Abstract
This study aimed to investigate the prevalence, antibiogram of Pseudomonas aeruginosa (P. aeruginosa), and the distribution of virulence genes (oprL, exoS, phzM, and toxA) and the antibiotic-resistance genes (blaTEM, tetA, and blaCTX-M). A total of 285 fish (165 Oreochromis niloticus and 120 Clarias gariepinus) were collected randomly from private fish farms in Ismailia Governorate, Egypt. The collected specimens were examined bacteriologically. P. aeruginosa was isolated from 90 examined fish (31.57%), and the liver was the most prominent infected organ. The antibiogram of the isolated strains was determined using a disc diffusion method, where the tested strains exhibited multi-drug resistance (MDR) to amoxicillin, cefotaxime, tetracycline, and gentamicin. The PCR results revealed that all the examined strains harbored (oprL and toxA) virulence genes, while only 22.2% were positive for the phzM gene. On the contrary, none of the tested strains were positive for the exoS gene. Concerning the distribution of the antibiotic resistance genes, the examined strains harbored blaTEM, blaCTX-M, and tetA genes with a total prevalence of 83.3%, 77.7%, and 75.6%, respectively. Experimentally infected fish with P. aeruginosa displayed high mortalities in direct proportion to the encoded virulence genes and showed similar signs of septicemia found in the naturally infected one. In conclusion, P. aeruginosa is a major pathogen of O. niloticus and C. gariepinus. oprL and toxA genes are the most predominant virulence genes associated with P. aeruginosa infection. The blaCTX-M, blaTEM, and tetA genes are the main antibiotic-resistance genes that induce resistance patterns to cefotaxime, amoxicillin, and tetracycline, highlighting MDR P. aeruginosa strains of potential public health concern.
Subject terms: Microbiology, Molecular biology, Diseases
Introduction
Pseudomonads are one of the most threatening fish pathogens that induce ulcerative syndrome and hemorrhagic septicemia1. Various bacterial pathogens affect a wide range of aquatic species and are responsible for considerable economic losses, worldwide. It was estimated that up to 50% of farms fish are lost due to bacterial infection before their marketing. The economic losses are mainly attributed to poor growth, high mortalities, and improper flesh quality2–5. Pseudomonas aeruginosa is a part of normal fish microbiota, however under stressful conditions such as malnutrition and overcrowding the bacteria have become highly opportunistic and pathogenic, causing serious illness including, hemorrhagic septicemia, gill necrosis, abdominal distension, splenomegaly, friable liver, and congested kidney6.
P. aeruginosa is an aerobic Gram-negative motile, non-spore-forming rods, catalase, and oxidase-positive7. Besides, this bacterium possesses many virulence-related determinants including cell-mediated and secreted virulence types. The cell-mediated types include; pilli, flagella, and lipopolysaccharide (LPS) which are commonly involved in bacterial colonization and motility, delivery of active proteins into the host cells, and establishment of persistent infections8. Likewise, the secreted virulence factors fortify the inflammatory processes, induce severe tissue damages, facilitate bacterial invasion and dissemination, and accelerate the progression of diseases9.
Indeed, the identification of Pseudomonas spp. is essential for accurate diagnosis, outbreaks prediction, and preventive and/or prophylactic measures implementation in aquaculture10. Like most of invading pathogens, phenotypic characterization of P. aeruginosa required protracted and complicated steps of morphological and biochemical identifications, which were intractable, time-wasting, and occasionally unreliable11. Therefore, the molecular-based detection of this pathogen could help to obtain a full idea about the ecological importance of such pathogens and rescind the shortcomings of conventional methods12. Most of the Pseudomonas species showed different resistance patterns against various antimicrobial agents13. Specifically, P. aeruginosa could develop innate and/or acquired resistance to several antimicrobial agents due to the active efflux of antibiotics and the permeability of its outer membrane9. Thus, the molecular typing of most inherited antibiotic-resistance genes should be performed to avoid the emergence of antibiotic-resistant strains that have a global health concern14.
Nowadays, serious attempts for further detection of P. aeruginosa were considered not only for its economic impact but also for its public health importance. Pseudomonas infection, generally induced by P. aeruginosa has public health concern and causes healthcare-associated illnesses for many consumers15. Pseudomonas species are presently cataloged as a food-borne illness that affects humans by consuming spoiled foods and ready-to-eat products, as well as manipulating the contaminated seafood16,17. Although few studies have claimed possible routes of disease transmission, people with weakened immune systems or those works with infected fish are usually at risk6. P. aeruginosa infection could occur in humans via the consumption of raw fish and its byproducts18. It is reported that the typical colonies of P. aeruginosa isolated from fish are closely related to those causing hospital-acquired pneumonia in humans17.
The application of the antimicrobial susceptibility testing is ultimately required for disease control. Globally, the extensive use of antimicrobial agents in the aquaculture sector, as well as transmission of multidrug-resistant bacterial pathogens between terrestrial and aquatic ecosystems have been reported19. The antibiotic resistance mechanism mostly occurs due to the transfer of R-Plasmid20. Various reports investigated the occurrence and transmission of antibiotic resistance between human and animal food21, with less attention to fish, so that fewer data are available about the antimicrobial resistance in fish22.
Several virulence factors are commonly associated with P. aeruginosa especially exotoxin A and exotoxin S that are controlled by a cell to cell signaling patterns, in addition, exotoxin A is responsible for the inhibition of protein-biosynthesis, while Exotoxin S is a bi-functional protein that enhances the ribosyltransferase and GTPase activity resulting in cell-apoptosis23. Furthermore, P. aeruginosa produces phenazine compounds, which are biologically active substances involved in bacterial competitiveness and virulence in both human and animal hosts24. The outer membrane proteins (L-lipoproteins) of P. aeruginosa are incriminated in the bacterial resistance to antiseptics and antibiotics25. The resistance of P. aeruginosa to the β-lactam antibiotics including penicillin (1st, 2nd, and 3rd generations) and cephalosporin (such as cefotaxime) is mainly attributed to the Extended Spectrum Beta-lactamases (ESBLs). blaCTX-M and blaTEM are the main Extended Spectrum β-lactamases genes that induce such type of resistance26.
In this context, the present study aimed to identify the phenotypic characteristics of P. aeruginosa strains, retrieved from two freshwater cultured fish species (Oreochromis niloticus and Clarias gariepinus) and their antibiogram. Furthermore, the genotypic characterization of the virulence genes: oprL; outer membrane lipoprotein-L, exoS; exotoxin S gene, phzM; phenazine-modifying gene, and toxA; exotoxin A gene as well as the antibiotic-resistance genes: blaTEM and blaCTX-M; ESBLs genes, and tetA; tetracycline resistance gene was performed to examine their potential for bacterial pathogenicity and probe their potential mechanisms of resistance to commercially available antibiotics.
Methods
Animal ethics
All methods were carried out following relevant guidelines and regulations. Handling of fish and all the experimental protocols carried out by well-trained scientists according to the guidelines of the Animal Ethics Review Committee of Suez Canal University. All used protocols are approved by Suez Canal University, Egypt.
Fish sampling
A total of 285 fish samples represented as 165 O. niloticus (70 apparently healthy and 95 moribunds) and 120 C. gariepinus (55 apparently healthy and 65 moribunds) were collected randomly; (5 sampling points at two months interval) from two private freshwater farms located at Ismailia Governorate, Egypt over the period of July 2018 to February 2019. The farms implemented semi-intensive culture system and applied the same management and treatment programs. Based on the clinical history, the farms routinely used different antibacterial agents such as amoxicillin, cefotaxime, tetracycline, norfloxacin, and gentamicin for regular prophylactic and treatment protocols. Fresh specimens of apparently healthy and moribund fish were transferred alive in aerated plastic bags to the laboratory of Microbiology, Faculty of Veterinary Medicine, Suez Canal University for further clinical and bacteriological examinations. Samples from liver, kidneys, and spleen were collected aseptically following the protocols of Yanong27.
