Abstract
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel coronavirus that emerged recently and has created a global pandemic. Symptomatic SARS-CoV-2 infection, termed coronavirus disease 2019 (COVID-19), has been associated with a host of symptoms affecting numerous organ systems, including the lungs, cardiovascular system, kidney, central nervous system, gastrointestinal tract, and skin, among others.
Objective
Although several risk factors have been identified as related to complications from and severity of COVID-19, much about the virus remains unknown. The host immune response appears to affect the outcome of disease. It is not surprising that patients with intrinsic or secondary immune compromise might be particularly susceptible to complications from SARS-CoV-2 infection. Pathogenic loss-of-function or gain-of-function heterozygous variants in nuclear factor-κB2 have been reported to be associated with either a combined immunodeficiency or common variable immunodeficiency phenotype.
Methods
We evaluated the functional consequence and immunologic phenotype of a novel NFKB2 loss of function variant in a 17-year-old male patient and describe the clinical management of SARS-CoV-2 infection in this context.
Results
This patient required a 2-week hospitalization for SARS-CoV-2 infection, including 7 days of mechanical ventilation. We used biologic therapies to avert potentially fatal acute respiratory distress syndrome and treat hyperinflammatory responses. The patient had an immunologic phenotype of B-cell dysregulation with decreased switched memory B cells. Despite the underlying immune dysfunction, he recovered from the infection with intense management.
Conclusions
This clinical case exemplifies some of the practical challenges in management of patients with SARS-CoV-2 infection, especially in the context of underlying immune dysregulation.
Key words: COVID-19, immunodeficiency, NF-κB, NF-κB pathway, NF-κB2, SARS-CoV-2
Abbreviations used: ACTH, Adrenocorticotropic hormone; CID, Combined immunodeficiency; CVID, Common variable immunodeficiency; COVID-19, Coronavirus disease 2019; CP, COVID-19 convalescent plasma; CRP, C-reactive protein; CT, Cycle threshold; DAVID, Deficit in anterior pituitary function and variable immune deficiency; FDA, US Food and Drug Administration; GOF, Gain-of-function; LOF, Loss-of-function; LRT, Lower respiratory tract; NF-κB, Nuclear factor-κB; NF-κB1, Nuclear factor-κB1; NF-κB2, Nuclear factor-κB2; NK, Natural killer; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; Treg, Regulatory T; XLA, X-linked agammaglobulinemia
Since its discovery more than 30 years ago, the nuclear factor κB (NF-κB) pathway has come to occupy a pivotal position in the function of the immune system and mediates a variety of cellular processes, including immune response, inflammation, cellular proliferation, and cell survival, among others.1 The NF-κB complex consists of 5 key transcription factors, including RelA, RelB, c-Rel, nuclear factor κB1 (NF-κB1), and nuclear factor κB2 (NF-κB2), which are regulated by the inhibitory IκB kinase complex.2 NF-κB1 and NF-κB2 are sequestered in the cytoplasm of the cell along with their inhibitory counterparts. The activation of NF-κB occurs through 1 of 2 pathways, the canonical and noncanonical, both of which are vital for immune regulation. The canonical pathway is triggered by stimulation through the T- and B-cell receptors, Toll-like receptors, and the TNF superfamily receptors. NF-κB1 is a component of the canonical pathway, and following degradation of the IκBα molecule in the proteasome, the cleaved p50 molecule transiently translocates to the nucleus, where it heterodimerizes with the p65/RelA molecule and the c-Rel molecule.3 The noncanonical pathway, of which NF-κB2 is a key player, involves activation through a specific set of receptors, including BAFF-R, CD40, and other TNF superfamily receptors family members, such as RANK. The NF-κB2 pathway involves processing of the p100 precursor molecule to form the p52 protein, which complexes with RelB and results in nuclear translocation. The canonical pathway of NF-κB is integral to the holistic function of the immune response, whereas the noncanonical pathway appears to play an adjunct role and is relevant to specific aspects of adaptive immunity. Patients with loss-of-function (LOF) variants in NFΚB2 have been described to have either a B-cell defect with a common variable immunodeficiency (CVID)-like or combined immunodeficiency (CID) phenotype.4, 5, 6 Although NF-κB plays a critical role in B-cell development and activation and was first identified in activated B cells, its role is not exclusive to B cells and it plays a significant role in multiple components of the immune system, both innate and adaptive.7
The newly described severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pathogen has rapidly spread worldwide, leading to a global pandemic and more than half a million deaths to date. The virus gains access to epithelial cells in the upper respiratory tract by initially binding to the angiotensin-converting enzyme 2 (ACE2) receptor.8 , 9 The early immune response to SARS-CoV-2 likely involves the innate immune system and sensing of viral RNA by Toll-like receptors and other pathogen recognition receptors, on the basis of parallels drawn from related viruses, such as SARS-CoV-1 and Middle East respiratory syndrome coronavirus (MERS-CoV).10 The activation of pathogen recognition receptors triggers various signaling cascades and production of cytokines. Type 1 interferons are key antiviral cytokines,11 , 12 but in addition to these, IL-1b, IL-6, IL-18, TNF-α, and other proinflammatory cytokines and chemokines are secreted. The immune dysregulation observed in patients with severe SARS-CoV-2 infection suggests that there is an imbalance between the host immune response and immune evasion tactics of the pathogen.13, 14, 15 The kinetics of the immune response and triggering of proinflammatory pathways leads to the observed “hyperinflammatory” phenotype or “cytokine storm” described in SARS-CoV-2 infection.16 , 17 Patients with acute SARS-Co-V-2 infection demonstrate pan-lymphopenia,18 which is more profound in patients with severe disease than in those with mild disease.
It is reasonable to postulate that in patients with primary immunodeficiencies and immune regulatory disorders, the range of disease from mild to severe would depend on the specific immune defect and immune response to the infection. Recently, a study of a small group of patients with humoral immune defects demonstrated that B cells might play a critical role in immune dysregulation in response to SARS-CoV-2 infection.19 In this study, patients with agammaglobulinemia due to absent B cells (X-linked agammaglobulinemia [XLA] or other causes of agammaglobulinemia) appeared to have a milder clinical course than did patients with CVID with hypogammaglobulinemia and dysregulated but detectable B cells, in whom the disease was more severe, necessitating multiple therapeutic agents and other life-saving measures. In a recent study, the use of a Bruton tyrosine kinase inhibitor was shown to improve markers of the inflammatory response in patients with coronavirus disease 2019 (COVID-19),20 which may represent a pharmacologic phenocopy of Bruton tyrosine kinase LOF in patients with XLA. In other patients with XLA however, the disease course appears to be prolonged, requiring adjunct therapy,21 which suggests that decreased mortality risk does not imply rapid recovery from infection. The data from patients with XLA and CVID suggests that dysregulated B cells might contribute to the pathology of SARS-CoV-2 infection and that the absence of B cells or low immunoglobulin levels in itself may not necessarily predicate high mortality but may still result in increased morbidity.
There are several reports in the literature on the association of NFΚB2 variants with either CVID or CID. NFΚB2 LOF variants located at the C-terminus of the protein and associated with an autosomal dominant–negative phenotype were shown to result in severe B-cell deficiency with hypogammaglobulinemia and alopecia.4 , 22 , 23 On the other hand, gain-of-function (GOF) variants in the NFKB2 gene resulted in a CID phenotype in some patients with impaired T-cell and B-cell function6 without endocrine anomalies or ectodermal dysplasia. This is in contrast to what has been observed in patients with the LOF variants, who have been reported to have endocrine anomalies manifesting as adrenocorticotropic hormone (ACTH) and growth hormone deficiencies, also called deficit in anterior pituitary function and variable immune deficiency (DAVID) syndrome.4 , 23 A recent cohort analysis by Klemann et al, which examined several patients with NF-κB2 revealed that adrenal insufficiency was noted in less than half of patients (21 of 50 [42%]).5 The penetrance and expressivity of NF-κB2 variants has been previously shown to be variable, depending on the specific type of genetic variant present, but in the Klemann study, the overall penetrance of disease was high, as only 2 of 50 patients were totally asymptomatic.5 Several of these patients had clear manifestations of CID with associated T-cell dysfunction and autoimmunity. Natural killer (NK) cell cytotoxic function has been reported to be impaired in NF-κB2 p52 haploinsufficiency24 and has also been shown to be adversely affected in SARS-CoV-2 infection with impaired degranulation and decreased production of key cytotoxic proteins and chemokines, as well as elevated serum IL-6 level.25, 26, 27 Here, we have described a patient with a novel NFKB2 LOF variant who developed severe SARS-CoV-2 infection requiring a multipronged approach to effectively manage the disease and its associated complications. We anticipate that these findings will be informative in the management of patients with an underlying intrinsic immunodeficiency, particularly affecting the B- and/or T-cell compartment.
Methods
The patient provided consent in accordance with an institutional review board–approved protocol (no. 00000015); in addition, specific consent was obtained for publication of the study findings. All dates have been written relative to first day of hospitalization (eg, day 1).
