Abstract
Background
Meyerozyma guilliermondii is a yeast which could be isolated from a variety of environments. The vka1 strain isolated and purified from the organic compost was found to have composting potential. To better understand the genes assisting the composting potential in this yeast, whole genome sequencing and sequence annotation were performed.
Results
The genome of M. guilliermondii vka1 strain was sequenced using a hybrid approach, on Illumina Hiseq-2500 platform at 100× coverage followed by Nanopore platform at 20× coverage. The de novo assembly using dual-fold approach had given draft genome of 10.8 Mb size. The genome was found to contain 5385 genes. The annotation of the genes was performed, and the enzymes identified to have roles in the degradation of macromolecules are discussed in relation to its composting potential. Annotation of the genome assembly of the related strains had revealed the unique biodegradation related genes in this strain. Phylogenetic analysis using the rDNA region has confirmed the position of this strain in the Ascomycota family. Raw reads are made public, and the genome wide proteome profile is presented to facilitate further studies on this organism.
Conclusions
Meyerozyma guilliermondii vka1 strain was sequenced through hybrid approach and the reads were de novo assembled. Draft genome size and the number of genes in the strain were assessed and discussed in relation to the related strains. Scientific insights into the composting potential of this strain are also presented in relation to the unique genes identified in this strain.
Keywords: Biodegradation, Genome annotation, Genome sequence, Illumina, NGS, Oxford Nanopore
Background
The yeast Meyerozyma guilliermondii (Wick.) Kurtzman and M. Suzuki, comb. nov. was first described as Endomycopsis guilliermondii by Wickerham [1]. The species was later placed in the genus Pichia, as Pichia guilliermondii [2] and recently renamed as Meyerozyma guilliermondii [3]. This ascomycetous yeast is widely distributed in the natural environment and forms part of the saprophytes on human skin and mucosal microflora. Meyerozyma caribbica (anamorph Candida fermentati) and M. guilliermondii (anamorph Candida guilliermondii) are two closely related species [4, 5]. This yeast has been an object of several studies, with a broad bibliography describing its multiple interesting properties and applications [6] and considered ubiquitous as they are found in deep-sea hydrothermal systems of the mid-Atlantic rift [7], wastewater treatment plants [8], maize wounds [9], and insect surfaces [10]. The yeast synthesizes large quantities of riboflavin [11] and has been extensively used in biotechnology industry.
M. guilliermondii is involved in xylitol production and polycyclic aromatic hydrocarbon degradation [12, 13]. It has been used industrially for the bioremediation purposes as well as for phosphate solubilizing [14, 15]. This yeast facilitates the process of tannin and saponin degradation also [16, 17].
In this study, we report the draft genome of the strain vka1 involved in the composting of organic waste. Hybrid sequencing followed by de novo assembly of the whole genome was performed. The objective of the study was to identify the number of genes in this strain and to find out and annotate the genes involved in the decomposing process, thus to give scientific back up for the biocomposting capability for this strain.
Methods
Yeast isolation and DNA extraction
The M. guilliermondii vka1 strain was isolated and purified from the organic compost at Kerala Agricultural University, India, following the standard protocol [18]. DNA was extracted according to the protocol by Dellaporta et al. [19]. For the extraction, pure culture of the yeast was isolated from the conical flask, frozen with liquid nitrogen in a mortar, ground to fine powder, and finally transferred to a tube containing extraction buffer (100 mM Tris pH 8.0, 50 mM EDTA pH 8.0, 500 mM NaCl, 10 mM mercaptoethanol and 1.25% SDS). After mixing, 5 M potassium acetate was added and incubated at 0 °C for 20 min. Supernatant obtained after centrifugation was poured into a clean tube where genomic DNA was allowed to precipitate with isopropanol for 30 min at – 20 °C. After centrifugation, pellet was resuspended in 50 mM Tris, 10 mM EDTA (pH 8.0) and transferred to a 20 μL tube where DNA was precipitated with 80% ethanol and the dried pellet was subsequently dissolved in 10 mM Tris, 1 mM EDTA (pH 8.0).
