Abstract
Leishmania donovani is a trypanosomatidic parasite and causes the lethal kala-azar fever, a neglected tropical disease. The Trypanosomatida are devoid of transcriptional gene regulation and rely on gene copy number variations and translational control for their adaption to changing conditions. To survive at mammalian tissue temperatures, L. donovani relies on the small heat shock protein HSP23, the loss of which renders the parasites stress sensitive and impairs their proliferation. Here, we analysed a spontaneous escape mutant with wild type-like in vitro growth. Further selection of this escape strains resulted in a complete reversion of the phenotype. Whole genome sequencing revealed a correlation between stress tolerance and the massive amplification of a six-gene cluster on chromosome 35, with further analysis showing over expression of the casein kinase 1.2 gene as responsible. In vitro phosphorylation experiments established both HSP23 and the related P23 co-chaperone as substrates and modulators of casein kinase 1.2, providing evidence for another crucial link between chaperones and signal transduction protein kinases in this early branching eukaryote.
Subject terms: Post-translational modifications, Parasitology, Chromosomes, DNA recombination, Post-translational modifications, Cell biology, Microbiology, Molecular biology, Systems biology, Infectious diseases, Parasitic infection
Introduction
Protozoan parasites of the genus Leishmania cause upwards of one million known infections of humans annually1, with an unknown number of non-symptomatic carriers. The resulting immune pathologies, summarily known as leishmaniasis, are among the most important of the neglected tropical diseases (NTDs) as defined by the WHO2. In addition, the leishmaniae share many important molecular features with the other Trypanosomatida, namely Trypanosoma cruzi and Trypanosoma brucei, the causative agents of the NTDs Chagas Disease and Human African Trypanosomiasis (HAT), respectively. The most severe form of leishmaniasis, visceral leishmaniasis, is caused by the closely related L. donovani and L. infantum and results in persistent fever, enlargement of spleen and liver, a depletion of antigen-presenting cells, anaemia and a concomitantly increased susceptibility to secondary infections. Without effective chemotherapy, this form of the disease is lethal. There is no effective vaccine or prophylaxis, and the chemotherapeutics against leishmaniasis are fraught with drawbacks such as toxic side effects, teratogenicity, high costs, inconsistent efficacy and/or existing or developing resistance3,4.
From the biological viewpoint, the most striking feature of trypanosomatidic protozoa is their complete lack of canonical, gene-specific regulation of transcription. Leishmania chromosomes consist of long, polycistronic transcription units of functionally non-related genes5–7 that are transcribed into polycistronic precursor RNAs. Precursor RNAs are then processed via trans-splicing coupled to polyadenylation to yield mature, monocistronic mRNAs8,9. This is reflected by the lack of genes for transcription regulator protein orthologs in the parasites' genomes. While differential RNA degradation and/or processing is likely responsible for fluctuating mRNA levels9–11, short-term regulation of gene expression likely relies on regulated translation12–15.
Another genetic feature of Leishmania is a natural, constitutive aneuploidy16–18, but also the ability to generate and maintain quantitative genetic diversity through intrachromosomal or episomal amplification of genes or gene clusters. Inverted and non-inverted repeat sequences of the SIDER family frame and facilitate such gene amplificates19,20, causing gene copy number variations21 in the parasite populations from which the best adapted variants may be selected under adverse conditions such as drug pressure22–25. This may also happen after artificial genetic changes in the laboratory. Spontaneous reversal of phenotypes was described for LPG2−/−- and HSP100−/−-null mutants of L. major26,27. Such phenotype reversals can even be mimicked by functional cloning using cosmid libraries of Leishmania genomic DNA28,29.
Leishmania parasites reside as flagellated, elongated promastigotes attached to the midgut walls of infected phlebotomine sandflies from where they are transmitted to mammals including humans. Taken up by host antigen-presenting cells, they undergo morphological stage conversion to non-flagellated, non-motile, ovoid amastigotes that reside within the phagosomes mainly of macrophages30,31. The sandfly stage is also characterised by a rapid, logarithmic growth while the amastigote has a reduced proliferation rate and can even enter quiescence32–34. Another consequence of the transmission from arthropod to mammalian hosts is the dual trigger of mammalian tissue temperatures and the acidified milieu inside macrophage phagosomes. Indeed, in vitro exposure of axenically cultivated promastigotes to both increased temperature (33–37° C, depending on the Leishmania species) and slightly acidic growth medium (pH 5–5.5) can trigger the morphological and metabolic shift to an amastigote-like cell35–37.
At the same time, the temperature shift also induces a classic cellular stress response, with increased synthesis of several heat shock protein families, including HSP100/ClpB38,39, HSP90 and HSP7012, a 60 kDa chaperonin (CPN60.2)40 and its likely co-chaperone CPN1041.
Another chaperone, the small heat shock protein 23 (HSP23) also shows a temperature-dependent expression, and HSP23 null mutants (HSP23−/−) indeed suffer a severe viability loss at mammalian tissue temperature (37 °C), impaired chemical stress tolerance and a reduced growth at permissive temperatures42. HSP23 belongs to the rather diverse small HSP family that are characterised primarily by the so-called α-crystallin domain, but has a unique overall domain structure compared with other small HSPs and is more closely related to the p23 co-chaperone of Leishmania which modulates the accessibility of the ATPase domain of the major chaperone HSP90 for HSP90 inhibitors43.
In this paper, we describe the spontaneous reversal of the temperature-sensitive (TS) phenotype of L. donovani HSP23−/− null mutants under in vitro selection at permissive and non-permissive temperatures. The escape variants display wild-type level temperature tolerance and in vitro proliferation rates and chemoresistance, due to a massive amplification of the casein kinase 1.2 (CK1.2) gene. We further show that HSP23, but also P23, are substrates of CK1.2, establishing yet another link44,45 between protein kinase-mediated signalling and the chaperone machinery of Leishmania.
Results
Emergence of an escape variant of HSP23−/−
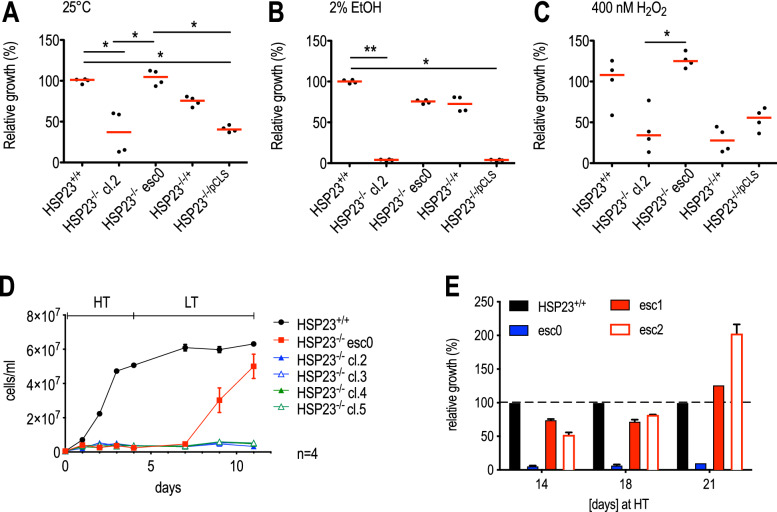
It was previously shown that the loss of HSP23 renders Leishmania donovani promastigotes incapable of surviving at mammalian tissue temperatures (hereafter referred to as high temperature (HT)) and more sensitive towards several stressors such as ethanol and acidic pH. Moreover, in vitro proliferation at 25 °C (hereafter referred to as low temperature (LT)) was also affected42. After approximately 25 in vitro passage cycles at LT, however, an escape population emerged with wild type-like growth phenotype which we designated HSP23−/− esc0. We also generated new HSP23−/− null mutants, following the same recombination strategy as used for the parental strain of esc042. The clone 2 was tested and still shows the original, growth impaired phenotype (Fig. 1A). As expected, HSP23−/− cl.2 growth can be restored by episomal expression of a HSP23 transgene (HSP23−/−/+), but not by the expression vector alone (HSP23−/−/pCLS).
Figure 1.
Phenotype comparison of HSP23−/− and HSP23−/− escape mutants under different culture conditions. 1✕106 cells were seeded into 7 ml of supplemented M199 + medium. Cell density on day 4 post seeding was then normalised to wild-type (HSP23+/+) cell density (set at 100%). Parasites were grown at LT (25 °C) (A), 2% EtOH at LT (B) or 400 nM H2O2 at LT (C). Differences were tested for significance using the Kruskal–Wallis test: *p < 0.05, **p < 0.01 (n = 4) (D) Heat selection of HSP23−/− esc0: 5✕105 cells/ml were seeded into 10 ml of M199 + medium and incubated for 4 days at HT (37 °C). The cultures were transferred back to LT and grown to mid-log phase for seven days. (E) Growth analysis of two heat selected (D) HSP23−/− mutant populations at HT, the non selected HSP23−/− esc0 population and HSP23+/+ wild type cells (set as 100%). Cells were diluted twice weekly to 5✕105 cells/ml. Error bars represent the standard error of the mean (SEM).
Very similar results were obtained under challenge with 2% ethanol. Here, growth of HSP23−/− cl.2 and HSP23−/−/pCLS was almost completely suppressed. By contrast, HSP23−/− esc0 and HSP23−/−/+ showed no significant impairment relative to wild type (HSP23+/+) (Fig. 1B).
We also tested the oxidative stress sensitivity of wild type (HSP23+/+) and mutant strains by monitoring in vitro growth under H2O2 challenge (400 nM) (Fig. 1C). Again, HSP23−/− esc0 was indistinguishable from wild type, while HSP23−/− cl.2 and HSP23−/−/pCLS were significantly affected, similar to the growth defects under standard culture conditions (25 °C, panel A). Surprisingly, the add-back strain, HSP23−/−/+, did not show restored growth under H2O2 resistance either, indicating a difference between add-back and escape strain.
Earlier work42 suggested that exposure of HSP23−/− to HT was lethal to the mutant. To assess the survival of a panel of HSP23−/− clones at elevated temperatures, we exposed wild type and null mutant clones to HT for 4 days, monitoring cell density daily (Fig. 1D). In keeping with earlier experiments, only wild type (HSP23+/+) showed growth at HT. Shifting back the cultures to LT, all HSP23−/− clones except for HSP23−/− esc0 showed no outgrowth, indicating that the 4-day exposure to HT had been indeed lethal to HSP23−/− clones 2 and 3. The HSP23−/− esc0 alone showed outgrowth starting at 2 days after the temperature downshift, indicating that a substantial number of promastigotes had survived the preselection.
