Abstract
Purpose
The present study established microRNA (miRNA) expression profiles for rat ovaries displaying polycystic ovary syndrome (PCOS) with insulin resistance and explored the underlying biological functions of differentially expressed miRNAs.
Methods
A PCOS with insulin resistance rat model was created by administering letrozole and a high-fat diet. Total RNA was extracted from the ovaries of PCOS with insulin resistance rats and normal rats. Three ovaries from each group were used to identify differentially expressed miRNAs by deep sequencing. A hierarchical clustering heatmap and volcano plot were used to display the pattern of differentially expressed miRNAs. Gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were conducted to explore the potential target genes of the differentially expressed miRNAs and identify their putative biological function. Nine of the differentially expressed miRNAs were selected for validation by Real-time Quantitative PCR (qRT-PCR).
Results
A total of 58 differentially expressed miRNAs were identified in the rat ovaries exhibiting PCOS with insulin resistance compared with control ovaries, including 23 miRNAs that were upregulated and 35 miRNAs that were downregulated. GO and KEGG pathway analyses revealed that the predicted target genes were related to metabolic processes, cellular processes, and metabolic pathways. Furthermore, qRT-PCR confirmed that miR-3585-5p and miR-30-5p were significantly upregulated and miR-146-5p was downregulated in the ovaries of PCOS with insulin resistance rats compared with the controls.
Conclusion
These results indicate that differentially expressed miRNAs in rat ovaries may be involved in the pathophysiology of insulin resistance in PCOS. Our study may be beneficial in establishing miRNAs as novel diagnostic and therapeutic biomarkers for insulin resistance in PCOS.
Keywords: MicroRNAs, High-throughput sequencing, Polycystic ovary syndrome, Insulin resistance
Introduction
Polycystic ovary syndrome (PCOS), which affects 6–20% of women during reproductive age, is one of the most common endocrine and metabolic disorders [1–3]. According to the Rotterdam criteria, PCOS may be diagnosed with at least two of the three following characteristics: anovulation, clinical and/or biochemical signs of hyperandrogenism and polycystic ovaries [4, 5]. PCOS is also associated with infertility, insulin resistance, obesity, glucose tolerance, dyslipidemia, type 2 diabetes and increased risk for cardiovascular disease [6]. Insulin resistance and hyperandrogenism are not only the two main characteristic features of PCOS, but also important pathogenic factors for PCOS [7]. Furthermore, there are approximately 40–70% of PCOS patients with insulin resistance. Insulin may promote luteinizing hormone release, increase androgen levels by ovarian theca cells, and inhibit the production of sex hormone binding protein by the liver, resulting in elevated levels of circulating free testosterone. Therefore, insulin synergizing with androgen could result in deteriorating anovulation, hyperandrogenism, hyperinsulinemia and infertility in PCOS patients. However, whether insulin resistance is the cause or the result of PCOS remains to be determined. Genetic studies and familial clustering of phenotypic features have indicated that PCOS is heritable [8–11]. Genome-wide association studies have found over 20 PCOS susceptibility genes; however, they account for < 10% of all PCOS cases [12–16]. Therefore, other mechanisms must play a role in the development of PCOS as genetic factors are responsible for only a small portion of the cases. One potential mechanism is an epigenetic process which could yield the same phenotypic characteristics as heritable genetics [17, 18].
MicroRNAs (miRNAs) are small, single-stranded noncoding RNAs consisting of 18–24 nucleotides in length. They regulate post-transcriptional gene expression by binding to the 3′ untranslated region of target RNAs [19]. MiRNAs are involved in many physiological processes including cell growth, differentiation, proliferation and metabolism [20]. Furthermore, several studies have shown that dysfunctional miRNAs are also associated with the pathological mechanism of PCOS [21]. Sang et al. identified highly expressed miRNAs in human follicular fluids [22]. In addition, Chen et al. found that miR-93 is upregulated in the adipose tissue of PCOS patients and women with insulin resistance [23]. In our previous study, we found differentially expressed circular RNAs (circRNAs) in the granulosa cells of reproductive-aged PCOS women [24]. However, there is no literature regarding miRNA expression profiling in the ovaries of PCOS patients with insulin resistance. In the present study, we identified differentially expressed miRNAs from PCOS ovaries in an insulin-resistant rat model and determined their putative function.
Materials and methods
Animals and treatment
All animal experiments were carried out based on the Guidelines for the Care and Use of Experimental Animals and approved by the Animal Ethics Committee of the Guangzhou University of Chinese Medicine (20180715002). Three-week-old female Sprague Dawley rats were purchased from the Animal Centre of Guangdong Province (Guangdong, China). Animals were fed adaptively for 1 week under conditions of 55–65% humidity, 21–22 °C temperature, and a 12 h light/12 h dark cycle. Twenty rats were randomly divided into two groups. Ten rats in the control group (CON) were fed a normal diet (Research Diet, D12450, 10% fat), while 10 rats in the treatment group were fed a high-fat diet (Research Diet, D12492, 60% fat) for 8 weeks (LEHF group). In addition, the con rats were gavaged with 1% carboxymethylcellulose (CMC) solution once daily, and LEHF rats were gavaged with 1 mg/kg letrozole in 1% CMC once daily for 21 days [25]. All rats were weighed each week.
