Abstract
Several machine learning (ML) algorithms have been increasingly utilized for cardiovascular disease prediction. We aim to assess and summarize the overall predictive ability of ML algorithms in cardiovascular diseases. A comprehensive search strategy was designed and executed within the MEDLINE, Embase, and Scopus databases from database inception through March 15, 2019. The primary outcome was a composite of the predictive ability of ML algorithms of coronary artery disease, heart failure, stroke, and cardiac arrhythmias. Of 344 total studies identified, 103 cohorts, with a total of 3,377,318 individuals, met our inclusion criteria. For the prediction of coronary artery disease, boosting algorithms had a pooled area under the curve (AUC) of 0.88 (95% CI 0.84–0.91), and custom-built algorithms had a pooled AUC of 0.93 (95% CI 0.85–0.97). For the prediction of stroke, support vector machine (SVM) algorithms had a pooled AUC of 0.92 (95% CI 0.81–0.97), boosting algorithms had a pooled AUC of 0.91 (95% CI 0.81–0.96), and convolutional neural network (CNN) algorithms had a pooled AUC of 0.90 (95% CI 0.83–0.95). Although inadequate studies for each algorithm for meta-analytic methodology for both heart failure and cardiac arrhythmias because the confidence intervals overlap between different methods, showing no difference, SVM may outperform other algorithms in these areas. The predictive ability of ML algorithms in cardiovascular diseases is promising, particularly SVM and boosting algorithms. However, there is heterogeneity among ML algorithms in terms of multiple parameters. This information may assist clinicians in how to interpret data and implement optimal algorithms for their dataset.
Subject terms: Computational biology and bioinformatics, Machine learning, Cardiovascular diseases
Introduction
Machine learning (ML) is a branch of artificial intelligence (AI) that is increasingly utilized within the field of cardiovascular medicine. It is essentially how computers make sense of data and decide or classify a task with or without human supervision. The conceptual framework of ML is based on models that receive input data (e.g., images or text) and through a combination of mathematical optimization and statistical analysis predict outcomes (e.g., favorable, unfavorable, or neutral). Several ML algorithms have been applied to daily activities. As an example, a common ML algorithm designated as SVM can recognize non-linear patterns for use in facial recognition, handwriting interpretation, or detection of fraudulent credit card transactions1,2. So-called boosting algorithms used for prediction and classification have been applied to the identification and processing of spam email. Another algorithm, denoted random forest (RF), can facilitate decisions by averaging several nodes. While convolutional neural network (CNN) processing, combines several layers and apples to image classification and segmentation3–5. We have previously described technical details of each of these algorithms6–8, but no consensus has emerged to guide the selection of specific algorithms for clinical application within the field of cardiovascular medicine. Although selecting optimal algorithms for research questions and reproducing algorithms in different clinical datasets is feasible, the clinical interpretation and judgement for implementing algorithms are very challenging. A deep understanding of statistical and clinical knowledge in ML practitioners is also a challenge. Most ML studies reported a discrimination measure such as the area under an ROC curve (AUC), instead of p values. Most importantly, an acceptable cutoff for AUC to be used in clinical practice, interpretation of the cutoff, and the appropriate/best algorithms to be applied in cardiovascular datasets remain to be evaluated. We previously proposed the methodology to conduct ML research in medicine6. Systematic review and meta-analysis, the foundation of modern evidence-based medicine, have to be performed in order to evaluate the existing ML algorithm in cardiovascular disease prediction. Here, we performed the first systematic review and meta-analysis of ML research over a million patients in cardiovascular diseases.
Methods
This study is reported in accordance with the Preferred Reporting Information for Systematic Reviews and Meta-Analysis (PRISMA) recommendations. Ethical approval was not required for this study.
Search strategy
A comprehensive search strategy was designed and executed within the MEDLINE, Embase, and Scopus databases from database inception through March 15, 2019. One investigator (R.P.) designed and conducted the search strategy using input from the study’s principal investigator (C.K.). Controlled vocabulary, supplemented with keywords, was used to search for studies of ML algorithms and coronary heart disease, stroke, heart failure, and cardiac arrhythmias. The detailed strategy is available from the reprint author. The full search strategies can be found in the supplementary documentation.
Study selection
Search results were exported from all databases and imported into Covidence9, an online systematic review tool, by one investigator (R.P.). Duplicates were identified and removed using Covidence's automated de-duplication functionality. The de-duplicated set of results was screened independently by two reviewers (C.K. and H.V.) in two successive rounds to identify studies that met the pre-specified eligibility criteria. In the initial screening, two investigators (C.K. and H.V.) independently examined the titles and abstracts of the records retrieved from the search via the Covidence portal and used a standard extraction form. Conflicts were resolved through consensus and reviewed by other investigators. We included abstracts with sufficient evaluation data, including methodology, the definition of outcomes, and an appropriate evaluation matrix. Studies without any kind of validation (external validation or internal validation) were excluded. We excluded reviews, editorials, non-human studies, letters without sufficient data.
Data extraction
We extracted the following information, if possible, from each study: authors, year of publication, study name, test types, testing indications, analytic models, number of patients, endpoints (CAD, AMI, stroke, heart failure, and cardiac arrhythmias), and performance measures ((AUC, sensitivity, specificity, positive cases (the number of patients who used the AI and were positively diagnosed with the disease), negative cases (the number of patients who used the AI and were negative with the AI test), true positives, false positives, true negatives, and false negatives)). CAD was defined as coronary artery stenosis > 70% using angiography or FFR-based significance. Cardiac arrhythmias included studies involving bradyarrhythmias, tachyarrhythmias, atrial, and ventricular arrhythmias. Data extraction was conducted independently by at least two investigators for each paper. Extracted data were compared and reconciled through consensus. In case studies which did not report positive and negative cases, we manually calculated by standard formulae using statistics available in the manuscripts or provided by the authors. We contacted the authors if the data of interest were not reported in the manuscripts or abstracts. The order of contact originated with the corresponding author, followed by the first author, and then the last author. If we were unable to contact the authors as specified above, the associated studies were excluded from the meta-analysis (but still included it in the systematic review). We also excluded manuscripts or abstracts without sufficient evaluation data after contacting the authors.
