Abstract
Influenza-specific antibody dependent cellular cytotoxicity (ADCC) antibodies have a broad cross reactivity and potential as an immune correlate for universal vaccines. Peptide-mapping for ADCC reactivity of H1-HA and H7-HA proteins from human serum samples identified high ADCC-inducing peptides in both the HA1 and HA2 regions. Vaccination of mice with single ADCC-peptides induced ADCC activity leading to partial protection from lethal influenza challenge, with increased survival, reduced viral loads, and reduced activation of NK cells in the lungs. Targeted vaccination strategies to elicit ADCC responses may provide an approach for universal vaccines.
Keywords: Influenza, ADCC, Antibodies, Peptide-mapping
1. Introduction
Current inactivated influenza vaccines (IIV) primarily work by targeting antibodies towards the Hemagglutinin (HA) head to block virus infection. However, these antibody responses are highly strain-specific and fallible due to antigenic drift or mismatch. Calls for improvement to the breadth of immune reactivity elicited by influenza vaccines has led to the research of additional immune correlates for protection and development of universal vaccine strategies.
Antibodies have a fragment antigen binding (Fab), which is antigen specific, and a constant fragment (Fc). The Fc domain mediates antibody effector functions due to Fab binding of cognate antigen, leading to cross linking of Fc receptors (FcR) on innate and adaptive immune cells [1]. FcR crosslinking of NK cells initiates Antibody dependent cellular cytotoxicity (ADCC) that leads to their activation (CD69+), degranulation (CD107a+) of cytotoxic granules and cytokine production (IFN-γ [2], and destruction of virus infected cells.
ADCC responses have shown a high level of cross-reactivity between seasonal and avian influenza viruses in the absence of virus neutralization [2], and increased responses correlate with reduced viral shedding during infection [1] and symptom severity [3]. Importantly, in adults cross-reactive ADCC antibodies are already present before the development of neutralizing antibody responses [4], reflecting their protective roles in the early phase of influenza infection. Influenza-specific ADCC responses are increased by a recent infection [3,5], but are not boosted by current inactivated influenza vaccines [6]. Therefore, new strategies need to be devised and assessed to stimulate the production of cross-reactive ADCC antibodies against influenza.
Both the HA head and the stem region contain broadly conserved epitopes, yet polyclonal serum has shown greater ADCC function to the HA-stem than recombinant HA1 proteins which predominantly represent the HA-head [5]. Broadly cross-reactive monoclonal antibodies targeting the conserved HA-stem [7], NP [8] and M2e [9] utilize Fc/FcR interactions for protection. Therefore, ADCC antibodies can potentially recognize more conserved epitopes than neutralizing antibodies [7], however there are limited reports on mapping ADCC-epitopes [10]. Identification of minimal epitopes is a major hurdle for the design of subunit and peptide-based vaccination. Subunit peptide-based vaccine approaches are an attractive target for universal vaccines, due to their stability, rapid production, and adaptability to sequence updates. Antibodies can recognize conformational or linear protein epitopes, from 2 to 85 amino acids in length, and the majority of B cell epitopes are 15 amino acid long based on identification from antigen-antibody complexes [11].
H7N9 avian influenza viruses have been a threat of pandemic emergence since 2012, and widespread vaccination of poultry in China since 2017 have diminished the circulation of H7N9 viruses. However, there has been many cases of human infection and mortality, and recruitment of cross-reactive ADCC antibodies have played an important role in survival from severe H7N9 infection [4]. Therefore, we aimed to map cross-reactive HA ADCC epitopes from both existing homotypic H1-HA and heterosubtypic H7-HA proteins to identify universal vaccine targets for stimulating ADCC responses and determine their protective potential.
