Abstract
Proteins created from recurrent fusion genes like CBFB-MYH11 are prevalent in acute myeloid leukemia (AML), often necessary for leukemogenesis, persistent throughout the disease course, and highly leukemia specific, making them attractive neoantigen targets for immunotherapy. A nonameric peptide derived from a prevalent CBFB-MYH11 fusion protein was found to be immunogenic in HLA-B*40:01+ donors. High-avidity CD8+ T cell clones isolated from healthy donors killed CBFB-MYH11+ HLA-B*40:01+ AML cell lines and primary human AML samples in vitro. CBFB-MYH11–specific T cells also controlled CBFB-MYH11+ HLA-B*40:01+ AML in vivo in a patient-derived murine xenograft model. High-avidity CBFB-MYH11 epitope–specific T cell receptors (TCRs) transduced into CD8+ T cells conferred antileukemic activity in vitro. Our data indicate that the CBFB-MYH11 fusion neoantigen is naturally presented on AML blasts and enables T cell recognition and killing of AML. We provide proof of principle for immunologically targeting AML-initiating fusions and demonstrate that targeting neoantigens has clinical relevance even in low–mutational frequency cancers like fusion-driven AML. This work also represents a first critical step toward the development of TCR T cell immunotherapy targeting fusion gene–driven AML.
Keywords: Immunology, Oncology
Keywords: Cancer immunotherapy, Leukemias, T cells
Introduction
Fusion genes represent essential early events in leukemic transformation in acute myeloid leukemia (AML), particularly in younger patients (1). The core-binding factor (CBF) leukemias are a prevalent subgroup of AML that arise from the leukemia-initiating gene fusions CBFB-MYH11 and RUNX1-RUNX1T1, which are produced by characteristic chromosomal translocations (2). T cell immunotherapy could be used to target such fusions. Protein products from cancer-specific mutations, including gene fusions, can be processed into short peptides and presented on the cell surface in the context of human leukocyte antigen (HLA), giving rise to T cell neoantigens with high specificity for malignant cells (3–5). The importance of neoantigens in antitumor immunity is evident from clinical successes with adoptive transfer of neoantigen-specific tumor-infiltrating lymphocytes (6–8) and neoantigen vaccines (9–11), and the contribution of neoantigen-specific responses to the efficacy of immune checkpoint blockade therapy (3) and graft-versus-malignancy responses after allogeneic hematopoietic cell transplantation (12). Effective immune responses against neoantigens present in the founding malignant clone can be curative. However, while neoantigens are well investigated in solid tumors, little is known about neoantigens in AML outside of the subset of the disease bearing mutations in nucleophosmin 1 (NPM1) (13–15), and AML fusions in particular have not been definitively identified as neoantigens (5).
A fusion between the CBFB and MYH11 genes (16) resulting from inv(16) and t(16;16) occurs in approximately half of patients with CBF AML, representing 12% of pediatric and 7% of adult AML overall (1, 17). This fusion is an essential leukemia-initiating event (18–20) that occurs in the founding clone (21), is stable across the disease course, and persists at relapse (22, 23). Among patients with de novo inv(16) or t(16;16) AML, specific chromosomal breakpoints are highly recurrent, with 80% to 90% of patients sharing the type A CBFB-MYH11 fusion variant (24). Despite being classified as a favorable-risk AML subtype (25), only 50% to 60% of patients with CBFB-MYH11+ AML are cured with intensive chemotherapy alone, and outcomes in older adults are particularly poor (26, 27). Concurrent signaling mutations also negatively impact prognosis (28). Deep molecular remissions are not always achieved with standard therapies but appear necessary to prevent subsequent relapse (29), highlighting an unmet need for novel and more effective treatments, especially for relapsed and chemotherapy-refractory disease. In contrast to neoantigens derived from unique patient-specific cancer mutations (4), CBFB-MYH11 is highly recurrent and could serve as a shared target for immunotherapy in patients with CBFB-MYH11+ AML. Moreover, targeting the clonal CBFB-MYH11 should prevent escape of leukemic subclones.
Some investigators have successfully used patient tumor (3) or peripheral blood (PB) (30, 31) as sources of neoantigen-specific T cells. However, in AML, lymphopenia and/or disease-related immune dysfunction (3, 32–36) hinder identification of neoantigen-specific T cells directly from patients. We (37) and others (38) have successfully identified antigen-specific T cells from the PB repertoire of healthy donors. Therefore, we used a reverse-immunology strategy employing in vitro stimulation of healthy HLA-typed donor CD8+ T cells to identify high-avidity T cells specific for an epitope derived from the CBFB-MYH11 type A fusion protein. We determined that the epitope these T cells recognize is naturally processed and presented from the CBFB-MYH11 type A fusion in primary AML samples. We demonstrated efficacy of CBFB-MYH11–specific T cells in vivo using a patient-derived xenograft (PDX) in a humanized mouse model that reliably engrafts CBF AML (39). Gene transfer of high-avidity T cell receptors (TCRs) conferred CBFB-MYH11–specific cytotoxicity to CD8+ T cells. Our data indicate that the CBFB-MYH11 fusion neoantigen is a promising target for T cell immunotherapy for individuals with CBFB-MYH11+ AML.
Results
Candidate peptide epitopes are predicted from a prevalent CBFB-MYH11 fusion.
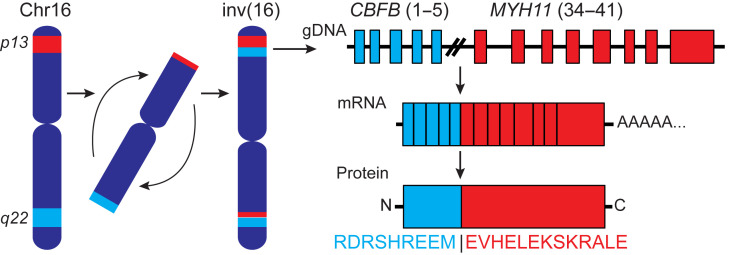
Variants of the CBFB-MYH11 fusion gene are created by different chromosomal breakpoints. The type A fusion (Figure 1), produced by joining exons 5 and 34 of CBFB and MYH11, respectively, is the predominant CBFB-MYH11 fusion (24). We evaluated the amino acid sequence spanning the fusion in silico to determine the probability that fusion-derived peptides would bind to one of 20 prevalent HLA class I molecules, including 5 HLA-A, 7 HLA-B, and 8 HLA-C alleles. Six candidate epitopes with binding affinities of less than 500 nM were predicted by at least 1 of the 3 HLA-binding prediction algorithms used (Table 1). Peptides with predicted IC50 ≤ 500 nM were identified only for HLA-B*40:01 and -B*44:02. The peptide REEMEVHEL had particularly high predicted binding affinity (<26 nM) to HLA-B*40:01 by all 3 algorithms.
Figure 1. Schematic of the type A CBFB-MYH11 fusion resulting from an inversion in chromosome 16.
In the type A fusion, exon 5 of CBFB is fused to exon 34 of MYH11, creating a fusion protein with the junctional amino acid sequence depicted.
Table 1. Candidate peptide epitopes derived from the CBFB-MYH11 type A fusion protein.
An immunogenic CBFB-MYH11 epitope is presented on HLA-B*40:01.
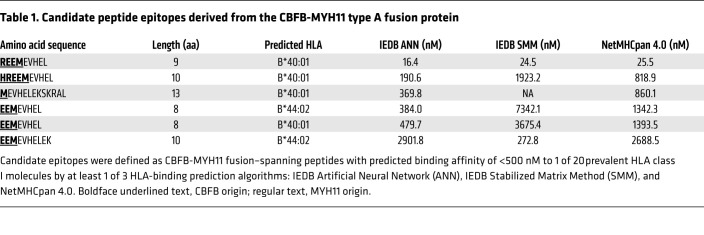
HLA-binding prediction algorithms effectively identify peptides with a high probability of binding to a particular HLA molecule. However, not all peptides predicted to bind to HLA actually do bind, and not all that bind are immunogenic (i.e., elicit T cell responses) (40, 41). Therefore, we directly tested the immunogenicity of candidate CBFB-MYH11 epitopes in vitro by stimulating CD8+ T cells from HLA-typed healthy donors with autologous mature DCs pulsed with a pool of candidate CBFB-MYH11 and control peptide epitopes. After 12–13 days of stimulation, we tested for specific lysis of peptide-pulsed autologous lymphoblastoid cell lines (LCLs) in 51Cr-release cytotoxicity assay (CRA). In 2 donors (D1 and D2, Figure 2A), we identified T cell lines that specifically lysed peptide-pulsed targets (D1, n = 1; D2, n = 8). As expected, most of the wells of T cells showed nonspecific reactivity or no reactivity. We subsequently determined that a subset of the wells contained T cells that specifically lysed target cells pulsed with control peptides known to be immunogenic (Figure 2A, black triangles). However, we also determined that some of the peptide-specific wells contained T cells that specifically lysed target cells pulsed with CBFB-MYH11 peptides (D1, n = 1; D2, n = 3; red triangles). We cloned the CBFB-MYH11–reactive T cells by limiting dilution, expanded them, and tested them for recognition of single peptides. The D1 T cell clone and 3 D2 clones, as well as 2 clones from a third donor (D3), were REEMEVHEL specific and did not recognize LCLs alone or with other peptides, including additional candidate peptides predicted to bind HLA-B*40:01 (Figure 2B and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI137723DS1) or a nonameric peptide known to bind HLA-B*40:01 (ref. 42 and Figure 2C). The clones’ lack of recognition of HLA-B*40:01+ LCLs in the absence of exogenous REEMEVHEL peptide indicated specific recognition of the peptide-HLA complex rather than alloreactivity to HLA-B*40:01. All 6 clones specifically lysed targets pulsed with as little as 1 ng/mL peptide in CRA, and 4 had high functional avidity (IC50 <1 ng/mL; Figure 2D). To further validate the immunogenicity of the epitope, we stimulated CD8+ T cells from an additional 3 donors and elicited REEMEVHEL-specific responses in all 3 tested (Supplemental Figure 1).
Figure 2. Highly avid CD8+ T cells recognizing an HLA-B*40:01–restricted epitope of CBFB-MYH11 can be isolated from healthy donors.