Based on this assumption, we compared two of the outgrowth populations with the original HSP23−/− esc0 strain and wild type for their ability to sustain growth at HT over a period of 21 days. The result of this experiment is shown in Fig. 1E; growth of the mutants is shown in [%] of wild type growth. While the original, not preselected strain (esc0) shows only marginal growth, the two populations (esc1, esc2) coming from the preselection shown in Fig. 1D show increasing vitality over the course of the experiment, reaching a relative growth exceeding that of wild type cells in the last days. Obviously, the preselection at HT followed by unimpeded growth at LT led to the spontaneous emergence of two fully temperature tolerant strains while the parent strain esc0 is not capable of growth at 37 °C. This indicates that esc0, while showing increased survival at 37 °C compared with the other HSP23-/- clones, required a recovery period to fully express temperature tolerance.
Genetic Adaptions of HSP23−/− Mutants
In lieu of transcriptional control Leishmania gene expression is often modulated by gene dosage through chromosomal aneuploidies18,46, gene copy number variations20,47 or selection of circular and linear amplicons19,48,49. In particular, such genomic alterations are frequently found in leishmaniae after challenge with stressful conditions such as drug pressure or environmental changes50,51.
To identify the genomic alterations that compensate for the lack of HSP23 at permissive and non-permissive temperatures, we performed whole genome sequencing of L. donovani 1S wild type selected at LT (n = 2) and HT (n = 2), HSP23−/− clone 2 (n = 1) and clone 3 (n = 1) selected at LT, HSP23−/− esc0 selected at LT (n = 2) and HSP23−/− esc1 (n = 1) and esc2 (n = 1) selected at HT. The sequence read data are available at https://www.ncbi.nlm.nih.gov/sra/PRJNA633969.
Firstly, we verified the accuracy of the HSP23 gene replacements for HSP23−/− cl.2, HSP23−/−cl.3, and HSP23−/− esc mutants (Supplementary Figs. S1, S2), finding the homologous recombination to have worked entirely as expected.
Using NGS sequence read coverage data, we analysed the aneuploidy patterns (Fig. S3, Table S2). Karyotype differences between HSP23+/+ and HSP23−/− clones and escape variants were observed. Full or mosaic aneuploidy was found for chromosomes 8 and 35 in HSP23−/− esc0, cl.2 and cl.3 at LT. Chromosomes 12 and 23 show a somy reduction in all HSP23−/− escape variants while chromosome 14 shows mosaic trisomy in three of four HSP23−/− escape samples.
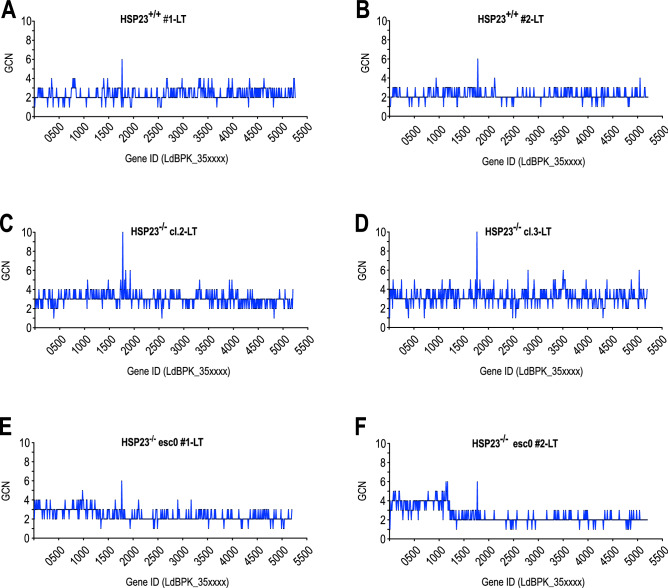
We next performed a genome-wide analysis of individual gene copy number variations for all strains subjected to whole genome sequencing. For the most part, changes of CNVs correlated with the observed somy changes (Table S2). However, the read depth analysis for chromosome 35 revealed striking discrepancies. As observed before, the HSP23−/− clones 2 and 3 that were maintained at LT are trisomic for chromosome 35 which is reflected in an overall 50% increase of gene copy numbers compared with the wild type samples (Fig. 2 A–D). For both samples of HSP23−/− esc0 maintained at LT we had observed a slight mosaic somy for chromosome 35 at 2.26 and 2.36 (Table S1). Gene copy number analysis, however, showed a selective amplification of a 0.55-megabase region at the 5′ end of chromosome 35 from nucleotide positions ~ 4500 bp to 554,500 bp and ~ 4500 bp to ~ 557,500 bp for sample 1 and sample 2, respectively. The observed amplifications encompass 124 and 126 genes, respectively, and result in 1.5- to twofold increase of gene copy numbers in these regions (Fig. 2E,F).
Figure 2.
Strain-specific gene copy number variations on chromosome 35 at permissive temperatures. Normalised copy number for each gene (#1-#530) on chromosome 35 is shown for (A, B) HSP23+/+, (C, D) HSP23−/−, and (E, F) HSP23−/− esc0. All strains were selected and grown at 25 °C (LT).
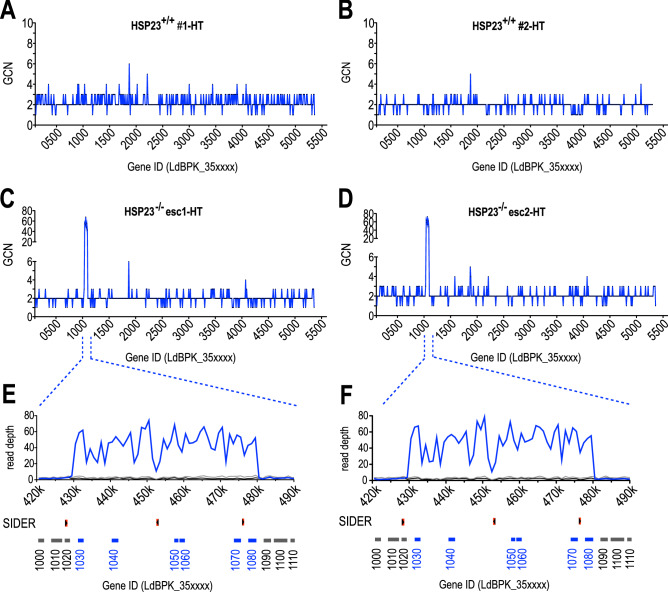
Analysis of the HSP23−/− esc1 and -esc2 strains, both survivors of a 4-day HT challenge and able to grow at HT (Fig. 1D,E), displayed a more specific and extreme amplification. Compared with wild type cells selected at HT (Fig. 3A,B), esc1 and esc2 both show a 43- to 73-fold gene copy number increase for a cluster of six genes on chromosome 35 (Fig. 3C,D). This 49-kb region (position 430,500–479,500) is delimited by SIDER2 elements19 and codes for casein kinase 1.2 (CK1.2) (LdBPK_351030), two CCCH-type zinc finger proteins (LdBPK_351040 and LdBPK_351060), a cupin domain-containing protein (LdBPK_351050), an undefined putative protein kinase (LdBPK_351070) and a hypothetical protein (LdBPK_351080) (Fig. 3E,F).
Figure 3.
Strain-specific gene copy number variations on chromosome 35 at non-permissive temperatures. Normalised copy numbers for indicated genes (LdBPK_35xxxx) on chromosome 35 is shown for (A, B) HSP23+/+-HT, (C, D) HSP23−/− esc1/2-HT, (E, F) enlargement of the amplified region on chromosome 35 between positions 420,000 and 490,000. Sequencing read coverage is shown for HSP23+/+ (LT, black), HSP23−/− esc1/2 (HT, blue) and HSP23−/− cl. 2/3 (LT, grey). Open reading frames and corresponding gene IDs are indicated in grey for non-amplified or blue for amplified CDSs; positions of SIDER2 repeat elements are indicated in red. LT:25 °C, HT:37 °C.
It has been previously demonstrated that karyotypic variation is associated with modulation of transcript abundance23. Thus, we questioned whether the observed amplification of the six-gene cluster on chromosome 35 in heat-resistant HSP23−/− esc1/2 also changes RNA and protein abundance. Indeed, qRT-PCR analysis confirmed that amplification of the gene cluster in HSP23−/− esc1/2 mutants results in a 10- to 41-fold increase of mRNA for the co-amplified genes (Table 1). With an anti-CK1.2 (LdBPK_351030) antibody available52, we performed Western blot analysis. Indeed, we observed a 2- to threefold up regulation of CK1.2 protein levels in the HSP23−/− esc1/2 mutants, confirming the correlation between DNA copy numbers, RNA levels and protein abundance (Fig. 4A,B).
Table 1.
Gene copy numbers (GCNs) and corresponding RNA levels of amplified candidates genes on chromosome 35 (LdBPK_351030-351080).
| Gene ID: LdBPK | 351030 | 351040 | 351050 | 351060 | 351070 | 351080 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GCN | RNA | GCN | RNA | GCN | RNA | GCN | RNA | GCN | RNA | GCN | RNA | |
| HSP23+/+ #1-LT | 3 | 1.028 ± 0.06 | 2 | 1.050 ± 0.06 | 3 | 1.029 ± 0.04 | 3 | 0.988 ± 0.03 | 2 | 0.989 ± 0.01 | 2 | 0.998 ± 0.01 |
| HSP23−/− esc0 #1 -LT | 4 | 1.445 ± 0.05 | 3 | 1.908 ± 0.07 | 4 | 1.798 ± 0.16 | 3 | 1.505 ± 0.19 | 5 | 1.64 ± 0.17 | 4 | 2.253 ± 0.17 |
| HSP23+/+ #1-HT | 2 | 0.915 ± 0.05 | 3 | 2.487 ± 0.32 | 3 | 1.120 ± 0.11 | 2 | 0.787 ± 0.11 | 2 | 0.953 ± 0.06 | 2 | 0.922 ± 0.15 |
| HSP23−/− esc1-HT | 56 | 15.55 ± 1.6 | 53 | 35.60 ± 1.71 | 68 | 29.53 ± 3.42 | 48 | 10.60 ± 0.86 | 55 | 28.03 ± 2.55 | 43 | 40.72 ± 1.42 |
| HSP23−/− esc2-HT | 63 | 13.11 ± 0.45 | 58 | 31.45 ± 1.28 | 73 | 30.95 ± 4.26 | 54 | 9.822 ± 1.05 | 62 | 26.44 ± 1.99 | 48 | 40.39 ± 6.05 |
RNA values are mean ± standard errors of the means (SEM). qRT-PCR data were obtained from three biological and two technical replicates each.