2. Results
2.1. Peptide mapping of ADCC activity for cross-reactivity
A high level of cross-reactivity has been reported for H7-HA proteins for ADCC activity in hemagglutinin inhibition (HAI) seronegative individuals [4]. Therefore, we sought to identify minimal epitope regions within the HA protein which could be attributed to ADCC cross-reactivity using overlapping peptide libraries for HA proteins from H1N1 (A/California/04/2009) and H7N9 (A/Shanghai/02/2013) viruses. A FACS based NK activation assay (Fig. 1A) was used to quantify ADCC responses (Supplementary Fig. 1AB), and IgG responses by standard ELISA for recombinant HA proteins and peptides (Fig. 1B). We assessed peptide ADCC responses in plasma collected before and after H1N1 pandemic infection (D1–3, Supplementary Fig. 1A). We found that recent H1N1 infection did not show a consistent pattern across donors (n = 3) of fold-change enrichment of ADCC responses for particular H1-HA or H7-HA peptides (Fig. 1D). To further assess ADCC responses at baseline before infection, we used pre H1N1 infection samples (Positive) from a household study and compared responses to household members who did not become infected (Negative) [3]. We did not find a difference in the profile of H1-HA targeted peptides between uninfected (Negative) and H1N1 infected (Positive) household contacts at baseline to account for acquisition of infection (Fig. 1E).
Fig. 1. HA peptide landscape for antibody binding and ADCC function.
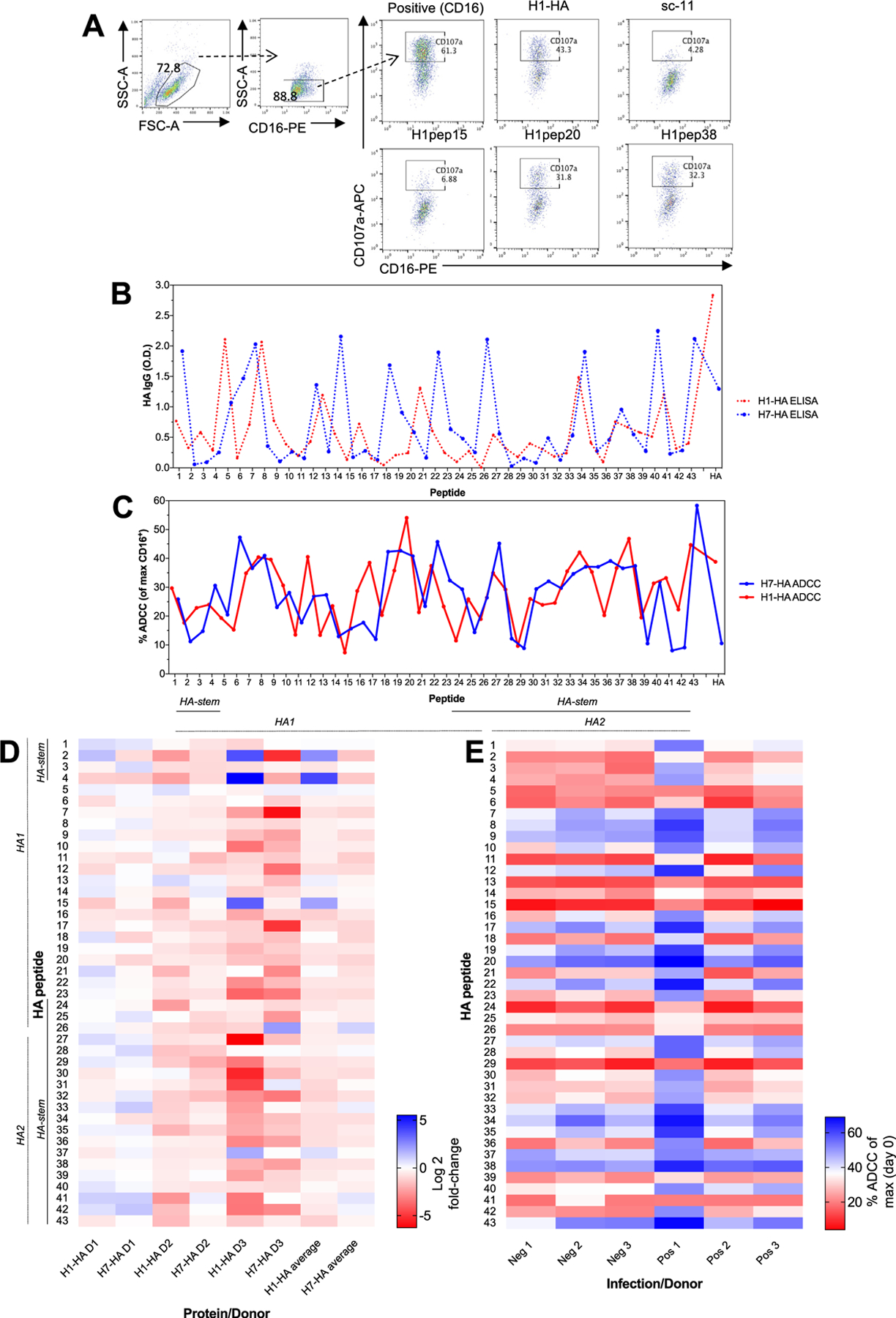
(A) A FACS based NK activation assay was used to assess ADCC antibody responses (representative FACS plots from Positive 1 donor). H1- and H7-HA peptides and full-proteins IgG levels (by ELISA, dotted lines (B)) and ADCC responses (plain lines (C)) (n = 15 human serums). Data represents the mean average. (D) Heat map of fold-change of post- versus pre-H1N1 infection ADCC responses for H1-HA and H7-HA peptides (values are represented as Log2). (E) Heat map of H1-HA peptide ADCC responses (% ADCC (of max CD16+) from (A) for uninfected negative donors (Neg 1–3) and infected positive donors (Pos 1–3). Experiment was repeated twice.