Flow cytometry
Whole blood cells and PBMCs were collected with EDTA and used for multiparametric immunophenotyping. A large panel of T- and B-cell subset markers and regulatory T (Treg) cells were assessed by flow cytometry at different time points: 10 months before SARS-CoV-2 infection, 4 days after hospital admission for SARS-CoV-2 infection (day 4), and at days 15 and 35 after hospitalization (day 15 and day 35). Briefly, 1 million cells, either from whole blood or PBMCs, were stained with markers for T-cell subsets, B- cell subsets, or Treg cells to identify naive and memory T cells and recent thymic emigrants (CD45RA, CD45RO, CD62L, CCR7, CD27, and CD31), activated T cells (CD25+ and HLA DR+), T-cell receptor expression (αβ vs γδ+), exhausted and senescent T cells (CD57, CD28, and PD-1), memory B-cell subsets (CD27, IgD, IgM, IgG, and IgA), early and late B-cell differentiation markers (CD27, CD10, CD24, CD38, CD21, and IgM), and Treg cells (CD4, CD25, and FOXP3). For analysis, 10,000 CD45+ lymphocytes for TBNK subset quantitation, 10,000 CD45+ lymphocytes for expanded T- and B-cell phenotyping, and 50,000 CD45+ lymphocytes for Treg cell quantitation were collected on a CytoFlex flow cytometer (Beckman Coulter, La Brea, Calif) with data acquisition using Kaluza C software (version 1.1, Beckman Coulter).
Diagnosis of SARS-CoV-2 infection by PCR
Clinical diagnostic assay
Detection of SARS-CoV-2 RNA from the patient’s nasopharyngeal, lower respiratory tract (LRT), and blood specimens was performed in the Clinical Microbiology Laboratory at Nationwide Children’s Hospital by using a modified US Centers for Disease Control and Prevention assay under US Food and Drug Administration (FDA) Emergency Use Authorization (In Vitro Diagnostics Emergency Use Authorization, High Complexity Molecular-Based Laboratory Developed Tests [https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-authorizations#covid19ivd]). This assay has an analytic sensitivity of 100% at 188 genomic copies/mL for nasopharyngeal specimens. Briefly, total nucleic acid was extracted by using the NucliSENS easyMag platform (bioMerieux, Durham, NC); 5 μL of the eluate was added to 20 μL of reaction mixture (4 μL of TaqPath-1-Step RT-qPCR Master Mix [ThermoFisher, Waltham, Mass], 1.5 μL of each primer/probe mix [Integrated DNA Technologies, Coralville, Iowa] and sterile distilled RNase-free water). The reaction was performed on QuantStudio 7 flex (Life Technologies, Grand Island, NY) under the following conditions: 25°C for 2 minutes, 50°C for 15 minutes, enzyme activation at 95°C for 2 minutes, and 45 cycles of 95°C for 3 seconds and 55°C for 30 seconds. This assay targets 2 regions of nucleocapsid gene of SARS-CoV-2, N1 and N2, as well as a human housekeeping gene (RNaseP), to assess the quality of the specimen. Viral load data for this assay are described as the “N1” RT-PCR or “N2” RT-PCR, which are reported as “true” cycle threshold (Ct) values and reflect the number of amplification cycles required for the fluorescent signal to cross the threshold (ie, higher viral loads are indicated by lower Ct values and vice versa).
Research diagnostic assay
To detect SARS-CoV-2 in the blood during the acute illness by using PCR, a research use–only PCR assay was developed. Briefly, 140 μL of whole blood was thawed at room temperature, and RNA was extracted by using a Qiagen QIAmp Viral RNA Kit (Qiagen USA, Germantown, Md). Reverse transcription of 10 μL of extracted RNA was completed by using a High Capacity cDNA Reverse Transcriptase Kit (Thermo Fisher, Waltham, Mass). The quantitative PCR was completed by using the StepOnePlus Real-Time PCR system (ThermoFisher) and IDT Primetime qPCR Probe assay (Integrated DNA Technologies, Coralville, Iowa) with N-gene–specific primers and probes (World Health Organization guidance document for laboratory testing for the 2019 novel coronavirus in suspected human cases [https://www.who.int/docs/default-source/coronaviruse/peiris-protocol-16-1-20.pdf?sfvrsn=af1aac73_4]). The viral load data for this assay are described as the “N” RT-PCR and are reported in cycle threshold (Ct) values.
Serologic testing for anti–SARS-CoV-2 antibodies
Serum samples from the patient and COVID-19 convalescent plasma (CP) donors were evaluated for qualitative anti–SARS-CoV-2 IgG against the viral nucleocapsid protein by using the EDI Novel Coronavirus COVID-19 IgG ELISA Kit (Epitope Diagnostics Inc, San Diego, Calif), which was clinically validated and performed at Mayo Clinic Laboratories. Samples with an antibody index value less than 1.01 were considered negative, those with a value of 1.01 to less than 1.21 were considered indeterminate, and those with values of 1.21 or higher were considered positive.
Because of the limited data available on performance of commercial SARS-CoV-2 antibody tests, frozen aliquots of matched serum samples from the patient were evaluated later for anti–SARS-CoV-2 antibodies by using the Emergency Use Authorization-authorized Abbott Architect SARS-CoV-2 IgG chemiluminescent microparticle immunoassay, which targets the viral nucleocapsid protein and is validated for clinical use at Nationwide Children’s Hospital per the manufacturer’s instructions. Samples were tested and interpreted per the manufacturer’s recommendations, with the signal-to-calibrator index value of 1.4 or more considered positive, whereas any value less than 1.4 was considered negative.
Serum IL-6 levels
Serum IL-6 was assessed at multiple time points by using the ProQuantum human IL-6 assay (ThermoFisher). Briefly, the ProQuantum assay uses 2 oligonucleotide labels, which bind a pair of antibodies targeted against IL-6. PCR amplification utilizes the proximity ligation assay principle, which measures the amount of fluorescent signal (which is proportional to the amount of DNA produced after each amplification cycle). The cytokine level was quantitated by using a recombinant IL-6 cytokine standard curve using the fluorescence threshold data at multiple time points.
NF-κB pathway protein expression studies
PBMCs isolated from sodium heparin–anticoagulated blood were stimulated with or without anti-CD3 antibody at 1 μg/mL (eBioscience, catalog no. 16-0037085) for 48 hours. Cell lysates were prepared and immunoblotted with anti–phospho-NF-κB2 (Cell Signaling Technology, catalog no. 164810, S866/870), anti–NF-κB2 (Cell Signaling Technology, catalog no. 1637359), and anti–phospho-NF-κBp65 (Cell Signaling Technology, catalog no. 163033, S536). T-cell blasts were generated from PBMCs by stimulation with anti-CD3 and anti-CD28 (1 μg/mL) in the presence of IL-2 (10 ng/mL) for 8 days. IL-2 was added every 2 or 3 days. Cell lysates were prepared and analyzed for full-length p100 and processed form, p52 of NF-κB2. Immunoblotting of β-actin (Cell Signaling Technology, catalog no. 164970) was used as a loading control. Images were acquired and analyzed with Image Studio software (LI-COR Biosciences, Lincoln, Neb). The data are shown as band intensities normalized to the β-actin control by densitometry. The control data are represented as means plus or minus SEMs from 3 healthy controls.