(A) Percentage specific lysis of peptide-pulsed targets for individual wells of CD8+ T cell lines after in vitro peptide stimulation of 2 HLA-B*40:01+ donors. Percent specific lysis was calculated as percent lysis of peptide-pulsed targets minus percent lysis of no-peptide targets to remove nonspecific reactivity to autologous target cells alone. Gray circles indicate wells with no peptide-specific reactivity. The single-peptide specificity of T cells in wells with peptide-specific lysis was subsequently determined: red triangles, REEMEVHEL-specific T cells; black triangles, T cells specific for control (known immunogenic, non–CBFB-MYH11) epitopes. Each plot depicts a single experiment. (B) Percentage lysis of HLA-B*40:01+ LCL targets with REEMEVHEL peptide (circles), pooled additional predicted HLA-B*40:01–binding peptides (squares), or no exogenous peptide (triangles) by REEMEVHEL-specific CD8+ T cell clones isolated from 3 healthy donors in 4-hour CRA. (C) Percentage lysis of HLA-B*40:01+ LCL targets with either REEMEVHEL peptide (circles), known HLA-B*40:01–binding peptide KECVLHDDL (diamonds), or no exogenous peptide (triangles) by REEMEVHEL-specific CD8+ T cell clones in 4-hour CRA. For B and C, peptides were used at 1000 ng/mL each; error bars are SD of 3 biological replicates. (D) Percentage lysis of targets pulsed with various concentrations of REEMEVHEL peptide by REEMEVHEL-specific CD8+ T cell clones. Mean and SEM of 3 technical replicates are shown. (E) Representative flow plots (from 3 experiments) of CBFB-MYH11/HLA-B*40:01 pHLA tetramer staining of REEMEVHEL-specific T cell clones and an irrelevant clone specific for an epitope (IPRAHNRLV) presented on HLA-B*07:02 (negative control). Cells are gated on live single CD4–CD8+ cells. (F) Percentage lysis of REEMEVHEL-pulsed LCLs (1000 ng/mL) with varying HLA types (including all class I HLA types of the 3 donors) by 6 REEMEVHEL-specific T cell clones.
All 6 T cell clones stained with REEMEVHEL/B*40:01 peptide/HLA (pHLA) tetramer (Figure 2E; gating strategy, Supplemental Figure 2), consistent with the predicted HLA restriction of the epitope. For additional corroboration, we tested clones for lysis of HLA-diverse LCL target cells pulsed with REEMEVHEL peptide. LCLs were chosen to represent the individual HLAs of the 3 donors (D1–D3; Supplemental Table 2) with minimal overlap across LCLs. Only HLA-B*40:01+ LCLs were lysed by clones, confirming the HLA-B*40:01 restriction (Figure 2F). Although all 3 donors shared both HLA-B*40:01 and -C*03:04 (Supplemental Table 2), and HLA-B*40:01+ LCLs were also HLA-C*03:04+, REEMEVHEL peptide–pulsed HLA-C*03:04+ HLA-B*40:01– LCLs were not lysed by clones. REEMEVHEL was also predicted to bind to HLA-B*40:02 (Supplemental Figure 3A). Two high-avidity clones lysed REEMEVHEL-pulsed HLA-B*40:02+ LCL targets at low levels (Supplemental Figure 3B), indicating that REEMEVHEL is also presented on HLA-B*40:02, but that clones preferentially recognize the epitope on HLA-B*40:01, reflecting their fine specificity. These results demonstrate that the predicted CBFB-MYH11 epitope is immunogenic and confirm its binding to HLA-B*40:01 and HLA-B*40:02.
CBFB-MYH11/B*40:01 epitope–specific T cell clones kill AML cell lines.
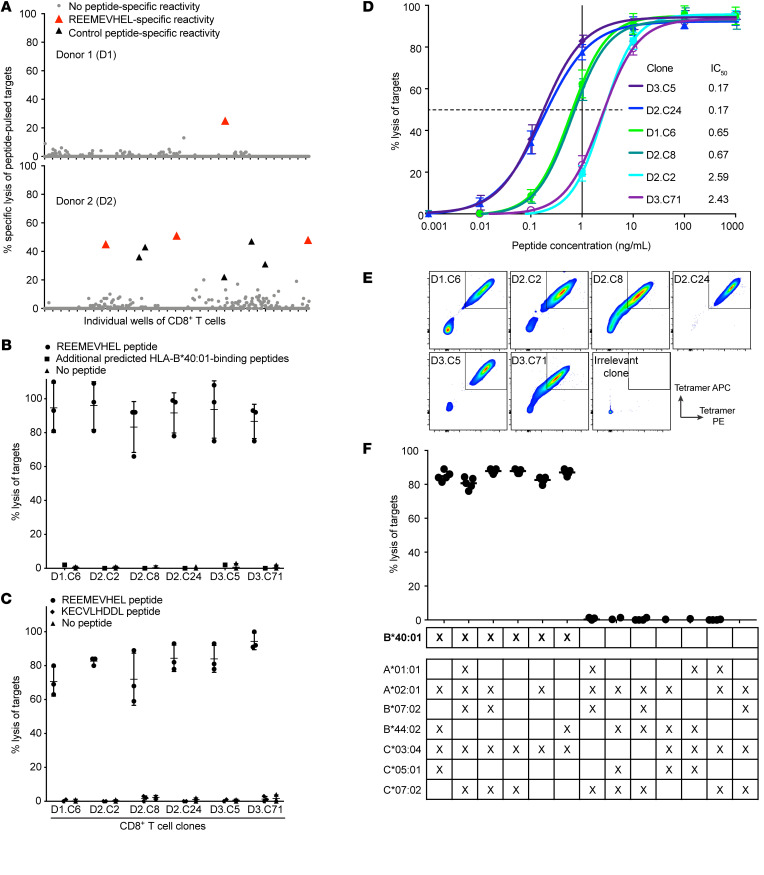
We next asked whether the REEMEVHEL epitope was processed from the CBFB-MYH11 protein and presented by endogenous antigen-presenting machinery in AML cell lines. To create CBFB-MYH11+ HLA-B*40:01+ AML cell line targets, we transduced naturally HLA-B*40:01+ NB-4 cells to express the full-length CBFB-MYH11 type A fusion protein, and naturally CBFB-MYH11+ ME-1 cells to express HLA-B*40:01 (Supplemental Figure 4, A and B, respectively), and determined that the transduced cell lines presented the REEMEVHEL epitope (Supplemental Figure 4C). We then evaluated the ability of the high-avidity T cell clone D2.C24 to eliminate CBFB-MYH11+ HLA-B*40:01+ targets in a flow cytometry–based cytotoxicity assay, which enables the assessment of T cell killing over hours to days (43, 44). The percentage survival of CBFB-MYH11+ NB-4 cells steadily decreased during coculture with D2.C24, while mock-transduced NB-4 cells were unchanged until increasing at 96 hours (Figure 3A, representative absolute counts; Figure 3B, representative flow plots for 1 experiment; and Figure 3C, summary percentage survival). D2.C24 also killed ME-1 cells transduced to express HLA-B*40:01. While HLA-B*40:01+ ME-1 cells were killed less efficiently than NB-4 cells, survival of HLA-B*40:01+ ME-1 cells declined over time in culture with D2.C24 (Figure 3D, representative absolute counts; Figure 3E, representative flow plots for 1 experiment; and Figure 3F, summary percentage survival), whereas survival of HLA-B*40:01– mock-transduced ME-1 cells was constant. These findings demonstrate that REEMEVHEL is endogenously processed and presented on HLA-B*40:01 in transduced AML cell lines, conferring susceptibility to killing by epitope-specific CD8+ T cells.
Figure 3. T cells specific for the HLA-B*40:01–restricted CBFB-MYH11 epitope kill AML cell lines.
(A) Absolute cell number of viable NB-4 cells either CBFB-MYH11–transduced (TD) (solid line) or mock-TD (dashed line) after coculture with high-avidity REEMEVHEL-specific clone D2.C24 from a representative experiment. (B) Representative flow plots for experiments shown in A and C, depicting viable single cells at time points after coculture. (C) Percentage survival of NB-4 cells, either WT (left) or transduced to express the full-length CBFB-MYH11 type A fusion (right), at time points after coculture with the D2.C24 T cell clone. (D) Absolute cell number of viable ME-1 cells either HLA-B*40:01–TD (solid line) or mock-TD (dashed line) and cocultured with clone D2.C24 from a representative experiment. (E) Representative flow plots for experiments shown in D and F, depicting viable single cells at time points after coculture. (F) Percentage survival of ME-1 cells, either WT (left) or transduced to express HLA-B*40:01 (right), at time points after coculture with the D2.C24 T cell clone. Viable cell numbers were assessed at varying time points by flow cytometry and percent survival calculated as described in Supplemental Methods. For C and F, colored bars indicate mean and error bars SD for 3–10 technical replicate samples. Two-sample unpaired 2-tailed t tests were performed to compare 0 hours with each subsequent time point.
CBFB-MYH11/B*40:01–specific T cell clones recognize and kill primary AML in vitro.
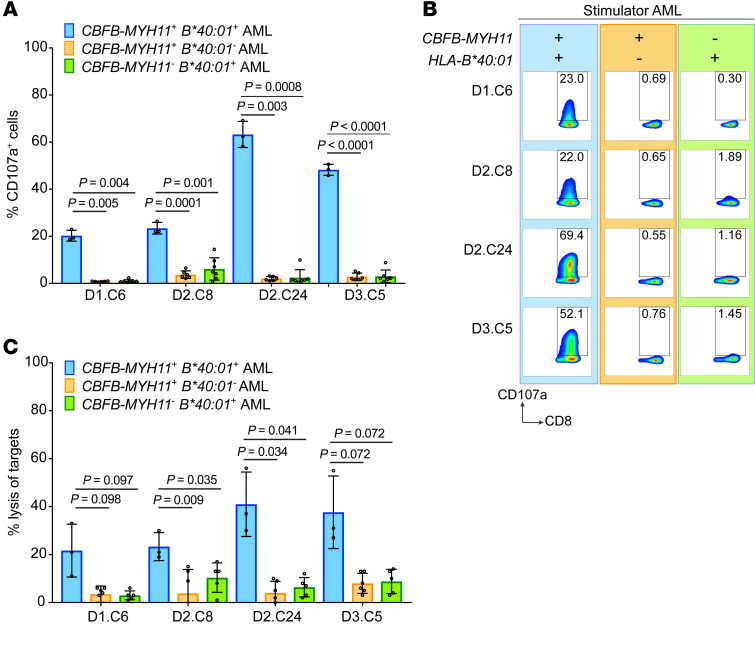
We then asked whether REEMEVHEL was also processed and presented on primary leukemic blasts obtained from patient PB at AML diagnosis. All 4 high-avidity REEMEVHEL-specific clones (D1.C6, D2.C8, D2.C24, and D3.C5) expressed CD107a in response to stimulation with CBFB-MYH11+ HLA-B*40:01+ primary AML samples from 3 different patients, but not in response to multiple different primary AML samples lacking either the fusion (n = 7) or the restricting HLA genotype (n = 6), reflecting specific degranulation and release of cytotoxic molecules in response to antigen naturally processed and presented on primary CBFB-MYH11+ HLA-B*40:01+ AML (Figure 4, A and B). Moreover, all 4 high-avidity clones lysed only CBFB-MYH11+ HLA-B*40:01+ primary AML samples from 3 different patients in a 4-hour CRA, but showed minimal lysis of primary AML samples lacking either the fusion (n = 6) or restricting HLA (n = 5) (Figure 4C). The observed levels of specific lysis of primary CBFB-MYH11+ HLA-B*40:01+ AML were comparable to what other groups testing antigen-specific T cells against primary AML have published (15, 45, 46). Taken together, these results indicate that REEMEVHEL is a recurrent, immunogenic CBFB-MYH11+ AML neoantigen that is naturally processed and presented on primary AML cells and that CBFB-MYH11+ HLA-B*40:01+ AMLs are killed by T cells specific for the fusion protein epitope REEMEVHEL.
