Abstract
The congenital sideroblastic anemias (CSAs) can be caused by primary defects in mitochondrial iron-sulfur (Fe-S) cluster biogenesis. HSCB (heat shock cognate B), which encodes a mitochondrial cochaperone, also known as HSC20 (heat shock cognate protein 20), is the partner of mitochondrial heat shock protein A9 (HSPA9). Together with glutaredoxin 5 (GLRX5), HSCB and HSPA9 facilitate the transfer of nascent 2-iron, 2-sulfur clusters to recipient mitochondrial proteins. Mutations in both HSPA9 and GLRX5 have previously been associated with CSA. Therefore, we hypothesized that mutations in HSCB could also cause CSA. We screened patients with genetically undefined CSA and identified a frameshift mutation and a rare promoter variant in HSCB in a female patient with non-syndromic CSA. We found that HSCB expression was decreased in patient-derived fibroblasts and K562 erythroleukemia cells engineered to have the patient-specific promoter variant. Furthermore, gene knockdown and deletion experiments performed in K562 cells, zebrafish, and mice demonstrate that loss of HSCB results in impaired Fe-S cluster biogenesis, a defect in RBC hemoglobinization, and the development of siderocytes and more broadly perturbs hematopoiesis in vivo. These results further affirm the involvement of Fe-S cluster biogenesis in erythropoiesis and hematopoiesis and define HSCB as a CSA gene.
Keywords: Genetics, Hematology
Keywords: Bone marrow, Genetic diseases, Mitochondria
Introduction
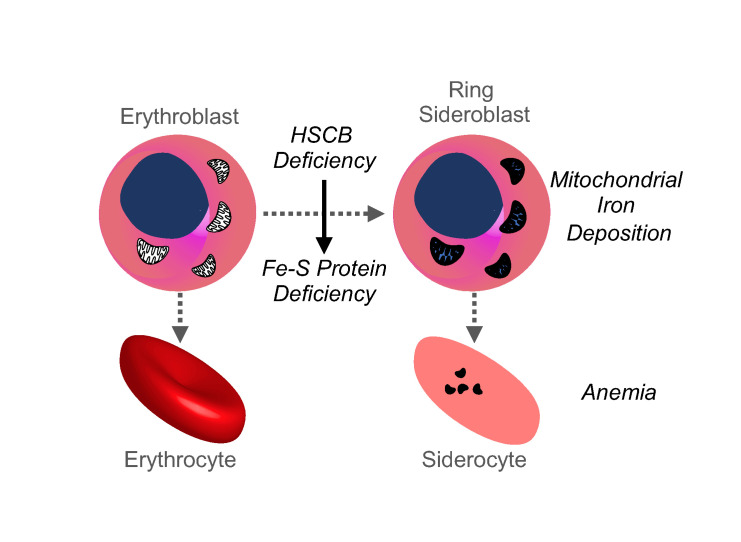
The congenital sideroblastic anemias (CSAs) are a group of uncommon inherited blood disorders characterized by abnormal iron accumulation in the mitochondria of erythroid cells. As the normal location of erythroid mitochondria is in the vicinity of the nucleus, the pathological iron deposits appear to ring the nucleus, resulting in the characteristic “ring sideroblast.” The mitochondrion is the nexus of iron metabolism in the erythroid progenitor, being the site of iron incorporation into protoporphyrin IX to form heme, and being essential for iron-sulfur (Fe-S) cluster biogenesis. Genes in multiple pathways contribute to the pathogenesis of CSA, including those involved in heme biosynthesis (ALAS2, SLC25A38), mitochondrial Fe-S cluster assembly and transport (ABCB7, GLRX5, HSPA9), mitochondrial protein synthesis (mtDNA deletions, TRNT1, LARS2, YARS2, PUS1), and mitochondrial energy metabolism (mtDNA deletions, MT-ATP6, SLC19A2, NDUFB11) (1). However, many cases of CSA remain genetically unexplained. The identification of the genetic defects responsible for CSA will assist in predicting clinical comorbidities and may guide the design of subtype-specific therapies, which are, unfortunately, limited at this time. More fundamentally, the mechanisms leading to the formation of the ring sideroblast and the physiopathology underlying these diseases are still not well understood.
Of the CSAs, those due to mitochondrial Fe-S cluster assembly defects are the rarest. Only 5 families have been reported with mutations in ATP binding cassette transporter subfamily B member 7 (ABCB7), 3 with glutaredoxin 5 (GLRX5) mutations, and 11 with heat shock protein family A member 9 (HSPA9; also known as mortalin) mutations (1). The mitochondrial Fe-S pathway can be subdivided into 3 steps: de novo assembly of the Fe-S cluster on a scaffold protein, release of the nascent cluster, and transfer and insertion of the cluster into target apoproteins (reviewed in refs. 2, 3). ABCB7 is a putative mitochondrial “Fe-S cluster exporter” involved in cytosolic Fe-S cluster protein maturation; HSPA9 and GLRX5 are involved in the scaffold release step. HSPA9 encodes a mitochondrial chaperone belonging to the HSP70 family of heat shock proteins. The essential cochaperone of HSPA9, heat shock cognate B (HSCB), also known as HSC20 (heat shock cognate protein 20), stimulates the ATPase activity of HSPA9 and facilitates the release of the newly formed 2-iron 2-sulfur [2Fe-2S] cluster from the scaffold protein, iron-sulfur cluster assembly enzyme (ISCU), and transfers it to GLRX5. GLRX5 is the immediate donor of [2Fe-2S] clusters to target apoproteins as well as to subsequent assembly factors that convert the [2Fe-2S] cluster to a [4Fe-4S] cluster. One of the mitochondrial recipient proteins is ferrochelatase (FECH), the last enzyme in heme biosynthesis, which is a dimer stabilized by 2 [2Fe-2S] clusters that are essential for its activity (4). Other targets include respiratory complexes I, II, and III, which incorporate structural and catalytic [2Fe-2S] and [4Fe-4S] clusters required for the electron transport chain. Disruption of mitochondrial Fe-S cluster assembly also impairs the maturation of cytosolic Fe-S proteins, including the bifunctional protein IRP1/ACO1 (iron regulatory protein 1/cytosolic aconitase). IRP1 and its functional ortholog, IRP2, control intracellular iron homeostasis by binding to mRNA stem-loops termed iron-responsive elements (IREs), located in either the 5′- or 3′-untranslated regions (UTRs) of genes relevant to iron metabolism, affecting mRNA translation or stability, respectively. In the setting of iron deficiency or cytosolic Fe-S deficiency, IRP1/ACO1 is an active IRP, favoring iron uptake and limiting its utilization. IRP2 is degraded in the presence of increased levels of cytosolic iron in an Fe-S cluster–dependent manner (5). Among those target mRNAs with 5′-IREs that are repressed by iron or Fe-S deficiency is the erythroid isoform of δ-aminolevulinate synthase (ALAS2), the first and rate-limiting enzyme of heme biosynthesis. Thus, Fe-S cluster biogenesis defects can influence multiple aspects of heme and mitochondrial respiratory protein function, all of which are implicated in CSA.
Given the requirement for HSCB in Fe-S cluster biogenesis, we hypothesized that HSCB mutations could cause CSA. Accordingly, we screened patients with genetically unexplained CSA, initially by targeted Sanger sequencing and subsequently by whole exome sequencing (WES), for rare HSCB variants. We identified one proband with biallelic variants: a frameshift variant, Thr87Asnfs*27, and an ETS transcription factor binding site variant in the promoter region. We established that the variants segregated with the disease in the family and that HSCB expression was reduced in the patient’s skin fibroblasts. We conducted experiments that validated the pathogenicity of the variants, including the effect of the promoter allele on HSCB expression. We also studied the impact of HSCB loss on erythropoiesis and hematopoiesis in cell culture and in vivo models, demonstrating its effect on Fe-S–dependent enzymes, oxidative phosphorylation, iron metabolism, and hemoglobinization in an erythroid context, including the evolution of siderocytes in a mouse model, affirming the causative nature of the mutations in this patient.
Results
Biallelic loss-of-function HSCB variants in a patient with CSA.