Quality assessment
We created the proposed guidance quality assessment of clinical ML research based on our previous recommendation (Table 1)6. Two investigators (C.K. and H.V.) independently assessed the quality of each ML study by using our proposed guideline to report ML in medical literature (Supplementary Table S1). We resolved disagreements through discussion amongst the primary investigators or by involving additional investigators to adjudicate and establish a consensus. We scored study quality as low (0–2), moderate (2.5–5), and high quality (5.5–8) as clinical ML research.
Table 1.
Proposed quality assessment of ML research for clinical practice.
| Algorithms |
|
Clarity of algorithms Propose new algorithms Select the proper algorithms Compare alternative algorithms |
| Resources |
|
Reliable database/center Number of database/centers Number of samples (patients/images) Type and diversity of data |
| Sufficient reported data |
|
Manuscript with sufficient supplementary information Letter or editor, short article, abstract Report baseline characteristics of patients |
| Ground truth |
|
Comparison to expert clinicians Comparison to validated clinical risk models |
| Outcome |
|
Assessment of outcome based on standard medical taxonomy External validation cohort |
| Interpretation |
|
Report both discrimination and calibration metrics Report one or more of the following: sensitivity, specificity, positive, negative cases, balanced accuracy |
Statistical analysis
We used symmetrical, hierarchical, summary receiver operating characteristic (HSROC) models to jointly estimate sensitivity, specificity, and AUC10. and denote the sensitivity and specificity of the ith study. is the variance of and is the variance of .
The HSROC model for study i fits the following
= and =1- . when no disease and for those with disease. And and follow normal distribution.
We conducted subgroup analyses stratified by ML algorithms. We assessed the performances of a subgroup-specific and statistical test of interaction among subgroups. We performed all statistical analyses using OpenMetaAnalyst for 64-bit (Brown University), R version 3.2.3 (Metafor and Phia packages), and Stata version 16.1 (Stata Corp, College Station, Texas). The meta-analysis has been reported in accordance with the Meta-analysis of Observational Studies in Epidemiology guidelines (MOOSE)11.
Results
Study search
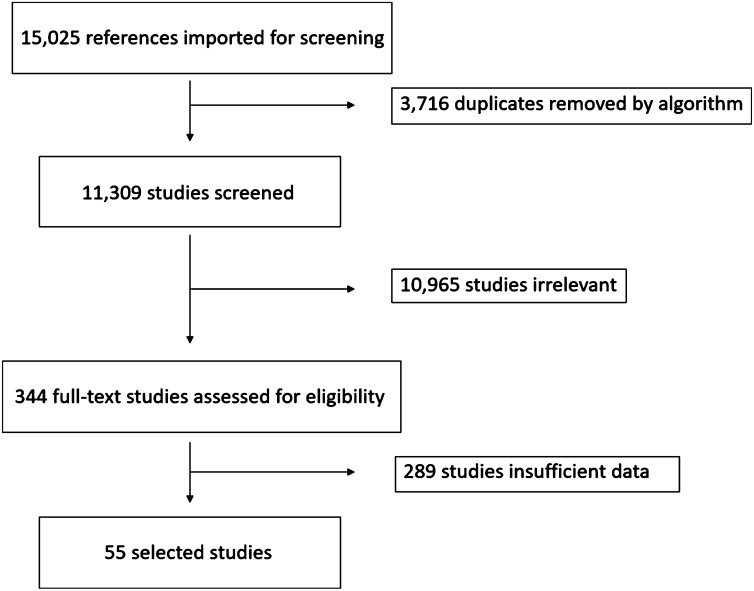
The database searches between 1966 and March 15, 2019, yielded 15,025 results. 3,716 duplicates were removed by algorithms. After the screening process, we selected 344 articles for full-text review. After full text and supplementary review, we excluded 289 studies due to insufficient data to perform meta-analytic approaches despite contacting corresponding authors. Overall, 103 cohorts (55 studies) met our inclusion criteria. The disposition of studies excluded after the full-text review is shown in Fig. 1.
Figure 1.
Study design. This flow chart illustrates the selection process for published reports.
Study characteristics
Table 2 shows the basic characteristics of the included studies. In total, our meta-analysis of ML and cardiovascular diseases included 103 cohorts (55 studies) with a total of 3,377,318 individuals. In total, 12 cohorts assessed cardiac arrhythmias (3,144,799 individuals), 45 cohorts are CAD-related (117,200 individuals), 34 cohorts are stroke-related (5,577 individuals), and 12 cohorts are HF-related (109,742 individuals). The characteristics of the included studies are listed in Table 2. We performed post hoc sensitivity analysis, excluding each study, and found no difference among the results.
Table 2.
