Abstract
Background:
The cardiomyocyte apoptosis is considered as one of major contributions to cardiac remodeling after myocardial infarction (MI). Numerous studies find that circular RNAs (circRNAs) play pivotal roles in a variety of biological functions. However, the role of circ_0068655 in MI and human induced pluripotent stem-derived cardiomyocytes (HCMs) remains unknown.
Methods:
The expression of circ_0068655, miR-498, and PRKC apoptosis WT1 regulator (PAWR) in human MI heart tissues and hypoxia subjected HCMs was evaluated with qRT-PCR and Western blot. The effects of circ_0068655 on hypoxia-induced apoptotic death and cell migration in HCMs were evaluated with qRT-PCR, cell viability, cell death ELISA (POD), and Caspase-3 activity assay, and Trans-well assay, respectively. Furthermore, luciferase assay, qRT-PCR, biotin-labeled miRNA pulldown assay, and Western blot were employed in the functional studies.
Results:
We found that the expression of circ_0068655 and PAWR was enhanced in MI patients and hypoxia subjected HCMs; by contrast, the expression of miR-498 decreased. Inhibited expression of circ_0068655 in HMCs counteracted hypoxia-induced apoptotic death and impaired cell migration, in sharp contrast to circ_0068655 knockdown. We identified that circ_0068655 sponged an endogenous miR-498 to sequester and inhibit its activity, leading to the increased PAWR expression.
Conclusions:
Our findings reveal that the expression of circ_0068655 can promote cardiomyocyte apoptosis through the modulation of miR-498-PAWR axis in vitro, which highlights the diagnostic and therapeutic value of circ_0068655 in patients with MI.
Keywords: Circular RNA, Myocardial infarction, MiR-498, PAWR, Cardiomyocyte apoptosis
Introduction
Myocardial infarction (MI) is a common cardiac emergency with a sudden loss of blood or oxygen supply [1]. Cardiomyocyte apoptosis, hypertrophy, and a series of inflammatory and immune responses are associated with MI [2, 3, 4], leading to chronic heart failure and cardiac remodeling [5, 6]. Even though the management of MI has been improved over the past several decades, exploring the pathogenesis of MI is crucial. The apoptosis of cardiomyocytes resulting from MI has been identified as an essential process in the progression to heart failure [7]. Thus, to discover novel molecular mechanisms withstanding MI-related apoptosis attracts widespread interest.
Circular RNAs (circRNAs) are characterized as a class of endogenous noncoding RNAs and have high stability and resistance against RNA exonuclease due to their covalently closed-loop structures [8]. Numerous circRNAs are found in human serum exosomes and their functions are identified, including binding to and regulating microRNA (miRNA) as a miRNA sponge, forming RNA–protein interaction complexes, regulating the parental gene expression, and sequestering proteins from their native subcellular localization [9–11]. Several circRNAs have been demonstrated that play the pivotal roles in cardiac development and physiology [12, 13]. Aberrant expression of these circRNAs is associated with heart failure, MI, and hypertrophy [13, 14]. Some studies demonstrated that the upregulation of CDR1as is associated with MI, and the downregulation of heart-related circular RNA (HRCR) is associated with hypertrophic cardiomyopathy [15, 16], suggesting the potential crucial role of circRNAs under these pathologic conditions.
In a most recent study, over 15,000 cardiac circRNAs within human heart tissues have been identified using purpose-designed bioinformatics tools [12, 17]. However, circ_0068655 has never been studied in cardiovascular disease studies, indicating the potential importance of circ_0068655 in MI mechanism study.
In the current study, we identified that circ_0068655 and the PRKC apoptosis WT1 regulator (PAWR) was upregulated in MI heart tissues and hypoxia-induced cell line. High expression level of circ_0068655 was associated with high apoptosis cell rate. In the functional studies, we found that circ_0068655 acted as an efficient miR-498 sponge and exerted its function by modulating the miR-498/PAWR axis, thereby, exacerbating cell apoptosis rate and reducing cell viability. These findings revealed the functions of a novel circRNA in MI-related apoptosis a suggested that circ_0068655 might be a therapeutic candidate for MI.
Materials and methods
Cell culture and treatment
Commercial human induced pluripotent stem-derived cardiomyocytes (HCMs) were maintained in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific, In., Waltham, MA, USA) composing of 10% fetal bovine serum (FBS, Sigma Aldrich, St. Louis, MO, USA). The cells were cultured in a hypoxia incubator (5% CO2, 94% N2, and 1% O2) for 3, 6, and 12 h.
Total RNA extraction and RNA expression
Total RNA was extracted using Trizol Reagent (Invitrogen), followed by cDNA synthesis with SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) according to the manufacturers’ introductions. The expression of these genes was quantified with RT-PCR using SYBR Green (CloudSeq Biotech Inc., Shanghai, China) technology on the ABI Prims 7900HT Sequence Detection System (Applied Biosystems, Carlsbad, CA, USA). The species-specific gene circ_00686552, miR-498, and PAWR sequences were customized designed and synthesized (Sangon Biotech, Co., Ltd., Shanghai, China) [18, 19]. U6 was used as an internal control for the expression of circ_00686552 and miR-498, and GAPDH was used as an internal control for the expression of PAWR. The primers were shown in Table 1.