Figure 4.
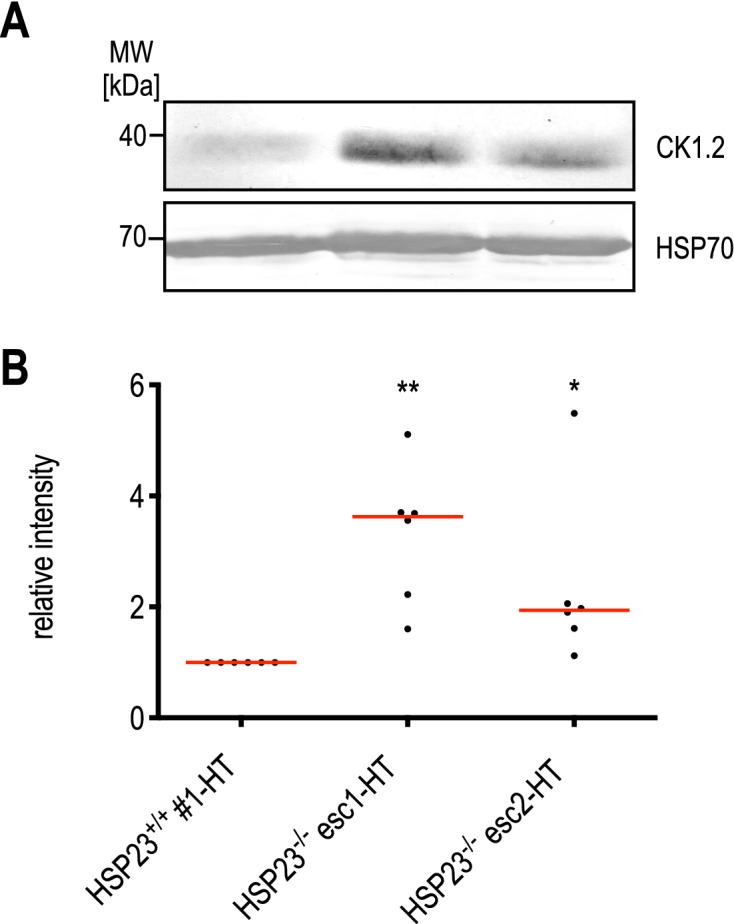
Abundance of CK1.2 in HSP23−/− esc1/2 mutants. (A) Western blot analysis of CK1.2 in HSP23 + / + and HSP23−/− esc1/2 using a rabbit anti-CK1.2 antibody (SY3535) (1:250). A chicken anti-HSP70 (1:5000) antibody was used as a loading control. (B) CK1.2 protein level was quantified using ImageJ Fiji Software (Version 2.0.0) and first normalised to Coomassie loading control and then to the HSP23 + / + (37 °C) sample. Statistical testing was carried out by Kruskal–Wallis test; *p < 0.05, **p < 0.01; (n = 6).
Functional Cloning of Stress Tolerance Marker
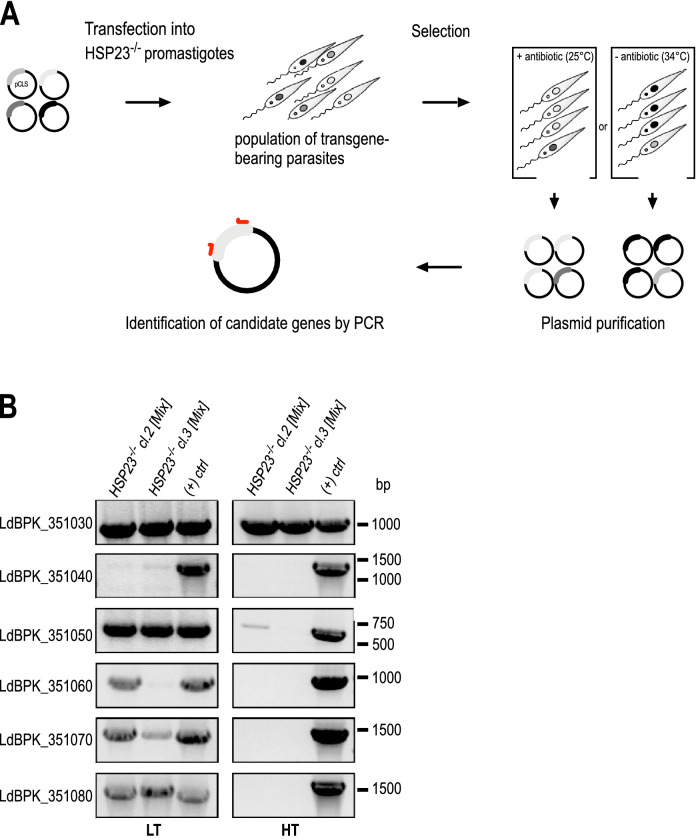
Next, we tested whether the escape mutant phenotype can be attributed to the observed amplification of the six-gene cluster on chromosome 35. We therefore co-transfected a mixture of six different over expression plasmids, harbouring the genes from the amplified cluster (LdBPK_351030—LdBPK_351080), into TS HSP23−/− lines. The transfected lines were selected either at LT under antibiotic selection or at HT without antibiotics, herein named as HSP23−/− [Mix]-LT and HSP23−/− [Mix]-HT, respectively (Fig. 5A). While empty vector-transfected cells did not survive the selection at non-permissive temperatures, two independent TS HSP23−/− clones transfected with the plasmid mixture recovered, confirming that over expression of genes from the amplified cluster causes the escape mutant phenotype.
Figure 5.
Selection of candidate genes in the HSP23−/− background at non-permissive temperatures (A) Schematic drawing of the transgene selection procedure. Six over expression plasmids harbouring LdBPK_351030 to LdBPK_351080 were co-transfected into HSP23−/− null mutants and selected either at LT with the selective antibiotic, or at HT, generating HSP23−/− [Mix]-LT or HSP23−/− [Mix]-HT cell populations. After selection, plasmids were isolated from the Leishmania populations by Phenol–Chloroform extraction. (B) Analytical PCR using gene-specific primers to test for the presence of candidate transgenes in the recovered plasmids. Transgene plasmid DNA was used as positive control (( +) ctrl). PCR products were analysed by agarose gel electrophoresis and ethidium bromide staining; the position of corresponding DNA size marker bands is shown on the right. The left panel shows the PCR products after LT selection, the right panel shows the results after HT selection.
To pinpoint the gene(s) mediating survival at HT, plasmid DNA was re-isolated from surviving populations, and PCR was performed with transgene-specific primers (Fig. 5B). While a complete set of plasmids was still recovered from HSP23−/− [Mix]-LT after LT selection (LTS), selection at HT (HTS) almost exclusively selected for the CK1.2 transgene, confirming a role of CK1.2 in reversing the TS phenotype of HSP23−/−. This was confirmed by qRT-PCR analysis, showing only increased abundance of CK1.2 RNA after HT selection, but not of the other transgenes (Fig. S4).
CK1.2 transgene is amplified under temperature selection
To further validate theses findings, we tested whether transfection of the CK1.2-encoding over expression plasmid alone can restore temperature tolerance to HSP23−/−. Indeed, ectopic over expression of CK1.2 alone abrogated characteristic heat-induced defects such as the aggregation and rounding of the normally elongated promastigotes, as visualised by anti-alpha-tubulin staining (Fig. 6B), but also restored tolerance to EtOH and H2O2-induced protein folding stress at LT (Fig. S5).
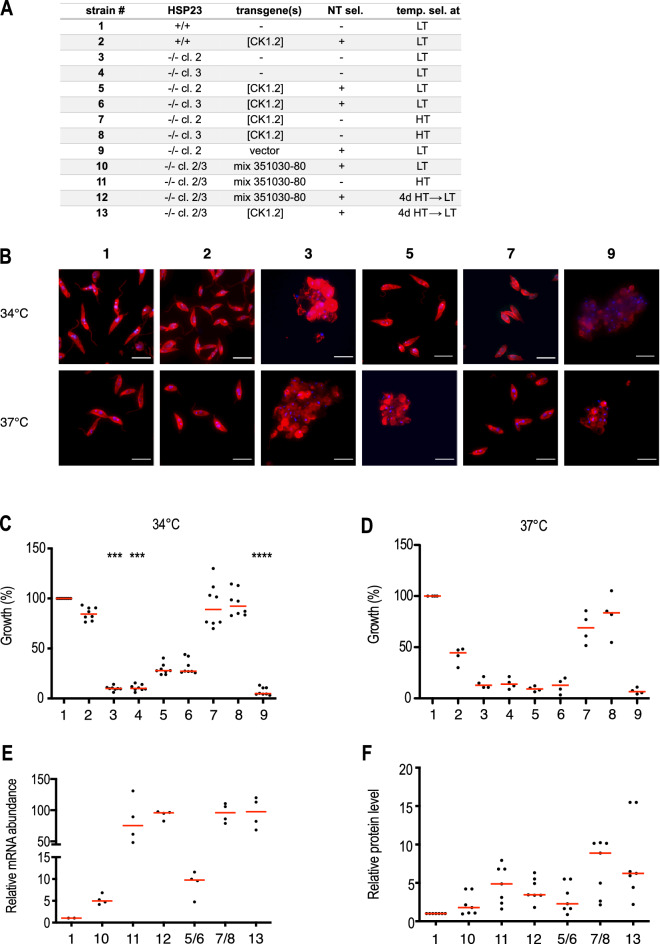
Figure 6.