The 18S rDNA gene of strain vka1 was PCR amplified and Sanger sequenced. Based on the sequence, the species of the yeast was identified. To confirm the species identity, five 18S rDNA gene sequences each from five related species (M. guilliermondii, M. caribbica, M. carpophila, Candida albicans, and C. amylolentus) were retrieved from NCBI GenBank and subjected to the cluster analysis along with our sequence, with 1000 bootstrap replications.
Sequencing and assembly
Genomic DNA was sequenced and de novo assembly of the reads was done. Sequencing was performed on Illumina Hiseq-2500 platform at 100× coverage followed by Nanopore sequencing at 20× coverage. Quality of the Illumina and Nanopore raw reads was assessed using FASTQC [20] and Poretools [21], respectively. The de novo assembly was performed using ABySS 2.2.4 [22]. Different k-mer lengths (40–190) were employed to optimize the assembly and the best k-mer value of 144 was chosen for the final output. The assembly was further conducted using SPAdes 3.11.1 [23] employing different k-mer lengths (21, 33, 55, 77, 99, and 111), setting the --cov cutoff parameter to auto, and using the --careful option.
Gene prediction and annotation
Gene prediction from the scaffolds and annotation of these genes were performed using the following procedure. The assembly was input in to Augustus [24] using Candida guilliermondii as the trained dataset. Predicted genes were further analyzed using BLAST+ [25] and classified into different pathways using KAAS (KEGG Automatic Annotation Server) [26]. BBH (bi-directional best hit), the best method for annotating complete genomes in KEGG, was employed for the prediction, with M. guilliermondii selected as the organism.
Analysis of genes in comparison with other strains
To understand the genes imparting the biodegradation capability to vka1 strain, its genome was compared with those of YLG18 and ATCC6260 strains. The assembled genome sequences, ASM975640v1 and ASM694215v1 of YLG18 and ATCC6260 strains, respectively, were downloaded in FASTA format from the SRA database of NCBI. The assembly was then annotated using the same pipeline detailed above.
Phylogenetic analysis
The rDNA region, comprising the complete sequence of 18S rRNA, ITS1, 5.8S rRNA, ITS2, and 28S rRNA, from vka1 strain was used as query in BLASTn and 10 sequences belonging to various genera of Ascomycota were retrieved. Sequences were then aligned using MAFFT v.7.0 using L-INS-i strategy [27] and the aligned sequences analyzed using MEGA X [28], employing NJ method with 1000 bootstrap replications. The evolutionary distances were computed using the Jukes-Cantor method.
Results
The yeast has been isolated and purified from the composted material and DNA was extracted using Dellaporta method. Purity of the DNA was good enough for sequencing. Analysis of the 18S rDNA sequence has shown that the organism is M. guilliermondii. Cluster analysis with the sequences of the related yeasts had further confirmed the identity of the strain.
The Illumina results had 10,514,805 reads and the Nanopore had 261,366 reads. Raw reads were of high quality and the genome features of the strain are summarized in Table 1. Reads are made available in the NCBI SRA database under the BioProject No. PRJNA598411. Although the genome was sequenced with a combination of long and short reads for obtaining larger number of scaffolds, only a paired-end short read library was employed in de novo assembly. The dual-fold assembly approach has used ABySS 2.2.4 followed by confirmation with SPAdes 3.11.1 assemblers. Different k-mer values were employed for obtaining the assembly and the best value was 144. The final assembly consisted of 36 scaffolds as predicted by the assemblers. After the final assembly, draft genome with 10.8 Mb size was obtained.
Table 1.
