Abstract
Introduction.
Despite a wealth of epidemiological evidence that cumulative parental lifetime stress experiences prior to conception are determinant of offspring developmental trajectories, there is a lack of insight on how these previous stress experiences are stored and communicated intergenerationally. Preconception experiences may impact offspring development through alterations in transcriptional regulation of the placenta, a major determinant of offspring growth and sex-specific developmental outcomes. We evaluated the lasting influence of maternal and paternal preconception stress (PCS) on the mid-gestation placenta and fetal brain, utilizing their transcriptomes as proximate readouts of intergenerational impact.
Methods.
To assess the combined vs. dominant influence of maternal and paternal preconception environment on sex-specific fetal development, we compared transcriptional outcomes using a breeding scheme of one stressed parent, both stressed parents, or no stressed parents as controls.
Results.
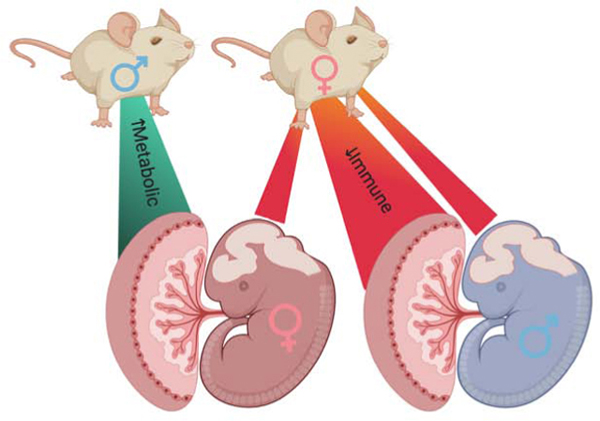
Interestingly, offspring sex affected the directionality of transcriptional changes in response to PCS, where male tissues showed a predominant downregulation, and female tissues showed an upregulation. There was also an intriguing effect of parental sex on placental programming where paternal PCS drove more effects in female placentas, while maternal PCS produced more transcriptional changes in male placentas. However, in the fetal brain, maternal PCS produced overall more changes in gene expression than paternal PCS, supporting the idea that the intrauterine environment may have a larger overall influence on the developing brain than it does on shaping the placenta.
Discussion.
Preconception experiences drive changes in the placental and the fetal brain transcriptome at a critical developmental timepoint. While not determinant, these altered transcriptional states may underlie sex-biased risk or resilience to stressful experiences later in life.
Graphical abstract
Lifetime stress and trauma, or the cumulative stressors experienced by an individual across their lifespan, is a predisposing factor for the development of chronic diseases, including neuropsychiatric disorders [1–4]. Neuropsychiatric disorders affect approximately 19.1% of adults in the United States [5]. Evidence from both rodent models and humans suggest that the physiological and psychological consequences of lifetime stress can be transmitted from parent to offspring, i.e. intergenerational transmission [5–7]. A mechanism by which maternal and paternal experiences prior to conception impact offspring development is through altering the transcriptional landscape in embryonic and extra-embryonic tissues, including the placenta [8–10]. Prenatal stress significantly alters sex-specific placental gene expression, driving sex differences in neurodevelopmental programming and offspring growth, metabolism, and behavior in adulthood [12–16]. Preconception trauma in women, and stress in female rodents, is associated with changes in offspring brain development, stress sensitivity, and neuropsychiatric outcomes [17–22]. Although studies have addressed the effects of maternal stress during gestation, little is known about the mechanisms by which maternal preconception experiences elicit such effects on offspring outcomes [17,23,24]. Recent studies have also demonstrated that paternal preconception experiences are transmitted to offspring via altered sperm content [25–28]. Specifically, paternal preconception stress programs offspring neurodevelopment and stress reactivity via sperm small-RNA content [29–33]. While maternal and paternal preconception contributions have been independently examined, an understanding of parental influences and driving forces on fetal development at the molecular level is lacking.
Conception involves a complex interplay of opposing maternal and paternal interests orchestrated at the level of the epigenome. Imprinting is the most widely recognized example of this “genetic-conflict hypothesis”, where paternally derived genes promote growth to maximize offspring fitness and maternally derived genes suppress growth to mitigate the energetic costs of gestation [34–36]. Post-conception, genetic-conflict is mediated at the placenta. As both the barrier and central integrator of signals between the maternal and fetal compartments, the placenta must meet the metabolic demands of the fetus and maintain maternal survival [35,37,38]. In response to a dynamic maternal milieu, broad placental transcriptional responses drive physiological adaptations to maintain support in a fetal sex-specific manner [39–44]. Inappropriate or ill-timed reactivity to perturbations in the maternal environment by the placenta can result in nutrient deficits and restricted resources to highly metabolic tissues, such as the fetal brain [14,45–47]. Proper placental development and function is a key determinant of gestational growth and long-term offspring health outcomes [48–57]. Sex-specific parental experiences prior to conception may affect the developing fetal and placental transcriptome, potentially altering the developmental trajectory of offspring in a fetal sex-specific manner.
In this study, adult male and female mice were exposed to four weeks of chronic stress. In order to examine the enduring effects of pre-conception stress (PCS), two weeks prior to breeding mice were returned to standard housing conditions. To determine potential independent and interacting effects of maternal and paternal PCS on sex-specific offspring development, we assessed global changes in the placental and fetal brain transcriptome at embryonic day 12.5 (E12.5). This timepoint coincides with maturation of the placenta and differentiation of the hypothalamus, critical events in tissues determining offspring growth and neurodevelopment [58,59]. We therefore evaluated changes in the placenta and fetal brain transcriptome as a proxy for the potential enduring effects of PCS on offspring development. While not determinant, such changes provide a critical snapshot of altered pathways that may underlie sex-biased risk or resilience later in life.
Materials and Methods
Animals
Mice for parental preconception stress studies were generated from one breeding cohort of virgin in-house bred C57BL/6:129 hybrid strain mice. Mice were housed in a 12 hr light/dark cycle and provided ad libitum access to food (Purina Rodent Chow; 28.1% protein, 59.8% carbohydrate, 12.1% fat) and water. All studies were performed according to experimental protocols approved by the University of Pennsylvania Institutional Animal Care and Use Committee and were conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
Preconception Stress
On postnatal day (PN) 28, male (n=32) and female (n=32) mice were weaned into pair-housing and randomly assigned to a control non-stressed group or to an experimental group that underwent 28 days of preconception stress (PCS), consistent with our paternal preconception stress model [29]. Briefly, seven different stressors were administered, one per day, and order was randomized across weeks of stress. Stressors included 36 hr constant light, 15 min exposure to fox odor (1:5000 2,4,5-trimethylthiazole; Acros Organics, Geel, Belgium), novel object (marbles) overnight, 15 min restraint in a 50 mL conical tube, multiple cage changes, novel 100 dB white noise overnight, and saturated bedding overnight.