Estrus cycle
The estrus cycle was examined daily. Depending on the results of microscopic analysis of a vaginal smear, the animals were flushed with physiological saline from the 12th day to the end of the experiment.
Blood and tissue sampling
All rats were anesthetized with isoflurane after overnight fasting. Blood samples were collected from the heart and the resulting serum samples were stored at – 80 °C. Serum levels of follicle stimulating hormone (FSH), luteinizing hormone (LH), estradiol (E2), testosterone (T), progesterone (P), fasting blood glucose (FBG) and fasting insulin (FINS) were measured using ELISA kits (Cloud-Clone Corp., Wuhan, China) according to the manufacturer’s protocol. The homeostasis model assessment for insulin resistance (HOMA-IR) was calculated as previously described using the following formula: HOMA-IR = (FBG × FINS) / 22.5 [26].
Ovary samples were quickly collected from the animals, and one ovary from each rat was fixed in 4% paraformaldehyde. The ovary tissue samples were then dehydrated, embedded in paraffin, and sectioned into 4 µm sections. Each section was stained with hematoxylin and eosin (H&E). The other rat ovary was stored at – 80 °C and used to establish miRNA expression profiles.
RNA-sequencing (RNA-seq) and bioinformatics analysis
RNA isolation and RNA sequencing were performed as previously described [24]. Briefly, total RNA was extracted from ovaries using TRIzol reagent according to the manufacturer’s instructions. The concentration of RNA was measured with a NanoDrop 2000 spectrophotometer. An RNA library was generated from total RNA using the NEB Next Ultra Directional RNA Library Prep Kit for Illumina (NEB, MA, USA). The RNA library quality and quantity were assessed with an Agilent 2100 Bioanalyzer and an ABI Step One Plus Real-Time PCR System, respectively. The RNA library was run on a HiSeq 2000 platform (Illumina, CA, USA) for sequence analysis. The clean reads were filtered from raw reads by FastQC and used for further bioinformatics analysis. The differentially expressed miRNAs were selected based on log2 (LEHF/CON) > 1 and P value < 0.05.
Hierarchical clustering heatmaps and volcano plots generated by R package version 1.0.8 software (https://cran.r-project.org/web/packages/pheatmap/) were used to display the differentially expressed miRNA patterns between the two groups. Gene ontology (GO) analysis was performed to explore the molecular function, cellular components and biological processes of the differentially expressed miRNAs (https://www.geneontology.org). The biological pathways of the differentially expressed miRNAs were further analyzed using the Kyoto Encyclopedia of Genes and Genomes (KEGG; https://www.genome.jp/kegg/) database.
Real-time quantitative PCR (qRT-PCR)
To verify the results of RNA sequencing, we measured relative miRNA expressions by qRT-PCR analysis using SYBR Premix Ex Taq II (Takara, China). The U6 gene was used as an internal control. The relative expression of miRNAs was calculated using the comparative Ct (2−ΔΔCT) method [27]. The sequences of primers used in this study are shown in Table 1.
Table 1.
The primer sequences in this study
| Gene name | Primer sequences (5′-3′) |
|---|---|
| rno-miR-3585-5p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACATGAAA |
| rno-miR-3585-5p-F | AACACGCTTCACAAGAAGGTG |
| rno-miR-200a-3p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACATCG |
| rno-miR-200a-3p-F | AACACGCTAACACTGTCTGGT |
| rno-miR-30a-5p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTTCCA |
| rno-miR-30a-5p-F | AACACGCTGTAAACATCCTCG |
| rno-miR-134-5p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCCCTC |
| rno-miR-134-5p-F | AACAGTGTGTGACTGGTTGAC |
| rno-miR-132-3p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCGACCA |
| rno-miR-132-3p-F | AACACGCTAACAGTCTACAGC |
| rno-miR-146b-5p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACAGCC |
| rno-miR-146b-5p-F | AACACGCTGAGAACTGAATTCC |
| rno-miR-21-5p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCAACA |
| rno-miR-21-5p-F | AAGCGACCTAGCTTATCAGACT |
| rno-miR-124-3p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGCATT |
| rno-miR-124-3p-F | TAAGGCACGCGGTGAA |
| rno-miR-122-5p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAAACA |
| rno-miR-122-5p-F | AACCGGTGGAGTGTGACAAT |
| rno-miR-375-3p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCACGC |
| rno-miR-375-3p-F | AACACGCTTTGTTCGTTCGG |
Statistical analysis
All values are presented as the mean ± SD. Comparisons between experimental and control groups were conducted by a Student’s t test. Data analyses were performed using SPSS 19.0 software (SPSS Inc., IL, USA). P value < 0.05 was considered statistically significant.