Characteristics of the included studies.
| First author | Analytic model | Sample | Indication | Imaging | Comparison | Database |
|---|---|---|---|---|---|---|
| Cardiac arrhythmias | ||||||
| Alickovic et al. (2016) | RF | 47 | Arrhythmia detection | ECG | Five ECG signal patterns from MIT-BIH (normal (N), Premature Ventricular Complex (PVC), Atrial Premature Contraction (APC), Right Bundle Branch Block (RBBB) and Left Bundle Branch Block (RBBB)) and four ECG patterns from St. -Petersburg Institute of Cardiological Technics 12lead Arrhythmia Database (N, APC, PVC and RBBB) | St. Petersburg and MIT-BIH database |
| Au-Yeung et al. (2018) | 5sRF, 10sRF, SVM | 788 | Ventricular arrhythmia | ICD Data | SCD-HeFT study | |
| Hill et al. (2018) | Logistic-linear regression, SVM, RF | 2,994,837 | Development of AF/flutter in gen pop | Clinical data | ML compared with conventional linear statistical methods | UK Clinical Practice Research Datalink (CPRD) between 01–01-2006 and 31–12-2016 was undertaken |
| Kotu et al. (2015) | k-NN, SVM, RF | 54 | arrhythmic risk stratification of post MI patients | Cardiac MRI | Low LVEF and Scar versus textural features of scar | Single center |
| Ming-Zher Poh et al. (2018) | CNN | 149,048 | AF | ECG | Several publicly accessible PPG repositories, including the MIMIC-III critical care database,11 the Vortal data set from healthy volunteers12 and the IEEE-TBME PPG Respiratory Rate Benchmark data set.1 | |
| Xiaoyan Xu et al. (2018) | CNN | 25 | AF | ECG | MIT-BIH Atrial Fibrillation database | MIT-BIH Atrial Fibrillation Database |
| Coronary artery disease | ||||||
| Araki et al. (2016) | SVM classifier with five different kernels sets | 15 | Plaque rupture prediction | IVUS | 40 MHz catheter utilizing iMap (Boston Scientific, Marlborough, MA, USA) with 2,865 frames per patient (42,975 frames) and (b) linear probe B-mode carotid ultrasound (Toshiba scCNNer, Japan) | Single center |
| Araki et al. (2016) | SVM combined with PCA | 19 | Coronary risk assessment | IVUS | Single center | |
| Arsanjani et al. (2013) | boosting algorithm | 1,181 | Perfusion SPECT in CAD | Perfusion SPECT | 2 experts, combined supine/prone TPD | Single center |
| BaumCNN et al. (2017) | Custom-built algorithm | 258 | ctFFR in detecting relevant lesions | Invasive FFR determination of relevant lesions | the MACHINE Registry | |
| Coenen (2018) | Custom-built algorithm | 351 | Invasive FFR / Computational flow dynamics based (CFD-FFR) | CT angiography | Invasive FFR / Computational flow dynamics based (CFD-FFR) | 5 centers in Europe, Asia, and the United States |
| Dey et al. (2015) | boosting algorithm | 37 | Coronary CTA in ischemic heart disease patients to predict impaired myocardial flow reserve | CCTA | Clinical stenosis grading | Single center |
| Eisenberg et al. (2018) | boosting algorithm | 1925 | MPI in CAD | SPECT | Human visual analysis | The ReFiNE registry |
| Freiman et al. (2017) | Custom-built algorithm | 115 | CCTA in coronary artery stenosis | CCTA | Cardiac image analysis | The MICCAI 2012 challenge |
| Guner et al. (2010) | CNN | 243 | Stable CAD | Myocardial perfusion SPECT (MPS) |
SPECT evaluation and human–computer interaction One expert reader who has 10 years of experience and six nuclear medicine residents who have two to four years of experience in nuclear cardiology took part in the study |
Single center |
| Hae et al. (2018) | Logistic-linear regression, SVM, RF, boosting algorithm | 1,132 | Prediction FFR in stable and unstable angina patients | FFR, CCTA | Single center | |
| Han et al. (2017) | Logistic-linear regression | 252 | Physiologically significant CAD | CCTA and invasive fractional flow reserve (FFR | The DeFACTO study | |
| Hu (Xiuhua) et al. (2018) | Custom-built algorithm | 105 | Intermediate coronary artery lesions | CCTA | CCTA-FFR vs Invasive angiography FFR | Single center |
| Hu et al. (2018) | Boosting algorithm | 1861 | MPI in CAD | SPECT | True early reperfusion | Multicenter REFINE SPECT registry |
| Wei et al. (2014) | Custom-built algorithm | 83 | Noncalcified plaques (NCPs) detection on CCTA | CCTA | Single center | |
| Kranthi et al. (2017) | Boosting algorithm | 85,945 | CCTA in CAD | CCTA | 66 available parameters (34 clinical parameters, 32 laboratory parameters) | Single center |
| Madan et al. (2013) | SVM | 407 | Urinary proteome in CAD | global proteomic profile analysis of urinary proteome | Indian Atherosclerosis Research Study | |
| Zellweger et al. (2018) | Custom-built algorithm | 987 | CAD evaluation | N/A | Framingham scores | The Ludwigshafen Risk and Cardiovascular Health Study (LURIC) |
| Moshrik Abd alamir et al. (2018) | Custom-built algorithm | 923 | ED patients with chest pain -CTA analysis | CT Angiography | Single center | |
| Nakajima et al. (2017) | CNN | 1,001 | Previous myocardial infarction and coronary revascularization | SPECT | Expert consensus interpretations | Japanese multicenter study |
| Song et al. (2014) | SVM | 208 | Risk prediction in ACS | N/A | Single center | |