Genome features of M. guilliermondii strain vka1
| Genome | Features | Value |
|---|---|---|
| M. guilliermondii strain vka1 | Reads |
1,05,14,805 (Illumina) 2,61,366 (Nanopore) |
| Scaffolds | 36 | |
| Max scaffold length | 14,99,707 | |
| Min scaffold length | 1270 | |
| Genome size | 10.8 Mb | |
| N50 | 5,67,598 | |
| No. of genes | 5385 |
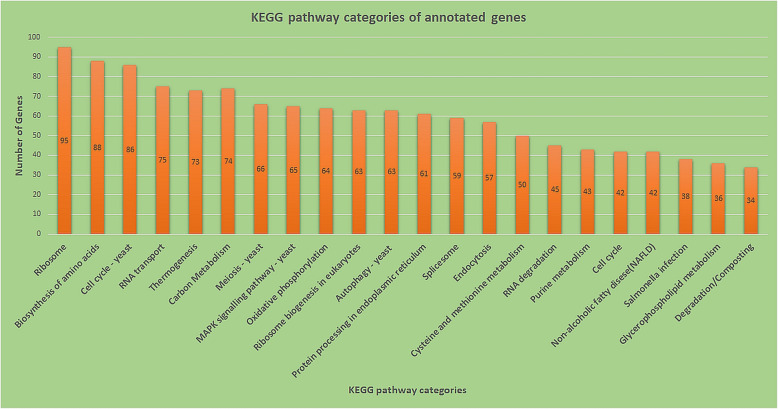
Augustus, a gene prediction program that uses a generalized hidden Markov model, was used for prediction of the genes. It has predicted 5385 genes from the vka1 strain (Table 1). KAAS annotation has classified the genes into 380 groups involved in diverse pathways (Fig. 1), of which 20 categories represented the enzymes involved in the degradation of macromolecules. The enzymes with roles in the biodegradation processes along with the list of other proteins identified in the genome are presented in Supplementary Table 1.
Fig. 1.
Major KEGG pathway categories of annotated genes from the M. guilliermondii strain vka1
Comparison of the genes identified from the vka1, YLG18, and ATCC6260 strains had revealed the unique biodegradation-related genes in vka1. The enzymes S-(hydroxymethyl)glutathione dehydrogenase, phenylacetate 2-hydroxylase, trimethyllysine dioxygenase, 4-aminobutyrate aminotransferase, and delta3-delta2-enoyl-CoA isomerase were found only in vka1 strain.
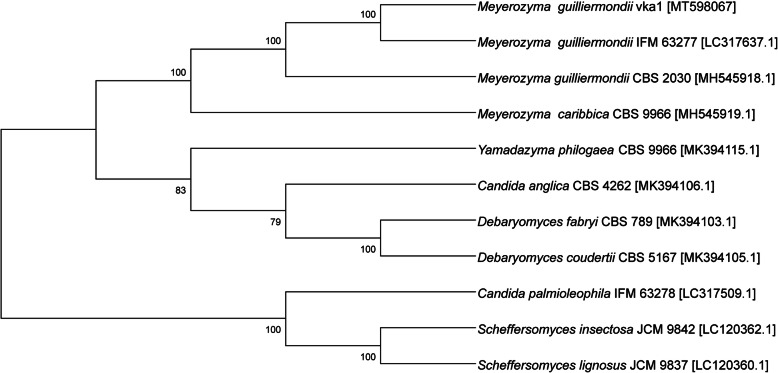
Phylogenetic analysis was carried out using the rDNA sequences of different genera belonging to Ascomycetes phylum. The phylogenetic tree had high bootstrap values for all the branches. Of the two major clades (Fig. 2), the first one had eight accessions divided into two sub-clusters. The first sub-cluster included Meyerozyma genus with 100% bootstrap support. Strain vka1 clustered along with M. guilliermondii strain IFM 63277. The second sub-cluster had two Debaryomyces accessions along with Candida anglica and Yamadazyma philogaea. Second major clade had three accessions, two belonging to Scheffersomyces, and one Candida palmioleophila accession forming an outgroup.
Fig. 2.
Phylogenetic relationships (cladogram) based on the sequences of rDNA region from selected genomes, inferred using the Neighbour Joining (NJ) method. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches
Discussion
The ascomycetous yeast M. guilliermondii is widely distributed in the environment including human body. This microbe is also exploited commercially in riboflavin and xylitol production [11]. However, it is well understood to have roles in degradation of macromolecules including polycyclic aromatic hydrocarbons [13], phosphates [15], tannins and saponins [16]. Due to these properties, it is well utilized in bioremediation [14].