Clinical and postmortem inspections
Fish were clinically examined for the presence of external and internal lesions according to Austin et al.28. Briefly, the fish were examined in a sterile manner using a three-line incision in the case of O. niloticus or a V-shaped incision in the case of C. gariepinus. Antiseptic treatment of the skin was performed using 70% ethyl alcohol. The incision was carried out with sharp pointed scissors, which were introduced into the anus in such manner that the intra-abdominal point remained steady while the cut was made in close contact with the ventral side to avoid internal tissue damage. The abdominal wall was removed; the internal organs were exposed and examined macroscopically for any gross abnormalities.
Isolation and identification of P. aeruginosa
A loopful sample from the collected internal organs was directly streaked onto cetrimide agar and MacConkey's agar (Oxoid, UK), and left incubated at 37 °C for 24 h under aerobic condition. The production of yellowish-green fluorescent pigment is commonly associated with Pseudomonads29. All suspected colonies were harvested and purified for phenotypic and biochemical characteristics. Briefly, all isolates were identified morphologically using Gram's stain and biochemically using various biochemical tests; catalase, oxidase, urease, indole, methyl red, Voges Proskauer, citrate utilization, H2S production, mannitol fermentation, and gelatin hydrolysis, as well as for their motility using hanging drop technique according to Mac Faddin30. Furthermore, the identified isolates were confirmed using a species-specific set of primers (PaF: 5′-GGGGGA TCTTCGGACCTCA-3′; PaSR: 5′-TCCTTAGAGTGCCCACCCG-3′) targeting 16S rRNA gene of P. aeruginosa as described elsewhere by Spilker et al.31.
Antimicrobial susceptibility testing
The susceptibility of the retrieved isolates to different commercial antimicrobial agents (Oxoid), including ofloxacin (5 µg), amoxicillin (10 µg), cefotaxime (30 µg), tetracycline (30 µg), levofloxacin (5 µg), gentamicin (10 µg), norfloxacin (10 µg), tobramycin (10 µg), and colistin sulfate (25 µg) was evaluated using a disc diffusion method32. The selected antimicrobial agents are representatives of the drugs used commonly in the aquaculture sector in Egypt and were selected according to the National Antimicrobial Resistance Monitoring System records. The test was performed using Muller Hinton agar plates (Oxoid, UK) and the plates were incubated at 37 °C for 24 h. The test was conducted according to the instructions of the Clinical Laboratory Standards Institute (CLSI) criteria33.
Molecular typing of the virulence and antibiotic-resistant genes of the isolated P. aeruginosa strains
DNA of purified strains was extracted using a silica-based membrane QIAamp DNA Mini kit (Catalogue no. 51304) according to the manufacturer’s instructions. Genomic DNA templates were quantified using Nanodrop (Nanodrop 1000, Thermo Scientific, UK), adjusted to 100 ng μL−1, and stored at − 20 °C until used for PCR. Ninety representative P. aeruginosa strains (the same strains that were tested for the antimicrobial susceptibility) were tested for the detection of virulence genes, four sets of primers targeting (oprL, exoS, phzM, and toxA) genes were selected based on the previous publications of Xu et al.34, Winstanley et al.35, Finnan et al.36, and Matar et al.37, respectively. Further, to verify the resistance of retrieved strains to the commercially available antibiotics, three sets of primers targeting blaTEM, tetA, and blaCTX-M genes were also selected according to Colom et al.38, Randall et al.39, and Fazeli et al.40, respectively. All primers supplied by (Metabion Company, Germany), and their oligonucleotides sequences and PCR conditions are given in Table 1. PCR reactions (25 μL) were amplified in T100 Gradient Thermocycler (Biometra, Jena, Germany) using EmeraldAmp GT PCR Master Mix (Code No. RR310A, Takara, Japan). A reaction with no template DNA was used as a negative control, while a virulent reference strain of P. aeruginosa (multidrug-resistant to amoxicillin, cefotaxime, tetracycline, and gentamicin), kindly provided by Animal Health Research Institute in Dokki, Cairo, Egypt, was used as positive control. The products were screened by horizontal 1.5% (w/v) agarose gel electrophoresis (AppliChem GmbH, Darmstadt, Germany) and then photographed.
Table 1.
List of oligonucleotide sequences and their PCR conditions used in the current study.
| Target gene | Primer sequences 5′-3' | Amplicon (bp) | PCR conditions | References | ||||
|---|---|---|---|---|---|---|---|---|
| No of cycles | Denaturation | Annealing | Extension | Final Extension | ||||
| oprL | ATG GAA ATG CTG AAA TTC GGC CTT CTT CAG CTC GAC GCG ACG | 504 | 40 | 96 ˚C for 1 min | 55 ˚C for 1 min | 72 ˚C for 1 min | 72˚C for 10 min | 34 |
| toxA | GACAACGCCCTCAGCATCACCAGC CGCTGGCCCATTCGCTCCAGCGCT | 396 | 30 | 94 ˚C for 1 min | 55 ˚C for 1 min | 72 ˚C for 1 min | 37 | |
| exoS | GCGAGGTCAGCAGAGTATCG TTCGGCGTCACTGTGGATGC | 118 | 36 | 94 ˚C for 30 s | 58 ˚C for 30 s | 68 ˚C for 1 min | 35 | |
| phzM | ATGGAGAGCGGGATCGACAG ATGCGGGTTTCCATCGGCAG | 875 | 30 | 94 °C for 30 s | 54 ˚C for 30 s | 72 ˚C for 1 min | 36 | |
| blaTEM | ATCAGCAATAAACCAGC CCCCGAAGAACGTTTTC | 516 | 32 | 94 °C for 30 s | 54 ˚C for 30 s | 72 ˚C for 1 min | 38 | |
| tetA | GGTTCACTCGAACGACGTCA CTGTCCGACAAGTTGCATGA | 576 | 30 | 94 ˚C for 1 min | 56 ˚C for 30 s | 72 ˚C for 1 min | 39 | |
| blaCTX-M |
ATGTGCAGYACCAGTAARGT TGGGTRAARTARGTSACCAGA |
593 | 35 | 95 ˚C for 45 s | 51 ˚C for 45 s | 72 ˚C for 1 min | 40 | |
Pathogenicity test
Acclimation period
A total of 150 healthy O. niloticus weighing 45 ± 3 g with no history of previous infections were collected randomly from Fish Research Center, Suez Canal University, Egypt, and left acclimated in a clean fiberglass tank of 1 m3 holding capacity for 15 days prior to the challenge. Oreochromis niloticus was selected as a model for the present study due to its local availability and ease of cultivation, handling, and transportation. The tank was filled with sand-filtered, UV-sterilized, dechlorinated tap water with an average salinity of 0.3 ± 0.1 g L−1. Dissolved oxygen was monitored at 5 ± 1 mg L−1 using automatic air suppliers (RINA, Genova, Italy), while the water temperature was maintained at 27 ± 0.52 °C. Tank pH was regulated at 7.5 and 13 h light/11 h dark cycle was adopted. Ammonia and nitrite were measured twice a week and never exceeded 0.05 and 0.25 mg L−1, respectively. The fish were fed two times daily (09:00 and 20:00 h) until visual satiety on a commercial pellet of 30% crude protein (Skretting, Alexandria, Egypt). The organic wastes and other debris were siphoned and 30% of the water was replaced daily to reduce the toxicity of ammonia. Fish that showed normal reflexes with no apparent lesions selected for the challenge trial.
