Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in Wuhan city of China in late December 2019 and identified as a novel coronavirus. Due to its contagious nature, the virus spreads rapidly and causes coronavirus disease 2019 (COVID-19). The global tally of COVID-19 was 28 million in early September 2020. The fears and stress associated with SARS-CoV-2 has demolished the socio-economic status worldwide. Researchers are trying to identify treatments, especially antiviral drugs and/or vaccines, that could potentially control the viral spread and manage the ongoing unprecedented global crisis. To date, more than 300 clinical trials have been conducted on various antiviral drugs, and immunomodulators are being evaluated at various stages of COVID-19. This review aims to collect and summarize a list of drugs used to treat COVID-19, including dexamethasone, chloroquine, hydroxychloroquine, lopinavir/ritonavir, favipiravir, remdesivir, tociluzimab, nitazoxanide and ivermectin. However, some of these drugs are not effective and their use has been suspended by WHO.
Keywords: 2019-nCoV, Coronavirus 2019, drugs, global tally, severe acute respiratory syndrome coronavirus 2, WHO
Introduction
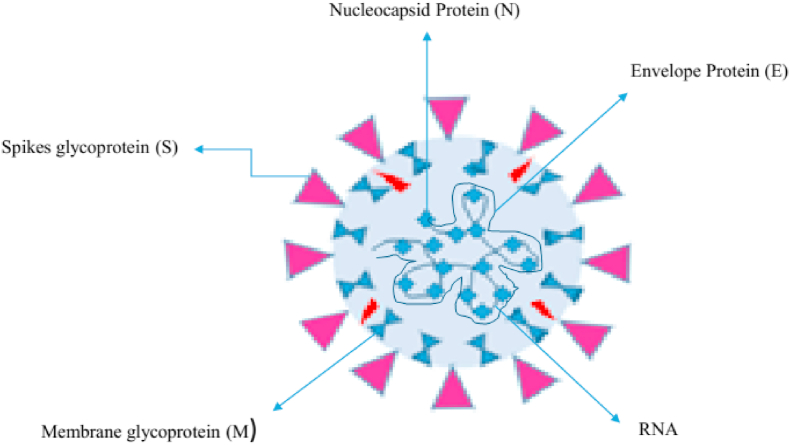
The recent emergence of the novel coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which caused an outbreak of coronavirus disease 2019 (COVID-19) in China, has brought about serious threats to public health worldwide. Coronaviruses belong to the Nidovirales order, which are large and positive-sense enveloped RNA viruses; divided into four genera: α, β, γ and δ. Since the start of the twenty-first century, two β-coronaviruses have caused epidemics of human fatal pneumonia. SARS-CoV-2 belongs to the genus β-coronavirus, which is comprised of crown-like, enveloped, positive-sense single-stranded RNA viruses (Fig. 1). In coronavirus particles, the nucleocapsid protein packages the genome RNA to form a helical nucleocapsid. Various factors are causing the situation to become more serious. First, there is a shortage of effective point-of-care testing assays for rapid, highly accurate identification of SARS-CoV-2-infected individuals. Moreover, asymptomatic and pre-asymptomatic SARS-CoV-2-infected patients are highly contagious, and there is a lack of appropriate detection assays. SARS-CoV emerged in 2002 and caused the disease SARS to spread to five continents, with a case fatality rate of 10% before it was contained in 2003 (more cases reported in 2004). In 2012, the appearance of Middle East respiratory syndrome coronavirus (MERS-CoV) in the Arabian Peninsula caused periodic epidemics in humans with a mortality rate of 35%. SARS-CoV and MERS-CoV are zoonotic viruses, and they cross-species barriers respectively through bat/palm fruit fly [1] and dromedary camels [2]. Seven coronaviruses have been identified that cause human diseases. Four viruses—229E, OC43, NL63 and HKU1—are ubiquitous and usually cause common cold-like symptoms in individuals with immunity [3]. These zoonotic viruses are endemic in the human population, account for up to 30% of mild respiratory tract infections, and can lead to serious complications or death in young children, and in aged and immunocompromised individuals [3,4].
Fig. 1.
Structural composition (proteins and genetic material) of severe acute respiratory syndrome coronavirus 2.
Nucleotide detection assay
During the early period of the SARS-CoV-2 outbreak, many SARS-CoV-2-infected individuals were identified promptly, due to the rapid development and widespread use of DNA-sequencing technology and the RT-PCR method [5].
Commercial nucleotide detection methods
Many molecular diagnostic companies and institutes are devoted to developing integrated, random-access, point-of-care molecular devices for fast and accurate diagnosis of SARS-CoV-2 infection [6]. As of 19 April 2020, the US Food and Drug Administration had approved more than 37 SARS-CoV-2 RNA detection kits (https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-authorizations).
Non-commercialized RNA detection methods
Many institutes are keen to adopt isothermal nucleic acid amplification technology for the point-of-care testing of SARS-CoV-2, avoiding the need for a highly expensive thermal cycler. Loop-mediated isothermal amplification (LAMP) may be the most promising alternative to PCR, as it offers several advantages, in terms of specificity, sensitivity, reaction efficiency and product yield. For example, a reverse transcription (RT)-LAMP assay has been developed, targeting Nsp3 for SARS-CoV-2 detection, of which the limit of detection was 100 copies per reaction [7]. Yu et al. prepared an RT-LAMP assay targeting ORF1ab and the spike gene, of which the limits of detection were 20 copies/reaction and 200 copies/reaction, respectively, within 60 min. The evaluation of a small clinical sample has demonstrated that some of the developed RT-LAMP assays have 100% specificity and 100% sensitivity [5].
Understanding cellular entry of SARS-CoV-2
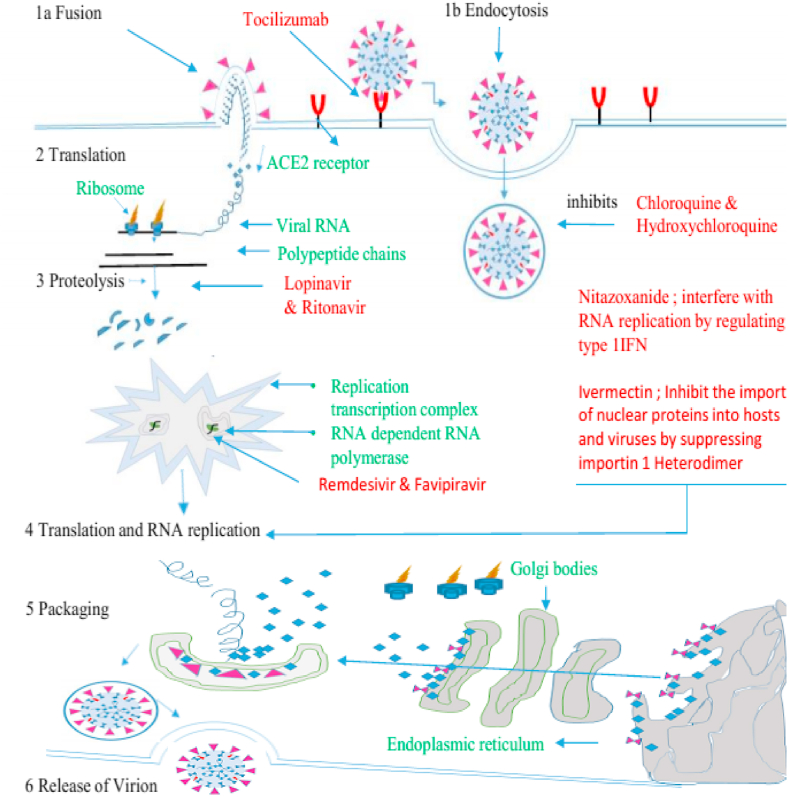
Coronaviruses use spikes of advanced glycoprotein (S) (Fig. 1) homotrimers to promote binding, fusion and entry to the host cell. These spikes are the core antigen existing on the viral surface and the target for neutralizing antibodies during the infection. As a result, glycoprotein (S) acts as a potential residue for vaccine design. Glycoprotein (S) synthesized as a single polypeptide of ∼1300 amino acids precursor belongs to class I viral fusion protein [8]. In most of the coronaviruses, the S protein is processed by host proteases and forms two subunits, named S1 and S2, that remain non-covalently bound in the pre-fusion conformation. The S1 N-terminal subunit contains four domains rich in β, respectively named as A, B, C and D, where the domain A or B serves as a receptor-binding domain in different coronaviruses. The S2 subunit transmembrane C-terminal is a metastable spring-loaded fusion mechanism [9]. During the entry process, the S2 subunit is further cleaved proteolytically at the S2 site instantly upstream of the fusion peptide [10]. The second cleavage step occurs for all coronaviruses and is believed to activate proteins for membrane fusion, which occurs through irreversible conformational changes [11,12]. Recently, cryo-electronic microscopy has led to the determination of the extracellular domain structure of the coronavirus S glycoprotein before and after fusion, providing an image of the beginning and end of the fusion reaction [9,13,14]. The entrance of coronavirus into the host cell is a complex process, necessitating several cellular factors. First, the attachment of the virus to the binding receptors, which enhances the cell surface density of the viral particles and/or promotes interaction with the fusion receptors. For instance, the binding of HCoV-OC43 and bovine coronavirus to N-acetyl-9-O-acetylneuraminic acid [15], binding of HCoV-NL63 and SARS-CoV to heparan sulphate proteoglycans [16,17] and binding of HCoV-HKU1 to O-acetylated sialic acid [18]. In some cases, the attachment of the viral particle to the host cell is redundant [2]. In other cases, the reduction of adhesion receptors primes to a lack of interaction between the virus and host cells, which leads to a significant decline in viral infectivity [17,19,20]. Coronaviruses use multiple fusion receptors, except for HCoV-NL63, which similar to SARS-CoV uses human angiotensin-converting enzyme 2, most α-coronaviruses use aminopeptidase N (CD13) to enter cells [21]. According to reports, HCoV-OC43 uses HLA class I molecules or sialic acid [22], MERS-CoV uses dipeptidyl peptidase 4 (DPP4 or CD26) [23] and the receptor for HCoV-HKU1 is still unknown [18]. Recognizing different receptors means not only different cell orientations, but also different internalization pathways. It is worth mentioning that the recent study also emphasizes the importance of other cytokines for viral tissue specificity, including tissue-specific proteases [24,25] (Fig.3).