Though Illumina sequencing platform offers better read accuracy, short read lengths is a concern [29]. On the other hand, Nanopore platform generates longer reads though comparatively error prone [30]. Method based on the combined analysis of short and long reads generated from different platforms is found to result in comprehensive genome characterization [31].
Several new de novo assembly tools have been developed recently to assemble short sequencing reads generated by next-generation sequencing platforms. Different assembly tools have their advantages and disadvantages and hence a dual-fold approach is reported to give better assemblies [32]. Thus, ABySS and SPAdes based de novo assembly was chosen in this study. The genome size shown by the assembly was slightly higher than 10.6 Mb in M. guilliermondii strain ATCC6260 and 10.64 Mb in P. perniciosus, both sequenced on Illumina HiSeq-2500 platform [33]. The gene calling algorithm Augustus has predicted 5385 genes from the vka1 strain. The number of genes obtained is more than that in a previous study which reported 5275 genes [34], but slightly lower than 5401 genes reported by De Marco et al. [33].
Bi-directional best hit method followed in this study is identified as the best method for annotating the complete genomes [35]. The annotation had shown that the genome accommodates many enzymes which are directly involved in the biodegradation processes, explaining the composting potential of this strain. The enzyme salicylate hydroxylase is identified to be involved in decarboxylative hydroxylation [36]. Similarly, urea carboxylase is involved in carboxylation in the degradation process [37]. Enzymes cyanamide hydratase [38], phenylacetate 2-hydroxylase [39, 40], aldehyde dehydrogenase [41], saccharopine dehydrogenase [42], sarcosine oxidase [43], dihydrolipoamide dehydrogenase [44], trimethyllysine dioxygenase [45], 2-oxoglutarate dehydrogenase [44], malonate-semialdehyde dehydrogenase [46], acyl-CoA dehydrogenase [47], hydroxymethylglutaryl-CoA synthase [48], 3-hydroxyisobutyryl-CoA hydrolase [49], 4-aminobutyrate aminotransferase [50], S-(hydroxymethyl)glutathione dehydrogenase [51], acyl-CoA oxidase [52], delta3-delta2-enoyl-CoA isomerase [53], beta-galactosidase [54], alpha-mannosidase [55], and hexosaminidase [56] are all well understood to have direct involvement in the macromolecule catabolism. This array of macromolecule degrading enzymes, including the unique genes identified, explains the composting potential of this strain. Future studies are required on these enzymes and on this yeast to understand the mechanisms.
The phylogenetic analysis was in agreement with the previous taxonomic studies on Meyerozyma and ascomycetes. The analysis had confirmed that vka1 strain belongs to M. guilliermondii and bootstrap value of 100% affirms this result. The genera Debaryomyces, Yamadazyma, and Candida which clustered into a single clade, with the bootstrap value of 83.0%, have previously been shown to be close relatives of Meyerozyma [3, 57, 58].
Conclusion
M. guilliermondii is an ascomycetous yeast universally distributed in the natural environment and having a wide range of industrial applications. The draft genome of a new strain, vka1, with potential for composting the organic wastes is being reported. Hybrid sequencing followed by dual-fold assembly was used to assess the genome size and the annotation has shown the total number of genes coded. Twenty categories of enzymes which are involved in the macromolecule degradation were identified, and this wide spectrum along with the unique enzymes identified explains the composing potential of this strain. The phylogenetic analysis has confirmed its position in the Ascomycota family.
Supplementary information
Additional file 1. Supplementary Table 1. Detailed results of KEGG (KAAS) analysis of Meyerozyma guilliermondii strain vka1 genome, showing the genes for the enzymes annotated and their respective categories (Categories given in boldface).
Acknowledgements
Not applicable
Abbreviations
- EDTA
Ethylenediaminetetraacetic acid
- ITS
Internal transcribed spacer
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MEGA
Molecular Evolutionary Genetics Analysis
- NCBI
National Center for Biotechnology Information
- SDS
Sodium dodecyl sulfate
Authors’ contributions
RV did data curation, draft genome assembly, gene finding, and genome annotation. DM has isolated the yeast, isolated genomic DNA, and sequenced. All authors have read and approved the final manuscript.