Challenge trial
It was performed according to the directive on the protection of animals used for scientific purposes and following the ethical approval sheet mentioned above. One hundred and twenty of acclimated O. niloticus were equally distributed into three groups; each contributed 2 glass aquaria of 80 L and 20 fish holding capacity. The trial was performed in duplicates (n = 2). The fish of the first group (C) were injected intraperitoneally (IP) with 100 µL of sterile phosphate buffer saline and served as a control, while the fish of the other groups (T1 and T2) were injected with 100 µL of the overnight culture of virulent P. aeruginosa strains (A and B, respectively) at a concentration of 3 × 107 cells mL−1 1. The isolates were selected based on the fact that all recovered isolates have identical biochemical and molecular properties. One of these isolates (A) harbored oprL, toxA, and phzM virulent genes, while the other (B) just encoded the oprL and toxA genes. For inocula preparation, bacteria were routinely cultured on tryptic soy broth (Oxoid) at 37 °C for 24 h. Thereafter, the growing bacteria were adjusted to the desired concentration using 0.5 McFarland standards and by the Helber counting chamber. The pathological lesions and cumulative deaths were recorded daily among experimental groups for 14 days post-challenge. Moribund and freshly dead fish were collected, examined immediately to verify the cause of death. Mortalities were considered only when the injected strain was recovered from the experimentally infected fish (Koch’s postulates).
Statistical analysis
The Chi-square was carried out to analyze the data to test the null hypothesis of various treatments using the statistical analysis software (SAS, Software version 9.4, SAS Institute, Cary, NC). The significance level was (P < 0.05).
Results
Clinical and postmortem findings
In the present study, a total of 285 fish samples represented as 165 O. niloticus and 120 C. gariepinus were collected randomly and examined. The clinical inspection revealed that 95 O. niloticus (95/165, 57.5%) and 65 C. gariepinus (65/120, 54.1%) were moribund and showed the signs of hemorrhagic septicemia. Regardless of the fish species, most of the naturally infected fish shared the same typical clinical signs, including hemorrhages on external body surfaces, mainly at the ventral aspect of the abdomen and around the vent (Fig. 1a). Others showed fins erosions, skin darkness, and detached scales (Fig. 1b). Internally, the infected fish showed typical signs of hemorrhagic septicemia represented by pale liver, necrotic gills, engorged spleen, and abdominal dropsy with reddish ascetic exudates (Fig. 2a,b).
Figure 1.
Clinical examination of naturally infected fish (a) catfish (Clarias gariepinus) showing irregular hemorrhages on external body surfaces, especially at the ventral part of the abdomen and around the vent (white arrows), (b) Nile tilapia (Oreochromis niloticus) showing scattered hemorrhagic spots (white arrows), detached scales (*), and fins erosion (black arrows).
Figure 2.
Post-mortem examination of naturally infected fish (a) Nile tilapia (Oreochromis niloticus) showing pale liver (black arrow), necrotic gills (white arrow), engorged spleen (*), serous fluid exudate, and congested kidney (#), (b) catfish (Clarias gariepinus) showing friable liver (#), congested kidneys (white arrows), and engorged spleen (*).
Bacteriological assay
The bacteriological examination revealed that all retrieved isolates were motile, Gram-negative bacilli, arranged in double or short chains. The colonies reacted positively to catalase, oxidase, nitrate reduction, gelatin hydrolysis, citrate utilization, and mannitol fermentation, while they were negative for H2S production, urease, Voges Proskauer, indole, and methyl red. Based on the morphological and biochemical characteristics, all isolates were identified as P. aeruginosa. The typical isolates of P. aeruginosa displayed large irregular colonies with a fruity odor and produced a yellowish-green fluorescent pigment on cetrimide agar (C.A) at 37 °C for 24 h. The bacteria grew on MacConkey's agar and showed flat, smooth, non-lactose fermenting colonies with regular edge and alligator skin like from the top view. Furthermore, all isolates were positive for the PCR amplification of species-specific 16S rRNA gene. The total prevalence of P. aeruginosa among the examined fish was 31.57%. The highest prevalence was recorded in O. niloticus (54/165, 32.72%) followed by C. gariepinus (36/120, 30%). None of the examined apparently healthy fish yielded P. aeruginosa, moreover, P. aeruginosa was isolated from 54 moribund O. niloticus (54/95, 56.8%) and 36 moribund C. gariepinus (36/65, 55.4%). The statistical analysis demonstrated that there was no significant difference in the prevalence of P. aeruginosa in O. niloticus and C. gariepinus (P > 0.05) (Table 2). Regarding the prevalence of P. aeruginosa in various infected organs, the liver was the most prominent infected organ, followed by the kidney and spleen (Table 3). The presence of P. aeruginosa in at least one organ of the fish was considered positive for the bacterium. The statistical analysis showed a significant difference in the prevalence of P. aeruginosa among the internal organs of the examined fish (P < 0.05).
Table 2.
Total prevalence of P. aeruginosa among the examined O. niloticus and C. gariepinus.
| Fish species | No of Examined fish | Positive for P. aeruginosa | |||
|---|---|---|---|---|---|
| No | % | Chi-square value | P-value | ||
| O. niloticus | 165 | 54 | 32.72 | 0.2392 | 0.6248 |
| C. gariepinus | 120 | 36 | 30 | ||
| Total | 285 | 90 | 31.57 | ||
Table 3.
Prevalence of P. aeruginosa in different organs of naturally infected O. niloticus and C. gariepinus.
| Fish species | No of isolates | Organs | Chi-square value | P-value | |||||
|---|---|---|---|---|---|---|---|---|---|
| Liver | Kidney | Spleen | |||||||
| No | % | No | % | No | % | ||||
| O. niloticus | 112 | 54 | 48.2 | 40 | 35.7 | 18 | 16.1 | 26.4643 | < .0001 |
| C. gariepinus | 84 | 36 | 42.8 | 28 | 33.3 | 20 | 23.9 | 6.0952 | 0.0475 |
| Total | 196 | 90 | 45.9 | 68 | 34.7 | 38 | 19.4 | ||
Antibiogram of the isolated P. aeruginosa strains
The antimicrobial susceptibility testing was conducted on 90 representative isolates of P. aeruginosa (one isolate/each infected fish; isolated from the liver). As shown in Table 4, the tested strains showed different resistance patterns to various antimicrobial agents (P < 0.0001). The examined strains were entirely sensitive to colistin sulfate (100%), highly sensitive to norfloxacin (88.89%), while exhibited remarkable resistance to amoxicillin (83.3%), cefotaxime (77.7%), tetracycline (75.6%), and gentamicin (67.6%). Additionally, out of them, 50 strains (50/90, 55.5%) showed multi-drug resistance to four antimicrobial agents; amoxicillin, cefotaxime, tetracycline, and gentamicin (Table 6).
Table 4.
Antimicrobial resistance profiles of P. aeruginosa strains (n = 90) retrieved from naturally infected O. niloticus and C. gariepinus.
| Antimicrobial agents | Concentration (µg) | Sensitive | Intermediate | Resistant | |||
|---|---|---|---|---|---|---|---|
| No | % | No | % | No | % | ||
| Ofloxacin | 5 | 63 | 70 | 19 | 21.11 | 8 | 8.89 |
| Amoxicillin | 10 | 0 | 0 | 15 | 16.7 | 75 | 83.3 |
| Cefotaxime | 30 | 5 | 5.6 | 15 | 16.7 | 70 | 77.7 |
| Tetracycline | 30 | 0 | 0 | 22 | 24.4 | 68 | 75.6 |
| Levofloxacin | 5 | 11 | 12.22 | 66 | 73.33 | 13 | 14.44 |
| Gentamicin | 10 | 0 | 0 | 29 | 32.2 | 61 | 67.6 |
| Norfloxacin | 10 | 80 | 88.89 | 6 | 6.67 | 4 | 4.44 |
| Tobramycin | 10 | 0 | 0 | 70 | 77.8 | 20 | 22.2 |
| Colistin sulfate | 25 | 90 | 100 | 0 | 0 | 0 | 0 |
| Chi-square value | 642.2441 | 276.3880 | 352.3856 | ||||
| P-value | < .0001 | < .0001 | < .0001 | ||||
Table 6.