Figure 4. The HLA-B*40:01–restricted CBFB-MYH11 epitope is a bona fide AML antigen.
Four high-avidity T cell clones were tested for recognition of CBFB-MYH11+ HLA-B*40:01+ primary AML, as well as primary AML lacking either the fusion or restricting HLA genotype. (A) Degranulation of T cell clones in response to primary AML was determined by measurement of T cell CD107a presentation in response to stimulation with primary AML (blue bars, CBFB-MYH11+ HLA-B*40:01+, n = 3 different primary AML samples; orange bars, CBFB-MYH11+ HLA-B*40:01–, n = 7; green bars, CBFB-MYH11– HLA-B*40:01+, n = 6). (B) Representative flow plots from one CD107a degranulation experiment are shown from left to right: CBFB-MYH11+ HLA-B*40:01+ AML (blue background), negative control stimulators CBFB-MYH11+ HLA-B*40:01– AML (orange background), and CBFB-MYH11– HLA-B*40:01+ AML (green background). (C) Lysis of primary AML by T cell clones was tested in a 4-hour CRA with effector/target (E:T) ratio of 20:1 (blue bars, CBFB-MYH11+ HLA-B*40:01+, n = 3; orange bars, CBFB-MYH11+ HLA-B*40:01–, n = 6; green bars, CBFB-MYH11– HLA-B*40:01+, n = 5). For A and C, mean and SD for each clone are shown are from a single experiment with 3–7 biological replicates for each group of AML samples. Statistics were calculated using unpaired 2-tailed t tests with Welch’s correction.
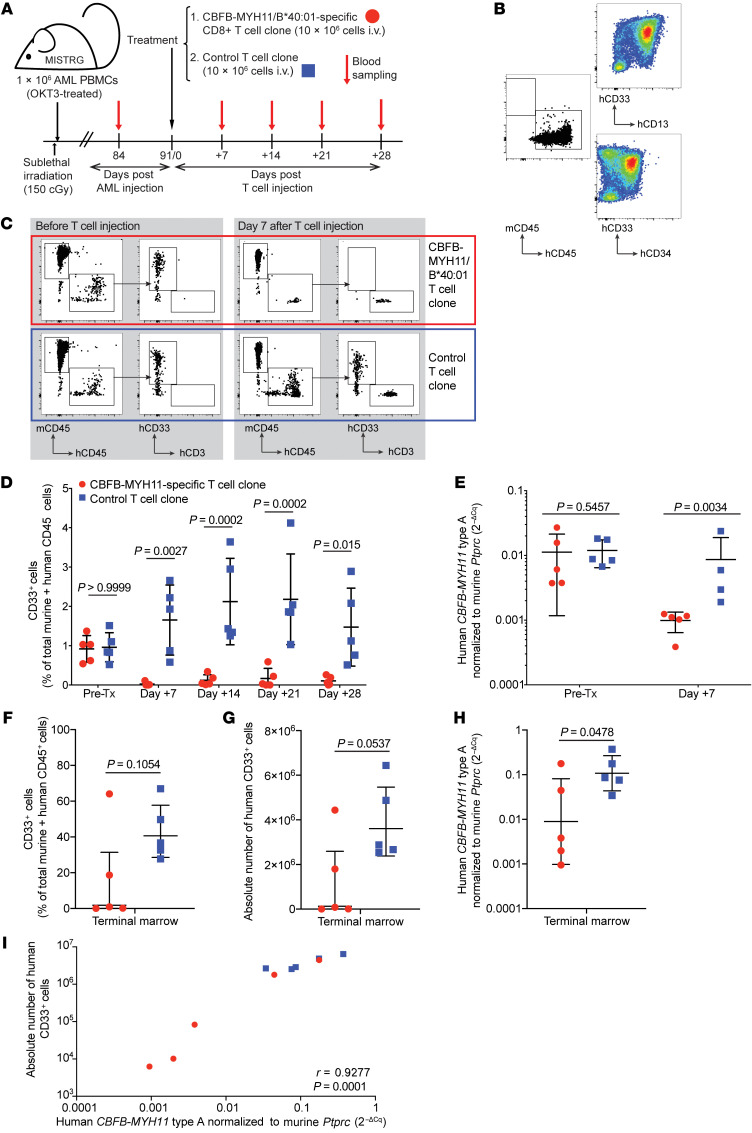
A high-avidity CBFB-MYH11/B*40:01–specific T cell clone controls AML in vivo.
We next evaluated the efficacy of the best-performing clone, D2.C24, in vivo. In contrast to other immunodeficient mouse strains, MISTRG (M-CSFh/h IL-3/GM-CSFh/hSIRPAh/hTPOh/h RAG2–/– IL2RG–/–) mice reliably engraft with CBFB-MYH11+ AML (39), providing a suitable model for in vivo evaluation of CBFB-MYH11–specific antileukemic activity. Therefore, we transplanted patient-derived primary CBFB-MYH11+ HLA-B*40:01+ AML into MISTRG mice (Figure 5A). Because many T cell immunotherapies appear to be most effective in the setting of remission or measurable residual disease (47), our objective was to model a similarly low disease burden state. Once engraftment with human CD45+ AML was established, mice were treated i.v. with D2.C24 clone or a control T cell clone. Control T cells were isolated by similar methods of in vitro stimulation from a healthy volunteer and were specific for a candidate neoantigen peptide presented by HLA-B*07:02, an allele for which AMLs used in these experiments were genotypically negative. We first engrafted a primary CBFB-MYH11+ HLA-B*40:01+ AML (AML1), which was positive for CD33 as well as CD34 and CD13 (Figure 5B). All mice underwent planned sacrifice 1 month after T cell injection; while we expected to see differences in leukemia burden in this time frame, given the low levels of disease, no survival differences were expected or observed. Mice treated with D2.C24 showed a significant reduction in PB human CD45+CD33+ cells by flow cytometry, beginning as early as 7 days after T cell infusion (representative flow plots in Figure 5C) and persisting 3 weeks later, as compared with mice treated with control clone (Figure 5D). Real-time quantitative PCR to detect the disease-specific CBFB-MYH11–encoding transcript also demonstrated significantly lower expression of fusion transcript, corresponding to AML burden, in mice 7 days after treatment with CBFB-MYH11–specific T cell clone as compared with mice treated with control T cells at the same time point (Figure 5E and Supplemental Figure 5). Human CD33+ cells were undetectable or present at a very low level in the terminal bone marrow analysis of 3 of the 5 mice treated with CBFB-MYH11/B*40:01–specific T cells (Figure 5, F and G), although the difference between the 2 groups did not reach statistical significance. Relative expression of CBFB-MYH11 type A transcripts was significantly lower in the bone marrow of mice treated with CBFB-MYH11/B*40:01–specific T cells (Figure 5H). Correlation between disease burden by flow cytometric measures and disease burden by molecular measures was high (r = 0.9277; Figure 5I).
Figure 5. CBFB-MYH11/B*40:01–specific T cells control AML in vivo in a PDX murine model.
(A) Experiment overview: Newborn, preconditioned MISTRG mice were injected intrahepatically with 1 × 106 PBMCs (OKT3-pretreated to prevent xenogeneic graft-versus-host disease) from HLA-B*40:01+ patients with active CBFB-MYH11+ AML. After 12 weeks of AML engraftment, mice received 10 × 106 CD8+ T cells i.v., either D2.C24 clone (high-avidity, CBFB-MYH11/B*40:01–specific) or a control clone (specific for a candidate neoantigen epitope, IPRAHNRLV, presented by HLA-B*07:02, for which this AML was genotypically negative), then were monitored by weekly PB sampling. (B) Primary AML PBMCs (AML1, 81% blasts) before OKT3 treatment and injection into mice were stained for myeloid markers using AML tracking in mice. (C) Representative flow plots of mice PB pretreatment (left) and 7 days after injection (right) with either CBFB-MYH11/B40:01–specific (red box) or control (blue box) T cell clones. (D) Summary of PB disease burden by flow cytometry after CBFB-MYH11/B40:01–specific (red circles) or control (blue squares) T cell treatment. Statistics were calculated using repeated-measures 2-way ANOVA. (E) Human CBFB-MYH11 type A transcript expression, normalized to murine CD45 (Ptprc) as 2–ΔCq, was assessed before and 7 days after administration of CBFB-MYH11/B40:01–specific or control T cells. (F–H) AML burden in terminal bone marrow as percentage (F) or absolute number (G) of human CD33+ cells, and relative CBFB-MYH11 type A transcript expression (H). (I) Correlation between marrow disease burden as measured by flow cytometry and real-time qPCR was determined by calculation of Pearson correlation coefficient. For all groups and time points, n = 5, except for control T cell–treated mice on day 7 (n = 4) owing to poor RNA yield from 1 sample. Except where noted, statistics were calculated using unpaired 2-tailed parametric t tests. Mean and SD are shown.
Overall, our observations indicated that targeting the CBFB-MYH11/B*40:01 epitope in vivo with the D2.C24 T cell clone had an antileukemic effect but did not lead to complete clearance of AML in all mice at 1 month. We hypothesized that leukemia persistence in some of the CBFB-MYH11/B*40:01–specific T cell–treated mice could reflect loss of CBFB-MYH11 neoantigen presentation by the AML, either by loss of HLA class I expression or by loss of REEMEVHEL peptide presentation. Surface expression of HLA class I on human CD45+ cells from terminal marrow was similar in mice treated with CBFB-MYH11/B*40:01–specific T cells versus control T cells (Supplemental Figure 6A), suggesting that altered HLA class I expression was not the mechanism of leukemia persistence. We then used an in vitro CD107a degranulation assay to test whether the D2.C24 T cell clone could recognize AML in terminal bone marrow samples from T cell treated–mice, which would indicate whether the persistent AML still presented REEMEVHEL peptide on HLA-B*40:01. Bone marrow samples from mice with persistent AML (as detected by flow cytometry and relative CBFB-MYH11 transcript expression) in both treatment groups induced degranulation of the CBFB-MYH11/B*40:01–specific D2.C24 T cell clone (Supplemental Figure 6B). The proportion of CD107a+ T cells was highest when cultured with marrow specimens containing higher percentages of human CD33+ cells (Supplemental Figure 6C). The bone marrow of 2 CBFB-MYH11/B*40:01–specific T cell–treated mice had very low-frequency events on flow cytometry, at the limit of detection of human CD33+ cells in our assay; these samples did not induce T cell degranulation above background levels, consistent with the observation that these 2 mice had completely cleared their disease and that no neoantigen remained. Since the persisting AML continued to present the neoantigen, we hypothesized that a T cell problem might have enabled leukemia persistence in a subset of animals. In support of this hypothesis, we observed that circulating human CD3+ T cells became undetectable in CBFB-MYH11/B*40:01–specific T cell–treated mice by day +14 but were detectable at all time points in mice treated with control epitope-specific T cells (Supplemental Figure 6D). Additionally, the absolute number of human CD45+CD3+ cells in terminal bone marrow was over a log lower in CBFB-MYH11/B*40:01–specific T cell–treated compared with control epitope-specific T cell–treated mice (Supplemental Figure 6E). These findings suggest that leukemia persistence in this experiment was likely related to inadequate T cell expansion and/or persistence. Lack of CBFB-MYH11/B*40:01–specific T cell persistence was antigen specific, as a similarly rapid drop in the control T cells was not observed, and likely resulted from the increased susceptibility of the clones, which had undergone multiple prior cycles of in vitro activation and expansion, to activation-induced cell death, exhaustion, senescence, or another process in the face of ongoing antigen exposure over weeks.