Characterisation of transgenic L. donovani strains. (A) List of transgenic parasite cell lines tested. NT sel = selected under nourseothricine; temp. sel. = temperature selection scheme. (B) 5✕105 cells/ml were seeded into 7 ml of M199 + medium and incubated for 4 days at 34 °C or 37 °C. On day 4, cells were stained with mouse anti-α-tubulin mAB (1:4000) and anti-mouse-IgG Alexa Fluor 594 (1: 500) and DAPI. Images were processed and merged using the ImageJ Fiji Software (Version 2.0.0). Scale bar:10 µm (C, D) Growth behaviour of transgenic L. donovani strains at 34 °C (n = 8) or 37 °C (n = 4). Cells were seeded at 5✕105/ml and grown for 4 days. Cell density was measured and normalised to wild type (HSP23+/+) cell density. Significance was tested using the Kruskal–Wallis test: ***p < 0.001, ****p < 0.0001. (E, F) CK1.2 expression levels were determined by qRT-PCR using gene-specific primers (E) and Western blotting using the anti-CK1.2 antibody (F). Data were normalised to the HSP23+/+ samples.
Moreover, in all tested strains (Fig. 6A), ectopic expression of CK1.2 strongly depended on the pre-selection condition: HT-selected lines were able to grow both at 34 °C and 37 °C and reached similar cell densities as HSP23−/− esc mutants (Fig. 6C,D). In contrast, only partial reversion of the TS phenotype was observed in LT-selected HSP23−/− [CK1.2] lines as judged by the comparably slow growth at 34 °C and failure to grow at 37 °C (Fig. 6C,D).
Based on these results, we reasoned that the compensation for the loss of HSP23 might correlate with CK1.2 expression levels. To test this hypothesis, we analysed CK1.2-encoding mRNA as well as protein levels in the over expressing lines selected under both permissive and non-permissive conditions. As suspected, HTS resulted in ~10–15-fold higher RNA and ~2.5-fold higher CK1.2 protein levels when compared to LTS (Fig. 6E,F). Further, shifting HSP23−/− [Mix]-LT and HSP23−/− [CK1.2]-LT lines to higher temperatures was associated with a 10–19-fold increase of CK1.2 mRNA levels and 2.6–3.9-fold protein levels. Continuous incubation of HSP23+/+ wild type cells at non-permissive temperatures or transfection of HSP23+/+ parasites with CK1.2 over expression plasmids with subsequent HTS did not result in comparable increases of CK1.2 mRNA or protein levels (data not shown). In addition, the increased mRNA as well as protein abundance observed for the heat-selected HSP23−/− esc1/2 populations was reversible once the cultures were transferred back to LT (Fig. S6).
In summary, our data demonstrate that i) there is a strong selective pressure for increased CK1.2 protein levels in HSP23−/− null mutants and ii) that the observed gene dosage changes are under tight, temperature-dependent selection.
HSP23 is a Casein Kinase 1.2 substrate in vitro
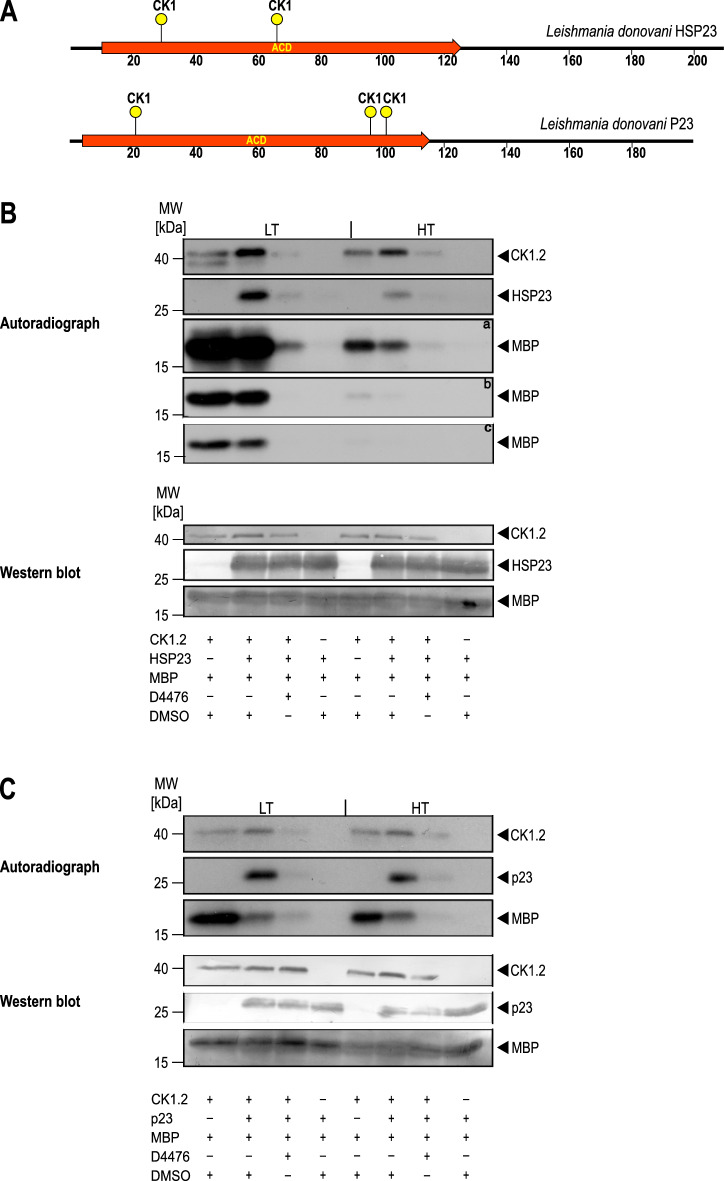
Casein kinase 1.2 is implicated in the heat shock response of Leishmania as an upstream protein kinase of HSP9045. Moreover, both HSP23 and the closely related, HSP90-modulating co-chaperone P2343 contain possible CK1 phosphorylation sites (Figs. 7A, S7). To elucidate possible enzyme–substrate interactions between CK1.2 and HSP23, but also P23, we performed in vitro phosphorylation assays with recombinantly expressed L. donovani CK1.2 using myelin basic protein (MBP), HSP23 and P23 as substrates and/or modulatory chaperones. The proteins were incubated for 30 min with [γ-32P]-ATP at LT and HT, separated by SDS-PAGE and visualised by autoradiography.
Figure 7.
In vitro kinase assays using [γ-32P]-ATP. (A) Schematic representation of the putative CK1 phosphorylation sites present in HSP23 and P23. (B, C) 0.25 µg recombinant Casein kinase 1.2 was incubated with 3 µg of myelin basic protein (MBP), 1.57 µg of recombinant HSP23 (B) or 1.3 µg P23 (C) in the presence of either 10 µM D4476 inhibitor or equivalent concentrations of DMSO. The reactions were incubated at LT (25 °C) or HT (37 °C) for 30 min and stopped by addition of loading buffer. A reaction without the kinase was included to test for possible autophosphorylation of substrates. Proteins were separated on a 12.5% SDS-PAGE gel followed by Western blot. The incorporation of [γ-32P]-ATP was monitored by autoradiography (upper panels). a = 1 week exposure; b = 24 h exposure, c = 12 h exposure of X-Ray films. The blot was probed with an anti-CK1.2 (SY3535), HSP23 and P23 antibody, or stained with Coomassie blue to evaluate loading (lower panels). The figure shows representative Western blots from a series (n = 4) of experiments.
Figure 7B shows the activity of CK1.2 in a reaction mix containing the kinase, MPB and HSP23. CK1.2 readily phosphorylates MBP at LT and at HT, with a slight drop of activity at HT. Adding HSP23 to the mix has a threefold effect: i) autophosphorylation of CK1.2 increases, ii) HSP23 becomes radiolabeled, and iii) MPB phosphorylation is reduced at higher temperature. We conclude that CK1.2 autophosphorylation and the concomitant reduction of activity is furthered by HSP23. In turn, HSP23 is a substrate of CK1.2. The latter activity is verified by the loss of HSP23 phosphorylation in the presence of the CK1-specific inhibitor, D4476.
We also tested the impact of the co-chaperone P23 on CK1.2 activity. As shown in Fig. 7C, P23 has a very similar impact to HSP23, furthering autophosphorylation of CK1.2 and reducing MBP phosphorylation. Like HSP23, P23 is a substrate for CK1.2-mediated phosphorylation, adding a third member of the chaperone family of proteins to the list of CK1.2 clients.
We conclude, that CK1.2 over expression has a likely effect on the phosphorylation status and possibly the activity of chaperones in Leishmania and its over expression by gene amplification may thus impact on the protein folding homeostasis in the parasite.
Discussion
Genetic compensation for the loss of HSP23 in Leishmania
Double-allele replacement of HSP23 in L. donovani results in a reduced growth at 25 °C, the temperature commonly associated with the promastigote stage, but also a loss of viability at temperatures > 30 °C42. Our results show that all HSP23−/− null mutants that we obtained carry a partial or complete amplification of chromosome 35, either as trisomy or as a duplication of the 5′-terminal third of this chromosome. A passage at 37 °C of the HSP23−/− esc0 clone, which overcame the growth reduction at 25 °C, even selected for the massive amplification of a six-gene region from the 5′ terminus of chromosome 35. As reported before16,23, changes in DNA copy numbers observed for the six-gene cluster on chromosome 35 correlated well with an increase of mRNA levels. However, upon removal of HTS, mRNA returned to basal levels indicating that the gDNA amplificates are lost from the selected populations in the absence of temperature selection, although this remains to be verified experimentally.
Our observations match the genomic changes associated with drug resistance where selective chromosomal somy changes appear early in the development of resistance and are followed, upon more stringent selection, by the appearance of extrachromosomal amplificates that are usually lost from the populations once the selection is removed18–20,53–55.
In recent years, remarkable progress has been made towards understanding the mechanisms underlying the high plasticity of the Leishmania genome. Circular or linear extrachromosomal episomes are commonly formed through rearrangements at the level of direct or inverted repeat sequences56,57. Furthermore, short interspersed degenerated retroposons (SIDERs) that are widely distributed through the entire Leishmania genome were shown to be involved in the regulation of gene expression at both the post-transcriptional (SIDER2) or translational (SIDER1) level58,59 but also in inducing DNA rearrangements19.
Indeed, a genome-wide bioinformatic analysis of the Leishmania genome19 described several SIDER sequences that either flanked or were present within the identified DNA region on chromosome 35 amplified in heat selected HSP23−/− esc mutants. More importantly, the amplification identified by NGS in the HSP23−/− esc1,2 mutants matched with a predicted putative product, thus highlighting the power of SIDER element-based prediction of DNA rearrangements and its importance in the response to changing stress conditions.