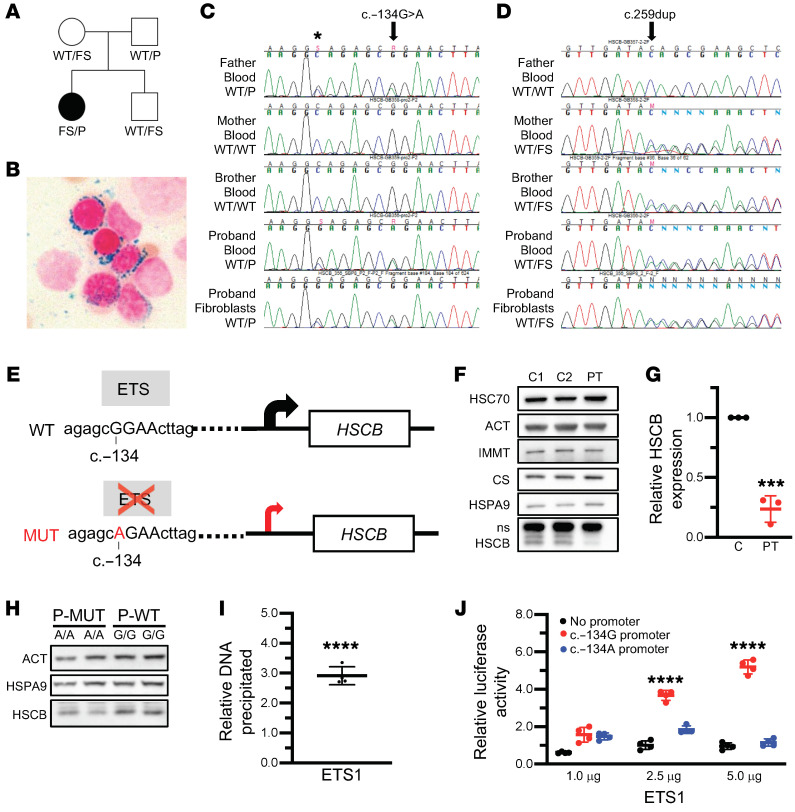
We evaluated 141 probands with CSA for HSCB mutations by Sanger sequencing and/or WES. Mutations in known CSA genes were subsequently identified in 71 of these patients by candidate gene sequencing or WES (data not shown). Three of the 141 patients carried rare (minor allele frequency < 0.005) variants in HSCB. In 2 of them, their CSA could be accounted for by mutations in other genes (ALAS2 and PUS1). One female patient (Figure 1A) carried a frameshift variant (NM_172002.4, c.259dup/p.Thr87Asnfs*27) not present in the Genome Aggregation Database (gnomAD; http://gnomad.broadinstitute.org) as well as a rare promoter variant (c.–134G>A/rs551244874; gnomAD minor allele frequency 9.69 × 10–5). The overall loss-of-function allele frequency for HSCB is 7.66 × 10–6 in gnomAD with no reported homozygotes for any loss-of-function allele, suggesting that null individuals are not viable. The HSCB variants segregate with the disease in the family and are present in the patient’s fibroblasts, confirming that neither variant is somatic (Figure 1, C and D). WES of the patient, her brother, and her parents revealed no mutations in other genes known to be associated with CSA.
Figure 1. HSCB expression is decreased by CSA patient-specific mutations.
(A) Pedigree of the family and HSCB variants: WT allele, c.–134A>C promoter variant (P), and c.259dup/p.Thr87Asnfs*27 frameshift allele (FS). (B) Perls Prussian blue iron staining of the proband’s bone marrow demonstrating characteristic ring sideroblasts. Original magnification, ×1000. (C and D) Sequencing traces of the P and FS variants, respectively. Asterisk indicates a common promoter polymorphism. (E) Representation of the HSCB promoter for the WT or P alleles. Predicted ETS binding site and the consequential reduction in transcription caused by the G/A substitution are indicated by the gray boxes and red arrow, respectively. (F) Representative Western blots of total protein extracts from skin fibroblasts from the proband (PT) and 2 unrelated female control fibroblast lines (C1 and C2). Actin (ACT) and heat shock cognate 70 (HSC70) were used as total protein controls, whereas IMMT (mitofilin) and citrate synthase (CS) controlled for mitochondrial content. (G) Quantification of PT and control (C) fibroblast HSCB expression. n = 3, unpaired t test, ***P < 0.001. (H) Western blot of CRISPR/Cas9–edited K562 clones homozygous for the WT (G/G) or the P allele (P-MUT A/A). (I) ChIP analysis of the HSCB promoter region in K562 cells using an ETS1-specific antibody demonstrating specific binding of ETS1 to the c.–134A region. Unpaired t test, ****P < 0.0001 vs. IgG alone. (J) Luciferase promoter activity in HeLa cells transfected with a promoterless luciferase expression construct or a similar plasmid containing the HSCB promoter with either the WT (c.–134G) or P (c.–134A) variants. Data are normalized to the activity of the promoterless construct cotransfected with 5 μg of the ETS1 plasmid. ANOVA, ****P < 0.0001 vs. no promoter.
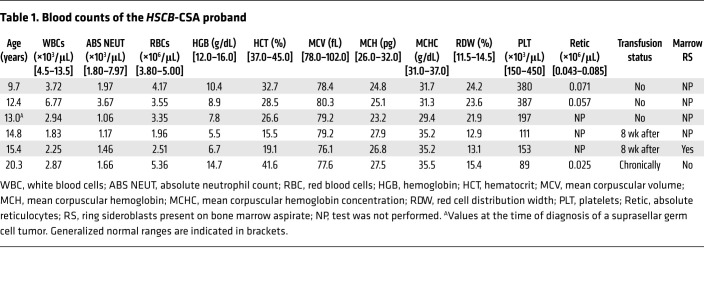
The proband was discovered to be anemic at the age of 10. At age 13, she was diagnosed with a suprasellar germ cell tumor that was treated with craniospinal radiation (Table 1). Because of persistent anemia and evolving pancytopenia, a bone marrow aspiration and biopsy were performed at age 15 years. Iron staining of the aspirate revealed ring sideroblasts (Figure 1B). She has received chronic transfusions since the age of 17. At age 20 years, at the time of this study, she had mild neutropenia and moderate thrombocytopenia. At the time of writing, at age 26 years, she was clinically stable. Given the history of radiation therapy, we compared the patient’s peripheral blood and skin fibroblast exome sequences and found no somatic variants indicative of a myelodysplastic syndrome, including those associated with acquired sideroblastic anemia (e.g., SF3B1 mutations; data not shown). Furthermore, X-inactivation studies performed on peripheral blood demonstrated that the patient’s hematopoiesis was not clonal (data not shown).
Table 1. Blood counts of the HSCB-CSA proband.
The mRNA transcribed from the HSCB coding sequence variant p.Thr87Asnfs*27 likely is degraded by nonsense-mediated decay and/or encodes a prematurely truncated, nonfunctional protein lacking the DNA-J domain, responsible for the interaction with HSPA9, and the C-terminal helix, critical for ISCU binding. We hypothesized that the promoter variant, which is located in a DNase-hypersensitive site in human CD34+ hematopoietic stem cells (Encode data accessed from http://genome.ucsc.edu/), adversely affects HSCB transcription. We used Genomatix MatInspector software to determine potential transcription factor binding sites in the region of the variant (Figure 1E). In silico, the c.–134G>A variant is predicted to disrupt the core sequence of a binding site for an E26 transformation–specific (ETS) DNA-binding factor. There are 28 human ETS transcription factors, each of which binds a conserved DNA motif (GGAA/T); surrounding sequences determine the binding specificity of different family members (6). To determine whether the patient’s variant alleles impaired HSCB expression, we measured HSCB protein levels by Western blot (Figure 1F). HSCB protein expression in the proband’s fibroblasts was about 75% less (24% ± 11% residual protein, t test, P < 0.001; Figure 1G) than in 2 unrelated skin fibroblast controls. Homozygous c.–134G>A mutant K562 human erythroleukemic cell lines expressed less HSCB protein than their control counterparts (~85%, n = 2 and n = 4 G/G c.–134A/A and G/G clones, respectively, t test, P = NS), suggesting the possibility of functional significance of this sequence variant (Figure 1H). We did not have access to CD34+ hematopoietic precursor cells from the proband, limiting our ability to evaluate the effect of the patient’s mutations on chromatin binding and/or gene expression and cellular phenotypes in primary erythroid cells. Consequently, to further test our hypothesis that the ETS binding site variant is responsible is the second, recessive allele, we performed chromatin immunoprecipitation of ETS1, one of the primary hematopoietically expressed ETS factors, in K562 cells and found that ETS1 does indeed occupy the HSCB promoter region (Figure 1I). We also examined the activity of the HSCB promoter c.–134G>A variant in a luciferase reporter assay in HeLa cells (Figure 1J). We found that expression of ETS1 produced a dose-dependent increase in luciferase activity in the WT c.–134G promoter construct that was attenuated in the c.–134A variant construct. Taken together, these results confirm that the HSCB sequence variants detected in the proband CSA patient collectively result in decreased HSCB protein expression.
Depletion of HSCB perturbs oxidative phosphorylation, iron metabolism, and hemoglobinization in K562 erythroleukemia cells.
