Box plots display median and interquartile range, and whiskers represent minimum and maximum values. P values less than 0.05 were considered significant: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Study approval.
All experimental protocols were reviewed and approved by the Veterinary Office of the canton of Zürich, Switzerland. Mice were treated in accordance with the guidelines of the Swiss Federal Veterinary Office.
Author contributions
MP performed experiments, analyzed data, and wrote the manuscript. GI and CAS performed experiments and analyzed data. AY, RB, ILD, NS, EYLC, and KH performed experiments. GT analyzed data. RH designed the study and performed experiments. DJS and FV designed the study, performed experiments, analyzed data, and wrote the manuscript. The order of the 2 first authors was determined by mutual agreement.
Supplementary Material
Acknowledgments
This study was supported by the Swiss National Science Foundation (MD-PhD scholarship, project 23530_183984 to MP, and MD-PhD scholarship 4221-06-2017 to RB), by the Swiss Federal Commission for Technology and Innovation (19300.1 PFLS-L to DJS), by the Promedica Foundation (to FV), by the Olga Mayenfisch Stiftung (to FV), and by the Novartis Foundation (to FV).
Version 1. 07/14/2020
In-Press Preview
Version 2. 09/14/2020
Electronic publication
Version 3. 10/01/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(10):5576–5590.https://doi.org/10.1172/JCI137282.
Contributor Information
Marc Pfefferlé, Email: marc.pfefferle@uzh.ch.
Giada Ingoglia, Email: giada.ingoglia@usz.ch.
Christian A. Schaer, Email: christian.schaer@usz.ch.
Ayla Yalamanoglu, Email: ayla.yalamanoglu@usz.ch.
Irina L. Dubach, Email: IrinaLea.Dubach@usz.ch.
Ge Tan, Email: ge.tan@fgcz.ethz.ch.
Emilio Y. López-Cano, Email: emilio.yanguez@fgcz.ethz.ch.
Nadja Schulthess, Email: nadja.schulthess@usz.ch.
Kerstin Hansen, Email: kerstin.hansen@usz.ch.
Rok Humar, Email: Rok.Humar@usz.ch.
Dominik J. Schaer, Email: dominik.schaer@usz.ch.
Florence Vallelian, Email: Florence.Vallelian@usz.ch.
References
- 1.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122(3):787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murray PJ. Macrophage polarization. Annu Rev Physiol. 2017;79:541–566. doi: 10.1146/annurev-physiol-022516-034339. [DOI] [PubMed] [Google Scholar]
- 3.Gautier EL, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13(11):1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buehler PW, Humar R, Schaer DJ. Haptoglobin therapeutics and compartmentalization of cell-free hemoglobin toxicity. Trends Mol Med. 2020;26(7):683–697. doi: 10.1016/j.molmed.2020.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Schaer DJ, Buehler PW, Alayash AI, Belcher JD, Vercellotti GM. Hemolysis and free hemoglobin revisited: exploring hemoglobin and hemin scavengers as a novel class of therapeutic proteins. Blood. 2013;121(8):1276–1284. doi: 10.1182/blood-2012-11-451229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baek JH, et al. Hemoglobin-driven pathophysiology is an in vivo consequence of the red blood cell storage lesion that can be attenuated in guinea pigs by haptoglobin therapy. J Clin Invest. 2012;122(4):1444–1458. doi: 10.1172/JCI59770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lei C, et al. Nitric oxide decreases acute kidney injury and stage 3 chronic kidney disease after cardiac surgery. Am J Respir Crit Care Med. 2018;198(10):1279–1287. doi: 10.1164/rccm.201710-2150OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gladwin MT, Kanias T, Kim-Shapiro DB. Hemolysis and cell-free hemoglobin drive an intrinsic mechanism for human disease. J Clin Invest. 2012;122(4):1205–1208. doi: 10.1172/JCI62972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belcher JD, et al. Heme triggers TLR4 signaling leading to endothelial cell activation and vaso-occlusion in murine sickle cell disease. Blood. 2014;123(3):377–390. doi: 10.1182/blood-2013-04-495887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh S, et al. Extracellular hemin crisis triggers acute chest syndrome in sickle mice. J Clin Invest. 2013;123(11):4809–4820. doi: 10.1172/JCI64578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vinchi F, et al. Hemopexin therapy improves cardiovascular function by preventing heme-induced endothelial toxicity in mouse models of hemolytic diseases. Circulation. 2013;127(12):1317–1329. doi: 10.1161/CIRCULATIONAHA.112.130179. [DOI] [PubMed] [Google Scholar]