The distribution of the multi-drug resistance patterns, antimicrobial resistance genes and virulence genes among the tested P. aeruginosa strains (n = 90).
| Number of isolates | Phenotypic resistance patterns | Antimicrobial resistance genes | Virulence genes |
|---|---|---|---|
| 41 | Amoxicillin, cefotaxime, tetracycline, gentamycin | blaTEM, blaCTX-M, tetA | oprL, exoS |
| 9 | Amoxicillin, cefotaxime, tetracycline, gentamycin | blaTEM, blaCTX-M, tetA | oprL, exoS, phzM |
| 11 | Amoxicillin, tobramycin, tetracycline, gentamycin | blaTEM, tetA | oprL, exoS, phzM |
| 5 | Amoxicillin, cefotaxime, tobramycin | blaTEM, blaCTX-M | oprL, exoS |
| 5 | Amoxicillin, cefotaxime, tetracycline | blaTEM, blaCTX-M, tetA | oprL, exoS |
| 4 | Amoxicillin, cefotaxime, tobramycin, ofloxacin, levofloxacin | blaTEM, blaCTX-M | oprL, exoS |
| 4 | Cefotaxime, ofloxacin, Norfloxacin | blaCTX-M | oprL, exoS |
| 2 | Cefotaxime, tetracycline | blaCTX-M, tetA | oprL, exoS |
| 9 | Levofloxacin | – | 0prL, exoS |
Molecular typing of the virulence and antibiotic-resistance genes of the isolated P. aeruginosa strains
Several antibiotic-resistance and virulence genes associated with a natural outbreak of P. aeruginosa were selected based on the previous publications and were examined in ninety representative P. aeruginosa strains (the same strains that were tested for the antimicrobial susceptibility). The results showed that all tested strains harbored oprL gene (100%) with specific amplicons size of 504 bp (Fig. 3, Table 5), as well as toxA gene (100%) with amplicons size of 396 bp (Fig. 4, Table 5), while 20 strains (22.2%) were positive to the phzM gene with fragments size of 875 bp (Fig. 5, Table 5). On the contrary, none of the tested P. aeruginosa strains were positive for exoS gene (Fig. 6, Table 5). The statistical analysis revealed a significant difference in the prevalence of various virulence genes among the tested strains (P < 0.0001).
Figure 3.
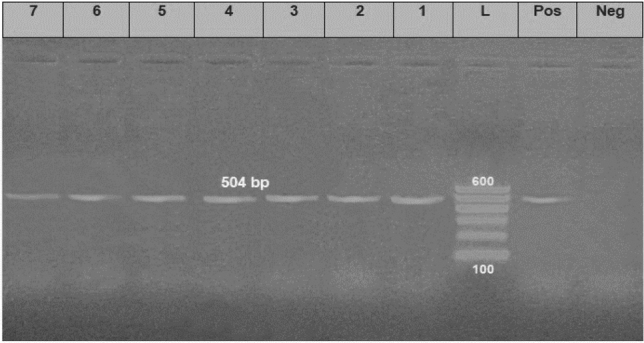
Electrophoretic pattern of primers targeting oprL gene. Lane L: 100 bp molecular weight ladder (Fermentas, Thermo Scientific, Germany); lane Pos: the positive control (Reference strain from Animal Health Research Institute in Dokki, Cairo, Egypt), and lane Neg: the negative control (template without DNA). Lanes 1–7: the specific DNA product (504 bp) amplified from representative isolates of P. aeruginosa.
Table 5.
The prevalence of virulence and antibiotic resistance genes among isolated P. aeruginosa strains from naturally infected O. niloticus and C. gariepinus.
| Gene function | Gene acronym | Prevalence | Statistical analysis | |
|---|---|---|---|---|
| No | % | |||
| Virulence genes | oprL | 90 | 100 |
Chi-square value = 297.0 P value < .0001 |
| toxA | 90 | 100 | ||
| exoS | 0 | 0 | ||
| phzM | 20 | 22.2 | ||
|
Antibiotic-resistance Genes |
blaTEM | 75 | 83.3 |
Chi-square value = 2.4415 P-value 0.2950 |
| tetA | 68 | 75.6 | ||
| blaCTX-M | 70 | 77.7 | ||
Figure 4.
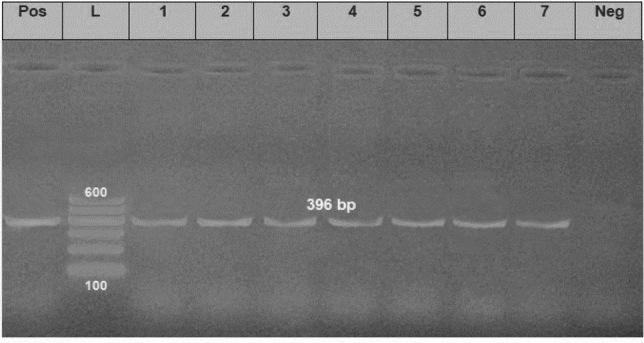
Electrophoretic pattern of primers targeting toxA gene. Lane L: 100 bp molecular weight ladder (Fermentas, Thermo Scientific, Germany); lane Pos: the positive control (Reference strain from Animal Health Research Institute in Dokki, Cairo, Egypt), and lane Neg: the negative control (template without DNA). Lanes 1–7: the specific DNA product (396 bp) amplified from representative isolates of P. aeruginosa.
Figure 5.
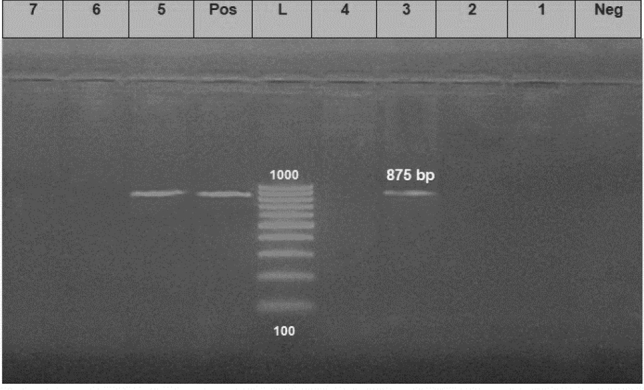
Electrophoretic pattern of primers targeting phzM gene. Lane L: 100 bp molecular weight ladder (Fermentas, Thermo Scientific, Germany); lane Pos: the positive control (Reference strain from Animal Health Research Institute in Dokki, Cairo, Egypt), and lane Neg: the negative control (template without DNA). Lanes 1–7: the specific DNA product (875 bp) amplified from representative strains of P. aeruginosa.
Figure 6.
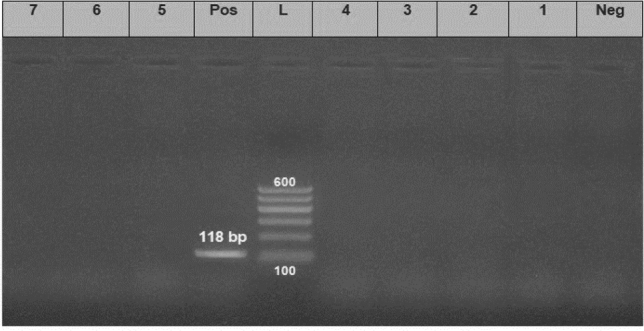
Electrophoretic pattern of primers targeting exoS gene. Lane L: 100 bp molecular weight ladder (Fermentas, Thermo Scientific, Germany); lane Pos: the positive control (Reference strain from Animal Health Research Institute in Dokki, Cairo, Egypt), and lane Neg: the negative control (template without DNA). Lanes 1–7: the specific DNA product (118 bp) amplified from representative strains of P. aeruginosa.