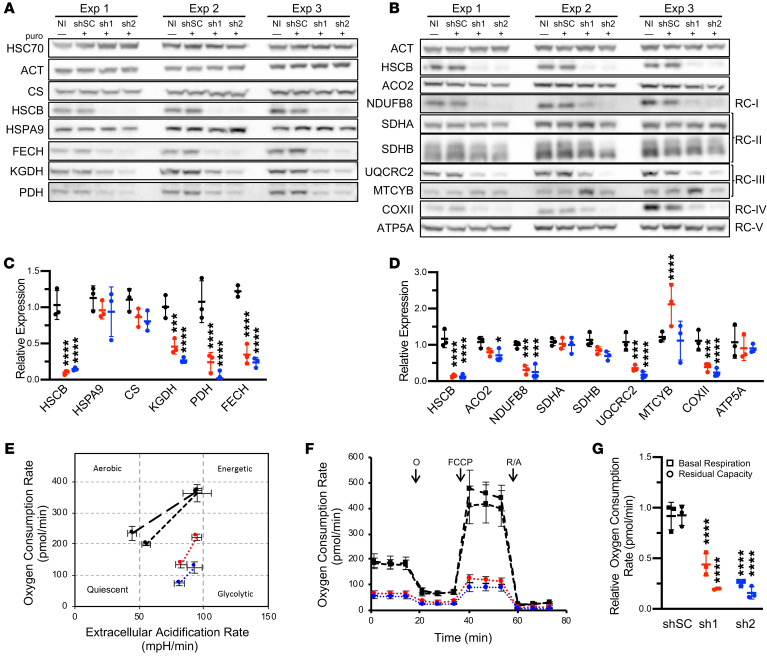
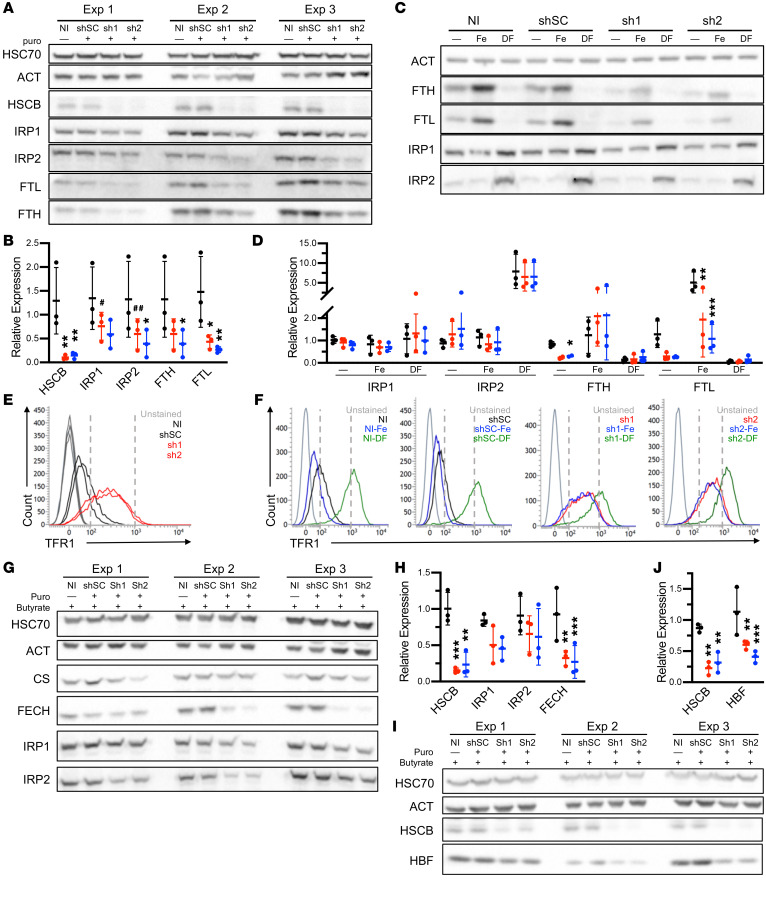
Previous characterization of mammalian HSCB has been performed in non-hematopoietic, non-erythroid cells (7–9), but the regulation of iron homeostasis, heme biosynthesis, and Fe-S cluster assembly is unique in erythroid cells. Moreover, germline defects in GLRX5 and HSPA9, 2 other proteins that mediate Fe-S cluster transfer, largely affect erythropoiesis (1). To determine the effects of HSCB in an erythroid system, we performed shRNA knockdown experiments in undifferentiated and differentiated K562 erythroleukemia cells. HSCB depletion to levels similar to those in the patient’s fibroblasts (Figure 2, A–D, and Supplemental Figure 1, A–C; supplemental material available online with this article; https://doi.org/10.1172/JCI135479DS1) did not affect HSPA9 expression, but did reduce expression of mitochondrial enzymes containing Fe-S clusters, such as FECH and mitochondrial aconitase (ACO2), as well as multiple respiratory complex (RC) proteins containing Fe-S cluster–dependent subunits. HSCB knockdown also impaired the lipoylation of the E2 subunits of the pyruvate and α-ketoglutarate dehydrogenase complexes, likely because lipoic acid biosynthesis requires an Fe-S enzyme (10). Commensurate with the reduction of tricarboxylic acid cycle enzymes and RC proteins, Seahorse extracellular flux analysis showed that HSCB-depleted K562 cells were less energetic, were more dependent on glycolysis, and had reduced oxidative phosphorylation capacity (Figure 2, E–G). These results are concordant with those previously observed in HeLa cells (8).
Figure 2. HSCB depletion decreases Fe-S protein expression and impairs respiration.
(A–D) Western blots of Fe-S cluster biogenesis, Fe-S, and lipoylated proteins (A) and respiratory proteins (B) in K562 erythroleukemia cells 10 days after control and HSCB-specific shRNA infection. Three independent experiments are shown. Western blots in A and B are quantified in C and D, respectively. Scrambled control (shSC) and HSCB‑specific shRNAs 1 (sh1) and 2 (sh2) are indicated by black, red, and blue, respectively. NI, noninfected. For all panels, mean expression ± SD is normalized to NI cells. ANOVA, *P < 0.05, ***P < 0.001, ****P < 0.0001 vs. shSC. (E–G) Seahorse Extracellular Flux (XF) Analyzer Cell Energy Phenotype analyses of shRNA-treated K562 cells. Data presented are representative of at least 3 independent experiments. NI and shSC-treated K562 cells are indicated by black points with long-dashed and short-dashed lines, respectively. sh1- and sh2-treated K562 cells are indicated by red and blue, respectively. (E) Cell Mito Stress Test energy maps of basal (left points) and stressed (right points) oxygen consumption versus extracellular acidification, which is a measure of glycolysis. (F) Seahorse extracellular flux analysis. Oligomycin (O), carbonyl cyanide p-trifluoromethoxy-phenylhydrazone (FCCP), and rotenone/antimycin A (R/A) allow assessment of basal ATP production, maximal respiration, and non-mitochondrial respiration, respectively. (G) Basal respiration and residual respiratory capacity. Activity normalized to NI cells. ANOVA, ****P < 0.001 vs. shSC. ACO2, aconitase 2; ATP5A, ATP synthase F1 subunit α; COXII, mitochondrially encoded cytochrome c oxidase II; CS, citrate synthase; FECH, ferrochelatase; HSPA9, heat shock protein family A (Hsp70) member 9; KGDH, α-ketoglutarate dehydrogenase; MTCYB, mitochondrially encoded cytochrome b; NDUFB8, NADH:ubiquinone oxidoreductase subunit B8; PDH, pyruvate dehydrogenase; RC, respiratory complex; SHDA and SDHB, succinate dehydrogenase A and B subunits; UQCRC2, ubiquinol–cytochrome c reductase core protein 2. E2 subunits of PDH and KGDH were detected with an anti–lipoic acid antibody.
We also investigated iron metabolism in HSCB-shRNA–treated cells. Interestingly, IRP protein expression was slightly decreased in erythroid HSCB-depleted cells (Figure 3A). This result was unexpected, as previous work had demonstrated increased IRP1 activity, but unaffected protein levels (9). Despite the reduced IRP1 and IRP2 protein levels, expression of IRP target proteins was consistent with increased IRP activity; both ferritin L and H chain expression was decreased, whereas cell surface TFR1 expression was increased (Figure 3, A, B, and E). We also determined that HSCB-depleted cells were able to respond to iron chelation and overload. We found that reduced expression of HSCB resulted in retained, but attenuated responses (Figures 3, C, D, and F).
Figure 3. HSCB deficiency affects iron metabolism and hemoglobinization in K562 cells.
(A) Western blots of 3 independent experiments of iron metabolism proteins in uninduced K562 cells infected with viruses encoding scrambled (shSC) and HSCB-specific shRNAs (sh1 and sh2) for 10 days. (B) Quantification of Western blots in A. shSC, sh1, and sh2 are indicated by black, red, and blue, respectively. (C and D) Western blots (C) and quantification (D) of iron metabolism proteins in uninduced K562 cells treated with shRNAs as in A and either iron (Fe) or the iron chelator desferrioxamine (DF), demonstrating reduced expression but retained response of iron-regulated proteins to iron concentrations. One representative blot of 3 independent experiments is shown. Two ferritin heavy chain (FTH) outliers were removed based on Grubbs’s test P < 0.05. (E and F) Transferrin receptor 1 (TFR1) cell surface expression by flow cytometry of cells in A and C, respectively. (G–J) K562 cells treated with shRNAs as in A and differentiated by exposure to sodium butyrate for 96 hours. Western blotting demonstrates alterations in iron metabolism proteins (G and H) and impaired hemoglobin F expression (I and J). Three independent experiments are shown. For all panels, mean expression ± SD is normalized to noninfected (NI) cells. ANOVA, *P < 0.05, **P < 0.01, ***P < 0.001, #P = 0.06, ##P = 0.07 vs. shSC.
To evaluate erythroid-specific phenotypes in the context of HSCB depletion, we repeated the analyses in K562 cells induced to differentiate by exposure to sodium butyrate for 96 hours. We found that depletion of HSCB impaired expression of FECH and fetal hemoglobin, the major hemoglobin expressed by K562 cells (Figure 3, G–J, and Supplemental Figure 1, D and E). These findings are consistent with reduced HSCB levels being responsible for the hypochromic microcytic anemia observed in the patient.
Knockdown of zebrafish hscb demonstrates an erythropoietic defect that is partially rescued by IRE-less alas2.