Funding
This work is part of the project Distributed Information Centre, supported by Department of Biotechnology, Ministry of Science and Technology, Government of India.
Availability of data and materials
The authors declare that sequence reads associated with this article are available in the NCBI SRA database under the BioProject No. PRJNA598411.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that the research was conducted with no commercial, personal, or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ravisankar Valsalan, Email: rsv518@gmail.com.
Deepu Mathew, Email: deepu.mathew@kau.in.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s43141-020-00074-2.
References
- 1.Wickerham LJ, Burton KA. A clarification of the relationship of Candida guilliermondii to other yeasts by a study of their mating types. J Bacteriol. 1954;68(5):594–597. doi: 10.1128/JB.68.5.594-597.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wickerham LJ. Validation of the species Pichia guilliermondii. J Bacteriol. 1966;92(4):1269. doi: 10.1128/JB.92.4.1269-.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurtzman CP, Suzuki M. Phylogenetic analysis of ascomycete yeasts that form coenzyme Q-9 and the proposal of the new genera Babjeviella, Meyerozyma, Millerozyma, Priceomyces, and Scheffersomyces. Mycoscience. 2010;51:2–14. doi: 10.1007/s10267-009-0011-5. [DOI] [Google Scholar]
- 4.Bai FY, Liang HY, Jia JH. Taxonomic relationships among the taxa in the Candida guilliermondii complex, as revealed by comparative electrophoretic karyotyping. Int J Syst Evol Microbiol. 2000;50(Pt 1):417–422. doi: 10.1099/00207713-50-1-417. [DOI] [PubMed] [Google Scholar]
- 5.Vaughan-Martini A, Kurtzman CP, Meyer SA, O'Neill EB. Two new species in the Pichia guilliermondii clade: Pichia caribbica sp. nov., the ascosporic state of Candida fermentati, and Candida carpophila comb. nov. FEMS Yeast Res. 2005;5(4-5):463–469. doi: 10.1016/j.femsyr.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Papon N, Savini V, Lanoue A, Simkin AJ, Creche J, Giglioli-Guivarc'h N, et al. Candida guilliermondii: biotechnological applications, perspectives for biological control, emerging clinical importance and recent advances in genetics. Curr Genet. 2013;59(3):73–90. doi: 10.1007/s00294-013-0391-0. [DOI] [PubMed] [Google Scholar]
- 7.Gadanho M, Sampaio J. Occurrence and diversity of yeasts in the mid-Atlantic ridge hydrothermal fields near the Azores Archipelago. Microb Ecol. 2005;50:408–417. doi: 10.1007/s00248-005-0195-y. [DOI] [PubMed] [Google Scholar]
- 8.Lahav R, Fareleira P, Nejidat A, Abeliovich A. The identification and characterization of osmotolerant yeast isolates from chemical wastewater evaporation ponds. Microb Ecol. 2002;43(3):388–396. doi: 10.1007/s00248-002-2001-4. [DOI] [PubMed] [Google Scholar]
- 9.Nout MJ, Platis CE, Wicklow DT. Biodiversity of yeasts from Illinois maize. Can J Microbiol. 1997;43(4):362–367. doi: 10.1139/m97-050. [DOI] [PubMed] [Google Scholar]
- 10.Suh SO, Blackwell M. Three new beetle-associated yeast species in the Pichia guilliermondii clade. FEMS Yeast Res. 2004;5(1):87–95. doi: 10.1016/j.femsyr.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 11.Protchenko OV, Boretsky Yu R, Romanyuk TM, Fedorovych DV. Oversynthesis of riboflavin by yeast Pichia guilliermondii in response to oxidative stress. Ukr Biokhim Zh. 2000;72(2):19–23. [PubMed] [Google Scholar]
- 12.Hesham AEL, Khan S, Liu X, Zhang Y, Wang Z, Yang M. Application of PCR-DGGE to analyse the yeast population dynamics in slurry reactors during degradation of polycyclic aromatic hydrocarbons in weathered oil. Yeast. 2006;23(12):879–887. doi: 10.1002/yea.1401. [DOI] [PubMed] [Google Scholar]
