Abstract
Background:
Intestinal organoids have evolved as potential molecular tools that could be used to study host-microbiome interactions, nutrient uptake, and drug screening. Gut epithelial barrier functions play a crucial role in health and diseases, especially in autoimmune diseases, such as inflammatory bowel diseases (IBDs), because they disrupt the epithelial mucosa and impair barrier function.
Methods:
In this study, we generated an in vitro IBD model based on dextran sodium sulfate (DSS) and intestinal organoids that could potentially be used to assess barrier integrity. Intestinal organoids were long-term cultivated and characterized with several specific markers, and the key functionality of paracellular permeability was determined using FITC-dextran 4 kDa. Intestinal organoids that had been treated with 2 µM DSS for 3 h were developed and the intestinal epithelial barrier function was sequentially evaluated.
Results:
The results indicated that the paracellular permeability represented epithelial characteristics and their barrier function had declined when they were exposed to FITC-dextran 4 kDa after DSS treatment. In addition, we analyzed the endogenous mRNA expression of pro-inflammatory cytokines and their downstream effector genes. The results demonstrated that the inflammatory cytokines genes significantly increased in inflamed organoids compared to the control, leading to epithelial barrier damage and dysfunction.
Conclusion:
The collective results showed that in vitro 3D organoids mimic in vivo tissue topology and functionality with minor limitations, and hence are helpful for testing disease models.
Electronic supplementary material
The online version of this article (10.1007/s13770-020-00278-0) contains supplementary material, which is available to authorized users.
Keywords: Intestinal stem cells, Organoids, IBD, Epithelial barrier, Ex vivo model
Introduction
The intestinal epithelial barrier has a fundamental physiological role in nutrient absorption and helps maintain host homeostasis. Simultaneously, it acts as the mechanistic immune barrier because it selectively controls the passage of biotic, xenobiotic, and pathogenic substances from the external environment to the luminal space [1]. Defects in the intestinal mucosa lead to impaired barrier function and a leaky gut. This increased epithelial barrier permeability is associated with a range of gastrointestinal (GI) tract diseases, such as inflammatory bowel diseases (IBD), irritable bowel syndrome (IBS), and colorectal cancer [2, 3].
Organoids have been successfully developed for various sets of organs, such as the intestine, kidney, lung, liver, pancreas, and brain, from a number of sources, e.g., embryonic stem cells (ESC), induced pluripotent stem cells (iPSC), and adult stem cells [4]. Intestinal organoids are isolated from the LGR5 expressing mature intestine crypts, and they possess close structural and functional similarities with original organs of the body [5]. Post-conditioning of the culture methods and media with suitable combinations of growth factors, such as epidermal growth factor (EGF), Noggin, R-spondin-1, and Wnt3A, allow the crypts to expand during long-term culturing [6]. Intestinal organoids have been successfully used in various stomach disease models and for detecting microbiome interactions. For example, the pathophysiological responses of Helicobacter pylori were elucidated by applying a microinjection technique to gastric enteroids [7]. A recent study demonstrated the applicability of organoids in regenerative therapeutic practice where engrafted colonoids (colon organoids) were able to cure EDTA induced colon injury in a mouse model [8].
Inflammatory bowel diseases, including ulcerative colitis (UC) and Crohn’s disease are idiopathic inflammatory diseases induced by various immune responses, genetic and environmental factors [9]. Although, accurate etiology of IBDs elusive, genome wide association studies uncovered few genetic factors susceptible to crohn’s disease [10]. Later studies identified that nucleotide-binding oligomerization domain containing 2 (NOD2), ATG16L1 and IRGM activates the immune responses contributes in elevated expression of pro-inflammatory cytokines, such as IL-6, TNF-α, and IL-1β, during infection [11]. There is also a rise in C-reactive protein (CRP) [12]. The up-regulation of pro-inflammatory cytokines associated with dysregulated barrier integrity and tight junction dynamics ultimately leads to IBD related pathogenesis [13]. Furthermore, chronic inflammation during acute stage IBD contributes to a variety of GI cancers, such as colorectal cancer, small bowel adenocarcinoma, and other extra intestinal cancers [14]. In previous studies, murine models were predominantly used to dissect the pathophysiology of IBD and chemical inducers. For example, dextran sodium sulfate (DSS), trinitrobenzene sulfonic acid (TNBS), and oxazolone were used to generate in vivo models [15, 16]. They succeeded as disease models, but ethical issues, and laborious and expensive procedures are major disadvantages of the in vivo models [17]. Consequently, in vitro 3D organoid models are now used as alternative research tools because they serve the same purpose as in vivo models [3]. Therefore, we used intestinal organoids to evaluate an inflammatory model and its effect on barrier function.
In this study, we maintained the intestinal organoids for more than 10 generations (> P10) by adopting procedures used in previous studies and serum free commercial media [18]. Furthermore, we evaluated the epithelial barrier function using a FITC-dextran 4 kDa permeability assay. Then we generated the inflammatory IBD intestinal organoid model by DSS induction and analyzed the endogenous expression of pro-inflammatory cytokines and their downstream effector genes. Finally, we tested barrier integrity under inflammatory conditions using a FITC-dextran 4 kDa permeability assay. A comprehensive summary of the experimental results showed that intestinal organoids are potential alternatives to in vivo disease models and demonstrated that barrier function declined in inflamed samples.
Materials and methods
Experimental animals
Five-week-old ICR mice were obtained from Japan SLC (Shizuoka, Japan) and were taken to Central Lab. Animal, Inc. (Seoul, Korea). All procedures, including animal handling, feeding, and sample collection, followed the standard operating protocols of the Animal Biotechnology Division at the National Institute of Animal Science, Korea.