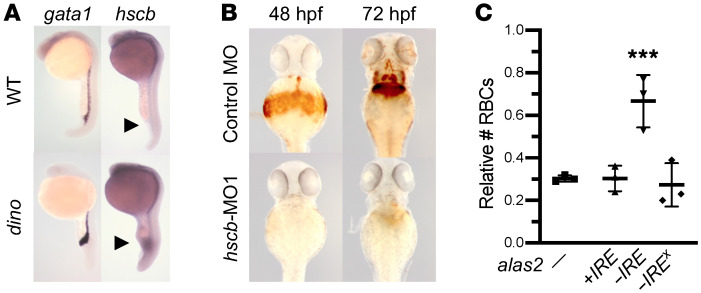
To further assess the role of HSCB in erythropoiesis, we examined hscb mRNA expression and the effect of hscb knockdown in zebrafish embryos. We found that hscb was expressed throughout the embryo and not disproportionately expressed in hematopoietic tissues (Figure 4A). However, morpholino knockdown of hscb resulted in an embryo with a grossly normal body plan, but a reduced number of erythrocytes and decreased hemoglobinization (Figure 4, B and C, and Supplemental Figure 2, A and B). Similar to HSCB-knockdown K562 cells, expression of the erythroid-specific form of the transferrin receptor was upregulated in hscb-morphant embryos, suggesting activation of IRPs (Supplemental Figure 2C). IRE-dependent suppression of ALAS2 translation likely contributes to the pathogenesis of CSAs related to Fe-S cluster deficiency (11). To determine whether this is also the case in HSCB deficiency, we coinjected hscb zebrafish embryo morphants with mRNAs encoding WT zebrafish alas2, WT alas2 with a mutated 5′-IRE, and alas2 with a premature stop codon also lacking the 5′-IRE. We found that only the IRE-less alas2 mRNA encoding the WT protein partially rescued the phenotype (Figure 4C), indicating that some of the HSCB deficiency erythroid phenotype can be attributed to an alas2–5′-IRE–dependent mechanism.
Figure 4. HSCB depletion in zebrafish embryos impairs hemoglobinization.
(A) Whole-mount in situ hybridization detects ubiquitous expression of hscb in zebrafish embryos. hscb is weakly expressed in blood islands (arrowheads) at 24 hours postfertilization (hpf). The signal in the ventral tissues is enhanced in the embryos from the dino mutant, which has a ventralized phenotype (29). gata1 is an erythroid transcription factor used to mark the blood islands. (B) hscb morpholino (MO1) induces anemia in zebrafish embryos. Reduced o-dianisidine (heme) staining is observed at both 48 hpf and 72 hpf in the morphant embryos. (C) IRE-independent expression of alas2 rescues the erythroid phenotype in hscb morphant embryos. Tg(globin-LCR:EGFP) transgenic zebrafish embryos were injected with a control morpholino, hscb-MO1 (–), hscb-MO1 + alas2 RNA with the 5′-IRE (+IRE), hscb-MO1 + alas2 RNA without 5′-IRE (–IRE), and hscb-MO1 + alas2 RNA without 5′-IRE but with a premature stop codon (–IREX). The effect of the hscb-MO1 alone and in combination with alas2 RNAs on the numbers of RBCs per embryo was quantified by flow cytometry. All data are normalized to embryos injected with the control morpholino alone. Data are expressed as mean ± SD of 3 independent experiments. ANOVA, ***P < 0.001 vs. hscb-MO1 only.
Hscb is an essential gene in mice, and its loss in hematopoietic tissues results in siderocytic anemia and loss of hematopoietic progenitors.
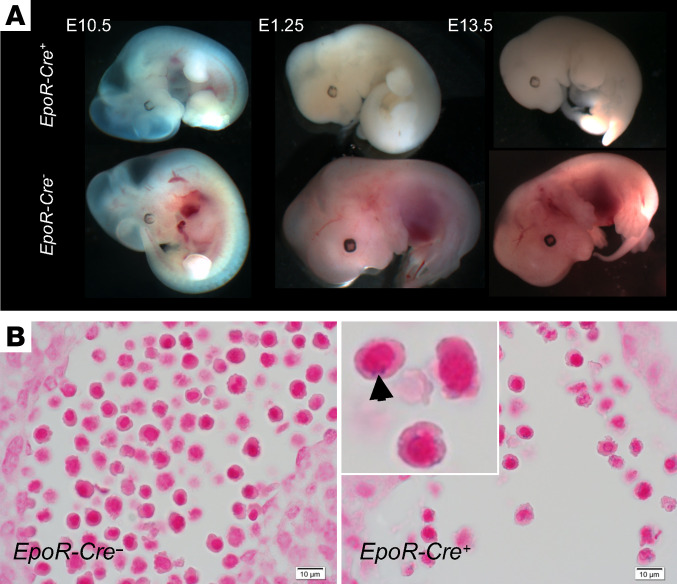
To examine the effect of HSCB deficiency in an intact mammal, we created null and conditionally targeted mouse Hscb alleles. Homozygous Hscb-null (Hscb–/–) embryos were not observed as early as embryonic day 7.5 (E7.5), indicating that germline Hscb deficiency is lethal in early embryogenesis. To determine the effect of Hscb loss on erythropoiesis, we bred the conditionally targeted allele (Hscbfl) with a transgenic line expressing Cre recombinase under the control of the erythropoietin receptor (Eportm1(EGFP/cre)UK, Epor-Cre), which is restricted to erythroblasts beginning at E10.5 (12). Hscbfl/– Epor-Cre+ animals uniformly died of anemia before E14.5 (Figure 5A). A subset of circulating nucleated erythrocytes containing coarse iron granules, typical of those seen in sideroblastic anemia, were observed only in Hscbfl/– Epor-Cre+ embryos at E11.5 (Figure 5B).
Figure 5. Prenatal lethality in mice lacking erythroid Hscb.
(A) Erythroid-specific deletion of Hscb with Epor-Cre results in death due to anemia in embryos before E14.5. Before E11.5, when the Epor-Cre becomes active, mutant embryos are somewhat hemoglobinized, whereas later embryos show extreme pallor. (B) Iron-stained histologic sections of E11.5 Hscbfl/– Epor-Cre+ animals demonstrate nucleated primitive RBCs containing coarse iron-positive granules, consistent with ring sideroblasts. Original magnification, ×400; inset, original magnification, ×2000.
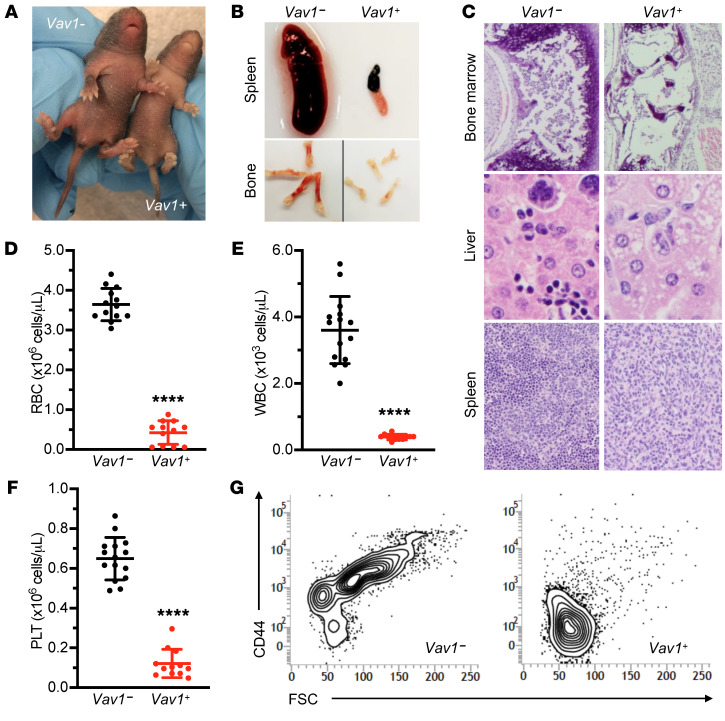
By contrast, Hscbfl/– animals carrying a Vav1-Cre transgene that leads to pan-hematopoietic deletion yielded grossly normal live-born mice in the expected Mendelian ratio. However, soon after birth, these animals began to exhibit growth delay and became pale (Figure 6A); most died of bacterial pneumonia between postnatal day 4 (P4) and P7 (Supplemental Figure 3). Hscbfl/– Vav1-Cre+ P7 pups had atrophic spleens, pale coloration of bones (Figure 6B), and extremely limited hematopoiesis in the bone marrow, liver, and spleen (Figure 6C and Supplemental Figure 3, C and D). Hscbfl/– Vav1-Cre+ were profoundly pancytopenic, with decreases in the RBC, WBC, and platelet counts (Figure 6, D–F). The hemoglobin and hematocrit were markedly decreased, whereas the mean corpuscular volume (MCV) was normal (data not shown). Terminal and stress erythropoiesis were assessed in marrow and spleen by flow cytometry (13). Ter119/CD44 staining showed a near-complete absence of early- and late-stage erythroblasts in the marrow (Figure 6G) and spleen (data not shown) of knockout animals, demonstrating that Hscb is required for murine normal and stress erythropoiesis.
Figure 6. Postnatal pancytopenia in mice with pan-hematopoietic loss of Hscb.