Breeding scheme
Consistent with previous experiments, following stress PCS mice were returned to standard housing for 14 days to recover from the acute effects of stress on reproductive behavior [25]. Control and PCS mice were then paired in a full factorial design resulting in the following breedings: non-stressed controls (C♂-C♀, n=8), PCS dams (C♂-S♀, n=8), PCS sires (S♂-C♀, n=8), and PCS dams and sires (S♂-S♀, n=8). Breeding pairs were housed together for a maximum of 4 nights, separated when a plug was observed, and litters were collected at embryonic day 12.5 (E12.5). All offspring were generated from one breeding cohort of PCS
Tissue Collection
Pregnant females were anesthetized and decapitated at E12.5. Placentas and fetal brains were collected from each litter, rapidly frozen in liquid nitrogen, and stored at −80°C. Tissues from one male and one female from each litter were used for further analyses.
RNA Sequencing and analysis
RNA from E12.5 placentas and fetal brains was isolated by RNeasy kit (Qiagen, Valencia, CA) and suspended in RNAse-free water. Libraries for RNA Sequencing were prepared using TruSeq Library Preparation Kit v2 (Illumina, San Diego, CA) according to the manufacturer’s protocol. Quantity and quality of libraries were assessed an Agilent 4200 TapeStation (Agilent Technologies, Wilmington, DE). Individually barcoded libraries were pooled by tissue and by sex, such that 3 control (C♂-C♀), 3 maternal PCS (C♂-S♀), 2 paternal PCS (S♂-C♀), and 6 biparental PCS (S♂-S♀) samples were sequenced on each Illumina NextSeq 500 (single-end 75bp) flow cell, to control for batch effects. Fastq files containing an average of 50 million reads were processed in the R environment using Kallisto (version 0.43.1) to perform pseudoalignment and abundance quantification of reads to the mouse transcriptome (Mus musculus v. 79). Gene set enrichment analysis (GSEAv2.0.7, Broad Institute, Cambridge, MA) of normalized counts was used to interpret broad patterns of gene expression and determine greater-than-chance enrichment of biological pathways in a threshold-free manner (i.e. without consideration for differential gene expression). Collections of c2 (curated: Kegg), and c5 (gene ontology) annotated gene sets were obtained from the Molecular Signature Database (MSigDBv3.0.1, Broad Institute, Cambridge, MA) available for use with GSEA software. Gene set permutations (1000) were computed in GSEA to determine FDR, nominal p value, and normalized enrichment score (NES) of each gene set. Significance thresholds were set at FDR ≤ 0.0025, p ≤ 0.01, and NES ≥ 1.6 to account for gene set permutation.
Results
Reproductive outcomes of parental PCS pairings
Adult male and female mice were paired in full factorial design two weeks after completion of the PCS paradigm to generate 4 pairing groups: non-stressed controls (C♂-C♀), PCS dams (C♂- S♀), PCS sires (S♂-C♀), and PCS dams and sires (S♂-S♀). Final viable litters fitting the criteria of successful mating two weeks post-stress and staged at E12.5, are described in Table 1. Despite variability in plug and pregnancy rates between groups, these differences were not statistically significant (χ2 (3, N=20) = 3.60 and χ2 (3, N=14) = 2.57, p > 0.05, respectively).
Table 1.
Summary of breeding outcomes.
| Pairings |
|||||
|---|---|---|---|---|---|
| C♂-C♀ (n=8) | C♂-S♀ (n=8) | S♂-C♀ (n=8) | S♂-S♀ (n=8) | P valueb | |
| Vaginal Plug Observed, n (%) | 3/8 (37.5) | 6/8 (75) | 3/8 (37.5) | 8/8 (100) | 0.31 |
| Pregnant/Plugged, n (%) | 3/3 (100) | 3/6 (50) | 2/3 (67) | 6/8 (75) | 0.46 |
| Litter Sizea | 10.33 ± 1.53 | 12 ± 1.00 | 10.5 ± 0.71 | 9.67 ± 3.98 | 0.96 |
| Males per Littera | 7.3 ± 2.08 | 4.3 ± 1.15 | 5 ± 1.41 | 5.3 ± 2.73 | 0.79 |
| Females per Littera | 3 ± 1.00 | 7.7 ± 1.53 | 5.5 ± 0.71 | 4.3 ± 2.07 | 0.51 |
Values represent litter characteristics at E12.5 expressed as mean ± SD
Pearson’s □2
Placental sex-specific transcriptomic responses to parental PCS
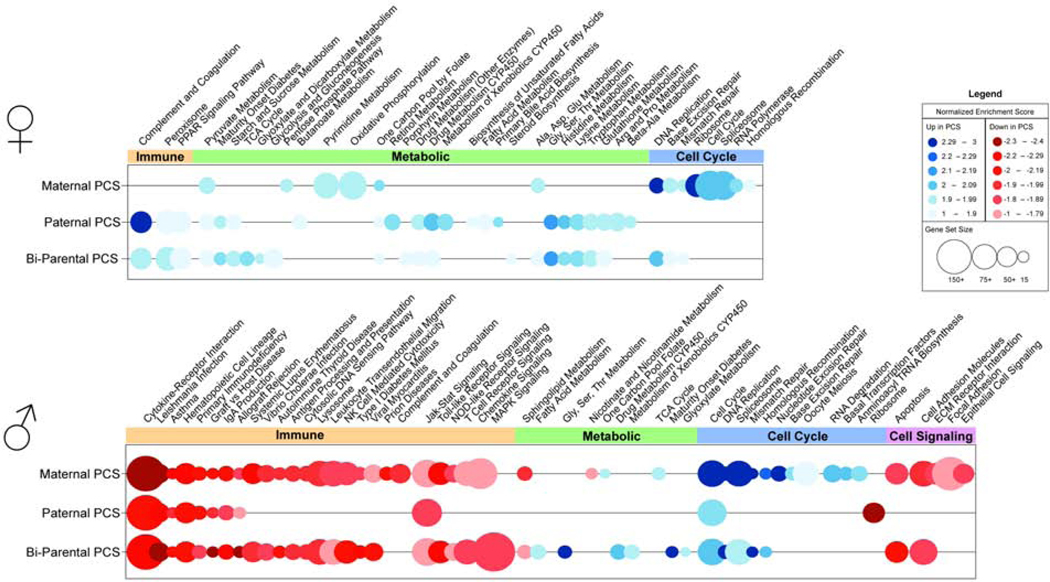
To examine how maternal and paternal preconception experience may be reflected at the level of the placenta by mid-gestation, we compared transcriptional profiles of male and female placentas from control (C♂-C♀), maternal PCS (C♂-S♀), paternal PCS (S♂-C♀), and bi-parental PCS (S♂-S♀) using RNA-sequencing. By GSEA analysis with enrichment criteria of FDR ≤0.0025 p ≤0.01 and NES ≥1.6, there was a main effect of PCS on female placental gene expression, irrespective of parental sex (Table 2), where gene sets involved in pyruvate and folate metabolism were especially enriched in PCS female placentas relative to controls (Fig 1 and Supplementary Table 1). In analyzing parent-specific contributions of PCS, we found that cell metabolism and cell cycling gene sets were increased in placentas of maternal PCS, but paternal PCS female placentas increased gene sets for carbohydrate, lipid, and amino acid metabolism. Bi-parental PCS female placentas were positively enriched for steroid biosynthesis and cell metabolism gene sets, specifically the TCA cycle and glycolysis/gluconeogenesis. Paternal effects were most similar to bi-parental PCS in female placentas where both increased expression of gene sets involved in PPAR signaling and complement and coagulation.
