Abstract
In this study, we evaluated and compared six SARS-CoV-2 serology kits including the Abbott SARS-CoV-2 IgG assay, Beckman Access SARS-CoV-2 IgG assay, OCD Vitros OCD Anti-SARS-CoV-2 Total antibody assay, Roche Elecsys Anti SARS-CoV-2 assay, Siemens SARS-CoV-2 Total assay, and cPass surrogate viral neutralising antibody assay. A total of 336 non-duplicated residual serum samples that were obtained from COVID-19 confirmed patients (n=173) on PCR and negative controls (n=163) obtained pre-December 2019 before the COVID-19 pandemic were used for the study. These were concurrently analysed on the different immunoassay platforms and correlated with clinical characteristics. Our results showed all assays had specificity ranging from 99.3% to 100.0%. Overall sensitivity across all days of symptoms, in descending order were OCD (49.1%, 95% CI 41.8–56.5%), cPass (44.8%, 95% CI 37.5–52.3%), Roche (41.6%, 95% CI 34.5–49.0%), Siemens (39.9%, 95% CI 32.9–47.3%), Abbott (39.8%, 95% CI 32.9–47.3%) and Beckman (39.6%, 95% CI 32.5–47.3%). Testing after at least 14 days from symptom onset is required to achieve AUCs greater than 0.80. OCD and cPass performed the best in terms of sensitivity for >21 days symptoms with 93.3% (95% CI, 73.5–99.2%) and 96.7% (95% CI, 82.8–99.9%), respectively. Both also shared the greatest concordance, kappa 0.963 (95% CI 0.885–1.0), p<0.001, and had the lowest false negative rates. Serology results should be interpreted with caution in certain cases. False negatives were observed in a small number of individuals with COVID-19 on immunosuppressive therapy, pauci-symptomatic or who received antiretroviral therapy. In conclusion, all assays exhibited excellent specificity and total antibody assays with spike protein configurations generally outperformed nucleocapsid configurations and IgG assays in terms of diagnostic sensitivity.
Key words: COVID-19, SARS-CoV-2, serology, concordance, evaluation
Introduction
A cluster of unexplained viral pneumonia cases first identified in Wuhan in December 2019 has now progressed to the global pandemic known as the coronavirus disease 2019 (COVID-19). Clinical features include a spectrum ranging from asymptomatic infection, mild acute respiratory illness, fever, and diarrhoea, to that of multi-organ failure requiring intensive care support.1 Presence of co-morbidities such as diabetes mellitus, hypertension, and cardiovascular disease confer poorer clinical outcomes and increased mortality.2 Laboratory markers such as elevated white blood cell count, neutrophil count, lactate dehydrogenase, alanine aminotransferase, aspartate aminotransferase, total bilirubin, creatinine, troponins, D-dimer and procalcitonin are associated with poorer prognosis for COVID-19 patients.3 The demand for laboratory testing, especially in a timely fashion, is crucial for clinical management and public health efforts to limit transmission. Currently, the gold-standard for diagnosing COVID-19 is detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA via real time reverse transcription polymerase chain reaction (rRT-PCR), targets of which may include a combination of nucleocapsid (N), envelope (E), RNA-dependent-RNA polymerase (RdRp), and open reading frame (orf1a and orf1b) genes.4
SARS-CoV-2 serological assays have both clinical and epidemiological uses. At the individual level, serological tests may be used to support clinical diagnosis by determining recent or previous infection, thereby contributing to quarantine measures, or by determining the immunological status in vaccinated individuals with a view for booster requirements. In addition, there have been suggestions regarding the utility of ‘immune passports’ with the use of seropositive status conferring potential immunity and allowing the individual to return to work or certified fit-to-travel.5 At a public health level, population screening with serology offers insights into epidemiology and immunity of the population. Also, seroprevalence data may allow government officials to more effectively implement interventions strategies such as lockdowns or targeted physical distancing measures. The advent of commercially available SARS-CoV-2 serological assays allows for more ubiquitous testing amongst laboratories. Nevertheless, there are currently limited head-to-head comparisons of test performance between the available testing platforms. The aim of our study is to compare six commercial serological tests with various assay design configurations directed against separate SARS-CoV-2 antigens, including a viral neutralisation antibody assay, and to inform users as to their clinical and interpretive differences.
Methods
Ethics approval for our study was given by the National Healthcare Group Domain Specific Review Board (NHG ROAM Reference Number: 2020/00337 and 2020/00407).
In our institution, we had access to a number of instruments used in the latest SARS-CoV-2 serological assays. For purposes of this study, we used the Abbott Architect i4000SR for the Abbott SARS-CoV-2 IgG (Abbott Diagnostics, USA), Roche Cobas E411 for testing the Elecsys Total Anti-SARS-CoV-2 antibody (Roche Diagnostics, Switzerland), UniCel DxI 800 for the Access SARS-CoV-2 IgG antibody assay (Beckman Coulter, USA), ADVIA Centaur XPT for Siemens SARS-CoV-2 Total (COV2T) assay (Siemens Healthineers, Germany), VITROS 3600 for the Vitros OCD Anti-SARS-CoV-2 Total antibody (Ortho Clinical Diagnostics, USA), and the Evolis Complete System (Biorad, USA) for the cPass SARS-CoV-2 surrogate viral neutralising antibody detection ELISA kit (Genscript, USA). A summary of the analysers used, the assay configuration and interpretation of the quantitative values are provided in Table 1 . These assays will subsequently be referred to as Abbott, Beckman, cPass, OCD, Roche, and Siemens assays.