(A) Postnatal day 7 (P7) Hscbfl/– ± Vav1-Cre littermates. Control pup (left) and runted, pale Vav1-Cre+ (right). (B) Anemia and lack of erythropoiesis are grossly appreciated by pallor of the long bones in the Hscbfl/– Vav1-Cre+ mutants. The spleens are also profoundly atrophic, reflecting a loss of all hematopoietic elements, including lymphocytes. (C) H&E-stained tissue sections of P7 Hscbfl/– Vav1-Cre+ mutant bone marrow (original magnification, ×100), liver (original magnification, ×400), and spleen (original magnification, ×100) show an extreme loss of hematopoietic elements. (D–F) Profound pancytopenia in P7 Hscbfl/– Vav1-Cre+ animals. In the mutants, hemoglobin (D), white blood cell count (E), and platelet count (F). t test, ****P < 0.0001. (G) Representative flow cytometry scattergrams of bone marrow gated on Ter119+ cells show an absence of CD44+ cells with higher forward scatter (FSC) typical of maturing erythroblasts in the Hscbfl/– ± Vav1-Cre+ animals (n = 3 in each group).
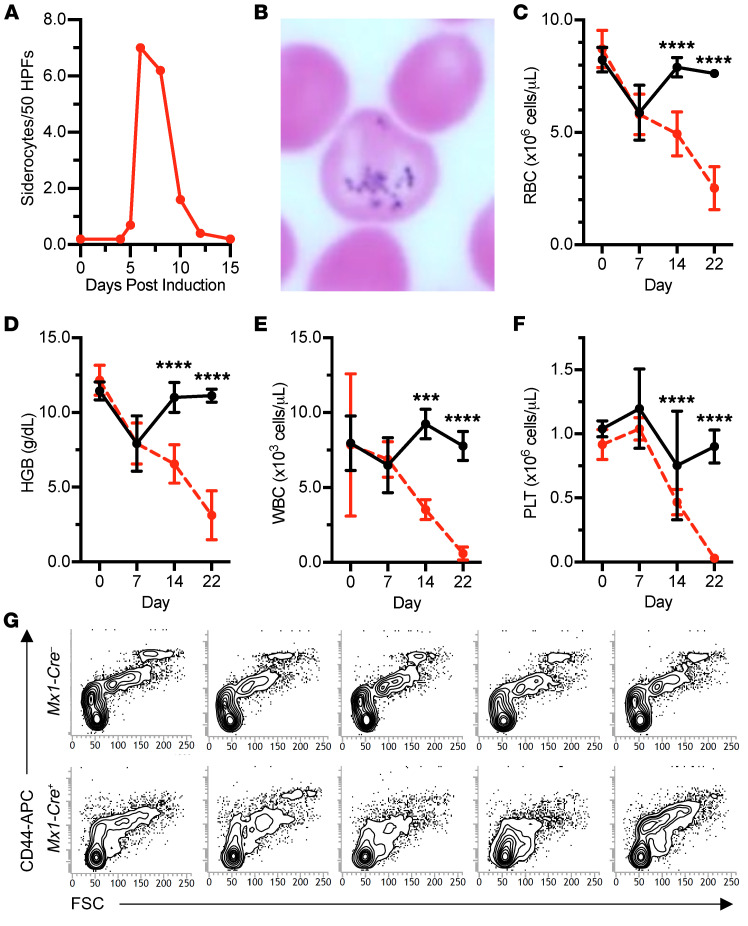
To further evaluate the role of Hscb in murine hematopoiesis, we transplanted E14.5 fetal liver hematopoietic stem cells from Hscbfl/– animals with or without an inducible Mx1-Cre transgene into lethally irradiated, congenic adult WT recipient mice. We allowed the chimeras to engraft for 8 weeks, after which Cre recombinase was induced by polyinosinic-polycytidylic acid [poly(I:C)] injections on alternate days for 5 days (3 doses), and quantitative and qualitative hematologic parameters were followed for 3 weeks (Supplemental Figure 4A). No quantitative differences were observed in the first week; however, beginning on day 5, and peaking on day 7, a transient population of siderocytes appeared in the peripheral blood of the recipients transplanted with Hscbfl/– Mx1-Cre+ hematopoietic stem cells (Figure 7, A and B). Thereafter, steep drops in the RBC count, hemoglobin, WBC count, and platelet count necessitated euthanasia of most mice by day 22 (Figure 7, C–F, and Supplemental Figure 4, B and C). Histologically, the bone marrow was nearly acellular, lacking any recognizable hematopoietic elements (data not shown). Flow cytometry analysis supported the absence of erythroid progenitors in the bone marrow (Figure 7G) and spleen (data not shown), confirming that steady-state and stress erythropoiesis are both affected by Hscb depletion.
Figure 7. Hematopoietic deletion of Hscb in the adult mouse results in a transient siderocytic anemia and bone marrow failure due to progenitor cell depletion.
(A–F) Time course of cytopenias following Mx1-Cre–induced Hscb deletion in hematopoietic stem cell transplant chimeras. Hscbfl/– ± Mx1-Cre fetal liver cells were transplanted into adult animals and allowed to engraft for 8 weeks. Thereafter, Hscb deletion was induced by injection with polyinosinic-polycytidylic acid [poly(I:C)]. (A and B) There was a transient population of siderocytes present only in the Mx1-Cre+ animals that peaked on day 7 after the initiation of induction. HPFs, high-power fields. Original magnification, ×1000. Beginning after day 7 of induction, Mx1-Cre+ animals experienced a precipitous drop in RBCs (C), hemoglobin (HGB) (D), WBCs (E), and platelets (PLT) (F). n = 5 in each group. Decreases in RBCs, HGB, and WBCs and an increase in PLT, observed immediately after induction (day 7), are likely related to poly(I:C)-induced cytokines. ANOVA, ***P < 0.001, ****P < 0.0001. (G) Flow cytometry scattergrams of bone marrow gated on Ter119+ cells show a decrease in CD44+ cells with higher forward scatter (FSC) typical of maturing erythroblasts in the MX1-Cre+ animals (n = 5 in each group).
Discussion
Here, in a very large cohort of patients with CSA, we identify an individual with 2 variants in the mitochondrial heat shock protein cochaperone HSCB. We demonstrate using patient-derived and induced-mutant cell lines that each variant decreases protein expression and that loss of HSCB reduces Fe-S cluster protein abundance, cellular respiration, and erythroid differentiation. Furthermore, studies in zebrafish hscb-morphant embryos and genetically engineered, Hscb-deficient mice indicate that HSCB is particularly important for erythropoiesis and can induce the accumulation of erythroid iron. In toto, this provides strong evidence that these mutations are causative of the sideroblastic phenotype in this patient. This result is congruent with the fact that mutations in other proteins, including HSPA9 — HSCB’s chaperone partner — as well as the mitochondrial glutaredoxin GLRX5 and ABCB7, involved in the transfer of nascent Fe-S clusters to recipient mitochondrial and cytosolic proteins, result in a CSA phenotype. As such, it further solidifies the understanding of this class of disorders.
Furthermore, we also noted that the patient developed moderate thrombocytopenia and leukopenia over time (Table 1). In this regard, we observed that complete Hscb loss leads to not only anemia but also bone marrow failure in mouse models. Flow cytometry experiments performed using bone marrow from both Vav1-Cre+ (Supplemental Figure 5) and Mx1-Cre+ (not shown) animals demonstrated that lineage-positive (Lin+) cells were dramatically decreased. Nonetheless, the Lin–Sca-1+Kit+ cell population persisted. It is unclear whether this result reflects the ability of these cells to persist in the absence of Hscb, or an effect related to the onset of Cre transgene expression.
The apparent rarity of the HSCB-related, and indeed the HSPA9- and GLRX5-related, anemias and their cell type selectivity may be a consequence of both the essential nature of HSCB in mammalian cells and a unique sensitivity or dependence of this pathway in erythroblasts. The early embryonic lethality of Hscb-null mouse embryos demonstrated here and the severe, respiratory-deficient phenotypes seen in yeast loss-of-function mutants (14) argue that a human null in any of these genes would be inviable. In the HSPA9 anemia, which is the most commonly described of Fe-S–associated CSAs, a variety of null alleles are present in trans of a functionally mild missense variant or a common variant (minor allele frequency ~30%–40%) at rs10117T, which is associated with reduced HSPA9 mRNA expression (15). Indeed, the relative frequency of this common variant may account for the more frequent occurrence of the HSPA9-associated disease; any null allele occurring in the population stands a good chance to be in trans of this low-expresser variant, resulting in sufficient loss of function to cause a mild sideroblastic phenotype. Similarly, we provide evidence here that reduced protein expression caused by a mutation in an ETS transcription factor binding site in the promoter of HSCB results in reduced protein expression in K562 cells. It is possible that a deficiency of ETS factor binding at this site contributes to the erythroid and hematopoietic restriction of the patient’s phenotype. Indeed, in contrast to fibroblasts from patients with GLRX5 deficiency due to missense mutations (16), we observed no abnormalities of Fe-S cluster protein expression or respiratory function in HSCB patient-derived fibroblasts (data not shown), suggesting that the c.–134A>G allele has little overall effect on HSCB expression in fibroblasts, or that fibroblasts are more tolerant than erythroid cells to Fe-S cluster loss.