- 13.Mussatto SI, Silva CJ, Roberto IC. Fermentation performance of Candida guilliermondii for xylitol production on single and mixed substrate media. Appl Microbiol Biotechnol. 2006;72(4):681–686. doi: 10.1007/s00253-006-0372-z. [DOI] [PubMed] [Google Scholar]
- 14.Carmona FJ, Pascual J, Felix F. Proceedings of 6th International Conference on Sustainable Solid Waste Management (NAXOS2018), 13-16 June 2018. Naxos Island: Poster; 2018. Targeted composting of olive mil wastes. Comparison between three-phase and two-phase waste composting; p. 32. [Google Scholar]
- 15.Ganapathy B, Yahya A, Ibrahim N. Bioremediation of palm oil mill effluent (POME) using indigenous Meyerozyma guilliermondii. Environ Sci Pollut Res. 2019;26(11):1113–11125. doi: 10.1007/s11356-019-04334-8. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Ying Y, Li X, Yao X. Changes in tannin and saponin components during co-composting of Camellia oleifera Abel shell and seed cake. PLoS ONE. 2020;15(3):e0230602. doi: 10.1371/journal.pone.0230602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Ying Y, Yao X, Huang W, Tao XJB. Degradations of tannin and saponin and changes in nutrition during co-composting of shell and seed cake of Camellia oleifera Abel. BioResources. 2020;15(2):2721–2734. doi: 10.1371/journal.pone.0230602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi GW, Um HJ, Kim Y, Kang HW, Kim M, Chung BW, Kim YH. Isolation and characterization of two soil derived yeasts for bioethanol production on cassava starch. Biomass Bioenergy. 2010;34(8):1223–1231. doi: 10.1016/j.biombioe.2010.03.019. [DOI] [Google Scholar]
- 19.Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: Version II. Plant Mol Biol Report. 1983;1(4):19–21. doi: 10.1007/BF02712670. [DOI] [Google Scholar]
- 20.Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. In: Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom, available at http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 21.Loman NJ, Quinlan AR. Poretools: a toolkit for analyzing nanopore sequence data. Bioinformatics. 2014;30(23):3399–3401. doi: 10.1093/bioinformatics/btu555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19(6):1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24(5):637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- 25.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35(Web Server issue):W182-185. 10.1093/nar/gkm321 [DOI] [PMC free article] [PubMed]
- 27.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20(4):1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kircher M, Heyn P, Kelso J. Addressing challenges in the production and analysis of illumina sequencing data. BMC Genomics. 2011;12(1):382. doi: 10.1186/1471-2164-12-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laver T, Harrison J, O’neill PA, Moore K, Farbos A, Paszkiewicz K, Studholme DJ. Assessing the performance of the oxford nanopore technologies minion. Biomol Detect Quantif. 2015;3:1–8. doi: 10.1016/j.bdq.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Au KF, Sebastiano V, Afshar PT, Durruthy JD, Lee L, Williams BA, van Bakel H, Schadt EE, Reijo-Pera RA, Underwood JG, Wong WH. Characterization of the human ESC transcriptome by hybrid sequencing. Proc Natl Acad Sci U S A. 2013;110(50):E4821–E4830. doi: 10.1073/pnas.1320101110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin Y, Li J, Shen H, Zhang L, Papasian CJ, Deng HW. Comparative studies of de novo assembly tools for next-generation sequencing technologies. Bioinformatics. 