Table 1.
Primer sequences in this study
| Primer | Forward primer | Reverse primer |
|---|---|---|
| U6 | AACGCTTCACGAATTTGCGT | CTCGCTTCGGCAGCACA |
| circ_00686552 | GCCCAGTACCAGTTCA | CCAGTATTTCCTGCTTT |
| miR-498 | TTTCAAGCCAGGGGGCGTTTTTC | GCTTCAAGCTCTGGAGGTGCTTTTC |
| PAWR | GCCGCAGAGTGCTTAGATGAG | GCAGATAGGAACTGCCTGGATC |
| Lentiviral-based shRNA targeting circ_0068655 sequences | ||
| circ_0068655 | ||
| shRNA #1 | ccggGTACCAGTTCAGCAAGTGTCTggatccAGACACTTGCTGAACTGGTACtttttg | aattcaaaaaGTACCAGTTCAGCAAGTGTCTggatccAGACACTTGCTGAACTGGTAC |
| shRNA#2 | ccggGCAAGTGTCTCTACTGTCAGAggatccTCTGACAGTAGAGACACTTGC tttttg | aattcaaaaaGTACCAGTTCAGCAAGTGTCTggatccAGACACTTGCTGAACTGGTAC |
| Circular RNA and the corresponding mutation of miR-498 binding site sequences | ||
| Circular RNA | ||
| circ_0068655 | ACGCTCGAGGTGCAAGTGAAAGTGT | ACGGCGGCCGCCACCAAATAATGGTTCTGGTTCCA |
| circ_0068655 binding site | TGTAGGAAAACATATGGTAGCCG | CGGCTACCATATGTTTTCCTACA |
| PAWR | ACGCTCGAGAATTTAAGGGATTTTCTTC | ACGGCGGCCGCTCCCAAAGTGCTGGGATTAC |
| PAWR binding site | TAATATATGAAAGGGAATAGCCT | AGGCTATTCCCTTTCATATATTA |
Bioinformatic analysis, vector construction and cell transfection
For circ_0068655 knockdown version, lentiviral-based shRNAs targeting circ_0068655 (Table 1) were synthesized and purchased from GeneChem (Shanghai, China). For circ_0068655 overexpression version, sequence was synthesized and amplified by PrimerSTAR Max DNA Polymerase Mix (Takara, Dalian, China). The PCR product was inserted into pLCDH vector and the vector contained a front circular frame and a back circular frame. For circ_0068655 mutation version, Circular RNA Interactome (http://circinteractome.nia.nih.gov) was used to predict the potential targeted sequences between circ_0068655 and miR-498. The miR-498 mimics were synthesized by GenePharma. The circ_0068655 sequence and circ_0068655 sequence with mutation of miR-498 binding site (Table 1) were synthesized using overlap extension PCR and subcloned into the luciferase reporter psiCHECK2 (Promega, Madison, WI, USA) and designated as circ_0068655-WT and circ_0068655-MT, respectively. The transfection efficiency of circ_0068655 and miR-498 was confirmed using qRT-PCR after HCMs were transfected with sh-Circ_0068655 and miR-498 mimics and inhibitor, respectively.
Additionally, PAWR has one putative 10-mer site that matches to the hsa_miR-498 seed region through Targetscan (http://www.targetscan.org/vert_71) predication. The PAWR sequence and PAWR sequence with mutation of miR-498 binding site (Table 1) were synthesized and subcloned into the luciferase reporter psiCHECK2 (Promega) and designated as PAWR 3′ UTR-WT and PAWR 3′ UTR-MT, respectively.
To evaluate the effects of circ_0068655 on hypoxia treated HCMs, the cells were transfected with sh-Circ_0068655. To reveal the relationship between circ_0068655 and miR-498, HCMs were co-transfected with sh-circ_0068655 and miR-498 inhibitor. The transfected HCMs were cultured in a hypoxia incubator for 12 h for cell viability, apoptotic cell rate, and migration and invasion evaluations. The gene and protein expression of PAWR in sh-Circ-0068655 and miR-498 co-transfection cells was evaluated using qRT-PCR and Western blot, respectively.
Cell viability assay
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT) assay was employed to evaluate the cell viability according to the manufacturer’s instruction. Briefly, transfected cells were seeded in 96-well plates with 5,000 cells/well and cultured in a hypoxia incubator for 12 h. MTT (20 µl) solution was added to culture medium and 200 µl dimethyl sulfoxide was used to dissolve the blue formazan after removing the medium. The spectrophotometric absorbance was measured by a microplate reader with a wavelength of 550 nm to reveal the cell viability. The cells without transfection and hypoxia treatment was used as a control group and the absorbance was normalized to the control group.