Concerning the distribution of the antibiotic resistance genes, 75 strains (83.3%) were positive for the blaTEM gene with a specific amplicon size of 516 bp, while 68 strains (75.6%) harboring tetA gene with a specific amplicon size of 576 bp, in addition, 70 strains (77.7%) harboring the blaCTX-M gene and gave specific amplicon size of 593 bp (Table 5, Figs. 7,8 and 9).
Figure 7.
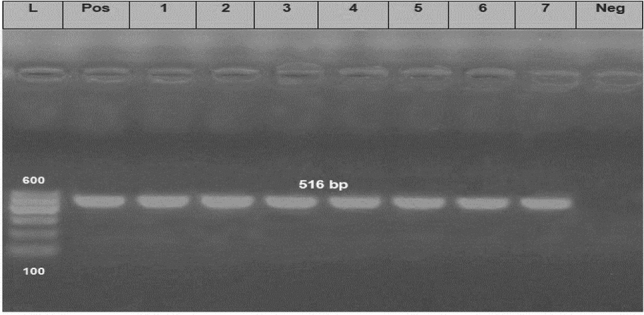
Electrophoretic pattern of primers targeting blaTEM gene. Lane L: 100 bp molecular weight ladder (Fermentas, Thermo Scientific, Germany); lane Pos: the positive control (Reference strain from Animal Health Research Institute in Dokki, Cairo, Egypt), and lane Neg: the negative control (template without DNA). Lanes 1–7: the specific DNA product (516 bp) amplified from representative isolates of P. aeruginosa.
Figure 8.
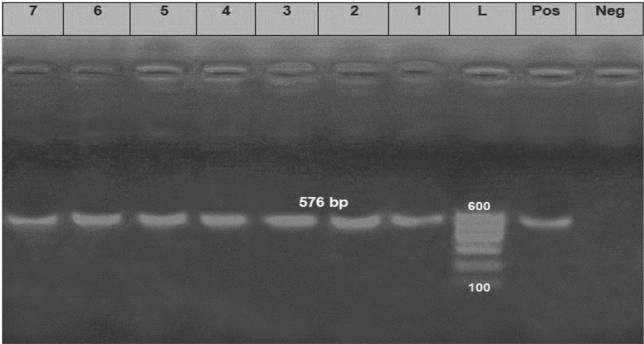
Electrophoretic pattern of primers targeting tetA gene. Lane L: 100 bp molecular weight ladder (Fermentas, Thermo Scientific, Germany); lane Pos: the positive control (Reference strain from Animal Health Research Institute in Dokki, Cairo, Egypt), and lane Neg: the negative control (template without DNA). Lanes 1–7: the specific DNA product (576 bp) amplified from representative strains of P. aeruginosa.
Figure 9.
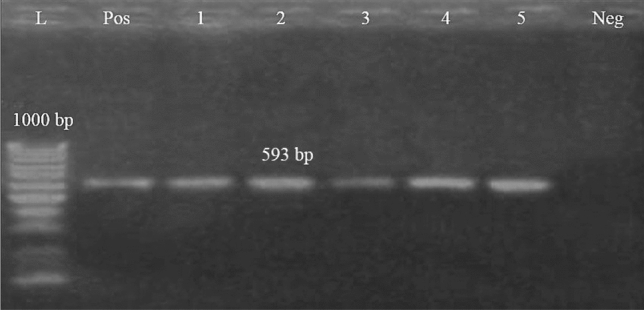
Electrophoretic pattern of primers targeting blaCTM gene. Lane L: 100 bp molecular weight ladder (Fermentas, Thermo Scientific, Germany); lane Pos: the positive control (Reference strain from Animal Health Research Institute in Dokki, Cairo, Egypt), and lane Neg: the negative control (template without DNA). Lanes 1–5: the specific DNA product (593 bp) amplified from representative strains of P. aeruginosa.
In the present study, as illustrated in Tables 4 and 6; the isolated strains exhibited a remarkable resistance to amoxicillin (83.3%), cefotaxime (77.7%) tetracycline (75.6%), and gentamicin (67.6%). Furthermore, Fifty strains (50/90, 55.5%) showed multi-drug resistance to four antimicrobial agents: amoxicillin, cefotaxime, tetracycline, and gentamicin and harbored blaTEM, blaCTX-M, and tetA antimicrobial resistance genes. Out of which, forty-one strains harbored two virulence genes (oprL and toxA), while nine strains harbored three virulence genes (oprL, toxA, and phzM). The statistical analysis revealed a non-significant difference in the prevalence of various antimicrobial-resistance genes among the tested strains (P > 0.05). The distribution of the multi-drug resistance patterns, antimicrobial resistance genes, and virulence genes among the tested P. aeruginosa strains (n = 90) illustrated in Table 6.
Pathogenicity (Challenge trial)
The clinical signs as well as fish morbidity and mortality were recorded in all experimental groups for 14 days post-challenge. The results demonstrated that the fish of the control group did not reveal any mortalities or pathological lesions, while those of the other groups displayed high mortalities and pathognomonic lesions of hemorrhagic septicemia, similar to those reported in naturally infected fish. It was noted that cumulative deaths were relatively associated with encoded virulence genes (Fig. 10), where the maximum mortality rate (87.5%) was recorded in T1 group, followed by T2 group (67.5%). Indeed, O. niloticus inoculated with a virulent strain A produced higher mortalities in a shorter time compared to those exposed to strain B. Approximately, 87.5% of infected fish in T1 group died within 7 days post-inoculation, while those of T2 group showed delayed mortality (67.5%) up to 12 days post-inoculation. Clinically, most of the experimentally infected fish showed exophthalmia, abdominal ascites, hemorrhagic gills, detached scales, and skin ulcers. The postmortem findings revealed that the challenged fish displayed typical signs of septicemia manifested by congested liver, enlarged spleen, and accumulation of serous bloody fluid in the abdominal cavity. In terms of bacteriology, P. aeruginosa was successfully isolated from skin ulcers and internal organs of dead and moribund fish, and the results confirmed that all isolates belong to P. aeruginosa based on biochemical characteristics and molecular typing.
Figure 10.
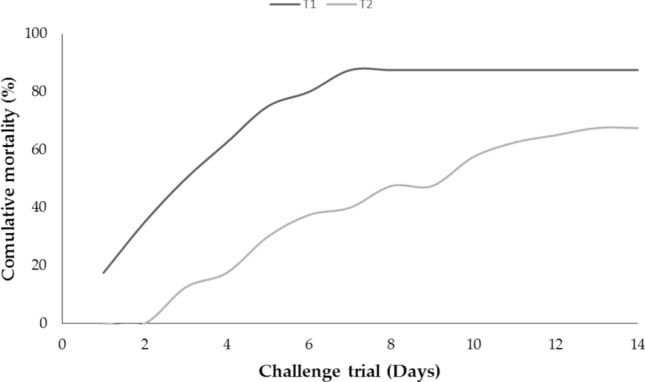
Cumulative mortality of O. niloticus subjected to intraperitoneal injection with 0.1 ml of virulent P. aeruginosa strain A (T1) and P. aeruginosa strain B (T2) at concentration of 3 × 107 cells mL-1.