Some peptides elicited more robust ADCC activities, indicating an ADCC epitope landscape. In HA1 ‘head’ region peptide HA-20 elicited the largest ADCC response (33.3 ± 20.3% versus an average of 17.3%). In the HA2 ‘stem’ region, the highest ADCC response (28.9%±22.6) was towards HApep38. These two peptides, HApep20 and HApep38 also had high levels of ADCC from H7-HA derived peptides (25.1%±20.8 and 23.1%±20.3 respectively, Fig. 1C).
Cross-reactivity between H1- and H7-HA peptides was observed for functional ADCC responses (blue and red solid lines, Fig. 1C), despite little sequence conservation (Supplementary Fig. 1), and we found 2 of 7 participants had high level H7-HA ADCC responses at >10% NK92 activation (Fig. 1C), consistent with other studies [12]. ADCC responses towards H1-HA and H7-HA peptides correlated in a representative donor (Positive 1) (R2 = 0.2655, p = 0.0004) (Supplementary Fig. 1C). Furthermore, the correlation between peptide ADCC responses was then assessed in all donors and was significant for high (HApep20, R2 = 0.9554, p < 0.0001 (Supplementary Fig. 1D) and HApep38 R2 = 0.8644, p < 0.0001, Supplementary Fig. 1E), or low-ADCC inducing peptides (HApep15, R2 = 0.9554, p < 0.0001, *Supplementary Fig. 1F). However, there was no correlation between H1-HA and H7-HA peptide IgG responses by ELISA (Supplementary Fig. 1G), or H1-HA for ADCC versus ELISA (Supplementary Fig. 1H). This suggests that ADCC antibodies represent a subset of HA-binding antibodies and their function is predominantly dependent on the Fc maturation rather than serum concentration, which is consistent with ADCC in the absence of HAI [1]. The detection of ADCC responses to some peptides in the absence of ELISA response may be attributable to other Ig isotypes or low affinity IgG resulting in a low level of ADCC activity which is below the limit of detection by ELISA.
2.2. Vaccination with ADCC-epitopes and mouse H1N1 challenge
We next investigated in mouse vaccination and homotypic challenge experiments to determine the protective capacity of single ADCC activating peptides (Supplementary Fig. 2a). Mice were vaccinated with (a) full H1-HA protein as a positive control, (b) high ADCC response peptides: H1pep20 from the HA1 region or H1pep38 from the HA2 region, low ADCC response peptide: H1pep15 from the HA1 region, and a scrambled version of H1pep20 as a negative control (sc11).
Vaccination with H1-HA, H1pep20 or H1pep38 elicited a detectable ADCC response to H1N1-infected cells (Fig. 2A), relative to convalescent H1N1 recovered serum, whilst H1pep15 and scrambled peptide vaccination did not. However, mouse vaccination with H1 peptides did not elicit cross-reactive H7N7 ADCC activity to H7N7 virus infected cells (Fig. 2A). Peptide vaccination did not elicit neutralising antibodies to H1N1 or H7N7 viruses, whilst H1-HA protein vaccination gave substantial H1N1 VNA titers (Supplementary Fig. 2B).
Fig. 2. Mouse vaccination with high ADCC activating peptides results in partial protection from influenza infection.