- 12.Vinchi F, et al. Hemopexin therapy reverts heme-induced proinflammatory phenotypic switching of macrophages in a mouse model of sickle cell disease. Blood. 2016;127(4):473–486. doi: 10.1182/blood-2015-08-663245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baek JH, et al. Iron accelerates hemoglobin oxidation increasing mortality in vascular diseased guinea pigs following transfusion of stored blood. JCI Insight. 2017;2(9):93577. doi: 10.1172/jci.insight.93577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deuel JW, et al. Hemoglobinuria-related acute kidney injury is driven by intrarenal oxidative reactions triggering a heme toxicity response. Cell Death Dis. 2016;7:e2064. doi: 10.1038/cddis.2015.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato GJ, et al. Sickle cell disease. Nat Rev Dis Primers. 2018;4:18010. doi: 10.1038/nrdp.2018.10. [DOI] [PubMed] [Google Scholar]
- 16.Reiter CD, et al. Cell-free hemoglobin limits nitric oxide bioavailability in sickle-cell disease. Nat Med. 2002;8(12):1383–1389. doi: 10.1038/nm1202-799. [DOI] [PubMed] [Google Scholar]
- 17.Schaer CA, et al. Mechanisms of haptoglobin protection against hemoglobin peroxidation triggered endothelial damage. Cell Death Differ. 2013;20(11):1569–1579. doi: 10.1038/cdd.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaer CA, et al. Haptoglobin preserves vascular nitric oxide signaling during hemolysis. Am J Respir Crit Care Med. 2016;193(10):1111–1122. doi: 10.1164/rccm.201510-2058OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Theurl I, et al. On-demand erythrocyte disposal and iron recycling requires transient macrophages in the liver. Nat Med. 2016;22(8):945–951. doi: 10.1038/nm.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaer DJ, Vinchi F, Ingoglia G, Tolosano E, Buehler PW. Haptoglobin, hemopexin, and related defense pathways-basic science, clinical perspectives, and drug development. Front Physiol. 2014;5:415. doi: 10.3389/fphys.2014.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaer DJ, Alayash AI, Buehler PW. Gating the radical hemoglobin to macrophages: the anti-inflammatory role of CD163, a scavenger receptor. Antioxid Redox Signal. 2007;9(7):991–999. doi: 10.1089/ars.2007.1576. [DOI] [PubMed] [Google Scholar]
- 22.Kaempfer T, et al. Extracellular hemoglobin polarizes the macrophage proteome toward Hb-clearance, enhanced antioxidant capacity and suppressed HLA class 2 expression. J Proteome Res. 2011;10(5):2397–2408. doi: 10.1021/pr101230y. [DOI] [PubMed] [Google Scholar]
- 23.Boyle JJ, et al. Coronary intraplaque hemorrhage evokes a novel atheroprotective macrophage phenotype. Am J Pathol. 2009;174(3):1097–1108. doi: 10.2353/ajpath.2009.080431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyle JJ, et al. Activating transcription factor 1 directs Mhem atheroprotective macrophages through coordinated iron handling and foam cell protection. Circ Res. 2012;110(1):20–33. doi: 10.1161/CIRCRESAHA.111.247577. [DOI] [PubMed] [Google Scholar]
- 25.Finn AV, et al. Hemoglobin directs macrophage differentiation and prevents foam cell formation in human atherosclerotic plaques. J Am Coll Cardiol. 2012;59(2):166–177. doi: 10.1016/j.jacc.2011.10.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haldar M, et al. Heme-mediated SPI-C induction promotes monocyte differentiation into iron-recycling macrophages. Cell. 2014;156(6):1223–1234. doi: 10.1016/j.cell.2014.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohyama M, et al. Role for Spi-C in the development of red pulp macrophages and splenic iron homeostasis. Nature. 2009;457(7227):318–321. doi: 10.1038/nature07472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vallelian F, et al. Glucocorticoid treatment skews human monocyte differentiation into a hemoglobin-clearance phenotype with enhanced heme-iron recycling and antioxidant capacity. Blood. 2010;116(24):5347–5356. doi: 10.1182/blood-2010-04-277319. [DOI] [PubMed] [Google Scholar]
- 29.Schaer CA, Schoedon G, Imhof A, Kurrer MO, Schaer DJ. Constitutive endocytosis of CD163 mediates hemoglobin-heme uptake and determines the noninflammatory and protective transcriptional response of macrophages to hemoglobin. Circ Res. 2006;99(9):943–950. doi: 10.1161/01.RES.0000247067.34173.1b. [DOI] [PubMed] [Google Scholar]