Table 2.
Summary of placental gene sets altered in parental PCS offspring.
| Female |
Male |
|||||
|---|---|---|---|---|---|---|
| Maternal | Paternal | Bi-parental | Maternal | Paternal | Bi-parental | |
| KEGG | 15 | 22 | 23 | 45 | 11 | 37 |
| BP | 25 | 41 | 14 | 286 | 4 | 95 |
Total gene sets with significantly increased or decreased expression in male and female PCS offspring placentas relative to controls. GSEA enrichment criteria were FDR ≤ 0.0025 p ≤ 0.01, and NES ≥ 1.6.
Figure 1.
Parental preconception stress dramatically reduces male offspring placental transcription. Bubble plots of gene sets identified by gene set enrichment analysis (GSEA) as significantly altered in female (top) and male (bottom) placentas of maternal (n=3), paternal (n=2), and bi-parental (n=6) PCS offspring relative to controls (n=3). Bubbles represent the normalized enrichment score (NES) of a particular gene set, where color indicates increased (blue) or decreased (red) enrichment, color intensity indicates magnitude of change, and bubble diameter represents gene set size. Differentially enriched gene sets, meeting criteria of FDR ≤ 0.0025 p ≤ 0.01, and NES ≥ 1.6, were grouped into functional categories spanning immunity (red), metabolism (green), cell cycling (blue), and cell signaling (purple). Gene set enrichment analysis (GSEA) of placentas at embryonic day 12.5 (E12.5) indicated a marked decrease in gene expression in placentas of male PCS offspring relative to controls, specifically in genes mediating immune processes. Female PCS offspring increased placental expression of gene sets mediating metabolic processes. A full list of enriched gene sets and statistics are detailed in Supplementary Tables 1 and 2.
In male placental tissue, gene expression was substantially altered by PCS regardless of parental sex (Table 2), where specifically there was a reduction in expression of gene sets involved in immune function and allograft rejection relative to control males (Fig 1 and Supplementary Table 2). In analyzing parent-specific contributions of PCS, we found that maternal PCS reduced expression of gene sets for complement and coagulation and the extracellular matrix, while increasing expression of cell metabolic and mRNA surveillance gene sets relative to control male placentas. Gene sets involved in MAPK signaling were reduced in expression, and gene sets regulating lipid, amino acid and carbohydrate metabolism increased expression in bi-parental PCS offspring placentas relative to control male placentas. Maternal effects were most similar to bi-parental PCS in male placentas where both reduced expression of gene sets involved in additional immune pathways, and increased expression of DNA repair gene sets relative to control male placentas.
Fetal brain sex-specific transcriptomic responses to parental PCS
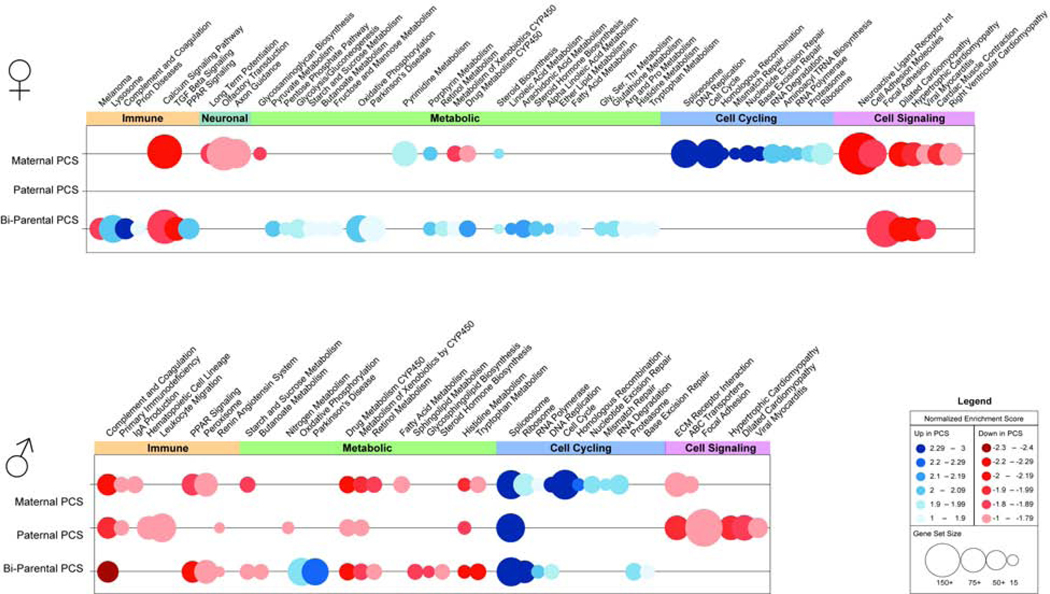
To examine PCS parent-of-origin effects reflected in the developing brain, transcriptional profiles of E12.5 brains were also determined by RNA-sequencing. GSEA analysis of the E12.5 brain transcriptome revealed a main effect of PCS, regardless of parental sex, on both male and female offspring fetal brain gene expression compared to offspring brains from non-stressed parents (Table 3). Specific effects of maternal PCS showed that female offspring brains increased gene expression of pathways related to DNA replication and repair, and nucleotide metabolism, but neuron and synapse development gene sets, such as calcium signaling, axon guidance, and long term potentiation were decreased in expression (Fig 2 and Supplementary Table 3). Bi-parental PCS female brains increased expression of complement and coagulation, PPAR signaling, and cellular metabolism gene sets, while decreasing expression of TGF-B, MAPK, and cell-cell signaling gene sets relative to control female brains. Relative to control offspring, no gene sets were significantly altered by paternal PCS in female brains. Bi-parental and maternal PCS female brains similarly decreased calcium signaling gene sets and increased steroid biosynthesis gene set expression.
Table 3.
Summary of fetal brain gene sets altered in parental PCS offspring.
| Female |
Male |
|||||
|---|---|---|---|---|---|---|
| Maternal | Paternal | Bi-parental | Maternal | Paternal | Bi-parental | |
| KEGG | 17 | 0 | 33 | 23 | 16 | 22 |
| BP | 235 | 0 | 22 | 204 | 22 | 66 |
Total gene sets with significantly increased or decreased expression in male and female fetal PCS offspring brains relative to controls. GSEA enrichment criteria were FDR ≤ 0.0025 p ≤ 0.01, and NES ≥ 1.6.
