Abstract
Objectives
Investigation whether in depth characterization of virus variant patterns can be used for epidemiological analysis of the first severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection clusters in Hamburg, Germany.
Methods
Metagenomic RNA-sequencing and amplicon-sequencing and subsequent variant calling in 25 respiratory samples from SARS-CoV-2 infected patients involved in the earliest infection clusters in Hamburg.
Results
Amplikon sequencing and cluster analyses of these SARS-CoV-2 sequences allowed the identification of the first infection cluster and five non-related infection clusters occurring at the beginning of the viral entry of SARS-CoV-2 in the Hamburg metropolitan region. Viral genomics together with epidemiological analyses revealed that the index patient acquired the infection in northern Italy and transmitted it to two out of 134 contacts. Single nucleotide polymorphisms clearly distinguished the virus variants of the index and other clusters and allowed us to track in which sequences worldwide these mutations were first described. Minor variant analyses identified the transmission of intra-host variants in the index cluster and household clusters.
Conclusions
SARS-CoV-2 variant tracing allows the identification of infection clusters and the follow up of infection chains occurring in the population. Furthermore, the follow up of minor viral variants in infection clusters can provide further resolution on transmission events indistinguishable at a consensus sequence level.
Keywords: Severe acute respiratory syndrome coronavirus 2 infection cluster, Viral genomics, Viral variants
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) first emerged in late 2019 in Wuhan, China [1]. Sequences of SARS-CoV-2 were immediately publicly available and online tools, such as GISAID (Global initiative on sharing all influenza data), allowed phylogenetic network analyses [[2], [3], [4]]. Genomic epidemiology of SARS-CoV-2 in different countries allowed conclusions to be drawn on major viral lineages, transmission chains and routes of infection [[5], [6], [7]]. Genome surveillance has been highly useful to monitor distinct geographical lineages. However, no notable association of genome sequence variation with transmissibility and pathogenicity has been reported with the exception of one mutation in the spike protein, D614G, contributing to higher transmissibility [8] and becoming a major global variant.
We used viral genomics and variant calling to describe the initial entry of SARS-CoV-2 into the metropolitan region of Hamburg by proven and suspected travel returnees in February and March 2020. We were able to trace back the putative geographical infection sites and characterized viral transmission chains in the earliest documented infection clusters in Hamburg.
Methods
Patients and SARS-CoV-2 diagnostics
The index patient (P0), the first case of coronavirus disease 2019 (COVID-19) in northern Germany, tested SARS-CoV-2-positive on 27 February 2020 after returning from a vacation in northern Italy. His spouse (P02) and a co-worker (P01) tested positive on 4 March and 2 March, respectively (see Supplementary file, Fig. S1). Subsequent samples from individuals involved in five unrelated clusters were collected between 1 March and 12 March following the test criteria of the Robert Koch Institute at that time (individuals with respiratory symptoms returning from high-risk regions or individuals with contacts to positively tested persons). Limited information on sampling date, gender, age and travel history was retrieved using a questionnaire.
Contact tracing was performed by public health officials (see Supplementary Table S1). SARS-CoV-2 molecular diagnosis was performed as described previously [9,10]. The local ethics committee approved the study (PV7306).
High throughput sequencing and variant calling
Detailed information on sequencing, bioinformatics and variant calling are described in the Supplementary material (Tables S2–S4). All metagenomic and amplicon-based sequences are publicly accessible at ENA, PRJEB38546.
Results
Details on the timelines of the clinical and laboratory findings of patients 0 (index patient), 1 and 2, forming the first known SARS-CoV-2 infection cluster in the Hamburg metropolitan region, are shown in the Supplementary material (Fig. S1). Infectious virus was cultured from the initial oropharyngeal swabs of all three patients including patient 2 [11], who was asymptomatic based on the WHO list of symptoms valid in March. Virus culture together with subgenomic RNAs detected by high throughput sequencing from the initial sample of P02 (see Fig. 1A and Supplementary material, Table S3) suggests active viral replication while asymptomatic. Of note, patient 2 developed only very mild symptoms, a mild cough, (see Supplementary material, Fig. S1).