Save for FECH mutations, which result in erythropoietic protoporphyria and an equivocal sideroblastic phenotype (17), it is unclear why mutations in specific Fe-S cluster recipient proteins or in proteins involved in earlier steps in Fe-S cluster biogenesis do not result in CSA. In particular, defects in the initial assembly of the Fe-S cluster tend to result in complex neurometabolic phenotypes. It may be that Fe-S clusters or their precursors are particularly toxic or accumulate in the iron- and heme-rich environment of the erythroblast mitochondrion. Alternatively, given the impact of Fe-S cluster deficiency on heme synthesis — mediated by both FECH deficiency and, through the hyperactivation of IRP1, IRE binding activity and concomitant ALAS2 downregulation — there may be insufficient heme at critical stages of erythroid maturation to sustain a heme-dependent network of differentiation and survival signals (18, 19). Heme deficiency is likely in part contributory, as this and other work has consistently shown that rendering ALAS2 expression IRE-independent has a salutary effect on Fe-S cluster–deficient erythropoiesis (11). Such a strategy could, in theory, be exploited therapeutically.
In conclusion, our work establishes HSCB as a mitochondrial Fe-S–associated CSA gene. Moreover, we confirm the aberrant activity of IRPs in this context and demonstrate that targeting the alas2-IRE, and releasing the repressive effect of IRPs on translation, can be sufficient to prevent the anemia. Finally, we showed that HSCB is not required only for erythropoiesis and hemoglobinization but also more generally for hematopoiesis.
Methods
Sequencing.
Except as noted, DNA was prepared from peripheral blood using the PureGene DNA isolation kit (Gentra). For Sanger sequencing, the HSCB promoter region (c.–250 to c.–1), exons, and intron-exon junctions were amplified and sequenced using the primers described in Supplemental Table 1 and analyzed with Sequencher 5.4.1 (Gene Codes Corp.). Paired-end whole exome sequencing (WES) libraries were prepared with the Agilent SureSelect V5 exon selection kit and sequenced to a median depth of ~100× with 100 bp paired-end reads on an Illumina NextSeq 2500 instrument. Data were processed on an internally validated pipeline and analyzed on the WuXi NextCODE genomics platform (www.wuxinextcode.com).
Cell culture.
Primary fibroblasts and HeLa cells were cultured in DMEM (11965-092, Gibco) and K562 cells in RPMI 1640 medium (11875-093, Gibco). All media were supplemented with 10% FBS (MilliporeSigma), 2 mM glutamine (Corning), and 100 IU/mL penicillin/100 μg/mL streptomycin (Corning) and described as “complete media.” Primary fibroblasts were cultured from a skin biopsy from the proband or discarded skin samples from unrelated healthy females. Fibroblasts were passaged, frozen, or harvested at ~80% confluence. For HSCB depletion experiments, K562 cells were infected with Mission Lentiviral Transduction particles encoding shRNA control (“shSC”; SHC002V) or shRNA directed against HSCB (SHCLNV, TRCN000013-4428, -4940, -8234 [“sh1”], -8487 [“sh2”], and -8909) (MilliporeSigma), according to the manufacturer’s instructions. Briefly, 1 × 105 cells were combined with 7.5 μL lentiviral particles. A noninfected well was used as a control. At 24 hours after infection, cells were diluted with complete media, and at 72 hours cells were washed 3 times in sterile 1× PBS and resuspended in complete media supplemented with puromycin (1 μg/mL; A11138-03, Gibco) for infected cells. Experiments were conducted between day 6 and day 21. Control and HSCB-depleted K562 cells were treated with 0.6 mM sodium butyrate (30342, MilliporeSigma) to induce hemoglobinization, 0.1 mM deferiprone (379409, MilliporeSigma) for iron chelation, and 200 μM iron–nitrilotriacetic acid (FeNTA; ferric iron, F7134, MilliporeSigma, and nitrilotriacetic acid, N9877, MilliporeSigma) for iron overload.
CRISPR/Cas9 gene editing.
K562 cells were edited by CRISPR/Cas9 technology according to established protocols (20). Briefly, an sgRNA adjacent to the promoter HSCB variant was selected using Deskgen software (https://www.deskgen.com/landing/) (g/AGAGCGGAACTTAGAACCC, where “g” is an additional nucleotide; activity score: 67; off-target score: 87). Annealed complementary sgRNAs were cloned in pSpCas9(BB)-2A-GFP (PX458, Addgene) using BbSI restriction enzyme and T4 ligase (New England Biolabs) and the clone sequence verified, propagated, and purified using PureLink HiPure Plasmid Filter Maxiprep Kit (Thermo Fisher Scientific), quantified (NanoDrop, Thermo Fisher Scientific), and resequenced. Two symmetric templates of reparation (ssODN) were designed and purchased (Integrated DNA Technologies): one to edit only the corresponding GGG protospacer adjacent motif (PAM) sequence (CTCAATGCCTCCTGGGAGTTGTAGTTTAGAAGGGAGAGCgGAACTTAGAACCCtttTTCCCCTCGGGTGATCCCGCCCCCTCGACTCCCCAGCCAATCAG, where “g” is the WT promoter variant and “ttt” the edited PAM; ssODN-PAM), and the other to edit the PAM site and the targeted locus (CTCAATGCCTCCTGGGAGTTGTAGTTTAGAAGGGAGAGCaGAACTTAGAACCCtttTTCCCCTCGGGTGATCCCGCCCCCTCGACTCCCCAGCCAATCAG, where “a” is the mutated promoter variant; ssODN-MUT). Genomatix software was used to analyze the impact of PAM site editing, and the TTT substitution was selected on the basis of the weakest impact predicted in silico. K562 cells (1 × 106) were nucleofected with 3 μg PX458-HSCB and 30 μM ssODNs (either the ssODN-PAM or ssODN-MUT) using an Amaxa Nucleofector 2b device (Lonza), the Cell Line Nucleofector Kit V, and program T-016 according to the manufacturer’s instructions. After nucleofection, the cells were resuspended in 0.5 mL of complete RPMI medium and transferred to a 6-well plate filled with 1.5 mL of RPMI medium supplemented with 1 μM SCR7, an inhibitor of DNA ligase IV and nonhomologous end joining (MilliporeSigma). After 24 hours, single GFP+ cells were sorted using a FACSAria III cell sorter (BD Biosciences) in 96-well U-well plates filled with 200 μL complete RPMI medium and expanded for 15 days. Wells containing single colonies were identified visually, and DNA was prepared with QuickExtract DNA Extraction Solution (Lucigen), amplified with target-specific primers, and sequenced and analyzed with Sequencher 5.4.1. Two independent clones homozygous for the PAM modification only and 2 independent clones homozygous for the PAM modification and the promoter variant were identified, expanded, and resequenced after 2 weeks in culture.
Western blotting.
Cell pellets were resuspended in Laemmli buffer 1× solution (62.5 mM Tris-HCl [pH 6.7], 10% glycerol, and 2% SDS final concentrations) and denatured for 5–10 minutes at 90°C. Protein content was quantified with the DC protein colorimetric assay (Bio-Rad) using serum albumin standards. For all K562 experiments, at least 3 independent infections were performed to obtain samples for immunoblotting. Fifteen to forty micrograms of total protein, supplemented with 40 mM dl-dithiothreitol (D9779, MilliporeSigma) and bromophenol blue, was loaded in TruPAGE 4%–20%, 4%–8%, or 12% precast acrylamide gels (MilliporeSigma) and run with TruPAGE SDS running buffer (PCG3001, MilliporeSigma). Transfers were performed on nitrocellulose membranes (1620112, Bio-Rad) using transfer buffer (100 mM glycine, 17 mM Tris base, 20% ethanol), and the efficiency of transfer was assessed by ponceau red staining. Membranes were cut according to the loaded ladder (Dual Color, Bio-Rad), and blocked at least 1 hour at room temperature with agitation in Tris-buffered saline buffer with 0.1% Tween 20 (TBST) supplemented with 5% nonfat dry milk. Membranes were incubated overnight with primary antibodies (Supplemental Table 2) at 4°C with agitation. Membranes were washed 30 minutes with TBST and incubated with secondary antibodies conjugated with horseradish peroxidase (Supplemental Table 3) for 1 hour, at room temperature with agitation. Membranes were washed again, and detection was performed on a ChemiDoc MP analyzer using ECL detection reagents of different sensitivities (Prime or Select, GE Healthcare) depending on the protein of interest. Membranes were stripped in TBST, reblotted with other primary antibodies, or stored at –20°C for subsequent reuse. ECL images were captured electronically with Image Lab 5.2.1 (Bio-Rad) and protein expression quantitated by densitometry using Image Studio Lite (LI-COR Biosciences).
HSCB promoter reporter assay.
The human minipromoter region of HSCB was amplified with 5′ primer 5′-AATTCTCGAGATGACTCACCGCGTGAGCCCACCTGGAGCC-3′ and 3′ primer 5′-AATTAAGCTTAGCAAAGAGAGCGTCTAACCAGACTAATGTTG-3′. The PCR product was digested with XhoI and HindIII and ligated into pGL3 Basic plasmid. The reporter plasmid with G>A SNP was generated by mutagenesis with primers 5′-TAGTTTAGAAGGGAGAGCAGAACTTAGAACCCGGGT-3′ and 5′-ACCCGGGTTCTAAGTTCtGCTCTCCCTTCTAAACTA-3′ using the QuikChange kit (Agilent). K562 cells were transfected with 9 μg of control plasmid (pGL3 Basic), WT reporter plasmid, and reporter plasmid with G>A SNP, together with 1 μg of Renilla luciferase plasmid (pRL-TK), with the Nucleofector kit (Lonza) according to the manufacturer’s protocol. The transfected cells were incubated for 48 hours, and the reporter activity was measured using the Dual Luciferase Assay System (Promega). The data were analyzed from 3 independent experiments.