| VanHouten et al. (2014) | Logistic-linear regression, RF | 20,078 | Risk prediction in ACS | N/A | Single center | |
| Xiao et al. (2018) | CNN | 15 | Ischemic ST change in ambulatory ECG | ECG | Long-Term ST Database (LTST database) from PhysioNet | |
| Yoneyama et al. (2017) | CNN | 59 | Detecting culprit coronary arteries | CCTA and myocardial perfusion SPECT | Single center | |
| Stroke | ||||||
| Abouzari et al. (2009) | CNN | 300 | SDH post-surgery outcome prediction | CT head | Single center | |
| Alexander Roederer et al.(2014) | Logistic-linear regression | 81 | SAH-Vasospasm prediction | Passively obtained clinical data | Single center | |
| Arslan et al. (2016) | Logistic-linear regression, SVM, boosting algorithm | 80 | Ischemic stroke | EMR | Single center | |
| Atanassova et al. (2008) | CNN | 54 | Major stroke | Diastolic BP | 2 CNNs compared | Single center |
| Barriera et al. (2018) | CNN | 284 | Stroke (ICH and ischemic stroke) | CT head | Stroke neurologists reading CT | Single center |
| Beecy et al. (2017) | CNN | 114 | Stroke | CT head | Expert consensus interpretations | Single center |
| Dharmasaroja et al. (2013) | CNN | 194 | Stroke/intracranial hemorrhage | CT head | Thrombolysis after ischemic stroke | Single center |
| Fodeh et al. (2018) | SVM | 1834 | Atraumatic ICH | EHR review | Single center | |
| Gottrup et al. (2005) | kNN, Custom-built algorithm | 14 | Acute ischemic stroke | MRI | Applicability of highly flexible instance-based methods | Single center |
| Ho et al. (2016) | SVM, RF, and GBRT models | 105 | Acute ischemic stroke | MRI | Classification models for the problem of unknown time-since-stroke | Single center |
| Knight-Greenfield et al. (2018) | CNN | 114 | Stroke | CT head | Expert consensus interpretations | Single center |
| Ramos et al. (2018) | SVM, RF, Logistic-linear regression, CNN | 317 | SAH | CT Head | Delayed cerebral ischemia in SAH detection | Single center |
| SÜt et al. (2012) | MLP neural networks | 584 | Stroke mortality | EMR data | Selected variables using univariate statistical analyses | N/A |
| Paula De Toledo et al. (2009) | Logistic-linear regression | 441 | SAH | CT Head | Algorithms used were C4.5, fast decision tree learner, partial decision trees, repeated incremental pruning to produce error reduction, nearest neighbor with generalization, and ripple down rule learner | Multicenter Register |
| Thorpe et al. (2018) | decision tree | 66 | Stroke | Transcranial Doppler | Velocity Curvature Index (VCI) vs Velocity Asymmetry Index (VAI) | Single center |
| Williamson et al. (2019) | BOOSTING algorithm, RF | 483 | Risk stratification in SAH | True poor outcomes | Single center | |
| Xie et al. (2019) | Boosting algorithm | 512 | Predict Patient Outcome in Acute Ischemic Stroke | CT Head and clinical parameters | Feature selections were performed using a greedy algorithm | Single center |
| Heart failure | ||||||
| Andjelkovic et al. (2014) | CNN | 193 | HF in congenital heart disease | Echocardiography | Single center | |
| Blecker et al. (2018) | Logistic-linear regression | 37,229 | ADHF | Early ID of patients at risk of readmission for ADHF | 4 algorithms tested | Single center |
| Gleeson et al. (2016) | Custom-built algorithm | 534 | HF | Echocardiography and ECG | Data mining was applied to discover novel ECG and echocardiographic markers of risk | Single center |
| Golas et al. (2018) | Logistic-linear regression, boosting algorithm, CNN | 11,510 | HF | EHR | Heat failure patients to predict 30 day readmissions | Several hospitals in the Partners Healthcare System |
| Mortazavi et al. (2016) | Random forests, boosting, combined algorithms or logistic regression | 1653 | HF | Surveys to hospital examinations | Tele-HF trial | |
| Frizzell et al | Random forest and gradient-boosted algorithms | 56,477 | HF | EHR | Traditional statistical methods | GWTG-HF registry |
| Kasper Rossing et al. (2016) | SVM | 33 | HFpEF | Urinary proteomic analysis | Heart failure clinic (Single center) | |
| Kiljanek et al. (2009) | RF | 1587 | HF | Clinical diagnosis | Development of congestive heart failure after NSTEMI | CRUSADE registry |
| Liu et al. (2016) | Boosting algorithm, Logistic-linear regression | 526 | HF | Medical data, blood test, and echocardiographic imaging | Predicting mortality in HF | Single center |
SVM support vector machine, RF random forest, CNN convolutional neural network, kNN k-nearest neighbors, PCA principal component analysis, GBRT gradient boosted regression trees, MLP multilayer perceptron, HER electronic health record, HF heart failure, HFpEF heart failure with preserved ejection fraction, ADHF acute decompensated heart failure, SAH subarachnoid hemorrhage, SDH subdural hematoma, ICH intracerebral hemorrhage, CAD coronary artery disease, ACS acute coronary syndrome, CCTA coronary computed tomography angiography, FFR fractional flow reserve, IVUS intravascular ultrasound, ICD implantable cardioverter-defibrillator, AF atrial fibrillation, ECG electrocardiogram.