Results
Clinical history relevant to NF-κB2 LOF
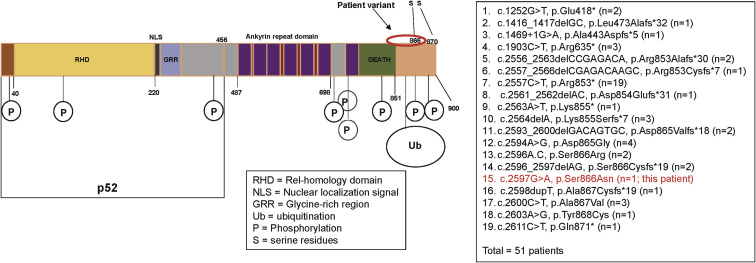
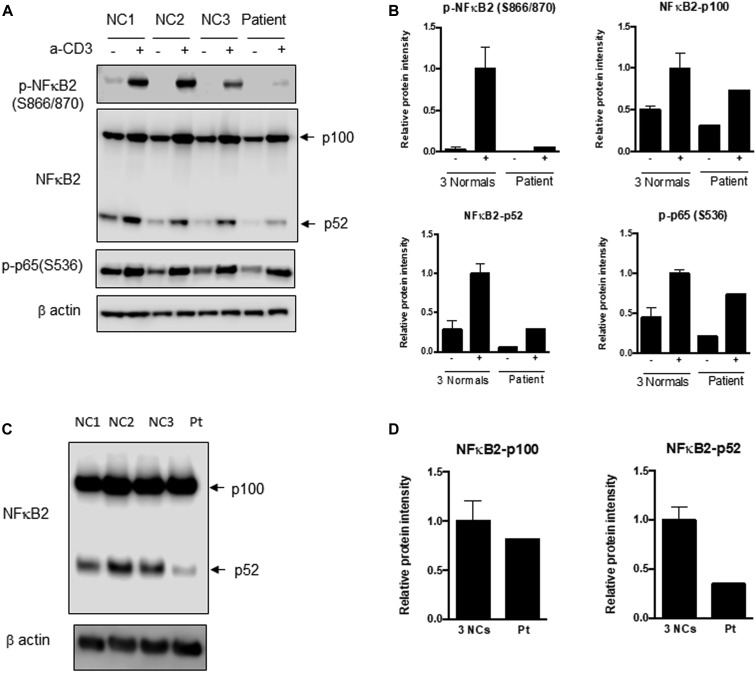
The male patient, who is currently 18 years old, initially presented at 8 years of age with recurrent upper respiratory infections, pneumonia, and recurrent orolabial herpes simplex, confirmed by PCR to be herpes simplex 1. Immunologic evaluation (Table I ) at that time revealed a primary hypogammaglobulinemia with an IgG level of 208 mg/dL and normal serum albumin level. B cells were present but at a low level, and the levels of total and switched memory B cells were markedly decreased. The patient mounted partial humoral immune responses to protein and polysaccharide vaccines. He did not have further clinical evidence of a cellular immune defect or endocrine dysfunction as reported in patients with DAVID syndrome. He developed multiple nevi, nail dystrophy, and a skin rash suggestive of cutaneous psoriasis, as well as alopecia areata that later progressed to alopecia totalis. His working diagnosis at the time was CVID with autoimmunity, and subcutaneous immunoglobulin replacement therapy every 2 weeks was initiated, resulting in clinical improvement of the severity and frequency of infections. When he was 16 years old, targeted next-generation sequencing performed with a 207-gene panel for inborn errors of immunity identified a variant of uncertain significance in the NFKB2 gene. The missense variant was in exon 23 of the NFKB2 gene, c. 2597G>A, p.Ser866Asn (NM_001077494.3, hg38). This Ser866Asn variant is in the extreme C-terminal domain of NFkB2 (p100) molecule, whereas the p52 cleaved protein consists mainly of the Rel-homology domain and the glycine-rich region but is truncated before the ankyrin repeat domain (Fig 1 ). It is a highly conserved nucleotide, but the Grantham distance is small (46 [range 0-215], suggesting insignificant physicochemical differences between serine and asparagine. It is not represented in the large population databases (1000 Genomes, gnomAD, and ESP), although it has been documented as a variant of uncertain significance previously in ClinVar (RCV000812720.1; this patient). In silico prediction tools such as SIFT, MutationTaster and PolyPhen2 categorize this variant as deleterious, disease-causing, and probably damaging, respectively. The patient’s parents were also tested, and his mother was found to be a germline mosaic for the NFKB2 variant. To determine the effect of this novel variant in the NFΚB2 gene on NF-κB2 processing subsequent to activation, immunoblotting was performed to assess the NF-κB pathway, as well as to determine whether the NF-κB2 protein was adequately phosphorylated at serines 866 and 870 after stimulation (Fig 2 , A-D). Visual assessment and densitometry revealed that the patient had obviously decreased levels of phospho-NF-κB2 after stimulation compared with the average levels in healthy controls after stimulation. Because NF-κB2 is a key component of the noncanonical pathway of NF-κB, the level of the active form, p52 molecule, derived from the precursor p100 protein was also assessed and found to be markedly decreased after stimulation compared with the controls in PBMCs and T-cell blasts (Fig 2, A and B and 2, C and D, respectively). The magnitude of decrease in phospho-NF-κB2 and p52 is more suggestive of a dominant negative effect for this variant rather than true haploinsufficiency (Fig 2, A-D). The quantitative data from densitometry suggest that despite the presence of a faint band for NF-κB2 on the Western blot, the amount of phosphorylated protein in the patient is less than 5% of the amount in healthy controls and similar to that observed in the unstimulated controls (Fig 2, A-D). This would contradict the expectation of a haploinsufficiency model, which would suggest that approximately 50% of the protein should be present. The same finding is also true for p52 protein level, which represents the sum of expression from the wild-type and mutant alleles, and this too is less than the expected half the normal protein level needed for haploinsufficiency (Fig 2, A-D). The noncanonical NF-κB pathway has been thought to be dependent on RelB interactions, whereas RelA has traditionally been regarded as essential to the canonical pathway. Transcription-activating heterodimers are typically formed between RelB and p52 of NF-κB2 and p50 and RelA (p65) for the NF-κB1 canonical pathway; however, other combinations of homodimers (p50 and p52) as well as p52-RelA and p50-RelB heterodimers have been reported, although these appear to have very specific functions.28 Other studies have also shown that there is cross talk between the canonical and noncanonical pathways, indicating that they are not mutually exclusive.29 The defective phosphorylation of NF-κB2 with decreased processing of p52 along with the patient's clinical phenotype indicate that this novel variant is indeed pathogenic and affects NF-κB signaling; however, the effects appear to be largely restricted to the B-cell compartment, with no overt evidence of a combined immunodeficiency or cellular immune dysfunction.
Table I.
Immunologic features of patient with NFκB2 LOF
| Patient age (y) | Data | Patient result | Reference interval or control result |
|---|---|---|---|
| 8 (at diagnosis) | IgG level (mg/dL) (L) | 208 mg/dL | 537-1,432 |
| IgA level (mg/dL) (L) | 10 mg/dL | 54-219 | |
| IgM level (mg/dL) (L) | 24 mg/dL | 26-112 | |
| Proportion of switched memory B cells (%) | 0.2 | Median: 20 | |
| Tetanus titers before/after DTaP booster (IU/mL) | <0.1/0.7 | ≥0.1 | |
| Diphtheria titers before/after DTaP booster (IU/mL) | <0.1/<0.1 | ≥0.1 | |
| Baseline pneumococcal antibody levels (μg/mL) | For 15 of 23 serotypes, ≥1.3 | For each serotype, ≥1.3 | |
| Pneumococcal 12 serotype avidity panel (AI units) | for 6 of 12, <0.3; for all <1.0 | 0.3-4.0 (reportable range) | |
| CD45+ ALC (cells/μL) | 2,091 | 1,500-6,800 | |
| CD3+ T cells (cells/μL) | 1,514 | 1,200-2,600 | |
| CD4+ T cells (cells/μL) | 1,222 | 650-1,500 | |
| CD8+ T cells (cells/μL) (L) | 259 | 370-1,100 | |
| CD19+ B cells (cells/μL) | 349 | 270-860 | |
| CD4/CD8 ratio | 4.7 | >0.9 | |
| 8 (at diagnosis) | Lymphocyte prolifer ation to mitogens∗ | Unstimulated: 121 PHA: 291,693 ConA: 350,462 PWM: 156,686 |
0-286 cpm ≥135,190 cpm ≥73,522 cpm ≥26,677 cpm |
| Lymphocyte proliferation to antigens∗ | Unstimulated: 111 Candida: 2,606 (low) Tetanus: 207 (low) |
≥15,289 cpm ≥4,761 cpm |
|
| 17 (after COVID-19 infection; HD 42 from onset of illness) | Lymphocyte proliferation to mitogens† | % CD3+ T cells to PHA: 84.2% % CD3+ T cells to PWM: 18.6% |
≥58.5% ≥3.5% |
| Lymphocyte proliferation to anti-CD3 panel† | % CD3+ T cell to soluble anti-CD3: 45.9% % CD3+ T cell to soluble anti-CD3+ anti-CD28: 66.9% % CD3+ T cell to soluble anti-CD3+ IL-2: 59.7% |
≥20.3% ≥44.6% ≥46.2% |
AI, Avidity index; ALC, absolute lymphocyte count; ConA, concanavalin A; cpm, counts per minute; DTaP, diphtheria and tetanus toxoids and acellular pertussis; HD, healthy donor; L, low; PWM, pokeweek mitogen.
Expressed as cpm of tritiated thymidine.
Expressed as percentage of proliferating CD3+ T cells relative to total CD3+ T cells.
Fig 1.
Schematic drawing of the NF-κB2 protein. This represents the protein domains of NF-κB2 for the full-length precursor and the processed p52 protein. The known NF-κB2 variants are listed along with this patient’s variant. A total of 50 patients with 18 variants have been described in the literature.5 This patient’s variant is p.Ser866Asn in the C-terminal region of the protein.
Fig 2.
Immunoblotting for NF-κB2, p52, and phosphorylated p65 (RelA). A, PBMCs from the patient (Pt) and healthy controls 1 to 3 (NC 1, NC 2, and NC3) were stimulated with soluble anti-CD3. Immunoblotting was performed for NF-κB2 precursor (p100) and the processed form (p52), phosphorylated NF-κB2, and phosphorylated p65 (RelA) of the canonical pathway in both stimulated and unstimulated samples. There is a distinct inability to phosphorylate NF-κB2, and the amount of processed p52 protein is substantially reduced in the patient sample following stimulation. B, Densitometric analysis of the immunoblot in (A). C, T-cell blasts were generated from PBMCs by stimulation with anti-CD3 and anti-CD28 with IL-2. Blast cell lysates were analyzed for p100 and p52 of NF-κB2. D, Bar graphs depict the relative expression levels of these proteins normalized to β-actin by densitometry. Compared with in normal controls, there is a small decrease in p100 but a remarkable reduction in the processed form, p52 protein, which correlates with the PBMC data. The extent of decrease in phospho-NF-κB2 and p52 is supportive of a complete deficiency for this variant.