Capspase-3 activity
An Apo-ONE Homogeneous Caspase-3/7 Assay kit (Promega, Madison, WI, USA) was used to evaluate the caspase-3 activity according to the manufacturer’s instruction. Briefly, hypoxia treated (12 h) cells were incubated with Caspase-Glo® 3/7 reagent for 3 h and the luminescence was measured in a plate-reading luminometer. The cells without transfection and hypoxia treatment was used as a control group in the circ_0068655 function study. In the circ_0068655/miR-498/PAWR axis function study, the cells transfected with negative vector were used as a control group. The absorbance was normalized to the control group.
Cell death ELISA (POD) assay
A cell death detection ELISA kit (Roche Applied Science, Indianapolis, IN, USA) was applied to measure the total quantity of mono- and oligonucleosomes according to the manufacturer’s instruction. In brief, the hypoxia treated cells were lysed and the lysates were incubated with anti-histone-biotin and anti-DNA-POD antibodies in a streptavidin-coated microplate for 90 min at room temperature. After removing the medium, 2, 2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic) acid substrate was added and absorbance was measured with a wavelength of 405 nm. The cells without transfection and hypoxia treatment was used as a control group in the circ_0068655 functional study. In the circ_0068655/miR-498/PAWR axis functional study, the cells transfected with negative vector were used as a control group. The absorbance was normalized to the control group.
Trans-well assay
To investigate the effects of circ_0068655 on hypoxia treated cells, two-chamber trans-well assay was applied according the previous report [20]. Briefly, transfected HCMs were seeded into the upper chamber of Corning chambers (Corning Inc., Corning, NY, USA) and the chambers were incubated in a hypoxia condition for 12 h. To evaluate the invaded cells, the upper chambers were coated with Matrigel (BD Bioscience, Franklin Lakes, NJ, USA) and incubated at 37 °C for 30 min before the cells were seeded. The migrated cells were stained with the crystal violet (Beyotime Biotechnology, Shanghai, China). The cells without transfection and hypoxia treatment was used as a control group in the circ_0068655 functional study. In the circ_0068655/miR-498/PAWR axis functional study, the cells transfected with negative vector were used as a control group. The relative migration and invasion were normalized to the control group.
Nuclear and cytoplasmic fraction isolation
The Ambion® PARIS™ system (Thermo Fisher Scientific) was used for the RNA and protein isolation from HCMs according to the manufacturer’s instruction. Briefly, HCMs was mixed with ice cold cell disruption buffer and incubated on ice for 10 min to completely lyse cells. The mixture was centrifuged at 4 °C with 500 × g for 5 min and the supernatant was collected as the cytoplasmic fraction into an RNase free tube for cytoplasmic RNA isolation. Ice cold cell disruption buffer was added to the formed pellet for the nuclear pellet lysis and collection for the nuclear RNA isolation. To isolate cytoplasmic fraction and nuclear lysate RNA, an equal volume of lysis/binding solution was added to the lysates at room temperature. Preheated elution solution was used to collect RNA and the RNA samples were used for qRT-PCR to confirm the circ_0068655 location.
Dual-luciferase reporter assay
For dual-luciferase reporter assay, sequences of circ_0068655-WT and PAWR 3′UTR-WT vectors with related reporters were transfected into HCMs and HEK-293T cells using Lipofectamine 2000 (Thermo Fisher Scientific). Mutated plasmids were used as controls. The cells were transfected with the related reporter plasmids and related miRNA or negative control. After 48 h transfection, dual-luciferase reporter assay system (Promega) was applied to evaluate luciferase activity according to the manufacturer’s instructions. Briefly, passive lysis buffer was added to perform lysis and luciferase assay buffer II containing luciferase assay substrate was added to measure firefly luciferase activity. Stop & Glo® reagent was added to the mixture to measure Renilla luciferase activity and firefly luciferase was used as a reporter gene for the normalized control.
Western blot
The treated cells were homogenized in NP-40 lysis buffer (1% NP-40, 20 mM Tris, 137 mM NaCl, 20% glycerol, 10 mM PMSF, 1 mM Na3VO4, 10 mM NaF, 2.5 mg/ml aprotinin, and 2.5 mg/ml leupeptin). Protein concentrations were determined with the Bicinchoninic Acid (BCA) Protein Assay Kit (Thermo Fisher Scientific), and 20–30 µg protein was denatured by heating at 95 °C for 10 min before loading into SDS-PAGE gel. The following dilutions of primary antibody were used: PAWR (1:1000; Cell Signaling, Danvers, MA, USA) and GAPDH (1:1000, Cell Signaling). The protein expression of PAWR were detected by an enhanced chemiluminescence detection system (Amersham Biosciences, Piscataway, NJ, USA).
Biotinylated-probe pull-down assay
Biotinylated-probe pull-down assay was performed as the previous described [21]. Biotin-labeled miR-498 was transfected into HCMs. The whole cell lysates were incubated with streptavidin-coupled magnetic beads (Thermo Fisher Scientific) for an additional 16 h at 4 °C with rotation. The bead-RNA complexes were harvested and washed, followed by DNase treatment and purification. The expression of circ_0068655 and PAWR was evaluated by qRT-PCR. All procedures were performed in the RNase-free conditions.
