Abstract
Aberrant activation of androgen receptor (AR) signaling occurs in patients treated with AR-targeted therapies, contributing to the development of castration-resistant prostate cancer (CRPC) and therapeutic resistance. Over the past decade, many AR variants (AR-Vs) have been identified in prostate cancer cell lines and clinical CRPC specimens. These AR-Vs lack the COOH-terminal ligand-binding domain (LBD), and may mediate constitutively active AR signaling acquired following AR-targeting therapies. AR splice variant-7 (AR-V7), one of the most well characterized AR-Vs, can be reliably measured in tissue and liquid biopsy specimens, and blood-based detection of AR-V7 is a reliable indicator of poor outcome to relatively novel hormonal therapies (NHT) such as abiraterone and enzalutamide in men with metastatic CRPC (mCRPC). Given the important clinical implication of AR-Vs, this short review will focus on studies addressing how AR-Vs are regulated in prostate cancer. With regard to the molecular origin of AR-Vs, it is established that expression of AR-Vs is highly correlated with androgen deprivation and suppression of AR signaling. Therapeutic targeting of the AR axis may result in active transcription of the AR gene, elevated activities of certain components of the mRNA splicing machinery, as well as AR genomic alterations, all of which may explain the molecular origin of AR-Vs. Although a unified hypothesis is currently lacking, existing data suggest that elevated expression of AR-Vs, which in general occurs quite specifically in a cellular environment where the canonical AR signaling is suppressed, is driven by both genomic and epigenomic features acquired in the development of CRPC.
Keywords: Androgen receptor, Androgen receptor splice variant-7, Castration-resistant prostate cancer
1. Introduction
In 1941, Huggins and Hodges [1] demonstrated the clinical benefit of hormonal manipulation in patients with metastatic prostate cancer, establishing prostate cancer as an androgen-dependent disease. Since then, androgen deprivation therapy (ADT) has been the mainstay of prostate cancer treatment. Androgens mediate the biological effects through the androgen receptor (AR). Given time, patients with advanced prostate cancer treated with ADT inevitably progress to castration-resistant prostate cancer (CRPC). In CRPC, AR expression is often increased, and AR activity is typically maintained under castrate levels of circulating testosterone (<50 ng/dL), implicating continued dependence of CRPC on AR signaling [[2], [3], [4]]. The importance of sustained AR activity in CRPC was further supported by the successful clinical development of more potent AR-targeting agents that either antagonize AR (enzalutamide, apalutamide, darolutamide) or further suppress extragonadal androgen synthesis (abiraterone) [[5], [6], [7], [8], [9], [10], [11], [12]]. Abiraterone blocks the synthesis of both adrenal and intra-tumoral androgens through inhibition of CYP17A activity [5,7], while AR antagonists bind to the AR ligand-binding domain (LBD) with higher affinity [6,8]. Importantly, these relatively novel agents demonstrated clinical benefits beyond metastatic CRPC (mCRPC), leading to approved use in earlier disease settings, including metastatic castration-sensitive prostate cancer (CSPC) [9] and non-metastatic CRPC [[10], [11], [12]].
Nearly all treated patients will inevitably develop acquired resistance to AR-targeting agents. Mechanisms of resistance can be broadly categorized into AR-dependent and AR-indifferent mechanisms [13]. Given the intent of this review, we will focus on AR signaling mediated by AR splice variants (AR-Vs) that lack the LBD, the target of all current AR-targeting agents. Specifically, we will focus on current knowledge and understanding concerning regulation of AR-V expression and molecular origin of AR-Vs in prostate cancer. Readers are directed to recent review articles covering other topics related to AR-Vs, including AR-V functional mechanisms and therapeutic implications [14,15].