- 30.Schaer DJ, Schaer CA, Schoedon G, Imhof A, Kurrer MO. Hemophagocytic macrophages constitute a major compartment of heme oxygenase expression in sepsis. Eur J Haematol. 2006;77(5):432–436. doi: 10.1111/j.1600-0609.2006.00730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knutson MD, Oukka M, Koss LM, Aydemir F, Wessling-Resnick M. Iron release from macrophages after erythrophagocytosis is up-regulated by ferroportin 1 overexpression and down-regulated by hepcidin. Proc Natl Acad Sci U S A. 2005;102(5):1324–1328. doi: 10.1073/pnas.0409409102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knutson MD, Vafa MR, Haile DJ, Wessling-Resnick M. Iron loading and erythrophagocytosis increase ferroportin 1 (FPN1) expression in J774 macrophages. Blood. 2003;102(12):4191–4197. doi: 10.1182/blood-2003-04-1250. [DOI] [PubMed] [Google Scholar]
- 33.Li N, et al. Nrf2 is a key transcription factor that regulates antioxidant defense in macrophages and epithelial cells: protecting against the proinflammatory and oxidizing effects of diesel exhaust chemicals. J Immunol. 2004;173(5):3467–3481. doi: 10.4049/jimmunol.173.5.3467. [DOI] [PubMed] [Google Scholar]
- 34.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 35.Cuadrado A, et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat Rev Drug Discov. 2019;18(4):295–317. doi: 10.1038/s41573-018-0008-x. [DOI] [PubMed] [Google Scholar]
- 36.Ishii T, et al. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J Biol Chem. 2000;275(21):16023–16029. doi: 10.1074/jbc.275.21.16023. [DOI] [PubMed] [Google Scholar]
- 37.Thimmulappa RK, et al. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J Clin Invest. 2006;116(4):984–995. doi: 10.1172/JCI25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linker RA, et al. Fumaric acid esters exert neuroprotective effects in neuroinflammation via activation of the Nrf2 antioxidant pathway. Brain. 2011;134(pt 3):678–692. doi: 10.1093/brain/awq386. [DOI] [PubMed] [Google Scholar]
- 39.Mills EL, et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature. 2018;556(7699):113–117. doi: 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singer BD, Chandel NS. Immunometabolism of pro-repair cells. J Clin Invest. 2019;129(7):2597–2607. doi: 10.1172/JCI124613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kovac S, Angelova PR, Holmström KM, Zhang Y, Dinkova-Kostova AT, Abramov AY. Nrf2 regulates ROS production by mitochondria and NADPH oxidase. Biochim Biophys Acta. 2015;1850(4):794–801. doi: 10.1016/j.bbagen.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kobayashi EH, et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat Commun. 2016;7:11624. doi: 10.1038/ncomms11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim SW, Lee HK, Shin JH, Lee JK. Up-down regulation of HO-1 and iNOS gene expressions by ethyl pyruvate via recruiting p300 to Nrf2 and depriving It from p65. Free Radic Biol Med. 2013;65:468–476. doi: 10.1016/j.freeradbiomed.2013.07.028. [DOI] [PubMed] [Google Scholar]
- 44.Olagnier D, et al. Activation of Nrf2 signaling augments vesicular stomatitis virus oncolysis via autophagy-driven suppression of antiviral immunity. Mol Ther. 2017;25(8):1900–1916. doi: 10.1016/j.ymthe.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olagnier D, et al. Nrf2 negatively regulates STING indicating a link between antiviral sensing and metabolic reprogramming. Nat Commun. 2018;9(1):3506. doi: 10.1038/s41467-018-05861-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weis S, et al. Metabolic adaptation establishes disease tolerance to sepsis. Cell. 2017;169(7):1263–1275.e14. doi: 10.1016/j.cell.2017.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Motterlini R, Otterbein LE. The therapeutic potential of carbon monoxide. Nat Rev Drug Discov. 2010;9(9):728–743. doi: 10.1038/nrd3228. [DOI] [PubMed] [Google Scholar]
- 48.Wandersee NJ, Birkenmeier CS, Gifford EJ, Mohandas N, Barker JE. Murine recessive hereditary spherocytosis, sph/sph, is caused by a mutation in the erythroid alpha-spectrin gene. Hematol J. 2000;1(4):235–242. doi: 10.1038/sj.thj.6200030. [DOI] [PubMed] [Google Scholar]