Table 1.
Summary of SARS-CoV-2 serology assay configuration, instruments, and interpretation
| Instrument | Assay name | SARS-CoV-2 serology assay configuration | Interpretation for signal values |
|---|---|---|---|
| Abbott Architect i4000SR | Abbott SARS-CoV-2 IgG antibody | Chemiluminescent immunoassay detecting IgG antibodies to nucleocapsid protein | <1.4: Non-reactive≥1.4: Reactive |
| Roche Cobas E411 | Elecsys Total Anti-SARS-CoV-2 antibody | Sandwich immunoassay detecting antibodies (IgM/IgG) to nucleocapsid protein | <1.0: Non-reactive≥1.0: Reactive |
| Beckman Unicel DxI 800 | Access SARS-CoV-2 IgG antibody | Chemiluminescent immunoassay detecting IgG specific to RBD of S1 protein | <0.8: Non-reactive≥0.8 to <1.0: Equivocal≥1.0: Reactive |
| ADVIA Centaur XPT | Siemens SARS-CoV-2 Total antibody | One-step sandwich immunoassay detecting antibodies (IgM/IgG) to RBD of S1 protein | <1.0: Non-reactive≥1.0: Reactive |
| VITROS 3600 | Vitros OCD Anti-SARS-CoV-2 Total antibody | Chemiluminescent immunoassay detecting antibodies (IgA, IgG, IgM) to S1 protein. | <1.0: Non-reactive≥1.0: Reactive |
| Evolis Complete System | cPass SARS-CoV-2 neutralising antibody ELISA | Blocking ELISA detection. HRP conjugated with recombinant RBD and human ACE-2 protein. Protein-protein interaction between HRP-RBD and ACE-2 are blocked by neutralising antibodies | Calculated as inhibition %<20%: Non-reactive≥20%: Reactive |
HRP, horseradish peroxidase; RBD, receptor binding domain.
We collected a total of 336 non-duplicated residual serum samples for the study that were obtained from COVID-19 confirmed patients (n=173) and negative controls (n=163) obtained pre-December 2019 before the COVID-19 pandemic. We prospectively selected samples between 30 March 2020 and 15 June 2020 from COVID-19 patients in our institution. When samples were collected from patients, we recorded the number of days following onset of symptoms. All had a minimum of one real time reverse transcription polymerase chain reaction (rRT-PCR) respiratory sample positive on our Cobas 6800 SARS-CoV-2 assay (Roche Diagnostics, Switzerland), or the A∗Star Fortitude kit (Accelerate Technologies, Singapore) with the cycle threshold (CT) value being lower than cut-off. This was the gold standard to which our results were compared. Samples were collected in serum separator tubes (Beckton Dickinson, USA), centrifuged at 3000 rpm for 8 min and after clinical testing, residual sera were collected in accordance with previously described laboratory protocols for COVID-19 sample handling.6 Days of symptoms were recorded based on first day of onset of COVID-19 symptoms, as documented by managing clinicians. Patients who were asymptomatic at the time of PCR testing were excluded. Archived serum samples taken from patients prior to December 2019 representing COVID-19 naivety were used as negative controls. These included healthy blood donors as well as patients with and without other positive serological tests: anti-extractable nuclear antigen antibodies (9); anti-glomerular basement membrane antibodies (4); anti-smooth muscle antibody (3); hepatitis A IgM (3); Epstein–Barr virus IgM (3); anti-intrinsic factor IgG (5); cytomegalovirus IgM (4); cytomegalovirus IgG (3); syphillis treponemal antibody (5); Epstein–Barr virus IgA (7); Leptospira IgM (3); hepatitis C antibody (9); hepatitis B surface antigen (7); hepatitis B e antigen (2); anti-double stranded DNA IgG (3); rubella IgM (4); antinuclear antibodies (3); hepatitis A IgG (3); dengue virus IgG (1); varicella zoster IgM (1); human immunodeficiency virus (8); and varicella zoster virus IgG (6). Prior to testing patients' sera, calibration was performed and quality controls were passed as per manufacturers' instructions.
Statistical analyses were performed using the Statistical Package for Social Sciences (SPSS) 25.0 (IBM, USA) with statistical significance determined at p<0.05. Sensitivity and specificity were expressed as percentages with their respective 95% confidence interval (CI). The relationship between the six immunoassays and their qualitative agreement were assessed using the inter-rater Gwet's AC1 kappa concordance to account for potential paradox effect with Cohen's kappa.7 Receiver operator characteristic (ROC) and area under the curve (AUC) values were analysed to determine the discriminative ability of the tests. Quantitative values of the signal range in both reactive and non-reactive samples were presented.
Results
Specificity
Specificities of all six assays were excellent. Five of the assays (Abbott, Roche, OCD, Siemens, cPass) had 100% specificity with no cross-reactivity in the negative control panel. Beckman had one serum sample which was positive (signal value 1.69) from a healthy volunteer. This was repeated and remained positive, thus giving a specificity of 99.3% (95% CI 95.7–99.9%). In general, the quantitative signals in this negative cohort except for Beckman, were distinctly lower than the threshold cut-off for each assay (Supplementary Table 1, Appendix A).