Statistical analysis
The data are presented as the mean ± standard deviation (SD). SPSS 13.0 software package was used to analyze the data. Three independent experiments were performed in all in vitro experiments. Comparisons between two groups were made using a two-tailed t test. When more than 2 groups were compared, a Shapiro–Wilk normality test was performed to asses for a normal distribution, and one-way ANOVA analysis was employed to determine the statistical significance. The level of significance was set at p < 0.05.
Results
Circ_0068655 was identified to be significantly up-regulated in MI and hypoxia-induced HCMs
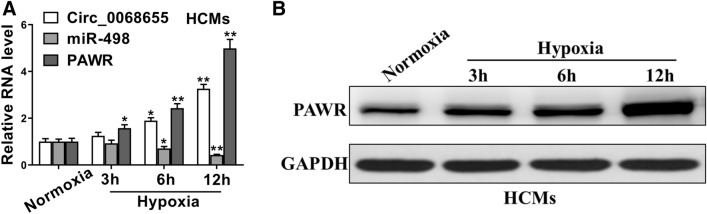
We found that the expression of circ_0068655 and PAWR in HCMs increased with hypoxia inducing duration increasing, while the expression of miR-498 showed an opposite trend (Fig. 1A). The protein expression of PAWR confirmed these findings (Fig. 1B). These results indicated that circ_0068655 may contribute to the negative effects on in vitro hypoxia-induced HCMs.
Fig. 1.
The dysregulation of circ_0068655, miR-498, and PAWR in cardiomyocytes subjected to hypoxic insult. A The expression of circ_0068655, miR-498, and PAWR in HCMs subjected to hypoxia treatment for 3, 6, and 12 h was evaluated by qRT-PCR. B The protein expression of PAWR in HCMs subjected to hypoxia treatment for 3, 6, and 12 h was evaluated by western blotting. The data are expressed as the mean ± SD. *p < 0.05; **p < 0.01
Circ_0068655 promoted hypoxia-induced cell injury
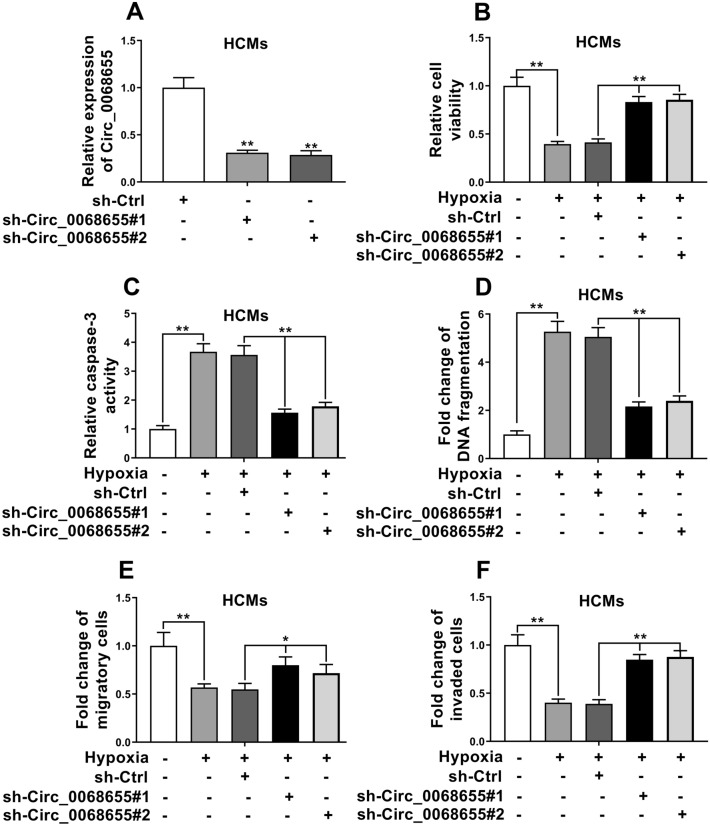
After transfected with sh-Circ_0068655, the expression of circ_0068655 decreased comparing to the control (Fig. 2A), indicating that the expression of circ_0068655 was suppressed due to the sh-Circ_0068655 transfection. After treated with hypoxia, the cells with lower circ_0068655 expression had significantly higher cell viability, migration, and invasion (Fig. 2), but lower apoptotic cell rate (Fig. 2C, D) versus the control group. These results suggested that knockdown the expression of circ_0068655 can prevent the cell injury induced by hypoxia.
Fig. 2.