Discussion
Pseudomonas aeruginosa is a ubiquitous pathogen and is one of the principal causes of septicemia in freshwater fish, resulting in tremendous economic losses in fish producing sectors worldwide41. In the present study, a total of 285 fish samples (165 O. niloticus and 120 C. gariepinus) were collected randomly from private freshwater farms at Ismailia Governorate, Egypt, for clinical and bacteriological examinations. The results of clinical inspection revealed that 95 O. niloticus (95/165, 57.5%) and 65 C. gariepinus (65/120, 54.1%) were moribund and exhibited the typical signs of hemorrhagic septicemia, skin ulcerations, and fin rots. Moreover, internally the infected fish showed pale liver, engorged spleen, and abdominal dropsy with reddish ascetic exudates. These findings agreed with those obtained by Eissa et al.1 and Magdy et al.42 who reported that the postmortem findings due to P. aeruginosa infection were ascites, hepatic and renal necrosis, and congestion of all internal organs. The disease induced by P. aeruginosa is mainly associated with exophthalmia, skin discoloration, hemorrhage, detached scales, and abdominal distension43. Concerning the phenotypic characteristics of P. aeruginosa, all the isolated strains exhibited the typical phenotypic characteristics, culture characters, and biochemical characteristics of P. aeruginosa. These findings are nearly similar to those obtained by Aprameya44.
In the present study, P. aeruginosa isolated from the examined fish with a prevalence of 31.57%. The highest prevalence recorded in O. niloticus (32.72%) followed by C. gariepinus (30%), in agreement with Magdy et al.42 who recorded that the prevalence of P. aeruginosa in O. niloticus and C. gariepinus was 34.4% and 27.5%, respectively. Interestingly, none of the apparently healthy fish yielded P. aeruginosa, while the bacteria recovered from all moribund O. niloticus and C. gariepinus with a total prevalence of 56.8% and 55.4%, respectively. Regarding the distribution of P. aeruginosa in various internal organs, the liver was the most prominent infected organ, followed by the kidney and spleen, which is consistent with those reported by Eissa et al.1. Variations in prevalence could be related to the geographical distribution, environmental factors, host susceptibility, and the season of sample collection. Regarding the antimicrobial susceptibility testing, the examined strains were sensitive to colistin sulfate (100%), followed by norfloxacin (88.89%), while showed remarkable resistance to amoxicillin (83.3%), cefotaxime (77.7%) tetracycline (75.6.8%), and gentamicin (67.6%). Furthermore, Fifty strains (50/90, 55.5%) showed multi-drug resistance to four antimicrobial agents: amoxicillin, cefotaxime, tetracycline, and gentamicin and harbored blaTEM, blaCTX-M, and tetA antimicrobial resistance genes. Out of which, forty-one strains harbored two virulence genes (oprL and toxA), while nine strains harbored three virulence genes (oprL, toxA, and phzM). These findings consistent with those recorded by Eid et al.45 and Nasreen et al.46 who reported the resistance to tetracycline and gentamycin. Globally, several antimicrobial agents are frequently used for the treatment and/ or prevention of fish bacterial diseases. The indiscriminate use of antibiotics, as well as the emerging antibiotic resistance genes, could result in the occurrence of multi-drug resistant (MDR) strains47–50. Therefore, the routine application of antibiotic sensitivity testing is significant to select the specifically effective antibiotic and overcome such a problem51–53.
In the present study, the PCR results revealed that all the tested strains were positive for oprL and toxA genes, in agreement with Kenneth54. L- Lipoproteins refer to the outer membrane proteins associated with P. aeruginosa that enable the microorganism to resist the antiseptics and variable antimicrobial agents. L- lipoproteins are restricted to Pseudomonads, so it could be a reliable target used in both identification and virulence determination of Pseudomonads in clinical specimens55. Exotoxin A is an extracellular product of virulent P. aeruginosa that is encoded by the toxA gene in their chromosome. It acts by inhibition of protein-biosynthesis in the host cell, resembling the action of diphtheria toxin56.
On the contrary, none of the tested strains was positive for the exoS gene, while 20 strains were positive to the phzM gene. These results agreed with those recorded by Nowroozi et al.57. The presence of the phzM gene in 22.2% of the examined strains indicates their ability to produce a phenazine toxin, which in turn enhances their survival rate and colonization to the host even under adverse environmental circumstances58,59. Concerning the distribution of the antibiotic-resistance genes: the examined strains harbored blaTEM, blaCTX-M, and tetA genes with a total prevalence of 83.3%, 77.7%, and 75.6%, respectively, in agreement with Ndi and Barton60, and Ishida et al.61. The presence of these genes explains the phenotypic resistance of the tested P. aeruginosa strains to cefotaxime, amoxicillin, and tetracycline62.
Regarding the pathogenicity trial, fish challenged with P. aeruginosa provided high mortality rates in direct proportion to the encoded virulence genes and showed similar signs of septicemia found in the naturally infected one. Our results are in a good agreement with Magdy et al.42 who noticed marked histopathological alterations in C. gariepinus experimentally challenged with P. aeruginosa. The present findings paralleled with those reported by Devakumar et al.63 who observed degenerative changes in all tissues (brain, ovary, liver, and gills) of crabs infected with P. aeruginosa. Likewise, Derwa et al.64 demonstrated that O. niloticus experimentally infected with P. aeruginosa clinically suffered from exophthalmia, scale losses, skin ulcerations, and external hemorrhages over all the body surfaces and at the base of fins, while, internally they showed a distended gall bladder, congestion, and enlargement of liver, spleen, and kidneys. The degenerative changes could be related to the fatal and subversive influence of bacterial toxins, enzymes, and bioactive extracellular components, which promote tissue damage and cell necrosis23. The elaboration of exotoxins: groups of proteins, is the most crucial factor in P. aeruginosa pathogenicity. The exotoxins are proven to induce liver necrosis, hemorrhage, and renal nephrosis65.
In conclusion, P. aeruginosa is one of the major recurrent emerging pathogens frequently isolated from O. niloticus and C. gariepinus. The recovery of multi-drug resistant (MDR) strains of P. aeruginosa gave a warning to the potential and proper use of antibiotics. The most frequent antibiotic-resistance genes associated with P. aeruginosa isolated from fish are blaTEM, tetA, and blaCTX-M genes that induced resistance patterns to amoxicillin, tetracycline, and cefotaxime, respectively. Routine application of the antimicrobial susceptibility testing is necessary to prevent the emergence of antibiotic-resistant strains of potential public health concern. oprL and toxA genes are the most predominant virulence genes associated with P. aeruginosa.
Acknowledgements
"The authors are grateful to the Researchers Supporting Project number (RSP-2020/53), King Saud University, Riyadh, Saudi Arabia".
Author contributions
A.M.A. designed the study and drafted the manuscript. A.M.A., M.A.M., F.M.Y., W.N.H., H.F.H., A.W.E., and M.H.A. carried out the experiments; A.M.A., M.A.M., E.S., A.W.E., H.F.H., and W.N.H. performed the data analysis, data accuracy, validation. A.M.A., W.N.H., H.F.H., M.A.M., and E.S. did the investigation, supervision, and the statistical analysis; A.M.A. and M.A.M. Writing, Review & Editing. All authors have read and agreed to the published version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Eissa N, El-Ghiet E, Shaheen A, Abbass A. Characterization of Pseudomonas species isolated from tilapia “Oreochromis niloticus” in Qaroun and Wadi-El-Rayan lakes, Egypt. Global Veterinaria. 2010;5:116–121. [Google Scholar]
- 2.Algammal AM, et al. Molecular typing, antibiogram and PCR-RFLP based detection of Aeromonas hydrophila complex isolated from Oreochromis niloticus. Pathogens. 2020;9:238. doi: 10.3390/pathogens9030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aly, S. M. A review of fish diseases in the Egyptian aquaculture sector: Working report. (2013).