Fig. 3.
Scheme of investigated antiviral against coronavirus disease 2019, their potential targets and mechanism of action, Names of drugs are in red, steps involved from fusion to release of virion are in black. ACE2, angiotensin-converting enzyme 2; RTC, replication transcription complex (RTC).
Emergence and global tally of SARS-CoV-2
In late December 2019, some local health centres in Wuhan, China, reported groups of patients with pneumonia with unfamiliar sources. These were epidemiologically related to a wholesale seafood and wet animal market by the European Centre for Disease Prevention and Control [26]. Soon after, unbiased sequencing technology was used to identify an unknown β-coronavirus from samples taken from individuals with pneumonia. Epithelial cells from the human respiratory tract were used to isolate a new coronavirus, initially called 2019-nCoV, which has formed an additional clade within the subgenus sarbecovirus, the subfamily Orthocoronavirinae. Unlike MERS-CoV and SARS-CoV, 2019-nCoV, now named SARS-CoV-2 by WHO, is the seventh member of the coronavirus family that infects humans.
Like other RNA viruses, SAR-CoV-2 has a high mutation rate, which is closely linked to increased virulence and virus evaluability [27]. Mutation in the S protein is of significant clinical and public health concern as it may alter a virus's tropism, including the adaptation of the virus to new hosts, or increase the virus's pathogenesis [28]. Recently Yao et al. accounted for a direct link of genetic mutation and variation of SARS-CoV-2 pathogenicity. They investigated 11 samples from SARS-CoV-2-infected individuals [29]. Interestingly six different mutations were detected in the S protein. Two of these mutations were single nucleotide variants that led to similar missense mutations [30].
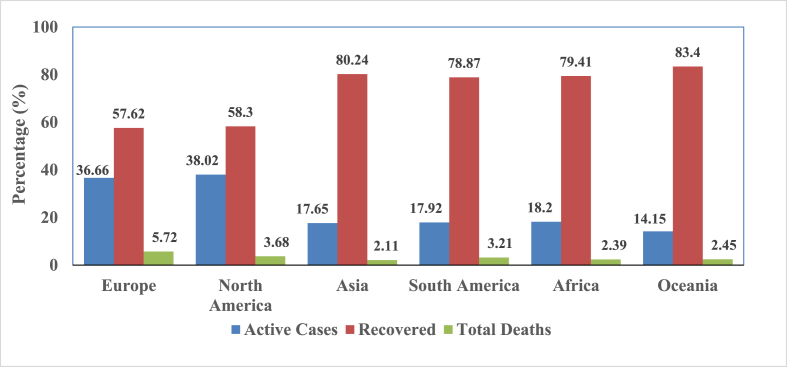
Importantly these isolates resulted in varied cytopathic effects and viral loads in Vero-E6 cells, which revealed that genetic mutation in SARS-CoV-2 causes substantial changes to enhance the pathogenicity of the virus [29,30]. With such vast genetic mutations, recurrent recombination of their genomes, and increased interface activity between humans and animals, new coronaviruses may emerge from time to time in humans [31,32]. By early September 2020, more than 28 million people had contracted COVID-19, and about 0.91 million had died. Despite substantial efforts by the global health community, the impact of COVID-19 in terms of morbidity and mortality rates remains high, especially in high-income countries compared with low-income countries. It was reported that in the first 5 months of the pandemic, the high-income and upper-middle-income countries had higher incidences and death rates per million compared with low-income and middle-income countries [33].
Fig. 2 demonstrates the continent-wise distribution, deaths and recoveries of COVID-19 up to 2 September 2020 (https://www.worldometers.info/coronavirus/).
Fig. 2.
Coronavirus disease 2019 continent-wise distribution.
Potential drugs against COVID-19
Currently, there is no specific and effective antiviral therapy available for COVID-19. However, most individuals with COVID-19 have a mild/asymptomatic or moderate course of the disease; in up to 5%–10% of individuals the course of the disease is severe and can lead to death in vulnerable sections of the community. The immediate need for effective drugs is crucial at this stage (www.covidreference.com) [34].
Enhanced supportive care remains the mainstay of treatment. More than 300 clinical trials for drugs have been conducted, some of which will be published in the coming months. WHO launched the Solidarity clinical trial for COVID-19 treatment to further evaluate remdesivir, hydroxychloroquine/chloroquine and lopinavir-ritonavir with and without interferon-β. However, various other antiviral agents and immunomodulators are being evaluated for use at various stages of COVID-19. The registry of international clinical trials can be found on the WHO website and at ClinicalTrials.gov [35]. Currently, it is strongly recommended to recruit patients to participate in ongoing trials because we cannot determine whether the benefits of most treatments outweigh the hazards, and these studies will provide much-needed evidence for the efficacy and safety of various treatments for COVID-19 [36].
Chloroquine phosphate
Chloroquine phosphate is an old drug used to treat malaria, it has been verified in a multicentre clinical trial conducted in China, with excellent efficacy and adequate safety in the treatment of COVID-19-related pneumonia [37]. Chloroquine phosphate was also used against the SARS-CoV and Zika virus epidemics as an important controlling agent. The role of chloroquine is to increase the pH of intracellular vacuoles and to change the protein degradation pathway by acid hydrolase in the lysosome, macromolecular production in the endosome, and post-translational protein modification in the Golgi. It also interferes with macrophages and other antigen-presenting cells, thus having an anti-rheumatic response [38]. Other studies reported that chloroquine has potential activity against various viruses by deceleration of the endosomal pH needed for virus–cell fusion, also interfering with cellular SARS-CoV receptor glycosylation [39]. The activity of chloroquine against viruses and as an anti-inflammatory agent might be one of the reasons for its effective efficacy to treat individuals with COVID-19 pneumonia. Over the last 70 years, chloroquine has been found to be a cost-effective and safe drug [37] and in China, chloroquine phosphate at a dose of 500 mg/day in numerous clinical trials has shown potential activity against COVID-19. Hence, it is postulated that chloroquine could be used as a possible treatment for individuals with COVID-19-related pneumonia [40].