Suppression of circ_0068655 alleviated hypoxia-induced cell injury. A The expression of circ_0068655 in HCMs transfected with shRNAs to knockdown circ_0068655 (sh-Circ_0068655#1, sh-Circ_0068655#2) or empty vector (sh-Ctrl) was determined by qRT-PCR. B Cell viability, C caspase-3 activity, D DNA fragmentation, E migration, and F invasion were evaluated by MTT assay, Caspase-3 activity assay, cell death ELISA (POD) assay, and trans-well assay in HCMs transfected with shRNAs against Circ_0068655 or empty vector (sh-Ctrl), respectively. The data are expressed as the mean ± SD. *p < 0.05; **p < 0.01
Circ_0068655 Sponged miR-498 in HCMs
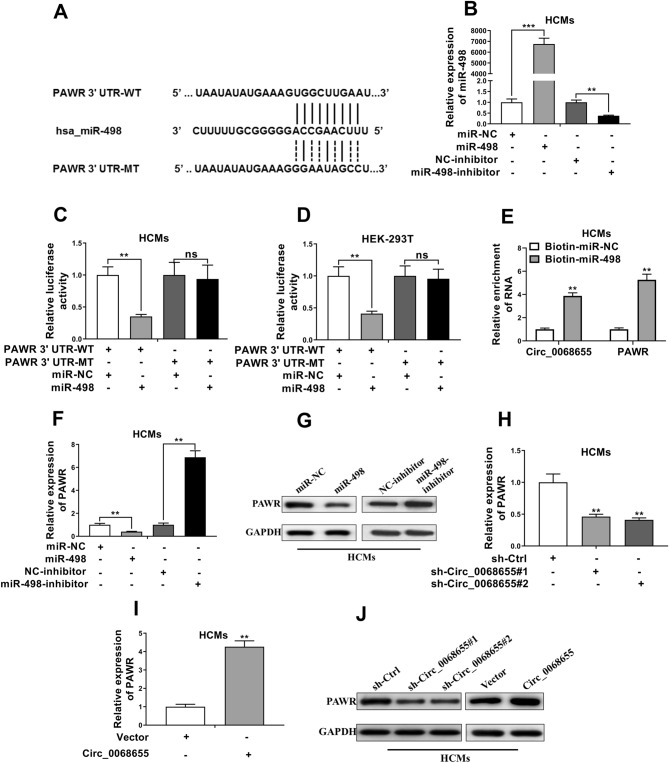
To observe cellular localization of circ_0068655, qRT-PCR was applied to analyze nuclear and cytoplasmic circ_0068655 RNA. Results showed that circ_0068655 transcript located in the cytoplasm (Fig. 3A). We found that circ_0068655 shares 8-mer bindings site of miR-498 by using Circular RNA Interactome (Fig. 3B). Subsequently, luciferase reporter assays were used to determine whether miR-498 directly targets circ_0068655. The luciferase activity of circ_0068655 luciferase reporters significantly reduced in the cells transfected with miR-498 mimics (Fig. 3C, D, indicating that miR-498 was the circ_0068655 associated miRNA in HCMs and HEK-293T cells. However, there was no significant difference between the cells transfected with mutant plasmid (Fig. 3C, D). Moreover, knockdown of circ_0068655 can promote miR-498 expression, while overexpression of circ_0068655 can suppress miR-498 expression (Fig. 3E, F). These results suggested that circ_0068655 directly sponged miR-498.
Fig. 3.
Circ_0068655 acted as a sponge to negatively regulate miR-498. A Levels of circ_0068655 in the nuclear and cytoplasmic fractions in HCMs were confirmed via qRT-PCR. B Graphical representation showing the predicted sites of miR-498 for binding to circ_0068655 and its corresponding mutation. Luciferase reporter assay for the interaction between miR-498 and circ_0068655 in C HCMs and D HEK-293T cells. E The expression of miR-498 in HCMs after transfected with shRNAs against circ_0068655 or empty vector (sh-Ctrl) was determined by qRT-PCR. F The expression of miR-498 in HCMs after transfected with circ_0068655 plasmid or empty vector (Vector) was determined by qRT-PCR. The data are expressed as the mean ± SD. *p < 0.05; **p < 0.01, ns: no significant difference
Circ_0068655 acted as a competing endogenous RNA to promote PAWR expression
We found that the 3′ UTR of PAWR shares the 10-mer binding site of miR-498 by using Targetscan (Fig. 4A). After transfected with miR-498 mimics, the expression of miR-498 in HCMs was greater versus the cells without transfection, while the expression was inhibited after the cells was transfected with miR-498 inhibitor (Fig. 4B). Luciferase reporter assays showed that miR-498 mimics reduced the luciferase activity of the PAWR with a wild-type vector and there was no significant effect in the cells with the mutant plasmid transfection in HCMs and HEK-293 T cells (Fig. 4C, D). Moreover, an RNA pull-down assay was performed by using biotin-labeled miR-498. We found that both of circ_0068655 and PAWR were pulled down by biotin-labeled miR-498 (Fig. 4E), suggesting both of circ_0068655 and PAWR had binding site with miR-498. The expression of PAWR in HCMs transfected with miR-498 mimics was suppressed, while the expression was enhanced in the cells transfected with miR-498 inhibitor (Fig. 4F, G), indicating that miR-498 can inhibit the expression of PAWR. Moreover, knockdown of circ_0068655 decreased the protein and mRNA levels of PAWR, while overexpression of circ_0068655 promoted PAWR expression (Fig. 4H–J). Taken together, these results suggested that circ_0068655 can enhance PAWR expression by competitively binding miR-498.