Over expression of Casein Kinase 1.2 rescues temperature tolerance in HSP23−/−
Two lines of evidence point at amplification of casein kinase 1.2 as the mechanism for restoring temperature tolerance to HSP23−/− mutants. Firstly, of the six genes found on the amplified gene cluster after HTS and transfected as transgenes into HSP23−/− clones 2 and 3, only LdBPK_351030 – CK1.2 was retained under HTS of the mixed population while LTS yielded all six transgenes. Secondly, in the CK1.2-over expressing HSP23−/− parasites, survival and growth under HTS correlated with CK1.2 mRNA and protein abundance, resulting in almost wild type-like temperature tolerance for HT-selected populations with 6–ninefold increased CK1.2 abundance. Shifting the LT-selected strains to HT quickly led to an increase of expression. In a similar way, HT-selected HSP23−/− esc strains that were transferred back to LT, showed a reduction of CK1.2 mRNA and protein suggesting that the observed gene amplification is reversible. We conclude that survival and growth of HSP23−/− mutants at temperatures matching mammalian tissue temperature is a function of CK1.2 expression. Conversely, cultivation at ambient temperature leads to a rapid reduction of over expression, indicating adverse effects of increased CK1.2 abundance in the absence of temperature stress.
Members of the casein kinase 1 family are highly conserved acidophilic serine/threonine kinases with reported roles in cell cycle60, circadian rhythm61–63, cell signalling, and membrane trafficking64. The Leishmania genome harbours six different Casein kinase isoforms with CK1.2 being the most highly expressed and abundant form65–67. CK1.2 was shown to display an ubiquitous localisation and was found in the cytoplasm, nucleus, the basal body and the flagellar pocket of L. donovani68, the major interface of the parasite with its environment and the origin of exosomes. Consistently, CK1.2 is known to be part of the protein load of immune-modulatory exosomes that are shed by Leishmania parasites in a temperature-dependent manner during the intracellular amastigote stage within the host cell69,70. This, and the fact that inhibition of CK1.2 with the specific inhibitor D4476 is cytotoxic in axenic amastigotes and reduces the parasite load of infected macrophages in vitro52, highlights the essential role of this protein kinase in stage differentiation and intracellular survival.
As reported previously for human CK1 enzymes71, known substrates of Leishmania CK1.2 include the major chaperones HSP70 and HSP90, the latter being essential for the rapidly growing promastigote and playing a pivotal role in the life cycle control of Leishmania donovani45,68,72,73.
CK1.2 phosphorylates HSP23 and P23 in vitro
Our results indicate that another two chaperone proteins, HSP23 and P23, are likely substrates of CK1.2. Indeed, both proteins contain several canonical consensus sequences (pSer/Thr-X-X-(X)-Ser/Thr) for CK1 protein kinases (Fig. 7A). P23 was already described to be phosphorylated by CK2 in other organisms74,75. Moreover, Leishmania HSP23 and P23 appear to modulate the CK1.2 activity and specificity in turn (Fig. 7B,C), increasing autophosphorylation activity and reducing MBP phosphorylation. The inverse correlation between autophosphorylation and substrate-specific activity is a known characteristic of CK1 in other systems76–78.
P23 is a known co-chaperone of HSP90 and binds to the middle domain of HSP90 with a higher affinity for the ATP-bound state. This interaction was shown to temporarily stabilise the closed conformation of HSP90 and at the same time slowing the ATPase cycle79–84. Such an inhibitory effect on the HSP90 ATPase activity was also described for P23 and, to a lesser degree, for HSP23 in Leishmania braziliensis85. Strong structural as well as functional conservation of HSP23 orthologs in Trypanosomatida (V. Adaui, C. Kröber-Boncardo, unpublished) suggests a similar modulating effect of the L. donovani HSP23 protein on HSP90 function.
The interaction between P23 and HSP90 can be inhibited by the HSP90 specific inhibitors geldanamycin (GA) and radicicol leading to the abrogation of a functional HSP90 cycle86,87. The loss of P23 in Leishmania, but also in other organisms, consistently results in hypersensitivity to those HSP90 inhibitors. The fact that over expression of HSP23 in L. donovani can compensate the loss of P23 under GA or radicicol induced HSP90 inhibition43 further supports data demonstrating an intrinsic co-chaperone activity of HSP23 on HSP90. The growth reduction phenotype observed for HSP23−/− null mutants may therefore be partly due to a deregulation of HSP90. Since HSP90 is also a known in vitro substrate of CK1.245, the over expression of CK1.2 in HSP23−/− escape variants might be one way to counteract the deregulation by altering the phosphorylation state of HSP90 itself or of its co-chaperones such as P23. Post-translational modifications of HSP90 are extremely important for the fine-tuning of the chaperone cycle and affect its conformation, ATPase activity, interaction with co-chaperones, the recruitment and activation of various client proteins, cell growth, and viability45,71,88.
A surprising result was obtained with a gene add-back line of HSP23−/− which showed restored in vitro growth at 37 °C and under 2% ethanol challenge, but was unable to proliferate under H2O2, while the complementation with CK1.2 alleviated H2O2-mediated growth inhibition. We suspect that the effects of HSP23−/− on growth under H2O2 mirror those at 25 °C standard culture conditions where gene add-back also had a moderate effect. This indicates that the strong growth defects under heat stress inducing conditions, i.e. HT or 2% ethanol, and the more moderate effects seen at LT and under oxidative stress may be due to different cellular pathways.
In summary, our data suggest a regulatory network comprising CK1.2, HSP23, P23 and HSP90. This is supported by the following results : i) HSP23, P23, and HSP90 are in vitro substrates of CK1.2 and ii) HSP23 and P23 affect CK1.2 activity but also HSP90 activity85.
Comparative phosphoproteome analysis of HSP23−/− esc mutants and CK1.2 over expressing lines under both permissive and non permissive temperatures may provide new insights into the functional network of these chaperones and their regulation by CK1.2. In addition, such studies may allow to identify novel clients and downstream signalling pathways of CK1.2 which may elucidate the effect of CK1.2 over expression in HSP23−/− null mutants and eventually shed light on its role in Leishmania temperature tolerance. Furthermore, the putative CK1.2 phosphorylation site(s) of HSP23 but also P23 can be mutated to assess the resulting phenotype in terms of in vitro growth, temperature- and chemotolerance.
Impact on target-specific drug development
Our data show that in spite of its importance for the parasite’s temperature tolerance, HSP23 may be a poor target for therapeutic intervention due to the prevalence of genetic variants that can overcome the loss of HSP23 function. The plasticity of the Leishmania genomes offers large ranges of genetic variants from which to select, both in the laboratory and in the wild, allowing them to overcome the loss or loss of function of single proteins. Thus, our data also highlight the risk of using genetically modified, live-attenuated Leishmania parasites89–91 for vaccine purposes, since supposedly attenuating genetic modifications may be easily reversed or compensated through selection from pre-existing, beneficial gene amplification variants within Leishmania populations.
Materials and methods
Reagents and chemicals
All reagents, unless otherwise stated, were purchased from Merck, Darmstadt, Germany or its Sigma-Aldrich subsidiary.
Leishmania culture conditions
Leishmania donovani strain 1S36 and mutants thereof were cultured in M199 + medium73 with the respective antibiotics at 25 °C and passaged every 3–4 days.
Electrotransfection of Leishmania parasites
Electrotransfection and selection was performed as described92. To isolate single clones from a transgenic parasite population, 0.5 cells per well were seeded in 96-well plates in a final volume of 200 µl M199 + medium supplemented with the respective antibiotics G418 (50 µg/ml, Roth, Karlsruhe, Germany), puromycin (25 µg/ml) and ClonNat (150 µg/ml, Werner Bioreagents, Jena, Germany) and 1 × Penicillin/Streptomycin. After 10–14 days of selection, emerging clones were transferred to culture flasks and further passaged under antibiotic selection.
Construction and preparation of recombinant DNA
LdBPK_340230 (HSP23), LdBPK_351030 (CK1.2), LdBPK_351040, LdBPK_351050, LdBPK_351060, LdBPK_351070 and LdBPK_351080 coding sequences were amplified from L. donovani 1S genomic DNA using specific primer pairs (Table S1) that introduce restriction sites as indicated. Fragments were subsequently ligated into the pCL2S vector93 predigested at the matching restriction sites. Plasmids were amplified in E. coli DH5α and purified by CsCl density gradient ultracentrifugation94.
For generation of the CK1.2 protein expression plasmid, the pJC45 plasmid40 was modified by introducing a KpnI restriction site into the multiple cloning site to create the pJC65 plasmid using primer Link-pJC65 + and Link-pJC65-. The open reading frame of Leishmania donovani CK1.2 was isolated from the pCL2S_LdBPK_351030 plasmid using KpnI and BamHI restriction sites and ligated between the matching sites in pJC65. A map of pJC65 is shown in Fig. S8.
Plasmid purification from Leishmania parasites
2–4 ✕ 109 log-phase promastigotes previously transfected with a mixture of over expression plasmids were harvested by centrifugation at 1250 ✕ g for 8 min at 4 °C. After three washing steps with 1✕ PBS, plasmids DNA was purified by alkaline lysis following the protocol for plasmid DNA mini preparation94. After phenol/chloroform/isoamylalcohol (25:24:1) extraction, plasmid DNA was precipitated by addition of 0.1 volume of 10 M ammonium acetate and 2.5 volumes 100% Ethanol. The pellet was dissolved for 24 h at 4 °C with Tris/EDTA buffer (pH 8.0). Next, 10 µl (approximately 100 ng) of isolated plasmid DNA was electroporated into E.coli XL one Blue Electroporation-Competent Cells (Agilent Technologies, Santa Clara, USA) cells using 0.1 cm gap Gene Pulser cuvettes (Bio Rad, Munich, Germany). Amplified DNA was isolated using the NucleoBond PC 20 kit (Machery-Nagel, Düren, Germany) following the manufacturer’s instructions, and eluted in 20 µl ultrapure ddH2O.