Hydroxychloroquine
Hydroxychloroquine, an aminoquinoline, is less toxic; with the N-diethyl chain of chloroquine substituted by an N-hydroxyethyl side chain. This alteration in the side chain makes the hydroxychloroquine more soluble than chloroquine. The antiviral activity of hydroxychloroquine is similar to that of chloroquine; by increasing the pH the hydroxychloroquine affects the activated immune cells and Toll-like receptor-mediated signal transduction, down-regulating Toll-like receptor expression and reducing the production of interleukin-6 (IL-6) [41]. Even though hydroxychloroquine and chloroquine have the same antimalarial action, hydroxychloroquine is preferred over chloroquine for its low ocular toxicity [42]. Prolonged treatment (at least 5 years) with hydroxychloroquine may cause retinopathy, which is a dose-limiting adverse effect of hydroxychloroquine, and a safe daily dose appears to correspond to 6.5 mg/kg of ideal body weight and 5.0 mg/kg of actual body weight [40]. Although in their activity against coronaviruses both chloroquine and hydroxychloroquine have the same theoretical application, the anti-coronaviral activity of chloroquine is supported by more clinical data [43]. Availability of hydroxychloroquine and chloroquine is low in some countries, and chloroquine has a more adverse effect than hydroxychloroquine. For instance, in individuals with COVID-19, chloroquine leads to prolongation of the QT interval when interacting with lopinavir/ritonavir. Consequently, it is crucial when unable to treat COVID-19 patients, to consider using hydroxychloroquine instead of chloroquine. In Iran, it was necessary to use hydroxychloroquine instead of chloroquine due to scarcity. Hydroxychloroquine hindrance by other antiviral agents, such as oseltamivir, lopinavir/ritonavir, ribavirin, and by interferons and intravenous immunoglobulins is currently under investigation. Million et al. conducted a comprehensive meta-analysis on the use of chloroquine derivatives in COVID-19 patients. It was observed that the combined therapy of hydroxychloroquine with azithromycin was more effective than hydroxychloroquine alone and the combined therapy was considered more important than monotherapy [44]. The synergistic approach was supported by an in vitro study and this combination should not be neglected in COVID-19 therapy [45]. Despite controversy as the virus spreads across borders, chloroquine derivatives are used by physicians on a large scale [46].
Remdesivir
The novel pro-drug remdesivir, a nucleotide analogue intracellularly activated to adenosine triphosphate analogue, which hinders the viral RNA polymerase, was initially developed as an antiviral drug against Ebola virus and Marburg virus infections. Remdesivir has potential activity against a broad range of virus families such as filoviruses (e.g. Ebola) and coronaviruses (e.g. SARS-CoV and MERS-CoV), displaying an effective therapeutic and prophylactic action against these coronaviruses in non-clinical models. An in vitro analysis of remdesivir demonstrated activity against SARS-CoV-2 in Vero E6 cells with an EC50 value of 1.76 μM, which suggests that in non-human primate models the working concentration is expected to be achieved [47]. According to the report, an individual with COVID-19 in the USA exhibited substantial improvement when treated with remdesivir intravenously [48], after which a rapid assessment was made to check the effectiveness and safety of remdesivir in hospitalized individuals with COVID-19. The group of patients severely affected by COVID-19 were treated with remdesivir, and it was found that among 53 individuals, 36 (68%) showed clinical improvement [49]. Because there are no placebos or active comparators in this study, it is difficult to draw any specific conclusions, and the determination of efficacy will require ongoing randomized, placebo-controlled results of remdesivir treatment. At present, four clinical trials recruiting patients in the USA and two other trials enrolled only in China have been registered on ClinicalTrials.gov: NCT04257656 (serious disease) and NCT04252664 (mild to moderate disease) [36]. Brouqui et al. have also compiled the critical data published on remdesivir use in COVID-19 patients. They found that no research supports convincingly the use of remdesivir in seriously ill patients. Treatment at the time of diagnosis is the key outcome for individuals with COVID-19; however, the adverse reaction associated with remdesivir sometimes leads to the interruption of treatment, and the intravenous route in this regard would probably restrict the use of remdesivir [50].
Favipiravir
The pro-drug favipiravir (6-fluoro-3-hydroxy-2-pyrazine carboxamide) is activated by intracellular phosphoribosylation to favipiravir ribofuranosyl-5B-triphosphate (Favipiravir-RTP), which acts as a substrate for RNA-dependent RNA polymerase (RdRp) and potentially inhibits the activity of RdRp in RNA viruses. Among several types of RNA viruses, the catalytic domain of RdRp is conserved, so the broad antiviral activity of favipiravir is supported by this mechanism of action. The active Favipiravir-RTP with an IC50 of 0.022 μg/mL hinders influenza virus RdRp, although up to 100 μg/mL does not affect the human DNA polymerase α, β, γ subunits. Additionally, favipiravir has a broad range of inhibitory effects on RNA viruses, e.g the arena-, bunya-, flavi- and filoviruses causing haemorrhagic fevers [51,52], and to some extent it is effective against Ebola virus [51,53,54]. Favipiravir is one of the possible drugs for COVID-19, as SARS-CoV-2 has been identified as a single-stranded RNA virus with an RdRp gene like SARS-CoV and MERS-CoV. However, there are no authentic in vitro and preclinical animal studies available. In an in vitro study, favipiravir inhibited SARS-CoV-2 in Vero E6 cells with an EC50 of 61.88 μM [48], but other studies showed that in vitro at concentrations under 100 μM favipiravir did not affect SARS-CoV-2 [55]. At present, the existing knowledge is not sufficient to endorse the use of favipiravir to treat COVID-19, and further research is needed.
Lopinavir and ritonavir
Lopinavir are protease inhibitors that are usually used to treat human immunodeficiency virus infection with and a booster drug ritonavir. Lopinavir and/or ritonavir have in vitro anti-coronaviral activity; they inhibit the proteases, which are important enzymes for polyprotein processing in coronaviruses. In vitro studies have reported that lopinavir can inhibit SARS-CoV, and the EC50 of lopinavir is acceptable. Against SARS-CoV-2, lopinavir showed efficacy in Vero E6 cells, with an estimated EC50 of 26.62 μM [56].
In Singapore, five individuals with COVID-19 were treated with lopinavir and ritonavir within 1–3 days of duration, but evidence of clinical use was ambiguous. Although fever developed within 1 to 3 days after the onset of lopinavir/ritonavir, in two patients disease progression was not prevented. The decrease in viral load indicated by the circulation threshold in lopinavir/ritonavir-treated and untreated individuals, and also nasopharyngeal swab results, were similar [57]. In a retrospective study of 134 COVID-19 patients, it was found that improving symptoms and reduced viral load were not significantly different among the lopinavir/ritonavir treatment group (n = 52), arbidol treatment group (n = 34) and control group (n = 48) [58].
Another study of 47 individuals with COVID-19 compared the standard of care (arbidol plus interferon-α inhaler) in five patients with combined treatment of lopinavir/ritonavir plus the standard of care (arbidol plus interferon-α inhaler) in 42 patients, and reported shorter times to normal body temperature and negative test in clinical samples for SARS-CoV-2 [59]. In another report, 44 patients with mild/moderate COVID-19 were randomly assigned to receive lopinavir/ritonavir (21 patients), arbidol (16 patients) or no antiviral drugs (7 patients as controls). There was no statistically significant difference in the fever reduction rate, cough relief rate, chest CT scan improvement rate, or clinical condition deterioration rate among the three groups (all p > 0.05). Overall, 5 (23.8%) patients in the lopinavir/ritonavir group experienced adverse events during follow up whereas no obvious adverse events occurred in the arbidol or control groups. The study concluded that lopinavir/ritonavir or arbidol monotherapy seemed to have little benefit in improving the clinical outcome of mild/moderate COVID-19, but lopinavir/ritonavir may cause more adverse events [60].