2011;27(15):2031–2037. doi: 10.1093/bioinformatics/btr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Marco L, Epis S, Capone A, Martin E, Bozic J, Crotti E, Ricci I, Sassera D. The genomes of four Meyerozyma caribbica isolates and novel insights into the Meyerozyma guilliermondii species complex. G3 (Bethesda) 2018;8(3):755–759. doi: 10.1534/g3.117.300316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan W, Zhang S, Wu M, Zhang W, Zhou J, Dong W, et al. The draft genome sequence of Meyerozyma guilliermondii strain YLG18, a yeast capable of producing and tolerating high concentration of 2-phenylethanol. 3. Biotech. 2019;9(12):441. doi: 10.1007/s13205-019-1975-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aoki-Kinoshita KF, Kanehisa M (2007) Gene annotation and pathway mapping in KEGG. In: Comparative Genomics, Bergman, N. H. (ed.), Methods in Molecular Biology vol. 396, ISBN 978-1-934115-37-4, Humana Press, pp. 71-91. doi: 10.1007/978-1-59745-515-2_6 [DOI] [PubMed]
- 36.You IS, Ghosal D, Gunsalus IC. Nucleotide sequence analysis of the Pseudomonas putida PpG7 salicylate hydroxylase gene (nahG) and its 3'-flanking region. Biochemistry. 1991;30(6):1635–1641. doi: 10.1021/bi00220a028. [DOI] [PubMed] [Google Scholar]
- 37.Kanamori T, Kanou N, Atomi H, Imanaka T. Enzymatic characterization of a prokaryotic urea carboxylase. J Bacteriol. 2004;186(9):2532–2539. doi: 10.1128/jb.186.9.2532-2539.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Biss M, Fu Y, Xu X, Moore SA, Xiao W. Two duplicated genes DDI2 and DDI3 in budding yeast encode a cyanamide hydratase and are induced by cyanamide. J Biol Chem. 2015;290(20):12664–12675. doi: 10.1074/jbc.M115.645408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mingot JM, Peñalva MA, Fernández-Cañón JM. Disruption of phacA, an Aspergillus nidulans gene encoding a novel cytochrome P450 monooxygenase catalyzing phenylacetate 2-hydroxylation, results in penicillin overproduction. J Biol Chem. 1999;274(21):14545–14550. doi: 10.1074/jbc.274.21.14545. [DOI] [PubMed] [Google Scholar]
- 40.Rodríguez-Sáiz M, Barredo JL, Moreno MA, Fernández-Cañón JM, Peñalva MA, Díez B. Reduced function of a phenylacetate-oxidizing cytochrome p450 caused strong genetic improvement in early phylogeny of penicillin-producing strains. J Bacteriol. 2001;183(19):5465–5471. doi: 10.1128/jb.183.19.5465-5471.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohta T, Tani A, Kimbara K, Kawai F. A novel nicotinoprotein aldehyde dehydrogenase involved in polyethylene glycol degradation. Appl Microbiol Biotechnol. 2005;68(5):639–646. doi: 10.1007/s00253-005-1936-z. [DOI] [PubMed] [Google Scholar]
- 42.Gaziola SA, Sodek L, Arruda P, Lea PJ, Azevedo RA. Degradation of lysine in rice seeds: Effect of calcium, ionic strength, S-adenosylmethionine and S-2-aminoethyl- l-cysteine on the lysine 2-oxoglutarate reductase-saccharopine dehydrogenase bifunctional enzyme. Physiol Plant. 2000;110(2):164–171. doi: 10.1034/j.1399-3054.2000.110204.x. [DOI] [Google Scholar]
- 43.Kim JM, Shimizu S, Yamada H. Sarcosine oxidase involved in creatinine degradation in Alcaligenes denitrificans subsp. denitrificans J9 and Arthrobacter spp. J5 and J11. Agric Biol Chem. 1986;50(11):2811–2816. doi: 10.1080/00021369.1986.10867841. [DOI] [Google Scholar]
- 44.Oppermann FB, Schmidt B, Steinbüchel A. Purification and characterization of acetoin:2,6-dichlorophenolindophenol oxidoreductase, dihydrolipoamide dehydrogenase, and dihydrolipoamide acetyltransferase of the Pelobacter carbinolicus acetoin dehydrogenase enzyme system. J Bacteriol. 1991;173(2):757–767. doi: 10.1128/jb.173.2.757-767.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Vlies N, Ofman R, Wanders RJ, Vaz FM. Submitochondrial localization of 6-N-trimethyllysine dioxygenase—implications for carnitine biosynthesis. FEBS J. 2007;274(22):5845–5851. doi: 10.1111/j.1742-4658.2007.06108.x. [DOI] [PubMed] [Google Scholar]