To confirm that the observed in vivo recognition of AML by our CBFB-MYH11/B*40:01–specific T cells was reproducible, we repeated the experiment in a second cohort of MISTRG mice engrafted with a different primary CBFB-MYH11+ HLA-B*40:01+ AML (AML2) over 5 months (Supplemental Figure 7A). In this experiment, mice engrafted with AML2 and treated with D2.C24 showed reduced percentages of PB CD45+CD33+ cells by flow cytometry at days 3 and 7 after T cell injection (Supplemental Figure 7B), and a significant difference in the absolute number of human CD33+ cells on terminal analysis of PB (Supplemental Figure 7C) and spleen (Supplemental Figure 7D) on day 10. No significant difference in the absolute number of human CD33+ cells in the terminal bone marrow was observed between groups (Supplemental Figure 7D), which may reflect inadequate time for the T cells to traffic to and clear bone marrow disease over the experimental time course.
Although the 2 leukemia samples behaved differently in xenografts, with AML1 engrafting at higher levels than AML2 (mean 0.89%, range 0.52%–1.37%, vs. mean 0.19%, range 0.01%–2.45%, respectively), an antileukemic effect of T cells targeting the CBFB-MYH11/B*40:01 neoantigen was seen in both experiments. Taken together, these data indicate that CBFB-MYH11/B*40:01–specific T cells have in vivo efficacy against primary CBFB-MYH11+ AML in a xenotransplantation model.
CBFB-MYH11/B*40:01–specific T cells recognize a unique leukemia-specific epitope.
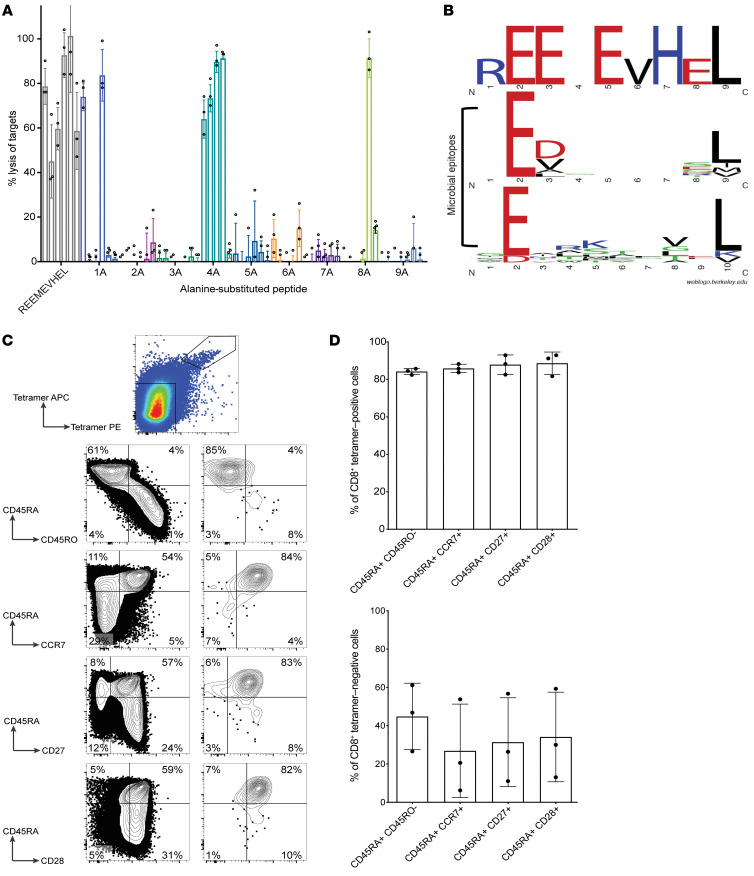
Having identified the CBFB-MYH11/B*40:01 neoantigen as a target for T cell immunotherapy, we next considered potential for cross-reactivity of CBFB-MYH11/B*40:01–specific T cells against similar epitopes and consequent toxicity, first by using alanine scanning across the epitope. None of the 6 T cell clones recognized peptides with alanine residues substituted at positions 2, 3, 5, 6, 7, or 9 (Figure 6A). Position 2 and the C-terminal residue (here, position 9) are known anchor residues (48) and, as expected, were predicted to be necessary for peptide binding to HLA-B*40:01 (Supplemental Table 3). Glutamic acids at positions 3 and 5, valine at position 6, and histidine at position 7 were essential for T cell recognition but do not affect predicted HLA binding. No sequences with ≥80% identity to the REEMEVHEL epitope or to sequences with X substituted in nonessential positions 1, 4, and/or 8 were identified in the nonredundant Basic Local Alignment Search Tool (BLAST) database of Homo sapiens protein sequences. The most similar peptide identified by BLAST (STEMEVHEL; derived from multiple isoforms of phosphatase and actin regulator 1 [PHACTR1]) was not recognized by any of the high-avidity clones (Supplemental Figure 8A) in CRA. A similar HLA-B*40:01–restricted epitope (RESEEESVSL) from the male minor histocompatibility antigen UTY (49) was also not recognized (Supplemental Figure 8B). As CBFB-MYH11/B*40:01–specific T cell responses might represent cross-reactivity to a microbial epitope, we aligned REEMEVHEL to 21 known HLA-B*40:01–restricted microbial T cell epitopes in the Immune Epitope Database, but found no overall similarity except as expected at anchor positions 2 and 9 (Figure 6B and ref. 48). In these studies, we did not find evidence that CBFB-MYH11/B*40:01–specific T cells were cross-reactive to similar epitopes.
Figure 6. CBFB-MYH11/B*40:01–specific CD8+ T cells recognize a unique epitope and have a naive phenotype in healthy donors.
(A) Percentage lysis by REEMEVHEL-specific T cell clones of HLA-B*40:01+ LCLs pulsed with individual peptides (at 1000 ng/mL) with alanine substitution at each position. Each bar represents percentage lysis of targets by one T cell clone from 3 technical replicate experiments with mean and SD shown. (B) Sequence logo of critical residues of the CBFB-MYH11/B*40:01 epitope for T cell clones compared with residues shared among known HLA-B*40:01–restricted microbial epitopes from IEDB. (C) Phenotypic evaluation of epitope-specific T cells isolated in healthy donors by CBFB-MYH11/B*40:01 tetramer enrichment of PBMCs from healthy donors and surface marker staining. Representative flow plots of viable CD8+ single cells from tetramer enrichment of donor D1 are shown (of 3 biological replicate experiments summarized in D). (D) Summary of phenotypic evaluation from C across 3 biological replicates (donors D1–D3); mean and SD are shown.
To further exclude the possibility that CBFB-MYH11/B*40:01–specific T cells represent cross-reactive memory T cells, we isolated unmanipulated epitope-specific CD8+ T cells from PBMCs of 3 HLA-B*40:01+ healthy donors (D1–D3) using pHLA tetramer enrichment. Tetramer-positive T cells from all 3 donors showed a naive CD45RA+CD45RO–CCR7+CD27+CD28+ phenotype (Figure 6, C and D; gating strategy, Supplemental Figure 9A), indicating that CBFB-MYH11/B*40:01–specific T cells in healthy donors were not memory T cells. Polyclonal T cell lines generated from sorted, expanded tetramer-positive CD8+ cells lysed HLA-B*40:01+ targets with but not without REEMEVHEL peptide (Supplemental Figure 9B), confirming their specificity. The naive immunophenotype of epitope-specific T cells in healthy donors suggests that the reproducible immunogenicity of the epitope was not due to cross-reactivity with preexisting memory responses.
CBFB-MYH11/B*40:01–specific TCRs are diverse and confer epitope-specific antileukemic cytotoxicity.
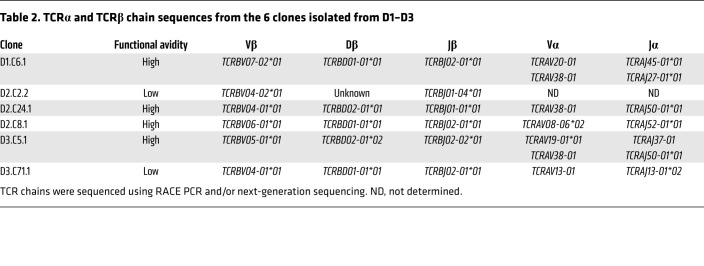
Adoptive transfer of T cells engineered to express antigen-specific TCRs can overcome quantitative and/or qualitative defects in natural antileukemic T cell immunity, which are known to exist in AML (32–36). Having determined that the CBFB-MYH11 neoantigen was a suitable AML target, we thus assessed the feasibility of transferring high-avidity CBFB-MYH11/B*40:01–specific TCRs as a first step toward immunotherapy development. To determine whether the epitope-specific TCRs shared common features, we sequenced the TCRα and TCRβ chains from all 6 T cell clones isolated from normal donors. The CBFB-MYH11/B*40:01–specific TCRs were diverse: no 2 clones isolated from a single donor shared the same Vβ region, and only 1 Vβ region (TCRBV04-01*01) was shared by 2 clones isolated from different donors (Table 2). To generate transgenic TCRs, we paired the TCRα and TCRβ chains of the 4 high-avidity clones (D1.C6, D2.C8, D2.C24, and D3.C5) in lentiviral vectors (LVs). Two α chains were identified in 2 clones (D1.C6 and D3.C5); the existence of T cell clones with 2 rearranged α chains has been previously described (50). For these clones, 2 transgenic TCRs (B1A1 and B1A2) were constructed.
Table 2. TCRα and TCRβ chain sequences from the 6 clones isolated from D1–D3.