In a randomized trial of 199 individuals with severe COVID-19, compared with standard treatment alone, lopinavir/ritonavir (400/100 mg) was added to standard treatment twice a day for 14 consecutive days and did not decrease clinical improvement time [61]. Lopinavir/ritonavir had a tendency to reduce mortality (19% versus 25%, respectively), and there was a greater difference in mortality within 12 days of symptom onset, although the difference was not statistically significant. The rate of SARS-CoV-2 decrease was similar between the group receiving lopinavir/ritonavir and the group not receiving lopinavir/ritonavir. The proportion of early discontinuation for with lopinavir/ritonavir treatment was 14%. The patients engaged in this study had a progressive infection and previously had considerable tissue damage (as shown by compromised lung function and a 25% death rate in the control group). Even highly active antibacterial agents have limited efficacy in advanced bacterial pneumonia [62].
Enhanced clinical recovery (from 16.0 to 17.0 days) and reduced fatality (from 19.0% to 27.1%) were detected in an ad hoc subgroup of patients who received treatment within 12 days of symptom onset, but not in patients who received treatment later [63]. In another study of 280 individuals with COVID-19, the time from onset to antiviral treatment was considered a risk factor for serious illness. Patients in the mild group received antiviral treatment earlier (Severe group 1.19 ± 0.45 days compared with 2.65 ± 1.06 days, p < 0.001) [62]. Whether early COVID-19 treatment with lopinavir/ritonavir has clinical applications is an important question that required further research.
Nitazoxanide
Nitazoxanide and its active metabolite tizoxanide have been shown to have effective in vitro activity against SARS-CoV-2 and MERS-CoV in Vero E6 cells, with EC50 of 2.12 μM and 0.92 μM, respectively. In addition to coronavirus, it also exhibits broad-spectrum antiviral activity against influenza virus, respiratory syncytial virus, parainfluenza virus, rotavirus and norovirus [63]. It is believed that this broad-spectrum antiviral activity is due to the following characteristics. The basis of its activity is to interfere with the host regulatory pathways associated with viral replication, rather than virus-specific pathways [64]. Nitazoxanide up-regulates the innate antiviral mechanism through extensive amplification of cytoplasmic RNA induction and the type I IFN pathway. Nitazoxanide interferes with viral infection by up-regulating the specific host mechanism of viral targeting to avoid host cellular defence [65].
Because of its broad-spectrum antiviral activity, nitazoxanide is being studied in clinical trials, including randomized controlled trials, to control influenza and other acute respiratory infections, although the results are either not encouraging or unavailable. However, the in vitro activity of nitazoxanide against SARS-CoV-2 is encouraging, it is clear that more data are needed to determine its role in the management of COVID-19 [55].
Ivermectin
Ivermectin is a broad-spectrum antiparasitic drug approved by the US Food and Drug Administration. In recent years, it has shown in vitro antiviral activity against various viruses. Initially, it was considered to be an inhibitor of the interaction of importin 1 heterodimer (accountable for nuclear importation of integrase protein) and human immunodeficiency virus 1. This confirms that ivermectin is responsible for inhibiting the nuclear import of integrase protein and human immunodeficiency virus type-1 replication. Other effects of ivermectin have been reported, other than the inhibition of the nuclear import of host and viral proteins. It has been shown to limit the infection of certain RNA viruses (including influenza virus, dengue fever virus and West Nile virus). Ivermectin has been similarly proven to be effective against the DNA virus pseudorabies virus both in vitro and in vivo and increased survival of pseudorabies virus-infected mice [66]. An in vitro study reported that ivermectin was found to be an inhibitor of SARS-CoV-2. A single addition to Vero-hSLAM cells 2 hours after their infection with SARS-CoV-2 can effectively reduce the viral RNA approximately 5000 times for 48 hours. The authors assumed that this may be accomplished by inhibiting importin-α/β1-mediated nuclear importation of viral proteins and that this inhibition would disrupt the immune escape mechanism of the virus [67]. Additional in vitro, in vivo and clinical trials are required to conclude its role in COVID-19 treatment.
Tocilizumab
Tocilizumab is a recombinant humanized monoclonal antibody IL-6 receptor inhibitor used to treat rheumatoid arthritis. Tocilizumab binds to the soluble and membrane-bound IL-6 receptors, ultimately binding to gp130 on the cell membrane, thereby inhibiting signal transduction and acting as a pro-inflammatory agent [56,68]. Similar to the alterations in SARS and MERS, the plasma of an individual with COVID-19 had high levels of the cytokines IL-6, IL-2, IL-7, IL-10, tumour necrosis factor-α, interferon-γ-inducible protein, macrophage inflammatory protein 1α, monocyte chemoattractant protein and granulocyte colony-stimulating factor, which suggests a cytokine storm [69], and is associated with the severity and prognosis of the disease. In autopsy samples from individuals who died from severe COVID-19, histological examination revealed bilateral diffuse alveolar injury with fibrinous mucus-like exudates. Mononuclear inflammatory lymphocytes were observed in both lungs [70]. Some studies have reported the immune properties of individuals with COVID-19 and found that T cells and inflammatory monocytes are highly activated, resulting in excessive production of cytokines and inflammatory storm. Among the cytokines, IL-6 is the significant cytokine that causes an inflammatory storm, which may lead to increased pulmonary alveolar–capillary blood gas exchange dysfunction, particularly impaired oxygen diffusion, which ultimately leads to pulmonary fibrosis and organ failure [71].
A recent study of 21 individual with COVID-19 treated with tocilizumab, who had a fever, dyspnoea, and CT image deterioration showed a remarkable improvement in body temperature, respiration and chest tightness after receiving tocilizumab treatment. Of the 21 infected individuals, 15 recovered from respiratory diseases, and two were withdrawn from the ventilator after 5 days of treatment [72].
Low number of lymphocytes is an important sign of severity and diagnosis in COVID-19 patients [73]. In a study with tocilizumab, 85% of the patients had low lymphocyte counts. Although in 52.6% of patients the lymphocyte counts had returned to normal after 5 days of treatment, and their C-reactive protein levels had also returned to normal. Although CT scanning showed lung turbidity in 90.5% of the patients, suggesting that it takes a long time to recover. However, in this study, there were no reports of adverse reactions and subsequent lung infections and 90.5% of patients displayed significant progress and good prognosis, together with two critical individuals. Hence, tocilizumab can be an efficient treatment for severe COVID-19, which may be due to the blocking of IL-6-related fever and inflammatory storm response [72].
Dexamethasone
Dexamethasone is a steroid that has been used in many diseases since the 1960s, including inflammatory disorders and certain cancers, to alleviate inflammation. Since 1977, it has been listed in multiple formulations on the WHO Model List of Essential Medicines, and is currently off-patent and available in most countries [74].
According to new studies, dexamethasone was the first agent to be proven to cause ‘significant reduction in mortality’ in individuals with COVID-19 who require oxygen or ventilation. As part of the random evaluation of the Oxford University's COVID-19 treatment (RECOVERY) trial, some patients are being treated with steroids. During the trial, a total of 2104 patients were randomly assigned to receive oral or intravenous dexamethasone, 6 mg once a day for 10 days, and were randomized with 4321 patients in the control group. Dexamethasone reduced the mortality of ventilated patients by 35% (rate 0.65, 95% CI 0.48–0.88; p 0.0003), whereas it decreased by 20% among patients who received oxygen only (0.80, 95% CI 0.67–0.96; p 0.0021). No benefit was seen in patients who do not require respiratory support (1.22, 95% CI 0.86–1.75; p 0.14) The Oxford University research on various drugs that may help against COVID-19 found that dexamethasone improves the survival of patients with COVID-19 who are receiving oxygen or using ventilators. For those receiving oxygen therapy, it saved the lives of one patient among 25 patients who were treated (reducing the number of deaths by about one-fifth). For patients using a ventilator, it saved the lives of eight patients (reducing deaths by approximately one-third). In particular, it seems to help avoid the damage caused by cytokine storms in the most severely affected patients https://www.recoverytrial.net [75]. Physicians must ensure the laboratory-confirmed SARS-CoV-2 status of patients and their medical history, and must consult the official recommendations of WHO for dexamethasone clinical use, because from time to time drugs are suspended and new drugs are approved. It was also explained in the study that dexamethasone does not help those who are mildly infected with the virus, indicating that it works by treating specific symptoms rather than the virus itself [76]. The researchers shared preliminary observations with WHO on the test findings and look forward to completing data analysis. WHO may coordinate a meta-analysis to improve our overall understanding of the intervention. In addition, WHO clinical guidance will be updated to reflect how and when the drug should be used in COVID-19 [74].