- 49.Mohandas N. Anemia lurking in introns. J Clin Invest. 2019;129(7):2655–2657. doi: 10.1172/JCI129443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chasis JA, Mohandas N. Erythroblastic islands: niches for erythropoiesis. Blood. 2008;112(3):470–478. doi: 10.1182/blood-2008-03-077883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xue L, Galdass M, Gnanapragasam MN, Manwani D, Bieker JJ. Extrinsic and intrinsic control by EKLF (KLF1) within a specialized erythroid niche. Development. 2014;141(11):2245–2254. doi: 10.1242/dev.103960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li W, et al. Identification and transcriptome analysis of erythroblastic island macrophages. Blood. 2019;134(5):480–491. doi: 10.1182/blood.2019000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lavin Y, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159(6):1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scott CL, et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat Commun. 2016;7:10321. doi: 10.1038/ncomms10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kurihara T, Warr G, Loy J, Bravo R. Defects in macrophage recruitment and host defense in mice lacking the CCR2 chemokine receptor. J Exp Med. 1997;186(10):1757–1762. doi: 10.1084/jem.186.10.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He H, Venema VJ, Gu X, Venema RC, Marrero MB, Caldwell RB. Vascular endothelial growth factor signals endothelial cell production of nitric oxide and prostacyclin through flk-1/KDR activation of c-Src. J Biol Chem. 1999;274(35):25130–25135. doi: 10.1074/jbc.274.35.25130. [DOI] [PubMed] [Google Scholar]
- 57.Halpern KB, et al. Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells. Nat Biotechnol. 2018;36(10):962–970. doi: 10.1038/nbt.4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwartz RE, et al. Multipotent adult progenitor cells from bone marrow differentiate into functional hepatocyte-like cells. J Clin Invest. 2002;109(10):1291–1302. doi: 10.1172/JCI15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huch M, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160(1-2):299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.MacParland SA, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun. 2018;9(1):4383. doi: 10.1038/s41467-018-06318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emmenegger U, Schaer DJ, Larroche C, Neftel KA. Haemophagocytic syndromes in adults: current concepts and challenges ahead. Swiss Med Wkly. 2005;135(21-22):299–314. doi: 10.4414/smw.2005.10976. [DOI] [PubMed] [Google Scholar]
- 64.Ingoglia G, et al. Line-selective macrophage activation with an anti-CD40 antibody drives a hemophagocytic syndrome in mice. Blood Adv. 2020;4(12):2751–2761. doi: 10.1182/bloodadvances.2020001624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun J, et al. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 2002;21(19):5216–5224. doi: 10.1093/emboj/cdf516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu Y, Liu Y, An W, Song J, Zhang Y, Zhao X. STING-mediated inflammation in Kupffer cells contributes to progression of nonalcoholic steatohepatitis. J Clin Invest. 2019;129(2):546–555. doi: 10.1172/JCI121842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baeck C, et al. Pharmacological inhibition of the chemokine CCL2 (MCP-1) diminishes liver macrophage infiltration and steatohepatitis in chronic hepatic injury. Gut. 2012;61(3):416–426. doi: 10.1136/gutjnl-2011-300304. [DOI] [PubMed] [Google Scholar]
- 68.Haukeland JW, et al. Systemic inflammation in nonalcoholic fatty liver disease is characterized by elevated levels of CCL2. J Hepatol. 2006;44(6):1167–1174. doi: 10.1016/j.jhep.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 69.Tosello-Trampont AC, Landes SG, Nguyen V, Novobrantseva TI, Hahn YS. Kuppfer cells trigger nonalcoholic steatohepatitis development in diet-induced mouse model through tumor necrosis factor-α production. J Biol Chem. 2012;287(48):40161–40172. doi: 10.1074/jbc.M112.417014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reid DT, Reyes JL, McDonald BA, Vo T, Reimer RA, Eksteen B. Kupffer cells undergo fundamental changes during the development of experimental NASH and are critical in initiating liver damage and inflammation. PLoS One. 2016;11(7):e0159524. doi: 10.1371/journal.pone.0159524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang W, et al. Depletion of liver Kupffer cells prevents the development of diet-induced hepatic steatosis and insulin resistance. Diabetes. 2010;59(2):347–357. doi: 10.2337/db09-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dahl M, et al. Protection against inhaled oxidants through scavenging of oxidized lipids by macrophage receptors MARCO and SR-AI/II. J Clin Invest. 2007;117(3):757–764. doi: 10.1172/JCI29968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Georgoudaki AM, et al. Reprogramming tumor-associated macrophages by antibody targeting inhibits cancer progression and metastasis. Cell Rep. 2016;15(9):2000–2011. doi: 10.1016/j.celrep.2016.04.084. [DOI] [PubMed] [Google Scholar]
