Recently, DNAJC7 was found to be associated with amyotrophic lateral sclerosis (ALS) in a single large-scale exome sequencing study.1 Multiple protein-truncating variants were detected in individuals with ALS that were absent in control subjects.1 DNAJC7 encodes a member of the DnaJ heat-shock protein family (HspP40), which functions in protein homeostasis, including protein folding and degradation.2 To validate the pathogenic role of DNAJC7 in ALS and further understand the relevant clinical features, we screened a Taiwanese ALS cohort for DNAJC7 mutations.
Methods
A consecutive series of 325 unrelated individuals (191 men and 134 women) with ALS diagnosed by the revised EL Escorial criteria 2015 were enrolled into the study.3 The average at disease onset was 54.3 (range 19–89) years. Seventy patients (21.5%) were affected by a bulbar-onset ALS, 39 patients (12%) had an ALS family history, 64 individuals carried a mutation in other ALS disease genes, such as SOD1, C9ORF72, TARDBP, or FUS, and 254 patients had an apparently sporadic ALS without any known mutation for ALS. Genomic DNA was extracted from peripheral blood samples. Mutation analyses of the coding region of DNAJC7 were performed by PCR amplification and Sanger sequencing. The amplicon sequences were compared with the reference DNAJC7 coding sequence (NM_003315.4). All participants provided a written informed consent, and this study was approved by the Institutional Review Board of Taipei Veterans General Hospital.
Results
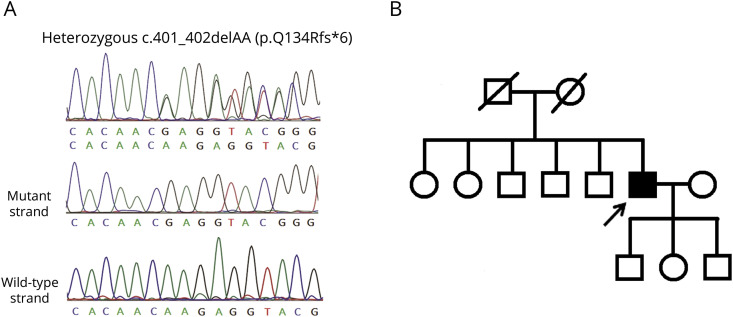
Mutational analysis of DNAJC7 in the 325 ALS patients revealed only one heterozygous truncating frameshift variant, c.401_402delAA (p.Q134Rfs*6) (figure, A), in one single individual with apparently sporadic ALS. Neither this variant nor other DNAJC7 loss-of-function (LOF) variant was found in the 1,517 ethnically matched control genomes in the Taiwan Biobank database (taiwanview.twbiobank.org.tw). The p.Q134Rfs*6 variant was also absent in the Genome Aggregation Database (gnomAD v2.1.1; gnomad.broadinstitute.org). The variant was predicted as a disease-causing mutation by 2 bioinformatics tools, MutationTaster (mutationtaster.org)4 and Combined Annotation Dependent Depletion (CADD v1.6; cadd.gs.washington.edu)5 with the CADD PHRED score 31.
Figure. The DNAJC7 mutation and pedigree of the patient with ALS.
(A) Sanger sequence traces of the DNAJC7 c.401_402delAA (p.Q134Rfs*6) mutation identified in the ALS patient in this study. The heterozygous frameshift mutations are clearly demonstrated by sequencing the TA-subcloned PCR fragments. (B) The pedigree structure of the ALS patient with the DNAJC7 mutation. Open symbol: unaffected; filled symbol: affected; symbol with diagonal line: deceased subjects; square: male; circle: female; arrow: the proband. ALS = amyotrophic lateral sclerosis.