Fig. 4.
PAWR competed with circ_0068655 for miR-498 binding sites. A Graphical representation showing the predicted miR-498 binding sites in the region of PAWR and its corresponding mutant sequence. B The expression of miR-498 in HCMs after transfected with miR-498 mimics and miR-498 inhibitor or their respective negative controls was determined by qRT-PCR. (C and D) Luciferase assays were performed in C HCMs and D HEK-293T cells co-transfected with wild-type or mutant psiCHECK2-PAWR 3’UTR and miR-498 mimics or miR-NC. E circ_0068655 and PAWR were pulled down and enriched with biotin labeled miR-498. F,G The gene and protein expression of PAWR in HCMs transfected with miR-498 mimics alone or miR-498 inhibitor was determined by F qRT-PCR and G western blotting, respectively. H PAWR expression in HCMs transfected with shRNAs against circ_0068655 or empty vector (sh-Ctrl) was determined by qRT-PCR. I PAWR expression in HCMs transfected with circ_0068655 plasmid or its respective empty vector was determined by qRT-PCR. J The protein expression of PAWR in HCMs transfected with plasmids as described above was evaluated by western blotting. The data are expressed as the mean ± SD, **p < 0.01; ***p < 0.001
Circ_0068655 suppressed cell viability and promoted apoptosis by sponging miR-498 and down-regulating PAWR expression
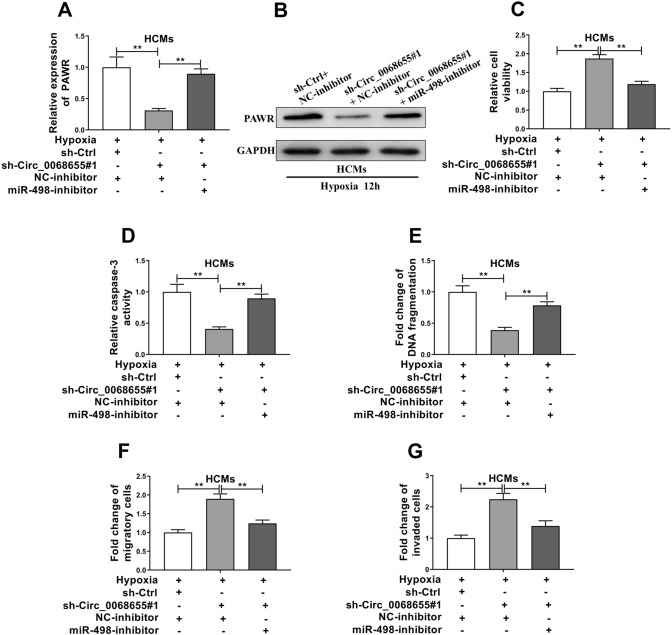
The gene and protein expression of PAWR was repressed in the cells with circ_0068655 siRNA transfection (Fig. 5A, B). Importantly, the inhibition of circ_0068655 siRNA on PAWR was attenuated by miR-498 inhibitor (Fig. 5A). The protein expression change of PAWR confirmed this finding (Fig. 5B). Therefore, we inferred that circ_0068655 might exert its function via modulation of the miR-498/PAWR axis. We further explored the biological function of circ_0068655 and miR-498 in HCMs. The MTT assay revealed that knockdown of circ_0068655 can significantly promote cell viability and cell migration and invasion, and repressed the apoptotic cell rate (Fig. 5C–G). In order to investigate whether circ_0068655 exerted it function through sponge activity of miR-498, we co-transfected miR-498 inhibitor and circ_0068655 siRNA in HCMs. The effect of circ_0068655 knockdown on cell viability, apoptotic cell rate, migration and invasion was reversed by the miR-498 inhibitor (Fig. 5C–G). These results indicate that circ_0068655 may exert its function via modulation of miR-498/PAWR axis.
Fig. 5.
Circ_0068655 exerted its function via modulation of the circ_0068655/miR-498/PAWR axis. HCMs were co-transfected with shRNA against circ_0068655, miR-498 inhibitor, empty vector, or negative control inhibitor, followed by hypoxia incubation for 12 h. The A RNA and B protein expression of PAWR, C cell viability, D caspase-3 activity, E DNA fragment, F relative migration, and G invasion were determined by qRT-PCR, western blotting, MTT assay, Caspase-3 activity assay, cell death ELISA (POD) assay, and trans-well assay, respectively. The data are expressed as the mean ± SD, **p < 0.01
Discussion
CircRNAs were found as a novel class of endogenous noncoding RNAs that have covalently closed-loop structures and have higher stability and resistance against RNA exonuclease comparing to linear RNA [14]. Because of the distinct properties, circRNAs attract widespread attention in various disease studies [14, 22, 23]. However, the mechanisms of circRNAs in cardiac pathophysiology are still unclear. Our current study showed detailed negative effects of circ_0068655 on HCMs. In MI tissues, circ_0068655 had higher expression and its overexpression promoted cardiomyocyte ischemia-related apoptotic death, whereas its down-regulation protected cardiomyocyte against hypoxic injury through targeting miR498-PAWR regulatory cascade.