Next generation sequencing
DNA libraries were created using the Nextera XT library kit (Illumina, San Diego, USA) and the Nextera XT index kit (Illumina, San Diego, USA) according to manufacturer’s instructions. Leishmania gDNA was isolated using the Bioline Isolate 2 Genomic DNA Kit according to the manufacturer's protocol and set to a final concentration of 0.5 ng/µl using the Qubit (3.0)-system (Thermo Fisher Scientific, Waltham, USA). 1 ng of gDNA was used in the subsequent transposase-mediated tagmentation step. After purification of the DNA library using the Agencourt AMPure XP kit (Beckman Coulter, Fullerton, USA), the size distribution of DNA fragments was determined using a Bioanalyzer (Agilent Technologies, Santa Clara, USA) and the high sensitivity DNA analysis kit (Agilent Technologies, Santa Clara, USA). Each sample was further diluted to 4 nM taking into account the average fragment size and DNA concentration. A DNA library pool was prepared mixing 5 µl of each diluted sample in a Lobind 1.5 mL tube (Eppendorf, Hamburg, Germany). To denature the DNA-library, 5 µl of the DNA pool was mixed with 5 µl of freshly prepared 0.2 N NaOH. After incubation for 5 min at RT, 990 µl of pre-chilled HT1 buffer was added resulting in a 20 pM DNA library. The DNA library was further diluted with HT1 buffer to 10 pM in a final volume of 600 µl and 6 µl of a denatured PhyX reference library was added. Prior to loading the sequencing cartridge MiSeq Reagent Kit v3 (600-cycle) the library was heat denatured (95 °C, 2 min) and incubated for 5 min on ice. Sequencing was carried out on a MiSeq sequencer (Illumina, San Diego, USA).
RNA extraction, cDNA synthesis and quantitative Real-time PCR (qRT-PCR)
Total cellular RNA was isolated from 5 ✕ 107 log-phase L. donovani parasites using the InviTrap Spin Cell RNA Mini Kit (Stratec, Birekenfeld, Germany) according to manufacturer‘s instructions. First strand cDNA synthesis from 800 ng of RNA was performed using the QuantiTect(R) Reverse Transcription Kit (Qiagen, Venlo, Netherlands) following the manufacturer's protocol.
For qRT-PCR, 1 µl of cDNA was included in a final volume of 20 µl using the Biozym Blue S'Green qPCR Kit (Biozym Scientific GmbH, Oldendorf, Germany). Reactions were run on a Rotor Gene real time PCR cycler (RG 6000, Corbett Research, Sydney, Australia) and analysed95. Gene expression levels were normalised to HSP23+/+-LT samples. qPCR primer sequences are listed in Table S1.
Western blotting
For each sample, 1 ✕ 107 parasites were dissolved in sample buffer and separated by SDS-PAGE, followed by semi-dry Western blot94. Primary antibody polyclonal anti-CK1.2 rabbit (SY3535) (1:250)52 or polyclonal anti-HSP70 IgY(1:500)38, anti-P23 IgY (1:500)43 and anti-HSP23 IgY (1:250)42 in blocking solution was used in conjunction with anti-rabbit-IgG::-AP (goat, 1:1000) or anti-chicken-IgY::AP (rabbit, 1:5000) secondary antibodies (Dianova, Hamburg, Germany) and developed using Nitro Blue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl phosphate39.
Immunofluorescence assays
1 ✕ 107 parasites were gently sedimented (10 min, 1000 ✕ g, 4 °C), washed once with PBS and suspended in 100 µl of PBS. A 40 µl aliquot was smeared on a glass microscopic slide, air-dried and fixed for 3 min in ice cold methanol. Slides were gently washed three times (0.1% Triton X-100 in PBS) and permeabilised for 15 min in 0.1% Triton X-100, 50 mM NH4Cl in PBS. Slides were then incubated in blocking solution (2% BSA, 0.1% Triton X-100 in PBS) for 1 h. Polyclonal anti-tubulin-IgG (mouse) (1:4000) was added in blocking solution and incubated for 1 h at RT. Cells were washed three times for at least 15 min and then incubated with anti-mouse Alexa Fluor(R) 594 IgG (goat, 1:250)) together with DAPI (1:50 in PBS). For microscopic visualisation, slides were covered with 20 µl of Mowiol and cover slips and left for 24 h at 4 °C. Fluorescence microscopy was carried out using an EVOS FL Auto Cell Imaging System (Thermo Fisher Scientific, Waltham, USA). Images were processed using the ImageJ, Fuji Software (Version 2.0.0).
Expression and purification of recombinant HSP23 and CK1.2 protein
The pJC65-CK1.2, pJC45-HSP2342 and pJC45-P2343 plasmids were transformed into the bacterial strain BL21(DE3) (pAPlacIQ)40 and soluble (His)10-tagged proteins were purified using HisBind Resin (Novagen, Madison, WI) as described previously96. The eluate was supplemented with 10% glycerol, shock-frozen and stored in aliquots at − 80 °C.
Casein kinase assay
CK1.2 Kinase in vitro activity testing was performed essentially as described45. Briefly, recombinant proteins were added to buffer A (40 mM HEPES (pH7.5), 130 mM KCl, 15 mM MgCl2, 0.5 mM 1,10-Phenanthroline, 1 mM Phenylmethylsulfonyl fluoride, 25 mM ß-glycerophosphate, 0.1 mM sodium orthovanadate, 1 mM EGTA, 1 mM dithiothreitol, 0.05 mM ATP and 10 µCi [γ-32P]-ATP (Hartmann Analytics, Braunschweig, Germany) in a final volume of 25 µl. Recombinant proteins and substrates were used in the following concentrations: recLdCK1.2: 0.25 µg (6 pmol), recLdHSP23: 1.57 µg (60 pmol), recLdP23: 1.3 µg (60 pmol), myelin basic protein (MBP): 3 µg (162 pmol). CK1.2 specific inhibitor D4476 was used at a concentration of 10 µM in the presence of 0.7% DMSO. Controls were performed with equivalent final concentrations of DMSO. The reactions were incubated for 30 min at 25 °C or 37 °C and stopped by adding 1 Vol of SDS-PAGE sample buffer94. The samples were heated to 95 °C for 10 min and run on a 12.5% SDS polyacrylamide gel. After Western blot onto a polyvinylidenefluoride (PVDF) membrane (GE Healthcare Life Sciences, Munich, Germany), 32P incorporation was detected by autoradiography (Fuji Super RX-N, Tokyo, Japan).
In silico procedures
In silico cloning, DNA and protein sequence analysis was performed using the MacVector software version 17.x. Numerical data were analysed using the Prism software (version 8, GraphPad Software LLC). Halftone images were optimised in toto for contrast and brightness using the ImageJ Fiji Software (Version 2.0.0)(NIH). Composite figures were assembled using the Intaglio software (Purgatory). The heat map was generated using the Heatmapper Online Tool97.
Bioinformatics and NGS analysis
Reads were aligned to TriTrypDB version 42 of L. donovani BPK using Bowtie298. Chromosome ploidy was determined by an iterative approach using an in-house program: initially, the coverage of each chromosome was estimated to be 2.0. The mean coverage of the positions in the 15th to 85th quantile and the ratio coverage/mean were calculated. Coverages were then updated according to these fractions and the procedure repeated until conversion. For gene coverages, a similar approach was used, with the difference that the copy number was normalised to the previously calculated chromosome ploidies.
Statistics
Statistical comparisons between groups were performed using one-way analysis of variance (ANOVA)/Kruskal–Wallis test with Dunn’s post test. Differences were considered significant at a level of p < 0.05.
Supplementary information
Acknowledgements
We thank our colleagues H. Zirpel and V. Adaui for many helpful discussions, Daniel Cardar for the use of the sequencing facility, and N. Rachidi for support with the casein kinase assays and for a gift of anti-CK1.2 antibody.
Author contributions
C. K.-B.: study concept, gene replacement, phenotype analyses, whole genome sequencing, in vitro selection, targeted over expression, in vitro phosphorylation analyses, manuscript preparation. S.L.: Bioinformatics analysis. C. B.: In vitro proliferation analyses, immune fluorescent microscopy, whole genome sequencing. J.C.: study concept, supervision and manuscript preparation.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
Whole genome sequencing data are available from: https://www.ncbi.nlm.nih.gov/sra/PRJNA633969.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-72724-x.
References
- 1.Alvar J, et al. Leishmaniasis worldwide and global estimates of its incidence. PLoS ONE. 2012;7:e35671. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.2WHO. Integrating Neglected Tropical Diseases into Global Health and Development: Fourth WHO Report on Neglected Tropical Diseases (2017).
- 3.Sundar S, Rai M. Treatment of visceral leishmaniasis. Expert Opin. Pharmacother. 2005;6:2821–2829. doi: 10.1517/14656566.6.16.2821. [DOI] [PubMed] [Google Scholar]
- 4.4Hendrickx, S., Maes, L., Croft, S. L. & Caljon, G. in Drug Resistance in Leishmania Parasites (eds Ponte-Sucre A. & Padrón-Nieves M.) (Springer, 2018).