Figure 2.
Maternal preconception stress shapes the offspring fetal brain transcriptome. Bubble plots of gene sets identified by gene set enrichment analysis (GSEA) as significantly altered in fetal brains of female (top) and male (bottom) offspring of maternal (n=3), paternal (n=2), and bi-parental (n=6) PCS relative to controls (n=3). Bubbles represent the normalized enrichment score (NES) of a particular gene set, where color indicates increased (blue) or decreased (red) enrichment, color intensity indicates magnitude of change, and bubble diameter represents gene set size. Differentially enriched gene sets, meeting criteria of FDR ≤ 0.0025 p ≤ 0.01, and NES ≥ 1.6, were grouped into functional categories spanning immunity (red), metabolism (green), cell cycling (blue), and cell signaling (purple). Gene set enrichment analysis (GSEA) of placentas at embryonic day 12.5 (E12.5) indicated a marked decrease in gene expression in placentas of male PCS offspring relative to controls, specifically in genes mediating immune Supplementary Tables 3 and 4.
In male brains, PCS increased expression of genes involved in generating the spliceosome, and decreased genes involved in complement and coagulation, CYP450 metabolism, and histidine metabolism relative to control males (Fig 2 and Supplementary Table 4). Maternal PCS increased cell cycle, DNA repair, and transcriptional machinery gene set expression, but decreased gene sets involved in humoral immunity, nutrient transport, and fatty acid metabolism. Paternal PCS reduced gene expression related to immune signaling, calcium signaling, and nitrogen metabolism. Bi-parental PCS increased expression of oxidative phosphorylation and proteolysis genes, while gene sets regulating steroid hormone biosynthesis were reduced in expression relative to controls. Bi-parental and maternal PCS both increased expression of DNA replication and transcription gene sets, and relatively reduced expression of complement and coagulation, extracellular matrix, PPAR signaling, and metabolism of amino acids, carbohydrates, and retinol gene sets compared to controls.
Discussion
Despite evidence that parental adverse life experiences prior to conception are a strong determinant of offspring neurodevelopmental outcomes, there is a lack of understanding as to how maternal or paternal lifetime stress interact to influence offspring development. Developmental trajectories are modulated in an offspring sex-specific manner through changes in the epigenetic and transcriptional patterns of placental tissues contributing to sex differences in fetal development [13,15,60,61]. In the current study, we probed for parent- and offspring-sex-specific changes in the placental and fetal brain transcriptome as a snapshot of mid-gestation, a key period of placental maturation and brain development, in response to MPS.
The placenta expresses the fetal genetic sex, which facilitates differences in male and female placental transcription, largely promoted by the X and Y chromosomes [10,50,62,63]. These divergent gene expression patterns alter placental function and responses to the maternal environment, ultimately driving sex differences in offspring developmental trajectories [12,16,64–66]. Consistent with previous studies, we found that offspring exhibited sex-specific changes to the transcriptional landscape of the mid-gestation placenta in response to parental preconception stress (PCS). Indeed, increased expression of immune-related genes was also observed in the male placenta of dams exposed to early preconception stress [67]. Although preconception paternal derived programming of female offspring outcomes [68–70] and preconception maternal derived effects on adult male offspring [19,71,72] have been documented, little is known about the mechanisms driving this parent- and offspring- sex specificity. Importantly, our data also demonstrated parental sex-specific effects on placental transcriptional changes, such that maternal and paternal PCS offspring differed in the magnitude, directionality, and functional categories of gene sets affected. The predominant effect of paternal preconception stress on the placenta was an upregulation in expression of 19 gene sets involved in metabolic signaling, specifically in the female placenta. When examined together with maternal effects, bi-parental PCS female placentas still shared a significant enrichment of gene sets involved in cellular and energy metabolism with paternal PCS placentas, suggesting that paternal life experiences dominate over maternal for his female offspring placenta. Paternal PCS shaping of female placental nutrient synthesis and metabolism at this dynamic time point in gestation aligns with the paternal interests in “genetic conflict theory”, altering placental nutrient supply, and ultimately determining intrauterine growth and metabolism of female offspring [34,73–75]. It is unclear why this effect is only apparent in the female placenta, but as we are studying a single timepoint in development, we may be observing a sex difference dependent on the rate or this particular stage of development.
In stark contrast to female placental outcomes, male offspring, regardless of parental sex, robustly reduced placental gene expression when compared to same sex controls. Specifically, maternal PCS had a much greater influence on the male placental transcriptome than paternal PCS, robustly altering the expression of 45 gene sets, relative to 11 with paternal PCS, where a majority of the gene sets were involved in immunity and immune signaling, suggesting a potential shift in placental immune environment at this mid-gestation time point in the male placenta. Bi-parental PCS males shared 21 of 37 altered placental gene sets with maternal PCS, further supporting the dominant role of maternal PCS on the male placental transcriptome. Changes in the complex and precise regulation of immunity at the maternal-fetal interface may underlie susceptibility or resilience to immune challenges during gestation, specifically in the male placenta, and have long term effects on the developmental trajectory of male offspring [76–80].
The preconception environment of the parent is a strong determinant of the offspring neurodevelopmental outcomes [19,53,72,81–86]. Transcriptional adaptations of the placenta drive sex-specific changes in communication and transport at the maternal:fetal interface that impact fetal brain development [10,50,62,66]. Given the parent and offspring sex-specific changes in placental gene expression, we also examined the E12.5 brain transcriptome. Maternal PCS drove transcriptional alterations in both male and female fetal brains relative to paternal PCS. There were no significantly enriched gene sets in paternal PCS females, suggesting that at this mid-gestational time point there is: (1) minimal transcriptional influence of paternal PCS the female fetal brain, or (2) transmission of a heterogenous signal that does not fall into distinct gene sets. However, maternal PCS only shared ~25% of enriched gene sets with bi-parental PCS, which suggests some influence of paternal PCS. Similar to the placenta, there was a significant reduction in gene expression of male PCS fetal brains, excepting gene sets involved in cell cycling. Maternal PCS male brains were enriched for 23 gene sets relative to 16 in paternal PCS, which suggests some influence of paternal PCS. Similar to the placenta, there was a significant reduction in gene expression of male PCS fetal brains, excepting gene sets involved in cell cycling. Maternal PCS male brains were enriched for 23 gene sets relative to 16 in paternal PCS. When accounting for 6 gene sets enriched in all PCS groups, there was a 50% overlap of gene sets altered in bi-parental and maternal PCS relative to only 6% in paternal PCS, supporting the dominant role of maternal PCS on gene expression changes in the fetal brain for both sexes.