ML algorithms and prediction of CAD
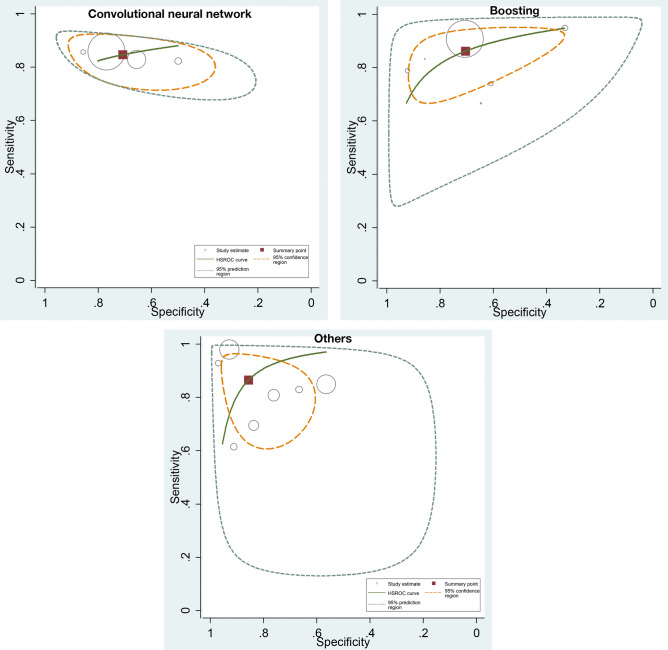
For the CAD, 45 cohorts reported a total of 116,227 individuals. 10 cohorts used CNN algorithms, 7 cohorts used SVM, 13 cohorts used boosting algorithm, 9 cohorts used custom-built algorithms, and 2 cohorts used RF. The prediction in CAD was associated with pooled AUC of 0.88 (95% CI 0.84–0.91), sensitivity of 0.86 (95% CI 0.77–0.92), and specificity of 0.70 (95% CI 0.51–0.84), for boosting algorithms and pooled of AUC 0.93 (95% CI 0.85–0.97), sensitivity of 0.87 (95% CI 0.74–0.94), and specificity of 0.86 (95% CI 0.73–0.93) for custom-built algorithms (Fig. 2).
Figure 2.
ROC curves comparing different machine learning models for CAD prediction. The prediction in CAD was associated with pooled AUC of 0.87 (95% CI 0.76–0.93) for CNN, pooled AUC of 0.88 (95% CI 0.84–0.91) for boosting algorithms, and pooled of AUC 0.93 (95% CI 0.85–0.97) for others (custom-built algorithms).
ML algorithms and prediction of stroke
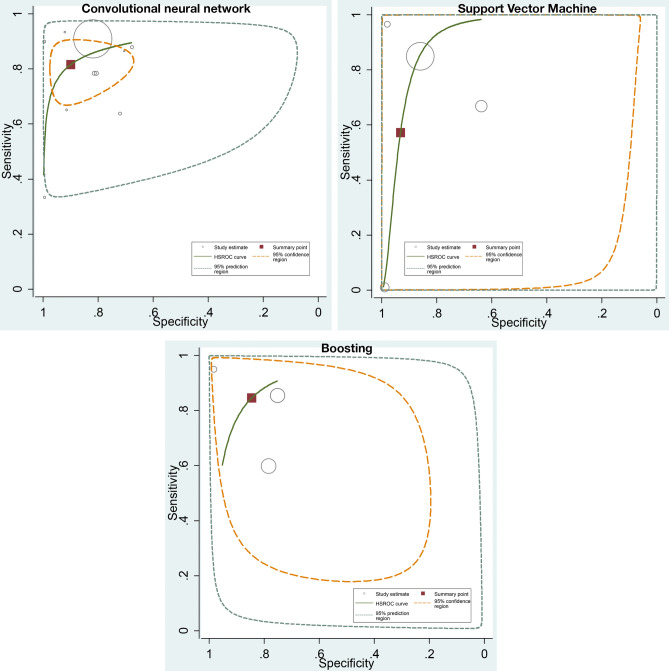
For the stroke, 34 cohorts reported a total of 7,027 individuals. 14 cohorts used CNN algorithms, 4 cohorts used SVM, 5 cohorts used boosting algorithm, 2 cohorts used decision tree, 2 cohorts used custom-built algorithms, and 1 cohort used random forest (RF). For prediction of stroke, SVM algorithms had a pooled AUC of 0.92 (95% CI 0.81–0.97), sensitivity 0.57 (95% CI 0.26–0.96), and specificity 0.93 (95% CI 0.71–0.99); boosting algorithms had a pooled AUC of 0.91 (95% CI 0.81–0.96), sensitivity 0.85 (95% CI 0.66–0.94), and specificity 0.85 (95% CI 0.67–0.94); and CNN algorithms had a pooled AUC of 0.90 (95% CI 0.83–0.95), sensitivity of 0.80 (95% CI 0.70–0.87), and specificity of 0.91 (95% CI 0.77–0.97) (Fig. 3).
Figure 3.
ROC curves comparing different machine learning models for stroke prediction. The prediction in stroke was associated with pooled AUC of 0.90 (95% CI 0.83–0.95) for CNN, pooled AUC of 0.92 (95% CI 0.81–0.97) for SVM algorithms, and pooled AUC of 0.91 (95% CI 0.81–0.96) for boosting algorithms.
ML algorithms and prediction of HF
For the HF, 12 cohorts reported a total of 51,612 individuals. 3 cohorts used CNN algorithms, 4 cohorts used logistic regression, 2 cohorts used boosting algorithm, 1 cohort used SVM, 1 cohort used in-house algorithm, and 1 cohort used RF. We could not perform analyses because we had too few studies (≤ 5) for each model.
ML algorithms and prediction of cardiac arrhythmias
For the cardiac arrhythmias, 12 cohorts reported a total of 3,204,837 individuals. 2 cohorts used CNN algorithms, 2 cohorts used logistic regression, 3 cohorts used SVM, 1 cohort used k-NN algorithm, and 4 cohorts used RF. We could not perform analyses because we had too few studies (≤ 5) for each model.