2. Summary of AR-Vs characterized so far
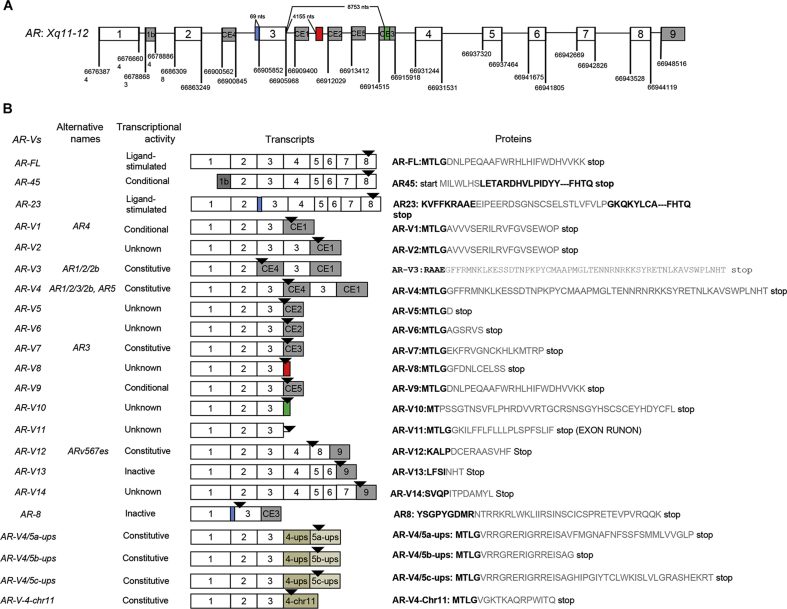
It is an established concept that aberrant AR activation occurs during the development of drug resistance to AR-targeting therapies. Specific types of AR alterations include AR gene amplification and overexpression [[16], [17], [18], [19], [20]], AR mutations [21], alterations leading to extragonadal (e.g., adrenal or intra-tumoral) androgen synthesis [[22], [23], [24], [25]], and emergence of constitutive active AR-Vs [[14], [15], [16]]. So far, more than 20 different AR-Vs have been identified (with mRNA sequences cloned) from both prostate cancer cell lines and clinical CRPC specimens (summarized in Fig. 1) [26]. We have previously categorized these AR-Vs into three different groups based on their transcriptional activity, including constitutively active AR-Vs (e.g., AR-V7, ARv567es), conditionally active AR-Vs (e.g., AR-V1, AR-V9), and inactive AR-Vs (e.g., AR-V13, AR-V14) [26]. Most of these AR-Vs have an intact transactivation N-terminal domain (NTD) and DNA-binding domain (DBD), followed by a variant-specific short peptide that replaces the C-terminal LBD [14,15,[27], [28], [29], [30]]. Relatively novel AR-Vs involving splicing between AR exon 3 and sequences from other segments of the genome (AR-V4/5a-ups, AR-V4/5b-ups, AR-V4/5c-ups, AR-V4-chr11) were identified in CRPC specimens recently, with all showing constitutive AR activity in vitro [31] (Fig. 1).
Figure 1.
Illustration of the AR splice variant transcript nomenclature, functional annotation, exon compositions and specific peptide sequences. (A) AR gene structure with canonical and cryptic exon splice junctions marked according to GRCh37/hg19 human genome sequences (not drawn to scale); (B) Nomenclature, functional annotation, exon compositions, and variant-specific mRNA (color matched to A) and peptide sequences (in gray). Modified from Reference [26] with addition of novel AR variants with unconventional exons (in gold) from structural rearrangements. AR, androgen receptor.
Among the AR-Vs identified so far, AR-V7 has been characterized the most extensively and continues to be the focus of studies on AR-Vs. AR-V7 mRNA and protein expression studies were first reported in 2009 [28]. It is now established that AR-V7 expression is specific to CRPC [32], negatively regulated by signaling mediated by the full-length AR (AR-FL) coexisting in the same cell [33,34], and may drive a progressive phenotype when canonical AR signaling is suppressed [33,35]. Although AR-V7 and AR-FL share common chromatin binding sites, AR-V7 also displayed distinct binding sites by interacting with different coregulators/repressors [35]. For example, key mediators such as HOXB13 and ZFX can mediate AR-V7 function [36,37], and AR-V7 prefers binding to open chromatin regions in prostate cancer cells [37,38]. It is, therefore, not surprising that AR-V7 drives a distinct transcriptional program in an AR-FL-indifferent manner in an earlier study [33]. Given the focus of this review, however, readers are directed to a few recent reviews and other published studies for detailed functional mechanisms and clinical implications concerning AR-Vs [14,15,26,30,31,33,34].