Sensitivity and AUC
We compared the qualitative results generated by each of the six assays. The overall sensitivity across all days of symptoms in descending ranking were: OCD (49.1%, 95% CI 41.8–56.5%), cPass (44.8%, 95% CI 37.5–52.3%), Roche (41.6%, 95% CI 34.5–49.0%), Siemens (39.9%, 95% CI 32.9–47.3%), Abbott (39.8%, 95% CI 32.9–47.3%) and Beckman (39.6%, 95% CI 32.5–47.3%). OCD and cPass performed the best in terms of sensitivity for >21 days group with 93.3% (95% CI, 73.5–99.2%) and 96.7% (95% CI, 82.8–99.9%), respectively. In the 14–20 days of symptoms group, OCD and Roche had equal highest sensitivity (88.5%, 95% CI 69.8–97.6%). For the 7–13 days of symptoms group, we found that OCD had the highest sensitivity (56.8%, 95% CI 39.5–72.9%), outperforming Beckman (29.7%, 95% CI 15.9–47.0%), p=0.019, and Siemens (32.4%, 95% CI 18–49.8%), p=0.035.
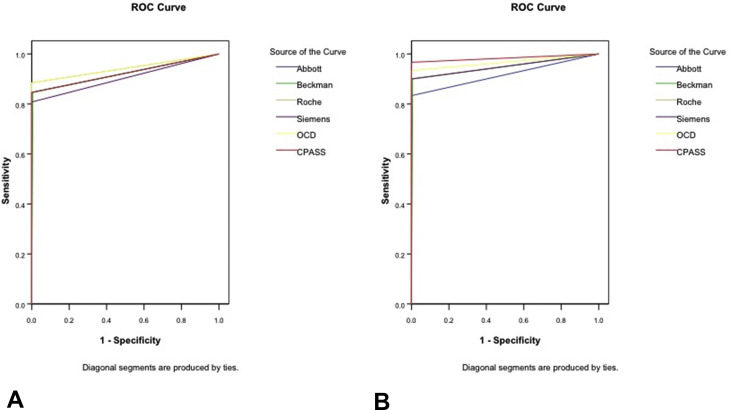
The discriminative ability of the binary/qualitative serology tests indicated that testing after at least 14 days from symptom onset is required to achieve AUCs of more than 0.80. Earlier testing was suboptimal with AUCs between 0.654 and 0.743 when testing was performed between days 7 and 13, and less then 0.60 when testing between days 1 and 6. Less than 10% of samples were positive when testing earlier than 7 days after symptom onset by any assay, suggesting that these assays have poor diagnostic capability in the early phase of symptoms for COVID-19. Comprehensive data are presented in Table 2 , and the ROC curves for binary discrimination shown in Fig. 1 .
Table 2.
Sensitivity of each immunoassay categorised by days of symptoms
| Days of symptoms |
Overall AUC [95% CI] | ||||
|---|---|---|---|---|---|
| 1–6 (n = 80) | 7–13a (n = 37) | 14–20 (n = 26) | 21–64 (n = 30) | ||
| Abbott | |||||
| n (%) | 7 (8.8) | 15 (40.5) | 22 (84.6) | 25 (83.3) | |
| 95% CI | 3.6–17.2 | 24.8–57.9 | 65.1–95.6 | 65.3–94.4 | |
| AUC [95% CI] | 0.533 [0.451–0.614]p = 0.420 | 0.700 [0.586–0.814]p < 0.001 | 0.917 [0.828–1.0]p < 0.001 | 0.914 [0.832–0.998]p < 0.001 | 0.692 [0.634–0.751]p < 0.001 |
| Beckmanb | |||||
| n (%) | 8 (10.0) | 11 (29.7) | 20 (76.9) | 26 (86.7) | |
| 95% CI | 4.4–18.8 | 15.9–47.0 | 56.3–91.0 | 69.3–96.2 | |
| AUC [95% CI] | 0.549 [0.467–0.632]p = 0.220 | 0.654 [0.549–0.769]p = 0.005 | 0.913 [0.824–1.0]p < 0.001 | 0.945 [0.879–1.0]p < 0.001 | 0.695 [0.636–0.753]p < 0.001 |
| Roche | |||||
| n (%) | 8 (10.0) | 14 (37.8) | 23 (88.5) | 27 (90.0) | |
| 95% CI | 4.4–18.8 | 22.4–55.2 | 69.8–97.6 | 73.5–97.9 | |
| AUC [95% CI] | 0.539 [0.458–0.621]p = 0.334 | 0.700 [0.586–0.814]p < 0.001 | 0.938 [0.859–1.0]p < 0.001 | 0.984 [0.883–1.0]p < 0.001 | 0.704 [0.647–0.762]p < 0.001 |
| OCD | |||||
| n (%) | 13 (16.3) | 21 (56.8) | 23 (88.5) | 27 (90.0) | |
| 95% CI | 8.9–26.2 | 39.5–72.9 | 69.8–97.6 | 73.5–97.9 | |
| AUC [95% CI] | 0.566 [0.484–0.648]p = 0.107 | 0.731 [0.664–0.879]p < 0.001 | 0.938 [0.859–1.0]p < 0.001 | 0.966 [0.912–1.0]p < 0.001 | 0.735 [0.670–0.791]p < 0.001 |
| Siemens | |||||
| n (%) | 9 (11.3) | 12 (32.4) | 21 (80.8) | 28 (93.3) | |
| 95% CI | 5.3–20.3 | 18.0–49.8 | 60.6–93.4 | 77.9–99.2 | |
| AUC [95% CI] | 0.553 [0.471–0.635]p = 0.197 | 0.671 [0.557–0.786]p = 0.002 | 0.896 [0.798–0.993]p < 0.001 | 0.984 [0.883–1.0]p < 0.001 | 0.698 [0.640–0.756]p < 0.001 |
| cPass | |||||
| n (%) | 8 (10.0) | 18 (48.6) | 22 (84.6) | 29 (96.7)c | |
| 95% CI | 4.4–18.8 | 31.9–65.6 | 65.1–95.6 | 82.8–99.9 | |
| AUC [95% CI] | 0.546 [0.464–0.625]p = 0.259 | 0.743 [0.633–0.853]p < 0.001 | 0.917 [0.828–1.0]p < 0.001 | 0.983 [0.944–1.0]p < 0.001 | 0.720 [0.663–0.776]p < 0.001 |
AUC, area under curve; CI, confidence interval.