Discussion
To the best of our knowledge, this is the first and largest novel meta-analytic approach in ML research to date, which drew from an extensive number of studies that included over one million participants, reporting ML algorithms prediction in cardiovascular diseases. Risk assessment is crucial for the reduction of the worldwide burden of CVD. Traditional prediction models, such as the Framingham risk score12, the PCE model13, SCORE14, and QRISK15 have been derived based on multiple predictive factors. These prediction models have been implemented in guidelines; specifically, the 2010 American College of Cardiology/American Heart Association (ACC/AHA) guideline16 recommended the Framingham Risk Score, the United Kingdom National Institute for Health and Care Excellence (NICE) guidelines recommend the QRISK3 score17, and the 2016 European Society of Cardiology (ESC) guidelines recommended the SCORE model18. These traditional CVD risk scores have several limitations, including variations among validation cohorts, particularly in specific populations such as patients with rheumatoid arthritis19,20. Under some circumstances, the Framingham score overestimates CVD risk, potentially leading to overtreatment20. In general, these risk scores encompass a limited number of predictors and omit several important variables. Given the limitations of the most widely accepted risk models, more robust prediction tools are needed to more accurately predict CVD burden. Advances in computational power to process large amounts of data has accelerated interest in ML-based risk prediction, but clinicians typically have limited understanding of this methodology. Accordingly, we have taken a meta-analytic approach to clarify the insights that ML modeling can provide for CVD research.
Unfortunately, we do not know how or why the authors of the analyzed studies selected the chosen algorithms from the large array of options available. Researchers/authors may have selected potential models for their databases and performed several models (e.g., running parallel, hyperparameter tuning) while only reporting the best model, resulting in overfitting to their data. Therefore, we assume the AUC of each study is based upon the best possible algorithm available to the associated researchers. Most importantly, pooled analyses indicate that, in general, ML algorithms are accurate (AUC 0.8–0.9 s) in overall cardiovascular disease prediction. In subgroup analyses of each ML algorithms, ML algorithms are accurate (AUC 0.8–0.9 s) in CAD and stroke prediction. To date, only one other meta-analysis of the ML literature has been reported, and the underlying concept was similar to ours. The investigators compared the diagnostic performance of various deep learning models and clinicians based on medical imaging (2 studies pertained to cardiology)21. The investigators concluded that deep learning algorithms were promising but identified several methodological barriers to matching clinician-level accuracy21. Although our work suggests that boosting models and support vector machine (SVM) models are promising for predicting CAD and stroke risk, further study comparing human expert and ML models are needed.
First, the results showed that custom-built algorithms tend to perform better than boosting algorithm for CAD prediction in terms of AUC comparison. However, there is significant heterogeneity among custom-built algorithms that do not disclose their details. The boosting algorithm has been increasingly utilized in modern biomedicine22,23. In order to implement in clinical practice, the essential stages of designing a model and interpretation need to be uniform24. For implementation in clinical practice, custom-built algorithms must be transparent and replicated in multiple studies using the same set of independent variables.
Second, the result showed that boosting algorithms and SVM provides similar pooled AUC for stroke prediction. SVMs and boosting shared a common margin to address the clinical question. SVM seems to perform better than boosting algorithms in patients with stroke perhaps due to discrete, linear data or a proper non-linear kernel that fits the data better with improved generalization. SVM is an algorithm designed for maximizing a particular mathematical function with respect to a given collection of data. Compared to the other ML methods, SVM is more powerful at recognizing hidden patterns in complicated clinical datasets2,25. Both boosting and SVM algorithms have been widely used in biomedicine and prior studies showed mixed results26–30. SVM seems to outperform boosting in image recognition tasks28, while boosting seems to be superior in omic tasks27. However, in subgroup analysis, using research questions or types of protocols or images showed no difference in algorithm predictions.
Third, for heart failure and cardiac arrhythmias, we could not perform meta-analytic approaches due to the small number of studies for each model. However, based on our observation in our systematic review, SVM seems to outperform other predictive algorithms in detecting cardiac arrhythmias, especially in one large study31. Interestingly, in HF, the results are inconclusive. One small study showed promising results from SVM32. CNN seems to outperform others, but the results are suboptimal33. Although we assumed all reported algorithms have optimal variables, technical heterogeneity exists in ML algorithms (e.g., number of folds for cross-validation, bootstrapping techniques, how many run time [epochs], multiple parameters adjustments). In addition, optimal cut off for AUC remained unclear in clinical practice. For example, high or low sensitivity/specificity for each test depends on clinical judgement based on clinically correlated. In general, very high AUCs (0.95 or higher) are recommended, and it is known that AUC 0.50 is not able to distinguish between true and false. In some fields such as applied psychology34, with several influential variables, AUC values of 0.70 and higher would be considered strong effects. Moreover, standard practice for ML practitioners recommended reporting certain measures (e.g., AUC, c-statistics) without optimal sensitivity and specificity or model calibration, while interpretation in clinical practice is challenging. For example, the difference in BNP cut off for HF patients could result in a difference in volume management between diuresis and IV fluid in pneumonia with septic shock.
Compared to conventional risk scores, most ML models shared a common set of independent demographic variables (e.g., age, sex, smoking status) and include laboratory values. Although those variables are not well-validated individually in clinical studies, they may add predictive value in certain circumstances. Head-to-head studies comparing ML algorithms and conventional risk models are needed. If these studies demonstrate an advantage of ML-based prediction, the optimal algorithms could be implemented through electronic health records (EHR) to facilitate application in clinical practice. The EHR implementation is well poised for ML based prediction since the data are readily accessible, mitigating dependency on a large number of variables, such as discrete laboratory values. While it may be difficult for physicians in resource-constrained practice settings to access the input data necessary for ML algorithms, it is readily implemented in more highly developed clinical environments.
To this end, the selection of ML algorithm should base on the research question and the structure of the dataset (how large the population is, how many cases exist, how balanced the dataset is, how many available variables there are, whether the data is longitudinal or not, if the clinical outcome is binary or time to event, etc.) For example, CNN is particularly powerful in dealing with image data, while SVM can reduce the high dimensionality of the dataset if the kernel is correctly chosen. While when the sample size is not large enough, deep learning methods will likely overfit the data. Most importantly, this study's intent is not to identify one algorithm that is superior to others.