2.1. AR-V transcription is regulated by AR-FL signaling
In CRPC, elevated AR expression may reflect AR self-regulation. Cai et al. [39] showed that AR binds to a site in intron 2 of the AR gene through recruitment of lysine-specific demethylase 1 (LSD1). This binding represses AR expression when androgen is present, suggesting a negative feedback mechanism of AR transcription. Therefore, it is possible that suppression of AR-FL signaling will abrogate LSD1 recruitment and release AR self-repression, leading to elevated AR transcription which in turn is generally coupled with occurrence of AR-Vs in CRPC. Although detailed mechanistic investigation may uncover additional key regulators, studies in multiple preclinical models have established that transcription of AR-Vs can be regulated by androgen signaling. For example, expression of AR-V7 in cell lines was shown to be dynamic, and can be rapidly suppressed by addition of androgens or induced by inhibition of AR-FL signaling (e.g., through androgen deprivation, anti-androgen enzalutamide, or AR-FL siRNA) [33,40], and AR-FL/AR-V expression can be dramatically upregulated upon castration in prostate cancer xenograft models (VCaP and LuCaP35) [34], or upon treatment with abiraterone in two CRPC xenografts (LuCAP35CR and LuCaP23CR) [41]. Increased expression of AR-FL/AR-V7 was also observed in other prostate cancer cells (e.g., LAPC4 and DuCaP cells) following long-term culture in the presence of enzalutamide [42]. Collectively, these findings from experimental models are consistent with the general expression pattern of AR-Vs that are more frequently and abundantly detected in CRPC than in CSPC [28,29,32,40,[43], [44], [45], [46], [47]]. Negative regulation of AR-V7 by androgen signaling was recently validated in a clinical trial of mCRPC patients treated with bipolar androgen therapies (BAT) [[48], [49], [50]], in which the majority of pre-treatment AR-V7-positive patients (determined by a blood-based test) turned AR-V7 negative following BAT [48]. In addition to prostate cancer, AR-FL and AR-V7 are known to be highly expressed in salivary duct carcinoma (SDC) collected before ADT [51]. Interestingly, three SDCs with the highest levels of AR-V7 were all from female patients in whom androgen levels were generally low and comparable to castrated males. Collectively, these findings suggest that suppression of androgen signaling may be a prerequisite for generation of constitutively active AR-Vs. In relation to this proposition, we previously postulated that generation of AR-V7 is coupled with active transcription from the AR gene locus [15,26]. Thus, the generation of AR-V7 in general will require super-active transcription from the AR locus.
2.2. The origin of AR-Vs at the genomic level
There are multiple examples of AR-Vs associated with or explained by AR genomic structural rearrangements (AR-GSRs). For example, CWR22Rv1 cells which express high level of AR-Vs harbor an approximately 35-kb intragenic duplication involving breakpoints at AR intron 2 and 3 [27,52]. In androgen-dependent CWR22Pc parental cells, only a rare subpopulation of cells harbors this AR-GSR. However, this AR-GSR is significantly enriched during castration, and its presence is associated with elevated expression of AR-Vs in this cell-based model of prostate cancer progression [52]. However, a recent study found that AR-V7 expression in the CWR22Rv1 was not affected following deletion of this duplicated exon 3 [53]. In addition, Li et al. [54] identified a ∼48-kb intragenic deletion within AR intron 1 in 20%–40% of CWR-R1 cells. Again, cells harboring this AR intron 1 deletion were enriched during castration, which was accompanied by elevated expression of AR-V7. Interestingly, single-cell cloning revealed that cells positive for the 48-kb intron deletion expressed high level of AR-V7 and displayed enzalutamide resistance. Conversely, deletion-negative cells expressed very low levels of AR-V7 and were sensitive to anti-androgen [55]. In these models, generation of AR-V7 is linked to AR-GSR. Nevertheless, the extent of AR-GSR as a driver of AR-V7 remains unclear, though it is likely AR-GSR may provide a permissive genomic context for the generation and regulation of AR-Vs including AR-V7.
A low-frequency constitutively active AR-V named ARv567es represents a relatively clear-cut example of AR-V expression driven by AR-GSR. An 8579-bp deletion of AR exons 5–7 was identified in the LuCaP86.2 xenograft model, providing possible explanation of the origin of high level of ARv567es expression in this model. In a different CRPC xenograft model, LuCaP136, a copy-neutral 8.7-kb inversion encompassing AR exon 5–7 was identified with exclusive expression of ARv567es [56]. Using TALEN-mediated genome engineering, Nyquist et al. [56] showed that rearrangement involving AR exon 5–7 such as deletion and inversion could result in the expression of ARv567es in cell line models. The extent of ARv567es expression explained by AR-GSR, however, remains unknown. It is worth noting that with respect to the detection of ARv567es in clinical specimens, potential measurement issues concerning PCR-based detection of ARv567es mRNA were thoroughly investigated. It was concluded that ARv567es is not a frequently expressed AR variant [57].