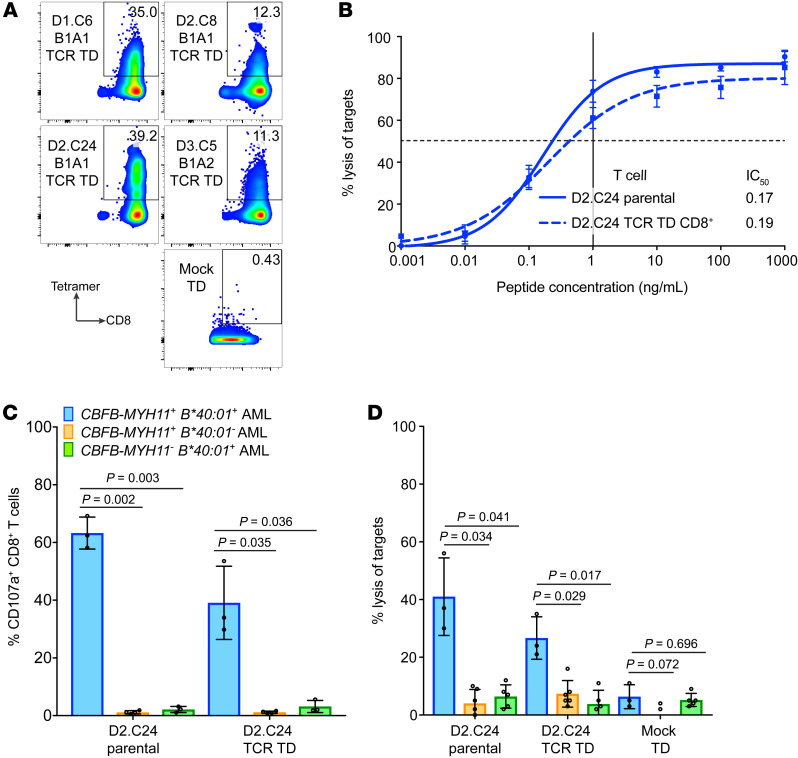
We successfully cloned 5 TCRs into LVs and transduced healthy donor CD8+ T cells. Four TCRs, representing all 4 parental clones, expressed well as measured by pHLA tetramer staining (Figure 7A). All 4 TCRs conferred CBFB-MYH11/B*40:01–specific function based on lysis of peptide-pulsed LCLs (Figure 7B and Supplemental Figure 10A). Donor CD8+ T cells transduced with the D2.C24 TCR showed particularly high functional avidity in peptide-titration CRA (Figure 7B). Additionally, when transduced into CD4+ T cells (Supplemental Figure 10B), the D2.C24 TCR showed evidence of some CD8 coreceptor-independent function in CD107a assay (Supplemental Figure 10, C and D) and in CRA (Supplemental Figure 10E), consistent with a high intrinsic affinity for the neoantigen (51, 52). D2.C24 TCR-transduced CD8+ T cells demonstrated cytolytic activity against CBFB-MYH11+ HLA-B*40:01+ primary AML samples (degranulation by CD107a assay, Figure 7C; and cytotoxicity by standard 4-hour CRA, Figure 7D) when tested against panels of primary samples from patients with active AML at diagnosis. Importantly, the TCR-transduced T cells did not degranulate with or lyse primary AML samples lacking either the fusion or restricting HLA genotype (Figure 7, C and D), indicating specific recognition of the CBFB-MYH11/B*40:01 epitope. These results indicate that CBFB-MYH11–specific TCR transfer confers epitope specificity and antileukemic cytotoxicity, and that the TCR-transduced CD8+ T cells can recognize CBFB-MYH11/B*40:01 epitope as it is naturally processed and presented on primary AML cells.
Figure 7. TCRs specific for the CBFB-MYH11/B*40:01 epitope confer antileukemic activity.
(A) Expression of transgenic TCRs in CD8+ T cells transduced (TD) with D1.C6 B1A1, D2.C8 B1A1, D2.C24 B1A1, and D3.C5 B1A2 TCR constructs or mock-TD is shown by staining with CBFB-MYH11/B*40:01 tetramer from a single experiment. (B) HLA-B*40:01+ LCLs pulsed with REEMEVHEL peptide at various concentrations were lysed in CRA by D2.C24 TCR-TD CD8+ T cells (dashed line) and D2.C24 parental clone (solid line). Mean and SEM for 3 technical replicates are shown. (C) Degranulation of D2.C24 TCR-TD CD8+ T cells and D2.C24 parental clone in response to primary AML was determined by measurement of T cell CD107a presentation in response to stimulation with primary AML (blue bars, CBFB-MYH11+ HLA-B*40:01+, n = 3 different primary AML samples; orange bars, CBFB-MYH11+ HLA-B*40:01–, n = 3; green bars, CBFB-MYH11– HLA-B*40:01+, n = 4). (D) Lysis of primary AML by D2.C24 TCR-TD CD8+ T cells, D2.C24 parental clone, and mock-TD CD8+ T cells was evaluated in a standard 4-hour CRA with E:T of 20:1 (CBFB-MYH11+ HLA-B*40:01+, n = 3 different AML samples; CBFB-MYH11+ HLA-B*40:01–, n = 6; CBFB-MYH11– HLA-B*40:01+, n = 5). For C and D, mean and SD are shown. Statistics were calculated using unpaired 2-tailed t tests with Welch’s correction.
Discussion
We have identified a CD8+ T cell epitope spanning the CBFB-MYH11 type A fusion protein that is naturally processed and presented on HLA-B*40:01 on primary AML blasts. To our knowledge, this epitope is the first reported neoantigen derived from an AML fusion protein. While fusion-derived CD8+ T cell neoantigens have been reported in other diseases, including chronic myeloid leukemia (53–56), acute lymphoblastic leukemia (57, 58), sarcomas (59, 60), and fusion-driven metastatic head and neck cancer (61), not all have been confirmed to be presented on primary tumor cells (5, 59, 60, 62). We elicited CD8+ T cell responses to the epitope in all 6 donors we tested, indicating that the epitope is consistently immunogenic. CD8+ T cells with high functional avidity for CBFB-MYH11/B*40:01 detected antigen that was processed from endogenous CBFB-MYH11 fusion protein and presented on HLA-B*40:01 on cell lines and primary AML cells. CBFB-MYH11/B*40:01–specific T cells showed epitope-specific antileukemic activity against primary AML cells in vitro as well as in vivo in a novel application of MISTRG PDX. We did not observe cross-reactivity of CBFB-MYH11/B*40:01–specific T cells against epitopes with similar amino acid sequences. We also observed no evidence of systemic toxicity in CBFB-MYH11–specific T cell–treated animals based on standard veterinary assessment in either murine experiment. As expected for de novo responses against a true neoantigen, unmanipulated epitope-specific CD8+ T cells isolated from healthy donor PBMCs had a predominantly naive phenotype. Taken together, our findings together provide proof of principle for AML fusion neoantigens as targets for T cell immunotherapy and indicate that the CBFB-MYH11/B*40:01 neoantigen is a suitable immunotherapeutic target for a subset of patients with AML.
Some key features of neoantigen quality for immunotherapeutic targeting have been postulated (63), including clonality, dissimilarity to self-antigens, similarity to microbial antigens, high protein expression, binding to HLA, and low likelihood that genetic abnormality yielding the neoantigen will be lost through deletion or transcriptional repression. The CBFB-MYH11 fusion–derived neoantigen has a number of these features. Our data indicate that the epitope lacks similarity to WT human proteins and binds to HLA-B*40:01. Moreover, as an essential leukemia-initiating event that occurs in a founding preleukemic clone (18–23), the fusion has high clonality and is unlikely to be lost without a significant cost to the leukemia’s fitness. As such, our approach targeting the essential CBFB-MYH11 fusion avoids the possibility that a subclone lacking the fusion will escape, in contrast to immunotherapies directed against an expendable surface marker (64), although other avenues of immune evasion such as downregulating HLA expression (44) or altering proteasomal processing of the epitope (65) are possible. We did not see evidence of AML escape from CBFB-MYH11–specific T cells through loss of HLA expression or presentation of the neoantigen in vivo.
While we were successful in stimulating CBFB-MYH11/B*40:01–specific T cell responses in blood from healthy donors, it is unclear whether the same might be true in patients. Neoantigen vaccines have shown clinical efficacy in solid tumors (9–11) in recent studies, but vaccines rely on the patient’s capacity to mount an effective immune response to the epitope or epitopes in the vaccine. We attempted to isolate and expand CBFB-MYH11/B*40:01–specific T cells ex vivo from PBMCs of patients with CBFB-MYH11+ HLA-B*40:01+ AML, but did not succeed in generating convincing data (data not shown); consequently, we cannot draw any conclusions about whether or not AML patients have preexisting T cell responses to the CBFB-MYH11/B*40:01 epitope. Others have been similarly unsuccessful in stimulating or detecting T cell responses against clearly immunogenic neoantigens in AML patients (15). Like other malignancies, AML appears to create an immunosuppressive microenvironment through the expression of inhibitory ligands (32), depletion of amino acids (33, 34), and other mechanisms (35, 36) that could preclude or inhibit T cell responses to AML neoantigens. Thus, we favor a transgenic TCR T cell approach over vaccination or ex vivo expansion, because adoptive transfer of autologous T cells genetically modified to express the CBFB-MYH11/B*40:01–specific TCR would circumvent numerical and/or functional defects in the T cell repertoire of patients with CBFB-MYH11+ AML.
In addition to overcoming deficiencies in naturally occurring antileukemia T cell responses, TCR transfer offers several other potential therapeutic benefits. TCR transfer enables patients to receive a product with a defined specificity and potency. After identifying a suitable target, like the CBFB-MYH11/B*40:01 neoantigen, a next key step in developing transgenic TCR T cell immunotherapy is identifying a suitable high-affinity antigen-specific TCR. Identifying the appropriate TCR for clinical translation may require screening numerous T cell clones specific for the target antigen (44, 45), since TCRs specific for the same antigen can have highly variable affinity, resulting in heterogeneous cytotoxic efficacy of the corresponding T cell clones. As expected, the CBFB-MYH11/B*40:01–specific T cell clones we identified varied in their functional avidity, with one T cell clone, D2.C24, demonstrating exceptional cytotoxic efficacy in all assays and its TCR showing strong therapeutic potential. Transgenic CD8+ T cells expressing D2.C24 would be expected to have similar cytotoxicity, greater proliferative capacity, and improved persistence as compared with the parental T cell clone. Before clinical translation, the transgenic TCR construct would be modified and optimized to further enhance the function of the product, for example, by including a CD8 costimulatory receptor to improve the function of CD4+ T cells engineered with a class I–restricted TCR and provide targeted help to epitope-specific CD8+ T cells (45, 66, 67). A safety switch could also be added to the transgene (45, 68) to allow for rapid removal of the transgenic T cells in the event of toxicity. To avoid immune escape by downregulated expression or loss of the restricting HLA allele, a multiplexed approach combining several transgenic T cell products with different specificities could be used to target CBFB-MYH11/B*40:01 together with other epitopes presented on diverse HLA molecules.