Several studies found that circRNAs had aberrant expression in various cardiovascular diseases [15, 16, 24, 25]. Upregulation of circ-Ttc3 may protect cardiomyocytes against the hypoxic injury [25]. In a mouse MI injury model, the overexpression of Cdr1as was associated with cardiac infarct size increase and cell apoptosis by sponging miR-7α [15]. Circ-HRCR can act as an endogenous miR-223 sponge and prevent cardiac hypertrophy and heart failure in a pressure overload-induced hypertrophic mouse model [16]. Herein, our findings provided a novel circRNA and demonstrated that the upregulation of circ-0068655 in cardiomyocytes can result in cell apoptosis and the downregulation can confer the potent resistance to hypoxic.
In the present study, we focused on the modulation on miRNA-protein sequestration in dissect the circ-0068655-mediated hypoxic injury. We found that circ_0068655 and PAWR had one potential 8-mer and 10-mer site that matches to the hsa_miR-498 seed region through online tools, respectively. We also found that circ_0068655 and PAWR had an opposite expression pattern comparing to miR-498 in human MI tissues. MiR-498 attracts numerous studies in various diseases and miR-498 showed contradictory effects. The overexpression of miR-498 in colon cancer cell lines could result in cell proliferation inhibition, while the expression of miR-498 can promote cell proliferation in Y79 cells [19, 26]. The comprehensive analysis of miR-498 in cardiac related diseases remains limited. In a recent study, it is demonstrated that the expression of miR-498 was upregulated in patients with congenital heart disease [27]. However, the role of miR-498 in MI is not clear. Our current study demonstrated that miR-498 may play a pivotal role to regulate normal heart functions. In our study we found that the expression of miR-498 decreased in MI tissues and hypoxia-induced HCMs, indicating miR-498 was positively linked to cardiomyocyte fate and sequestration of miR-498 by circ_0068655 deteriorate the hypoxia-induced apoptosis.
In the previous report, PAWR was associated with inhibition of angiogenesis that was enhanced upon hypoxia and acute MI [28, 29]. Our findings were consistent with these reports and the expression of PAWR increased in hypoxia-induced HCMs. Because of the endogenous miR-498, the PAWR expression was inhibited and the inhibition could be reversed when miR-498 inhibitor was applied. Our findings provided additional insight that PAWR is one of miR-498′s targets with clear evidence. Of note, we observed that the 3′ UTR of PAWR shares the binding sites of miR-498 matched in circ_0068655 by using bioinformatics. High PAWR expression in cardiac diseases and tumors often correlates with cell viability, apoptosis, and cell invasion [18, 29]. Therefore,
we further identified whether circ_0068655 acts as a sponge of miR-498 to promote PAWR expression in HCMs. Theoretically, circ_0068655 acted as a sponge to control miR-498 expression, with the result that increased expression of circ_0068655 should decrease miR-498 and promote PAWR expression. We demonstrated that PAWR is a direct target of miR-498 in HCMs. Moreover, we confirmed that circ_0068655 regulates PAWR expression through miR-498. The effect of circ_0068655 knockdown on HCMs can be reversed by the application of miR-498 inhibitor. These results revealed that circ_0068655 promotes HCM apoptosis by sponging miR-498.
In conclusion, we show that circ_0068655 plays a pivotal role in MI, and the circ_0068655-miR-498-PAWR regulatory axis underlies the cardiac regulation. These findings provide new insights for understanding the pathogenesis of MI-related cardiomyocyte apoptosis.
Acknowledgements
The study was supported by Key projects of Hebei Science and technology support plan (16277707D); Hebei provincial government funded specialty capacity building and specialty leader training Project (LS201808); Hebei Science and technology plan Project (19277757D).