The correlation of AR-GSRs and generation of novel AR-Vs has been reported in clinical specimens in recent studies [31,58]. Henzler et al. [31] conducted deep sequencing analysis of 30 autopsy samples (from 15 mCRPC patients). AR-GSRs were found to be specific to CRPC. The AR-GSRs identified in this study were diverse and generally not recurrent between different patients and specimens. In general, tumors with high level of AR-GSRs are more likely to display expression of AR-Vs. However, elevated AR-V7 expression may occur without evidence of AR-GSR, and no AR splicing alterations of any kind was observed in some tumors with AR-GSR. In the Henzler study, splicing between AR exon 3 and upstream exons was detected in one patient harboring 379 kb AR genomic tandem duplication. Novel CTD-truncated AR-Vs were identified from this patient and all of these newly characterized AR-Vs (AR-V4/5a-ups, AR-V4/5b-ups, AR-V4/5c-ups, AR-V4-chr11) (Fig. 1) process constitutively active AR transcriptional activity. De Laere et al. [58] performed low-pass whole-genome sequencing and targeted sequencing for AR on chromosome X and identified AR structural variants using cell-free DNA extracted from plasma. All four structural variants were detected, including deletion overlapping the AR-LBD, inversion of the entire AR-LBD, tandem duplication within intron 1 and fusion of AR-LBD with segments from other chromosomes. However, potentially novel AR-V transcripts were not cloned and characterized in the De Laere study [58]. Similar to the Henzler study [31], the majority of the AR-GSR positive patients in the De Laere study expressed AR-Vs with rare exceptions. Results from these studies using clinical specimens suggesting a correlation between AR-GSRs and AR-Vs expression, yet a clear-cut one-to-one correlation between AR-GSR and AR-Vs were only demonstrated in a few patients. Because both AR-GSRs and AR-Vs are specific to CRPC and can occur concurrently or independently, whether there is a general causal relationship remains to be determined.
2.3. Regulation of AR-Vs by alternative splicing
Given that the production of AR-Vs can be regulated at the transcriptional level, the general splicing machinery may also play a key role [59]. For AR-V7, RNA splicing involves inclusion of cryptic exon 3 (CE3) within the canonical intron 3 [28]. CE3 possesses the canonical splice site as well as 3′ polyadenylation signal sequences, and is considered to be the last exon of the AR-V7 transcript. Not surprisingly, a number of splicing factors have been found to participate in the inclusion of CE3, i.e., AR-V7 generation. For example, the heterogeneous nuclear ribonucleoproteins (hnRNP) were shown to be involved in AR-V generation in a few studies. Nadiminty et al. [60] showed downregulation of hnRNPA1 and hnRNPA2, both regulated by NF-kB, decreased the expression of AR-V7 in CWR22Rv1 and VCaP cells. The recruitment of hnRNPA1 and hnRNPA2 to AR-Vs including AR-V7 splicing sites were enhanced in enzalutamide resistant CRPC cell lines, and could be inhibited by chemical inhibition of hnRNPA1 [61]. Other hnRNPs such as hnRNPH1, hnRNPL and hnRNPF were also reported to physically interact with AR CE3 [[62], [63], [64]]. Liu et al. [65] demonstrated that two RNA splicing factors, U2AF65 and ASF/SF2, were recruited to AR pre-mRNA near the AR-V7 splicing sites. Interestingly, silencing of U2AF65 and ASF/SF2 only downregulated the AR-V7 RNA level, but not the AR-FL level. The expression of AR-Vs can also be regulated by multiple RNA-binding proteins. Knockdown of DDX39A and DDX39B, which belong to the DExD/H box family of ATP-dependent RNA-helicases, significantly downregulated AR-V7 mRNA and protein levels in CWR22Rv1 and VCaP cells, without affecting AR-FL mRNA levels [66]. Another RNA-binding protein Sam68 (encoded by KHDRBS1) controls the inclusion of CE3 and AR-V7 mRNA expression in RNA-binding-dependent manner [67]. Takayama et al. [68] also reported that the expression of AR-Vs in prostate cancer was affected by RNA-binding protein PSF. PSF enhances various splicing factors to promote AR expression and splicing. A recent study identified SF3B2 as a key splicing factor required for AR-V7 generation (Kawamura, Cancer Research, In Press). In addition, disruption of the polyadenylation signal in AR CE3 can also inhibit expression of AR-Vs, and silencing of cleavage and polyadenylation specificity factor CPSF1 inhibited mRNA expression of AR-Vs and abrogated androgen-independent growth in CWR22Rv1 and LNCaP95 cells [69].
3. Conclusion
The generation of constitutively active AR-Vs lacking the LBD is one of the major mechanisms contributing to CRPC and resistance to AR-targeting therapies. Certain AR-Vs such as AR-V7 are compatible with blood-based detection and have shown clinical utility in treatment and patient selection. In this review, we focused on current data concerning the molecular origin and regulation of AR-Vs in CRPC. AR-Vs could be generated independently or synergistically through AR genomic rearrangement or aberrant AR pre-mRNA splicing dynamics. However, a key prerequisite is suppression of AR signaling mediated by the canonical full-length AR. For AR-V7, production of its mature RNA and functional protein is a dynamic and reversible process through a general mechanism independent of structural alterations in the AR gene, and may require super-active AR transcription driven by genomic and/or epigenomic features acquired in CRPC or in a low-androgen environment. These conclusions are currently supported by data derived from both preclinical investigations and clinically oriented studies. Moving forward, the deeper genomic characterization of CRPC patients will provide more information on the molecular origin of AR-Vs. This in-depth understanding will help to guide the clinical development of therapeutic strategies, including agents with AR-V-specific activities, in AR variant-driven CRPC.