Results
Animal model of PCOS with insulin resistance
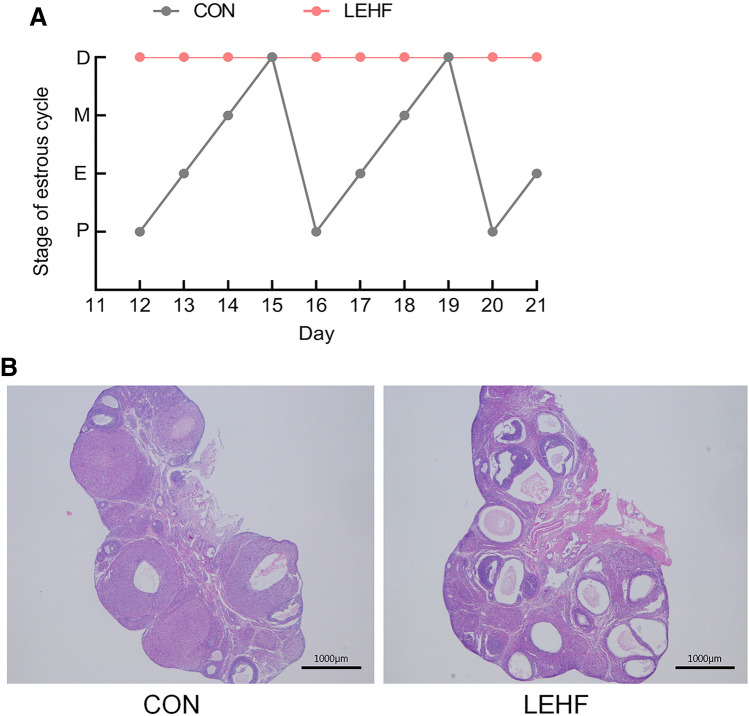
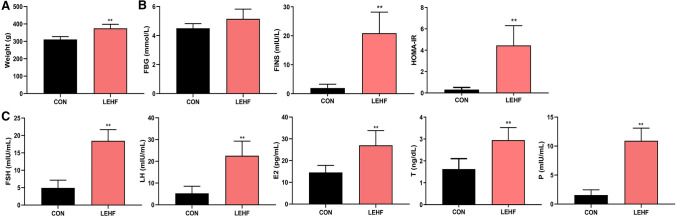
We used letrozole and a high-fat diet to establish a rat model with a PCOS and insulin-resistance phenotype. The CON rats exhibited a normal estrus cycle, whereas the LEHF rats had a disrupted estrous cycle (Fig. 1a). H&E staining results showed that the ovary in the CON group had follicles and corpora lutea at various stages of development, a normal theca and granulosa cell layer and no ovarian cysts. In the LEHF group, the ovary exhibited atretic antral follicles, more large cystic follicles with a thinner granulosa cell layer and fewer corpora lutea (Fig. 1b). Compared with the CON group, the body weight, FINS and HOMA-IR were significantly increased in rats from the LEHF group (Fig. 2a, b). Moreover, rats in the LEHF group exhibited a dramatic increase in the LH, FSH, E2, T and P levels (Fig. 2c). Taken together, these results indicated that we successfully established a rat PCOS model with insulin resistance.
Fig.1.
Change in estrous cycles and morphology of ovaries of rats from the LEHF-induced PCOS with insulin resistance and the groups. a Changes in estrous cycle between the LEHF and CON groups. b Morphological changes in the rat ovarian tissues as detected by H&E. LEHF letrozole and high-fat diet group, D diestrus, P proestrus, E estrus, M metestrus
Fig.2.
Changes in blood sugar and hormone levels between the LEHF and CON groups. a Comparison of weight between the two groups. b The values of FBG, FINS and HOMA-IR were determined for the two groups. c The LH, FSH, E2, T and P levels were determined for the two groups. *P < 0.05; **P < 0.01. FBG fasting blood glucose, FINS fasting insulin, HOMA-IR homeostasis model assessment of insulin resistance, FSH follicle stimulating hormone, LH luteinizing hormone, E2 estradiol, T testosterone, P progesterone
Differentially expressed miRNA profiles in the ovary of the LEHF and CON groups
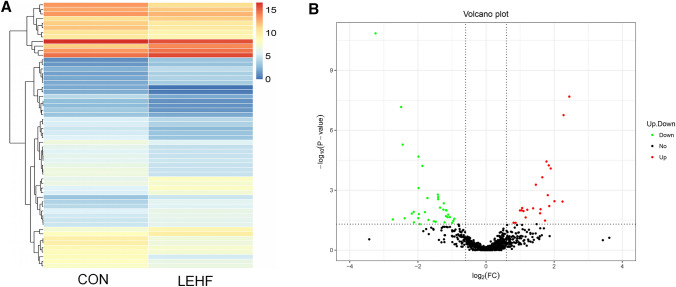
We conducted high-throughput sequencing to establish miRNA expression profiles for three ovary samples from the LEHF and CON groups. A total of 767 miRNAs were detected in the ovary samples. Among these, 58 differentially expressed miRNAs were found between the two groups (fold change ≥ 2 and P value < 0.05). Among the differentially expressed miRNAs, 23 were upregulated, while 35 were significantly downregulated in the LEHF group. The list of the top 10 differentially expressed miRNAs identified by deep sequencing is shown in Table 2. A hierarchical clustering heatmap (Fig. 3a) indicated that the expression of miRNAs in the LEHF group was significantly different from that of the CON group. A volcano plot (Fig. 3b) also displayed the pattern of upregulated and downregulated miRNAs.