Immunohistochemistry
The small intestine segments were fixed in 10% neutral-buffered formalin (Sigma-Aldrich, St Louis, MO, USA) after strong washing with ice cold phosphate-buffered saline (PBS). The segments were subsequently embedded in a paraffin block and the paraffin-embedded intestinal tissue was vertically and horizontally sectioned at a thickness of 3–5 μm. The sections were then deparaffinized in xylene, rehydrated with water via a graded alcohol series, and then processed prior to hematoxylin and eosin (Merck, Darmstadt, Germany) staining. In the immunohistochemical analysis, the sections were permeabilized with 0.1% Triton X-100 in PBS for 5 min and incubated with 0.1% normal goat serum for 1 h to block nonspecific binding after antigen retrieval by boiling the sections in a sodium citrate buffer solution. The samples were incubated with appropriate dilutions of primary antibodies, such as LGR5 anti-mouse Clone-OTI2A2 (Origene Technologies, Rockville, MD, USA), 36/E-cadherin anti-rabbit (BD biosciences, East Rutherford, NJ, USA), Bmi1 anti-rabbit ployclonal (Abcam, Cambridge, MA, USA), and mucin2 (F-2) anti-mouse (Santa Cruz Biotechnology, Santa Cruz, CA, USA) over night at 4 °C. After washing, the samples were reacted with anti-mouse and anti-rabbit secondary antibodies coupled to Alexa Fluor 488 and-594 (Molecular Probes/Life technologies, Waltham, MA, USA), respectively, for 1 h at room temperature. These fluorescent samples were counterstained with diamidino-2-phenylindole (DAPI). The images were captured using an Olympus X100 confocal microscope (Olympus, Tokyo, Japan).
Isolation and culturing of murine small intestinal organoids
Before the murine small intestine was isolated, the matrigel basement membrane matrix (Corning, New York, NY, USA) was thawed on ice and pre-heated in a 24-well (Corning) plate that had been placed inside a CO2-incubator (5% CO2, 37 °C). The mice were euthanized by cervical dislocation and the small intestine was obtained by making a V-shaped cut into the peritoneal cavity. The crypt culture protocol was adapted from StemCell Technologies (Vancouver, BC, Canada). Briefly, a piece of mouse small intestine (~ 5 cm) was isolated as shown in Figure S1. It was then washed with PBS after it had been opened longitudinally to remove the villi. The prepared intestine was diced into 2–4 mm pieces and transferred to a 50 mL tube containing 30 mL ice-cold PBS. The solution was then pipetted gently with a 10 mL pipette and the supernatant was discarded after the solution had settled down. This was repeated until the supernatant was clear, which was approximately 15 times. Then, 30 mL of enzyme free cell disassociation buffer (StemCell Technologies) was added and the tube was gently shaken at room temperature for 20 min. The sample was pipetted again to release the crypts, and then 5% FBS was added to the PBS and the sample was passed through a 70 mm cell strainer into another 50 mL tube to collect the crypts. The suspension was spun down at 300 × g for 5 min and the pellets were resuspended in 10 mL of 5% FBS that contained advanced Dulbecco’s modified Eagle medium/F12 (Gibco, Waltham, MA, USA) supplemented with penicillin/streptomycin. The washing cycles were repeated twice. The pellet was resuspended in 2 mL IntestiCult mouse organoid growth medium (StemCell Technologies) and the number of cells were counted using a hemocytometer under an inverted microscope.
A total seeding population of 1000–1500 crypts per 100 µL of matrigel were prepared and carefully placed in the middle of a 24 well plate (Corning). The matrigel started polymerizing after 2–3 min, after which the plate was moved to an incubator for further incubation at 37 °C and 5% CO2. The matrigel dome had fully polymerized after 20 min. Then, 900 µL IntestiCult organoid growth medium was gently added through the walls. The medium was replaced every 3 days and sub-cultivated once a week. The number of organoids was recorded in triplicate every week and the wells were sub-divided into three parts for cultivation.
Immunofluorescent staining
The organoids were maintained in 24-well plates (Corning) until maturation. The fixation media was aspirated in the wells, and the organoids were washed thoroughly with cold PBS and incubated in neutrally buffered 4% paraformaldehyde (Sigma-Aldrich) for 30 min at room temperature. Then the organoids were permeabilized in buffer containing 0.5% Triton-X100 (v/v) (Sigma-Aldrich) in PBS for 30 min at room temperature. The blocking step was performed using 3% bovine serum albumin (BSA) in PBS for 1 h at room temperature. The organoids were thoroughly rinsed with PBS and incubated overnight at 4 °C with the appropriate primary antibodies, such as LGR5 anti-mouse Clone-OTI2A2 (OriGene Technologies, Rockville, MD, USA), 36/E-cadherin anti-rabbit (BD biosciences), Bmi1 anti-rabbit polyclonal (Abcam), chromogranin A anti-rabbit polyclonal (Abcam), F-actin anti-rabbit polyclonal (Abcam), Cytokeratin 19 anti-rabbit polyclonal (Abcam), and mucin 2 (F-2) anti-mouse (Santacruz Biotech), at their appropriate dilutions. The marker gene expressions were detected by incubating the samples with corresponding secondary antibodies coupled to AlexaFluor-488 and 594 (Molecular Probes/Life Technologies) for 1 h at room temperature. These fluorescent samples were counterstained with diamidino-2-phenylindole (DAPI) and mounted on glass slides using ProLong Gold antifade (Life technologies, Waltham, MA, USA) mounting medium. The images were captured under an Olympus X100 confocal microscope (Olympus).
Epithelial barrier permeability assay using FITC-dextran 4 kDa
The epithelial barrier function was tested by diluting powdered FITC-dextran 4 kDa (Sigma-Aldrich) in nuclease free water, which resulted in a 1 mg/mL working solution. The organoids were placed in 24 well plates (Corning) and allowed to grow until fully developed into crypt and villi like structures. Then, 25 ng/mL of FITC-dextran 4 kDa was added to each well and the plate was incubated under normal growth conditions. The permeability was observed using luminal absorption and recorded for more than 60 min at 10 min intervals under a Leica CTR6000 fluorescent microscope (Leica, Wentzler, Germany).
Inflammatory model generation using DSS
The in vitro organoid IBD model was generated using 500 kDa DSS (Sigma-Aldrich) diluted in DNase and RNase free ultra-pure distilled water (Invitrogen, Carlsbad, CA, USA) to achieve a 1 mg/mL final concentration. Matured intestinal organoids that had been grown in 24 well plates (Corning) were used to test for inflammation and barrier integrity. Testing concentrations of 1 µM, 2 µM, or 5 µM DSS were added to each well for 60 min to determine the dose. The treatments were repeated three times and the plate was incubated under normal growth conditions. Figure S2 shows that the organoids were examined for inflammation at 30 min, 60 min, and 180 min. The images were captured using a Leica microscope and the bright field (BF) option. We then used the inflamed organoids to repeat the barrier permeability assay in the presence of FITC-dextran 4 kDa. The barrier integrity was estimated, and the results recorded.