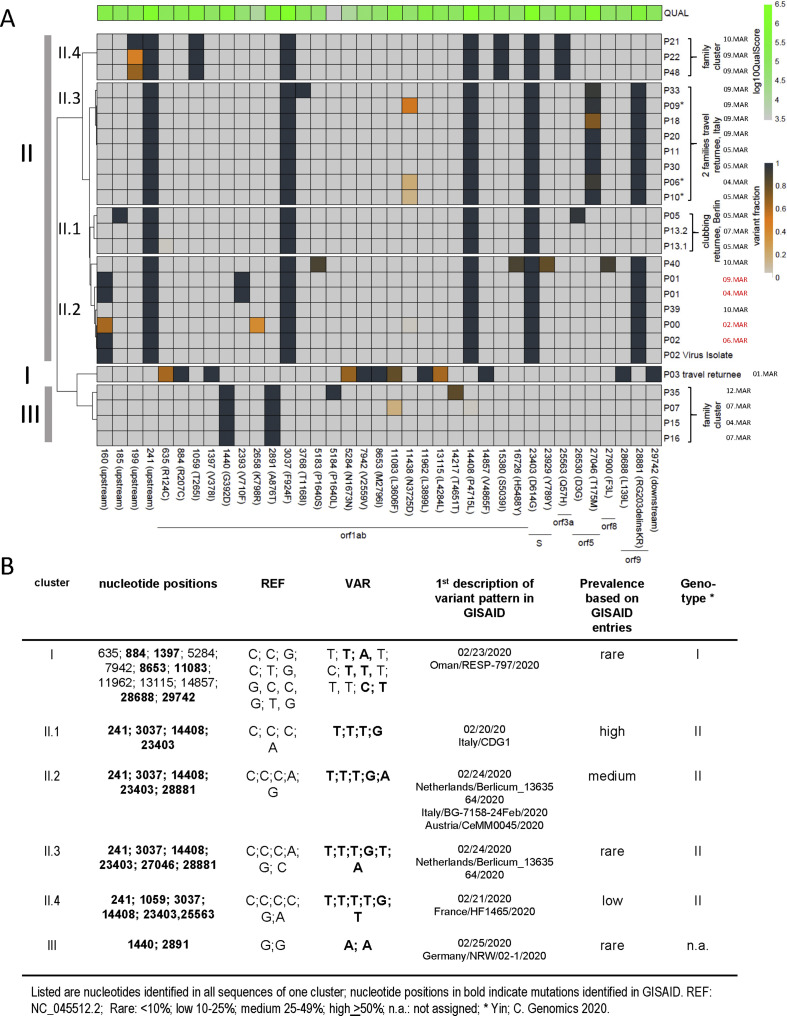
Within the 25 samples, we identified 37 single nucleotide polymorphisms (SNPs) (Fig. 1 a) with 2–12 fixed mutations per genome. In addition, 30% of the genomes showed one or two sub-consensus, minor variants. The sequences, based on SNPs, clustered in distinct variant patterns (I, II.1-II.4 and III). Clustering is mostly driven by SNPs located in the leader region (nt 241), nsp3 (nt 3037), RNApol (nt 14,408) and spike (nt 23,403; D614G). These co-occurring mutations have been recently described as one major variant occurring in Europe [8], referred to as genotype II [4]. Sub-patterns, II.2-II.4 are defined by additional SNPs, with different frequencies according to GISAID entries (Fig. 1b).
Fig. 1.
(a) Clustering of viral variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) sequences recovered from the index patient, patient 1, patient 2 and 19 SARS-CoV-2 sequences from respiratory swabs collected in the same time period in comparison to the reference sequence, NC_045,521. Nucleotide positions are indicated at the bottom. Only variants with sufficient coverage (>10) and single nucleotide polymorphism (SNP) present in more than 33% of all reads in at least one sample are included. I–III summarizes sequence patterns as defined by SNPs. The frequency of variants is indicated by the heat map ranging from grey (reference), yellow to dark blue (variant). The quality score per individual site is indicated at the top. ∗ indicates members within one family. Sampling dates are indicated on the right with the sampling date of the cases in the index cluster labelled in red. (b) Overview of individual SNPs defining pattern I, II (II.1-II.4) and III with sequence variation in comparison to the reference sequence.
We used the SNP information in the context of GISAID sequences to reproduce the geographic origins of the viral sequences (Fig. 1b). SNPs defining pattern II.1 (returnees from clubbing in Berlin) were first described in a sequence sampled in Italy. Within pattern II.2, the index cluster is defined by an additional SNP, nt 160, a variant not described at that time. This mutation in combination with details from questionnaires and time course of positivity strongly suggests that P01 and P02 acquired SARS-CoV-2 from P0, who was initially infected in northern Italy (see Supplementary material, Fig. S1). Sequences from P39 and P40, a couple returning from Italy, which cluster in II.2, do not show sequence variation at position 160.
Pattern II.3 and II.4, each separate from II.2 by one additional SNP. Pattern II.3 represents sequences from two families returning from a joint vacation in Italy. SNPs in II.3 were first described in sequences simultaneously sampled in the Netherlands, Italy and Austria. Sequences in pattern II.4 are derived from related travel returnees; characteristic SNPs in II.4 have been first reported in a sequence from France.
Pattern III, representing a family cluster, is defined by SNPs first reported in a sequence from North-Rhine Westphalia, Germany. Finally, pattern I, a sequence from a travel returnee from the Middle East shows 12 SNPs with six of them being previously reported in a sequence sampled in Oman.
Based on amplicon-sequencing, we defined minority variants, sub-consensus viral populations, at 14 positions with ten resulting in synonymous mutations.
Interestingly, in two family clusters, we identified minority variants allowing us to follow transmission events. In cluster II.4, sequences of P22 and P48 display multiple variants at nt 199 while P21 showed only one variant. The observed variant fractions are suggestive of transmission from P22 or P48 to P21 or alternatively infection from a common source and P21 has lost one variant. The transmission of intra-host variants with variant frequencies of the founding population in the recipient closely matching the donor suggests a high viral load exposure or a loose bottleneck in transmission events.