Table 2.
The list of the top ten differentially expressed miRNA identified by the deep sequencing
| miRNA_ID | CON | LEHF | Log2 fold change (LEHF/CON) | Up-down-regulation (LEHF/CON) | P value | FDR |
|---|---|---|---|---|---|---|
| rno-miR-509-3p | 46.38242401 | 252.6640226 | 2.445570126 | Up | 2.07E-08 | 7.94E-06 |
| rno-miR-547-3p | 3018.488892 | 14629.32712 | 2.276965017 | Up | 1.75E-07 | 3.36E-05 |
| rno-miR-547-5p | 74.34761696 | 254.435207 | 1.774939909 | Up | 3.67E-05 | 0.004016012 |
| rno-miR-201-5p | 43.16871548 | 154.8170877 | 1.842506645 | Up | 5.72E-05 | 0.005207537 |
| rno-miR-201-3p | 15.06913716 | 56.21809675 | 1.8994378 | Up | 8.21E-05 | 0.006294168 |
| rno-miR-3585-5p | 159.4036017 | 501.4001728 | 1.653278266 | Up | 0.000226462 | 0.015790557 |
| rno-miR-652-3p | 303.3559719 | 837.049714 | 1.464301597 | Up | 0.000532474 | 0.034033945 |
| rno-miR-200c-3p | 4.111515619 | 14.45494958 | 1.813821362 | Up | 0.001779502 | 0.090991889 |
| rno-miR-224-3p | 3.472553993 | 13.9860925 | 2.00992392 | Up | 0.003568794 | 0.142581276 |
| rno-miR-3596b | 1.989015013 | 9.43033931 | 2.245255516 | Up | 0.003717895 | 0.142581276 |
| rno-miR-132-5p | 369.5014638 | 38.99406342 | − 3.244253674 | Down | 1.42E-11 | 1.09E-08 |
| rno-miR-132-3p | 702.1346736 | 124.6887802 | − 2.49341612 | Down | 6.83E-08 | 1.75E-05 |
| rno-miR-146b-5p | 10354.12271 | 1898.054539 | − 2.447611968 | Down | 5.26E-06 | 0.000806138 |
| rno-miR-212-5p | 109.3275286 | 27.6345511 | − 1.984111634 | Down | 2.08E-05 | 0.002654303 |
| rno-miR-292-5p | 308.9159765 | 84.59455768 | − 1.868577729 | Down | 6.11E-05 | 0.005207537 |
| rno-miR-291a-3p | 98.98330743 | 25.07270548 | − 1.98106757 | Down | 0.000785138 | 0.046323121 |
| rno-miR-183-5p | 1512.932233 | 567.7884137 | − 1.413922053 | Down | 0.001674859 | 0.090991889 |
| rno-miR-3553 | 2817.78701 | 1062.511993 | − 1.40708344 | Down | 0.002209313 | 0.105908965 |
| rno-miR-1298 | 83.26407405 | 25.28723408 | − 1.719284907 | Down | 0.002478859 | 0.111840282 |
| rno-miR-219a-2-3p | 65.61026414 | 24.74143541 | − 1.406992327 | Down | 0.002836267 | 0.120856493 |
Fig.3.
Differentially expressed miRNAs were identified by deep sequencing. A hierarchical clustering heatmap (a) and a volcano plot (b) were used to display the differentially expressed miRNA patterns between the two groups
Functional analysis of differentially expressed miRNAs
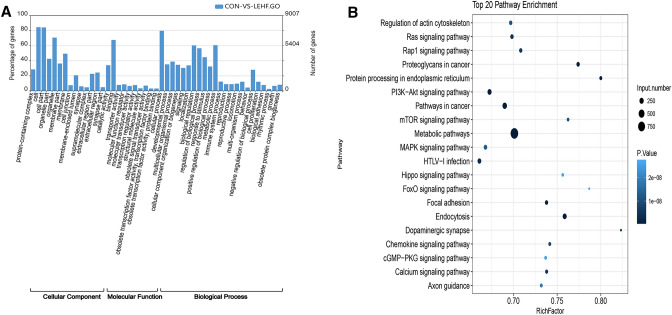
Ovary dysfunction and abnormal follicular development are two characteristics of PCOS that are aggravated by insulin resistance. To gain insight into the underlying mechanism of PCOS with insulin resistance, GO analysis was conducted to identify the biological processes, cellular components and molecular functions of the differentially expressed miRNAs (Fig. 4a). We discovered that the most enriched GO term for the cellular component was cell apart (GO:0044464), the most enriched term for the molecular function was cellular process (GO:0009987) and the most enriched term for the biological process was metabolic process (GO:0008152). Moreover, the metabolic pathway (path: rno01100) was the most enriched pathway in the KEGG database (Fig. 4b). The biological functions and pathways for the differentially expressed miRNAs are closely related to the development of insulin resistance in PCOS.