Quantitative RT-PCR
The non-treated control and the DSS treated organoids were washed with ice cold PBS and then incubated in Cultrex organoid harvesting solution (Trevigen, Gaithersburg, MD, USA) for 10 min in an incubator. The dislocated organoids were gently pipetted to release them from the matrix membrane. They were then centrifuged at 300 × g for 5 min and the supernatant was carefully discarded without disturbing the pellet. The pellet was thoroughly washed with PBS and spinned down for downstream processing. Total RNA in the test samples was isolated using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. The RNA quality was checked by agarose gel electrophoresis and the RNA quantity was determined by spectrophotometry at 260 nm [19]. The cDNA was synthesized from the RNA using the Superscript III First-Strand Synthesis System (Invitrogen). It was then serially diluted five-fold and quantitatively equalized for PCR amplification. Quantitative RT-PCR was performed using a technique utilized in a previous study [20], and the results were used to relatively quantify the expressions of the genes influenced by the prepared cDNA. The primers used in this study were designed using primer express software (Applied Biosystems, Foster City, CA, USA) using sequences from the GenBank database and these are shown in Table 1. Each test sample was run in triplicate.
Table 1.
Primers used in gene expression analysis by qRT-PCR to study the effects of intestinal inflammatory bowel diseases
| Gene name | Forward | Reverse |
|---|---|---|
| 18 s rRNA | CGCTGAGCCAGTCAGTGTAG | CCATCCAATCGGTAGTAGCG |
| IL6 | CCACGGCCTTCCCTACTTC | TTGGGAGTGGTATCCTCTGTGA |
| TNF-alpha | CACAAGATGCTGGGACAGTGA | TCCTTGATGGTGGTGCATGA |
| CXCL-1 | GCAGACCATGGCTGGGATT | CCTGAGGGCAACACCTTCAA |
| CLDN1 | TGACCGCTCAGGCCATCTAC | CTGCCCGGTGCTTTGC |
| MMP10 | GCACCCTCAGGGACCAACTTA | CAGGGAGTGGCCAAGTTCA |
| MMP3 | CCCCTGATGTCCTCGTGGTA | GCACATTGGTGATGTCTCAGGTT |
| STAT1 | GACCACGCCTTTGGGAAGTA | TCGCTTAGGGTCGTCAAGCT |
| Ubiquitin D | GGCCCAGGTGAAAGAGATGA | CACAACCTGCTTCTTAGGGATCA |
| NOS2 | GGATCTTCCCAGGCAACCA | CAATCCACAACTCGCTCCAA |
| Lysozyme | GGGACTCCTCCTGCTTTCTGT | GGCCAACTCACAACGATTGTAG |
Statistical analysis
The differential expression of genes among the control and treatment groups was compared by two-way ANOVA followed by the Bonferroni post hoc test and FITC-dextran intensities of early (30 min) and late stages (180 min) of IBD model were analyzed by t test using GraphPad V 5.0 software (San Diego, CA, USA). The differences were considered statistically significant at p < 0.05.
Results
Immunohistochemical analysis of intestinal tissue using intestinal stem cell markers
Mouse small intestine was tested for expression of epithelial stem cell markers. Initially, intestine tissue sections were subjected to a microscopic anatomy analysis using hematoxylin (purple) and eosin (pink) histological staining to identify any possible inflammation prior to crypt isolation. Vertical and horizontal sections of the intestine were illustrated at two different levels. These were an overview (100 μm), which showed the whole anatomy of the sections, and a detailed view (50 μm), which visualized the stem cell niche in mucosa (Figure S1). The detailed view showed the integral structures of the intestinal epithelium gland, such as crypts at the bottom and the finger shaped villi. The crypt structures were composed of crypt base columnar cells, which were combined with goblet and paneth cells (Figure S1), and the apical side of the villus region showing the microvilli (purple). There were no traces of inflammation identified in the intact intestine. Furthermore, immunofluorescent analysis of the small intestine tissue showed that stem cell markers involved in stemness and proliferation were expressed in the basolateral region. The fluorescent stained crypts showed markers specific to cell types that had distinct functions, such as leucine-rich repeat containing G protein–coupled receptor 5 (LGR5), a key gene required for stemness that is expressed in columnar crypt cells; goblet cells, which were detected by mucin2; proliferation related marker Bmi1 polycomb ring finger oncogenes (Bmi1), which were found in +4 cells adjacent to the paneth cells; and adhering junctions, which were identified by E-cadherin (Fig. 1).
Fig. 1.