Limitations
Although the performance of ML-based algorithms seems satisfactory, it is far from optimal. Several methodological barriers can confound results and increase heterogeneity. First, technical parameters such as hyperparameter tuning in algorithms are usually not disclosed to the public, leading to high statistical heterogeneity. Indeed, heterogeneity measures the difference in effect size between studies. Therefore, in the present study, heterogeneity is inevitable as several factors can lead to this (e.g., fine-tuning models, hyperparameter selection, epochs). It is also a not good indicator to use as, in our HSROC model, we largely controlled the heterogeneity. Second, the data partition is also arbitrary because of no standard guidelines for utilization. In the present study, most included studies use 80/20 or 70/30 for training and validation sets. In addition, since the sample size for each type of CVD is small, the pooled results could potentially be biased. Third, feature selection methodologies, and techniques are arbitrary and heterogeneous. Fourth, due to the ambiguity of custom-built algorithms, we could not classify the type of those algorithms. Fifth, studies report different evaluation matrices (e.g., some did not report positive or negative cases, sensitivity/specificity, F-score, etc.). We did not report the confusion matrix for this meta-analytic approach as it required aggregation of raw numbers from studies without adjusting for difference between studies, which could result in bias. Instead, we presented pooled sensitivity and specificity using the HSROC model. Although ML algorithms are robust, several studies did not report complete evaluation metrics such as positive or negative cases, Beyes, bias accuracy, or analysis in the validation cohort since there are many ways to interpret the data depending on the clinical context. Most importantly, some analyses did not correlate with the clinical context, which made it more difficult to interpret. The efficacy of meta-analysis is to increase the power of the study by using the same algorithms. In addition, clinical data are heterogeneous and usually imbalanced. Most ML research did not report balanced accuracy, which could mislead the readers. Sixth, we did not register the analysis in PROSPERO. Finally, some studies reported only the technical aspect without clinical aspects, likely due to a lack of clinician supervision.
Conclusion
Although there are several limitations to overcome to be able to implement ML algorithms in clinical practice, overall ML algorithms showed promising results. SVM and boosting algorithms are widely used in cardiovascular medicine with good results. However, selecting the proper algorithms for the appropriate research questions, comparison to human experts, validation cohorts, and reporting of all possible evaluation matrices are needed for study interpretation in the correct clinical context. Most importantly, prospective studies comparing ML algorithms to conventional risk models are needed. Once validated in that way, ML algorithms could be integrated with electronic health record systems and applied in clinical practice, particularly in high resources areas.
Supplementary information
Author contributions
C.K., H.H., S.B., Z.W., K.W.J., R.P., H.Z., S.K., B.N., T.K., U.B., J.L.H., W.T. had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: C.K., H.H., K.W.J., Z.W. Acquisition of data: C.K., H.H., R.P., H.J., T.K. Analysis and interpretation of data: B.N., Z.W. Drafting of the manuscript: C.K., H.H., S.B., U.B., J.L.H., T.W. Critical revision of the manuscript for important intellectual content: T.W., Z.W. Study supervision: C.K., T.W.
Funding
There was no funding for this work.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-72685-1.
References
- 1.Noble WS. Support vector machine applications in computational biology. Kernel Methods Comput. Biol. 2004;71:92. [Google Scholar]
- 2.Aruna S, Rajagopalan S. A novel SVM based CSSFFS feature selection algorithm for detecting breast cancer. Int. J. Comput. Appl. 2011;31:20. [Google Scholar]
- 3.Lakhani P, Sundaram B. Deep learning at chest radiography: Automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017;284:574–582. doi: 10.1148/radiol.2017162326. [DOI] [PubMed] [Google Scholar]
- 4.Yasaka K, Akai H. Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: A preliminary study. Radiology. 2018;286:887–896. doi: 10.1148/radiol.2017170706. [DOI] [PubMed] [Google Scholar]
- 5.Christ PF, Elshaer MEA, Ettlinger F, et al. Automatic Liver and Lesion Segmentation in CT Using Cascaded Fully Convolutional Neural Networks and 3D Conditional Random Fields. International Conference on Medical Image Computing and Computer-Assisted Intervention. Berlin: Springer; 2016. pp. 415–423. [Google Scholar]
- 6.Krittanawong C, Johnson KW, Rosenson RS, et al. Deep learning for cardiovascular medicine: A practical primer. Eur. Heart J. 2019;40:2058–2073. doi: 10.1093/eurheartj/ehz056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krittanawong C, Zhang H, Wang Z, Aydar M, Kitai T. Artificial intelligence in precision cardiovascular medicine. J. Am. Coll. Cardiol. 2017;69:2657–2664. doi: 10.1016/j.jacc.2017.03.571. [DOI] [PubMed] [Google Scholar]
- 8.Krittanawong C, Bomback AS, Baber U, Bangalore S, Messerli FH, Wilson Tang WH. Future direction for using artificial intelligence to predict and manage hypertension. Curr. Hypertens. Rep. 2018;20:75. doi: 10.1007/s11906-018-0875-x. [DOI] [PubMed] [Google Scholar]
- 9.Covidence systematic review software. Melbourne AVHIAawcoAD.