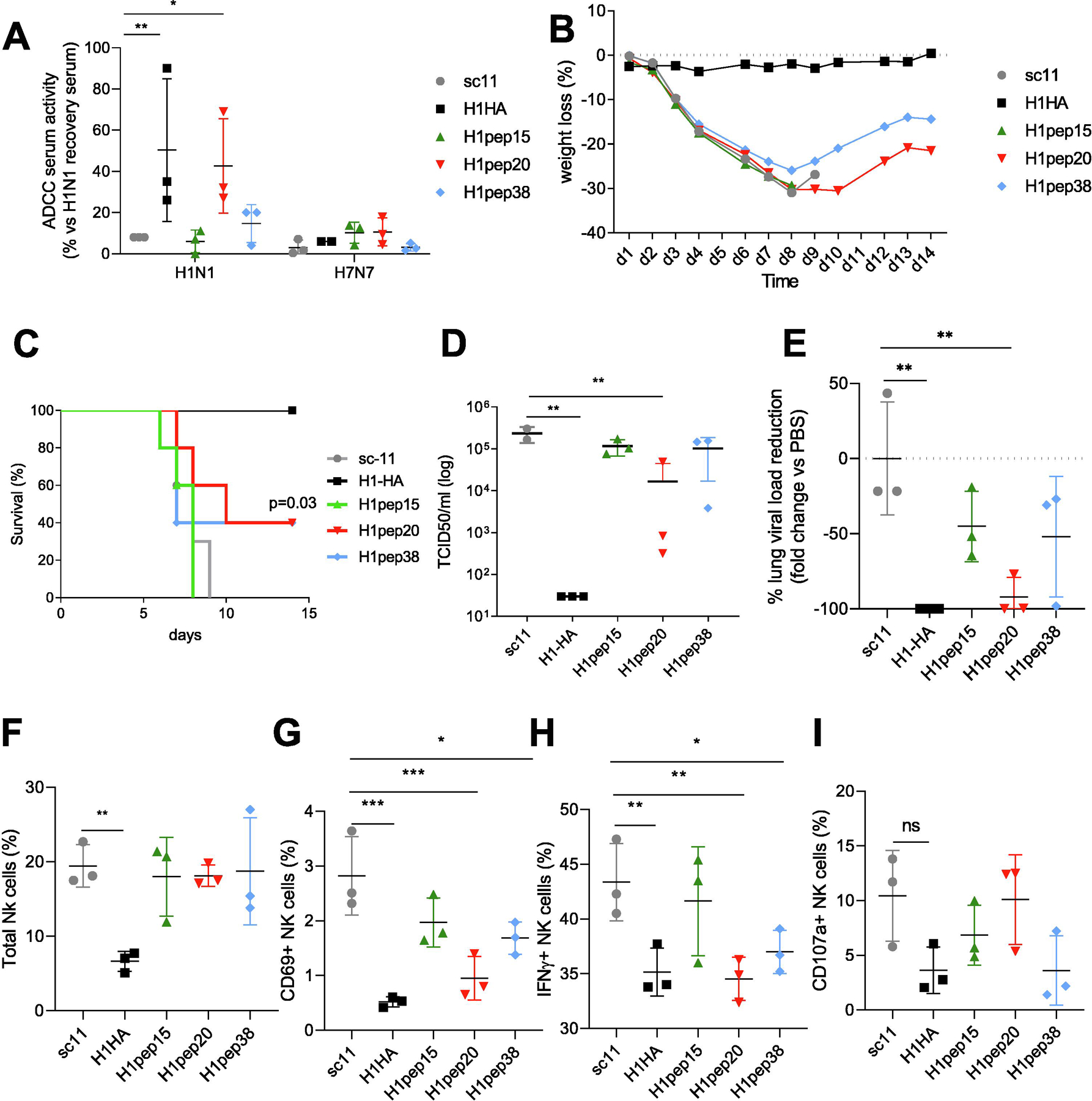
(A) Serum ADCC activity (by NFAT signaling of luciferase reporter NK cells) to H1N1-infected target cells 21 days post-vaccine. Day 14 wt loss (B) and survival (C) of vaccinated mice after lethal H1N1 challenge. TCID50 lung viral titers (D) and fold reduction of lung viral loads (E) at day 7 versus scrambled vaccinated mice. NK cell response in lungs at day 7 post infection for total NK cells (F), CD69+ NK cells (G), IFN-γ+ NK cells (H) and CD107a+ NK cells (I) (see gating FACS plots in Supplementary Fig. 2C). Data represents the mean average and SD. For (A) survival curves were compared for statistical significance with Log-rank Mantel-Cox test (n = 5 per group). For (B–I) data were compared for statistical significance by one-way ANOVA with Dunnett’s multiple comparison test versus scrambled or PBS group, (n = 3 per group) *p < 0.05, **p < 0.01, ***p < 0.005, experiments were repeated twice
Upon lethal H1N1 challenge of peptide vaccinated mice, vaccination with H1pep20 and H1pep38 peptides provided significant (p = 0.03) partial protection (40% survival), whilst H1pep15 and scrambled peptides did not, and full H1-HA protein provided 100% homologous protection (Fig. 2B–C). Furthermore, H1pep20 peptide vaccinated mice also had significantly less lung virus titers than those treated with scrambled peptide (92% fold change, p = 0.0068) (Fig. 2D).