Targets for neoantigen-specific TCR immunotherapies exist on a spectrum, ranging from highly recurrent neoantigens shared by numerous patients to personal neoantigens unique to a single patient’s disease. The CBFB-MYH11/B*40:01 neoantigen falls in the middle of that spectrum and would apply to about 1% of patients with AML (~200 patients per year) in the United States, based on the overall prevalence of the CBFB-MYH11 fusion (~10%) and the restricting HLA-B*40:01 (~4%–30% depending on ethnic background; ref. 69). Applicability would be even broader in regions like Asia where HLA-B*40:01 is more prevalent. Development of transgenic TCR immunotherapy for less prevalent neoantigens is becoming increasingly feasible (70, 71), partly because of the growing number of new nonviral technologies for rapid therapeutic gene transfer, including nanoparticles (72), RNA electroporation (73), and gene editing (74–76). Illustrating this fact are 2 clinical trials of personalized neoantigen-specific TCR-engineered T cell immunotherapy for the treatment of solid tumors (ClinicalTrials.gov NCT03970382, NCT03412877) that are currently enrolling. We envision a semipersonalized TCR T cell immunotherapy approach to employ CBFB-MYH11/B*40:01 and other well-characterized neoantigens as targets, and their corresponding TCRs as immunotherapeutics. After thorough preclinical evaluation of each TCR for safety and efficacy, they could serve as tools within a “toolbox” from which one or more TCRs could be selected for individual patients based on the specific profile of their disease.
We conclusively demonstrate that the leukemia-initiating CBFB-MYH11 fusion itself gives rise to a neoantigen with potential as an immunotherapeutic target, provide proof of principle for AML fusion–directed T cell immunotherapy, and propose the development of a semipersonalized TCR T cell immunotherapy strategy targeting fusion neoantigens in AML, including the CBFB-MYH11/B*40:01 T cell epitope and other similar fusion neoantigens that will be characterized in ongoing studies. Our results emphasize that even in a very-low-mutation disease like CBF- and fusion-driven AML (1, 77), neoantigens are a viable therapeutic target so long as the disease harbors at least 1 immunogenic fusion or mutation. CBFB-MYH11 and similar fusions that are early and essential events in AML and other malignancies are optimal targets that should enable elimination of the earliest founding clone with minimal risk of escape due to loss of the target protein or of on-target, off-tumor toxicity.
Methods
Human samples.
Blood samples were obtained from healthy volunteer donors and patients with AML. PBMCs were isolated from whole blood by Ficoll-Hypaque (PerkinElmer) density gradient centrifugation or from nonmobilized apheresis product. PBMCs were cryopreserved in RPMI 1640 supplemented with 20% human serum and 10% DMSO in vapor-phase liquid nitrogen in aliquots until use.
HLA-binding prediction analysis and peptides.
Immune Epitope Database (IEDB) Artificial Neural Network (ANN) (78–81), IEDB Stabilized Matrix Method (SMM) (82, 83), and NetMHCpan 4.0 (82, 84, 85) were used to predict peptide binding to 20 prevalent HLA molecules (details in Supplemental Methods). Peptides were defined as candidate epitopes if they had predicted IC50 less than 500 nM for any HLA by ≥1 algorithm. Control and CBFB-MYH11 peptides were synthesized using standard Fmoc chemistry (GenScript), reconstituted to a stock concentration of 10 mg/mL in DMSO, and stored at –20°C in aliquots until use.
Immunogenicity screening and identification of CBFB-MYH11–specific CD8+ T cells.
CD8+ T cells from HLA-typed volunteer PBMCs were highly purified by immunomagnetic bead depletion of CD8– cells (CD8+ T cell isolation kit, Miltenyi Biotec). Autologous dendritic cells (DCs) generated from monocytes by a modified fast DC protocol as described (37) were used as antigen-presenting cells. For each immunogenicity screen, a minimum of 10 × 106 CD8+ T cells were plated at 3 × 104 to 6 × 104 T cells per well in 96-well plates, along with autologous mature DCs in a T cell/DC ratio of 30:1. A total of up to 7 plates were used in each experiment. Before coculture with CD8+ T cells, DCs were incubated for 2 hours at 37°C with CBFB-MYH11 and control (non–CBFB-MYH11) peptides based on donor HLA type. Each peptide was used at a final concentration of 1 μg/mL in culture medium for the incubation. After incubation with peptides, DCs were then irradiated and washed before coculturing with T cells. Cultures were supplemented with IL-12 (10 ng/mL) at initiation and IL-15 (10 ng/mL) at day 7. Split-well 51Cr-release cytotoxicity assays (CRAs) were performed on day 12 to 13, using autologous lymphoblastoid cell lines (LCLs) with or without peptide as target cells. A well was considered positive if it exhibited greater than 20% lysis of peptide-pulsed LCLs and lysis of peptide-pulsed LCLs was at least 2-fold higher than lysis of LCLs without peptide. Peptide-specific T cells were cloned by limiting dilution using OKT3, IL-2, and feeder cells, then screened by split-well CRA on day 11 to 13. A clone was considered positive if it exhibited greater than 20% lysis of peptide-pulsed LCLs and lysis of peptide-pulsed LCLs was at least 5-fold higher than lysis of LCLs without peptide. Positive clones were expanded using OKT3, IL-2, and feeder cells (86), and their specificity and functional avidity were evaluated by peptide/HLA (pHLA) tetramer staining and functional assays (CRA and flow cytometry–based cytotoxicity assays) as described in Supplemental Methods.
Patient-derived xenografts.
PBMCs from HLA-B*40:01+ patients with active CBFB-MYH11+ AML were engrafted in immunodeficient MISTRG mice following an established protocol (87–90). Briefly, newborn mice were sublethally irradiated (150 cGy). AML PBMCs (1 × 106) were injected intrahepatically after treatment with anti-CD3 OKT3 antibody to in vivo–deplete human T cells that would cause xenogeneic graft-versus-host disease (91). Engraftment was confirmed 12–20 weeks later by PB flow cytometry. Freshly expanded T cells (10 × 106) from either an epitope-specific or a control CD8+ T cell clone were injected i.v. on a randomized basis 1 week later, and mice were subsequently monitored weekly by PB flow cytometry. The planned endpoint for both experiments was 1 month after T cell injection; the second experiment was terminated prematurely because of COVID-19–related closure of laboratory research at our center. The control T cell clone was specific for the peptide IPRAHNRLV presented on HLA-B*07:02 and was isolated from PBMCs from an HLA-B*07:02+ healthy donor using a method of in vitro stimulation identical to that used for D2.C24, followed by limiting-dilution cloning and rapid expansion. Real-time quantitative PCR (qPCR) was performed on cDNA generated from total RNA extracted from select PB samples to examine expression of CBFB-MYH11 type A transcript (Ipsogen kit, QIAGEN) and murine Ptprc (CD45; kit from Integrated DNA Technologies). Additional details of real-time qPCR and relative quantitation are in Supplemental Methods.
Tetramer enrichment.
CBFB-MYH11–specific T cells were enriched from PBMCs from healthy donors using pHLA tetramers based on methods adapted from Moon and coworkers (92) and detailed in Supplemental Methods. Enriched T cells were then characterized by flow cytometry, sorted, and expanded using OKT3, IL-2, and feeder cells (86).
TCR sequencing, transfer into lentiviral vectors, and transduction of T cells.
CBFB-MYH11/B*40:01–specific TCRβ and TCRα chains were sequenced by next-generation sequencing (Adaptive Biotechnologies) and by rapid amplification of complementary DNA ends (RACE) PCR as detailed in Supplemental Methods. TCRs were constructed by pairing the sequences encoding the dominant TCRα and TCRβ chains in each CBFB-MYH11/B*40:01–specific T cell clone and including cysteine modifications and codon optimization as previously described (45). When 2 dominant TCRα chains were identified from a single clone, 2 TCRs were assembled (e.g., B1A1 and B1A2) and tested. TCR constructs were synthesized by GeneArt (Life Technologies) and cloned into the pRRLSIN.cPPT.MSCV.WPRE lentiviral vector (LV) that included the RQR8 selection marker (93) by restriction digestion and ligation.
CD8+ or CD4+ T cells immunomagnetically purified from normal donor PBMCs were activated with Dynabeads Human T-Activator CD3/CD28 (Thermo Fisher Scientific) in 50 IU/mL IL-2 for 24 hours, then transduced with LV supernatant (Supplemental Methods). Four days after transduction, T cells were stained with CBFB-MYH11/B*40:01 pHLA tetramer and anti-CD8 mAb. CBFB-MYH11/B*40:01 tetramer–positive CD8+ T cells were sorted to greater than 95% purity, expanded (86), then evaluated by flow cytometry and functional assays.
Study approval.
Blood samples from healthy volunteer donors and patients with AML were obtained after written informed consent in accordance with the Declaration of Helsinki to participate in research protocols approved by the Institutional Review Board of the Fred Hutchinson Cancer Research Center (FHCRC). Samples from AML patients were obtained through the FHCRC/University of Washington Hematopoietic Diseases Repository (protocol 1690). Animal experiments were approved by Institutional Animal Care and Use Committee of the FHCRC (protocol 50941).
Author contributions
MAB designed and performed experiments, analyzed data, and wrote the manuscript. M. Bleakley designed experiments and wrote the manuscript. KAF, KBW, MEC, and CC performed experiments, analyzed data, and contributed figures and/or text to the manuscript. TMC, RGD, M. Brault, JS, and TMO performed experiments. KG and EE collected essential patient samples. SM provided content expertise for CBF AML. AR designed and analyzed murine experiments and edited the manuscript.
Supplementary Material
Acknowledgments
We thank the patients and healthy donors who provided the samples used in these studies. We also thank Jenny Lill, Ana Dios-Esponera, Howell Moffett, Jim Boonyaratanakornkit, Blair DeBuysscher, Felecia Wagener, Joshua Veatch, Brenda Jesernig, Nicholas Culores, and Aili Carmichael for technical assistance. We thank Ted Gooley and Jenna Voutsinas for their assistance with statistical analyses. We thank Deborah Banker for editorial assistance. We acknowledge Yale University, the University of Zürich, and Regeneron Pharmaceuticals, where MISTRG mice were generated thanks to the financial support of the Bill and Melinda Gates Foundation. This work was supported by Hyundai Hope on Wheels (TE 6705) and by a Stand Up To Cancer Innovative Research Grant (SU2C-AACR-IRG 14-17). Stand Up To Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C. MAB received additional support from a National Cancer Institute Paul Calabresi Career Development Award for Clinical Oncology (5 K12 CA076930-18) and grants from the Rally Foundation for Childhood Cancer Research and Alex’s Lemonade Stand–Foundation for Childhood Cancer. MISTRG studies were additionally supported by funding from the Fred Hutch Immunotherapy Integrated Research Center. This research was also supported by Shared Resources (Comparative Medicine, Flow Cytometry, and Experimental Histopathology) of the Fred Hutch/University of Washington Cancer Consortium (P30 CA015704).