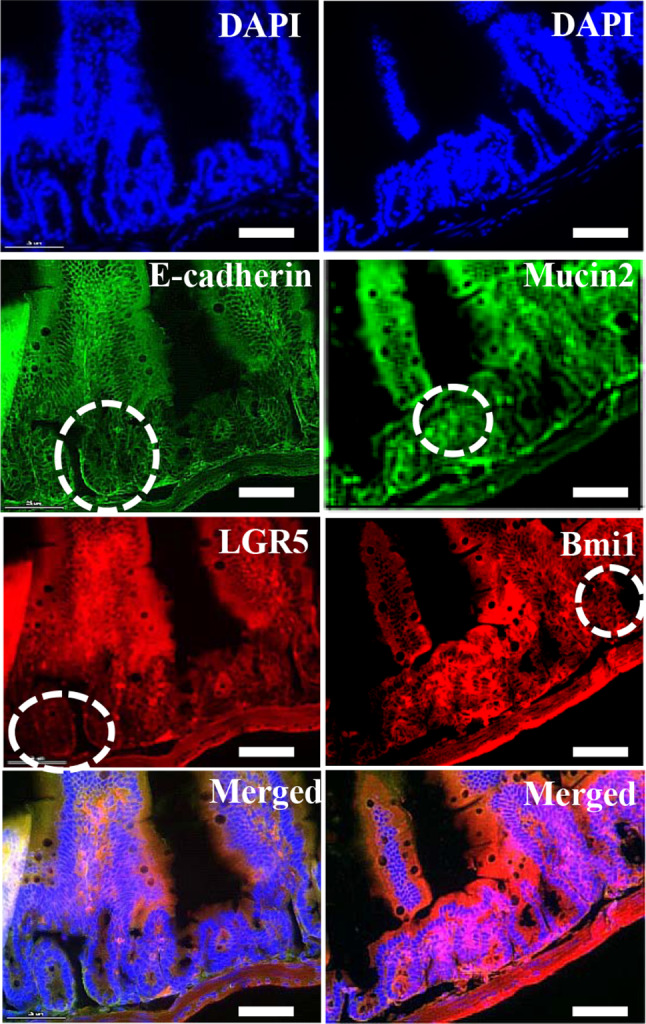
Immunohistochemical staining of mouse small intestine. Intestinal tissue expressed stem cell specific markers LGR5 (Red), Bmi1 (red), and mucin2 (green), together with adhere junctions specific E-cadherin (green) and counter stain DAPI (blue) were expressed in mouse small intestine and highlighted the positive cells with circles (white). Scale bar: 25 μm
Intestinal organoids showed consistent growth in the long-term cultures
The recapitulating capacity of the organoids was measured by long-term culturing. The mouse intestinal organoids were stably grown for more than 10 generations in low adhesive 24 well plates that contained mouse IntestiCult serum free culture media. The organoids in the long-term cultures showed distinct crypt and villus structures (branched structures) surrounding the lumen in the P2, P5, and P10 generations (Figure S3A, upper panel). Furthermore, Figure S3A also shows the growth stages of the organoid cultures and organoid population in the basement membrane (middle panel). Subsequent detailed structures represent organoid propagation from day 1 to the fully grown structure on day 10 (middle and lower panels). Following the general protocol and suggestions by IntestiCult manufacturer, StemCell Technologies, each well was sub-cultured into three wells based on the number of organoids present in the membrane. The organoid populations from the 100 μL matrix dome in each generation (n = 3 wells) were collected and used to construct a growth chart, which showed the generation passage number on the X-axis and organoid numbers per well on the Y-axis (Figure S3B). The combined results for the intact structures of the intestinal organoids at the P2, P5, and P10 passages and the growth chart indicated the recapitulating capacity of the crypt cells. Finally, the growth data showed that in each generation, an average of 203.7 organoid structures were present per basement matrix dome (Figure S3B). At this density, the organoids showed healthy and consistent growth.
Characterization and barrier permeability evaluation of intestinal organoids
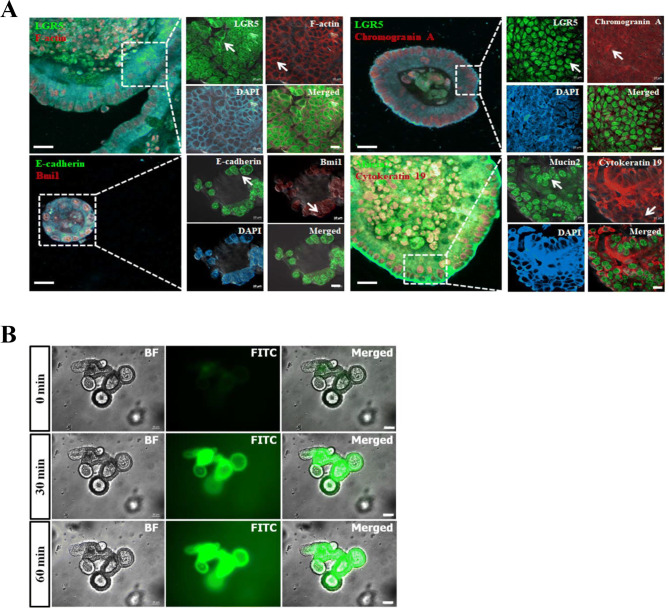
The organoids were cultured for 10 generations (P10) and evaluated for stemness and proliferation related genes. Their barrier function was assessed using FITC-dextran 4 kDa. The spatial expression of various epithelial markers was identified in respected cell types on the organoids (Fig. 2A). Columnar cells expressing LGR5 (green), goblet cells secreting mucin2 (green), which contributes to epithelial barrier integrity, proliferation marker Bmi1 (red), adhere junctions marker E-cadherin (green), matured enterocytes, goblet cells expressing cytokeratin 19 (red), enteroendocrine cells representing chromogranin A, and cytoskeleton representing F-actin are shown in Fig. 2A. The concomitant expression of intestinal epithelial genes in organoids derived from single intestinal crypts mimicked the topology of an in vivo intact intestine. Furthermore, the stable expression of stemness contributing genes, such as LGR5 and Bmi1, in long-term cultures showed the persistent recapitulating capacity of organoids under optimal conditions.
Fig. 2.
Characterization of intestinal organoids and evaluation of barrier function. A Immunofluorescent detection of intestinal organoids with Alexa Fluor 488 and 594 conjugated antibodies for stem cells and proliferation related markers, overview illustrating the morphology of individual organoids and specific region with dashed square (white) was highlighted. Detailed view of specific region was showing the spatial expression of LGR5 (green), Bmi1 (red), E-cadherin (green), mucin2 (green), chromogranin A (red) and DAPI (blue) in the different cell types. Positive cells expressing respected cell specific markers were identified with arrows (white). Scale bar: 20 μm and 10 μm. B Intestinal organoids were treated with FITC-dextran 4 kDa to assess barrier function. Organoids were indicated the bright field (grey), FITC (green), and merged, overlay panels showing at early point 0 min empty luminal space, following at 30 min and 60 min displaying the absorption of FITC-dextran 4 kDa (green) into the luminal space. The FITC was detected by excitation at 488 nm. Scale bar: 50 μm
The integrity and functionality of the epithelial barrier was determined by treating the intestinal organoids with FITC-dextran 4 kDa at 25 ng/mL and then recording the concentration. The fluorescent detection results showed that there was luminal translocation of FITC-dextran 4 kDa at 520 nm. As illustrated in Fig. 2B, no considerable traces of FITC-dextran 4 kDa were observed after a few minutes of treatment (upper row), but there was a significant rise in concentration at the 30 min time point (middle row) [21]. Concentration of FITC was maintained constantly beyond the 60 min as shown in Fig. 2B (bottom row). Slow diffusion of FITC-dextran 4 kDa indicated the presence of a mucous layer, which plays a major role in barrier function and nutrient absorption. The mucosal barrier function results showed that there was mucin2 secretion by goblet cells. Furthermore, the organoid functional testing results showed that their properties were similar to the in vivo gut absorption properties.