Clinical history relevant to SARS-CoV-2 infection
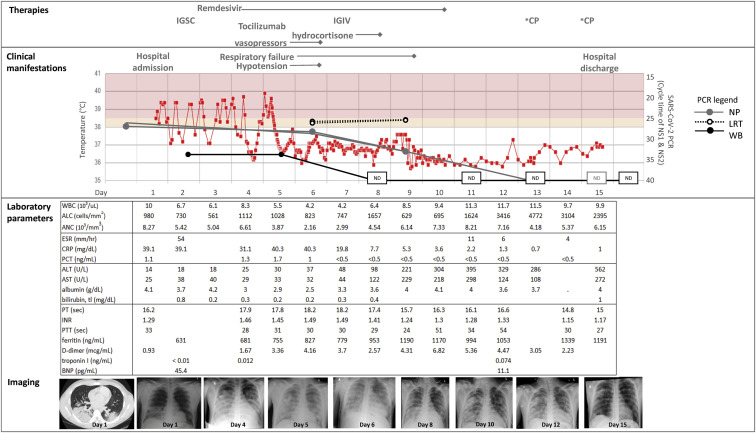
At age 17 years, the patient contacted his primary care provider 4 days before admission (day –4) with a 1-week history of nasal congestion and cough; he was prescribed amoxicillin for presumed sinusitis. The following day, he developed fever and anosmia. One day before admission, he was evaluated in the emergency department, where he underwent viral testing for SARS-CoV-2; however, the test result was not immediately available and he was discharged from the emergency department. He returned the following day with worsening fever and respiratory distress, by which time the SARS-CoV-2 PCR result was positive. He was hospitalized because of tachypnea, tachycardia, and hypoxemia, with an oxygen saturation level of 95% mm Hg. His laboratory test and microbiologic diagnostic results (Fig 3 ) were notable for lymphopenia (an absolute lymphocyte count of 980 cells/mm3), hyponatremia (131 mmol/L), mildly elevated d-dimer and ferritin levels, and elevated C-reactive protein and procalcitonin levels. The initial chest radiograph revealed a normal heart size but bilateral lower lobe ground glass opacities without effusion, for which antibiotics were initiated. He had persistent fever with tachypnea, dyspnea, chest pain, and hypoxemia requiring oxygen supplementation via nasal cannula. A computed tomography scan of the chest revealed consolidative pneumonia in the lung bases, scattered nodular densities with ground glass characteristics, and reactive mediastinal lymph nodes. On the basis of the computed tomography scan, an empiric course of antibiotics and antifungals was given to treat a possible opportunistic infection. An ACTH stimulation test performed on hospital day 2 (day 2) showed normal adrenal function. Because the patient missed his maintenance dose of replacement immunoglobulin (subcutaneous Ig) on account of his acute illness, he received a dose on day 2, which was approximately 2.5 weeks from his last maintenance dose. The patient was not eligible for national clinical trials of the antiviral drug remdesivir because of his age (<18 years), but a patient emergency investigational new drug application was approved by the FDA and treatment (200 mg intravenously once, then 100 mg intravenously every 24 hours) was initiated on day 3. On day 4, the patient’s respiratory distress, hypoxemia, lymphopenia, and inflammatory markers continued to worsen, and he was intubated and transferred to the pediatric intensive care service. He developed premature ventricular contractions, intermittent bigeminy, and nonspecific ST and T wave abnormalities, with normal cardiac enzyme levels and no evidence of myocarditis on echocardiogram. On day 5, because of concern for progression to a cytokine release syndrome, including ongoing fevers, new hypotension requiring inotropic support and stress-dose hydrocortisone, and increasing levels of inflammatory markers (C-reactive protein [CRP], IL-6, and d-dimer), a single dose of the IL-6 receptor antagonist tocilizumab (8 mg/kg) was administered. With these interventions, the patient's fever resolved and his CRP and procalcitonin levels began to improve. The patient received a dose (40 g) of intravenous immunoglobulin on day 7. Thanks to stabilization of his blood pressure, inotropic support was discontinued on day 6, and improvement in his respiratory status led to extubation on day 11.
Fig 3.
Clinical course of SARS-CoV-2 infection. Key clinical, laboratory, and radiologic parameters are depicted. ALC, absolute lymphocyte count; ALT, alanine transaminase; ANC, absolute neutrophile count; AST, aspartate transaminase; BNP, brain natiuretic peptide; ESR, erythrocyte sedimentation rate; IGIV, intravenous IgG; IGSC, subcutaneous IgG; INR, international normalized ratio; NP, natiuretic peptide; PT, prothrombin time; PTT, partial thromboplastin time; WB, whole blood; WBC, whole blood cell.
On day 8, the patient developed a transaminitis of unclear etiology. Doppler ultrasound demonstrated patent hepatic veins with no evidence of thrombi but abnormally decreased phasicity of flow, suggesting venous outflow resistance. His liver enzyme levels continued to rise; therefore, no more than the single dose of tocilizumab was given, and remdesivir was discontinued after the eighth dose of therapy (day 10). The patient's liver enzyme levels peaked on day 16 after hospitalization (alanine transaminase level 622 U/L) and improved thereafter. He was discharged from the hospital on day 15.
At the time of intubation on day 4, efforts were initiated to obtain COVID-19–positive CP because of the patient’s known humoral immunodeficiency and the absence of anti–SARS-COV-2 antibodies in contemporary commercial immunoglobulin preparations used for his maintenance treatments. Because of his age, he was ineligible to participate in national adult clinical trials assessing the safety and efficacy of COVID-19 CP.30 A second emergency investigational new drug was FDA-approved for directed-donor donation, and several eligible local donors were identified. The patient received 5 doses (2 U/dose) of CP in total, which were administered every other day starting on day 13, with 3 doses administered after discharge even though the patient was improving clinically before CP administration, suggesting that the overall clinical management was effective even without CP. At more than 60 days since his discharge, the patient is doing well without symptom recrudescence.
SARS-CoV-2 RNA detection
The PCR results from the different specimen types at each time point are shown in Table II . The nasopharyngeal swab was positive for SARS-CoV-2 on the day before hospitalization (day –1). Viral RNA was also detectable from LRT specimens (day 6) and whole blood samples (days 2 and 5). Over the course of remdesivir treatment, his SARS-CoV-2 PCR cycle threshold (Ct) values increased (indicating decreased viral burden) according to his nasopharyngeal and whole blood specimens but did not change across 2 LRT specimens (Fig 3). SARS-CoV-2 RNA was no longer detected from the patient's blood beginning on day 11. The PCR results from his nasopharyngeal samples on days 13 to 18 were negative, but as discussed in the clinical history, the PCR result was again positive on day 20, with Ct values for nucleocapsid targets of at least 35, suggesting low-level nonviable viral RNA present in the specimens. The nasopharyngeal PCR subsequently became inconclusive (1 of 2 targets detected) on day 22 and thereafter became repeatedly negative (days 28 and 35), indicating RNA clearance from the upper respiratory tract.
Table II.
SARS-CoV-2 RNA detection and SARS-CoV-2 IgG antibody testing in patient specimens and convalescent plasma
| Collection date∗ | SARS-CoV-2 PCR |
SARS-CoV-2 IgG |
||||||
|---|---|---|---|---|---|---|---|---|
| Patient (serum samples) |
CP donors (plasma samples) |
|||||||
| PCR specimen type | Target N (Ct value)† | Target N1 (Ct value) | Target N2 (Ct value) | Result | Send-out laboratory (EDI) IgG antibody index | In-house (Abbott Architect) IgG antibody index | Send-out laboratory (EDI) IgG antibody index | |
| Day –1 (day before hospitalization) | NP | 26.99 | 26.02 | DET | ||||
| Day 2 | Blood† | 33.66 | DET | |||||
| Day 4 | Serum | <1.01 | 0.69 | |||||
| Day 5 | Blood† | 33.73 | ||||||
| Day 6 | LRT | 25.63 | 25.67 | DET | ||||
| Day 6 | NP | 28.18 | 28.61 | DET | ||||
| Day 7 | ||||||||
| Day 8 | Blood† | ND | ND | |||||
| Day 9 | LRT | 25.27 | 25.38 | DET | ||||
| Day 9 | NP | 32.98 | 32.98 | DET | ||||
| Day 10 | ||||||||
| Day 11 | NP, blood† | ND | ND | 3.24 | 7.58 | |||
| Day 13 | NP | ND | ND | ND | 3.27 | 8.13 | 2.9 (CP donor 1)‡ | |
| Day 13 | Blood† | ND | ND | |||||
| Day 15 (day of discharge) | NP | ND | ND | ND | 2.72 | 7.68 | 2.8 (CP donor 2) | |
| Day 16 | Blood | ND | ND | ND | ||||
| Day 16 | NP | ND | ND | ND | ||||
| Day 18 | NP | ND | ND | ND | 1.49 (CP donor 3) | |||
| Day 18 | Blood | ND | ND | ND | ||||
| Day 20 | NP | 35.68 | 35.15 | DET | 1.51 (CP donor 4) | |||
| Day 20 | Blood† | ND | ND | 2.85 | 6.86 | |||
| Day 22 | NP | ND | 39.6 | INC | 2.69 | 6.83 | 3.84 (CP donor 5) | |
| Day 28 | NP | ND | ND | ND | ||||
| Day 35 | NP | ND | ND | ND | ||||
DET, Detected; EDI, Epitope Diagnostics Inc; INC, inconclusive; ND, not detected; NP, natiuretic peptide.