Chromatin immunoprecipitation and quantitative PCR.
Chromatin immunoprecipitation (ChIP) was performed using the Pierce Agarose ChIP Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol with slight modifications. Briefly, 1 × 107 K562 cells were cross-linked with 1% formaldehyde for 10 minutes at room temperature. One hundred microliters of prepared nuclei were digested with 6 units of micrococcal nuclease for 15 minutes at 37°C. Immunoprecipitation was completed by incubation of the digested chromatin with 2 μg of control IgG or ETS1 antibody (Santa Cruz Biotechnology) overnight at 4°C. The IP products were eluted, and DNA was purified with a DNA clean-up column (QIAGEN) according to the manufacturer’s protocol. Precipitated DNA encoding the HSCB promoter region was quantified by quantitative PCR with iQ SYBR Green Supermix (Bio-Rad) using primers 5′-CCCTGCCGCCGGACCAATGAGGAGCAGC-3′ and 5′-GCAAAGAGAGCGTCTAACCAGACTAATGTTG-3′.
Functional metabolic profiling.
Briefly, experiments were conducted on shRNA-noninfected and shRNA-infected K562 cells on day 6 or 7 on a Seahorse XFe96 extracellular flux analyzer. Seahorse XF Cell Energy Phenotype and Mito Stress Test kits were used according to the supplier’s recommendation (Agilent). K562 cells were counted and loaded onto a Cell-Tak–pretreated (1206D69, Corning) 96-well cell culture microplate on the day of assay following the manufacturer’s instructions. An optimal cell density of 7.5 × 104 cells per well, FCCP concentration of 0.25 μM, and oligomycin concentration of 1.0 μM were determined empirically. At a minimum, triplicates were performed for each sample on each plate, and 3 replicate experiments were conducted on different days. Oxygen consumption rate and extracellular acidification rate were quantified with Seahorse Wave software.
Flow cytometry.
All washing steps as well as antibody incubations were performed in U-bottom plates at 4°C in a sterile PBS supplemented with 2% FBS. Unstained cells were used as negative controls, and single-color-stained cells were used to set compensation thresholds. A BD FACSverse analyzer was used to acquire and analyze the data. Each analysis was conducted at least 3 times on independent samples. Human CD71-FITC antibody (130-098-781, Miltenyi Biotec) was used to determine cell surface expression of transferrin receptor 1. Ter119-FITC/CD44-APC (BioLegend 116206/BD Biosciences 559250) staining was used to assess mouse erythroblast maturation using a strategy described by others (13). Lineage-APC (51-9003632, BD Biosciences), Ly-6A/E–PE-Cy7 (Sca-1; 255981-82, Invitrogen), and CD117-PE (c-Kit; 12-1172-83, eBioscience) antibodies were used to identify hematopoietic stem cells (LSK: lineage negative, Sca-1 positive, c-Kit positive). For mouse experiments, bone marrow and spleen were harvested as described below and filtered with 70-μm cell strainers. Before LSK staining, RBCs were lysed using RBC lysis buffer (QIAGEN) for 5 minutes at room temperature.
Zebrafish.
Zebrafish were maintained and manipulated according to standard procedures (21, 22). AB, dino (dintm84) (23), and a Tg(globin-LCR:EGFP) transgenic line (24) were used in this study. The Tg(globin-LCR:EGFP) line expresses a GFP reporter under the control of the locus control region (LCR) enhancer of a zebrafish globin gene (24).
Zebrafish in situ hybridization.
Digoxigenin-labeled cRNA probes were synthesized using a DIG RNA labeling kit (Roche). Whole-mount in situ hybridization (WISH) was carried out as previously described (25). Heterozygous dino zebrafish were mated with each other to produce a pool of WT and homozygous mutant embryos for WISH analysis.
Morpholino injections.
Two splice-blocking antisense morpholinos for hscb were obtained from Gene Tools. The morpholino sequences were 5′-CACAATACAGTCCATTTACCTGTGA-3′ for MO1 and 5′-CTTCGCTGAGGAAAGACAAACACAT-3′ for MO2, complementary to the exon 2–intron 2 and intron 1–exon 2 boundaries of hscb pre-mRNA, respectively. The standard control MO from Gene Tools (5′-CCTCTTACCTCAGTTACAATTTATA-3′) was used as a negative control. The morpholinos were injected into 1-cell-stage zebrafish embryos at concentrations of 0.5 mM (MO1 and control MO) and 0.1 mM (MO2). The knockdown efficiency was confirmed by quantitative reverse transcription PCR using the following primers: hscb, 5′-CAGCACAGTTACAGTCAACA-3′ and 5′-GGAGAACTCTTTGCAGCTTCAC-3′; hprt, 5′-CAGTTAGCTTGTCAGAGTTGGACG-3′ and 5′-GTCCACCCATGCCTTTCATTG-3′; tfr1a, 5′-AATCGCATTATGAGGGTGGAA-3′ and 5′-GGGAGACACGTATGGAGAGAGC-3′.
o-Dianisidine staining and quantification of erythrocytes.
After MO injections, the morphant embryos were harvested at 48–72 hours postfertilization (hpf) for o-dianisidine staining, following the method described previously (26). The morphant embryos from the Tg(globin-LCR:EGFP) line were also harvested at 72 hpf for quantification of erythrocytes by FACS. The cells were disaggregated and sequentially filtered through 70-μm and 40-μm cell strainers. The percentage of GFP+ erythroid cells was analyzed by FACS using the FACSVantage SE machine (BD Biosciences).
Overexpression of alas2 in zebrafish embryos.
The constructs of alas2 with IRE in the 5′-UTR, alas2 with 5′-IRE (CAGUGC) deleted (–IRE), and the IRE-deleted alas2 with an engineered premature stop codon (–IREX) at amino acid residue 26 (Tyr26X) were described previously (11, 27). RNAs correlating to these alas2 variants were synthesized using SP6 mMessage mMachine (Ambion). The RNA (150 ng/μL) was injected into 1-cell-stage zebrafish embryos together with hscb-MO1 (0.5 mM). At 72 hpf, the morphant embryos were harvested for quantification of erythrocytes by FACS analysis.
Hscb-deficient mice.
Mice heterozygous for an Hscb conditional allele (Hscbfl/+) were derived from conditionally targeted mouse embryonic stem cells obtained from the Knockout Mouse Project (KOMP) Consortium. A constitutive null allele (Hscb+/–) was obtained by breeding of the conditional allele to the B6.Cg-Tg(CMV-Cre)1Cgn/J line. Both alleles were backcrossed to C57BL/6J for more than 10 generations each. Dissection of timed matings showed that Hscb–/– animals die before E7.5 (data not shown). Hscb+/– animals were bred individually to the hematopoietic-specific B6.Cg-Tg(Vav1-Cre)1Graf (Vav1-Cre), the erythroid-specific Eportm1(EGFP/cre)UK (EpoR-Cre), and the temporally inducible B6.Cg-Tg(Mx1-Cre)1Cgn/J (Mx1-Cre) Cre recombinase lines, which were subsequently bred to animals homozygous for the conditional allele (Hscbfl/fl). All mouse genotypes were determined using the Transnetyx automated genotyping platform.
Fetal liver stem cell transplants using Hscbfl/– ± Mx1-Cre donors were performed as previously described (28). After 8 weeks, the animals underwent induction of Mx1-Cre with poly(I:C). Each animal received 300 μg of poly(I:C) on alternate days for 5 days, totaling 3 injections. Retro-orbital blood was collected for complete blood counts (CBCs) on days 0, 7, 14, and 22 and diluted 1:4 in PBS for CBCs. Tail nicks were performed to generate blood smears on days 4, 5, 6, 8, 9, 10, and 12 to monitor for the presence of siderocytes in circulation. CBCs were performed on 37.5 μL of blood obtained from a neck vein from 7-day-old Hscb Vav1-Cre pups and diluted 1:8 in PBS.
Histology.
P7 pups were fixed whole in 10% buffered formalin for 2 weeks and embedded in paraffin. Deparaffinized sections of tissue were stained with H&E, Perls Prussian blue iron stain, or CD71 antibody (Tfr1) in the Boston Children’s Hospital, Department of Pathology Histology Laboratory. Images were acquired using either a ×10/0.30 or a ×4/0.13 objective lens on a BX51 microscope equipped with a DP71 Digital Camera employing Olympus MicroSuite FIVE Imaging Software. May-Grünwald-Giemsa–stained (MilliporeSigma) peripheral blood smears were photographed using a ×100/1.3 objective lens.
Statistics.
All statistical analyses and graphical representations were performed using GraphPad Prism (version 7.03). One-way ANOVA was performed with multiple comparisons and the uncorrected Fisher’s least significant difference test unless otherwise specified in the figure legend. Data are presented as mean ± SD. A P value of less than 0.05 was considered statistically significant.
Study approval.
Written informed consent was obtained from all participants under Boston Children’s Hospital (BCH) human subjects research protocol 06-12-053. Animal experiments were approved by the Animal Care and Use Committee at BCH.