ADCC antibodies have been shown to have a protective role in human infections, but high levels of activated NK cells at the site of infection are also associated with immunopathology [2], therefore we assessed in vivo NK cell activation in the lung (Fig. 2F–I, Supplementary Fig. 2C). H1-HA vaccinated mice had a significant reduction of more than 60% of total NK cells in the lung compared to scrambled peptide vaccinated mice (19.43%±2.84 for sc-11 mice versus mean of 6.63%±1.35 for H1-HA mice, p = 0.002, Fig. 2F). Similarly, H1-HA protein, H1pep20 and H1pep38 peptide vaccinated mice also had significantly reduced activated (CD69+) and cytokine production (IFN-γ+) by NK cells compared to scrambled peptide vaccinated mice (Fig. 2G–H). Whilst CD107a+ NK cells were similar across groups directly ex vivo (Fig. 2I). Furthermore, antibody responses at day 7 of infection were high for IgG homologous peptides in the serum Supplementary Fig. 2D) but not cross-reactive for H7-HA, nor significantly increased as IgA at the site of infection (BAL) (Supplementary Fig. 2E). Only H1-HA vaccination elicited high levels of H1-HA-specific serum IgG and secretory BAL IgA responses compared to scramble peptide vaccinated mice, and again responses were not cross-reactive in mice for H7-HA (Supplementary Fig. 2BD).
Immunopathology in the lung was assessed by protein leakage to the BAL, by total protein concentration (Supplementary Fig. 2F), pro-inflammatory IL-6 (Supplementary Fig. 2G) and histopathology scoring (Supplementary Fig. 2H). H1-HA protein vaccination significantly reduced total protein (Supplementary Fig. 2F) and IL-6 concentration (Supplementary Fig. 2G) in the BAL, whilst H1pep38 vaccination also reduced lung IL-6 levels compared to scrambled peptide vaccinated. Histopathology scoring revealed a significant reduction of lung pathology in H1-HA group but also in H1pep20 and H1pep38 vaccinated groups versus PBS (Supplementary Fig. 2H). H1-HA vaccination also altered the balance of macrophage responses, with reduced inflammatory monocytes and consequently increased alveolar macrophages (Supplementary Fig. 2IJ), whilst peptide vaccination had no impact on macrophage and monocyte recruitment to the lung. Therefore, ADCC-peptide vaccination provided partial protection against influenza infection to reduce lung viral loads, NK cell activation, and inflammation.
3. Discussion
In this study, we tested ADCC responses to HA-peptides in human serum to map cross-reactive HA regions and determine the protective potential of ADCC activating HA antibodies by mouse vaccination and challenge. To identify broadly cross-reactive ADCC antibodies, we first mapped linear HA epitopes that corresponded to high ADCC activity in the serum of influenza-infected subjects and used newly identified peptides for vaccination in mice. We found that ADCC epitopes can be found through-out the HA protein, in both the HA1 and HA2 regions, in the absence of neutralisation or with low antibody concentration, and human serum has many cross-reactive H7-HA responses. H7-peptides caused a greater level of ADCC reactivity than whole H7-HA protein, which may be due to greater antibody recognition. This could be exploited in universal vaccines to do better than nature and increase ADCC activity. Furthermore, we did not find a difference in the peptide landscapes for ADCC responses between household subjects who became infected versus those who did not, therefore the total magnitude of pre-existing ADCC responses remains an important correlate of protection.