Dextran sodium sulfate induced damage to the epithelial barrier
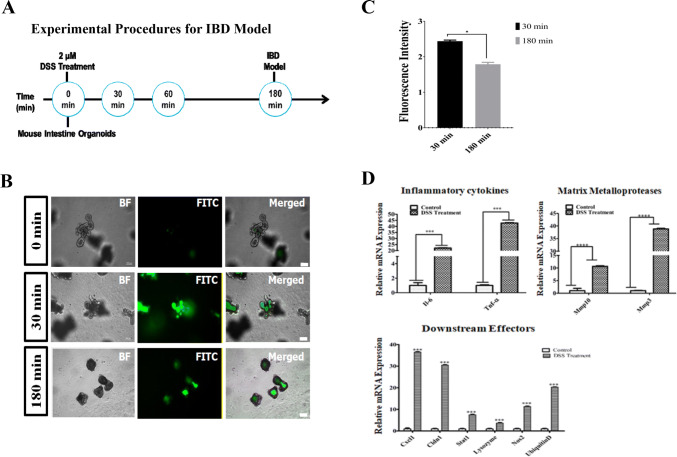
Dextran sodium sulfate (DSS) treatment damaged the organoids, which led to inflammation. Figure 3A is a schematic diagram that shows the in vitro organoid based inflammatory model development procedure when DSS was used. It also illustrates the lethal concentration and time taken to generate the model. In a two-step method, the first dose was determined by treating organoids with 1 μM, 2 μM and 5 μM of DSS in each well for 1 h. Results of inflamed organoids were showed an optimal concentration i.e. 2 μM to generate an IBD model (Figure S2A). Following, time scale findings showed the time needed to induce epithelial breach (Figure S2B). The lethal dose of 2 µM DSS systematically disrupted the organoid structure and caused morphological change (Fig. 3A). The DSS also caused a small amount of matrix membrane damage. Consequently, the absorption capability of the organoid epithelial barrier decreased, which was confirmed by the change in the luminal FITC-dextran 4 kDa concentration. The comparative results showed that the FITC-dextran 4 kDa treatment at 3 h showed reduced trans-location to the lumen compared to 30 min post inflammation (Fig. 3B). Furthermore, quantitative analysis also denoted the significant drift in FITC-dextran 4 kDa concentration at 180 min compared to 30 min of post DSS treatment (Fig. 3C). The 1 µM DSS concentration did not induce damage in the intestinal organoids at the 180 min time point, and no significant damage can be seen in Figure S2A. The 5 µM DSS concentration was considered highly cytotoxic because acute epithelial cell death was observed at 1 h post treatment and the FITC levels in the lumen were lower (Figure S2B).
Fig. 3.
Dextran sodium sulfate (DSS) induced barrier injury and qRT-PCR evaluation. A Schematic diagram showing development of the inflammatory model. B The DSS inflammatory organoid model loss of barrier function using FITC-dextran 4 kDa uptake. At the beginning of the experiment (0 min), no damage was found in organoid and observed FITC uptake (upper row). Following, no significant decline in FITC concentration (middle row) after 30 min of DSS treatment, but the FITC concentration was notably lower in the luminal space of the organoids after 180 min of treatment (bottom row). Scale bar: 100 μm. C Graph showing the quantitative difference in FITC intensity at 30 min and 180 min of post DSS treatment, data were normalized using bright field image. D The qRT-PCR analyses showing higher expression of pro-inflammatory cytokines TNF-α and IL-6 in the DSS treatment (left panel). Similarly, matrix metalloproteases MMP10 and MMP3 expressions were higher due to inflammation of the treated sample compared to normal conditions (middle panel). Downstream effectors of inflammation, such as chemokine receptor ligand CXCL1, reactive oxygen species marker (NOS2), proteome degrading ubiquitin D, tight junction protein CLDN1, and paneth cell marker lysozyme, are significantly more highly expressed in the treated sample. The values are the means plus the standard error of mean (S.E.M), and a p value < 0.05 (*) (< 0.0001***) was significant
Dextran sodium sulfate activated inflammatory signaling related to genes in the IBD model
A qRT-PCR study was used to assess cytokine signaling related to genes. The DSS caused the damaged organoids to become inflamed and increased cytokine signaling. The expressions of molecular markers involved in IBD pathogenesis were identified and the initial results showed that the inflammatory cytokines were up-regulated, particularly IL-6 and TNF-α. The TNF-α expression increased very significantly (42.78-fold increase) compared to the other tested genes in DSS inflamed organoids. Similarly, cytokine gene IL-6 was also more highly expressed (22.09-fold increase) in the treated samples, and its expression was very significantly greater than the control (Fig. 3D). Downstream effectors of TNF-α mediated inflammation were also significantly over-expressed (Fig. 3D). Inflammation caused by TNF-α and IL-6 stimulated the various signaling macro molecules, such as matrix metallopeptidases MMP10 (10.63 fold increase), MMP3 (38.8 fold increase), chemokine ligand CXCL1 (36.5 fold increase), junction protein CLDN1 (30.5 fold increase), and proteome degrading ubiquitin D (20.3 fold increase) in the DSS treated organoids (Fig. 3D). However, the free radical agent NOS2 (11.3-fold increase) and the signal transducer and activator of transcription 1 (STAT1) (7.49-fold increase) genes showed only a moderate increase in expression. The paneth cell marker gene, lysozyme, showed a significant rise in expression, but this was lower than the other genes. The expression patterns suggested that systemic inflammation played a crucial role in the disintegration of the organoid structure, which contributed to subsequent epithelial barrier dysfunction.