- 10.Rutter CM, Gatsonis CA. A hierarchical regression approach to meta-analysis of diagnostic test accuracy evaluations. Stat. Med. 2001;20:2865–2884. doi: 10.1002/sim.942. [DOI] [PubMed] [Google Scholar]
- 11.Stroup DF, Berlin JA, Morton SC, et al. Meta-analysis of observational studies in epidemiology: A proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000;283:2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 12.Wilson PW, D'Agostino RB, Levy D, Belanger AM, Silbershatz H, Kannel WB. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97:1837–1847. doi: 10.1161/01.cir.97.18.1837. [DOI] [PubMed] [Google Scholar]
- 13.Goff DC, Jr, Lloyd-Jones DM, Bennett G, et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: A report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. J. Am. Coll. Cardiol. 2014;63:2935–2959. doi: 10.1016/j.jacc.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conroy RM, Pyörälä K, Fitzgerald AP, et al. Estimation of ten-year risk of fatal cardiovascular disease in Europe: The SCORE project. Eur. Heart J. 2003;24:987–1003. doi: 10.1016/s0195-668x(03)00114-3. [DOI] [PubMed] [Google Scholar]
- 15.Hippisley-Cox J, Coupland C, Vinogradova Y, et al. Predicting cardiovascular risk in England and Wales: Prospective derivation and validation of QRISK2. BMJ (Clinical research ed) 2008;336:1475–1482. doi: 10.1136/bmj.39609.449676.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenland P, Alpert JS, Beller GA, et al. 2010 ACCF/AHA guideline for assessment of cardiovascular risk in asymptomatic adults: A report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2010;122:e584–636. doi: 10.1161/CIR.0b013e3182051b4c. [DOI] [PubMed] [Google Scholar]
- 17.Hippisley-Cox J, Coupland C, Brindle P. Development and validation of QRISK3 risk prediction algorithms to estimate future risk of cardiovascular disease: Prospective cohort study. BMJ (Clinical research ed) 2017;357:j2099. doi: 10.1136/bmj.j2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piepoli MF, Hoes AW, Agewall S, et al. 2016 European Guidelines on cardiovascular disease prevention in clinical practice: The Sixth Joint Task Force of the European Society of Cardiology and Other Societies on Cardiovascular Disease Prevention in Clinical Practice (constituted by representatives of 10 societies and by invited experts) Developed with the special contribution of the European Association for Cardiovascular Prevention & Rehabilitation (EACPR) Eur. Heart J. 2016;37:2315–2381. doi: 10.1093/eurheartj/ehw106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kremers HM, Crowson CS, Therneau TM, Roger VL, Gabriel SE. High ten-year risk of cardiovascular disease in newly diagnosed rheumatoid arthritis patients: A population-based cohort study. Arthritis Rheum. 2008;58:2268–2274. doi: 10.1002/art.23650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Damen JA, Pajouheshnia R, Heus P, et al. Performance of the Framingham risk models and pooled cohort equations for predicting 10-year risk of cardiovascular disease: A systematic review and meta-analysis. BMC Med. 2019;17:109. doi: 10.1186/s12916-019-1340-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Faes L, Kale AU, et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: A systematic review and meta-analysis. Lancet Digit. Health. 2019;1:e271–e297. doi: 10.1016/S2589-7500(19)30123-2. [DOI] [PubMed] [Google Scholar]
- 22.Mayr A, Binder H, Gefeller O, Schmid M. The evolution of boosting algorithms. From machine learning to statistical modelling. Methods Inf. Med. 2014;53:419–427. doi: 10.3414/ME13-01-0122. [DOI] [PubMed] [Google Scholar]
- 23.Buhlmann P, Gertheiss J, Hieke S, et al. Discussion of "the evolution of boosting algorithms" and "extending statistical boosting". Methods Inf. Med. 2014;53:436–445. doi: 10.3414/13100122. [DOI] [PubMed] [Google Scholar]
- 24.Natekin A, Knoll A. Gradient boosting machines, a tutorial. Front. Neurorobot. 2013;7:21–21. doi: 10.3389/fnbot.2013.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noble WS. What is a support vector machine? Nat. Biotechnol. 2006;24:1565–1567. doi: 10.1038/nbt1206-1565. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H, & Gu C. Support vector machines versus Boosting.
- 27.Ogutu JO, Piepho HP, Schulz-Streeck T. A comparison of random forests, boosting and support vector machines for genomic selection. BMC Proc. 2011;5(Suppl 3):S11. doi: 10.1186/1753-6561-5-S3-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun T, Wang J, Li X, et al. Comparative evaluation of support vector machines for computer aided diagnosis of lung cancer in CT based on a multi-dimensional data set. Comput. Methods Programs Biomed. 2013;111:519–524. doi: 10.1016/j.cmpb.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 29.Huang M-W, Chen C-W, Lin W-C, Ke S-W, Tsai C-F. SVM and SVM ensembles in breast cancer prediction. PLoS One. 2017;12:e0161501–e0161501. doi: 10.1371/journal.pone.0161501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caruana R, Karampatziakis N, & Yessenalina A. An empirical evaluation of supervised learning in high dimensions. In Proceedings of the 25th International Conference on Machine Learning: ACM, 2008, 96–103.
- 31.Hill NR, Ayoubkhani D, Lumley M, et al. Machine learning to detect and diagnose atrial fibrillation and atrial flutter (AF/F) using routine clinical data. Value Health. 2018;21:S213. [Google Scholar]
- 32.Rossing K, Bosselmann HS, Gustafsson F, et al. Urinary proteomics pilot study for biomarker discovery and diagnosis in heart failure with reduced ejection fraction. PLoS One. 2016;11:e0157167. doi: 10.1371/journal.pone.0157167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Golas SB, Shibahara T, Agboola S, et al. A machine learning model to predict the risk of 30-day readmissions in patients with heart failure: A retrospective analysis of electronic medical records data. BMC Med. Inform. Decis. Mak. 2018;18:44. doi: 10.1186/s12911-018-0620-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rice ME, Harris GT. Comparing effect sizes in follow-up studies: ROC Area, Cohen's d, and r. Law Hum Behav. 2005;29:615–620. doi: 10.1007/s10979-005-6832-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.