In summary, these findings demonstrate that parental PCS experience may shape offspring development in a parent- and offspring- sex specific manner. At mid-gestation, the fetal brain transcriptome appeared markedly altered by maternal PCS in both males and females. However, we saw far greater parental and offspring sex-specific outcomes in the placental transcriptome. Additionally, these data outline the sex-difference in transcriptional response to parental PCS; with male offspring suppressing and female offspring enriching immune and metabolic gene sets in both the placenta and the brain. Given the tight regulation of immunity and metabolism during development, these transcriptional adaptations, while not determinant, may suggest sex-specific shifts in utilization of energy resources and protection from the external environment. As these data are limited to transcriptional readouts, further studies are needed to understand the ultimate effects of these developmental events. This study supports the growing appreciation for the impact of parental lifetime stress and preconception adverse experiences on offspring development, possibly underlying sex differences in long-term health risk and resilience.
Supplementary Material
Highlights.
Preconception stress (PCS) alters offspring development in a parental and fetal sex-specific manner.
Maternal PCS markedly decreased immune-related genes in the male placenta.
Paternal PCS altered metabolic-related genes in the female placenta.
Acknowledgements
This work was supported by the National Institutes of Health grants ES028202, MH108286, HD097093 (TLB), and F32HD101301 (YMC).
Footnotes
Declarations of interest: none.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Sledjeski EM, Speisman B, Dierker LC, Does number of lifetime traumas explain the relationship between PTSD and chronic medical conditions? Answers from the National Comorbidity Survey-Replication (NCS-R), J. Behav. Med 31 (2008) 341–349. doi: 10.1007/s10865-008-9158-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sotiropoulos I, Catania C, Pinto LG, Silva R, Pollerberg GE, Takashima A, Sousa N, Almeida OFX, Stress acts cumulatively to precipitate Alzheimer’s disease-like tau pathology and cognitive deficits, J. Neurosci 31 (2011) 7840–7847. doi: 10.1523/JNEUROSCI.0730-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Berntson J, Patel JS, Stewart JC, Number of recent stressful life events and incident cardiovascular disease: Moderation by lifetime depressive disorder, J. Psychosom. Res 99 (2017) 149–154. doi: 10.1016/j.jpsychores.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Juster RP, McEwen BS, Lupien SJ, Allostatic load biomarkers of chronic stress and impact on health and cognition, Neurosci. Biobehav. Rev 35 (2010) 2–16. doi: 10.1016/j.neubiorev.2009.10.002. [DOI] [PubMed] [Google Scholar]
- [5].2018 NSDUH Annual National Report | CBHSQ Data, (n.d.). https://www.samhsa.gov/data/report/2018-nsduh-annual-national-report (accessed May 8, 2020).
- [6].Yao Y, Robinson AM, Zucchi FC, Robbins JC, Babenko O, Kovalchuk O, Kovalchuk I, Olson DM, Metz GA, Ancestral exposure to stressepigenetically programs preterm birth risk and adverse maternal and newbornoutcomes, BMC Med. 12 (2014) 121. doi: 10.1186/S12916-014-0121-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Franklin TB, Russig H, Weiss IC, Gräff J, Linder N, Michalon A, Vizi S, Mansuy IM, Epigenetic Transmission of the Impact of Early Stress Across Generations, Biol. Psychiatry 68 (2010) 408–415. doi: 10.1016/J.BIOPSYCH.2010.05.036. [DOI] [PubMed] [Google Scholar]
- [8].Bale TL, Lifetime stress experience: Transgenerational epigenetics and germ cell programming, Dialogues Clin. Neurosci 16 (2014) 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fowden AL, Coan PM, Angiolini E, Burton GJ, Constancia M, Imprinted genes and the epigenetic regulation of placental phenotype, Prog. Biophys. Mol. Biol 106 (2011) 281–288. doi: 10.1016/j.pbiomolbio.2010.11.005. [DOI] [PubMed] [Google Scholar]
- [10].Gabory A, Roseboom TJ, Moore T, Moore LG, Junien C, Placental contribution to the origins of sexual dimorphism in health and diseases: sex chromosomes and epigenetics, Biol. Sex Differ 4 (2013) 5. doi: 10.1186/2042-6410-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nugent BM, Bale TL, The omniscient placenta: Metabolic and epigenetic regulation of fetal programming, Front. Neuroendocrinol 39 (2015) 28–37. doi: 10.1016/J.YFRNE.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mao J, Zhang X, Sieli PT, Falduto MT, Torres KE, Rosenfeld CS, Contrasting effects of different maternal diets on sexually dimorphic gene expression in the murine placenta, Proc. Natl. Acad. Sci. U. S. A 107 (2010) 5557–5562. doi: 10.1073/pnas.1000440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Howerton CL, Morgan CP, Fischer DB, Bale TL, O-GlcNAc transferase (OGT) as a placental biomarker of maternal stress and reprogramming of CNS gene transcription in development, Proc. Natl. Acad. Sci. U. S. A 110 (2013) 5169–5174. doi: 10.1073/pnas.1300065110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Howerton CL, Bale TL, Targeted placental deletion of OGT recapitulates the prenatal stress phenotype including hypothalamic mitochondrial dysfunction., Proc. Natl. Acad. Sci. U. S. A 111 (2014) 9639–44. doi: 10.1073/pnas.1401203111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gabory A, Ferry L, Fajardy I, Jouneau L, Gothié JD, Vigé A, Fleur C, Mayeur S, Gallou-Kabani C, Gross MS, Attig L, Vambergue A, Lesage J, Reusens B, Vieau D, Remacle C, Jais JP, Junien C, Maternal Diets Trigger Sex-Specific Divergent Trajectories of Gene Expression and Epigenetic Systems in Mouse Placenta, PLoS One. 7 (2012). doi: 10.1371/journal.pone.0047986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bronson SL, Chan JC, Bale TL, Sex-Specific Neurodevelopmental Programming by Placental Insulin Receptors on Stress Reactivity and Sensorimotor Gating, Biol. Psychiatry 82 (2017) 127–138. doi: 10.1016/j.biopsych.2016.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Moog NK, Buss C, Entringer S, Shahbaba B, Gillen DL, Hobel CJ, Wadhwa PD, Maternal Exposure to Childhood Trauma Is Associated during Pregnancy with Placental-Fetal Stress Physiology, Biol. Psychiatry 79 (2016) 831–839. doi: 10.1016/j.biopsych.2015.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yehuda R, Daskalakis NP, Bierer LM, Bader HN, Klengel T, Holsboer F, Binder EB, Holocaust Exposure Induced Intergenerational Effects on FKBP5 Methylation, Biol. Psychiatry 80 (2016) 372–380. doi: 10.1016/j.biopsych.2015.08.005. [DOI] [PubMed] [Google Scholar]