Version 1. 08/24/2020
Electronic publication
Version 2. 10/01/2020
Print issue publication
Funding Statement
Stand Up To Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C.
M.A.B. received additional support from a National Cancer Institute Paul Calabresi Career Development Award for Clinical Oncology.
Footnotes
MEC’s and RGD’s present address is: Kite Pharmaceuticals, Santa Monica, California, USA.
JS’s present address is: Children’s Hospital of Los Angeles, Los Angeles, California, USA.
Conflict of interest: M. Bleakley is a founder and Scientific Advisory Board member of HighPassBio and a Scientific Advisory Board member of Orca Bio, and has also received compensation from Miltenyi Biotec for presentations at conferences and corporate symposia. The Fred Hutchinson Cancer Research Center has filed a provisional patent application (62/616,261) and a PCT application (PCT/US2019/013323) naming M. Bleakley and MAB as inventors, covering applications of T cell immunotherapy for CBF acute myeloid leukemia. Both applications have lapsed, and as a result, the TCRs are no longer subject to patent rights. RGD and MEC are currently employed by Kite Pharmaceuticals
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(10):5127–5141.https://doi.org/10.1172/JCI137723.
Contributor Information
Melinda A. Biernacki, Email: mbiernac@fredhutch.org.
Kimberly A. Foster, Email: kafoster@fredhutch.org.
Kyle B. Woodward, Email: kbwoodwa@fredhutch.org.
Michael E. Coon, Email: michael.e.coon@gmail.com.
Carrie Cummings, Email: ccumming@fredhutch.org.
Tanya M. Cunningham, Email: tmcunnin@fredhutch.org.
Robson G. Dossa, Email: robsondossa@yahoo.com.br.
Michelle Brault, Email: mbrault@fredhutch.org.
Jamie Stokke, Email: jstokke@chla.usc.edu.
Tayla M. Olsen, Email: tolsen@fredhutch.onmicrosoft.com.
Kelda Gardner, Email: gardnerk@uw.edu.
Elihu Estey, Email: eestey@seattlecca.org.
Soheil Meshinchi, Email: smeshinc@fhcrc.org.
Anthony Rongvaux, Email: rongvaux@fredhutch.org.
Marie Bleakley, Email: mbleakle@fredhutch.org.
References
- 1.Bolouri H, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med. 2018;24(1):103–112. doi: 10.1038/nm.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer. 2002;2(7):502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- 3.Yarchoan M, Johnson BA, Lutz ER, Laheru DA, Jaffee EM. Targeting neoantigens to augment antitumour immunity. Nat Rev Cancer. 2017;17(4):209–222. doi: 10.1038/nrc.2016.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer. 2014;14(2):135–146. doi: 10.1038/nrc3670. [DOI] [PubMed] [Google Scholar]
- 5.Biernacki MA, Bleakley M. Neoantigens in hematologic malignancies. Front Immunol. 2020;11:121. doi: 10.3389/fimmu.2020.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tran E, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science. 2015;350(6266):1387–1390. doi: 10.1126/science.aad1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tran E, et al. T-cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med. 2016;375(23):2255–2262. doi: 10.1056/NEJMoa1609279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zacharakis N, et al. Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat Med. 2018;24(6):724–730. doi: 10.1038/s41591-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sahin U, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547(7662):222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 10.Ott PA, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547(7662):217–221. doi: 10.1038/nature22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keskin DB, et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature. 2019;565(7738):234–239. doi: 10.1038/s41586-018-0792-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajasagi M, et al. Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood. 2014;124(3):453–462. doi: 10.1182/blood-2014-04-567933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greiner J, et al. Immune responses against the mutated region of cytoplasmatic NPM1 might contribute to the favorable clinical outcome of AML patients with NPM1 mutations (NPM1mut) Blood. 2013;122(6):1087–1088. doi: 10.1182/blood-2013-04-496844. [DOI] [PubMed] [Google Scholar]
- 14.Narayan R, et al. Acute myeloid leukemia immunopeptidome reveals HLA presentation of mutated nucleophosmin. PLoS One. 2019;14(7):e0219547. doi: 10.1371/journal.pone.0219547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Lee DI, et al. Mutated nucleophosmin 1 as immunotherapy target in acute myeloid leukemia. J Clin Invest. 2019;129(2):774–785. doi: 10.1172/JCI97482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu P, et al. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261(5124):1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 17.Grimwade D, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116(3):354–365. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 18.Castilla LH, et al. The fusion gene Cbfb-MYH11 blocks myeloid differentiation and predisposes mice to acute myelomonocytic leukaemia. Nat Genet. 1999;23(2):144–146. doi: 10.1038/13776. [DOI] [PubMed] [Google Scholar]
- 19.Kuo YH, et al. Cbf beta-SMMHC induces distinct abnormal myeloid progenitors able to develop acute myeloid leukemia. Cancer Cell. 2006;9(1):57–68. doi: 10.1016/j.ccr.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 20.Zhao L, et al. KIT with D816 mutations cooperates with CBFB-MYH11 for leukemogenesis in mice. Blood. 2012;119(6):1511–1521. doi: 10.1182/blood-2011-02-338210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McHale CM, et al. Prenatal origin of childhood acute myeloid leukemias harboring chromosomal rearrangements t(15;17) and inv(16) Blood. 2003;101(11):4640–4641. doi: 10.1182/blood-2003-01-0313. [DOI] [PubMed] [Google Scholar]
- 22.Parkin B, et al. Clonal evolution and devolution after chemotherapy in adult acute myelogenous leukemia. Blood. 2013;121(2):369–377. doi: 10.1182/blood-2012-04-427039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sood R, et al. Somatic mutational landscape of AML with inv(16) or t(8;21) identifies patterns of clonal evolution in relapse leukemia. Leukemia. 2016;30(2):501–504. doi: 10.1038/leu.2015.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schnittger S, Bacher U, Haferlach C, Kern W, Haferlach T. Rare CBFB-MYH11 fusion transcripts in AML with inv(16)/t(16;16) are associated with therapy-related AML M4eo, atypical cytomorphology, atypical immunophenotype, atypical additional chromosomal rearrangements and low white blood cell count: a study on 162 patients. Leukemia. 2007;21(4):725–731. doi: 10.1038/sj.leu.2404531. [DOI] [PubMed] [Google Scholar]
- 25.Döhner H, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Appelbaum FR, et al. The clinical spectrum of adult acute myeloid leukaemia associated with core binding factor translocations. Br J Haematol. 2006;135(2):165–173. doi: 10.1111/j.1365-2141.2006.06276.x. [DOI] [PubMed] [Google Scholar]
- 27.Vasu S, et al. Ten-year outcome of patients with acute myeloid leukemia not treated with allogeneic transplantation in first complete remission. Blood Adv. 2018;2(13):1645–1650. doi: 10.1182/bloodadvances.2017015222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Itzykson R, et al. Clonal interference of signaling mutations worsens prognosis in core-binding factor acute myeloid leukemia. Blood. 2018;132(2):187–196. doi: 10.1182/blood-2018-03-837781. [DOI] [PubMed] [Google Scholar]
- 29.Yin JA, O’Brien MA, Hills RK, Daly SB, Wheatley K, Burnett AK. Minimal residual disease monitoring by quantitative RT-PCR in core binding factor AML allows risk stratification and predicts relapse: results of the United Kingdom MRC AML-15 trial. Blood. 2012;120(14):2826–2835. doi: 10.1182/blood-2012-06-435669. [DOI] [PubMed] [Google Scholar]
- 30.Gros A, et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med. 2016;22(4):433–438. doi: 10.1038/nm.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Veatch JR, et al. Tumor-infiltrating BRAFV600E-specific CD4+ T cells correlated with complete clinical response in melanoma. J Clin Invest. 2018;128(4):1563–1568. doi: 10.1172/JCI98689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang L, Gajewski TF, Kline J. PD-1/PD-L1 interactions inhibit antitumor immune responses in a murine acute myeloid leukemia model. Blood. 2009;114(8):1545–1552. doi: 10.1182/blood-2009-03-206672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mussai F, et al. Acute myeloid leukemia creates an arginase-dependent immunosuppressive microenvironment. Blood. 2013;122(5):749–758. doi: 10.1182/blood-2013-01-480129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukuno K, et al. Expression of indoleamine 2,3-dioxygenase in leukemic cells indicates an unfavorable prognosis in acute myeloid leukemia patients with intermediate-risk cytogenetics. Leuk Lymphoma. 2015;56(5):1398–1405. doi: 10.3109/10428194.2014.953150. [DOI] [PubMed] [Google Scholar]
- 35.Buggins AG, et al. Microenvironment produced by acute myeloid leukemia cells prevents T cell activation and proliferation by inhibition of NF-kappaB, c-Myc, and pRb pathways. J Immunol. 2001;167(10):6021–6030. doi: 10.4049/jimmunol.167.10.6021. [DOI] [PubMed] [Google Scholar]
- 36.van Galen P, et al. Single-cell RNA-Seq reveals AML hierarchies relevant to disease progression and immunity. Cell. 2019;176(6):1265–1281.e24. doi: 10.1016/j.cell.2019.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bleakley M, et al. Leukemia-associated minor histocompatibility antigen discovery using T-cell clones isolated by in vitro stimulation of naive CD8+ T cells. Blood. 2010;115(23):4923–4933. doi: 10.1182/blood-2009-12-260539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strønen E, et al. Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science. 2016;352(6291):1337–1341. doi: 10.1126/science.aaf2288. [DOI] [PubMed] [Google Scholar]
- 39.Ellegast JM, et al. inv(16) and NPM1mut AMLs engraft human cytokine knock-in mice. Blood. 2016;128(17):2130–2134. doi: 10.1182/blood-2015-12-689356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robbins PF, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19(6):747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yadav M, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515(7528):572–576. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 42.Mommaas B, et al. Identification of a novel HLA-B60-restricted T cell epitope of the minor histocompatibility antigen HA-1 locus. J Immunol. 2002;169(6):3131–3136. doi: 10.4049/jimmunol.169.6.3131. [DOI] [PubMed] [Google Scholar]
- 43.Jedema I, van der Werff NM, Barge RM, Willemze R, Falkenburg JH. New CFSE-based assay to determine susceptibility to lysis by cytotoxic T cells of leukemic precursor cells within a heterogeneous target cell population. Blood. 2004;103(7):2677–2682. doi: 10.1182/blood-2003-06-2070. [DOI] [PubMed] [Google Scholar]