The dates are indicated as days starting with hospital admission (day 1).
Whole blood PCR performed with research SARS-CoV-2 PCR assay.
The CP donor antibody test results are shown for the day of infusion.
Flow cytometric immunophenotyping
Quantitative analysis for lymphocyte subsets (T, B, and NK cells) was performed at 4 different time points. The first blood sample was collected 10 months before the patient's SARS-CoV-2 infection and served as a baseline for comparison with samples collected and examined during his COVID-19 illness. During the COVID-19 infection, the relative frequency (percentage) of various lymphocyte subsets demonstrated increased distribution of CD4+ T cells and a decreased proportion of NK cells (Table III ). However, the absolute quantitation (cells/μL) was remarkable in that there was pan-lymphopenia observed during the patient's hospitalization for SARS-CoV-2 at day 4 (Table III). By day 15, his CD45+ lymphocyte and CD3+, CD4+, and CD8+ T-cell counts rebounded to normal levels. The CD4/CD8 ratio was relatively skewed only in the acute infection sample (taken on day 4). He had documented B-cell lymphopenia at 10 months before his hospitalization for COVID-19 (Table III). During and after his hospitalization, his B-cell count further decreased compared with his preinfection assessment result (Table III). His levels of cytotoxic NK cells (CD16++CD56+) were also decreased, and although they plummeted at the time of the infection, they rebounded to baseline levels after infection.
Table III.
Immunophenotyping analysis
| Cellular subset | 10 Months prior | Day 4 | Day 15 | Day 20 | Day35 | Control data, mean; 5%-95% CI |
|---|---|---|---|---|---|---|
| CD45+ ALC | 2,741 | 407 | 2,394 | 2,291 | 1,732; 971-2,497 | |
| CD3+ T cells (%) | 89.1 | 87.3 | 90.3 | 92 | 75.57; 61.28-84.29 | |
| CD3+ T cells (cells/μL) | 2,443 | 355 | 2,160 | 2,107 | 1,308; 677-1,936 | |
| CD4+ T cells (%) | 68.8 | 73.9 | 66.8 | 56.4 | 48; 36-60 | |
| CD4+ T cells (cells/μL) | 1,887 | 301 | 1,600 | 1,293 | 818; 471-1,194 | |
| CD8+ T cells (%) | 18.8 | 12.2 | 22 | 34.2 | 24; 16-32 | |
| CD8+ T cells (cells/μL) | 515 | 50 | 526 | 783 | 416; 181-715 | |
| CD4–CD8– DNT cells (%) | 1.4 | 1 | 1 | 1.1 | 3; 1-9 | |
| CD19+ B cells (%) | 5.3 | 10.8 | 2.7 | 3.2 | 10.96; 5.49-21.23 | |
| CD19+ B cells (cells/μL) | 145 | 44 | 64 | 72 | 192; 67-392 | |
| CD16/56+ NK cells (%) | 1.7 | 1 | 2.6 | 2.5 | 9.70; 3.18-20.14 | |
| CD56/16+ NK cells (cells/μL) | 47 | 4 | 63 | 58 | 166; 49-376 | |
| CD4/CD8 ratio | 3.7 | 6 | 3 | 1.7 | 2.17; 1.16-3.59 | |
| Naive B cells (% of CD19+ B cells) | 81.77 | 72.9 | 71.0; 56.0-92.0 | |||
| CD19+CD27+ memory B cells + plasmablasts (% CD19+ B cells) | 3.83 | 1.87 | 30.0; 11.0-49.0 | |||
| Marginal zone B cells (% CD19+ B cells) | 1.62 | 0.37 | 9.9; 1.8-21.0 | |||
| Switched memory B cells (% CD19+ B cells) | 0.18 | 0.75 | 23.0; 6.8-53.0 |
| Control data (2 independent experiments) | ||||||
|---|---|---|---|---|---|---|
| FOXP3+ (% CD4+) | 4.23 | 4.41 | 6.2 | 7.11; 10.95 | ||
| CD25hi (% Treg cells) | 1.26 | 1.62 | 3.55 | 4.67; 5.18 | ||
| CD25+CD127low/– (% Treg cells) | 4.46 | 4.14 | 6.95 | 6.20; 9.06 | ||
| FOXP3+CD127 low/– (% Treg cells) | 3.68 | 3.53 | 5.53 | 6.56; 10.52 | ||
| FOXP3+CD25+ (% Treg cells) | 3.48 | 3.43 | 5.9 | 6.51; 9.89 | ||
| FOXP3 MFI (from CD25+CD127 low/–) | 3,871 | 4,835 | 5,965 | 4,706; 10,719 | ||
| CD25 MFI (from FOXP3+) | 2,023 | 2,204 | 4,861 | 2,296; 3,529 | ||
| CD127 MFI (from FOXP3+) | 5,439 | 6,751 | 5,405 | 6,069; 5,234 |
ALC, Absolute lymphocyte count; DNT, double-negative T; NK, natural killer.
The dates of assessment are provided relative to day 1 of hospitalization.
Detailed T-cell subset phenotyping results for naive, memory, activated, exhausted, and senescent T cells were unremarkable (data not shown) and did not show any substantive differences from those of healthy controls, with the exception of the acute T-cell lymphopenia related to his infection and hospitalization, from which he recovered. Similar findings in certain T-cell subsets have also been described previously, although there is variability in the patient cohorts and time points studied. Assessment of T-cell function 5 weeks after hospitalization showed normal T-cell proliferation to mitogens, antigens, and stimulation with anti-CD3 (soluble) alone or with other costimulants (anti-CD28 and IL-2), further confirming a lack of a cellular immune defect in T cells, as has been described in some other patients with NF-κB2 variants (Table I).
B-cell subset phenotyping, on the other hand, showed a persistent decrease in total memory and particularly in switched memory B cells, consistent with his underlying humoral immunodeficiency related to the heterozygous NFΚB2 C-terminus LOF variant (Table III). The marginal zone B cells (CD19+CD27+IgM+IgD+) also showed an almost 4-fold decrease from before infection to during infection. The decrease in level of switched memory B cells also correlates with the patient's hypogammaglobulinemia, for which he was receiving treatment with replacement immunoglobulin therapy. Although there was a decline in total CD19+CD20+ B-cell count during the early stages of acute infection, his B-cell count did not rebound after recovery from infection, suggesting an intrinsic defect in the B-cell compartment, contrasting with his T-cell count, which improved rapidly following recovery from infection.
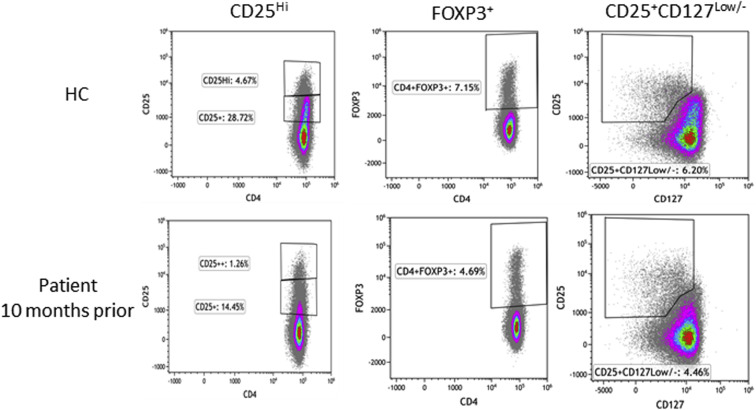
Treg cell subsets were quantified in PBMCs from 3 time points: baseline, 10 months before infection, day 4 of COVID-19 hospitalization, and after recovery from COVID-19 infection (day 18) (Table III and see Fig E1 [in this article's Online Repository at www.jacionline.org]). The patient had a decreased frequency (percentage) of CD25high CD4+ T cells compared with experimental healthy controls at baseline and during acute infection, although it was modestly improved after clinical recovery. Similar findings were observed for FOXP3+ Treg cells and CD25+CD127low/– Treg cells, with the postrecovery sample showing the highest proportion of these subsets compared with the baseline or acute infection samples, with normalization at posthospital day 20 for the CD25+CD127low/– Treg cells. Analysis of the median fluorescence intensity for FOXP3, CD25, and CD127 in the CD25+CD127low/– and FOXP3+ subsets respectively revealed comparable expression relative to that in healthy controls (Table III). This result suggests that there is no impairment in expression of FOXP3, CD25, or CD127 proteins on Treg subsets; however, the frequency (percentage) of these cell populations is reduced, although there has been quantitative recovery in the posthospitalization sample.
Fig E1.