- 5.Myler PJ, et al. Leishmania major Friedlin chromosome 1 has an unusual distribution of protein-coding genes. Proc. Natl. Acad. Sci. USA. 1999;96:2902–2906. doi: 10.1073/pnas.96.6.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Myler PJ, et al. Genomic organization and gene function in Leishmania. Biochem. Soc. Trans. 2000;28:527–531. doi: 10.1042/bst0280527. [DOI] [PubMed] [Google Scholar]
- 7.Ivens AC, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LeBowitz JH, Smith HQ, Rusche L, Beverley SM. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes. Dev. 1993;7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- 9.Clayton C, Shapira M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol. Biochem. Parasitol. 2007;156:93–101. doi: 10.1016/j.molbiopara.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Argaman M, Aly R, Shapira M. Expression of heat shock protein 83 in Leishmania is regulated post- transcriptionally. Mol. Biochem. Parasitol. 1994;64:95–110. doi: 10.1016/0166-6851(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 11.Zilka A, Garlapati S, Dahan E, Yaolsky V, Shapira M. Developmental regulation of HSP83 in Leishmania: transcript levels are controlled by the efficiency of 3? RNA processing and preferential translation is directed by a determinant in the 3' UTR. J. Biol. Chem. 2001;11:11. doi: 10.1074/jbc.M108271200. [DOI] [PubMed] [Google Scholar]
- 12.Brandau S, Dresel A, Clos J. High constitutive levels of heat-shock proteins in human-pathogenic parasites of the genus Leishmania. Biochem. J. 1995;310:225–232. doi: 10.1042/bj3100225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clos J, Brandau S, Hoyer C. Chemical stress does not induce heat shock protein synthesis in Leishmania donovani. Protist. 1998;149:167–172. doi: 10.1016/S1434-4610(98)70021-5. [DOI] [PubMed] [Google Scholar]
- 14.David M, et al. Preferential translation of Hsp83 in Leishmania requires a thermosensitive polypyrimidine-rich element in the 3' UTR and involves scanning of the 5' UTR. RNA. 2010;16:364–374. doi: 10.1261/rna.1874710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bifeld E, et al. Ribosome profiling reveals HSP90 inhibitor effects on stage-specific protein synthesis in Leishmania donovani. mSystems. 2018 doi: 10.1128/mSystems.00214-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leprohon P, et al. Gene expression modulation is associated with gene amplification, supernumerary chromosomes and chromosome loss in antimony-resistant Leishmania infantum. Nucl. Acids Res. 2009;37:1387–1399. doi: 10.1093/nar/gkn1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lachaud L, et al. Constitutive mosaic aneuploidy is a unique genetic feature widespread in the Leishmania genus. Microbes Infect. 2014;16:61–66. doi: 10.1016/j.micinf.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Dumetz F, et al. Modulation of aneuploidy in Leishmania donovani during adaptation to different in vitro and in vivo environments and its impact on gene expression. MBio. 2017 doi: 10.1128/mBio.00599-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ubeda JM, et al. Genome-wide stochastic adaptive DNA amplification at direct and inverted DNA repeats in the parasite Leishmania. PLoS Biol. 2014;12:e1001868. doi: 10.1371/journal.pbio.1001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laffitte MN, Leprohon P, Papadopoulou B, Ouellette M. Plasticity of the Leishmania genome leading to gene copy number variations and drug resistance. F1000Res. 2016;5:2350. doi: 10.12688/f1000research.9218.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iantorno SA, et al. Gene expression in Leishmania is regulated predominantly by gene dosage. mBio. 2017 doi: 10.1128/mBio.01393-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grondin K, Haimeur A, Mukhopadhyay R, Rosen BP, Ouellette M. Co-amplification of the gamma-glutamylcysteine synthetase gene gsh1 and of the ABC transporter gene pgpA in arsenite-resistant Leishmania tarentolae. EMBO J. 1997;16:3057–3065. doi: 10.1093/emboj/16.11.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ubeda JM, et al. Modulation of gene expression in drug resistant Leishmania is associated with gene amplification, gene deletion and chromosome aneuploidy. Genome Biol. 2008;9:R115. doi: 10.1186/gb-2008-9-7-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukherjee B, et al. Antimony-resistant but not antimony-sensitive Leishmania donovani up-regulates host IL-10 to overexpress multidrug-resistant protein 1. Proc. Natl. Acad. Sci. USA. 2013;110:E575–582. doi: 10.1073/pnas.1213839110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patino LH, et al. Major changes in chromosomal somy, gene expression and gene dosage driven by Sb(III) in Leishmania braziliensis and Leishmania panamensis. Sci. Rep. 2019;9:9485. doi: 10.1038/s41598-019-45538-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spath GF, Lye LF, Segawa H, Turco SJ, Beverley SM. Identification of a compensatory mutant (lpg2-REV) of Leishmania major able to survive as amastigotes within macrophages without LPG2-dependent glycoconjugates and its significance to virulence and immunization strategies. Infect. Immun. 2004;72:3622–3627. doi: 10.1128/IAI.72.6.3622-3627.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reiling L, et al. Spontaneous recovery of pathogenicity by Leishmania major hsp100-/- alters the immune response in mice. Infect. Immun. 2006;74:6027–6036. doi: 10.1128/IAI.00773-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turco S, Descoteaux A, Ryan K, Garraway L, Beverley S. Isolation of virulence genes directing GPI synthesis by functional complementation of Leishmania. Braz. J. Med. Biol. Res. 1994;27:133–138. [PubMed] [Google Scholar]
- 29.Reiling L, Chrobak M, Schmetz C, Clos J. Overexpression of a single Leishmania major gene is sufficient to enhance parasite infectivity in vivo and in vitro. Mol. Microbiol. 2010;76:1175–1190. doi: 10.1111/j.1365-2958.2010.07130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Killick-Kendrick R. The life-cycle of Leishmania in the sandfly with special reference to the form infective to the vertebrate host. Ann. Parasitol. Hum. Comp. 1990;65:37–42. doi: 10.1051/parasite/1990651037. [DOI] [PubMed] [Google Scholar]
- 31.Bogdan C, Gessner A, Solbach W, Rollinghoff M. Invasion, control and persistence of Leishmania parasites. Curr. Opin. Immunol. 1996;8:517–525. doi: 10.1016/S0952-7915(96)80040-9. [DOI] [PubMed] [Google Scholar]
- 32.Kloehn J, Saunders EC, O'Callaghan S, Dagley MJ, McConville MJ. Characterization of metabolically quiescent Leishmania parasites in murine lesions using heavy water labeling. PLoS Pathog. 2015;11:e1004683. doi: 10.1371/journal.ppat.1004683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McConville MJ, Saunders EC, Kloehn J, Dagley MJ. Leishmania carbon metabolism in the macrophage phagolysosome—feast or famine? F1000Res. 2015;4:938. doi: 10.12688/f1000research.6724.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jara M, et al. Tracking of quiescence in Leishmania by quantifying the expression of GFP in the ribosomal DNA locus. Sci. Rep. 2019;9:18951. doi: 10.1038/s41598-019-55486-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bates PA. Complete developmental cycle ofLeishmania mexicanain axenic culture. Parasitology. 1994;108:1–9. doi: 10.1017/S0031182000078458. [DOI] [PubMed] [Google Scholar]
- 36.Barak E, et al. Differentiation of Leishmania donovani in host-free system: analysis of signal perception and response. Mol. Biochem. Parasitol. 2005;141:99–108. doi: 10.1016/j.molbiopara.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 37.Rosenzweig D, et al. Retooling Leishmania metabolism: from sand fly gut to human macrophage. FASEB J. 2008;22:590–602. doi: 10.1096/fj.07-9254com. [DOI] [PubMed] [Google Scholar]
- 38.Hübel A, Brandau S, Dresel A, Clos J. A member of the ClpB family of stress proteins is expressed during heat shock in Leishmania spp. Mol. Biochem. Parasitol. 1995;70:107–118. doi: 10.1016/0166-6851(95)00012-P. [DOI] [PubMed] [Google Scholar]
- 39.Krobitsch S, et al. Leishmania donovani heat shock protein 100: characterization and function in amastigote stage differentiation. J. Biol. Chem. 1998;273:6488–6494. doi: 10.1074/jbc.273.11.6488. [DOI] [PubMed] [Google Scholar]
- 40.Schlüter A, et al. Expression and subcellular localization of Cpn60 protein family members in Leishmania donovani. Biochim. Biophys. Acta. 2000;1491:65–74. doi: 10.1016/S0167-4781(00)00028-2. [DOI] [PubMed] [Google Scholar]
- 41.Zamora-Veyl FB, Kroemer M, Zander D, Clos J. Stage-specific expression of the mitochondrial co-chaperonin of Leishmania donovani, CPN10. Kinetoplastid. Biol. Dis. 2005;4:3. doi: 10.1186/1475-9292-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hombach A, Ommen G, MacDonald A, Clos J. A small heat shock protein is essential for thermotolerance and intracellular survival of Leishmania donovani. J. Cell Sci. 2014;127:4762–4773. doi: 10.1242/jcs.157297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hombach A, Ommen G, Sattler V, Clos J. Leishmania donovani P23 protects parasites against HSP90 inhibitor-mediated growth arrest. Cell Stress Chaperones. 2015;20:673–685. doi: 10.1007/s12192-015-0595-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaur P, Garg M, Hombach-Barrigah A, Clos J, Goyal N. MAPK1 of Leishmania donovani interacts and phosphorylates HSP70 and HSP90 subunits of foldosome complex. Sci. Rep. 2017;7:10202. doi: 10.1038/s41598-017-09725-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hombach-Barrigah A, et al. Leishmania donovani 90 kD heat shock protein: impact of phosphosites on parasite fitness, infectivity and casein kinase affinity. Sci. Rep. 2019;9:5074. doi: 10.1038/s41598-019-41640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sterkers Y, Crobu L, Lachaud L, Pages M, Bastien P. Parasexuality and mosaic aneuploidy in Leishmania: alternative genetics. Trends Parasitol. 2014;30:429–435. doi: 10.1016/j.pt.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 47.Clayton CE. Life without transcriptional control? From fly to man and back again. Embo J. 2002;21:1881–1888. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beverley SM. Gene amplification in Leishmania. Annu. Rev. Microbiol. 1991;45:417–444. doi: 10.1146/annurev.mi.45.100191.002221. [DOI] [PubMed] [Google Scholar]