Significant difference in sensitivity between OCD and Beckman at 7–13 days of symptoms, where OCD performed better by 27.1% (95% CI 5.4–48.8; p = 0.019). Significant difference in sensitivity between OCD and Siemens at 7–13 days of symptoms, where OCD performed better by 24.4% (95% CI 2.4–46.3; p = 0.035).
Day 1–6: 1 equivocal and 3 insufficient omitted; Day 7–13 and Day 14–20: 1 equivocal and 1 insufficient omitted; Day ≥ 21: 1 equivocal omitted.
The 1 non-reactive subject was also non-reactive for all the other 5 tests. With n = 29, the 100% sensitivity has a lower 95% CI of 88.0%.
Fig. 1.
ROC curves (A) for day 14–20, and (B) >20 days of symptoms.
Quantitative signal values in COVID-19 patients
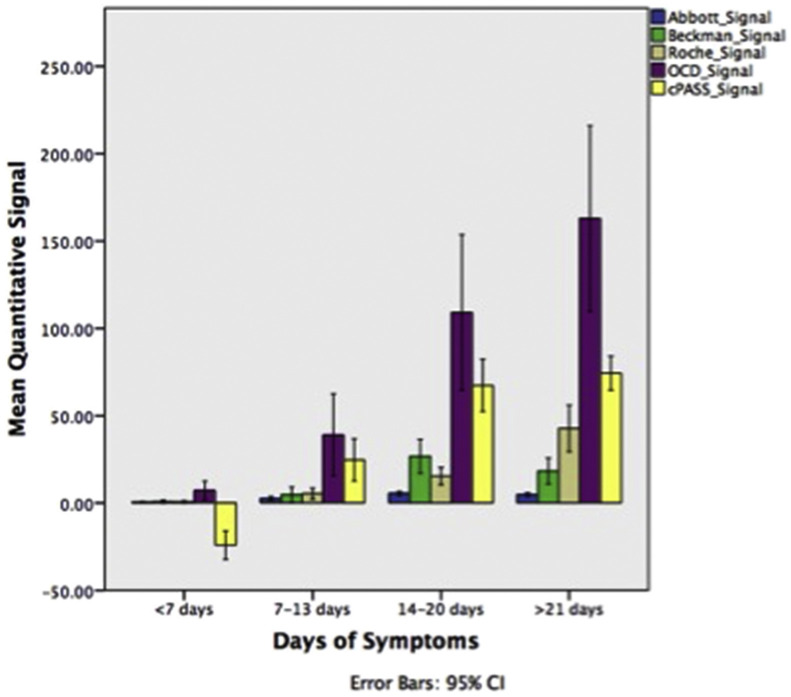
Quantitative signal values may be useful for estimating SARS-CoV-2 antibody titres in patients. For our cohort of patients we documented the minimum to maximum ranges of the reactive and non-reactive cases' signal values for each assay. We then demonstrated that the mean quantitative signals generally increased for COVID-19 patients across all assays, in tandem with increasing number of days from symptom onset (Fig. 2 ). One-way ANOVA comparing the mean signals between the days of symptoms was different (p<0.001) across five assays. The Siemens assay was not analysed due to the number of cases recorded as >10 and <0.05 (i.e., beyond analytical measuring range) and hence impractical for mean signal calculations. All five assays showed a significant difference (p<0.05) in mean quantitative values between >14 days and <14 days using Tukey's posthoc honest significant difference (HSD) test, in line with our ROC data described above (Supplementary Tables 2 and 3, Appendix A).
Fig. 2.
Quantitative signals generated for each assay.
Concordance between assays
We were interested in concordance between the assays, particularly when compared against a viral neutralisation antibody assay. Kappa concordance, categorised by days of symptoms and plotted against the qualitative result, is summarised in Table 3 . Data showed that for >21 days of symptoms, cPass and OCD had the greatest concordance between the assays with kappa 0.963 (95% CI 0.885–1.0), p<0.001. In the 14–20 days group, cPass agreed most with Abbott, with kappa 1.0 (95% CI 0.868–1.0), p<0.001, followed by both OCD and Roche which had similar kappa of 0.950 (95% CI 0.843–1.0), p<0.001. cPass also correlated well with Siemens for all days, kappa 0.942 (95% CI 0.909–0.974), p<0.001.