- [19].Bock J, Poeschel J, Schindler J, Börner F, Shachar-Dadon A, Ferdman N, Gaisler-Salomon I, Leshem M, Braun K, Poeggel G, Transgenerational sex-specific impact of preconception stress on the development of dendritic spines and dendritic length in the medial prefrontal cortex, Brain Struct. Funct 221 (2016) 855–863. doi: 10.1007/s00429014-0940-4. [DOI] [PubMed] [Google Scholar]
- [20].Huang Y, Shi X, Xu H, Yang H, Chen T, Chen S, Chen X, Chronic Unpredictable Stress Before Pregnancy Reduce the Expression of Brain-Derived Neurotrophic Factor and N-Methyl-D-Aspartate Receptor in Hippocampus of Offspring Rats Associated with Impairment of Memory, Neurochem. Res 35 (2010) 1038–1049. doi: 10.1007/s11064010-0152-0. [DOI] [PubMed] [Google Scholar]
- [21].Zaidan H, Leshem M, Gaisler-Salomon I, Prereproductive stress to female rats alters corticotropin releasing factor type 1 expression in ova and behavior and brain corticotropin releasing factor type 1 expression in offspring, Biol. Psychiatry 74 (2013) 680–687. doi: 10.1016/j.biopsych.2013.04.014. [DOI] [PubMed] [Google Scholar]
- [22].Zaidan H, Gaisler-Salomon I, Prereproductive stress in adolescent female rats affects behavior and corticosterone levels in second-generation offspring, Psychoneuroendocrinology. 58 (2015) 120–129. doi: 10.1016/j.psyneuen.2015.04.013. [DOI] [PubMed] [Google Scholar]
- [23].Stroud LR, Papandonatos GD, Parade SH, Salisbury AL, Phipps MG, Lester BM, Padbury JF, Marsit CJ, Prenatal major depressive disorder, placenta glucocorticoid and serotonergic signaling, and infant cortisol response, Psychosom. Med 78 (2016) 979–990. doi: 10.1097/PSY.0000000000000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tian X, Anthony K, Neuberger T, Diaz FJ, Preconception Zinc Deficiency Disrupts Postimplantation Fetal and Placental Development in Mice1, Biol. Reprod 90 (2014). doi: 10.1095/biolreprod.113.113910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rodgers AB, Morgan CP, Bronson SL, Revello S, Bale TL, Paternal stress exposure alters sperm microRNA content and reprograms offspring HPA stress axis regulation., J. Neurosci 33 (2013) 9003–12. doi: 10.1523/JNEUROSCI.0914-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, Bock C, Li C, Gu H, Zamore PD, Meissner A, Weng Z, Hofmann HA, Friedman N, Rando OJ, Paternally Induced Transgenerational Environmental Reprogramming of Metabolic Gene Expression in Mammals, Cell. 143 (2010) 1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Zhang Y, Qian J, Duan E, Zhai Q, Zhou Q, Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder, Science (80-. ). 351 (2016) 397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- [28].Lambrot R, Xu C, Saint-Phar S, Chountalos G, Cohen T, Paquet M, Suderman M, Hallett M, Kimmins S, Low paternal dietary folate alters the mouse sperm epigenome and is associated with negative pregnancy outcomes, Nat. Commun 4 (2013) 1–13. doi: 10.1038/ncomms3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chan JC, Morgan CP, Adrian Leu N, Shetty A, Cisse YM, Nugent BM, Morrison KE, Jašarević E, Huang W, Kanyuch N, Rodgers AB, Bhanu NV, Berger DS, Garcia BA, Ament S, Kane M, Neill Epperson C, Bale TL, Reproductive tract extracellular vesicles are sufficient to transmit intergenerational stress and program neurodevelopment, Nat. Commun 11 (2020) 1–13. doi: 10.1038/s41467-020-15305-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, Song L, Carone BR, Ricci EP, Li XZ, Fauquier L, Moore MJ, Sullivan R, Mello CC, Garber M, Rando OJ, Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals, Science (80-. ). 351 (2016) 391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Rodgers AB, Morgan CP, Leu NA, Bale TL, Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress, Proc. Natl. Acad. Sci 112 (2015) 13699–13704. doi: 10.1073/pnas.1508347112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Cunningham AM, Walker DM, Nestler EJ, Paternal transgenerational epigenetic mechanisms mediating stress phenotypes of offspring, Eur. J. Neurosci (2019) ejn.14582. doi: 10.1111/ejn.14582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gapp K, van Steenwyk G, Germain PL, Matsushima W, Rudolph KLM, Manuella F, Roszkowski M, Vernaz G, Ghosh T, Pelczar P, Mansuy IM, Miska EA, Alterations in sperm long RNA contribute to the epigenetic inheritance of the effects of postnatal trauma, Mol. Psychiatry (2018) 1–13. doi: 10.1038/s41380-018-0271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Haig D, Transfers and transitions: Parent-offspring conflict, genomic imprinting, and the evolution of human life history, Proc. Natl. Acad. Sci. U. S. A 107 (2010) 1731–1735. doi: 10.1073/pnas.0904111106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Haig D, Placental hormones, genomic imprinting, and maternal-fetal communication, J. Evol. Biol 9 (1996) 357–380. doi: 10.1046/j.1420-9101.1996.9030357.x. [DOI] [Google Scholar]
- [36].Spencer HG, Feldman MW, Clark AG, Weisstein AE, The Effect of Genetic Conflict on Genomic Imprinting and Modification of Expression at a Sex-Linked Locus, Genetics. 166 (2004) 565–579. doi: 10.1534/genetics.166.1.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Moore T, Review: Parent-offspring conflict and the control of placental function, in: Placenta, 2012. doi: 10.1016/j.placenta.2011.11.016. [DOI] [PubMed] [Google Scholar]
- [38].Haig D, Evolutionary conflicts in pregnancy and calcium metabolism - A review, Placenta. 25 (2004). doi: 10.1016/j.placenta.2004.01.006. [DOI] [PubMed] [Google Scholar]
- [39].Sandovici I, Hoelle K, Angiolini E, Constância M, Placental adaptations to the maternal–fetal environment: implications for fetal growth and developmental programming, Reprod. Biomed. Online. 25 (2012) 68–89. doi: 10.1016/J.RBMO.2012.03.017. [DOI] [PubMed] [Google Scholar]