Author contributions
Drafting of manuscript: Yezi Zhu.
Critical revision of the manuscript: Jun Luo.
Conflicts of interest
Dr. Luo reports personal fees from Sun Pharma, Janssen, Tolero, and Sanofi, research funding to his institution from Orion, Astellas, Sanofi, Constellation, ECOG, Pandomedx, and Gilead. Dr. Luo is an inventor of a technology that has been licensed to A&G, Tokai, and Qiagen.
Acknowledgement
Some of the work associated with the review paper was supported by National Institutes of Health Grants (R01 CA185297 and P30 CA006973), Department of Defense Prostate Cancer Research Program Grants (W81XWH-15-2-0050), Johns Hopkins Prostate SPORE Grant (P50 CA058236), and the Prostate Cancer Foundation.
Footnotes
Peer review under responsibility of Second Military Medical University.
References
- 1.Huggins C., Hodges C.V. Studies on prostatic cancer: I. The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. J Urol. 2002;168:9–12. doi: 10.1016/s0022-5347(05)64820-3. 1941. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y., Sawyers C.L., Scher H.I. Targeting the androgen receptor pathway in prostate cancer. Curr Opin Pharmacol. 2008;8:440–448. doi: 10.1016/j.coph.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Attard G., Cooper C.S., de Bono J.S. Steroid hormone receptors in prostate cancer: a hard habit to break? Canc Cell. 2009;16:458–462. doi: 10.1016/j.ccr.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Tran C., Ouk S., Clegg N.J., Chen Y., Watson P.A., Arora V. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–790. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Bono J.S., Logothetis C.J., Molina A., Fizazi K., North S., Chu L. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scher H.I., Fizazi K., Saad F., Taplin M.E., Sternberg C.N., Miller K. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 7.Ryan C.J., Smith M.R., de Bono J.S., Molina A., Logothetis C.J., de Souza P. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med. 2013;368:138–148. doi: 10.1056/NEJMoa1209096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beer T.M., Armstrong A.J., Rathkopf D.E., Loriot Y., Sternberg C.N., Higano C.S. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med. 2014;371:424–433. doi: 10.1056/NEJMoa1405095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fizazi K., Tran N., Fein L., Matsubara N., Rodriguez-Antolin A., Alekseev B.Y. Abiraterone plus prednisone in metastatic, castration-sensitive prostate cancer. N Engl J Med. 2017;377:352–360. doi: 10.1056/NEJMoa1704174. [DOI] [PubMed] [Google Scholar]
- 10.Hussain M., Fizazi K., Saad F., Rathenborg P., Shore N., Ferreira U. Enzalutamide in men with nonmetastatic, castration-resistant prostate cancer. N Engl J Med. 2018;378:2465–2474. doi: 10.1056/NEJMoa1800536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fizazi K., Shore N., Tammela T.L., Ulys A., Vjaters E., Polyakov S. Darolutamide in nonmetastatic, castration-resistant prostate cancer. N Engl J Med. 2019;380:1235–1246. doi: 10.1056/NEJMoa1815671. [DOI] [PubMed] [Google Scholar]
- 12.Smith M.R., Saad F., Chowdhury S., Oudard S., Hadaschik B.A., Graff J.N. Apalutamide treatment and metastasis-free survival in prostate cancer. N Engl J Med. 2018;378:1408–1418. doi: 10.1056/NEJMoa1715546. [DOI] [PubMed] [Google Scholar]
- 13.Watson P.A., Arora V.K., Sawyers C.L. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Canc. 2015;15:701–711. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo J., Attard G., Balk S.P., Bevan C., Burnstein K., Cato L. Role of androgen receptor variants in prostate cancer: report from the 2017 Mission Androgen Receptor Variants meeting. Eur Urol. 2018;73:715–723. doi: 10.1016/j.eururo.2017.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antonarakis E.S., Armstrong A.J., Dehm S.M., Luo J. Androgen receptor variant-driven prostate cancer: clinical implications and therapeutic targeting. Prostate Cancer Prostatic Dis. 2016;19:231–241. doi: 10.1038/pcan.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson D., Van Allen E.M., Wu Y.M., Schultz N., Lonigro R.J., Mosquera J.M. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;162:454. doi: 10.1016/j.cell.2015.06.053. [DOI] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takeda D.Y., Spisak S., Seo J.H., Bell C., O'Connor E., Korthauer K. A somatically acquired enhancer of the androgen receptor Is a noncoding driver in advanced prostate cancer. Cell. 2018;174:422–432. doi: 10.1016/j.cell.2018.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quigley D.A., Dang H.X., Zhao S.G., Lloyd P., Aggarwal R., Alumkal J.J. Genomic hallmarks and structural variation in metastatic prostate cancer. Cell. 2018;175:889. doi: 10.1016/j.cell.2018.10.019. [DOI] [PubMed] [Google Scholar]