Fig.4.
Gene ontology (GO) and KEGG pathway analysis for predicted targets of the differentially expressed miRNAs
Validation of candidate miRNAs by qRT-PCR
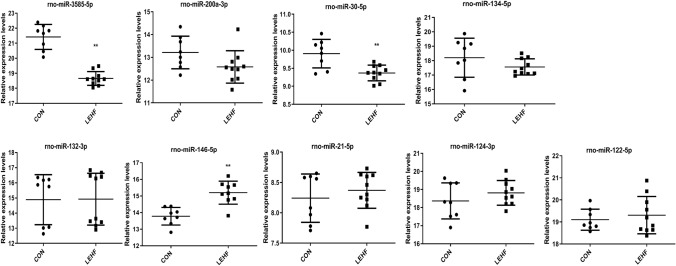
According to the literature and the fold-change of the deep sequencing results, four upregulated miRNAs (miR-3585-5p, miR-200-3p, miR-30-5p and miR-134-3p) and five downregulated miRNAs (miR-132-3p, miR-146-5p, miR-21-5p, miR-124-3p and miR-122-5p) were selected for validation using qRT-PCR [28, 29]. As shown in Fig. 5, qRT-PCR confirmed that compared with the CON group, miR-3585-5p and miR-30-5p were significantly upregulated, while miR-146-5p was downregulated in the rat ovaries of the LEHF group. These results were consistent with the sequencing results. However, two upregulated miRNAs (miR-200-3p and miR-134-3p) and four downregulated miRNAs (miR-132-3p, miR-21-5p, miR-124-3p and miR-122-5p) were not significantly expressed or inconsistently expressed as compared with the sequencing results.
Fig.5.
Validation of the miRNAs in the rat ovaries between the two groups by qRT-PCR. The expression of four upregulated miRNAs (miR-3585-5p, miR-200-3p, miR-30-5p and miR-134-3p) and five downregulated miRNAs (miR-132-3p, miR-146-5p, miR-21-5p, miR-124-3p and miR-122-5p) identified by deep sequencing were determined. *P < 0.05; **P < 0.01
Discussion
PCOS is a complex heterogeneous disorder characterized by hyperandrogenism and insulin resistance [30]. Insulin resistance can cause infertility and increased androgen levels, which exacerbate PCOS traits [31, 32]. In addition, the insulin-sensitizing drug metformin can improve menstrual disorder, promote ovulation, and elevate birth rate in PCOS, especially when accompanied by insulin resistance [33, 34]. Therefore, there should be a strong relationship between PCOS and insulin resistance. However, the underlying mechanism of insulin resistance in PCOS remains unclear. In the present study, we successfully established a model for PCOS with insulin resistance in rats by administrating letrozole and a high-fat diet to the rats. Letrozole is an aromatase inhibitor that blocks the conversion of androgen to estrogen. This results in elevated androgen levels, which is the main pathogenetic mechanism occurring in PCOS. Pubertal female rats were continuously treated with letrozole, inducing increased LH and T levels, abnormal estrous cycle, anovulation, increased ovarian weight, cysts, atretic follicles and no CL in adulthood [35, 36]. Furthermore, a high-fat diet in rats induces abnormal metabolism, including increased body weight, impaired glucose tolerance, and insulin resistance [37]. Thus, letrozole and a high-fat diet in rats contain both the reproductive and metabolic features associated with PCOS and insulin resistance.
Several miRNA, long-non-coding-RNA and circRNA expression profiling studies of PCOS have been conducted using different human components including serum, adipose tissue, uterine tissue, and follicular fluid [24, 38, 39]. Studies have implicated miRNAs in the pathogenesis of PCOS, including increased androgen [38, 39]. However, the exact role of miRNAs in the pathophysiology of PCOS with insulin resistance remains unclear.
The terms biological process, cellular component, and molecular function were predicted by a GO analysis of the differentially expressed miRNAs. Of these, the most significantly enriched term was metabolic process, indicating that these differentially expressed miRNAs participate in metabolism. Likewise, the most enriched pathway from the KEGG pathway analysis was also metabolism. It is known that insulin resistance, obesity, type 2 diabetes mellitus and dyslipidemia are common features of PCOS [1–3, 40]. Moreover, insulin resistance is an important characteristic of PCOS and contributes to the development of PCOS. Thus, these aberrantly expressed miRNAs may be the cause of insulin resistance in PCOS, though elucidating the underlying mechanism awaits further study.