Author contributions
JB, SSB, MMH, DRC, and MDF ascertained and phenotyped patients, processed patient samples, and maintained the clinical research database. DRC and AKS performed and interpreted gene/exome sequencing experiments. AC and SD performed fibroblast experiments. CG performed HSCB promoter reporter and CHIP experiments in HeLa cells. SD performed K562 CRISPR/Cas9 HSCB promoter editing and analyses. SD designed and performed most of the shRNA K562 cell studies. AC performed Seahorse experiments. AC, DRC, PJS, DL, and C Cao derived or maintained the Hscb-null mice. AC, DRC, and SD phenotyped the mice. DRC and MDF performed bone marrow transplantations. SD directly supervised AC and DL. C Chen, GJHS, NCH, and BHP designed, performed, or oversaw zebrafish experiments. MDF and SD reviewed and interpreted all the data. AC, MDF, and SD wrote the manuscript. PJS edited the manuscript. All authors reviewed the final manuscript except BHP; BHP died on December 28, 2017, before the completion of the manuscript. The work was presented with his consent during the 59th American Society of Hematology Annual Meeting and Exposition (2017).
Supplementary Material
Acknowledgments
The authors acknowledge the participation of the index patient and her family as well as all of the congenital sideroblastic anemia patients participating in our research. Ronald Mathieu from the Boston Children’s Hospital Division of Hematology/Oncology Harvard Stem Cell Institute provided expert flow cytometry support. Mouse transgenic services were provided by the Boston Children’s Hospital Center for Developmental Hematopoiesis Mouse Embryonic Stem Cell and Gene Targeting Core, a National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Cooperative Center of Excellence in Hematology. Nancy Chamberlin is acknowledged for critical review of the manuscript. This work was supported by the NIDDK (R01 DK087992, R24 DK094746) and an American Society of Hematology Bridge Grant.
Version 1. 07/07/2020
In-Press Preview
Version 2. 08/31/2020
Electronic publication
Version 3. 10/01/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(10):5245–5256.https://doi.org/10.1172/JCI135479.
Contributor Information
Andrew Crispin, Email: crispin.andrew@gmail.com.
Chaoshe Guo, Email: chaoshe.guo@bbctg.com.cn.
Caiyong Chen, Email: chency@zju.edu.cn.
Dean R. Campagna, Email: dean.campagna@childrens.harvard.edu.
Paul J. Schmidt, Email: Paul.Schmidt@childrens.harvard.edu.
Chang Cao, Email: chang.cao@childrens.harvard.edu.
Anoop K. Sendamarai, Email: anoops@gmail.com.
Gordon J. Hildick-Smith, Email: ghildicksmith@gmail.com.
Nicholas C. Huston, Email: nicholas.huston@yale.edu.
Jeanne Boudreaux, Email: jeanne.boudreaux@choa.org.
Sylvia S. Bottomley, Email: sbottomley34@gmail.com.
Matthew M. Heeney, Email: Matthew.Heeney@childrens.harvard.edu.
Barry H. Paw, Email: bpaw@rics.bwh.harvard.edu.
Mark D. Fleming, Email: mark.fleming@childrens.harvard.edu.
Sarah Ducamp, Email: sarah.ducamp@childrens.harvard.edu.
References
- 1.Ducamp S, Fleming MD. The molecular genetics of sideroblastic anemia. Blood. 2019;133(1):59–69. doi: 10.1182/blood-2018-08-815951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rouault TA, Maio N. Biogenesis and functions of mammalian iron-sulfur proteins in the regulation of iron homeostasis and pivotal metabolic pathways. J Biol Chem. 2017;292(31):12744–12753. doi: 10.1074/jbc.R117.789537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braymer JJ, Lill R. Iron-sulfur cluster biogenesis and trafficking in mitochondria. J Biol Chem. 2017;292(31):12754–12763. doi: 10.1074/jbc.R117.787101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crooks DR, Ghosh MC, Haller RG, Tong WH, Rouault TA. Posttranslational stability of the heme biosynthetic enzyme ferrochelatase is dependent on iron availability and intact iron-sulfur cluster assembly machinery. Blood. 2010;115(4):860–869. doi: 10.1182/blood-2009-09-243105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H, et al. FBXL5 regulates IRP2 stability in iron homeostasis via an oxygen-responsive [2Fe2S] cluster. Mol Cell. 2020;78(1):31–41.e5. doi: 10.1016/j.molcel.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Findlay VJ, LaRue AC, Turner DP, Watson PM, Watson DK. Understanding the role of ETS-mediated gene regulation in complex biological processes. Adv Cancer Res. 2013;119:1–61. doi: 10.1016/B978-0-12-407190-2.00001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim KS, Maio N, Singh A, Rouault TA. Cytosolic HSC20 integrates de novo iron-sulfur cluster biogenesis with the CIAO1-mediated transfer to recipients. Hum Mol Genet. 2018;27(5):837–852. doi: 10.1093/hmg/ddy004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maio N, Kim KS, Singh A, Rouault TA. A single adaptable cochaperone-scaffold complex delivers nascent iron-sulfur clusters to mammalian respiratory chain complexes I-III. Cell Metab. 2017;25(4):945–953.e6. doi: 10.1016/j.cmet.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uhrigshardt H, Singh A, Kovtunovych G, Ghosh M, Rouault TA. Characterization of the human HSC20, an unusual DnaJ type III protein, involved in iron-sulfur cluster biogenesis. Hum Mol Genet. 2010;19(19):3816–3834. doi: 10.1093/hmg/ddq301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ollagnier-De Choudens S, et al. Iron-sulfur center of biotin synthase and lipoate synthase. Biochemistry. 2000;39(14):4165–4173. doi: 10.1021/bi992090u. [DOI] [PubMed] [Google Scholar]
- 11.Wingert RA, et al. Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis. Nature. 2005;436(7053):1035–1039. doi: 10.1038/nature03887. [DOI] [PubMed] [Google Scholar]
- 12.Heinrich AC, Pelanda R, Klingmüller U. A mouse model for visualization and conditional mutations in the erythroid lineage. Blood. 2004;104(3):659–666. doi: 10.1182/blood-2003-05-1442. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, et al. Quantitative analysis of murine terminal erythroid differentiation in vivo: novel method to study normal and disordered erythropoiesis. Blood. 2013;121(8):e43–e49. doi: 10.1182/blood-2012-09-456079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voisine C, et al. Jac1, a mitochondrial J-type chaperone, is involved in the biogenesis of Fe/S clusters in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2001;98(4):1483–1488. doi: 10.1073/pnas.98.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitz-Abe K, et al. Congenital sideroblastic anemia due to mutations in the mitochondrial HSP70 homologue HSPA9. Blood. 2015;126(25):2734–2738. doi: 10.1182/blood-2015-09-659854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye H, et al. Glutaredoxin 5 deficiency causes sideroblastic anemia by specifically impairing heme biosynthesis and depleting cytosolic iron in human erythroblasts. J Clin Invest. 2010;120(5):1749–1761. doi: 10.1172/JCI40372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rademakers LH, Koningsberger JC, Sorber CW, Baart de la Faille H, Van Hattum J, Marx JJ. Accumulation of iron in erythroblasts of patients with erythropoietic protoporphyria. Eur J Clin Invest. 1993;23(2):130–138. doi: 10.1111/j.1365-2362.1993.tb00752.x. [DOI] [PubMed] [Google Scholar]
- 18.Tanimura N, et al. Mechanism governing heme synthesis reveals a GATA factor/heme circuit that controls differentiation. EMBO Rep. 2016;17(2):249–265. doi: 10.15252/embr.201541465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doty RT, et al. Single-cell analyses demonstrate that a heme-GATA1 feedback loop regulates red cell differentiation. Blood. 2019;133(5):457–469. doi: 10.1182/blood-2018-05-850412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawrence C, Adatto I, Best J, James A, Maloney K. Generation time of zebrafish (Danio rerio) and medakas (Oryzias latipes) housed in the same aquaculture facility. Lab Anim (NY) 2012;41(6):158–165. doi: 10.1038/laban0612-158. [DOI] [PubMed] [Google Scholar]
- 22. Westerfield M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio). University of Oregon Press; 2000. [Google Scholar]
- 23.Haffter P, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- 24.Ganis JJ, et al. Zebrafish globin switching occurs in two developmental stages and is controlled by the LCR. Dev Biol. 2012;366(2):185–194. doi: 10.1016/j.ydbio.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen C, et al. Snx3 regulates recycling of the transferrin receptor and iron assimilation. Cell Metab. 2013;17(3):343–352. doi: 10.1016/j.cmet.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaw GC, et al. Mitoferrin is essential for erythroid iron assimilation. Nature. 2006;440(7080):96–100. doi: 10.1038/nature04512. [DOI] [PubMed] [Google Scholar]
- 27.Chung J, et al. Iron regulatory protein-1 protects against mitoferrin-1-deficient porphyria. J Biol Chem. 2014;289(11):7835–7843. doi: 10.1074/jbc.M114.547778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohgami RS, et al. nm1054: a spontaneous, recessive, hypochromic, microcytic anemia mutation in the mouse. Blood. 2005;106(10):3625–3631. doi: 10.1182/blood-2005-01-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammerschmidt M, et al. Mutations affecting morphogenesis during gastrulation and tail formation in the zebrafish, Danio rerio. Development. 1996;123:143–151. doi: 10.1242/dev.123.1.143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.