- 20.Viswanathan S.R., Ha G., Hoff A.M., Wala J.A., Carrot-Zhang J., Whelan C.W. Structural alterations driving castration-resistant prostate cancer revealed by linked-read genome sequencing. Cell. 2018;174:433–447. doi: 10.1016/j.cell.2018.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisermann K., Wang D., Jing Y., Pascal L.E., Wang Z. Androgen receptor gene mutation, rearrangement, polymorphism. Transl Androl Urol. 2013;2:137–147. doi: 10.3978/j.issn.2223-4683.2013.09.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montgomery R.B., Mostaghel E.A., Vessella R., Hess D.L., Kalhorn T.F., Higano C.S. Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Canc Res. 2008;68:4447–4454. doi: 10.1158/0008-5472.CAN-08-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai C., Balk S.P. Intratumoral androgen biosynthesis in prostate cancer pathogenesis and response to therapy. Endocr Relat Canc. 2011;18:R175–R182. doi: 10.1530/ERC-10-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai C., Chen S., Ng P., Bubley G.J., Nelson P.S., Mostaghel E.A. Intratumoral de novo steroid synthesis activates androgen receptor in castration-resistant prostate cancer and is upregulated by treatment with CYP17A1 inhibitors. Canc Res. 2011;71:6503–6513. doi: 10.1158/0008-5472.CAN-11-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mostaghel E.A., Page S.T., Lin D.W., Fazli L., Coleman I.M., True L.D. Intraprostatic androgens and androgen-regulated gene expression persist after testosterone suppression: therapeutic implications for castration-resistant prostate cancer. Canc Res. 2007;67:5033–5041. doi: 10.1158/0008-5472.CAN-06-3332. [DOI] [PubMed] [Google Scholar]
- 26.Lu C., Luo J. Decoding the androgen receptor splice variants. Transl Androl Urol. 2013;2:178–186. doi: 10.3978/j.issn.2223-4683.2013.09.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dehm S.M., Schmidt L.J., Heemers H.V., Vessella R.L., Tindall D.J. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Canc Res. 2008;68:5469–5477. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu R., Dunn T.A., Wei S., Isharwal S., Veltri R.W., Humphreys E. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Canc Res. 2009;69:16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu R., Isaacs W.B., Luo J. A snapshot of the expression signature of androgen receptor splicing variants and their distinctive transcriptional activities. Prostate. 2011;71:1656–1667. doi: 10.1002/pros.21382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Z., Yang X., Sun F., Jiang R., Linn D.E., Chen H. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Canc Res. 2009;69:2305–2313. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henzler C., Li Y., Yang R., McBride T., Ho Y., Sprenger C. Truncation and constitutive activation of the androgen receptor by diverse genomic rearrangements in prostate cancer. Nat Commun. 2016;7:13668. doi: 10.1038/ncomms13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharp A., Coleman I., Yuan W., Sprenger C., Dolling D., Rodrigues D.N. Androgen receptor splice variant-7 expression emerges with castration resistance in prostate cancer. J Clin Invest. 2019;129:192–208. doi: 10.1172/JCI122819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu R., Lu C., Mostaghel E.A., Yegnasubramanian S., Gurel M., Tannahill C. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Canc Res. 2012;72:3457–3462. doi: 10.1158/0008-5472.CAN-11-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watson P.A., Chen Y.F., Balbas M.D., Wongvipat J., Socci N.D., Viale A. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci U S A. 2010;107:16759–16765. doi: 10.1073/pnas.1012443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cato L., de Tribolet-Hardy J., Lee I., Rottenberg J.T., Coleman I., Melchers D. ARv7 represses tumor-suppressor genes in castration-resistant prostate cancer. Canc Cell. 2019;35:401–413. doi: 10.1016/j.ccell.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai L., Tsai Y.H., Wang P., Wang J., Li D., Fan H. ZFX mediates non-canonical oncogenic functions of the androgen receptor splice variant 7 in castration-resistant prostate cancer. Mol Cell. 2018;72:341–354. doi: 10.1016/j.molcel.2018.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Z., Wu D., Thomas-Ahner J.M., Lu C., Zhao P., Zhang Q. Diverse AR-V7 cistromes in castration-resistant prostate cancer are governed by HoxB13. Proc Natl Acad Sci U S A. 2018;115:6810–6815. doi: 10.1073/pnas.1718811115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He Y., Lu J., Ye Z., Hao S., Wang L., Kohli M. Androgen receptor splice variants bind to constitutively open chromatin and promote abiraterone-resistant growth of prostate cancer. Nucleic Acids Res. 2018;46:1895–1911. doi: 10.1093/nar/gkx1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai C., He H.H., Chen S., Coleman I., Wang H., Fang Z. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Canc Cell. 2011;20:457–471. doi: 10.1016/j.ccr.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X., Morrissey C., Sun S., Ketchandji M., Nelson P.S., True L.D. Androgen receptor variants occur frequently in castration resistant prostate cancer metastases. PloS One. 2011;6 doi: 10.1371/journal.pone.0027970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mostaghel E.A., Marck B.T., Plymate S.R., Vessella R.L., Balk S., Matsumoto A.M. Resistance to CYP17A1 inhibition with abiraterone in castration-resistant prostate cancer: induction of steroidogenesis and androgen receptor splice variants. Clin Canc Res. 2011;17:5913–5925. doi: 10.1158/1078-0432.CCR-11-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoefer J., Akbor M., Handle F., Ofer P., Puhr M., Parson W. Critical role of androgen receptor level in prostate cancer cell resistance to new generation antiandrogen enzalutamide. Oncotarget. 2016;7:59781–59794. doi: 10.18632/oncotarget.10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hornberg E., Ylitalo E.B., Crnalic S., Antti H., Stattin P., Widmark A. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PloS One. 2011;6 doi: 10.1371/journal.pone.0019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welti J., Rodrigues D.N., Sharp A., Sun S., Lorente D., Riisnaes R. Analytical validation and clinical qualification of a new immunohistochemical assay for androgen receptor Splice variant-7 protein expression in metastatic castration-resistant prostate cancer. Eur Urol. 2016;70:599–608. doi: 10.1016/j.eururo.2016.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu Y., Sharp A., Anderson C.M., Silberstein J.L., Taylor M., Lu C. Novel Junction-specific and quantifiable in situ detection of AR-V7 and its clinical correlates in metastatic castration-resistant prostate cancer. Eur Urol. 2018;73:727–735. doi: 10.1016/j.eururo.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antonarakis E.S., Lu C., Luber B., Wang H., Chen Y., Zhu Y. Clinical significance of androgen receptor splice variant-7 mRNA detection in circulating tumor cells of men with metastatic castration-resistant prostate cancer treated with first- and second-line abiraterone and enzalutamide. J Clin Oncol. 2017;35:2149–2156. doi: 10.1200/JCO.2016.70.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scher H.I., Lu D., Schreiber N.A., Louw J., Graf R.P., Vargas H.A. Association of AR-V7 on circulating tumor cells as a treatment-specific biomarker with outcomes and survival in castration-resistant prostate cancer. JAMA Oncol. 2016;2:1441–1449. doi: 10.1001/jamaoncol.2016.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teply B.A., Wang H., Luber B., Sullivan R., Rifkind I., Bruns A. Bipolar androgen therapy in men with metastatic castration-resistant prostate cancer after progression on enzalutamide: an open-label, phase 2, multicohort study. Lancet Oncol. 2018;19:76–86. doi: 10.1016/S1470-2045(17)30906-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schweizer M.T., Antonarakis E.S., Wang H., Ajiboye A.S., Spitz A., Cao H. Effect of bipolar androgen therapy for asymptomatic men with castration-resistant prostate cancer: results from a pilot clinical study. Sci Transl Med. 2015;7:269ra2. doi: 10.1126/scitranslmed.3010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Isaacs J.T., D'Antonio J.M., Chen S., Antony L., Dalrymple S.P., Ndikuyeze G.H. Adaptive auto-regulation of androgen receptor provides a paradigm shifting rationale for bipolar androgen therapy (BAT) for castrate resistant human prostate cancer. Prostate. 2012;72:1491–1505. doi: 10.1002/pros.22504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang R.K., Zhao P., Lu C., Luo J., Hu R. Expression pattern of androgen receptor and AR-V7 in androgen deprivation therapy naive salivary duct carcinomas. Hum Pathol. 2019;84:173–182. doi: 10.1016/j.humpath.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y., Alsagabi M., Fan D., Bova G.S., Tewfik A.H., Dehm S.M. Intragenic rearrangement and altered RNA splicing of the androgen receptor in a cell-based model of prostate cancer progression. Canc Res. 2011;71:2108–2117. doi: 10.1158/0008-5472.CAN-10-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawamura N., Nimura K., Saga K., Ishibashi A., Kitamura K., Nagano H. SF3B2-mediated RNA splicing drives human prostate cancer progression. Canc Res. 2019;79:5204–5217. doi: 10.1158/0008-5472.CAN-18-3965. [DOI] [PubMed] [Google Scholar]