We established miRNA expression profiles from the ovaries of PCOS rats with insulin resistance along with control rats. A total of 58 differentially expressed miRNAs in the PCOS ovaries exhibited significant changes compared with the control group, including 23 that were upregulated and 35 that were downregulated. Of note, we found that miR-3585-5p and miR-30-5p were significantly upregulated, and miR-146-5p was downregulated in the LEHF group, which was consistent with the deep sequencing results. MiR-30 was significantly upregulated in ovaries from PCOS with insulin resistance rats which is consistent with previous studies [28, 41, 42]. However, miR-146 was significantly upregulated in serum from PCOS patients compared with that of healthy women, which is inconsistent with our findings [28, 42]. This difference may be explained by the natural heterogeneity of PCOS as well as the different sequencing method employed. Interestingly, the expression of miR-146 was significantly decreased in peripheral blood mononuclear cells from type-2-diabetes patients compared with control subjects, and miR-146 was negatively correlated to insulin resistance [43]. This indicates that miR-146 may play a key role in the pathogenesis of insulin resistance in PCOS, and miR-146 may represent a novel diagnostic marker or therapeutic target for insulin resistance in PCOS. Therefore, miR-146 should be further investigated in this context.
However, this study has some limitations. In view of the methods and results we have got, this paper should be described as a pilot study, and this study is not large enough to do justice to the heterogeneity of PCOS. But these results might provide some assistance for further study of insulin resistance in PCOS. Therefore, we will make further experiments to study insulin resistance in PCOS and the comprehensive experiments will be obtained and reported in future.
In conclusion, we found 58 differentially expressed miRNAs in a rat model of PCOS with insulin resistance by deep sequencing analysis and verified the differential expression of miR-146-5p, miR-3585-5p and miR-30-5p by qRT-PCR. Furthermore, miR-146 may represent a novel diagnostic marker and therapeutic target for insulin resistance in PCOS. Additional studies are needed to identify the underlying mechanism of miRNA function in the development of PCOS with insulin resistance.
Author contributions
CZ: Project development, Data collection, Data analysis, Manuscript writing. CY: Data collection, Data analysis. ZL: Data collection, Data analysis. HP: Data collection, Data analysis. KL: Data collection. HM: Project development, Funding acquisition, Data analysis, Manuscript editing.
Funding
This work was supported by National Natural Science Foundation of China (81574013).
Compliance with ethical standards
Conflict of interest
The authors declare no potential conflicts of interest.
Ethical approval
All animal experiments were conducted under the principles of the Guidelines to the Care and Use of Experimental Animals and approved by the animal Ethics Committee of Guangzhou University of Chinese Medicine.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Escobarmorreale HF. Polycystic ovary syndrome: definition, aetiology, diagnosis and treatment. Nat Rev Endocrinol. 2018;14(5):270–284. doi: 10.1038/nrendo.2018.24. [DOI] [PubMed] [Google Scholar]
- 2.Pasquali R, Gambineri A. New perspectives on the definition and management of polycystic ovary syndrome. J Endocrinol Invest. 2018;41(10):1123–1135. doi: 10.1007/s40618-018-0832-1. [DOI] [PubMed] [Google Scholar]
- 3.Walters KA, Gilchrist RB, Ledger WJ, Teede HJ, Handelsman DJ, Campbell RE. New perspectives on the pathogenesis of PCOS: neuroendocrine origins. Trends Endocrinol Metab. 2018;29(12):841–852. doi: 10.1016/j.tem.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Azziz R, Carmina E, Dewailly D, Diamantikandarakis E, Escobarmorreale HF, Futterweit W, Janssen OE, Legro RS, Norman RJ, Taylor A. Criteria for defining polycystic ovary syndrome as a predominantly hyperandrogenic syndrome: an androgen excess society guideline. J Clin Endocrinol Metab. 2006;91(11):4237–4245. doi: 10.1210/jc.2006-0178. [DOI] [PubMed] [Google Scholar]
- 5.The Rotterdam ESHRE/ASRM-sponsored, PCOS, consensus, workshop, group Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome (PCOS) Hum Reprod. 2004;19(1):41–47. doi: 10.1093/humrep/deh098. [DOI] [PubMed] [Google Scholar]
- 6.Ricardo A, Enrico C, ZiJiang C, Andrea D, Joop L. Polycystic ovary syndrome. Nat Rev Dis Primers. 2016;2:16057. doi: 10.1038/nrdp.2016.57. [DOI] [PubMed] [Google Scholar]