Table 3.
Kappa concordance between assays
| Beckman | Roche | Siemens | OCD | cPass | |
|---|---|---|---|---|---|
| All days | |||||
| Abbott | 0.929 (0.900–0.960) <0.001 |
0.938 (0.904–0.972) <0.001 |
0.934 (0.899–0.909) <0.001 |
0.912 (0.870–0.953) <0.001 |
0.923 (0.884–0.961) <0.001 |
| Beckman | 0.936 (0.907–0.965) <0.001 |
0.951 (0.925–0.976) <0.001 |
0.921 (0.889–0.954) <0.001 |
0.925 (0.894–0.957) <0.001 |
|
| Roche | 0.948 (0.917–0.979) <0.001 |
0.916 (0.876–0.956) <0.001 |
0.927 (0.890–0.965) <0.001 |
||
| Siemens | 0.921 (0.883–0.961) <0.001 |
0.942 (0.909–0.975) <0.001 |
|||
| OCD | 0.930 (0.893–0.967) <0.001 |
||||
| Days 1–6 | |||||
| Abbott | 0.906 (0.835–0.976) <0.001 |
0.925 (0.856–0.994) <0.001 |
0.939 (0.877–1.0) <0.001 |
0.904 (0.823–0.985) <0.001 |
0.925 (0.856–0.994) <0.001 |
| Beckman | 0.919 (0.853–0.984) <0.001 |
0.946 (0.892–1.0) <0.001 |
0.918 (0.851–0.984) <0.001 |
0.905 (0.835–0.976) <0.001 |
|
| Roche | 0.954 (0.899–1.0) <0.001 |
0.919 (0.844–0.993) <0.001 |
0.908 (0.831–0.986) <0.001 |
||
| Siemens | 0.934 (0.868–1.0) <0.001 |
0.954 (0.899–1.0) <0.001 |
|||
| OCD | 0.887 (0.798–0.975) <0.001 |
||||
| Days 7–13 | |||||
| Abbott | 0.774 (0.613–0.935) <0.001 |
0.742 (0.516–0.969) <0.001 |
0.647 (0.388–0.908) <0.001 |
0.568 (0.290–0.846) <0.001 |
0.626 (0.362–0.890) <0.001 |
| Beckman | 0.775 (0.614–0.935) <0.001 |
0.840 (0.701–0.979) <0.001 |
0.673 (0.488–0.857) <0.001 |
0.740 (0.570–0.910) <0.001 |
|
| Roche | 0.702 (0.461–0.943) <0.001 |
0.515 (0.225–0.805) <0.001 |
0.681 (0.433–0.929) <0.001 |
||
| Siemens | 0.519 (0.229–0.810) <0.001 |
0.687 (0.441–0.933) <0.001 |
|||
| OCD | 0.731 (0.500–0.961) <0.001 |
||||
| Days 14–20 | |||||
| Abbott | 0.913 (0.788–1.0) <0.001 |
0.950 (0.843–1.0) <0.001 |
0.946 (0.831–1.0) <0.001 |
0.950 (0.843–1.0) <0.001 |
1.0 (0.868–1.0) <0.001 |
| Beckman | 0.872 (0.721–1.0) <0.001 |
0.870 (0.717–1.0) <0.001 |
0.872 (0.721–1.0) <0.001 |
0.914 (0.788–1.0) <0.001 |
|
| Roche | 0.896 (0.737–1.0) <0.001 |
0.903 (0.755–1.0) <0.001 |
0.950 (0.843–1.0) <0.001 |
||
| Siemens | 0.896 (0.737–1.0) <0.001 |
0.946 (0.831–1.0) <0.001 |
|||
| OCD | 0.950 (0.843–1.0) <0.001 |
||||
| Days ≥21 | |||||
| Abbott | 0.885 (0.749–1.0) <0.001 |
0.913 (0.783–1.0) <0.001 |
0.913 (0.783–1.0) <0.001 |
0.874 (0.717–1.0) <0.001 |
0.837 (0.660–1.0) <0.001 |
| Beckman | 0.963 (0.881–1.0) <0.001 |
0.963 (0.881–1.0) <0.001 |
0.927 (0.819–1.0) <0.001 |
0.892 (0.763–1.0) <0.001 |
|
| Roche | 1.0 (0.884–1.0) <0.001 |
0.961 (0.787–1.0) <0.001 |
0.924 (0.809–1.0) <0.001 |
||
| Siemens | 0.961 (0.787–1.0) <0.001 |
0.924 (0.809–1.0) <0.001 |
|||
| OCD | 0.963 (0.885–1.0) <0.001 |
||||
Values are kappa (95% CI), p value.