Discussion
Intestinal organoids are evolving as potential alternatives to animal models when studying human GI tract diseases [22]. In the present study, we used mouse intestinal stem cell (ISC) generated organoids to study epithelial barrier integrity in acute inflammatory conditions. We isolated the crypts from the small intestines of ICR mice and cultivated them using the scaffold-based method. The basement matrix membrane was long-term cultured (> P10) using pre-conditioned media. We achieved a consistent growth rate of 203.7 organoids per cell on a 100 μL dome and preserved the recapitulating capacity (Figures S3A and S3B) [6]. Additionally, as shown in Figs. 1 and 2A, comparative studies of stained intestines and organoids showed that they had similar expressions of epithelium specific markers, such as LGR5, Bmi1, mucin2, and E-cadherin. This suggested that there was a physiological similarity between the intestinal tissue and organoids [6, 23, 24]. Along with common epithelial markers, additional genes, such as enteroendocrine expressing chromogranin A and cytokeratin 19, were tested in organoids and characterized after they had been cultured for ten generations (P10) [25]. The results showed that the organoids closely mimicked the in vivo organ physiology and remained indefinitely intact under controlled culture conditions [6].
In the present study, we did not investigate molecular mechanisms. Instead, we focused on model development and applicability by studying physical parameters, such as morphology and permeability. Paracellular permeability and nutrient absorption are the primary roles of the in vivo epithelial barrier, and their analogous functions were measured in in vitro by the FITC-dextran 4 kDa uptake assay. The results demonstrated that there was regulated absorption of FITC-dextran for 60 min post incubation and that the FITC-dextran reduced cell permeability when it reached its maximum capacity, which meant that it was not possible to maintain cell homeostasis. This and previous studies showed that gut barrier integrity plays a major role in health and disease, and this study also demonstrated that intestinal organoids have a gut functionality that is similar to in vivo models [26].
The chemical inducer DSS has been widely used to create inflammatory models that can be used to study IBD etiology in mice [16]. Besides the in vivo studies, recent organoids were developed from DSS treated mouse intestines to study various genetic manifestations including IBD [27]. While, the organoids developed from pluripotent stem cells are using to study IBD in human models, it indicating the significance of ex vivo models in disease dissection [28]. The high molecular weight DSS (500 kDa) was used to generate the inflammatory model, which was different to previous in vivo studies [29, 30]. The success of the model was confirmed by the organoid morphological changes caused by necrosis (Figure S2) and barrier disintegration, which was shown by the reduction in FITC-dextran 4 kDa translocation in the luminal cavity [31]. Organoids at 180 min post treatment showed severe damage to their basal layer, which led to structure disassociation. This revealed the pathology of barrier dysfunction or a leaky gut. These conditions were due to tight junctions (TJ) dislocation, which is the key event in IBD pathogenesis [32]. Similarly, in in vivo IBD infections increases the gut permeability several folds in response to high magnitude of inflammation and luminal contents interacts with intestinal serosa. Furthermore, recent study proved the permeability was observed on either side and it was confirmed by increased concentrations of FITC-dextran in plasma as well as blood-to-stool ratio [33].
Although the exact molecular mechanism behind barrier disintegrity was not verified by this study, the causative inflammatory signals were studied. Previous high throughput studies related to IBD showed that the inflammatory signals produced by DSS treated organoids positively correlated with the IBD models. Endogenous mRNA expression studies have revealed that the significant up-regulation of pro-inflammatory cytokines, chemokines, and transcription factors influenced barrier function during chronic inflammation [34]. After analyzing IBD pathogenesis, its influence on gut barrier function, and the results from previous studies, we selected downstream TNF-α and IL-6 signaling network genes, such as MMP3, MMP10, CXCL1, NOS2, CLDN1, lysozyme, and ubiquitin D, to test whether pro-inflammatory compounds increased barrier dysfunction [35–38]. In conjunction with previous studies, the qRT-PCR analysis showed that the higher TNF-α expressions led to the dysregulation of matrix metalloproteases [39]. Additionally, comparisons between previous studies and the current results suggested that the concomitant suppression of pro-inflammatory cytokines TNF-α, and IL-6 could be a potential therapeutic solution to IBD related pathogenesis [40].
In conclusion, the organoids offer a potentially less complicated opportunity to study disease models compared to in vivo methods. The results clearly showed the differences in barrier function under inflammatory conditions compared to normal conditions. Additionally, the magnitude of the molecular signals in organoids and in vivo IBD models were similar, which showed that the technique was robust. Apart from these advantages, organoids showed higher sensitivity to DSS treatment because necrosis was induced within 3 h after treatment. However, it is difficult to study the counter mechanism used by inflamed tissues to reduce inflammation because of the lack of a macro environment for the mucosal lymphoid tissue in organoids. However, organoids deliver precise disease mechanisms with minimal limitations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1. Isolation of intestinal crypts. A ~5-week-old ICR mouse was used to extract the intestinal stem cells (left upper panel). A section of the small intestine was obtained from the euthanized mouse and washed thoroughly with PBS under aseptic conditions (left lower panel). Histological H and E staining of vertical and horizontal sections from the small intestine showing the presence of crypts (oval structures at the bottom) and villi (finger like structures). The purple spots show nuclei and the epithelium is stained pink. Scale bar: 100 μm (right upper panel) and 50 μm (right middle and lower panels). (PPT 6462 kb)
Figure S2. In vitro intestinal organoid IBD model generation using DSS. (A) The organoids were treated with the chemical agent DSS at 1 µM, 2 µM, or 5 µM per well (upper panel) to identify and optimize the lethal dose needed to induce inflammation. After incubation for 1 h, inflammation was observed in the 2 µM and 5 µM wells, but not in 1 µM wells. (B) Time course data showing the effects of the 2 µM lethal dose at 30 min, 60 min, and 180 min post-treatment (lower panel). There was minimal damage at 30 min post-treatment, but the organoids were highly inflamed and had begun to disintegrate at 180 min. There was also damage to the basement membrane, magnitude of inflammation was indicated in circles using designated colors yellow (damage) and red (lethal) in the figure. Scale bar: 100 μm (PPT 6462 kb)
Figure S3. Culturing and measuring the growth rate of mouse intestinal organoids in long-term cultures. (A) Long term culturing of intestinal organoids for up to 10 generations (P2-P10). Middle and lower panels show various developmental stages. The organoids can be spheroidal (round shaped), show budding (spheroids with extension), and have mature villus and crypt like structures (branched structures). (B) Growth rate graph showing the number of organoids/well (mean n = 3 wells) growing in a 100 µL matrigel dome on a 24 well plate. Scale bar: 100 μm (PPT 6462 kb)
Acknowledgement
This work was supported with the National Institute of Animal Sciences (Grant No. PJ01422201), Rural Development Administration (RDA), Korea.