- [40].Sferruzzi-Perri AN, Higgins JS, Vaughan OR, Murray AJ, Fowden AL, Placental mitochondria adapt developmentally and in response to hypoxia to support fetal growth., Proc. Natl. Acad. Sci. U. S. A 116 (2019) 1621–1626. doi: 10.1073/pnas.1816056116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Coan PM, Angiolini E, Sandovici I, Burton GJ, Constância M, Fowden AL, Adaptations in placental nutrient transfer capacity to meet fetal growth demands depend on placental size in mice, J. Physiol 586 (2008) 4567–4576. doi: 10.1113/jphysiol.2008.156133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Napso T, Yong HEJ, Lopez-Tello J, Sferruzzi-Perri AN, The Role of Placental Hormones in Mediating Maternal Adaptations to Support Pregnancy and Lactation, Front. Physiol 9 (2018) 1091. doi: 10.3389/fphys.2018.01091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Schoenfelder S, Mifsud B, Senner CE, Todd CD, Chrysanthou S, Darbo E, Hemberger M, Branco MR, Divergent wiring of repressive and active chromatin interactions between mouse embryonic and trophoblast lineages, Nat. Commun 9 (2018) 1–10. doi: 10.1038/s41467-018-06666-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hemberger M, Hanna CW, Dean W, Mechanisms of early placental development in mouse and humans, Nat. Rev. Genet 21 (2020) 27–43. doi: 10.1038/s41576-019-0169-4. [DOI] [PubMed] [Google Scholar]
- [45].Logue OC, George EM, Bidwell GL, Preeclampsia and the brain: neural control of cardiovascular changes during pregnancy and neurological outcomes of preeclampsia., Clin. Sci. (Lond). 130 (2016) 1417–34. doi: 10.1042/CS20160108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wallingford MC, Benson C, Chavkin NW, Chin MT, Frasch MG, Placental Vascular Calcification and Cardiovascular Health: It Is Time to Determine How Much of Maternal and Offspring Health Is Written in Stone, Front. Physiol 9 (2018) 1044. doi: 10.3389/fphys.2018.01044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hart B, Morgan E, Alejandro EU, Nutrient sensor signaling pathways and cellular stress in fetal growth restriction, J. Mol. Endocrinol 62 (2019) R155–R165. doi: 10.1530/JME18-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Roos S, Powell TL, Jansson T, Placental mTOR links maternal nutrient availability to fetal growth., Biochem. Soc. Trans 37 (2009) 295–8. doi: 10.1042/BST0370295. [DOI] [PubMed] [Google Scholar]
- [49].Fowden AL, Sferruzzi-Perri AN, Coan PM, Constancia M, Burton GJ, Placental efficiency and adaptation: endocrine regulation., J. Physiol 587 (2009) 3459–72. doi: 10.1113/jphysiol.2009.173013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Rosenfeld CS, Sex-Specific Placental Responses in Fetal Development, Endocrinology. 156 (2015) 3422–3434. doi: 10.1210/en.2015-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Delhaes F, Giza SA, Koreman T, Eastabrook G, McKenzie CA, Bedell S, Regnault TRH, de Vrijer B, Altered maternal and placental lipid metabolism and fetal fat development in obesity: Current knowledge and advances in non-invasive assessment, Placenta. 69 (2018) 118–124. doi: 10.1016/j.placenta.2018.05.011. [DOI] [PubMed] [Google Scholar]
- [52].Dimasuay KG, Boeuf P, Powell TL, Jansson T, Placental responses to changes in the maternal environment determine fetal growth, Front. Physiol 7 (2016). doi: 10.3389/fphys.2016.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Muller CL, Anacker AM, Rogers TD, Goeden N, Keller EH, Forsberg CG, Kerr TM, La Wender C, Anderson GM, Stanwood GD, Blakely RD, Bonnin A, Veenstra-Vanderweele J, Impact of Maternal Serotonin Transporter Genotype on Placental Serotonin, Fetal Forebrain Serotonin, and Neurodevelopment, Neuropsychopharmacology. 42 (2017) 427–436. doi: 10.1038/npp.2016.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Rashid CS, Bansal A, Simmons RA, Oxidative stress, intrauterine growth restriction, and developmental programming of type 2 diabetes, Physiology. 33 (2018) 348–359. doi: 10.1152/physiol.00023.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Bansal A, Rashid C, Simmons RA, Impact of Fetal Programming on Mitochondrial Function and Susceptibility to Obesity and Type 2 Diabetes, in: Mitochondria Obes. Type 2 Diabetes, Elsevier, 2019: pp. 325–345. doi: 10.1016/b978-0-12-811752-1.00014-6. [DOI] [Google Scholar]
- [56].Paquette AG, Chu T, Wu X, Wang K, Price ND, Sadovsky Y, Distinct communication patterns of trophoblastic miRNA among the maternal-placental-fetal compartments, Placenta. 72–73 (2018) 28–35. doi: 10.1016/j.placenta.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Chang G, Mouillet J-F, Mishima T, Chu T, Sadovsky E, Coyne CB, Parks WT, Surti U, Sadovsky Y, Expression and trafficking of placental microRNAs at the fetomaternal interface, FA SEB J. 31 (2017) 2760–2770. doi: 10.1096/fj.201601146R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Chen VS, Morrison JP, Southwell MF, Foley JF, Bolon B, Elmore SA, Histology Atlas of the Developing Prenatal and Postnatal Mouse Central Nervous System, with Emphasis on Prenatal Days E7.5 to E18.5, Toxicol. Pathol 45 (2017) 705–744. doi: 10.1177/0192623317728134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Malassine A, Frendo J-L, Evain-Brion D, A comparison of placental development and endocrine functions between the human and mouse model, Academic.Oup.Com. (n.d.). doi: 10.1093/humupd/dmg043. [DOI] [PubMed] [Google Scholar]
- [60].Muralimanoharan S, Gao X, Weintraub S, Myatt L, Maloyan A, Sexual dimorphism in activation of placental autophagy in obese women with evidence for fetal programming from a placenta-specific mouse model, Autophagy. 12 (2016) 752–769. doi: 10.1080/15548627.2016.1156822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Gong S, Sovio U, Aye IL, Gaccioli F, Dopierala J, Johnson MD, Wood AM, Cook E, Jenkins BJ, Koulman A, Casero RA, Constância M, Charnock-Jones DS, Smith GC, Placental polyamine metabolism differs by fetal sex, fetal growth restriction, and preeclampsia, JCI Insight. 3 (2018). doi: 10.1172/jci.insight.120723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kalisch-Smith JI, Simmons DG, Dickinson H, Moritz KM, Review: Sexual dimorphism in the formation, function and adaptation of the placenta, Placenta. 54 (2017) 10–16. doi: 10.1016/J.PLACENTA.2016.12.008. [DOI] [PubMed] [Google Scholar]
- [63].Cvitic S, Longtine MS, Hackl H, Wagner K, Nelson MD, Desoye G, Hiden U, The Human Placental Sexome Differs between Trophoblast Epithelium and Villous Vessel Endothelium, PLoS One. 8 (2013) e79233. doi: 10.1371/journal.pone.0079233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Gallou-Kabani C, Gabory A, Tost J, Karimi M, Mayeur S, Lesage J, Boudadi E, Gross MS, Taurelle J, Vigé A, Breton C, Reusens B, Remacle C, Vieau D, Ekström TJ, Jais J-P, Junien C, Sex- and Diet-Specific Changes of Imprinted Gene Expression and DNA Methylation in Mouse Placenta under a High-Fat Diet, PLoS One. 5 (2010) e14398. doi: 10.1371/journal.pone.0014398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Osei-Kumah A, Smith R, Jurisica I, Caniggia I, Clifton VL, Sex-specific differences in placental global gene expression in pregnancies complicated by asthma., Placenta. 32 (2011) 570–8. doi: 10.1016/j.placenta.2011.05.005. [DOI] [PubMed] [Google Scholar]