Treg cell subset quantitation. The gating strategy for Treg quantitation is depicted for a representative healthy control (HC) and the patient. From CD4+ T cells, the frequency (percentage) of CD25high T cells (CD25hi), FOXP3+ Treg cells, and CD25+CD127low/– Treg cells were gated by using CD25, FOXP3, and CD127.
IL-6 measurement
Serum was collected and analyzed for IL-6 concentrations on days 2, 4, 5, 10, 18, and 22 after diagnosis and hospitalization with SARS-CoV-2 infection and compared with a preinfection/prehospitalization sample (10 months prior); the comparison showed significant increases over baseline and compared with samples from healthy controls at days 2, 4, and 5 (IL-6 levels of 117.5, 146, and 68.4 pg/mL, respectively) during active infection. The patient received a single dose (8 mg/kg) of a recombinant humanized mAb against the IL-6R, tocilizumab, on day 4. A posttocilizumab sample collected on day 10, which was within the half-life of the drug, showed an apparently paradoxic increase (to 612.5 pg/mL) in the level of serum IL-6. The IL-6 level gradually declined over the course of the next 2 weeks (day 8 and day 2 levels of 96.5 and 49.6 pg/mL respectively) but was still approximately 4 times higher than the healthy control data measured during that period. The early measurement of serum IL-6 affected the timing of administration of tocilizumab and correlated with other measures of inflammation (Fig 3). The decline in IL-6 levels after the initial spike from administration of tocilizumab correlated with a recent report on better prognosis in patients with COVID-19 with lower IL-6 levels after treatment.31
Assessment of antibody levels to SARS-CoV-2
The patient was discharged from the hospital 2 weeks after initial admission for SARS-Co-V-2 infection. The rationale for administration of CP was to mitigate the severity of his acute and critical illness as well as to provide passive immunity against viral recrudescence or reinfection given his underlying humoral immunodeficiency and the presumed lack of anti–SARS-CoV-2 antibodies in contemporary replacement immunoglobulin products. CP donors were identified, and the patient received 5 doses of CP starting at day 3. Assessment for SARS-CoV-2 antibodies was performed by using a clinically validated qualitative IgG antibody test at multiple time points, including days 4, 11, 13, and 15 during hospitalization and posthospital days 20 and 22 after discharge. Although the sample from day 4 showed no SARS-CoV-2 IgG antibodies, serologic evidence of antibodies was observed starting at day 11 before receipt of CP as well as after administration of CP (Table II). The positive antibody detection by 2 independent tests reduces the possibility of false-positive detection. The IgG antibody levels to SARS-CoV-2 were monitored through posthospital day 22.
Discussion
With the recent global outbreak of SARS-CoV-2, the concern for high-risk patients, such as those with underlying immune defects or other comorbidities, has been high. The spectrum of immunologic anomalies associated with NFκB2 gene defects is varied and can manifest primarily as a humoral immunodeficiency (CVID-like) or a combined immunodeficiency, affecting both T- and B-cell compartments.5 Some of the NF-κB2–deficient patients with the CVID-like phenotype also had endocrine defects with ACTH deficiency and nail dystrophy, described as DAVID syndrome.32 NFΚB2 gene defects can present with LOF in an autosomal dominant manner because of a nonprocessed form of mutant p100 leading to p52 dominant negative or haploinsufficient states, and these appear to be highly penetrant, although there is variable expressivity.4 , 23 , 33 , 34 Most of the LOF variants have been described in the C-terminal region and affect either the serine phosphorylation sites 866 and 870 or the ubiquitination site at amino acid 855. All of these variants affect the p100 precursor protein, creating a molecule that cannot be processed and thus reducing p52 nuclear translocation and heterodimer formation, with RelB. In contrast to the LOF variants, GOF NF-κB variants are largely clustered in the glycine-rich domain (Fig 1) or the ankyrin repeat domain. The GOF variants are likely to show incomplete penetrance with variable expressivity. As can be expected, heterozygous GOF variants result in constitutive activation with increased nuclear translocation of p52/RelB heterodimers and also in increased canonical NF-κB pathway activity, illustrating that NF-κB signaling is not confined to either the canonical or noncanonical pathway but is instead an integrated and interactive signaling mechanism.
NF-κB2 protein expression and its phosphorylation following activation (Fig 2) was used to establish that the novel LOF variant was indeed likely to have an impact on NF-κB signaling through both the noncanonical and canonical pathways owing to a significant decrease in phospho-NF-κB2 and generation of the processed form, p52. This finding is consistent with other variants reported at position 866, which as previously mentioned is a critical serine phosphorylation site. This patient had all the clinical features associated with LOF variants (Table I) with the exception of endocrinopathy (central adrenal insufficiency) associated with DAVID syndrome, and he had normal adrenal function documented during the acute illness. Despite onset in childhood, life-threatening infections are rarely reported. This was exemplified by the clinical history of this patient up to this point (ie, in his late teenage years), when he exhibited a life-threatening infection, in contrast to most teenagers who contract SARS-CoV-2 infection and remain either asymptomatic or only mildly symptomatic. He, however, has alopecia and nail dystrophy consistent with features seen in other patients. Although he did have a history of 1 episode of H1N1 pneumonia, he did not experience any opportunistic infections such as Pneumocystis jiroveci pneumonia.
This patient showed evidence of severe COVID-19 disease, as discussed previously, and his therapeutic regimen included compassionate use remdesivir,35 intensive cardiorespiratory support, tocilizumab, and CP (Fig 3). During the critical phase of illness, a lymphocyte subset quantitation (Table III) was performed; it revealed stark pan-lymphopenia, which has been reported in other patients with COVID-19 as well.18 , 36 When compared specifically with the patient's pre–COVID-19 immunophenotyping result, this result indicated that his lymphopenia was COVID-19–related rather than due to his underlying immunodeficiency. The post–hospital discharge rebound of the patient's T-cell counts to the pre–COVID-19 baseline from the hospital provided further evidence of this (Table III). Further immunophenotyping of T- and B-cell subsets was unrevealing, in contrast to the findings in certain other reports,36 although not all of the markers used were identical between this study and the published reports. Also, there may be differences in various immune cell populations, depending on the timing of analysis from onset of infection, age of patient, clinical status, and other factors. However, the numbers of switched memory B cells were decreased, as one would expect to find in patients carrying heterozygous NFΚB2 C-terminus LOF variants (Table III). Analysis of the patient's Treg cells revealed no intrinsic defects in Treg subset numbers compared with in the controls, especially after recovery (Table III and see Fig E1).
With evidence of respiratory failure in conjunction with CVID, it was imperative to determine whether biologicals should be used to control the cytokine storm reported in patients with COVID-19.13 , 37 , 38 IL-6 level was measured at several time points both during and following hospitalization with COVID-19, and it was elevated early on, which was informative from a clinical management standpoint. Data from other centers have variable cutoffs that define elevated IL-6 and trigger the use of biologic therapy. There are data to suggest that an IL-6 level of 80 pg/mL is associated with respiratory failure and more severe disease.39 Some centers have used lower cutoffs in protocols or clinical trials, whereas other centers do not have a fixed cutoff for IL-6 level at which blocking therapy is initiated. Tocilizumab has been used empirically in several centers to treat the hyperinflammatory response in COVID-19.40 , 41 Some centers assess serum IL-6 levels to determine timing of drug administration, whereas others use inflammatory markers, such as CRP and ferritin to determine the indication for tocilizumab. A study using tocilizumab reported clinical improvement in patients with COVID-19 who are receiving the drug.41 Although analogies can be drawn between the cytokine storm seen in COVID-19 and cytokine release syndrome in patients following chimeric antigen receptor T-cell therapy,42 whether such analogies are viable or valid, especially in intrinsically immunocompromised patients, is unclear. The decision to treat with biologic therapy should be decided not solely by a single biomarker but by several biomarkers of inflammation along with clinical context. In patients with COVID-19, IL-6 has been shown to be a particularly useful biomarker, in correlation with other biomarkers in predicting severe disease and reduced cytotoxicity, and thus it represents a useful therapeutic target with drugs, such as tocilizumab.25 , 43 In this patient, the IL-6 levels peaked at day 4 but began trending down (although this was not known before analysis) before the first and only dose of tocilizumab was administered (8 mg/kg). The half-life of the drug at this dose is approximately 13 days, and therefore, additional samples were collected after administration of tocilizumab at 3 different time points. A notable but not unusual finding was that IL-6 level was increased after tocilizumab was administered. This finding has been reported previously in patients with rheumatoid arthritis and Castleman disease, in which settings elevated IL-6 levels persisted for several days and has been attributed to saturation of the IL-6 receptors by the drug.44 In our patient, the highest IL-6 level was seen immediately after administration of tocilizumab, with levels gradually decreasing over the course of 2 weeks, although at that time point it had not returned to baseline (either before COVID-19 for the patient or comparable to the level in healthy controls). Tocilizumab can bind either the membrane-bound or soluble form of the IL-6 receptor and prevent IL-6/IL-6R from dimerizing with membrane-bound gp130, inhibiting downstream signaling. Because IL-6 can bind either membrane-bound IL-6R inducing “classic” signaling (which is primarily anti-inflammatory) or soluble IL-6R (thereby forming a complex with gp130 and mediating proinflammatory trans-signaling),45 it seems unlikely that IL-6 is competing with tocilizumab, which binds with high affinity to the membrane-bound or soluble form of the receptor, to generate circulating immune complexes. In fact, the levels of IL-6 after tocilizumab appear to be prognostically informative in COVID-19, and they may actually reflect the level of IL-6 production.31 In addition, in vitro studies in our laboratory with “spiked” IL-6 levels in healthy control serum samples to which tocilizumab has been added does not show any increase in IL-6 levels, indicating that the origin of the IL-6 is endogenous. Therefore, serum IL-6 levels after tocilizumab administration should be used for prognostication and for determining the necessity for additional dosing of tocilizumab, especially after the half-life of the drug has been reached. However, it would be reasonable to suggest that the use of additional doses of tocilizumab be based on multiple factors rather than on serum IL-6 levels alone. It is critical to note that when serum or plasma cytokine measurements are performed the results are often not comparable across immunoassay platforms, and thus, all serial monitoring of cytokines should be performed within the same laboratory and on the same platform and should account for the presence of heterophile antibodies, which might cause spurious elevation of cytokines, especially in multiplex assays.46 , 47