- 4.Enany ME, et al. Molecular typing and evaluation of Sidr honey inhibitory effect on virulence genes of MRSA strains isolated from catfish in Egypt. Pak. J. Pharm. Sci. 2018;31:1856–1870. [PubMed] [Google Scholar]
- 5.El-Sayed M, Algammal A, Abouel-Atta M, Mabrok M, Emam A. Pathogenicity, genetic typing, and antibiotic sensitivity of Vibrio alginolyticus isolated from Oreochromis niloticus and Tilapia zillii. Rev. Med. Vet. 2019;170:80–86. [Google Scholar]
- 6.Ardura A, Linde AR, Garcia-Vazquez E. Genetic detection of Pseudomonas spp. in commercial amazonian fish. Int. J. Environ. Res. Public Health. 2013;10:3954–3966. doi: 10.3390/ijerph10093954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uğur A, Ceylan Ö, Aslım B. Characterization of Pseudomonas spp. from seawater of the southwest coast of Turkey. J. Biodivers. Environ. Sci. 2012;6:15–23. [Google Scholar]
- 8.Zeng, L. Pseudomonas aeruginosa pathogenicity and antibiotic resistance, University of Florida, (2004).
- 9.Mesquita CS, Soares-Castro P, Santos PM, Mendez-Vilas A. Pseudomonas aeruginosa: phenotypic flexibility and antimicrobial resistance. Microbial pathogens and strategies for combating them: science, technology and education. 2013;1:650–665. [Google Scholar]
- 10.Fadel A, Mabrok M, Aly S. Epizootics of Pseudomonas anguilliseptica among cultured seabream (Sparus aurata) populations: Control and treatment strategies. Microb. Pathog. 2018;121:1–8. doi: 10.1016/j.micpath.2018.04.021. [DOI] [PubMed] [Google Scholar]
- 11.Serracca L, et al. Vibrio virulence genes in fishes collected from estuarine waters in Italy. Lett. Appl. Microbiol. 2011;53:403–408. doi: 10.1111/j.1472-765X.2011.03119.x. [DOI] [PubMed] [Google Scholar]
- 12.Tripathy S, et al. Characterisation of Pseudomonas aeruginosa isolated from freshwater culture systems. Microbiol. Res. 2007;162:391–396. doi: 10.1016/j.micres.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 13.Quinn PJ, et al. Veterinary Microbiology and Microbial Disease. Hoboken: Wiley; 2011. [Google Scholar]
- 14.Dalmasso A, et al. Development of a PCR assay targeting the rpoA gene for the screening of Vibrio genus. Food Anal. Methods. 2009;2:317. doi: 10.1007/s12161-009-9089-9. [DOI] [Google Scholar]
- 15.Zilberberg, M. D. & Shorr, A. F. Seminars in Respiratory and Critical Care Medicine. 010–015 (© Thieme Medical Publishers).
- 16.Gram, L. & Huss, H. H. The microbiological safety and quality of food 472–506 (Aspen Publishers, 2000).
- 17.Gram L, et al. Food spoilage—interactions between food spoilage bacteria. Int. J. Food Microbiol. 2002;78:79–97. doi: 10.1016/S0168-1605(02)00233-7. [DOI] [PubMed] [Google Scholar]
- 18.Novotny L, Dvorska L, Lorencova A, Beran V, Pavlik I. Fish: a potential source of bacterial pathogens for human beings. A review. Vet. Med.–Czech. 2006;49:343–358. doi: 10.17221/5715-VETMED. [DOI] [Google Scholar]
- 19.Cabello FC. Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ. Microbiol. 2006;8:1137–1144. doi: 10.1111/j.1462-2920.2006.01054.x. [DOI] [PubMed] [Google Scholar]
- 20.Sørum, H. in Antimicrobial resistance in bacteria of animal origin 213–238 (American Society of Microbiology, 2006).
- 21.van den Bogaard AE, Stobberingh EE. Epidemiology of resistance to antibiotics: links between animals and humans. Int. J. Antimicrob. Agents. 2000;14:327–335. doi: 10.1016/S0924-8579(00)00145-X. [DOI] [PubMed] [Google Scholar]
- 22.Biyela P, Lin J, Bezuidenhout C. The role of aquatic ecosystems as reservoirs of antibiotic resistant bacteria and antibiotic resistance genes. Water Sci. Technol. 2004;50:45–50. doi: 10.2166/wst.2004.0014. [DOI] [PubMed] [Google Scholar]
- 23.Strateva T, Mitov I. Contribution of an arsenal of virulence factors to pathogenesis of Pseudomonas aeruginosa infections. Ann. Microbiol. 2011;61:717–732. doi: 10.1007/s13213-011-0273-y. [DOI] [Google Scholar]
- 24.Mavrodi DV, et al. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 2001;183:6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikbin V, et al. Molecular identification and detection of virulence genes among Pseudomonas aeruginosa isolated from different infectious origins. Iran. J. Microbiol. 2012;4:118. [PMC free article] [PubMed] [Google Scholar]
- 26.Peymani A, Naserpour-Farivar T, Zare E, Azarhoosh K. Distribution of blaTEM, blaSHV, and blaCTX-M genes among ESBL-producing P. aeruginosa isolated from Qazvin and Tehran hospitals, Iran. J. Prev. Med. Hygiene. 2017;58:E155. [PMC free article] [PubMed] [Google Scholar]
- 27.Yanong, R. P. Seminars in Avian and Exotic Pet Medicine. 89–105 (Elsevier).
- 28.Austin, B. & Austin, D. Characteristics of the pathogens: Gram-negative bacteria. Bacterial Fish Pathogens: Diseases of Farmed and Wild Fish, 81–150 (2007).
- 29.Lamont IL, Martin LW. Identification and characterization of novel pyoverdine synthesis genes in Pseudomonas aeruginosa. Microbiology. 2003;149:833–842. doi: 10.1099/mic.0.26085-0. [DOI] [PubMed] [Google Scholar]
- 30.Mac Faddin, J. F. Media for isolation-cultivation-identification-maintenance of medical bacteria. (Williams & Wilkins, 1985).
- 31.Spilker T, Coenye T, Vandamme P, LiPuma JJ. PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J. Clin. Microbiol. 2004;42:2074–2079. doi: 10.1128/JCM.42.5.2074-2079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karatuna O, Yagci A. Analysis of quorum sensing-dependent virulence factor production and its relationship with antimicrobial susceptibility in Pseudomonas aeruginosa respiratory isolates. Clin. Microbiol. Infect. 2010;16:1770–1775. doi: 10.1111/j.1469-0691.2010.03177.x. [DOI] [PubMed] [Google Scholar]
- 33.CLSI, C. Performance standards for antimicrobial susceptibility testing. Clinical Lab Standards Institute (2016).