Mouse serum from peptide vaccination did not have ADCC cross-reactivity between H1N1 and H7N7 or H7N9 viruses that was observed in human serum. This may be due to the naïve background of mice prior to single peptide vaccination and their lack of previous diverse exposures to group 1 and 2 influenza viruses, especially H3N2 viruses, unlike humans. Therefore our mouse challenge experiments focused on homotypic H1N1 reactivity and influenza challenge to demonstrate the protective potential of ADCC responses themselves.
Vaccination of mice with ADCC activating peptides provided moderate protection, in terms of survival, viral loads and lung inflammation from homologous influenza virus challenge. Whilst peptide vaccination did not achieve the level of protection afforded by full H1-HA protein vaccination, it did demonstrate the protective potential of ADCC responses which has also been evident in human studies [1,3]. A limitation of our study is the reductionist approach of using linear peptides to identify antibody epitopes, as conformational epitopes may also play a role in influenza-ADCC responses. In addition, as only partial protection was achieved in our model, additional immune correlates targeted in conjunction with ADCC responses or combined mosaic ADCC peptide vaccination approaches could be used in the future using targets such as the NA, NP, or M2e proteins.
FcR maturation can increase antibody effector functions such as ADCC due to changes in antibody isotype, IgG subclass and glycosylation. We found ADCC function and increased protection by H1pep20 vaccination, and further dissection of antibodies generated is needed to attribute protection to ADCC function by Fc maturation as no single factor can determine Fc maturation in polyclonal serum. It is only by dissection of monoclonal antibodies and alteration of the Fc region by IgG subclass and glycan additions with the same Fab that these effector functions can be attributed to particular characteristics of an antibody. Furthermore, we did not determine the contribution of IgG subclass to ADCC effector function, which is likely attributable to IgG2a/b in mice.
NK cells can have different functions in the lung during influenza virus infection depending on the virus dose and timing of response. NK cell depletion in mice can lead to worsening of morbidity and mortality from mild influenza virus infection and therefore NK cells are necessary for protection, however NK cells can also be responsible for enhanced morbidity and mortality during more severe influenza infection by driving an extreme inflammatory reaction [2]. We observed increased ADCC, BAL IgA and VNA antibodies by H1-HA protein vaccination coincided with reduced total NK cell recruitment during infection and lowered virus replication and inflammation. We also observed reduced activation (CD69 and IFN-γ) of NK cells by H1-HA, H1pep20 and H1pep38 vaccination corresponding to improved protection and virus load reduction.
Other groups have also evaluated the protective function of ADCC antibodies by peptide mapping. Srivastava et al., identified the E1 peptide: 92–117 and E2 peptide: 124–159 amino acids of H1-HA protein as ADCC-epitopes on the HA head domain using serum samples from 6 H1N1-infected patients [13], but observed exaggerated inflammatory cell infiltration in the lungs alveolar damage and increased mortality of E1 peptide-vaccinated mice [13,14]. Peptide E1 and E2 correspond to HApep8 and HApep10/pep11 peptides respectively in our study, of which HApep8 had high ADCC and ELISA binding in cohort of serum but not to the same level as HApep20 and HApep38, whilst HApep10 and HApep11 did not correspond to high ADCC activity. In our hands peptide vaccinated mice displayed lower viral loads and inflammation and were therefore a promising vaccination platform. Overall ADCC-epitopes are present in both the HA1 and HA2 regions, and single ADCC-activating peptides provided partial protection from lethal influenza challenge, therefore representing a possible target in future combination vaccination strategies.
4. Materials and methods
4.1. Human subjects
Serum samples (n = 15) were collected from multiple sources to assess ADCC cross-reactivity in the community (pooled), before exposure and acquisition of infection (household study), and before and after pandemic infection (Red Cross donors) (Supplementary Table 1). Samples were from 3 sources: (1) pooled immune serum from Hong Kong Red Cross Blood Transfusion Service, pool 1 (n = 5), pool 2 (n = 5), (2) baseline samples from a household infection study [3], with infected index cases (Inf 1) and household contacts sampled, positive donors (Pos 1, 2, 3) became infected (RT-PCR confirmed) after exposure and negative donors were not infected (Neg 1, 2, 3), 3) blood donors (Donor 1–3, D1–3) at the Hong Kong Red Cross Blood Transfusion Service before and after the 2009 pandemic season with HAI confirmed H1N1 (A/California/04/2009) infection (pre and post serum for D1, D2, D3) [15]. The use of human samples were approved by the Institutional Review Board of the University of Hong Kong.