- 74.Kazankov K, et al. The role of macrophages in nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Nat Rev Gastroenterol Hepatol. 2019;16(3):145–159. doi: 10.1038/s41575-018-0082-x. [DOI] [PubMed] [Google Scholar]
- 75.Rivera CA, Adegboyega P, van Rooijen N, Tagalicud A, Allman M, Wallace M. Toll-like receptor-4 signaling and Kupffer cells play pivotal roles in the pathogenesis of non-alcoholic steatohepatitis. J Hepatol. 2007;47(4):571–579. doi: 10.1016/j.jhep.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Garcia-Martinez I, et al. Hepatocyte mitochondrial DNA drives nonalcoholic steatohepatitis by activation of TLR9. J Clin Invest. 2016;126(3):859–864. doi: 10.1172/JCI83885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang X, et al. Macrophage p38α promotes nutritional steatohepatitis through M1 polarization. J Hepatol. 2019;71(1):163–174. doi: 10.1016/j.jhep.2019.03.014. [DOI] [PubMed] [Google Scholar]
- 78.Svendsen P, et al. Antibody-directed glucocorticoid targeting to CD163 in M2-type macrophages attenuates fructose-induced liver inflammatory changes. Mol Ther Methods Clin Dev. 2017;4:50–61. doi: 10.1016/j.omtm.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boyle JJ, et al. Heme induces heme oxygenase 1 via Nrf2: role in the homeostatic macrophage response to intraplaque hemorrhage. Arterioscler Thromb Vasc Biol. 2011;31(11):2685–2691. doi: 10.1161/ATVBAHA.111.225813. [DOI] [PubMed] [Google Scholar]
- 80.Kim YC, Masutani H, Yamaguchi Y, Itoh K, Yamamoto M, Yodoi J. Hemin-induced activation of the thioredoxin gene by Nrf2. A differential regulation of the antioxidant responsive element by a switch of its binding factors. J Biol Chem. 2001;276(21):18399–18406. doi: 10.1074/jbc.M100103200. [DOI] [PubMed] [Google Scholar]
- 81.Alam J, et al. Heme activates the heme oxygenase-1 gene in renal epithelial cells by stabilizing Nrf2. Am J Physiol Renal Physiol. 2003;284(4):F743–F752. doi: 10.1152/ajprenal.00376.2002. [DOI] [PubMed] [Google Scholar]
- 82.Kapetanaki MG, Gbotosho OT, Sharma D, Weidert F, Ofori-Acquah SF, Kato GJ. Free heme regulates placenta growth factor through NRF2-antioxidant response signaling. Free Radic Biol Med. 2019;143:300–308. doi: 10.1016/j.freeradbiomed.2019.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Habib A, Finn AV. The role of iron metabolism as a mediator of macrophage inflammation and lipid handling in atherosclerosis. Front Pharmacol. 2014;5:195. doi: 10.3389/fphar.2014.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Joseph SB, Castrillo A, Laffitte BA, Mangelsdorf DJ, Tontonoz P. Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat Med. 2003;9(2):213–219. doi: 10.1038/nm820. [DOI] [PubMed] [Google Scholar]
- 85.Vallelian F, et al. Proteasome inhibition and oxidative reactions disrupt cellular homeostasis during heme stress. Cell Death Differ. 2015;22(4):597–611. doi: 10.1038/cdd.2014.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Santoro AM, Lo Giudice MC, D’Urso A, Lauceri R, Purrello R, Milardi D. Cationic porphyrins are reversible proteasome inhibitors. J Am Chem Soc. 2012;134(25):10451–10457. doi: 10.1021/ja300781u. [DOI] [PubMed] [Google Scholar]
- 87.Figueiredo RT, et al. Characterization of heme as activator of Toll-like receptor 4. J Biol Chem. 2007;282(28):20221–20229. doi: 10.1074/jbc.M610737200. [DOI] [PubMed] [Google Scholar]
- 88.Vallelian F, et al. Revisiting the putative role of heme as a trigger of inflammation. Pharmacol Res Perspect. 2018;6(2):e00392. doi: 10.1002/prp2.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474(7351):337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cabral F, et al. Purification of hepatocytes and sinusoidal endothelial cells from mouse liver perfusion. J Vis Exp. 2018;(132):56993. doi: 10.3791/56993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40(10):4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.