False negative rates and patient clinical characteristics
We determined the false negative rate (FNR) of each assay, using the remaining five assays as a composite reference standard (Table 4 ). The lower the percentage, the better the performance of the assay in question. For samples taken >21 days after symptom onset, cPass had the lowest FNR (0%, 95% CI 0.0–11.6%) followed by OCD (3.3%, 95% CI 0.08–17.2%). There was a significant difference of 13.3% (95% CI 1.1–25.4%), p=0.039, between Abbott and cPass. For 14–20 days, OCD and Roche had equal lowest FNR (3.8%, 95% CI 0.10–19.6%). OCD again demonstrated the lowest FNR in the 7–13 days group (8.1%, 95% CI 1.7–21.9%), and this was statistically significant when compared against Siemens (32.4%, 95% CI 18–49.8%), p=0.009 as well as Beckman (31.4%, 95% CI 15.9–47.0%), p=0.010.
Table 4.
False negative rate of each assay
| Assay | Days of symptoms |
|||
|---|---|---|---|---|
| 1–6 (n=80) | 7–13 (n=37) | 14–20 (n=26) | 21–64 (n=30) | |
| Abbott | 7 (8.8)b 3.6–17.2 |
9 (24.3) 11.8–41.2 |
2 (7.7) 0.94–25.1 |
4 (13.3)b 3.8–30.7 |
| Beckmana | 3 (3.9) 0.9–10.6 |
11 (31.4)d 15.9–47.0 |
2 (8.3) 0.94–25.1 |
2 (6.7) 0.82–22.1 |
| Roche | 6 (7.5) 2.8–15.6 |
10 (27.0)e 13.8–44.1 |
1 (3.8) 0.10–19.6 |
2 (6.7) 0.82–22.1 |
| OCD | 1 (1.3)c 0.03–6.8 |
3 (8.1)c 1.7–21.9 |
1 (3.8) 0.10–19.6 |
1 (3.3) 0.08–17.2 |
| Siemens | 5 (6.3) 2.1–14.0 |
12 (32.4)b 18.0–49.8 |
3 (11.5) 2.4–30.1 |
2 (6.7) 0.82–22.1 |
| cPass | 6 (7.5) 2.8–15.6 |
6 (16.2) 6.2–32.0 |
2 (7.7) 0.94–25.1 |
0 (0.0)c 0.0–11.6 |
| Significant differences | ||||
| (b–c) | 7.5% 95% CI 0.8–14.2 p=0.030 |
24.3% 95% CI 6.8–41.8 p=0.009 |
13.3% 95% CI 1.1–25.4 p=0.039 |
|
| (d–c) | 23.3% 95% CI 4.4–38.7 p=0.010 |
|||
| (e–c) | 18.9% 95% CI 2.1–35.7 p=0.033 |
|||
Values are n (%), 95% CI.
Day 1–6: 1 equivocal and 3 insufficient omitted; Day 7–13 and Day 14–20: 1 equivocal and 1 insufficient omitted; Day ≥21: 1 equivocal omitted.
See significant differences section of table.
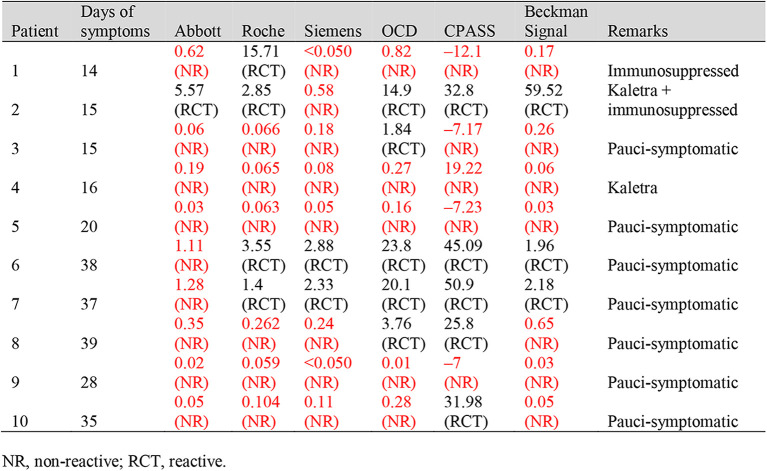
The clinical characteristics and signal values were also reviewed for COVID-19 patients with non-reactive results even for samples taken 14 days after symptom onset. In the 14–20 days group, the poorest performer was Siemens with five non-reactives as compared to the best performer, OCD, with three non-reactives. Patient 1 had end-stage renal disease as a result of IgA nephropathy, previously undergoing renal transplant and was on mycophenolate (Myfortic) immunosuppressant at the time of serology testing. Patients 2 and 4 had significant lymphopenia, required intubation with a course of intensive care unit stay, and were both administered lopinavir-ritonavir (Kaletra). Patient 2 was given hydrocortisone as well. The other two patients in this group were pauci-symptomatic. In the >21 days group, poorest performer Abbott had five non-reactives compared to only one non-reactive in the best performer cPass. All of these five patients were pauci-symptomatic and presented only with a mild acute respiratory illness, dry cough or sore throat. These data are summarised in Table 5 .
Table 5.