- 44.Paulson KG, et al. Acquired cancer resistance to combination immunotherapy from transcriptional loss of class I HLA. Nat Commun. 2018;9(1):3868. doi: 10.1038/s41467-018-06300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dossa RG, et al. Development of T-cell immunotherapy for hematopoietic stem cell transplantation recipients at risk of leukemia relapse. Blood. 2018;131(1):108–120. doi: 10.1182/blood-2017-07-791608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doubrovina E, Carpenter T, Pankov D, Selvakumar A, Hasan A, O’Reilly RJ. Mapping of novel peptides of WT-1 and presenting HLA alleles that induce epitope-specific HLA-restricted T cells with cytotoxic activity against WT-1(+) leukemias. Blood. 2012;120(8):1633–1646. doi: 10.1182/blood-2011-11-394619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chapuis AG, et al. T cell receptor gene therapy targeting WT1 prevents acute myeloid leukemia relapse post-transplant. Nat Med. 2019;25(7):1064–1072. doi: 10.1038/s41591-019-0472-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Falk K, et al. Peptide motifs of HLA-B58, B60, B61, and B62 molecules. Immunogenetics. 1995;41(2-3):165–168. doi: 10.1007/BF00182333. [DOI] [PubMed] [Google Scholar]
- 49.Vogt MH, et al. UTY gene codes for an HLA-B60-restricted human male-specific minor histocompatibility antigen involved in stem cell graft rejection: characterization of the critical polymorphic amino acid residues for T-cell recognition. Blood. 2000;96(9):3126–3132. [PubMed] [Google Scholar]
- 50.Padovan E, Casorati G, Dellabona P, Meyer S, Brockhaus M, Lanzavecchia A. Expression of two T cell receptor alpha chains: dual receptor T cells. Science. 1993;262(5132):422–424. doi: 10.1126/science.8211163. [DOI] [PubMed] [Google Scholar]
- 51.Choi EM, et al. High avidity antigen-specific CTL identified by CD8-independent tetramer staining. J Immunol. 2003;171(10):5116–5123. doi: 10.4049/jimmunol.171.10.5116. [DOI] [PubMed] [Google Scholar]
- 52.Johnson LA, et al. Gene transfer of tumor-reactive TCR confers both high avidity and tumor reactivity to nonreactive peripheral blood mononuclear cells and tumor-infiltrating lymphocytes. J Immunol. 2006;177(9):6548–6559. doi: 10.4049/jimmunol.177.9.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bocchia M, et al. Specific binding of leukemia oncogene fusion protein peptides to HLA class I molecules. Blood. 1995;85(10):2680–2684. doi: 10.1182/blood.V85.10.2680.bloodjournal85102680. [DOI] [PubMed] [Google Scholar]
- 54.Yotnda P, et al. Cytotoxic T cell response against the chimeric p210 BCR-ABL protein in patients with chronic myelogenous leukemia. J Clin Invest. 1998;101(10):2290–2296. doi: 10.1172/JCI488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bornhäuser M, et al. Prophylactic transfer of BCR-ABL-, PR1-, and WT1-reactive donor T cells after T cell-depleted allogeneic hematopoietic cell transplantation in patients with chronic myeloid leukemia. Blood. 2011;117(26):7174–7184. doi: 10.1182/blood-2010-09-308569. [DOI] [PubMed] [Google Scholar]
- 56.Clark RE, et al. Direct evidence that leukemic cells present HLA-associated immunogenic peptides derived from the BCR-ABL b3a2 fusion protein. Blood. 2001;98(10):2887–2893. doi: 10.1182/blood.V98.10.2887. [DOI] [PubMed] [Google Scholar]
- 57.Yotnda P, et al. Cytotoxic T cell response against the chimeric ETV6-AML1 protein in childhood acute lymphoblastic leukemia. J Clin Invest. 1998;102(2):455–462. doi: 10.1172/JCI3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zamora AE, et al. Pediatric patients with acute lymphoblastic leukemia generate abundant and functional neoantigen-specific CD8+ T cell responses. Sci Transl Med. 2019;11(498):eaat8549. doi: 10.1126/scitranslmed.aat8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van den Broeke LT, Pendleton CD, Mackall C, Helman LJ, Berzofsky JA. Identification and epitope enhancement of a PAX-FKHR fusion protein breakpoint epitope in alveolar rhabdomyosarcoma cells created by a tumorigenic chromosomal translocation inducing CTL capable of lysing human tumors. Cancer Res. 2006;66(3):1818–1823. doi: 10.1158/0008-5472.CAN-05-2549. [DOI] [PubMed] [Google Scholar]
- 60.Worley BS, et al. Antigenicity of fusion proteins from sarcoma-associated chromosomal translocations. Cancer Res. 2001;61(18):6868–6875. [PubMed] [Google Scholar]
- 61.Yang W, et al. Immunogenic neoantigens derived from gene fusions stimulate T cell responses. Nat Med. 2019;25(5):767–775. doi: 10.1038/s41591-019-0434-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Popovic J, Li LP, Kloetzel PM, Leisegang M, Uckert W, Blankenstein T. The only proposed T-cell epitope derived from the TEL-AML1 translocation is not naturally processed. Blood. 2011;118(4):946–954. doi: 10.1182/blood-2010-12-325035. [DOI] [PubMed] [Google Scholar]
- 63.McGranahan N, Swanton C. Neoantigen quality, not quantity. Sci Transl Med. 2019;11(506):eaax7918. doi: 10.1126/scitranslmed.aax7918. [DOI] [PubMed] [Google Scholar]
- 64.Salter AI, Pont MJ, Riddell SR. Chimeric antigen receptor-modified T cells: CD19 and the road beyond. Blood. 2018;131(24):2621–2629. doi: 10.1182/blood-2018-01-785840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ebstein F, et al. Exposure to Melan-A/MART-126-35 tumor epitope specific CD8(+)T cells reveals immune escape by affecting the ubiquitin-proteasome system (UPS) Sci Rep. 2016;6:25208. doi: 10.1038/srep25208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hung K, Hayashi R, Lafond-Walker A, Lowenstein C, Pardoll D, Levitsky H. The central role of CD4(+) T cells in the antitumor immune response. J Exp Med. 1998;188(12):2357–2368. doi: 10.1084/jem.188.12.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sommermeyer D, et al. Chimeric antigen receptor-modified T cells derived from defined CD8+ and CD4+ subsets confer superior antitumor reactivity in vivo. Leukemia. 2016;30(2):492–500. doi: 10.1038/leu.2015.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Di Stasi A, et al. Inducible apoptosis as a safety switch for adoptive cell therapy. N Engl J Med. 2011;365(18):1673–1683. doi: 10.1056/NEJMoa1106152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Middleton D, Jones AR, McCabe A. The Allele Frequency Net Database. http://allelefrequencies.net Updated July 10, 2019. Accessed July 10, 2020.
- 70.Jacoby K, et al. Abstract 4783: Highly efficient, non-viral precision genome engineering for the generation of personalized neoepitope-specific adoptive T cell therapies. Cancer Res. 2019;79:4783 [Google Scholar]
- 71.Yamamoto TN, Kishton RJ, Restifo NP. Developing neoantigen-targeted T cell-based treatments for solid tumors. Nat Med. 2019;25(10):1488–1499. doi: 10.1038/s41591-019-0596-y. [DOI] [PubMed] [Google Scholar]
- 72.Smith TT, et al. In situ programming of leukaemia-specific T cells using synthetic DNA nanocarriers. Nat Nanotechnol. 2017;12(8):813–820. doi: 10.1038/nnano.2017.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Campillo-Davo D, et al. Efficient and non-genotoxic RNA-based engineering of human T cells using tumor-specific T cell receptors with minimal TCR mispairing. Front Immunol. 2018;9:2503. doi: 10.3389/fimmu.2018.02503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roth TL, et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature. 2018;559(7714):405–409. doi: 10.1038/s41586-018-0326-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bailey SR, Maus MV. Gene editing for immune cell therapies. Nat Biotechnol. 2019;37(12):1425–1434. doi: 10.1038/s41587-019-0137-8. [DOI] [PubMed] [Google Scholar]
- 76.Stadtmauer EA, et al. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020;367(6481):eaba7365. doi: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Faber ZJ, et al. The genomic landscape of core-binding factor acute myeloid leukemias. Nat Genet. 2016;48(12):1551–1556. doi: 10.1038/ng.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim Y, et al. Immune epitope database analysis resource. Nucleic Acids Res. 2012;40(Web Server issue):W525–W530. doi: 10.1093/nar/gks438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nielsen M, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12(5):1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, Nielsen M. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Res. 2008;36(Web Server issue):W509–W512. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics. 2016;32(4):511–517. doi: 10.1093/bioinformatics/btv639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jurtz V, Paul S, Andreatta M, Marcatili P, Peters B, Nielsen M. NetMHCpan-4.0: improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol. 2017;199(9):3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sidney J, et al. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res. 2008;4:2. doi: 10.1186/1745-7580-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hoof I, et al. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61(1):1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nielsen M, Andreatta M. NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med. 2016;8(1):33. doi: 10.1186/s13073-016-0288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Riddell SR, Greenberg PD. The use of anti-CD3 and anti-CD28 monoclonal antibodies to clone and expand human antigen-specific T cells. J Immunol Methods. 1990;128(2):189–201. doi: 10.1016/0022-1759(90)90210-M. [DOI] [PubMed] [Google Scholar]
- 87.Rongvaux A, et al. Development and function of human innate immune cells in a humanized mouse model. Nat Biotechnol. 2014;32(4):364–372. doi: 10.1038/nbt.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Traggiai E, et al. Development of a human adaptive immune system in cord blood cell-transplanted mice. Science. 2004;304(5667):104–107. doi: 10.1126/science.1093933. [DOI] [PubMed] [Google Scholar]
- 89.Saito Y, et al. Peripheral blood CD34+ cells efficiently engraft human cytokine knock-in mice. Blood. 2016;128(14):1829–1833. doi: 10.1182/blood-2015-10-676452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Song Y, et al. A highly efficient and faithful MDS patient-derived xenotransplantation model for pre-clinical studies. Nat Commun. 2019;10(1):366. doi: 10.1038/s41467-018-08166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wunderlich M, Brooks RA, Panchal R, Rhyasen GW, Danet-Desnoyers G, Mulloy JC. OKT3 prevents xenogeneic GVHD and allows reliable xenograft initiation from unfractionated human hematopoietic tissues. Blood. 2014;123(24):e134–e144. doi: 10.1182/blood-2014-02-556340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moon JJ, et al. Tracking epitope-specific T cells. Nat Protoc. 2009;4(4):565–581. doi: 10.1038/nprot.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Philip B, et al. A highly compact epitope-based marker/suicide gene for easier and safer T-cell therapy. Blood. 2014;124(8):1277–1287. doi: 10.1182/blood-2014-01-545020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.