- 34.Xu J, Moore JE, Murphy PG, Millar BC, Elborn JS. Early detection of Pseudomonas aeruginosa–comparison of conventional versus molecular (PCR) detection directly from adult patients with cystic fibrosis (CF) Ann. Clin. Microbiol. Antimicrob. 2004;3:21. doi: 10.1186/1476-0711-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winstanley C, et al. Genotypic and phenotypic characteristics of Pseudomonas aeruginosa isolates associated with ulcerative keratitis. J. Med. Microbiol. 2005;54:519–526. doi: 10.1099/jmm.0.46005-0. [DOI] [PubMed] [Google Scholar]
- 36.Finnan S, Morrissey JP, O’gara F, Boyd EF. Genome diversity of Pseudomonas aeruginosa isolates from cystic fibrosis patients and the hospital environment. J. Clin. Microbiol. 2004;42:5783–5792. doi: 10.1128/JCM.42.12.5783-5792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matar GM, Ramlawi F, Hijazi N, Khneisser I, Abdelnoor AM. Transcription levels of pseudomonas aeruginosa exotoxin A gene and severity of symptoms in patients with otitis externa. Curr. Microbiol. 2002;45:350–354. doi: 10.1007/s00284-002-3703-z. [DOI] [PubMed] [Google Scholar]
- 38.Colom K, et al. Simple and reliable multiplex PCR assay for detection of bla TEM, bla SHV and bla OXA–1 genes in Enterobacteriaceae. FEMS Microbiol. Lett. 2003;223:147–151. doi: 10.1016/S0378-1097(03)00306-9. [DOI] [PubMed] [Google Scholar]
- 39.Randall L, Cooles S, Osborn M, Piddock L, Woodward MJ. Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the UK. J. Antimicrob. Chemother. 2004;53:208–216. doi: 10.1093/jac/dkh070. [DOI] [PubMed] [Google Scholar]
- 40.Fazeli N, Momtaz H. Virulence gene profiles of multidrug-resistant Pseudomonas aeruginosa isolated from Iranian hospital infections. Iran. Red Crescent Med. J. 2004;16:1–10. doi: 10.5812/ircmj.15722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts RJ. Fish Pathology. Hoboken: Wiley; 2012. [Google Scholar]
- 42.Magdy I, El-Hady M, Ahmed H, Elmeadawy S, Kenwy A. A contribution on Pseudomonas aeruginosa infection in African catfish (Clarias gariepinus) Res. J. Pharm. Biol. Chem. Sci. 2014;5:575–588. [Google Scholar]
- 43.Altinok I, Kayis S, Capkin E. Pseudomonas putida infection in rainbow trout. Aquaculture. 2006;261:850–855. doi: 10.1016/j.aquaculture.2006.09.009. [DOI] [Google Scholar]
- 44.Aprameya IV. Non-fermenting Gram-negative bacilli (NFGNB) other than Pseudomonas. J. Acad. Clin. Microbiol. 2013;15:59. doi: 10.4103/0972-1282.124588. [DOI] [Google Scholar]
- 45.Eid H, El Tabiy A, Fathy S. Prevalence and molecular characterization of Pseudomonas species isolated from fish markets in port-said. Suez Canal Vet. Med. J. SCVMJ. 2016;21:1–12. doi: 10.21608/scvmj.2016.62742. [DOI] [Google Scholar]
- 46.Nasreen M, Sarker A, Malek M, Ansaruzzaman M, Rahman M. Prevalence and resistance pattern of Pseudomonas aeruginosa isolated from surface water. Adv. Microbiol. 2015;5:74. doi: 10.4236/aim.2015.51008. [DOI] [Google Scholar]
- 47.Abd El-Baky RM, Farhan SM, Ibrahim RA, Mahran KM, Hetta HF. Antimicrobial resistance pattern and molecular epidemiology of ESBL and MBL producing Acinetobacter baumannii isolated from hospitals in Minia, Egypt. Alexandria J. Med. 2020;56:4–13. doi: 10.1080/20905068.2019.1707350. [DOI] [Google Scholar]
- 48.Abd El-Baky RM, et al. Prevalence and some possible mechanisms of colistin resistance among multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa. Infect. Drug Resist. 2020;13:323. doi: 10.2147/IDR.S238811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Algammal AM, Enany ME, El-Tarabili RM, Ghobashy MOI, Helmy YA. Prevalence, antimicrobial resistance profiles, virulence and enterotoxin-determinant genes of MRSA isolated from subclinical bovine mastitis samples in Egypt. Pathogens. 2020;9:1–11. doi: 10.3390/pathogens9050362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farhan SM, Ibrahim RA, Mahran KM, Hetta HF, El-Baky RMA. Antimicrobial resistance pattern and molecular genetic distribution of metallo-β-lactamases producing Pseudomonas aeruginosa isolated from hospitals in Minia, Egypt. Infect. Drug Resist. 2019;12:2125–2131. doi: 10.2147/IDR.S198373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Enany ME, et al. The occurrence of the multidrug resistance (MDR) and the prevalence of virulence genes and QACs resistance genes in E. coli isolated from environmental and avian sources. AMB Express. 2019;9:192. doi: 10.1186/s13568-019-0920-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Algammal AM, et al. Genes encoding the virulence and the antimicrobial resistance in enterotoxigenic and shiga-toxigenic E. coli isolated from diarrheic calves. Toxins (Basel) 2020;12:383. doi: 10.3390/toxins12060383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.El-Mokhtar MA, Hetta HF. Ambulance vehicles as a source of multidrug-resistant infections: a multicenter study in Assiut City, Egypt. Infect. Drug Resist. 2018;11:587. doi: 10.2147/IDR.S151783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kenneth, T. Todar's online textbook of bacteriology. Bacterial Protein Toxins (2011).
- 55.Remans K, Vercammen K, Bodilis J, Cornelis P. Genome-wide analysis and literature-based survey of lipoproteins in Pseudomonas aeruginosa. Microbiology. 2010;156:2597–2607. doi: 10.1099/mic.0.040659-0. [DOI] [PubMed] [Google Scholar]
- 56.Aljebory IS. PCR detection of some virulence genes of Pseudomonas aeruginosa in Kirkuk city, Iraq. J. Pharm. Sci. Res. 2018;10:1068–1071. [Google Scholar]
- 57.Nowroozi J, Sepahi AA, Rashnonejad A. Pyocyanine biosynthetic genes in clinical and environmental isolates of Pseudomonas aeruginosa and detection of pyocyanine’s antimicrobial effects with or without colloidal silver nanoparticles. Cell J. (Yakhteh) 2012;14:7. [PMC free article] [PubMed] [Google Scholar]
- 58.Bradbury RS, Roddam L, Merritt A, Reid DW, Champion AC. Virulence gene distribution in clinical, nosocomial and environmental isolates of Pseudomonas aeruginosa. J. Med. Microbiol. 2010;59:881–890. doi: 10.1099/jmm.0.018283-0. [DOI] [PubMed] [Google Scholar]
- 59.Cezairliyan B, et al. Identification of Pseudomonas aeruginosa phenazines that kill Caenorhabditis elegans. PLoS Pathog. 2013;9:1–9. doi: 10.1371/journal.ppat.1003101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ndi O, Barton M. Resistance determinants of Pseudomonas species from aquaculture in Australia. J. Aquac. Res. Dev. 2012;3:119. [Google Scholar]
- 61.Ishida, Y. et al. Molecular analysis of antimicrobial resistance in Gram-negative bacteria isolated from fish farms in Egypt. J. Vet. Med. Sci. 1002020148–1002020148 (2010). [DOI] [PubMed]
- 62.Cho J-K, Kim J-H, Kim J-M, Park C-K, Kim K-S. Antimicrobial resistance and distribution of resistance gene in Enterobacteriaceae and Pseudomonas aeruginosa isolated from dogs and cats. Korean J. Vet. Serv. 2013;36:171–180. doi: 10.7853/kjvs.2013.36.3.171. [DOI] [Google Scholar]
- 63.Devakumar D, Jayanthi J, Ragunathan M. Herbal alternate to Pseudomonas aerginosa infection in a freshwater crab, OZIOTELPHUSA SENEX SENEX. Int. J. Biol. Pharmacy Allied Sci. (IJBPAS). 2013;2:2142–2147. [Google Scholar]
- 64.Derwa H, AbdElWahab M, Kamal K. Role of some biological pollutants in relation to disease occurrence in Oreochromis Niloticus. Suez Canal Vet. Med. J. SCVMJ. 2017;22:247–257. doi: 10.21608/scvmj.2017.62179. [DOI] [Google Scholar]
- 65.Liu PV. Extracellular toxins of Pseudomonas aeruginosa. J. Infect. Dis. 1974;130:S94–S99. doi: 10.1093/infdis/130.Supplement.S94. [DOI] [PubMed] [Google Scholar]