Clinical characteristics of patients with false negative serology
Discussion
The SARS-CoV-2 virus is an enveloped RNA consisting of 4 structural proteins known as the spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins. S and N proteins in particular, have been the focus of several targets for serological assays on the basis that previous studies have indicated that they are the most immunogenic antigens.8 , 9 Walls and colleagues have shown coronavirus S protein mediates viral entry into host cells. The S protein consists of two functional subunits namely the S1 subunit which binds to the host cell receptor and S2 subunit which leads to fusion of viral and cellular membranes.10 The S1 subunit comprises the receptor binding domain (RBD) which attaches to the human angiotensin-converting-enzyme 2 (ACE-2), initiating entry into the host cell.11 The N protein, on the other hand, has been described to be highly immunogenic and expressed in large quantities during acute infection of SARS-CoV-2.12 Western blot studies also indicate that the N protein is a potent viral antigen for SARS-CoV-2 on the basis of IgG, IgA and IgM antibodies directed against N protein in COVID-19 positive sera.13
To our knowledge, our study is the first which investigated and compared immunoassays with different design configurations against the S1 subunit, RBD protein, and N antigen, including that of a surrogate viral neutralising antibody (NAb) assay. NAbs typically target the S1-RBD, inhibiting the binding of RBDs to their receptors and hence preventing S2-mediated membrane fusion or entry into the host cell. As a result, viral infection of the host is prevented and presence of such antibodies may confer a status of true immunity.14 Our results show that specificity for all six assays are excellent, and the one false reactive case in Beckman could be potential cross-reactivity of a common coronavirus in the healthy volunteer. In light of the limited sensitivity in the early stages of illness, antibody detection is not sufficiently reliable to act as a routine diagnostic test for COVID-19. This is supported by our previous work and current AUC results, which suggest that serological testing should generally be considered only 14 days after onset of symptoms.15 , 16 If testing is performed prior to this, users should consider the high possibility of false negative results. In this study, the best performer in terms of sensitivity across all days and also at >14 days of symptoms was OCD followed closely by cPass. This is unsurprising because both assay designs are towards the S1 spike protein and they also correlated well based on kappa's concordance. OCD in particular is a total antibody assay directed at IgA, IgM and IgG, hence its diagnostic accuracy outperformed assays raised against just IgG or IgG/IgM. As a a mucosal targeting virus, SARS-CoV-2 generates secretory IgA inducing strong mucosal immunity in the host. Yu and colleagues also showed that IgA antibody produced the earliest seroconversion amongst all antibodies, and was significantly greater than that of IgM in patients with both severe and non-severe illness.17 It would be valuable for vendors to consider providing quantitative signals for individual components to a total antibody assay in the diagnostic evaluation COVID-19. Perhaps disappointingly, Beckman's RBD assay design did not significantly correlate well with cPass but did have a better correlation with Abbott's assay which could be due to both of these assays only targeting IgG. Our cPass NAB results exhibited the best sensitivity in the cohort of patients >21 days, where only one case who was pauci-symptomatic demonstrated negative neutralising antibodies. These results corroborate with findings from Brouwer's group who showed potent neutralising antibody production in a convalescent cohort of patients and served as a marker of immunity.18 Interestingly, quantitative values for cPass ranged from a negative inhibition percentage of –81.1% to that of +91.1%. The negative values may be explained in part by reflecting statistical variation around ‘zero’ inhibition, or true biological enhancements where non-specific antibodies cross-react and bind to the receptors, decreasing binding avidity and hence inhibition.19
Our data provide important considerations for serology testing, and caveats for clinical use. Some patients who had been immunocompromised, given steroids, or some antivirals (two of our patients received a protease inhibitor antiretroviral) may exhibit a down-regulated immune response which manifests as a negative result in some of the immunoassays. Furthermore, pauci-symptomatic patients who were COVID-19 positive on rRT-PCR more frequently failed to have a positive serology on multiple immunoassays, even during the convalescent phase. This suggests that patients with mild symptoms may not display a strong immune response or antibody production, and clinicians need to interpret such cases with caution. Our finding are supported by the study by Long et al. which showed that the virus specific IgG levels in the asymptomatic group were significantly lower compared to symptomatic COVID-19 patients.20
The strength of our study includes using the same cohort of unique, non-duplicate COVID-19 patients' sera to compare performance head-to-head. Certain validation studies use the same patient but collect their sera at different time points for the calculation of sensitivity. This potentially leads to a positive skew in reporting data as an early seroconversion patient likely continues to exhibit reactive serology at later timepoints. A recent study by Public Health England showed the comparison between Abbott, Diasorin, Roche and Siemens, although this only accounted for convalescent patients (≥20 days of symptoms).21 Our study has similar specificity characteristics as described, but further categorises our patient cohort into four different time points, from <7 days to ≥21 days, which provides more detailed information about practical testing and their respective AUCs. Certain limitations in our study were that we did not have archived sera from patients with previous coronaviruses such as SARS-CoV-1 and MERS-CoV to test for cross-reactivity. As this study was approved only for residual sera, low volume samples were insufficient for certain assays to be performed, and these patients were excluded from the final data analysis. Lastly, our COVID-19 positive patients diagnosed via rRT-PCR were all assumed to be true positives only.