COVID-19 detection methods
Worldwide research approaches have been adopted to respond to the rapidly growing COVID-19 pandemic based on testing capabilities, public health resources and the spread of viruses among populations.
Molecular based laboratory testing
Currently, the US CDC recommended strategy for identifying individuals with COVID-19 is to test respiratory samples to diagnose the presence of one or several specific nucleic acid targets for SARS-CoV-2 [77]. Nasopharyngeal samples are the first choice for swab-based SARS-CoV-2 testing, but samples of the middle turbinate, oropharynx or anterior nostrils are also acceptable [78].
Flocking swabs are the best choice for sample collection; it is best to use a swab with a plastic shaft or aluminium. Cotton swabs containing calcium alginate should be avoided as it may affect PCR testing. After collecting the sample, it should be transferred to a universal transport medium to preserve the viral nucleic acid. Samples collected from endotracheal aspirates, sputum and bronchoalveolar lavage fluid can also be sent directly to the laboratory for processing, which may be more sensitive than the upper respiratory tract specimens [79]. Samples are subjected to RNA extraction, followed by target-specific nucleic acid detection through qualitative RT-PCR. The CDC and WHO have developed the most widely used SARS-CoV-2 detection method. The kit designed by CDC contains PCR primer probes for the two regions (N1 and N2) of the viral nucleocapsid gene, but the WHO PCR primer probes are different, and probe the SARS-CoV-2 envelope (E) gene and RNA-dependent RNA polymerase (RdRP) [77].
Both detection methods have high detection sensitivity and specificity for SARS-CoV-2 and have limited cross-reactivity with other circulating coronavirus strains, and both have a circulation threshold of <40 as a positive standard (https://www.fda.gov/media/135659/download). In another report, SARS-CoV-2 was studied by inspecting a strong correlation between Ct and sample infectivity in a cell culture model. According to that study, infected individuals are can be detected up to 20 days after the onset of symptoms using PCR; however, isolation of the virus is not possible after day 8 because of high viral loads of approx. 105 RNA copies per mL. RT-PCR can show that patients with a Ct value ≥ 34 can be discharged. It has been suggested that the sensitivity of amplification on gene E detection would be less than on gene N. Moreover, the correlation between the viral RNA load and cell culture model for each centre can be performed independently [80].
Antigen-detection-based tests
Diagnosis approaches that identify antigens for respiratory syncytial virus infection or influenza virus infection directly from clinical samples through immunoassays have been commercialized for decades; they are capable of showing results in a few minutes and have low complexity [81]. Current methods for identification of influenza and respiratory syncytial viruses suffer from suboptimal sensitivity to rule out disease [82,83], and SARS-CoV-2 may have the same problem. Experiments with specific guidance for proper interpretation need to be introduced. Test prototypes for other new coronaviruses have not yet obtained permission and regulatory approval is still in progress [84,85] (www.finddx.org/covid-19/pipeline on 23 March 2020). Monoclonal antibodies against the SARS-CoV-2 nucleocapsid protein have been developed and may form the basis of future rapid antigen detection tests [86].
Serology
Serological tests, such as ELISA, can detect antibodies against SARS-CoV-2 (such as IgG, IgM and IgA) from clinical specimens, which may not be as complicated as molecular tests and may be used in some cases for diagnosis [87]. However, at the onset of symptoms, when the risk of viral shedding and transmission seems to be highest, their use in the diagnosis of acute infections may be limited [78]. Antibody response to viral infection may take several days to be reliably detected [87]. A negative result cannot sort out SARS-CoV-2 infection, especially in people who have recently been exposed to the virus. Antibodies that cross-react with non-SARS-CoV-2 coronavirus proteins are also a possible problem, and the results of past or current contact with other human coronaviruses are encouraging [88].
In patients with advanced disease complications during drug therapy, serological testing may be more important, because RT-PCR may be false negative as the virus decreases over time [89]. For epidemiological research, continuous monitoring, vaccine research and risk assessment of potential medical personnel, it is necessary to develop serological tests that can accurately assess previous SARS-CoV-2 infection and immunity. In some countries/regions, immunoassays are already on the market, but their diagnostic reliability and optimal usage are still unclear.
Radiographical analysis
The image features reported in COVID-19 are variable and uncertain, and there is a great overlap with the image features of SARS and MERS. Early evidence suggests that initial chest imaging will show abnormalities in at least 85% of patients and that, initially, 75% of patients will have bilateral lung involvement, most often occurring in the sub-pleural and peripheral areas as ground-glass appearance with distortion and convergence. Older age and gradual merger can predict a poorer prognosis. In addition to the acute phase, CT monitoring of individuals recovering from COVID-19 is recommended to assess long-term or permanent lung injury, including fibrosis, such as in SARS and MERS [90]. An analysis of five people who were initially diagnosed with COVID-19 with a negative viral swab test showed the importance of early CT results for disease diagnosis. This study suggests that the presence of typical CT findings in people suspected of having the virus may be useful for initial screening [91]. However, early reports indicate that preliminary imaging examinations can show normal findings in 15% of people, so standard chest imaging examinations cannot eliminate infections. Because chest imaging of individuals with COVID-19 is an important part of patient care, further research is needed to expand the understanding of imaging results during disease. SARS and MERS results indicate that people recovering from COVID-19 should be followed to check for signs of repeated lung injury (i.e. interlobular trapped air, thickening or fibrosis). Preventive measures may play a key role in reducing the spread of disease from person to person in the hospital. Radiologists should pay attention to all procedures and techniques to reduce the risk of infection by medical staff and patients [73].
Vaccines updates
SARS-CoV-2 has caused a continuing public health crisis, but because of the novelty of the virus, there is currently no SARS-CoV-2-specific therapy or vaccine.
Researchers are working with different technologies, some of which have not previously been used in licensed vaccines. Six or more groups of volunteers have been injected with preparations; others have begun testing in animals (https://www.nature.com/articles/d41586-020-01221-y).Research teams from different universities and companies around the world have produced more than 118 SARS-CoV-2 vaccines. The 18 candidate vaccines currently in clinical evaluation (Table 1) (Table 2) and 129 pre-clinical candidate vaccines included inactivated vaccines, recombinant subunit vaccines, 203 nucleic acid vaccines, adenovirus vector vaccines and recombinant influenza viruses 204 carrier vaccine (https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines).
Table 1.
Profile of antivirals investigated for COVID-19 treatment in clinical trials or in vitro studies. Source: WHO, adapted from landscape analysis 17 February 2020 https://www.who.int/blueprint/priority-diseases/key
| Group | Drugs | Structure and PubChem CID | Route of administration | Current use | Mechanism of action | EC50 value in Vero E6 cell | Being tested? |
|---|---|---|---|---|---|---|---|
| Inhibitors of viral RNA polymerase/RNA synthesis | Remdesivir | PubChem CID:121304016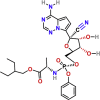
|
Intravenous | COVID-19 | A prodrug, adenosine nucleotide analogue, which acts as an RdRp inhibitor | 1.76 μM | Approved |
| Favipiravir | PubChem CID:492405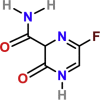
|
Intravenous | influenza | A prodrug, guanosine nucleotide analogue, which acts as an RdRp inhibitor | 61.88 μM | Yes | |
| Inhibitors of viral protein synthesis | Lopinavir/ritonavir | PubChem CID:11979606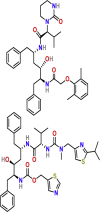
|
Orally | HIV | Inhibition of protease | 26.62 μM | Yes |
| Viral entry inhibitors | Hydroxychloroquine | PubChem CID:3652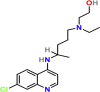
|
Orally | Antimalarial | Increase the endosomal pH required by the virus/Cell fusion, and interference Glycosylation of SARS-CoV (ACE-2) cell receptor | 4.51 μM | yes |
| Chloroquine | PubChem CID:2719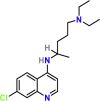
|
Orally | Antimalarial | 2.71 μM | |||
| Immunomodulatory | Nitazoxanide | PubChem CID:41684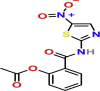
|
Orally | Anti-parasitic and antiviral | Interference involves viral replication, amplification of cytoplasmic RNA sensing, and host regulation pathways of type I interferon | 2.12 μM | yes |
| Ivermectin | PubChem CID:6321424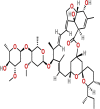
|
Orally | Anti-parasitic | Inhibit the import of nuclear proteins into hosts and viruses by suppressing importin 1 Heterodimer | NA | yes | |
| Immunomodulatory | Tocilizumab | NA | Intravenously | Rheumatoid arthritis; COVID-19 | inhibit signal transduction by specifically binding to sIL-6R and mIL-6R | NA | Approved |
| Anti-inflammatory | Dexamethasone | 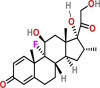 |
Intravenously and orally | Anti-inflammatory | Act as an inflammatory agent | NA | yes |
Note: Not applicable- NA.