Compliance with ethical standards
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethical statement
There are no animal experiments carried out for this article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Benjamin EJ, Virani SS, Callaway CW, Chamberlain AM, Chang AR, Cheng S, et al. Heart disease and stroke statistics-2018 update: a report from the American Heart Association. Circulation. 2018;137:e67–492. doi: 10.1161/CIR.0000000000000558. [DOI] [PubMed] [Google Scholar]
- 2.Shimizu I, Minamino T. Physiological and pathological cardiac hypertrophy. J Mol Cell Cardiol. 2016;97:245–262. doi: 10.1016/j.yjmcc.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Shinde AV, Frangogiannis NG. Fibroblasts in myocardial infarction: a role in inflammation and repair. J Mol Cell Cardiol. 2014;70:74–82. doi: 10.1016/j.yjmcc.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teringova E, Tousek P. Apoptosis in ischemic heart disease. J Trans Med. 2017;15:87-87. doi: 10.1186/s12967-017-1191-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gabriel-Costa D. The pathophysiology of myocardial infarction-induced heart failure. Pathophysiology. 2018;25:277–284. doi: 10.1016/j.pathophys.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 6.Schirone L, Forte M, Palmerio S, Yee D, Nocella C, Angelini F, et al. A review of the molecular mechanisms underlying the development and progression of cardiac remodeling. Oxid Med Cell Longev. 2017;2017:3920195. doi: 10.1155/2017/3920195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Empel VP, Bertrand AT, Hofstra L, Crijns HJ, Doevendans PA, De Windt LJ. Myocyte apoptosis in heart failure. Cardiovasc Res. 2005;67:21–29. doi: 10.1016/j.cardiores.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32:309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Armakola M, Higgins MJ, Figley MD, Barmada SJ, Scarborough EA, Diaz Z, et al. Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models. Nat Genet. 2012;44:1302–1309. doi: 10.1038/ng.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 11.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 12.Kim YY, Min H, Kim H, Choi YM, Liu HC, Ku SY. Differential MicroRNA expression profile of human embryonic stem cell-derived cardiac lineage cells. Tissue Eng Regen Med. 2017;14:163–169. doi: 10.1007/s13770-017-0051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M, Ding W, Sun T, Tariq MA, Xu T, Li P, et al. Biogenesis of circular RNA s and their roles in cardiovascular development and pathology. FEBS J. 2018;285:220–32. doi: 10.1111/febs.14191. [DOI] [PubMed] [Google Scholar]
- 14.Fan X, Weng X, Zhao Y, Chen W, Gan T, Xu D. Circular RNAs in cardiovascular disease: an overview. Biomed Res Int. 2017;2017:5135781. doi: 10.1155/2017/5135781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geng HH, Li R, Su YM, Xiao J, Pan M, Cai XX, et al. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One. 2016;11:e0151753. doi: 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 17.Tan WL, Lim BT, Anene-Nzelu CG, Ackers-Johnson M, Dashi A, See K, et al. A landscape of circular RNA expression in the human heart. Cardiovasc Res. 2016;113:298–309. doi: 10.1093/cvr/cvw250. [DOI] [PubMed] [Google Scholar]
- 18.Yang K, Shen J, Chen SW, Qin J, Zheng XY, Xie LP. Upregulation of PAWR by small activating RNAs induces cell apoptosis in human prostate cancer cells. Oncol Rep. 2016;35:2487–2493. doi: 10.3892/or.2016.4582. [DOI] [PubMed] [Google Scholar]
- 19.Yang L, Wei N, Wang L, Wang X, Liu QH. miR-498 promotes cell proliferation and inhibits cell apoptosis in retinoblastoma by directly targeting CCPG1. Childs Nerv Syst. 2018;34:417–422. doi: 10.1007/s00381-017-3622-8. [DOI] [PubMed] [Google Scholar]
- 20.Guo Z, Zhou F, Song W, Yu L, Yan W, Yin L, et al. Suppression of microRNA-101 attenuates hypoxia-induced myocardial H9c2 cell injury by targeting DIMT1-Sp1/survivin pathway. Eur Rev Med Pharmacol Sci. 2018;22:6965–6976. doi: 10.26355/eurrev_201810_16167. [DOI] [PubMed] [Google Scholar]
- 21.Yu X, Liu X, Wang R, Wang L. Long non-coding RNA NEAT1 promotes the progression of hemangioma via the miR-361-5p/VEGFA pathway. Biochem Biophys Res Commun. 2019;512:825–831. doi: 10.1016/j.bbrc.2019.03.084. [DOI] [PubMed] [Google Scholar]
- 22.Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rong D, Sun H, Li Z, Liu S, Dong C, Fu K, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8:73271–73281. doi: 10.18632/oncotarget.19154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai L, Qi B, Wu X, Peng S, Zhou G, Wei Y, et al. Circular RNA Ttc3 regulates cardiac function after myocardial infarction by sponging miR-15b. J Mol Cell Cardiol. 2019;130:10–22. doi: 10.1016/j.yjmcc.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 26.Gopalan V, Smith RA, Lam AK. Downregulation of microRNA-498 in colorectal cancers and its cellular effects. Exp Cell Res. 2015;330:423–428. doi: 10.1016/j.yexcr.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 27.Smith T, Rajakaruna C, Caputo M, Emanueli C. MicroRNAs in congenital heart disease. Ann Transl Med. 2015;3:333. doi: 10.3978/j.issn.2305-5839.2015.12.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Messmer-Blust A, An X, Li J. Hypoxia-regulated angiogenic inhibitors. Trends Cardiovasc Med. 2009;19:252–256. doi: 10.1016/j.tcm.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nukala SB, Regazzoni L, Aldini G, Zodda E, Tura-Ceide O, Mills NL, et al. Differentially expressed proteins in primary endothelial cells derived from patients with acute myocardial infarction. Hypertension. 2019;74:947–956. doi: 10.1161/HYPERTENSIONAHA.119.13472. [DOI] [PubMed] [Google Scholar]