- 54.Li Y., Hwang T.H., Oseth L.A., Hauge A., Vessella R.L., Schmechel S.C. AR intragenic deletions linked to androgen receptor splice variant expression and activity in models of prostate cancer progression. Oncogene. 2012;31:4759–4767. doi: 10.1038/onc.2011.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y., Chan S.C., Brand L.J., Hwang T.H., Silverstein K.A., Dehm S.M. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Canc Res. 2013;73:483–489. doi: 10.1158/0008-5472.CAN-12-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nyquist M.D., Li Y., Hwang T.H., Manlove L.S., Vessella R.L., Silverstein K.A. TALEN-engineered AR gene rearrangements reveal endocrine uncoupling of androgen receptor in prostate cancer. Proc Natl Acad Sci U S A. 2013;110:17492–17497. doi: 10.1073/pnas.1308587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bernemann C., Humberg V., Thielen B., Steinestel J., Chen X., Duensing S. Comparative analysis of AR variant AR-V567es mRNA detection systems reveals eminent variability and questions the role as a clinical biomarker in prostate cancer. Clin Canc Res. 2019;25:3856–3864. doi: 10.1158/1078-0432.CCR-18-4276. [DOI] [PubMed] [Google Scholar]
- 58.De Laere B., van Dam P.J., Whitington T., Mayrhofer M., Diaz E.H., Van den Eynden G. Comprehensive profiling of the androgen receptor in liquid biopsies from castration-resistant prostate cancer reveals novel intra-AR structural variation and splice variant expression patterns. Eur Urol. 2017;72:192–200. doi: 10.1016/j.eururo.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 59.Paschalis A., Sharp A., Welti J.C., Neeb A., Raj G.V., Luo J. Alternative splicing in prostate cancer. Nat Rev Clin Oncol. 2018;15:663–675. doi: 10.1038/s41571-018-0085-0. [DOI] [PubMed] [Google Scholar]
- 60.Nadiminty N., Tummala R., Liu C., Lou W., Evans C.P., Gao A.C. NF-kappaB2/p52:c-Myc:hnRNPA1 pathway regulates expression of androgen receptor splice variants and enzalutamide sensitivity in prostate cancer. Mol Canc Ther. 2015;14:1884–1895. doi: 10.1158/1535-7163.MCT-14-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tummala R., Lou W., Gao A.C., Nadiminty N. Quercetin Targets hnRNPA1 to overcome enzalutamide resistance in prostate cancer cells. Mol Canc Ther. 2017;16:2770–2779. doi: 10.1158/1535-7163.MCT-17-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fei T., Chen Y., Xiao T., Li W., Cato L., Zhang P. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc Natl Acad Sci U S A. 2017;114:E5207–E5215. doi: 10.1073/pnas.1617467114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang Y., Jia D., Kim H., Abd Elmageed Z.Y., Datta A., Davis R. Dysregulation of miR-212 promotes castration resistance through hnRNPH1-mediated regulation of AR and AR-V7: implications for racial disparity of prostate cancer. Clin Canc Res. 2016;22:1744–1756. doi: 10.1158/1078-0432.CCR-15-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fan L., Zhang F., Xu S., Cui X., Hussain A., Fazli L. Histone demethylase JMJD1A promotes alternative splicing of AR variant 7 (AR-V7) in prostate cancer cells. Proc Natl Acad Sci U S A. 2018;115:E4584–E4593. doi: 10.1073/pnas.1802415115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu L.L., Xie N., Sun S., Plymate S., Mostaghel E., Dong X. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene. 2014;33:3140–3150. doi: 10.1038/onc.2013.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakata D., Nakao S., Nakayama K., Araki S., Nakayama Y., Aparicio S. The RNA helicase DDX39B and its paralog DDX39A regulate androgen receptor splice variant AR-V7 generation. Biochem Biophys Res Commun. 2017;483:271–276. doi: 10.1016/j.bbrc.2016.12.153. [DOI] [PubMed] [Google Scholar]
- 67.Stockley J., Markert E., Zhou Y., Robson C.N., Elliott D.J., Lindberg J. The RNA-binding protein Sam68 regulates expression and transcription function of the androgen receptor splice variant AR-V7. Sci Rep. 2015;5:13426. doi: 10.1038/srep13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takayama K.I., Suzuki T., Fujimura T., Yamada Y., Takahashi S., Homma Y. Dysregulation of spliceosome gene expression in advanced prostate cancer by RNA-binding protein PSF. Proc Natl Acad Sci U S A. 2017;114:10461–10466. doi: 10.1073/pnas.1706076114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Etten J.L., Nyquist M., Li Y., Yang R., Ho Y., Johnson R. Targeting a single alternative polyadenylation site coordinately blocks expression of androgen receptor mRNA splice variants in prostate cancer. Canc Res. 2017;77:5228–5235. doi: 10.1158/0008-5472.CAN-17-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]