4.2. Recombinant proteins and peptides
HA influenza recombinant proteins, H1-HA (A/California/04/2009), H7-HA (A/Shanghai/2/2013), and gp120, were purchased commercially (SinoBiological, Beijing China). An overlapping peptide library of 43 linear peptides at 25 amino acids in length with 12 amino acids overlap were synthesized based on the protein sequence of H1-HA (A/California/04/2009) and H7-HA (A/Shanghai/2/2013) (Genscript, Piscataway, USA) (Supplementary Table 2). Amino acid similarity between H1 and H7 peptides was assessed by alignment using Water (EMBOSS, EMBL-EBI).
4.3. Human ADCC peptide mapping
We used a protein plate-bound NK ADCC CD107a degranulation assay [3] using CD16+ NK92 cells (Fox Chase Cancer Center, Philadelphia, USA). Briefly, blocked coated (40 µg/peptide or protein) immunosorbent plates (Nunc, Roskilde, Denmark) were bound by heat inactivated (56 °C for 30 mins) human immune serum (1:20), and incubated with 1 × 105 NK92 cells for 5 h, stained for anti-human CD16-PE, CD107a-APC (Biolegend, San Diego, USA), and assessed by flow cytometry on an Attune (Invitrogen, Carlsbad, USA). Responses were normalised to % of maximum CD107a+ of CD16 stimulated positive controls and background subtracted. CD16 coated wells determine the maximum threshold of NK92 activation. Results are calculated by % CD107a+ values minus paired serum background gp120, divided by the maximum CD107a+ value from CD16 coated wells, i.e.: (sample - gp120/CD16)*100 = % ADCC (of max CD16+).
4.4. Antibody ELISA for human and mouse samples
To assess influenza-specific antibodies human and mouse samples was probed by ELISA [3]. Briefly, blocked immunosorbent plates coated with recombinant proteins H1-HA, H7-HA (80 ng/ml), and overlapping peptides (80 ng/ml), were bound by immune serum (1:100) or BAL supernatant (1:2), and detected by anti-mouse or human IgG-HRP, or anti-mouse IgA-HRP (Invitrogen).
4.5. Mice vaccination and challenge model
Female BALB/c mice (6 weeks old) were vaccinated twice 3 weeks apart via the sub-cutaneous route with H1-HA protein or peptides (H1pep15, H1pep20, H1pep38 or scrambled) (10 µg in 50ul PBS plus 50ul Addavax (Invivogen, San Diego, USA) and challenged with H1N1 (A/California/04/2009, 20LD50, 1.04E3TCI-D50/25ul) influenza virus 3 weeks later (Supplementary Fig. 2A), monitored for survival, and lung viral titers were determined by standard TCID50.
Clotted serum (MiniCollect, Greiner BioOne, Kremsmünster, Austria) was collected at day 21 post-vaccination or day 7 post-infection. To evaluate cellular immunity, cells were sampled at day 7 post-infection from Bronchoalveolar lavage (BAL), lung, and mediastinal LN (mLN). The total protein concentration in BAL supernatants were measured by BCA Protein Assay kit (Thermo-Fisher, Waltham, USA) according to manufacturer’s instructions. All animal studies were approved by CULATR, The University of Hong Kong.
4.6. ADCC for murine samples
Mouse ADCC Reporter Bioassay was conducted according to manufacturer’s instructions (Promega, Madison, USA), with 25ul of 1:20 diluted heat inactivated mouse serum, added to 12.5x103 Raji cells at 16 h post-H1N1 infection (MOI4) and 75 × 103 Jurkat reporter cells (6:1 Effector:Target ratio).
4.7. Immune cell profiling
Immune cells were stained for viability (Zombie-Violet live/dead) (Biolegend), FcR blocked (anti-CD16/CD32, BD Bioscience, Franklin lakes, USA), and stained with one of two panels (all Biolegend). Cocktail 1 for Macrophages: anti-mouse F4/80-PE, MHC-II/I-AE-PerCPCy5.5, Ly-6G-APC, CD11c-FITC. Cocktail 2 for NK cells: anti-mouse CD3-BV510, NKp46/CD335-PE-TexasRed, CD107a-PE, CD69-BV605, IFN-γ-PerCPCy5.5.