- 49.Haimeur A, Ouellette M. Gene amplification in Leishmania tarentolae selected for resistance to sodium stibogluconate. Antimicrob. Agents Chemother. 1998;42:1689–1694. doi: 10.1128/AAC.42.7.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Downing T, et al. Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res. 2011;21:2143–2156. doi: 10.1101/gr.123430.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rogers MB, et al. Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res. 2011;21:2129–2142. doi: 10.1101/gr.122945.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rachidi N, et al. Pharmacological assessment defines Leishmania donovani casein kinase 1 as a drug target and reveals important functions in parasite viability and intracellular infection. Antimicrob. Agents Chemother. 2014;58:1501–1515. doi: 10.1128/AAC.02022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mannaert A, Downing T, Imamura H, Dujardin JC. Adaptive mechanisms in pathogens: universal aneuploidy in Leishmania. Trends Parasitol. 2012;28:370–376. doi: 10.1016/j.pt.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 54.Cuypers B, et al. Integrated genomic and metabolomic profiling of ISC1, an emerging Leishmania donovani population in the Indian subcontinent. Infect. Genet. Evol. 2018;62:170–178. doi: 10.1016/j.meegid.2018.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dumetz F, et al. Molecular preadaptation to antimony resistance in Leishmania donovani on the Indian Subcontinent. mSphere. 2018 doi: 10.1128/mSphere.00548-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grondin K, Papadopoulou B, Ouellette M. Homologous recombination between direct repeat sequences yields P-glycoprotein containing amplicons in arsenite resistant Leishmania. Nucl. Acids Res. 1993;21:1895–1901. doi: 10.1093/nar/21.8.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Navarro M, et al. Inverted repeat structure and homologous sequences in the LD1 amplicons of Leishmania spp. Mol. Biochem. Parasitol. 1994;68:69–80. doi: 10.1016/0166-6851(94)00147-2. [DOI] [PubMed] [Google Scholar]
- 58.Bringaud F, et al. Members of a large retroposon family are determinants of post-transcriptional gene expression in Leishmania. PLoS Pathog. 2007;3:1291–1307. doi: 10.1371/journal.ppat.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith M, Bringaud F, Papadopoulou B. Organization and evolution of two SIDER retroposon subfamilies and their impact on the Leishmania genome. BMC Genom. 2009;10:240. doi: 10.1186/1471-2164-10-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson AE, Chen JS, Gould KL. CK1 is required for a mitotic checkpoint that delays cytokinesis. Curr. Biol. 2013;23:1920–1926. doi: 10.1016/j.cub.2013.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fan MM, Zhang H, Hayden MR, Pelech SL, Raymond LA. Protective up-regulation of CK2 by mutant huntingtin in cells co-expressing NMDA receptors. J. Neurochem. 2008;104:790–805. doi: 10.1111/j.1471-4159.2007.05016.x. [DOI] [PubMed] [Google Scholar]
- 62.Meng QJ, et al. Setting clock speed in mammals: the CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron. 2008;58:78–88. doi: 10.1016/j.neuron.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eng GWL, Virshup DM. Site-specific phosphorylation of casein kinase 1 delta (CK1delta) regulates its activity towards the circadian regulator PER2. PLoS ONE. 2017;12:e0177834. doi: 10.1371/journal.pone.0177834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Knippschild U, et al. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell Signal. 2005;17:675–689. doi: 10.1016/j.cellsig.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 65.Allocco JJ, et al. Inhibitors of casein kinase 1 block the growth of Leishmania major promastigotes in vitro. Int J Parasitol. 2006;36:1249–1259. doi: 10.1016/j.ijpara.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 66.Silverman JM, et al. Proteomic analysis of the secretome of Leishmania donovani. Genome Biol. 2008;9:R35. doi: 10.1186/gb-2008-9-2-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paape D, Barrios-Llerena ME, Le Bihan T, Mackay L, Aebischer T. Gel free analysis of the proteome of intracellular Leishmania mexicana. Mol. Biochem. Parasitol. 2010;169:108–114. doi: 10.1016/j.molbiopara.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 68.Martel D, et al. The low complexity regions in the C-terminus are essential for the subcellular localisation of Leishmania casein kinase 1 but not for its activity. bioRxiv. 2020 doi: 10.1101/2020.02.28.969741. [DOI] [Google Scholar]
- 69.Silverman JM, et al. Leishmania exosomes modulate innate and adaptive immune responses through effects on monocytes and dendritic cells. J. Immunol. 2010;185:5011–5022. doi: 10.4049/jimmunol.1000541. [DOI] [PubMed] [Google Scholar]
- 70.Silverman JM, et al. An exosome-based secretion pathway is responsible for protein export from Leishmania and communication with macrophages. J. Cell Sci. 2010;123:842–852. doi: 10.1242/jcs.056465. [DOI] [PubMed] [Google Scholar]
- 71.Muller P, et al. C-terminal phosphorylation of Hsp70 and Hsp90 regulates alternate binding to co-chaperones CHIP and HOP to determine cellular protein folding/degradation balances. Oncogene. 2013;32:3101–3110. doi: 10.1038/onc.2012.314. [DOI] [PubMed] [Google Scholar]
- 72.Wiesgigl M, Clos J. Heat shock protein 90 homeostasis controls stage differentiation in Leishmania donovani. Mol. Biol. Cell. 2001;12:3307–3316. doi: 10.1091/mbc.12.11.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hombach A, Ommen G, Chrobak M, Clos J. The Hsp90-Sti1 Interaction is critical for Leishmania donovani proliferation in both life cycle stages. Cell Microbiol. 2013;15:585–600. doi: 10.1111/cmi.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kobayashi T, et al. Regulation of cytosolic prostaglandin E synthase by phosphorylation. Biochem. J. 2004;381:59–69. doi: 10.1042/BJ20040118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tosoni K, et al. The p23 co-chaperone protein is a novel substrate of CK2 in Arabidopsis. Mol. Cell Biochem. 2011;356:245–254. doi: 10.1007/s11010-011-0969-0. [DOI] [PubMed] [Google Scholar]
- 76.Graves PR, Roach PJ. Role of COOH-terminal phosphorylation in the regulation of casein kinase I delta. J. Biol. Chem. 1995;270:21689–21694. doi: 10.1074/jbc.270.37.21689. [DOI] [PubMed] [Google Scholar]
- 77.Cegielska A, Gietzen KF, Rivers A, Virshup DM. Autoinhibition of casein kinase I epsilon (CKI epsilon) is relieved by protein phosphatases and limited proteolysis. J. Biol. Chem. 1998;273:1357–1364. doi: 10.1074/jbc.273.3.1357. [DOI] [PubMed] [Google Scholar]
- 78.Gietzen KF, Virshup DM. Identification of inhibitory autophosphorylation sites in casein kinase I epsilon. J. Biol. Chem. 1999;274:32063–32070. doi: 10.1074/jbc.274.45.32063. [DOI] [PubMed] [Google Scholar]
- 79.Fang Y, Fliss AE, Rao J, Caplan AJ. SBA1 encodes a yeast hsp90 cochaperone that is homologous to vertebrate p23 proteins. Mol. Cell Biol. 1998;18:3727–3734. doi: 10.1128/mcb.18.7.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Felts SJ, Toft DO. p23, a simple protein with complex activities. Cell Stress Chaperones. 2003;8:108–113. doi: 10.1379/1466-1268(2003)008<0108:paspwc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Richter K, Walter S, Buchner J. The Co-chaperone Sba1 connects the ATPase reaction of Hsp90 to the progression of the chaperone cycle. J. Mol. Biol. 2004;342:1403–1413. doi: 10.1016/j.jmb.2004.07.064. [DOI] [PubMed] [Google Scholar]
- 82.McLaughlin SH, et al. The co-chaperone p23 arrests the Hsp90 ATPase cycle to trap client proteins. J. Mol. Biol. 2006;356:746–758. doi: 10.1016/j.jmb.2005.11.085. [DOI] [PubMed] [Google Scholar]
- 83.Chua CS, Low H, Goo KS, Sim TS. Characterization of Plasmodium falciparum co-chaperone p23: its intrinsic chaperone activity and interaction with Hsp90. Cell Mol. Life Sci. 2010;67:1675–1686. doi: 10.1007/s00018-010-0275-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Echeverria PC, et al. The Hsp90 co-chaperone p23 of Toxoplasma gondii: identification, functional analysis and dynamic interactome determination. Mol. Biochem. Parasitol. 2010;172:129–140. doi: 10.1016/j.molbiopara.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Batista FA, et al. Identification of two p23 co-chaperone isoforms in Leishmania braziliensis exhibiting similar structures and Hsp90 interaction properties despite divergent stabilities. FEBS J. 2015;282:388–406. doi: 10.1111/febs.13141. [DOI] [PubMed] [Google Scholar]
- 86.Stebbins CE, et al. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell. 1997;89:239–250. doi: 10.1016/S0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 87.Forafonov F, et al. p23/Sba1p protects against Hsp90 inhibitors independently of its intrinsic chaperone activity. Mol. Cell. Biol. 2008;28:3446–3456. doi: 10.1128/MCB.02246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mollapour M, Neckers L. Post-translational modifications of Hsp90 and their contributions to chaperone regulation. Biochem. Biophys. Acta. 1823;648–655:2012. doi: 10.1016/j.bbamcr.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dey R, et al. Live attenuated Leishmania donovani p27 gene knockout parasites are nonpathogenic and elicit long-term protective immunity in BALB/c mice. J. Immunol. 2013;190:2138–2149. doi: 10.4049/jimmunol.1202801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Elikaee S, et al. Leishmania major p27 gene knockout as a novel live attenuated vaccine candidate: protective immunity and efficacy evaluation against cutaneous and visceral leishmaniasis in BALB/c mice. Vaccine. 2019;37:3221–3228. doi: 10.1016/j.vaccine.2019.04.068. [DOI] [PubMed] [Google Scholar]
- 91.Zabala-Penafiel A, Todd D, Daneshvar H, Burchmore R. The potential of live attenuated vaccines against cutaneous Leishmaniasis. Exp. Parasitol. 2020;210:107849. doi: 10.1016/j.exppara.2020.107849. [DOI] [PubMed] [Google Scholar]
- 92.Zirpel H, Clos J. Gene replacement by homologous recombination. Methods Mol. Bio.l. 1971;169–188:2019. doi: 10.1007/978-1-4939-9210-2_8. [DOI] [PubMed] [Google Scholar]
- 93.Bartsch K, Hombach-Barrigah A, Clos J. Hsp90 inhibitors radicicol and geldanamycin have opposing effects on Leishmania Aha1-dependent proliferation. Cell Stress Chaperones. 2017;22:729–742. doi: 10.1007/s12192-017-0800-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Green MR, Sambrook J, Sambrook J. Molecular Cloning: A Laboratory Manual. 4. New York: Cold Spring Harbor Laboratory Press; 2012. [Google Scholar]
- 95.Bifeld E, et al. Geographical sequence variation in the Leishmania major virulence factor P46. Infect. Genet. Evol. 2015;30:195–205. doi: 10.1016/j.meegid.2014.12.029. [DOI] [PubMed] [Google Scholar]
- 96.Clos J, Brandau S. pJC20 and pJC40—two high-copy-number vectors for T7 RNA polymerase-dependent expression of recombinant genes in Escherichia coli. Prot. Expression Purif. 1994;5:133–137. doi: 10.1006/prep.1994.1020. [DOI] [PubMed] [Google Scholar]
- 97.Babicki S, et al. Heatmapper: web-enabled heat mapping for all. Nucl. Acids Res. 2016;44:W147–153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole genome sequencing data are available from: https://www.ncbi.nlm.nih.gov/sra/PRJNA633969.