Compliance with ethical standards
Conflicts of interest
The authors declare no conflicts of interest.
Ethical statement
The experimental use of mouse was performed after receiving approval of the Institutional Animal Care and Use Committee (IACUC) of National Institute of Animal Science (NIAS-2019-366), Korea.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chelakkot C, Ghim J, Ryu SH. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp Mol Med. 2018;50:103. doi: 10.1038/s12276-018-0126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deuring JJ, de Haar C, Kuipers EJ, Peppelenbosch MP, van der Woude CJ. The cell biology of the intestinal epithelium and its relation to inflammatory bowel disease. Int J Biochem Cell Biol. 2013;45:798–806. doi: 10.1016/j.biocel.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 3.Kraiczy J, Zilbauer M. Intestinal epithelial organoids as tools to study epigenetics in gut health and disease. Stem Cells Int. 2019;2019:7242415. doi: 10.1155/2019/7242415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 5.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 6.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 7.McCracken KW, Catá EM, Crawford CM, Sinagoga KL, Schumacher M, Rockich BE, et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature. 2014;516:400–404. doi: 10.1038/nature13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jee JH, Lee DH, Ko J, Hahn S, Jeong SY, Kim HK, et al. Development of collagen-based 3D matrix for gastrointestinal tract-derived organoid culture. Stem Cells Int. 2019;2019:8472712. doi: 10.1155/2019/8472712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ko JK, Auyeung KK. Inflammatory bowel disease: etiology, pathogenesis and current therapy. Curr Pharm Des. 2014;20:1082–1096. doi: 10.2174/13816128113199990416. [DOI] [PubMed] [Google Scholar]
- 10.Verstockt B, Smith KG, Lee JC. Genome-wide association studies in Crohn’s disease: past, present and future. Clin Transl Immunology. 2018;7:e1001. doi: 10.1002/cti2.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang YZ, Li YY. Inflammatory bowel disease: pathogenesis. World J Gastroenterol. 2014;20:91–99. doi: 10.3748/wjg.v20.i1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westbrook AM, Szakmary A, Schiestl RH. Mechanisms of intestinal inflammation and development of associated cancers: lessons learned from mouse models. Mutat Res. 2010;705:40–59. doi: 10.1016/j.mrrev.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hibiya S, Tsuchiya K, Hayashi R, Fukushima K, Horita N, Watanabe S, et al. Long-term inflammation transforms intestinal epithelial cells of colonic organoids. J Crohns Colitis. 2017;11:621–630. doi: 10.1093/ecco-jcc/jjw186. [DOI] [PubMed] [Google Scholar]
- 14.DeVoss J, Diehl L. Murine models of inflammatory bowel disease (IBD): challenges of modeling human disease. Toxicol Pathol. 2014;42:99–110. doi: 10.1177/0192623313509729. [DOI] [PubMed] [Google Scholar]
- 15.Scheiffele F, Fuss IJ. Induction of TNBS colitis in mice. Curr Protoc Immunol. 2002;Chapter 15:Unit 15.19. [DOI] [PubMed]
- 16.Kiesler P, Fuss IJ, Strober W. Experimental models of inflammatory bowel diseases. Cell Mol Gastroenterol Hepatol. 2015;1:154–170. doi: 10.1016/j.jcmgh.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bredenoord AL, Clevers H, Knoblich JA. Human tissues in a dish: the research and ethical implications of organoid technology. Science. 2017;355:eaaf9414. doi: 10.1126/science.aaf9414. [DOI] [PubMed] [Google Scholar]
- 18.Sato T, Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340:1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- 19.Lee BR, Kim H, Park TS, Moon S, Cho S, Park T, et al. A set of stage-specific gene transcripts identified in EK stage X and HH stage 3 chick embryos. BMC Dev Biol. 2007;7:60. doi: 10.1186/1471-213X-7-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rallabandi HR, Yang H, Jo YJ, Lee HC, Byun SJ, Lee BR. Identification of female specific genes in the W chromosome that are expressed during gonadal differentiation in the chicken. Korean J Poult Sci. 2019;46:287–296. doi: 10.5536/KJPS.2019.46.4.287. [DOI] [Google Scholar]
- 21.Pearce SC, Al-Jawadi A, Kishida K, Yu S, Hu M, Fritzky LF, et al. Marked differences in tight junction composition and macromolecular permeability among different intestinal cell types. BMC Biol. 2018;16:19. doi: 10.1186/s12915-018-0481-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sala FG, Kunisaki SM, Ochoa ER, Vacanti J, Grikscheit TC. Tissue-engineered small intestine and stomach form from autologous tissue in a preclinical large animal model. J Surg Res. 2009;156:205–212. doi: 10.1016/j.jss.2009.03.062. [DOI] [PubMed] [Google Scholar]
- 23.Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A. 2012;109:466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pastula A, Middelhoff M, Brandtner A, Tobiasch M, Höhl B, Nuber AH, et al. Three-dimensional gastrointestinal organoid culture in combination with nerves or fibroblasts: a method to characterize the gastrointestinal stem cell niche. Stem Cells Int. 2016;2016:3710836. doi: 10.1155/2016/3710836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagatake T, Fujita H, Minato N, Hamazaki Y. Enteroendocrine cells are specifically marked by cell surface expression of claudin-4 in mouse small intestine. PLoS One. 2014;9:e90638. doi: 10.1371/journal.pone.0090638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altay G, Larrañaga E, Tosi S, Barriga FM, Batlle E, Fernández-Majada V, et al. Self-organized intestinal epithelial monolayers in crypt and villus-like domains show effective barrier function. Sci Rep. 2019;9:10140. doi: 10.1038/s41598-019-46497-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitt M, Schewe M, Sacchetti A, Feijtel D, van de Geer WS, Teeuwssen M, et al. Paneth cells respond to inflammation and contribute to tissue regeneration by acquiring stem-like features through SCF/c-Kit signaling. Cell Rep. 2018;24:2312–28.e7. [DOI] [PubMed]