The use of CP has been described in a few studies of treatment of severe disease in patients with COVID-19.48, 49, 50 Because this patient had a humoral immunodeficiency and was receiving immunoglobulin replacement therapy, it seemed logical to use SARS-CoV-2 CP to provide passive therapeutic support to help clear the virus; therefore, he was given 5 doses of CP (Table II). A recent study of anti–SARS-CoV-2 antibodies in 149 convalescent patients revealed that very few of these individuals had high levels of neutralizing antibodies, but the presence of antibodies directed against the receptor-binding domain of the virus was present in many of these patients with antiviral activity, albeit at low levels, suggesting that strategies to increase the concentrations of such antibodies would be effective in design of a vaccine.51 The practical utility of administering CP has been limited by the inability to obtain quantitative assessment of SARS-CoV-2–specific IgG antibodies, particularly those with neutralizing capability, to determine the best donors for CP. However, the introduction of the IMMUNO-COV test, which can measure neutralizing antibodies to the SARS-CoV-2 spike glycoprotein by using a pseudovirus, or other types of neutralizing antibody tests will substantially facilitate this aspect of measurement of the serologic immune response to SARS-CoV-2 infection as it becomes increasingly available in diagnostic laboratories.52
Interestingly, assessment of IgG antibodies to SARS-CoV-2 before CP infusion showed a positive result, which was unexpected but perhaps not altogether surprising. The most plausible explanation for this phenomenon is that the patient was capable of mounting a partial humoral immune response (Table I), similar to other patients carrying heterozygous NF-κB2 C-terminus LOF variants,4 , 5 , 22 especially to neoantigens, such as SARS-CoV-2, despite the very low switched memory B cells in their blood. In addition, there may be switched memory B cells in secondary lymphoid organs, which cannot be assessed by peripheral analysis, and the patient's intact CD4+ T-cell compartment may likely provide adequate help to his B cells for antibody production. Most commercial immunoglobulin products are expected to contain little to no antibodies to SARS-CoV-2 in the current preparations, so it is unlikely that the replacement immunoglobulin accounted for this observed positive antibody response. Both SARS-CoV-2 serologic assays demonstrated 100% specificity and no cross-reactivity with seasonal coronaviruses, and therefore this does not account for the SARS-CoV-2 IgG antibody pre-CP. A recent study identified SARS-CoV-2–specific CD4+ and CD8+ T cells not only in a large proportion of the 20 patients who were recovering from COVID-19 infection but also in approximately half of the “unexposed” individuals (n = 20), which was suggested to be due to cross-reactivity with other more common coronaviruses.53 , 54 However, T-cell cross-reactivity does not necessarily have to correlate with antibody cross-reactivity.
It is unclear whether the quality (affinity and avidity) of the antibody response in these patients with NF-κB2 is the same as in healthy individuals, and it is likely that these “endogenous” antibodies may be relatively short-lived. Several days after the infusion, the index (ratio of signal to cutoff) of IgG antibodies appeared to be decreasing. In contrast to several other patients reported in the literature, the patient displayed evidence of clinical improvement even before initiation of CP therapy, although it may have had an adjunctive effect in consolidating recovery.
The patient tested negative for SARS-CoV-2 by PCR at least 3 times after discharge from the hospital, but a nasopharyngeal sample tested positive again at day 22. However, this positive result was not accompanied by any clinical manifestations. A positive PCR test after multiple negative PCR results has been previously reported.55 , 56 Also, Ct values greater than 25 to 30 have not been associated with culturable virus, further substantiating the hypothesis that this represents noninfectious viral RNA.57 Because this result was not associated with new clinical disease, further action was not taken. Serial monitoring of virus along with other clinical parameters may be useful for prognosis, as viral persistence has been shown to be correlative with severe disease.58 However, mild and transient viral positivity after recovery from clinical disease does not appear to correlate with adverse clinical outcomes based on this patient. Interestingly, in 1 study, asymptomatic individuals (n = 37) appeared to shed virus for a longer duration than symptomatic individuals, and they also produced SARS-CoV-2 IgG antibodies as well as neutralizing antibodies relatively early compared with patients with active disease.59
In summary, we have characterized the pathogenic impact of a novel heterozygous NFKB2 LOF C-terminus variant, which resulted in a CVID-like phenotype in this patient. An important lesson learned from this case is that although patients with inborn errors of immunity are predisposed to infection with higher morbidity and mortality in the context of such infection, patients with milder immunodeficiency (ie, partially preserved antibody response, intact T-cell function) can survive infections such as SARS-CoV-2 when treated with use of a multidisciplinary approach to clinical management and aggressive mitigation strategies, including monitoring of biomarkers and early utilization therapies, including biologics and immunomodulators when indicated.
Clinical implications.
A patient’s novel C-terminal loss-of-function variant in NFKB2 resulted in severe COVID-19 and required aggressive management of the primary and secondary immune dysregulation, resulting in a positive outcome.
Acknowledgments
The authors acknowledge the technical and clinical staff of the Diagnostic Immunology Laboratory of Nationwide Children's Hospital, who assisted with the flow cytometry assays.
Footnotes
This study was funded by the Department of Pathology and Department of Laboratory Medicine, Nationwide Children's Hospital, Columbus, Ohio.
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
Appendix
References
- 1.Oeckinghaus A., Ghosh S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol. 2009;1 doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Courtois G. The NF-kappaB signaling pathway in human genetic diseases. Cell Mol Life Sci. 2005;62:1682–1691. doi: 10.1007/s00018-005-5031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Q., Lenardo M.J., Baltimore D. 30 Years of NF-kappaB: A blossoming of relevance to human pathobiology. Cell. 2017;168:37–57. doi: 10.1016/j.cell.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen K., Coonrod E.M., Kumanovics A., Franks Z.F., Durtschi J.D., Margraf R.L., et al. Germline mutations in NFKB2 implicate the noncanonical NF-kappaB pathway in the pathogenesis of common variable immunodeficiency. Am J Hum Genet. 2013;93:812–824. doi: 10.1016/j.ajhg.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klemann C., Camacho-Ordonez N., Yang L., Eskandarian Z., Rojas-Restrepo J.L., Frede N., et al. Clinical and immunological phenotype of patients with primary immunodeficiency due to damaging mutations in NFKB2. Front Immunol. 2019;10:297. doi: 10.3389/fimmu.2019.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuehn H.S., Niemela J.E., Sreedhara K., Stoddard J.L., Grossman J., Wysocki C.A., et al. Novel nonsense gain-of-function NFKB2 mutations associated with a combined immunodeficiency phenotype. Blood. 2017;130:1553–1564. doi: 10.1182/blood-2017-05-782177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baltimore D. NF-kappaB is 25. Nat Immunol. 2011;12:683–685. doi: 10.1038/ni.2072. [DOI] [PubMed] [Google Scholar]
- 8.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and Is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tay M.Z., Poh C.M., Renia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makris S., Paulsen M., Johansson C. Type I interferons as regulators of lung inflammation. Front Immunol. 2017;8:259. doi: 10.3389/fimmu.2017.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Welsh R.M., Bahl K., Marshall H.D., Urban S.L. Type 1 interferons and antiviral CD8 T-cell responses. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi Y., Wang Y., Shao C., Huang J., Gan J., Huang X., et al. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27:1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lipworth B., Chan R., Lipworth S., RuiWen Kuo C. Weathering the cytokine storm in susceptible patients with severe SARS-CoV-2 infection. J Allergy Clin Immunol Pract. 2020;8:1798–1801. doi: 10.1016/j.jaip.2020.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siddiqi H.K., Mehra M.R. COVID-19 illness in native and immunosuppressed states: a clinical-therapeutic staging proposal. J Heart Lung Transplant. 2020;39:405–407. doi: 10.1016/j.healun.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F., Nie J., Wang H., Zhao Q., Xiong Y., Deng L., et al. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J Infect Dis. 2020;221:1762–1769. doi: 10.1093/infdis/jiaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quinti I., Lougaris V., Milito C., Cinetto F., Pecoraro A., Mezzaroma I., et al. A possible role for B cells in COVID-19? Lesson from patients with agammaglobulinemia. J Allergy Clin Immunol. 2020;146:211–213. doi: 10.1016/j.jaci.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roschewski M., Lionakis M.S., Sharman J.P., Roswarski J., Goy A., Monticelli M.A., et al. Inhibition of Bruton tyrosine kinase in patients with severe COVID-19. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abd0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyts I., Bucciol G., Quinti I., Neven B., Fischer A., Seoane E., et al. Coronavirus disease 2019 in patients with inborn errors of immunity: an international study. J Allergy Clin Immunol. 2020 Sep 24 doi: 10.1016/j.jaci.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee C.E., Fulcher D.A., Whittle B., Chand R., Fewings N., Field M., et al. Autosomal-dominant B-cell deficiency with alopecia due to a mutation in NFKB2 that results in nonprocessable p100. Blood. 2014;124:2964–2972. doi: 10.1182/blood-2014-06-578542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brue T., Quentien M.H., Khetchoumian K., Bensa M., Capo-Chichi J.M., Delemer B., et al. Mutations in NFKB2 and potential genetic heterogeneity in patients with DAVID syndrome, having variable endocrine and immune deficiencies. BMC Med Genet. 2014;15:139. doi: 10.1186/s12881-014-0139-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lougaris V., Tabellini G., Vitali M., Baronio M., Patrizi O., Tampella G., et al. Defective natural killer-cell cytotoxic activity in NFKB2-mutated CVID-like disease. J Allergy Clin Immunol. 2015;135:1641–1643. doi: 10.1016/j.jaci.2014.11.038. [DOI] [PubMed] [Google Scholar]
- 25.Mazzoni A., Salvati L., Maggi L., Capone M., Vanni A., Spinicci M., et al. Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J Clin Invest. 2020;130:4694–4703. doi: 10.1172/JCI138554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martinez-Colon G.J., McKechnie J.L., et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smale S.T. Dimer-specific regulatory mechanisms within the NF-kappaB family of transcription factors. Immunol Rev. 2012;246:193–204. doi: 10.1111/j.1600-065X.2011.01091.x. [DOI] [PubMed] [Google Scholar]
- 29.Shih V.F., Tsui R., Caldwell A., Hoffmann A. A single NFkappaB system for both canonical and non-canonical signaling. Cell Res. 2011;21:86–102. doi: 10.1038/cr.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joyner M, Wright RS, Fairweather D, Senefeld J, Bruno K, Klassen S, et al. Early safety indicators of COVID-19 convalescent plasma in 5,000 patients [e-pub ahead of print]. medRxiv https://doi.org/10.1101/2020.05.12.20099879. Accessed June 19, 2020.