- 7.Macut D, Bjekić-Macut J, Rahelić D, Doknić M. Insulin and the polycystic ovary syndrome. Diabetes Res Clin Pract. 2017;130:163–170. doi: 10.1016/j.diabres.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 8.Vink JM, Sadrzadeh S, Lambalk CB, Boomsma DI. Heritability of polycystic ovary syndrome in a Dutch twin-family study. J Clin Endocrinol Metab. 2006;91(6):2100–2104. doi: 10.1210/jc.2005-1494. [DOI] [PubMed] [Google Scholar]
- 9.Andrea D. Perspectives in polycystic ovary syndrome: from hair to eternity. J Clin Endocrinol Metab. 2016;101(3):759–768. doi: 10.1210/jc.2015-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abbott DH, Bacha F. Ontogeny of polycystic ovary syndrome and insulin resistance in utero and early childhood. Fertil Steril. 2013;100(1):2–11. doi: 10.1016/j.fertnstert.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luca L, Rebecca SS, Copperman AB, Salehi HS, Yi Z, Zhang W, Yaron T, Nathan K. Intrauterine reprogramming of the polycystic ovary syndrome: evidence from a pilot study of cord blood global methylation analysis. Front Endocrinol. 2017;8:352. doi: 10.3389/fendo.2017.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Day FR, Hinds DA, Tung JY, Stolk L, Styrkarsdottir U, Saxena R, Bjonnes A, Broer L, Dunger DB, Halldorsson BV. Causal mechanisms and balancing selection inferred from genetic associations with polycystic ovary syndrome. Nat Commun. 2015;6:8464. doi: 10.1038/ncomms9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geoffrey HM, Margrit U, David E, et al. Genome-wide association of polycystic ovary syndrome implicates alterations in gonadotropin secretion in European ancestry populations. Nat Commun. 2015;6:7502. doi: 10.1038/ncomms8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Z, Zhao H, He L, Shi Y, Qin Y, Shi Y, Li Z, You L, Zhao J, Liu J. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet. 2011;43(1):55–59. doi: 10.1038/ng.732. [DOI] [PubMed] [Google Scholar]
- 15.Shi Y, Zhao H, Shi Y, Cao Y, Yang D, Li Z, Zhang B, Liang X, Li T, Chen J. Genome-wide association study identifies eight new risk loci for polycystic ovary syndrome. Nat Genet. 2012;44(9):1020–1025. doi: 10.1038/ng.2384. [DOI] [PubMed] [Google Scholar]
- 16.Azziz R. New insights into the genetics of polycystic ovary syndrome. Nat Rev Endocrinol. 2016;12(2):74–75. doi: 10.1038/nrendo.2015.230. [DOI] [PubMed] [Google Scholar]
- 17.Heerwagen MJR, Miller MR, Barbour LA, Friedman JE. Maternal obesity and fetal metabolic programming: a fertile epigenetic soil. Am J Physiol Regul Integr Comp Physiol. 2010;299(3):R711–R722. doi: 10.1152/ajpregu.00310.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Azziz R. PCOS in 2015: new insights into the genetics of polycystic ovary syndrome. Nat Rev Endocrinol. 2016;12(2):183–183. doi: 10.1038/nrendo.2016.9. [DOI] [PubMed] [Google Scholar]
- 19.Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16(7):421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 20.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 21.Baiqi C, Ping Xu, Jing W, Chunping Z. The role of MiRNA in polycystic ovary syndrome (PCOS) Gene. 2019;706:91–96. doi: 10.1016/j.gene.2019.04.082. [DOI] [PubMed] [Google Scholar]
- 22.Sang Q, Yao Z, Wang H, Feng R, Wang H, Zhao X, Xing Q, Li J, Lin H, Wu L. Identification of microRNAs in human follicular fluid: characterization of microRNAs that govern steroidogenesis in vitro and are associated with polycystic ovary syndrome in vivo. J Clin Endocrinol Metab. 2013;798(7):3068–3079. doi: 10.1210/jc.2013-1715. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y-H, Heneidi S, Lee J-M, Layman LC, Stepp DW, Gamboa GM, Chen B-S, Chazenbalk G, Azziz R. miRNA-93 inhibits GLUT4 and is overexpressed in adipose tissue of polycystic ovary syndrome patients and women with insulin resistance. Diabetes. 2013;62(7):2278–2286. doi: 10.2337/db12-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang C, Liu J, Lai M, Li J, Zhan J, Wen Q, Ma H. Circular RNA expression profiling of granulosa cells in women of reproductive age with polycystic ovary syndrome. Arch Gynecol Obstet. 2019;300(2):431–440. doi: 10.1007/s00404-019-05129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kafali H, Iriadam M, Ozardal I, Demir N. Letrozole-induced polycystic ovaries in the rat: a new model for cystic ovarian disease. Arch Med Res. 2004;35(2):103–108. doi: 10.1016/j.arcmed.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 26.Hanley AJG, Williams K, Stern MP, Haffner SM. Homeostasis model assessment of insulin resistance in relation to the incidence of cardiovascular disease: the San Antonio Heart Study. Diabetes Care. 2002;25(7):1177–1184. doi: 10.2337/diacare.25.7.1177. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR. Method. 2002;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Motta AB. Epigenetic marks in polycystic ovary syndrome. Curr Med Chem. 2019;26:1–17. doi: 10.2174/0929867326666191003154548. [DOI] [PubMed] [Google Scholar]