- 46.Goodwin GW, Rougraff PM, Davis EJ, Harris RA. Purification and characterization of methylmalonate-semialdehyde dehydrogenase from rat liver. Identity to malonate-semialdehyde dehydrogenase. J Biol Chem. 1989;264(25):14965–14971. [PubMed] [Google Scholar]
- 47.Corydon TJ, Bross P, Jensen TG, Corydon MJ, Lund TB, Jensen UB, et al. Rapid degradation of short-chain Acyl-CoA Dehydrogenase variants with temperature-sensitive folding defects occurs after import into mitochondria. J Biol Chem. 1998;273(21):13065–13071. doi: 10.1074/jbc.273.21.13065. [DOI] [PubMed] [Google Scholar]
- 48.Shinohara M, Sato N, Kurinami H, Takeuchi D, Takeda S, Shimamura M, et al. Reduction of brain β-amyloid (Aβ) by Fluvastatin, a Hydroxymethylglutaryl-CoA Reductase inhibitor, through increase in degradation of amyloid precursor protein C-terminal fragments (APP-CTFs) and Aβ clearance. J Biol Chem. 2010;285(29):22091–22102. doi: 10.1074/jbc.M110.102277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan Y, Yang J, Gong Y, Li X, Hu H. 3-Hydroxyisobutyryl-CoA hydrolase involved in isoleucine catabolism regulates triacylglycerol accumulation in Phaeodactylum tricornutum. Philos Trans R Soc Lond Ser B Biol Sci. 2017;372(1728):20160409. doi: 10.1098/rstb.2016.0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Large PJ, Robertson A. The subcellular location of 4-aminobutyrate aminotransferase in Candida boidinii and its probable role in the breakdown of putrescine and spermidine. Yeast. 1988;4(2):149–153. doi: 10.1002/yea.320040209. [DOI] [Google Scholar]
- 51.Koopman F, Wierckx N, de Winde JH, Ruijssenaars HJ. Identification and characterization of the furfural and 5-(hydroxymethyl)furfural degradation pathways of Cupriavidus basilensis HMF14. Proc Natl Acad Sci U S A. 2010;107(11):4919. doi: 10.1073/pnas.0913039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dommes V, Baumgart C, Kunau WH. Degradation of unsaturated fatty acids in peroxisomes. Existence of a 2,4-dienoyl-CoA reductase pathway. J Biol Chem. 1981;256(16):8259–8262. [PubMed] [Google Scholar]
- 53.Palosaari PM, Hiltunen JK. Peroxisomal bifunctional protein from rat liver is a trifunctional enzyme possessing 2-enoyl-CoA hydratase, 3-hydroxyacyl-CoA dehydrogenase, and delta 3, delta 2-enoyl-CoA isomerase activities. J Biol Chem. 1990;265(5):2446–2449. [PubMed] [Google Scholar]
- 54.Kitamikado M, Ito M, Li YT. Isolation and characterization of a keratan sulfate-degrading endo-beta-galactosidase from Flavobacterium keratolyticus. J Biol Chem. 1981;256(8):3906–3909. [PubMed] [Google Scholar]
- 55.De Gasperi R, Daniel PF, Warren CD. A human lysosomal alpha-mannosidase specific for the core of complex glycans. J Biol Chem. 1992;267(14):9706–9712. [PubMed] [Google Scholar]
- 56.Conzelmann E, Sandhoff K. AB variant of infantile GM2 gangliosidosis: deficiency of a factor necessary for stimulation of hexosaminidase A-catalyzed degradation of ganglioside GM2 and glycolipid GA2. Proc Natl Acad Sci U S A. 1978;75(8):3979. doi: 10.1073/pnas.75.8.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459(7247):657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kurtzman CP, Robnett CJ. Relationships among genera of the Saccharomycotina (Ascomycota) from multigene phylogenetic analysis of type species. FEMS Yeast Res. 2013;13(1):23–33. doi: 10.1111/1567-1364.12006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary Table 1. Detailed results of KEGG (KAAS) analysis of Meyerozyma guilliermondii strain vka1 genome, showing the genes for the enzymes annotated and their respective categories (Categories given in boldface).
Data Availability Statement
The authors declare that sequence reads associated with this article are available in the NCBI SRA database under the BioProject No. PRJNA598411.