Table 2.
List of candidate vaccines currently under clinical trials
| Platform | Type of candidate vaccine | Developer | Coronavirus target | The current stage of clinical evaluation/regulatory status Coronavirus candidate | Same platform for non-coronavirus candidates |
|---|---|---|---|---|---|
| Non-replicating viral vector | ChAdOx1-S | University of Oxford/AstraZeneca | SARS-CoV-2 | Phase 3 ISRCTN89951424 Phase2b/3 2020-001228-32 Phase 1/2 PACTR202006922165132 2020-001072-15 |
MERS, influenza, TB, Chikungunya, Zika, MenB, plague |
| Non- Replicating Viral Vector | Adenovirus Type 5 Vector | CanSino Biological Inc./Beijing Institute of Biotechnology | SARS-CoV-2 | Phase 2 ChiCTR2000031781 Phase 1 ChiCTR2000030906 |
Ebola |
| RNA | LNP- encapsulated mRNA |
Moderna/NIAID | SARS-CoV-2 | Phase 2 NCT04405076 Phase 1 NCT04283461 |
multiple candidates |
| DNA | DNA plasmid vaccine with electroporation | Inovio Pharmaceuticals/International Vaccine Institute | SARS-CoV-2 | Phase 1/2 NCT04447781NCT04336410 | multiple candidates |
| Inactivated | Inactivated | Wuhan Institute of Biological Products/Sinopharm |
SARS-CoV-2 | Phase 1/2 ChiCTR2000031809 | |
| Inactivated | Inactivated | Beijing Institute of Biological Products/Sinopharm | SARS-CoV-2 | Phase 1/2 ChiCTR2000032459 | |
| Inactivated | Inactivated + alum | Sinovac | SARS-CoV-2 | Phase 1/2 NCT04383574NCT04352608 | SARS |
| Protein Subunit | Full-length recombinant SARS CoV-2 glycoprotein nanoparticle vaccine adjuvanted with Matrix M |
Novavax | SARS-CoV-2 | Phase 1/2 NCT04368988 | RSV; CCHF, HPV, VZV, EBOV |
| RNA | 3 LNP-mRNAs | BioNTech/Fosun Pharma/Pfizer | SARS-CoV-2 | Phase 1/2 2020-001038-36 NCT04368728 |
|
| Inactivated | Inactivated | Institute of Medical Biology , Chinese Academy of Medical Sciences |
SARS-CoV-2 | Phase 1 NCT04412538 | |
| DNA | DNA Vaccine (GX-19) |
Genexine Consortium | SARS-CoV-2 | Phase 1 NCT04445389 |
|
| Non- Replicating Viral Vector |
Adeno-based | Gamaleya Research Institute | SARS-CoV-2 | Phase 1 NCT04436471 NCT04437875 |
|
| Protein Subunit | Native like Trimeric subunit Spike Protein vaccine | Clover Biopharmaceuticals Inc./GSK/Dynavax | SARS-CoV-2 | Phase 1 NCT04405908 | HIV, REV Influenza |
| Protein Subunit | Adjuvanted recombinant protein (RBD-Dimer) | Anhui Zhifei Longcom Biopharmaceutical/Institute of Microbiology, Chinese Academy of Sciences |
SARS-CoV-2 | Phase 1 NCT04445194 | MERS |
| Protein Subunit | Recombinant spike protein with Advax™ Adjuvant |
Vaxine Pty Ltd/Medytox | SARS-CoV-2 | Phase 1 NCT04453852 | |
| RNA | LNP-nCoVsaRNA | Imperial College London | SARS-CoV-2 | Phase 1 ISRCTN17072692 |
EBOV; LASV, MARV, Inf (H7N9), RABV |
| RNA | mRNA | Curevac | SARS-CoV-2 | Phase 1 NCT04449276 | RABV, LASV, YFV; MERS, InfA, ZIKV, DENV, NIPV |
| RNA | mRNA | People's Liberation Army (PLA) Academy of Military Sciences/Walva × Biotech. |
SARS-CoV-2 | Phase 1 ChiCTR2000034112 |
Abbreviations: CCHF, Crimean–Congo haemorrhagic fever virus; DENV, dengue virus; EBOV, Ebola virus; HIV, human immunodeficiency virus; HPV, human papillomavirus; Inf (H7N9), influenza virus H7N9; LASV, Lassa mammarenavirus; MARV, Marburg virus; MenB, meningitis B; MERS, Middle East respiratory syndrome; NIPV, Nipah virus; RABV, rabies virus; REV, reticuloendothelial virus; RSV, respiratory syncytial virus; SARS, severe acute respiratory syndrome; TB, tuberculosis; VZV, varicella zoster virus.
Conclusion and perspective
There is an intensifying health crisis around the globe caused by COVID-19, mainly distressing economically developed countries and responsible for high morbidity and mortality rates. An escalating threat to global COVID-19 control mandates the development and application of new tools. At the forefront, the diagnosis of SARS-COV-2 from clinical samples must be rapid and accurate. Urgency is required for investigations to unravel the complex pathways that hamper not only the clinical application of available drugs but also the development of new anti-COVID-19 treatments. The absence of effective drugs is the prime reason for the rapid spread of COVID-19. In this review, we discuss drugs used against COVID-19 and their mechanisms of action, both those used initially and those drugs that are still in use, such as chloroquine, hydroxychloroquine, lopinavir/ritonavir, favipiravir, remdesivir, tociluzimab, nitazoxanide and ivermectin [92]. We also discuss diagnostic procedure such as molecular based tests, serology, radiography and antigen base detection and vaccines. Some of the drugs are not effective and have been suspended by WHO. New knowledge along with detailed understanding of COVID-19's mechanisms would be tremendously helpful in the development of new, highly effective anti-COVID-19 drugs. Studying coronaviruses will assist in understanding the principles of cross-species transmission and adaptation to humans and prepare for possible outbreaks of zoonotic diseases in the future and their treatment.
From the perspective of the current pandemic, designing effective therapies for SARS-CoV-2 is another challenge for scientists. Although many antiviral treatments against SARS and MERS have been reported to have strong in vitro activity, there are limited options with clinical potential. Treatment options are now available that can be used clinically during the ongoing SARS-CoV-2 epidemic. Some of the broad-spectrum antiviral drugs may be effective against SARS-CoV-2, and the current scenario of COVID-19 around the globe provides an opportunity to test them. The design and development of new broad-spectrum antiviral drugs that could potentially target all coronaviruses may be the only treatment option to prevent re-emerging and circulating coronaviruses. Moreover, new data on the clinical characteristics, treatment options and outcomes of COVID-19 appear constantly, and doctors working in patient care should keep up to date with the latest information on this issue. Finally, people should practice good hygiene, frequently washing their hands, maintaining social distances and wearing masks.
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
Not required.
Acknowledgement
AK appreciates the support provided by the Chinese Scholarship Council to conduct this study.