4.8. Histopathology scoring of lung tissues
Inflammation was assessed by histopathology scoring of H&E stained lung sections from day 7 post infection for inflammation in the airway (score 0–4), vascular (score 0–4) and parenchyma (score 0–5, ×10 magnification) for a total score up to 13 [16]. Scoring was blinded and averaged from 2 independent assessments.
4.9. Statistics
Results represent the mean ± stdev of three to five mice per group. Statistical significance was compared between vaccine groups and negative-control group using a standard Student t test (unless indicated) on Graphpad Prism Software v8, *p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.001.
Supplementary Material
Acknowledgments
We would like to thank Marianne Weyhmuller and Kerry Campbell at Fox Chase Cancer Center for supply of the NK92 cells, and Yizhuo Wang for technical assistance. This project utilised an Invitrogen Attune cytometer assisted by the Pasteur Foundation Asia.
Funding statement
This project was supported by the Hong Kong Health and Medical Research Fund (15141052 and 14130672) and National Institutes of Allergy and Infectious Diseases, National Institutes of Health (USA) (contract HHSN272201400006C).
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.vaccine.2020.07.008.
References
- [1].Jegaskanda S et al. Generation and protective ability of influenza virus-specific antibody-dependent cellular cytotoxicity in humans elicited by vaccination, natural infection, and experimental challenge. J Infect Dis 2016;214(6): 945–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Zhou G, Juang SW, Kane KP. NK cells exacerbate the pathology of influenza virus infection in mice. Eur J Immunol 2013;43(4):929–38. [DOI] [PubMed] [Google Scholar]
- [3].Valkenburg SA et al. Cross-reactive antibody-dependent cellular cytotoxicity antibodies are increased by recent infection in a household study of influenza transmission. Clin Transl Immunology 2019;8(11):e1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Vanderven HA et al. Fc functional antibodies in humans with severe H7N9 and seasonal influenza. JCI Insight 2017;2(13):92750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].de Vries RD et al. Influenza virus-specific antibody dependent cellular cytoxicity induced by vaccination or natural infection. Vaccine 2017;35 (2):238–47. [DOI] [PubMed] [Google Scholar]
- [6].Jegaskanda S et al. Standard trivalent influenza virus protein vaccination does not prime antibody-dependent cellular cytotoxicity in macaques. J Virol 2013;87(24):13706–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].DiLillo DJ et al. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat Med 2014;20(2):143–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].LaMere MW et al. Contributions of antinucleoprotein IgG to heterosubtypic immunity against influenza virus. J Immunol 2011;186(7):4331–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].El Bakkouri K et al. Universal vaccine based on ectodomain of matrix protein 2 of influenza A: Fc receptors and alveolar macrophages mediate protection. J Immunol 2011;186(2):1022–31. [DOI] [PubMed] [Google Scholar]
- [10].Ellebedy AH et al. Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proc Natl Acad Sci U S A 2014;111(36):13133–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kringelum JV et al. Structural analysis of B-cell epitopes in antibody:protein complexes. Mol Immunol 2013;53(1–2):24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jegaskanda S et al. Induction of H7N9-cross-reactive antibody-dependent cellular cytotoxicity antibodies by human seasonal influenza a viruses that are directed toward the nucleoprotein. J Infect Dis 2017;215(5):818–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Srivastava V et al. Identification of dominant antibody-dependent cell-mediated cytotoxicity epitopes on the hemagglutinin antigen of pandemic H1N1 influenza virus. J Virol 2013;87(10):5831–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ye ZW et al. Antibody-dependent cell-mediated cytotoxicity epitopes on the hemagglutinin head region of pandemic H1N1 influenza virus play detrimental roles in H1N1-infected mice. Front Immunol 2017;8:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Valkenburg SA et al. Preexisting antibody-dependent cellular cytotoxicity-activating antibody responses are stable longitudinally and cross-reactive responses are not boosted by recent influenza exposure. J Infect Dis 2016;214 (8):1159–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Horvat JC et al. Neonatal chlamydial infection induces mixed T-cell responses that drive allergic airway disease. Am J Respir Crit Care Med 2007;176 (6):556–64. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