- 28.Dotti I, Salas A. Potential use of human stem cell-derived intestinal organoids to study inflammatory bowel diseases. Inflamm Bowel Dis. 2018;24:2501–2509. doi: 10.1093/ibd/izy275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitajima S, Takuma S, Morimoto M. Histological analysis of murine colitis induced by dextran sulfate sodium of different molecular weights. Exp Anim. 2000;49:9–15. doi: 10.1538/expanim.49.9. [DOI] [PubMed] [Google Scholar]
- 30.Okayasu I, Hatakeyama S, Yamada M, Ohkusa T, Inagaki Y, Nakaya R. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology. 1990;98:694–702. doi: 10.1016/0016-5085(90)90290-H. [DOI] [PubMed] [Google Scholar]
- 31.Bardenbacher M, Ruder B, Britzen-Laurent N, Schmid B, Waldner M, Naschberger E, et al. Permeability analyses and three dimensional imaging of interferon gamma-induced barrier disintegration in intestinal organoids. Stem Cell Res. 2019;35:101383. doi: 10.1016/j.scr.2019.101383. [DOI] [PubMed] [Google Scholar]
- 32.Michielan A, D’Incà R. Intestinal permeability in inflammatory bowel disease: pathogenesis, clinical evaluation, and therapy of leaky gut. Mediators Inflamm. 2015;2015:628157. doi: 10.1155/2015/628157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaworska K, Konop M, Bielinska K, Hutsch T, Dziekiewicz M, Banaszkiewicz A, et al. Inflammatory bowel disease is associated with increased gut-to-blood penetration of short-chain fatty acids: a new, non-invasive marker of a functional intestinal lesion. Exp Physiol. 2019;104:1226–1236. doi: 10.1113/EP087773. [DOI] [PubMed] [Google Scholar]
- 34.Dobre M, Milanesi E, Mănuc TE, Arsene DE, Ţieranu CG, Maj C, et al. Differential intestinal mucosa transcriptomic biomarkers for Crohn’s disease and ulcerative colitis. J Immunol Res. 2018;2018:9208274. doi: 10.1155/2018/9208274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobayashi K, Arimura Y, Goto A, Okahara S, Endo T, Shinomura Y, et al. Therapeutic implications of the specific inhibition of causative matrix metalloproteinases in experimental colitis induced by dextran sulphate sodium. J Pathol. 2006;209:376–383. doi: 10.1002/path.1978. [DOI] [PubMed] [Google Scholar]
- 36.Kawamoto A, Nagata S, Anzai S, Takahashi J, Kawai M, Hama M, et al. Ubiquitin D is upregulated by synergy of notch signalling and TNF-α in the inflamed intestinal epithelia of IBD patients. J Crohns Colitis. 2019;13:495–509. doi: 10.1093/ecco-jcc/jjy180. [DOI] [PubMed] [Google Scholar]
- 37.Dhillon SS, Mastropaolo LA, Murchie R, Griffiths C, Thöni C, Elkadri A, et al. Higher activity of the inducible nitric oxide synthase contributes to very early onset inflammatory bowel disease. Clin Transl Gastroenterol. 2014;5:e46. doi: 10.1038/ctg.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giles EM, Sanders TJ, McCarthy NE, Lung J, Pathak M, MacDonald TT, et al. Regulation of human intestinal T-cell responses by type 1 interferon-STAT1 signaling is disrupted in inflammatory bowel disease. Mucosal Immunol. 2017;10:184–193. doi: 10.1038/mi.2016.44. [DOI] [PubMed] [Google Scholar]
- 39.Armaka M, Apostolaki M, Jacques P, Kontoyiannis DL, Elewaut D, Kollias G. Mesenchymal cell targeting by TNF as a common pathogenic principle in chronic inflammatory joint and intestinal diseases. J Exp Med. 2008;205:331–337. doi: 10.1084/jem.20070906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandborn WJ, Hanauer SB. Antitumor necrosis factor therapy for inflammatory bowel disease: a review of agents, pharmacology, clinical results, and safety. Inflamm Bowel Dis. 1999;5:119–133. doi: 10.1097/00054725-199905000-00008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Isolation of intestinal crypts. A ~5-week-old ICR mouse was used to extract the intestinal stem cells (left upper panel). A section of the small intestine was obtained from the euthanized mouse and washed thoroughly with PBS under aseptic conditions (left lower panel). Histological H and E staining of vertical and horizontal sections from the small intestine showing the presence of crypts (oval structures at the bottom) and villi (finger like structures). The purple spots show nuclei and the epithelium is stained pink. Scale bar: 100 μm (right upper panel) and 50 μm (right middle and lower panels). (PPT 6462 kb)
Figure S2. In vitro intestinal organoid IBD model generation using DSS. (A) The organoids were treated with the chemical agent DSS at 1 µM, 2 µM, or 5 µM per well (upper panel) to identify and optimize the lethal dose needed to induce inflammation. After incubation for 1 h, inflammation was observed in the 2 µM and 5 µM wells, but not in 1 µM wells. (B) Time course data showing the effects of the 2 µM lethal dose at 30 min, 60 min, and 180 min post-treatment (lower panel). There was minimal damage at 30 min post-treatment, but the organoids were highly inflamed and had begun to disintegrate at 180 min. There was also damage to the basement membrane, magnitude of inflammation was indicated in circles using designated colors yellow (damage) and red (lethal) in the figure. Scale bar: 100 μm (PPT 6462 kb)
Figure S3. Culturing and measuring the growth rate of mouse intestinal organoids in long-term cultures. (A) Long term culturing of intestinal organoids for up to 10 generations (P2-P10). Middle and lower panels show various developmental stages. The organoids can be spheroidal (round shaped), show budding (spheroids with extension), and have mature villus and crypt like structures (branched structures). (B) Growth rate graph showing the number of organoids/well (mean n = 3 wells) growing in a 100 µL matrigel dome on a 24 well plate. Scale bar: 100 μm (PPT 6462 kb)