Contributor Information
A. Khan, Email: asaf2018@lzu.edu.cn.
W. Sajjad, Email: wasim.sajjad71@yahoo.com.
References
- 1.Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haagmans B.L., Al Dhahiry S.H., Reusken C.B., Raj V.S., Galiano M., Myers R. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. 2014;14:140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isaacs D., Flowers D., Clarke J.R., Valman H.B., MacNaughton M.R. Epidemiology of coronavirus respiratory infections. Arch Dis Child. 1983;58:500–503. doi: 10.1136/adc.58.7.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu F., Yan L., Wang N., Yang S., Wang L., Tang Y. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin Infect Dis Off Publ Infect Dis Soc Am. 2020;71(15):793–798. doi: 10.1093/cid/ciaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loeffelholz M.J., Tang Y.W. Laboratory diagnosis of emerging human coronavirus infections—the state of the art. Emerg Microbe. Infect. 2020;9:747–756. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park G.S., Ku K., Baek S.H., Kim S.J., Kim S.I., Kim B.T. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) J Mol Diagn. 2020;22:729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walls A.C., Tortorici M.A., Bosch B.J., Frenz B., Rottier P.J.M., DiMaio F. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531(7592):114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burkard C., Verheije M.H., Wicht O., van Kasteren S.I., van Kuppeveld F.J., Haagmans B.L. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathogens. 2014;10 doi: 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci USA. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walls A.C., Tortorici M.A., Snijder J., Xiong X., Bosch B.J., Rey F.A. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc Natl Acad Sci USA. 2017;114:11157–11162. doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shang J., Zheng Y., Yang Y., Liu C., Geng Q., Tai W. Cryo-electron microscopy structure of porcine deltacoronavirus spike protein in the prefusion state. J Virol. 2018;92(4) doi: 10.1128/JVI.01556-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vlasak R., Luytjes W., Spaan W., Palese P. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc Natl Acad Sci USA. 1988;85:4526–4529. doi: 10.1073/pnas.85.12.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lang J., Yang N., Deng J., Liu K., Yang P., Zhang G. Inhibition of SARS pseudovirus cell entry by lactoferrin binding to heparan sulfate proteoglycans. PloS One. 2011;6 doi: 10.1371/journal.pone.0023710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milewska A., Zarebski M., Nowak P., Stozek K., Potempa J., Pyrc K. Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. J Virol. 2014;88:13221–13230. doi: 10.1128/JVI.02078-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang X., Dong W., Milewska A., Golda A., Qi Y., Zhu Q.K. Human coronavirus HKU1 spike protein uses O-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J Virol. 2015;89:7202–7213. doi: 10.1128/JVI.00854-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plochmann K., Horn A., Gschmack E., Armbruster N., Krieg J., Wiktorowicz T. Heparan sulfate is an attachment factor for foamy virus entry. J Virol. 2012;86:10028–10035. doi: 10.1128/JVI.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Connell B.J., Lortat-Jacob H. Human immunodeficiency virus and heparan sulfate: from attachment to entry inhibition. Front Immunol. 2013;4:385. doi: 10.3389/fimmu.2013.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn J.H., Li W., Choe H., Farzan M. Angiotensin-converting enzyme 2: a functional receptor for SARS coronavirus. Cell Mol Life Sci. 2004;61:2738–2743. doi: 10.1007/s00018-004-4242-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins A.R. HLA class I antigen serves as a receptor for human coronavirus OC43. Immunol Invest. 1993;22:95–103. doi: 10.3109/08820139309063393. [DOI] [PubMed] [Google Scholar]
- 23.Raj V.S., Mou H., Smits S.L., Dekkers D.H., Müller M.A., Dijkman R. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci USA. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kido H., Okumura Y., Yamada H., Le T.Q., Yano M. Proteases essential for human influenza virus entry into cells and their inhibitors as potential therapeutic agents. Curr Pharmaceut Des. 2007;13:405–414. doi: 10.2174/138161207780162971. [DOI] [PubMed] [Google Scholar]
- 26.ECDC. European Centre for Disease Prevention and Control . ECDC; Solna: 2020. Cluster of pneumonia cases caused by a novel coronavirus, Wuhan, China. [Google Scholar]
- 27.Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16(8) doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94(7) doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao H., Lu X., Chen Q., Xu K., Chen Y., Cheng L. 2020. Patient-derived mutations impact pathogenicity of SARS-CoV-2. 2020. CELL-D-20-01124. [Google Scholar]
- 30.Abdullahi I.N., Emeribe A.U., Ajayi O.A., Oderinde B.S., Amadu D.O., Osuji A.I. Implications of SARS-CoV-2 genetic diversity and mutations on pathogenicity of the COVID-19 and biomedical interventions. J Taibah Univ Med Sci. 2020;15:258–264. doi: 10.1016/j.jtumed.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong G., Liu W., Liu Y., Zhou B., Bi Y., Gao G.F. MERS, SARS, and Ebola: the role of super-spreaders in infectious disease. Cell Host Microbe. 2015;18:398–401. doi: 10.1016/j.chom.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabawoon W. medRxiv; 2020. Differences by country-level income in COVID-19 cases, deaths, case-fatality rates, and rates per million population in the first five months of the pandemic. 07.13.20153064. [Google Scholar]
- 34.Surek A., Ferahman S., Gemici E., Dural A.C., Donmez T., Karabulut M. 2020. Effects of COVID-19 pandemic on general surgical emergencies: are some emergencies really urgent? Level 1 Trauma center experience.10.21203/rs.3.rs-37618/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Solidarity” clinical trial for COVID-19 treatments. 2020. int/emergencies/diseases/novel-coronavirus-/global-research-on-novel-coronavirus—ncov/solidarity-clinical-trial-for-covid-19-treatments WHOJWHOSrGWAfhww. [Google Scholar]
- 36.McCreary E.K., Pogue J.M. Coronavirus disease 2019 treatment: a review of early and emerging options. Open Forum Infect Dis. 2020;7(4):ofaa105. doi: 10.1093/ofid/ofaa105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao J., Tian Z., Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends. 2020;14:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 38.Tortorici M.A., Walls A.C., Lang Y., Wang C., Li Z., Koerhuis D. Structural basis for human coronavirus attachment to sialic acid receptors. Nat Struct Mol Biol. 2019;26:481–489. doi: 10.1038/s41594-019-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plantone D., Koudriavtseva T. Current and future use of chloroquine and hydroxychloroquine in infectious, immune, neoplastic, and neurological diseases: a mini-review. Clin Drug Invest. 2018;38:653–671. doi: 10.1007/s40261-018-0656-y. [DOI] [PubMed] [Google Scholar]
- 40.Savarino A., Boelaert J.R., Cassone A., Majori G., Cauda R. Effects of chloroquine on viral infections: an old drug against today's diseases? Lancet Infect Dis. 2003;3:722–727. doi: 10.1016/S1473-3099(03)00806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jorge A.M., Melles R.B., Zhang Y., Lu N., Rai S.K., Young L.H. Hydroxychloroquine prescription trends and predictors for excess dosing per recent ophthalmology guidelines. Arthritis Res Ther. 2018;20:133. doi: 10.1186/s13075-018-1634-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan Y.W., Yam W.K., Sun J., Chu J.J.H. An evaluation of chloroquine as a broad-acting antiviral against hand, foot and mouth disease. Antivir Res. 2018;149:143–149. doi: 10.1016/j.antiviral.2017.11.017. [DOI] [PubMed] [Google Scholar]