- 29.Chen B, Xu P, Wang J, Zhang C. The role of MiRNA in polycystic ovary syndrome (PCOS) Gene. 2019;706:91–96. doi: 10.1016/j.gene.2019.04.082. [DOI] [PubMed] [Google Scholar]
- 30.Mayer SB, Evans WS, Nestler JE. Polycystic ovary syndrome and insulin: our understanding in the past, present and future. Womens Health. 2015;11(2):137–149. doi: 10.2217/whe.14.73. [DOI] [PubMed] [Google Scholar]
- 31.Dewailly D, Andersen CY, Balen AH, Broekmans FJM, Dilaver N, Fanchin R, Griesinger G, Kelsey T, La Marca A, Lambalk CB. The physiology and clinical utility of anti-Müllerian hormone in women. Human Reprod Update. 2014;20(3):370–385. doi: 10.1093/humupd/dmt062. [DOI] [PubMed] [Google Scholar]
- 32.Evanthia DK, Andrea D. Insulin resistance and the polycystic ovary syndrome revisited: an update on mechanisms and implications. Endocr Rev. 2012;33(6):981–1030. doi: 10.1210/er.2011-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sivalingam VN, Myers J, Nicholas S, Balen AH, Crosbie EJ. Metformin in reproductive health, pregnancy and gynaecological cancer: established and emerging indications. Human Reprod Update. 2014;20(6):853–868. doi: 10.1093/humupd/dmu037. [DOI] [PubMed] [Google Scholar]
- 34.Underdal M, Othelie S, Solhild O, Ingrid H, Hogetveit K. Does metformin treatment during pregnancy modify the future metabolic profile in women with PCOS? J Clin Endocrinol Metab. 2018;103(6):2408–2413. doi: 10.1210/jc.2018-00485. [DOI] [PubMed] [Google Scholar]
- 35.Kauffman AS, Thackray VG, Ryan GE, Tolson KP, Glidewell-Kenney CA, Semaan SJ, Poling MC, Iwata N, Breen KM, Duleba AJ. A novel letrozole model recapitulates both the reproductive and metabolic phenotypes of polycystic ovary syndrome in female mice. Biol Reprod. 2015;93(3):69. doi: 10.1095/biolreprod.115.131631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caldwell ASL, Middleton LJ, Jimenez M, Desai R, McMahon AC, Allan CM, Handelsman DJ, Walters KA. Characterization of reproductive, metabolic, and endocrine features of polycystic ovary syndrome in female hyperandrogenic mouse models. Endocrinology. 2014;155(8):3146–3159. doi: 10.1210/en.2014-1196. [DOI] [PubMed] [Google Scholar]
- 37.Hancock CR, Han D, Chen M, Terada S, Yasuda T, Wright DC, Holloszy JO. High-fat diets cause insulin resistance despite an increase in muscle mitochondria. Proc Natl Acad Sci USA. 2008;105(22):7815–7820. doi: 10.1073/pnas.0802057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li D, Li C, Xu Y, Xu D, Li H, Gao L, Chen S, Fu L, Xu X, Liu Y. Differential expression of microRNAs in the ovaries from letrozole-induced rat model of polycystic ovary syndrome. DNA Cell Biol. 2016;35(4):177–183. doi: 10.1089/dna.2015.3145. [DOI] [PubMed] [Google Scholar]
- 39.Fu L, Xu Y, Li D, Dai X, Xu X, Zhang J, Ming H, Zhang X, Zhang G, Ma Y. Expression profiles of mRNA and long noncoding RNA in the ovaries of letrozole-induced polycystic ovary syndrome rat model through deep sequencing. Gene. 2018;657:19–29. doi: 10.1016/j.gene.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 40.Dewailly D, Robin G, Peigne M, Decanter C, Pigny P, Catteaujonard S. Interactions between androgens, FSH, anti-Müllerian hormone and estradiol during folliculogenesis in the human normal and polycystic ovary. Human Reprod Update. 2016;22(6):709–724. doi: 10.1093/humupd/dmw027. [DOI] [PubMed] [Google Scholar]
- 41.Chen Z, Ou H, Wu H, Wu P, Mo Z. Role of microRNA in the pathogenesis of polycystic ovary syndrome. DNA Cell Biol. 2019;38(8):754–762. doi: 10.1089/dna.2019.4622. [DOI] [PubMed] [Google Scholar]
- 42.Long W, Zhao C, Ji C, Ding H, Cui Y, Guo X, Shen R, Liu J. Characterization of serum microRNAs profile of PCOS and identification of novel non-invasive biomarkers. Cell Physiol Biochem. 2014;33(5):1304–1315. doi: 10.1159/000358698. [DOI] [PubMed] [Google Scholar]
- 43.Balasubramanyam M, Aravind S, Gokulakrishnan K, Prabu P, Sathishkumar C, Ranjani H, Mohan V. Impaired miR-146a expression links subclinical inflammation and insulin resistance in type 2 diabetes. Mol Cell Biochem. 2011;351(1):197–205. doi: 10.1007/s11010-011-0727-3. [DOI] [PubMed] [Google Scholar]