- [66].Nugent BM, O’Donnell CM, Epperson CN, Bale TL, Placental H3K27me3 establishes female resilience to prenatal insults, Nat. Commun 9 (2018) 2555. doi: 10.1038/s41467-018-04992-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Bronson SL, Bale TL, Prenatal Stress-Induced Increases in Placental Inflammation and Offspring Hyperactivity Are Male-Specific and Ameliorated by Maternal Antiinflammatory Treatment, Endocrinology. 155 (2014) 2635–2646. doi: 10.1210/en.2014-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].McPherson NO, Owens JA, Fullston T, Lane M, Preconception diet or exercise intervention in obese fathers normalizes sperm microRNA profile and metabolic syndrome in female offspring, Am. J. Physiol. Metab 308 (2015) E805–E821. doi: 10.1152/ajpendo.00013.2015. [DOI] [PubMed] [Google Scholar]
- [69].Chambers TJG, Morgan MD, Heger AH, Sharpe RM, Drake AJ, High-fat diet disrupts metabolism in two generations of rats in a parent-of-origin specific manner, Sci. Rep 6 (2016) 1–11. doi: 10.1038/srep31857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Chang RC, Skiles WM, Chronister SS, Wang H, Sutton GI, Bedi YS, Snyder M, Long CR, Golding MC, DNA methylation-independent growth restriction and altered developmental programming in a mouse model of preconception male alcohol exposure, Epigenetics. 12 (2017) 841–853. doi: 10.1080/15592294.2017.1363952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Srinivasan M, Katewa SD, Palaniyappan A, Pandya JD, Patel MS, Maternal high-fat diet consumption results in fetal malprogramming predisposing to the onset of metabolic syndrome-like phenotype in adulthood, Am. J. Physiol. - Endocrinol. Metab 291 (2006). doi: 10.1152/ajpendo.00078.2006. [DOI] [PubMed] [Google Scholar]
- [72].Jabbar S, Chastain LG, Gangisetty O, Cabrera MA, Sochacki K, Sarkar DK, Preconception alcohol increases offspring vulnerability to stress, Neuropsychopharmacology. 41 (2016) 2782–2793. doi: 10.1038/npp.2016.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Brett K, Ferraro Z, Yockell-Lelievre J, Gruslin A, Adamo K, Brett KE, Ferraro ZM, Yockell-Lelievre J, Gruslin A, Adamo KB, Maternal–Fetal Nutrient Transport in Pregnancy Pathologies: The Role of the Placenta, Int. J. Mol. Sci 15 (2014) 16153–16185. doi: 10.3390/ijms150916153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Haffner S, Temprosa M, Crandall J, Fowler S, Goldberg R, Horton E, Marcovina S, Mather K, Orchard T, Ratner R, Barrett-Connor E, Diabetes Prevention Program Research Group, Intensive lifestyle intervention or metformin on inflammation and coagulation in participants with impaired glucose tolerance., Diabetes. 54 (2005) 1566–72. http://www.ncbi.nlm.nih.gov/pubmed/15855347 (accessed July 23, 2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Connor KL, Kibschull M, Matysiak-Zablocki E, Nguyen TTTN, Matthews SG, Lye SJ, Bloise E, Maternal malnutrition impacts placental morphology and transporter expression: an origin for poor offspring growth, J. Nutr. Biochem 78 (2020) 108329. doi: 10.1016/j.jnutbio.2019.108329. [DOI] [PubMed] [Google Scholar]
- [76].Shanks N, Lightman SL, The maternal-neonatal neuro-immune interface: are there long-term implications for inflammatory or stress-related disease?, J. Clin. Invest 108 (2001) 1567–73. doi: 10.1172/JCI14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Regal JF, Gilbert JS, Burwick RM, The complement system and adverse pregnancy outcomes., Mol. Immunol 67 (2015) 56–70. doi: 10.1016/j.molimm.2015.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Christian LM, Psychoneuroimmunology in pregnancy: Immune pathways linking stress with maternal health, adverse birth outcomes, and fetal development, Neurosci. Biobehav. Rev 36 (2012) 350–361. doi: 10.1016/J.NEUBIOREV.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Lewis EL, Barila G, Brown AG, Porrett PM, Elovitz MA, Dynamic changes in immune populations at the fetomaternal interface during term and preterm birth, J. Immunol 200 (2018). [Google Scholar]
- [80].Leavey K, Grynspan D, Cox BJ, Both “canonical” and “immunological” preeclampsia subtypes demonstrate changes in placental immune cell composition, Placenta. 83 (2019) 53–56. doi: 10.1016/j.placenta.2019.06.384. [DOI] [PubMed] [Google Scholar]
- [81].Class QA, Abel KM, Khashan AS, Rickert ME, Dalman C, Larsson H, Hultman CM, Långström N, Lichtenstein P, D’Onofrio BM, Offspring psychopathology following preconception, prenatal and postnatal maternal bereavement stress, Psychol. Med 44 (2014) 71–84. doi: 10.1017/S0033291713000780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Dietz DM, Laplant Q, Watts EL, Hodes GE, Russo SJ, Feng J, Oosting RS, Vialou V, Nestler EJ, Paternal transmission of stress-induced pathologies, Biol. Psychiatry 70 (2011) 408–414. doi: 10.1016/j.biopsych.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Korgan AC, O’Leary E, King JL, Weaver ICG, Perrot TS, Effects of paternal high-fat diet and rearing environment on maternal investment and development of defensive responses in the offspring, Psychoneuroendocrinology. 91 (2018) 20–30. doi: 10.1016/j.psyneuen.2018.02.010. [DOI] [PubMed] [Google Scholar]
- [84].Li J, Olsen J, Vestergaard M, Obel C, Attention-deficit/hyperactivity disorder in the offspring following prenatal maternal bereavement: A nationwide follow-up study in Denmark, Eur. Child Adolesc. Psychiatry 19 (2010) 747–753. doi: 10.1007/s00787-010-0113-9. [DOI] [PubMed] [Google Scholar]
- [85].Braun K, Bock J, Wainstock T, Matas E, Gaisler-Salomon I, Fegert J, Ziegenhain U, Segal M, Experience-induced transgenerational (re-)programming of neuronal structure and functions: Impact of stress prior and during pregnancy, Neurosci. Biobehav. Rev (2016). doi: 10.1016/j.neubiorev.2017.05.021. [DOI] [PubMed] [Google Scholar]
- [86].Huang Y, Shi X, Xu H, Yang H, Chen T, Chen S, Chen X, Chronic unpredictable stress before pregnancy reduce the expression of brain-derived neurotrophic factor and N-methyl-D-aspartate receptor in hippocampus of offspring rats associated with impairment of memory, Neurochem. Res 35 (2010) 1038–1049. doi: 10.1007/s11064-010-0152-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.