- 43.Sahraei Z., Shabani M., Shokouhi S., Saffaei A. Aminoquinolines against coronavirus disease 2019 (COVID-19): chloroquine or hydroxychloroquine. Int J Antimicrob Agents. 2020;55:105945. doi: 10.1016/j.ijantimicag.2020.105945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Million M., Gautret P., Colson P., Roussel Y., Dubourg G., Chabriere E. 2020. Clinical efficacy of chloroquine derivatives in COVID-19 infection: comparative meta-analysis between the big data and the real world; p. 100709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andreani J., Le Bideau M., Duflot I., Jardot P., Rolland C., Boxberger M. In vitro testing of combined hydroxychloroquine and azithromycin on SARS-CoV-2 shows synergistic effect. Microb Pathog. 2020;145:104228. doi: 10.1016/j.micpath.2020.104228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colson P., Rolain J.-M., Raoult D.J. Chloroquine for the 2019 novel coronavirus SARS-CoV-2. Int J Microb Agents. 2020;55(3):105923. doi: 10.1016/j.ijantimicag.2020.105923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A. Compassionate use of remdesivir for patients with severe Covid-19. N Engl J Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brouqui P., Giraud-Gatineau A., Raoult D. Elsevier; Amsterdam: 2020. Remdesivir investigational trials in COVID-19: a critical reappraisal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Du Y.X., Chen X.P. Favipiravir: pharmacokinetics and concerns about clinical trials for 2019-nCoV infection. Clin Pharmacol Ther. 2020;108(2):242–247. doi: 10.1002/cpt.1844. [DOI] [PubMed] [Google Scholar]
- 52.Furuta Y., Komeno T., Nakamura T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc Jpn Acad Ser B, Phys Biol Sci. 2017;93:449–463. doi: 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bai C.Q., Mu J.S., Kargbo D., Song Y.B., Niu W.K., Nie W.M. Clinical and virological characteristics of Ebola virus disease patients treated with favipiravir (T-705)-Sierra Leone, 2014. Clin Infect Dis. 2016;63:1288–1294. doi: 10.1093/cid/ciw571. [DOI] [PubMed] [Google Scholar]
- 54.Correction Experimental treatment with favipiravir for Ebola virus disease (the JIKI trial): a historically controlled, single-arm proof-of-concept trial in Guinea. PLoS Med. 2016;13 doi: 10.1371/journal.pmed.1001967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choy K.T., Wong A.Y., Kaewpreedee P., Sia S.F., Chen D., Hui K.P.Y. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir Res. 2020;178:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davies R., Choy E. Clinical experience of IL-6 blockade in rheumatic diseases—implications on IL-6 biology and disease pathogenesis. Semin Immunol. 2014;26:97–104. doi: 10.1016/j.smim.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 57.Young B.E., Ong S.W.X., Kalimuddin S., Low J.G., Tan S.Y., Loh J. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 2020;323:1488–1494. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gautret P., Lagier J.C., Parola P., Hoang V.T., Meddeb L., Sevestre J. Clinical and microbiological effect of a combination of hydroxychloroquine and azithromycin in 80 COVID-19 patients with at least a six-day follow up: a pilot observational study. Travel Med Infect Dis. 2020;34:101663. doi: 10.1016/j.tmaid.2020.101663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ye X.T., Luo Y.L., Xia S.C., Sun Q.F., Ding J.G., Zhou Y. Clinical efficacy of lopinavir/ritonavir in the treatment of Coronavirus disease 2019. Eur Rev Med Pharmacol Sci. 2020;24:3390–3396. doi: 10.26355/eurrev_202003_20706. [DOI] [PubMed] [Google Scholar]
- 60.Chen C.Y., Wang F.L., Lin C.C. Chronic hydroxychloroquine use associated with QT prolongation and refractory ventricular arrhythmia. Clin Toxicol. 2006;44:173–175. doi: 10.1080/15563650500514558. [DOI] [PubMed] [Google Scholar]
- 61.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N Engl J Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baden L.R., Rubin E.J. Covid-19 - the search for effective therapy. N Engl J Med. 2020;382:1851–1852. doi: 10.1056/NEJMe2005477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Şimşek Yavuz S., Ünal S. Antiviral treatment of COVID-19. Turk J Med Sci. 2020;50(Si-1):611–619. doi: 10.3906/sag-2004-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rossignol J.F. Nitazoxanide, a new drug candidate for the treatment of Middle East respiratory syndrome coronavirus. J Infect Public Health. 2016;9:227–230. doi: 10.1016/j.jiph.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jasenosky L.D., Cadena C., Mire C.E., Borisevich V., Haridas V., Ranjbar S. The FDA-approved oral drug nitazoxanide amplifies host antiviral responses and inhibits Ebola virus. iScience. 2019;19:1279–1290. doi: 10.1016/j.isci.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir Res. 2020;178:104787. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ketkar H., Yang L., Wormser G.P., Wang P. Lack of efficacy of ivermectin for prevention of a lethal Zika virus infection in a murine system. Diagn Microbiol Infect Dis. 2019;95:38–40. doi: 10.1016/j.diagmicrobio.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaly L., Rosner I. Tocilizumab—a novel therapy for non-organ-specific autoimmune diseases. Best Practice Research Clin Rheumatol. 2012;26:157–165. doi: 10.1016/j.berh.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 69.Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Resp Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou Y., Fu B., Zheng X., Wang D., Zhao C., qi Y. 2020. Aberrant pathogenic GM-CSF+ T cells and inflammatory CD14+CD16+ monocytes in severe pulmonary syndrome patients of a new coronavirus; p. 2020. 02.12.945576. [Google Scholar]
- 72.Xu X., Han M., Li T., Sun W., Wang D., Fu B. Effective treatment of severe COVID-19 patients with tocilizumab. Proc Natl Acad Sci USA. 2020;117(20):10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.WHO . WHO; Geneva: 16 June 2020. WHO, Adapted from WHO welcomes preliminary results about dexamethasone use in treating critically ill COVID-19 parents. [Google Scholar]
- 75.Theraphy R. 2020. Adapted from Recovery trails, randaomised evaluation of COVID-19 therapy, registered under CTN50189673. [Google Scholar]
- 76.BBC n. British broadcasting coporation (BBC) news.
- 77.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.La Scola B., Le Bideau M., Andreani J., Hoang V.T., Grimaldier C., Colson P. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Prendergast C., Papenburg J. Rapid antigen-based testing for respiratory syncytial virus: moving diagnostics from bench to bedside? Future Microbiol. 2013;8:435–444. doi: 10.2217/fmb.13.9. [DOI] [PubMed] [Google Scholar]
- 82.Chartrand C., Tremblay N., Renaud C., Papenburg J. Diagnostic accuracy of rapid antigen detection tests for respiratory syncytial virus infection: systematic review and meta-analysis. J Clin Microbiol. 2015;53:3738–3749. doi: 10.1128/JCM.01816-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Merckx J., Wali R., Schiller I., Caya C., Gore G.C., Chartrand C. Diagnostic accuracy of novel and traditional rapid tests for influenza infection compared with reverse transcriptase polymerase chain reaction: a systematic review and meta-analysis. Ann Intern Med. 2017;167:394–409. doi: 10.7326/M17-0848. [DOI] [PubMed] [Google Scholar]
- 84.Lau S.K., Woo P.C., Wong B.H., Tsoi H.W., Woo G.K., Poon R.W. Detection of severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in sars patients by enzyme-linked immunosorbent assay. J Clin Microbiol. 2004;42:2884–2889. doi: 10.1128/JCM.42.7.2884-2889.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen Y., Chan K.H., Kang Y., Chen H., Luk H.K., Poon R.W. A sensitive and specific antigen detection assay for Middle East respiratory syndrome coronavirus. Emerg Microb Infect. 2015;4:e26. doi: 10.1038/emi.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sheridan C. Fast, portable tests come online to curb coronavirus pandemic. Nat Biotechnol. 2020;38:515–518. doi: 10.1038/d41587-020-00010-2. [DOI] [PubMed] [Google Scholar]
- 87.Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Patrick D.M., Petric M., Skowronski D.M., Guasparini R., Booth T.F., Krajden M. An outbreak of human coronavirus OC43 infection and serological cross-reactivity with SARS coronavirus. Can J Infect Dis Med Microbiol. 2006;17:330–336. doi: 10.1155/2006/152612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hosseiny M., Kooraki S., Gholamrezanezhad A., Reddy S., Myers L. Radiology perspective of coronavirus disease 2019 (COVID-19): lessons from severe acute respiratory syndrome and middle east respiratory syndrome. Am J Roentgenol. 2020;214:1078–1082. doi: 10.2214/AJR.20.22969. [DOI] [PubMed] [Google Scholar]
- 91.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020:200343. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kakakhel M.A., Wu F., Khan T.A., Feng H., Hassan Z., Anwar Z., Faisal S., Ali I., Wang W. The first two months epidimiological study of COVID-19, related public health preparedness, and response to the ongoing epidemic in Pakistan. N Microb N Infect. 2020;37:100734. doi: 10.1016/j.nmni.2020.100734. [DOI] [PMC free article] [PubMed] [Google Scholar]