The gentleman carrying the DNAJC7 mutation had an initial symptom of left hand weakness at age 61 years. Neurologic examinations at age 62, approximately 1 year after the disease onset, revealed tongue atrophy with fasciculation, weakness and atrophy with fasciculation in the bilateral upper extremities (muscle strength of 2–3 of 5 according to the Medical Research Council scale), a mild degree of weakness of the left hip flexor (4/5), diminished deep tendon reflexes, and normal cognitive function. The symptoms deteriorated rapidly in the following 6 months, and the patient developed bilateral lower limbs weakness and respiratory distress requiring noninvasive ventilation support. The Amyotrophic Lateral Sclerosis Functional Rating Scale-revised (ALSFRS-R) scores were 38 at age 62 and dropped to 13 six months later.6 The patient did not carry any other mutation related to ALS and denied having a family history of ALS (figure, B).
Discussion
We identified a DNAJC7 LOF mutation, p.Q134Rfs*6, in a patient with apparently sporadic ALS. Its pathogenicity was supported by the following findings. First, DNAJC7 p.Q134Rfs*6 is absent in the 1,517 ethnically matched Taiwanese control genomes and the gnomAD. Second, the mutation was predicted as pathogenic by Mutation Taster and CADD programs. Furthermore, DNAJC7 p.Q134Rfs*6 is a truncating frameshift mutation, which putatively results in a truncated, often unstable protein product that compromises DNAJC7 functions. Similar DNAJC7 LOF mutation, p.R156*, has been demonstrated with significantly reduced protein production in vitro.1
The DNAJC7 p.Q134Rfs*6 mutation was identified in one of the 254 unexplained sporadic ALS patients (0.4%). In another recent study, DNAJC7 protein-truncating variants were identified in 8 out of 5,095 (0.16%) ALS patients.1 These findings suggest that DNAJC7 mutations are not a common cause of ALS.
The clinical features of ALS associated with DNAJC7 LOF mutations remain elusive. Our patient had a typical spinal-onset ALS but a rapidly progressive disease course with 35 points decline of the ALSFRS-R scores during the 18 months after disease onset. According to the PRO-ACT database, the average rate of the ALSFRS-R decline of the ALS patients was 1.02 points per month.7 Lack of ALS family history of the patient suggests that DNAJC7 may be a risk gene or mendelian disease gene with reduced penetrance for ALS. Regrettably, we cannot approach the patient's siblings and parents, and this is a limitation of the present study. Further studies are warranted to elucidate the role and phenotypic features of DNAJC7 mutations in ALS.
In conclusion, we identified a patient carrying a DNAJC7 p.Q134Rfs*6 mutation and suffering from a rapidly progressive spinal-onset ALS. The present study underlines the pathogenic role of DNAJC7 LOF mutation in ALS.
Acknowledgment
The authors would like to thank the patients who participated in this study.
Appendix. Authors
Study funding
This study was supported by the grants from the Ministry of Science and Technology, Taiwan (107-2314-B-075-014-MY3), Taipei Veterans General Hospital (V109C-060), Taiwan Motor Neuron Disease Association, and Brain Research Center, National Yang-Ming University from the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan.
Disclosure
The authors report no disclosures relevant to the manuscript. Go to Neurology.org/NG for full disclosures.
References
- 1.Farhan SMK, Howrigan DP, Abbott LE, et al. Exome sequencing in amyotrophic lateral sclerosis implicates a novel gene, DNAJC7, encoding a heat-shock protein. Nat Neurosci 2019;22:1966–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci 2006;63:2560–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ludolph A, Drory V, Hardiman O, et al. A revision of the El Escorial criteria—2015. Amyotroph Lateral Scler Frontotemporal Degener 2015;16:291–292. [DOI] [PubMed] [Google Scholar]
- 4.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 2014;11:361–362. [DOI] [PubMed] [Google Scholar]
- 5.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014;46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cedarbaum JM, Stambler N, Malta E, et al. The ALSFRS-R: a revised ALS functional rating scale that incorporates assessments of respiratory function. BDNF ALS Study Group (Phase III). J Neurol Sci 1999;169:13–21. [DOI] [PubMed] [Google Scholar]
- 7.Atassi N, Berry J, Shui A, et al. The PRO-ACT database: design, initial analyses, and predictive features. Neurology 2014;83:1719–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]