- 31.Quartuccio L, Sonaglia A, Pecori D, Peghin M, Fabris M, Tascini C, et al. Higher levels of IL-6 early after tocilizumab distinguish survivors from nonsurvivors in COVID-19 pneumonia: a possible indication for deeper targeting of IL-6. J Med Virol https://doi.org/10.1002/jmv.26149. Accessed August 11, 2020. [DOI] [PMC free article] [PubMed]
- 32.Quentien M.H., Delemer B., Papadimitriou D.T., Souchon P.F., Jaussaud R., Pagnier A., et al. Deficit in anterior pituitary function and variable immune deficiency (DAVID) in children presenting with adrenocorticotropin deficiency and severe infections. J Clin Endocrinol Metab. 2012;97:E121–E128. doi: 10.1210/jc.2011-0407. [DOI] [PubMed] [Google Scholar]
- 33.Ramakrishnan K.A., Rae W., Barcenas-Morales G., Gao Y., Pengelly R.J., Patel S.V., et al. Anticytokine autoantibodies in a patient with a heterozygous NFKB2 mutation. J Allergy Clin Immunol. 2018;141:1479–1482.e6. doi: 10.1016/j.jaci.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 34.Willmann K.L., Klaver S., Dogu F., Santos-Valente E., Garncarz W., Bilic I., et al. Biallelic loss-of-function mutation in NIK causes a primary immunodeficiency with multifaceted aberrant lymphoid immunity. Nat Commun. 2014;5:5360. doi: 10.1038/ncomms6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., et al. Compassionate use of remdesivir for patients with severe Covid-19. N Engl J Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thevarajan I., Nguyen T.H.O., Koutsakos M., Druce J., Caly L., van de Sandt C.E., et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat Med. 2020;26:453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herold T., Jurinovic V., Arnreich C., Lipworth B.J., Hellmuth J.C., Bergwelt-Baildon M.V., et al. Elevated levels of IL-6 and CRP predict the need for mechanical ventilation in COVID-19. J Allergy Clin Immunol. 2020;146:128–136.e4. doi: 10.1016/j.jaci.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo P., Liu Y., Qiu L., Liu X., Liu D., Li J. Tocilizumab treatment in COVID-19: a single center experience. J Med Virol. 2020;92:814–818. doi: 10.1002/jmv.25801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu X., Han M., Li T., Sun W., Wang D., Fu B., et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proc Natl Acad Sci U S A. 2020;117:10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee D.W., Gardner R., Porter D.L., Louis C.U., Ahmed N., Jensen M., et al. Current concepts in the diagnosis and management of cytokine release syndrome. Blood. 2014;124:188–195. doi: 10.1182/blood-2014-05-552729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vultaggio A, Vivarelli E, Virgili G, Lucenteforte E, Bartoloni A, Nozzoli C, et al. Prompt predicting of early clinical deterioration of moderate-to-severe COVID-19 patients: usefulness of a combined score using IL-6 in a preliminary study [e-pub ahead of print]. J Allergy Clin Immunol Pract https://doi.org/10.1016/j.jaip.2020.06.013. Accessed August 15, 2020. [DOI] [PMC free article] [PubMed]
- 44.Nishimoto N., Terao K., Mima T., Nakahara H., Takagi N., Kakehi T. Mechanisms and pathologic significances in increase in serum interleukin-6 (IL-6) and soluble IL-6 receptor after administration of an anti-IL-6 receptor antibody, tocilizumab, in patients with rheumatoid arthritis and Castleman disease. Blood. 2008;112:3959–3964. doi: 10.1182/blood-2008-05-155846. [DOI] [PubMed] [Google Scholar]
- 45.Rose-John S. IL-6 trans-signaling via the soluble IL-6 receptor: importance for the pro-inflammatory activities of IL-6. Int J Biol Sci. 2012;8:1237–1247. doi: 10.7150/ijbs.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knight V, Long T, Meng QH, Linden MA, Rhoads DD. Variability in the laboratory measurement of cytokines: a longitudinal summary of a College of American Pathologists proficiency testing survey [e-pub ahead of print]. Arch Pathol Lab Med https://doi.org/10.5858/arpa.2019-0519-CP. Accessed May 10, 2020. [DOI] [PubMed]
- 47.Martins T.B., Pasi B.M., Litwin C.M., Hill H.R. Heterophile antibody interference in a multiplexed fluorescent microsphere immunoassay for quantitation of cytokines in human serum. Clin Diagn Lab Immunol. 2004;11:325–329. doi: 10.1128/CDLI.11.2.325-329.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Casadevall A., Pirofski L.A. The convalescent sera option for containing COVID-19. J Clin Invest. 2020;130:1545–1548. doi: 10.1172/JCI138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci U S A. 2020;117:9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., et al. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vandergaast R, Carey T, Reiter S, Lech P, Gnanadurai C, Tesfay M, et al. Development and validation of IMMUNO-COV: a high-throughput clinical assay for detecting antibodies that neutralize SARS-CoV-2 [e-pub ahead of print]. bioRxiv https://doi.org/10.1101/2020.05.26.117549. Accessed June 12, 2020.
- 53.Braun J, Loyal L, Frentsch M, Wendisch D, Georg P, Kurth F, et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19 [e-pub ahead of print]. Nature https://doi.org/10.1038/s41586-020-2598-9. Accessed July 8, 2020. [DOI] [PubMed]
- 54.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sethuraman N., Jeremiah S.S., Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020;323:2249–2251. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- 56.Yuan J, Kou S, Liang Y, Zeng J, Pan Y, Liu L. PCR assays turned positive in 25 discharged COVID-19 patients [e-pub ahead of print]. Clin Infect Dis https://doi.org/10.1093/cid/ciaa398. Accessed August 1, 2020. [DOI] [PMC free article] [PubMed]
- 57.Bullard J, Dust K, Funk D, Strong JE, Alexander D, Garnett L, et al. Predicting infectious SARS-CoV-2 from diagnostic samples [e-pub ahead of print]. Clin Infect Dis https://doi.org/10.1093/cid/ciaa638. Accessed August 1, 2020. [DOI] [PMC free article] [PubMed]
- 58.Chang Zhao P., Zhang D., Dong J.H., Xu Z., Yang G., et al. Persistent viral presence determines the clinical course of the disease in COVID-19. J Allergy Clin Immunol Pract. 2020;8:2585–2591. doi: 10.1016/j.jaip.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Long Q.X., Tang X.J., Shi Q.L., Li Q., Deng H.J., Yuan J., et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]