Similarly, in cluster II.3 with two families returning from a joint vacation, sequences from P06, P09 and P10 from the same household show intra-host variants at position 11,438 indicative of high infection dose exposure during viral transmission from a common source to all three patients or between these individuals. With sampling 4–5 days apart (Figs. 1a), P06 and P10 might have had the same infection source, and one of them transmitted the virus to P09.
Discussion
We here report the genomic analysis of the index cluster and five unrelated family/household clusters occurring at the beginning of the pandemic in the metropolitan region of Hamburg, Germany. Our analysis created important insights into transmission events and molecular epidemiology of SARS-CoV-2. Based on comparative SNP analysis from our study and additional SARS-CoV-2 sequences from GISAID, we identified different virus clusters and variant patterns, which allowed us to discern the first infection clusters in Hamburg and to propose intra-cluster transmission routes. Transmission of SARS-CoV-2 from the index patient occurred to only two out of 132 contacts with whom prolonged and unprotected interaction took place.
The overall mutation frequency was relatively low in our analysis when compared with a previous report [12]. These differences may be explained by our variant calling approach, employing >33% variant frequencies as cut-off compared with 5% [12].
By following the transmission of intra-host variants, we propose high-dose transmission of SARS-CoV-2 occurring in these clusters. In contrast to our study, no evidence of transmission of intra-host variants has been described in a previous study [12], whereas in another study identical minor variants were identified in household and workplace clusters [13].
Overall, our study describes the initial SARS-CoV-2 clusters occurring in Hamburg and emphasizes that virus variant tracing can provide further resolution for the identification of transmission events, for example in clusters that are indistinguishable at the consensus sequence level.
Author information
SP, RK, TG, AG, MA, JK, ML and NF designed the study. SP, TG, MC, DI, DN and NF performed the literature search. SP, RK, TG, RS, MA, JK, ML and NF wrote the manuscript. SP, RK, TG, MC, DN, JO, SK, LO, KP, DI, JK, ML and NF collected the data. TG, AG and JH performed the bioinformatics analysis. SP, TG, MC, DI, ML and NF generated the figures and tables.
Transparency declaration
We declare that there are no competing interests.
Acknowledgements
We thank Svenja Reucher and Kerstin Reumann for excellent technical support and Uwe Ganz for graphical illustration. We gratefully acknowledge the authors, originating and submitting laboratories of the sequences from GISAID's EpiFlu™ Database1 on which part of this research is based. This work is partly funded by support of the Federal Ministry of Health, Germany (HPI-COVID-19).
Editor: M Cevik
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2020.09.034.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang 7B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data—from vision to reality. Euro Surveill. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang X., Wu C., Li X., Song Y., Yao X., Wu X. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. 2020 doi: 10.1093/nsr/nwaa036. nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin C. Genotyping coronavirus SARS-CoV-2: methods and implications. Genomics. 2020;112:3588–3596. doi: 10.1016/j.ygeno.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Candido D.S., Claro I.M., de Jesus J.G., Souza W.M., Moreira F.R.R., Dellicour S. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science. 2020;369:1255–1260. doi: 10.1126/science.abd2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng X., Gu W., Federman S., du Plessis L., Pybus O.G., Faria N.R. Genomic surveillance reveals multiple introductions of SARS-CoV-2 into Northern California. Science. 2020;369:582–587. doi: 10.1126/science.abb9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gudbjartsson D.F., Helgason A., Jonsson H., Magnusson O.T., Melsted P., Norddahl G.L. Spread of SARS-CoV-2 in the Icelandic population. N Engl J Med. 2020;382:2302–2315. doi: 10.1056/NEJMoa2006100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfefferle S., Reucher S., Norz D., Lutgehetmann M. Evaluation of a quantitative RT-PCR assay for the detection of the emerging coronavirus SARS-CoV-2 using a high throughput system. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.9.2000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolfel R., Corman V., Guggemos W., Seilmaier M., Zange S., Mueller M. Virological assessment of hospitalized cases of coronavirus disease 2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 11.Pfefferle S., Huang J., Nörz D., Indenbirken D., Oestereich L., Guenther S. Complete genome sequence of a SARS-CoV-2 strain isolated in Northern Germany. Microbiol Resource Announcement. 2020;9 doi: 10.1128/MRA.00520-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen Z., Xiao Y., Kang L., Ma W., Shi L., Zhang L. Genomic diversity of SARS-CoV-2 in coronavirus disease 2019 patients. Clin Infect Dis. 2020;71:713–720. doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lythgoe K.A., Hall M., Ferretti L., de Cesare M., MacIntyre-Cockett G., Trebes A. Shared SARS-CoV-2 diversity suggests localised transmission of minority variants. bioRxiv. 2020 doi: 10.1101/2020.05.28.1189928. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.