Future directions should revolve around testing of rRT-PCR together with serology which will improve overall diagnostic accuracy for COVID-19. The use of CT-values from PCR and quantitative signals generated by serological assays may provide utility regarding infective status and immunity of the patient. Quantitative signal values and their interpretations should be provided by vendors, as reports have shown that greater titres of SARS-CoV-2 antibodies in the early phase of COVID-19 translate to more deleterious outcomes for the patient.22 In such scenarios, serology may provide insight into prognosis and appropriate clinical management. Importantly, it has been suggested that neutralising antibodies directed against SARS-CoV-2 S and RBD proteins confer immunity upon the host and these have been the focus of several vaccine trials.14 Neutralising antibodies potentially trigger immunopathogenic and pro-inflammatory events in the host, hence serological testing may serve as a useful indicator whether immunisation has been acquired.23
Taken together, our report shows excellent specificity for all assays although sensitivities were poorer when compared to the manufacturer's claims. Spike protein designed immunoassays generally outperformed nucleocapsid target immunoassays. Clinicians should interpret negative serology results with caution if the patient is immunocompromised, given antiretrovirals, pauci-symptomatic or has had symptoms less than 14 days.
Acknowledgements
We would like to thank Temasek Holdings Pte Ltd for sponsoring the laboratory testing kits used in this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pathol.2020.09.007.
Conflicts of interest and sources of funding
PT has received grants paid to the National University Hospital from Roche (CP40617, MV40618), Johnson & Johnson (63623872FLZ3001, 63623872FLZ2002), Sanofi Pasteur (H-030-014), GlaxoSmithKline, and Shionogi (1601T0831). All other authors have no conflicts of interest to declare. No external funding was received for this study.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guan W.J., Liang W.H., Zhao Y., et al. Comorbidity and its impact on 1590 patients with COVID-19 in China: a nationwide analysis. Eur Respir J. 2020;55:2000547. doi: 10.1183/13993003.00547-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lippi G., Plebani M. Laboratory abnormalities in patients with COVID-2019 infection. Clin Chem Lab Med. 2020;58:1131–1134. doi: 10.1515/cclm-2020-0198. [DOI] [PubMed] [Google Scholar]
- 4.Hong K.H., Lee S.W., Kim T.S., et al. Guidelines for laboratory diagnosis of coronavirus disease 2019 (COVID-19) in Korea. Ann Lab Med. 2020;40:351–360. doi: 10.3343/alm.2020.40.5.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryant J.E., Azman A.S., Ferrari M.J., et al. Serology for SARS-CoV-2: apprehensions, opportunities, and the path forward. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abc6347. [DOI] [PubMed] [Google Scholar]
- 6.Tan S.S., Yan B., Saw S., et al. Practical laboratory considerations amidst the COVID-19 outbreak: early experience from Singapore. J Clin Pathol. 2020; Mar 20 doi: 10.1136/jclinpath-2020-206563. [DOI] [PubMed] [Google Scholar]
- 7.Zec S., Soriani N., Comoretto R., Baldi I. High agreement and high prevalence: the paradox of Cohen's kappa. Open Nurs J. 2017;11:211–218. doi: 10.2174/1874434601711010211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luchsinger L.L., Ransegnola B., Jin D., et al. Serological analysis of New York City COVID19 convalescent plasma donors. medRxiv. 2020; Jun 9 doi: 10.1101/2020.06.08.20124792. [DOI] [Google Scholar]
- 9.Tai W., He L., Zhang X., et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shang J., Ye G., Shi K., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dutta N.K., Mazumdar K., Gordy J.T. The nucleocapsid protein of SARS-CoV-2: a target for vaccine development. J Virol. 2020;94:e00647-20. doi: 10.1128/JVI.00647-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng W., Liu G., Ma H., et al. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem Biophys Res Commun. 2020;527:618–623. doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chew K.L., Tan S.S., Saw S., et al. Clinical evaluation of serological IgG antibody response on the Abbott Architect for established SARS-CoV-2 infection. Clin Microbiol Infect. 2020;26:1256. doi: 10.1016/j.cmi.2020.05.036. e9–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan S.S., Saw S., Chew K.L., et al. Comparative clinical evaluation of the Roche Elecsys and Abbott SARS-CoV-2 serology assays for COVID-19. Arch Pathol Lab Med. 2020; Sep 9 doi: 10.5858/arpa.2020-0499-SA. [DOI] [PubMed] [Google Scholar]
- 17.Yu H.Q., Sun B.Q., Fang Z.F., et al. Distinct features of SARS-CoV-2-specific IgA response in COVID-19 patients. Eur Respir J. 2020;56:2001526. doi: 10.1183/13993003.01526-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brouwer P.J.M., Caniels T.G., van der Straten K., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu X., Gilbert P.B., Hioe C.E., Zolla-Pazner S., Self S.G. Statistical approaches to analyzing HIV-1 neutralizing antibody assay data. Stat Biopharm Res. 2012;4:1–13. doi: 10.1080/19466315.2011.633860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long Q.X., Liu B.Z., Deng H.J., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 21.Public Health England . Jul 2020. Evaluation of sensitivity and specificity of 4 commercially available SARS-CoV-2 antibody immunoassays.http://www.gov.uk/government/publications/covid-19-head-to-head-laboratory-evaluation-of-4-commercial-serological-assays cited Aug 2020. [Google Scholar]
- 22.French M.A., Moodley Y. The role of SARS-CoV-2 antibodies in COVID-19: healing in most, harm at times. Respirology. 2020;25:680–682. doi: 10.1111/resp.13852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klasse P.J., Moore J.P. Antibodies to SARS-CoV-2 and their potential for therapeutic passive immunization. Elife. 2020;9 doi: 10.7554/eLife.57877. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.